Abstract
Purpose:
Treatment effect or radiation necrosis after stereotactic radiosurgery (SRS) for brain metastases is a common phenomenon often indistinguishable from true progression. Radiomics is an emerging field that promises to improve on conventional imaging. In this study, we sought to apply a radiomics-based prediction model to the problem of diagnosing treatment effect after SRS.
Methods and Materials:
We included patients in the Johns Hopkins Health System who were treated with SRS for brain metastases who subsequently underwent resection for symptomatic growth. We also included cases of likely treatment effect in which lesions grew but subsequently regressed spontaneously. Lesions were segmented semiautomatically on preoperative T1 postcontrast and T2 fluid-attenuated inversion recovery magnetic resonance imaging, and radiomic features were extracted with software developed in-house. Top-performing features on univariate logistic regression were entered into a hybrid feature selection/classification model, IsoSVM, with parameter optimization and further feature selection performed using leave-one-out cross-validation. Final model performance was assessed by 10-fold cross-validation with 100 repeats. All cases were independently reviewed by a board-certified neuroradiologist for comparison.
Results:
We identified 82 treated lesions across 66 patients, with 77 lesions having pathologic confirmation. There were 51 radiomic features extracted per segmented lesion on each magnetic resonance imaging sequence. An optimized IsoSVM classifier based on top-ranked radiomic features had sensitivity and specificity of 65.38% and 86.67%, respectively, with an area under the curve of 0.81 on leave-one-out cross-validation. Only 73% of cases were classifiable by the neuroradiologist, with a sensitivity of 97% and specificity of 19%.
Conclusions:
Radiomics holds promise for differentiating between treatment effect and true progression in brain metastases treated with SRS. A predictive model built on radiomic features from an institutional cohort performed well on cross-validation testing. These results warrant further validation in independent datasets. Such work could prove invaluable for guiding management of individual patients and assessing outcomes of novel interventions.
Summary
Treatment effect after stereotactic radiosurgery for brain metastases is a common phenomenon often indistinguishable from true progression. We applied a radiomics-based prediction model to discriminate treatment effect from true progression after stereotactic radiosurgery in an institutional cohort, with excellent performance on leave-one-out cross-validation. With further validation in independent datasets, such work could prove invaluable for guiding management of individual patients and assessing outcomes of novel interventions.
Introduction
Because of its excellent efficacy, short treatment time, and favorable toxicity profile compared with whole brain radiation (WBRT), stereotactic radiosurgery (SRS) is increasingly favored for patients with a limited number of brain metastases (1). There remains, however, a significant minority of cases in which brain metastases may appear to progress despite treatment, with local failure rates upward of 27% to 33% in some randomized trials (2–4). However, many lesions grow only transiently after SRS, and increasing tumor size is not always consistent with local progression (5). Treatment effects in irradiated brain tumors have been well described and involve a range of effects including inflammation, vascular injury, and necrosis (6). For SRS, necrosis may play a particularly prominent role, with radionecrosis rates after SRS as high as 26% (7).
At this time, the gold standard for distinguishing true progression from necrosis after SRS remains pathologic determination with surgery; conventional magnetic resonance imaging (MRI) is largely unhelpful in such cases (Fig. 1).
Fig. 1.
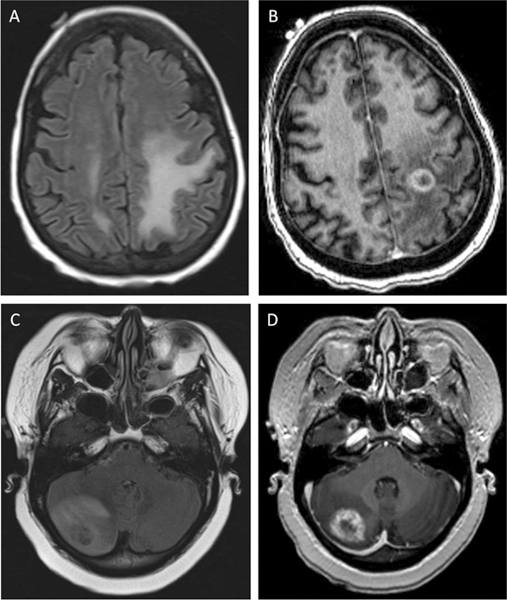
Progression and treatment effect of brain metastases on magnetic resonance imaging. Top: T2 Fluid-attenuated inversion recovery (FLAIR) (A) and T1 post-contrast (B) images for a patient with treatment effect of a left prefrontal metastasis from non-small cell lung cancer. Bottom: T2 FLAIR (C) and T1 postcontrast (D) images for a patient with true progression of a right cerebellar metastasis (breast primary) after stereotactic radiotherapy. Both cases were confirmed by surgical pathology.
Radiomics involves extracting quantitative texture features from medical images. Texture features are used as potential biomarkers for incorporation with other clinical markers for improved decision-making (8, 9). Radiomic features include a collection of shape and first- or higher order features describing gray-level textures that may not be readily recognized by conventional visual inspection. These types of radiomic features create high-dimensional information that can be analyzed and leveraged using machine learning algorithms for improved diagnostic accuracy.
In this study, we investigated whether a predictive model, based on radiomic features extracted from conventional brain MRI, could distinguish true progression from treatment effect in an institutional cohort of patients treated with SRS for brain metastases.
Methods and Materials
Patient data
The study was approved by the Johns Hopkins institutional review board. Electronic medical records of patients with brain metastases who underwent SRS or fractionated stereotactic radiation in 1 to 5 fractions at a single institution from June 2003 through September 2017 were reviewed to identify patients in whom treated lesions grew on imaging to the point of requiring surgical resection for symptom relief. We also identified patients without pathologic confirmation in whom treated lesions grew to a concerning extent after SRS but subsequently regressed, as well as patients who stabilized spontaneously without further intervention. For patients who underwent resection, we retrieved images from the preprocedure brain MRI performed at our institution, typically 1 day before or the morning of surgery. For the small set of patients without pathologic confirmation but with presumed treatment effect on imaging, we retrieved images from the study that showed the maximum size of the treated lesion. We excluded patients for whom the acquired MRI had substantial motion artifact or did not include T1 postcontrast (T1c) and T2 fluid-attenuated inversion recovery (FLAIR) sequences. MRI acquisition parameters varied across studies because of the long timespan over which patients were treated and followed in this study (Table E1; available online at https://doi.org/10.1016/jAjrobp.2018.05.041). For patients who underwent resection, surgical pathology was used to determine whether the lesion represented true malignant progression. Samples with a mix of tumor and necrosis were uniformly labeled as true progression to match our institutional practice of proceeding with adjuvant treatment (typically reirradiation or brachytherapy) if any component of active tumor were found.
Image segmentation, registration, and radiomic feature extraction
A pictorial schema of the steps required for creation of the prediction model based on radiomic features is shown in Fig. 2. As an initial step, the T2 FLAIR sequence for each lesion was rigidly registered to the T1c sequence based on bony anatomy, using the Velocity AI commercial software platform (Version 3.2.1, Varian Medical Systems, Palo Alto, CA).
Fig. 2.
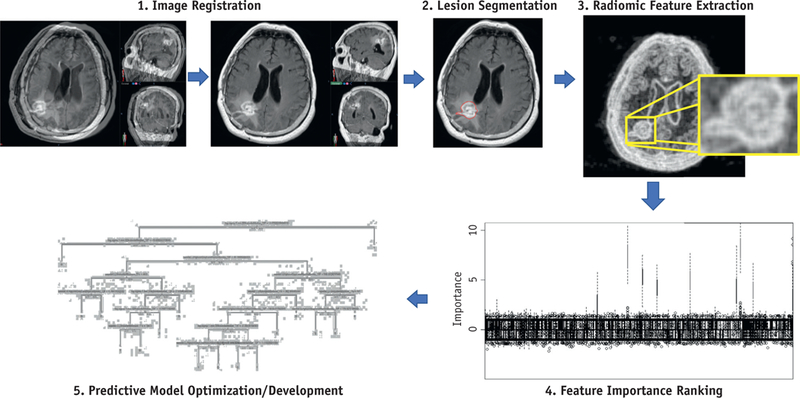
Analysis with radiomics. Major steps in radiomics include (1) image registration, (2) lesion segmentation, (3) radiomic feature extraction, (4) determination of the importance of each feature and ranking features accordingly, and (5) predictive modeling with a machine learning algorithm trained on an optimal set of features and parameters.
Semiautomatic segmentation of the single largest-diameter axial slice of each lesion on T1c sequence was applied using a multiparametric, deep learning, tissue signature model built using stacked sparse autoencoders (10). For a random subset of 10 cases, multiparametric, deep learning, tissue signature model segmentation showed high similarity (mean Dice similarity index = 0.86) compared with fully manual segmentations of the same cases. The single largest-diameter slice was chosen to correspond with standard clinical analysis using criteria from Response Evaluation Criteria in Solid Tumors.
The resulting regions of interest and associated T1c and T2 FLAIR images were exported as files in Digital Imaging and Communications in Medicine format for radiomic feature extraction using in-house software developed with MATLAB (The MathWorks, Natick, MA). We extracted 51 radiomic features from each T1c or FLAIR imaging sequence, divided into 5 categories: (1) first-order statistics (14 features), (2) gray level co-occurrence matrix (GLCM, 18 features), (3) gray level run length matrix (GLRLM, 11 features), (4) neighborhood gray tone difference matrix (NGTDM, 5 features), and (5) morphologic features (3 features). A comprehensive listing of extracted features is provided in Table E2 (available online at https://doi.org/10.1016/j.ijrobp.2018.05.041).
In broad terms, first-order features describe the statistics of voxel intensity distributions within the regions of interest, whereas second-order features, such as GLCM, GLRLM, and NGTDM, seek to quantify textural features (9). GLCM and GLRLM were evaluated in 4 directions (0°, 45°, 90°, and 135°) with values averaged to achieve rotational invariance, and the GLCM parameter, d, was set to 1 voxel in all directions. We used 64 equally sized bins for first-order statistics, and gray levels were quantized into 64 levels for GLCM, GLRLM, and NGTDM calculations. Both binning and gray levels were systematically varied between 8 and 512 (specifically 8, 16, 32, 64, 128, 256, and 512), and 64 was empirically chosen in each case. Of note, to preserve the maximal amount of information from original source data, images were not normalized before processing.
Feature selection and prediction modeling
As a next step, summary statistics for the radiomic values (mean and standard error of the mean) were computed for the 2 outcome classes and compared using an unpaired Student t test. Associations between the radiomic metrics and the final diagnosis were assessed on univariate logistic regression analysis, with radomic features ranked according to predictive performance using logistic regression as assessed by the area under the curve (AUC) for the receiver operating characteristic curve.
Further feature selection and prediction modeling were performed using the IsoSVM algorithm as applied to the top-performing radiomic features from univariate logistic regression (11). IsoSVM is a hybrid feature selection and classification algorithm based on the Isomap and Support Vector Machine (SVM) algorithms. By definition, SVM trains a classification model that creates a linear hyperplane to best separate the 2 groups in question. This concept can be extended for linearly inseparable data using kernel methods that map the data into a linearly separable space. We implemented IsoSVM to use the Isomap kernel to perform this nonlinear transformation because it has been shown that Isomap outperforms radial basis function, linear, quadratic, and cubic kernels (11).
Imbalance in the number of true progression and treatment effect cases was overcome by setting a separate misclassification penalty for the 2 classes. The optimal ratio of the misclassification penalty along with other IsoSVM parameters (nearest neighborhood and dimensionality) were optimized along with final selection of radiomic features for the model using grid search and leave-one-out cross-validation (LOOCV). Finally, 10-fold cross-validation was performed to assess the overall performance of the optimized IsoSVM model by AUC. To avoid bias resulting from unfortunate partitioning of the data, 10-fold cross-validation was performed 100 times with random initiali-zation of 10 disjoint groups. Statistical significance was set at P ≤ .05.
Neuroradiologist interpretation
All cases and associated clinical reports were reviewed by a board-certified neuroradiologist (D.L.) with 16 years of postfellowship experience; the neuroradiologist classified the lesions as representing either treatment effect or cancer progression based on imaging description and clinical interpretation but was blinded to final histopathologic and clinical outcome. Lesions with a high degree of diagnostic uncertainty were classified as indeterminate or mixed. Interpretation of other MRI sequences, such as perfusion, was permitted when available, but for conventional images, classification was mainly based on lesion growth over time.
Results
We identified 82 individual brain metastases with apparent progression after SRS across 66 patients who met our inclusion criteria. Of the 82 lesions, 77 underwent surgical resection for histopathologic diagnosis, but 5 lesions (in 5 different patients) were presumed to be a treatment effect based on subsequent spontaneous regression. Of 82 lesions, 52 (63%) were confirmed to be true progression based on surgical pathology, and the remaining 30 (37%) were labeled as treatment effect.
Patient and radiation treatment characteristics are summarized in Table 1. The most common primary histology was non-small cell lung cancer, followed by melanoma, breast, and small cell lung cancer. Other primary histologies included renal cell, endometrial, ovarian, thyroid, colorectal, and sarcoma. SRS was delivered using a range of treatment machines, including Gamma Knife, Cyber-Knife, and linear accelerator—based systems. Consistent with our institutional practice, brain metastases were most commonly treated to 18 to 20 Gy in 1 fraction. For larger tumors or sensitive locations, a hypofractionated fractionated stereotactic radiation approach was used, often 24 Gy in 3 fractions or 25 Gy in 5 fractions. For lesions that had undergone some form of reirradiation by the time of resection, there were 3 instances of prior SRS to the same location, 11 cases of prior/additional WBRT, and 4 cases of both WBRT and prior SRS. The great majority of lesions (64 of 82 [78%]) experienced only a single course of SRS before resection.
Table 1.
Tumor and treatment characteristics
| Characteristics | Data |
|---|---|
| Median patient age at SRS (range), y | 56.5 (29–86) |
| No. of lesions treated | 82 |
| Primary histology | |
| Non-small cell lung cancer | 28 |
| Melanoma | 21 |
| Breast | 16 |
| Small cell lung cancer | 6 |
| Other | 11 |
| SRS machine | |
| CyberKnife | 58 |
| Gamma knife | 8 |
| Linear accelerator | 16 |
| Treatment characteristics | |
| Median total dose (range), Gy | 20 (14–25) |
| Median fractions (range) | 1 (1–5) |
| No. treated with WBRT (%) | 15 (18.3) |
| No. with prior same-site SRS (%) | 7 (8.5) |
| Median time from SRS to resection (range), d* | 302 (21–1351) |
Abbreviations: SRS = stereotactic radiosurgery; WBRT = whole brain radiation therapy.
For lesions with radiographic pseudoprogression without pathology, time interval was from SRS until date of maximum size.
The 10 top-ranked radiomic features according to univariate logistic regression performance (AUC) are listed in Table 2. These included 6 features from T1c and 4 from T2 FLAIR.
Table 2.
Top 10 radiomic features according to performance on univariate logistic regression models
| Rank | Radiomic feature | Mean value in treatment effect (standard error) |
Mean value in true progression (standard error) |
P value* | AUC† |
|---|---|---|---|---|---|
| 1 | T1c minimum | 190.63 (17.12) | 139.42 (17.71) | .04 | 0.70 |
| 2 | FLAIR NGTDM coarseness | 0.05 (0.01) | 0.03 (0.00) | .03 | 0.69 |
| 3 | T1c NGTDM texture strength | 116.30 (13.68) | 82.34 (9.49) | .05 | 0.68 |
| 4 | FLAIR kurtosis | 3.09 (0.26) | 3.53 (0.15) | .15 | 0.67 |
| 5 | T1c NGTDM coarseness | 0.04 (0.00) | 0.03 (0.00) | .04 | 0.65 |
| 6 | T1c GLRLM gray level nonuniformity | 594.39 (70.12) | 842.25 (75.11) | .02 | 0.65 |
| 7 | T1c fractal dimension | 1.37 (0.03) | 1.43 (0.02) | .08 | 0.65 |
| 8 | FLAIR minimum | 4107.00 (353.47) | 3130.63 (337.94) | .05 | 0.65 |
| 9 | T1c GLRLM run percentage | 0.41 (0.03) | 0.48 (0.02) | .04 | 0.63 |
| 10 | FLAIR NGTDM texture strength | 338.99 (93.26) | 203.51 (31.36) | .18 | 0.63 |
Abbreviations: FLAIR = fluid-attenuated inversion recovery; GLRLM = gray level run-length matrix; NGTDM = neighborhood gray tone difference matrix; T1c = T1 post contrast.
P value is for difference in the means according to unpaired Student’s t test.
AUC is area under the receiver operating characteristic curve for performance of the radiomic feature on univariate logistic regression.
The optimal set of hyperparameters for the IsoSVM feature transformation and classification model, obtained using an LOOCV-based grid search, were Isomap neigh-borhood parameter k = 45, dimensionality d = 9, and misclassification ratio = 4:1 (treatment effect:true progression). The grid search for radiomic feature subsets (Top 5, 10, 15, .All) ultimately selected only the top 5 radiomic features (first 5 features in Table 2) for input into the optimal IsoSVM model. The optimized IsoSVM classifier had sensitivity and specificity of 65.38% and 86.67%, respectively, with an AUC of 0.81 on LOOCV (Fig. 3). On 10-fold cross-validation, the optimal IsoSVM model had sensitivity and specificity of 60.48% and 86.97%, respectively, with an AUC of 0.79. We also explored the value of combining radiomic features with a set of clinical features, including age, sex, and tumor or treatment characteristics (Table E3; available online at https://doi.org/10.1016/j-ijrobp.2018.05.041), but the clinical features ranked poorly on univariate logistic regression, and the performance of IsoSVM on LOOCV remained at 0.81. No clinical features were selected for use in classification.
Fig. 3.
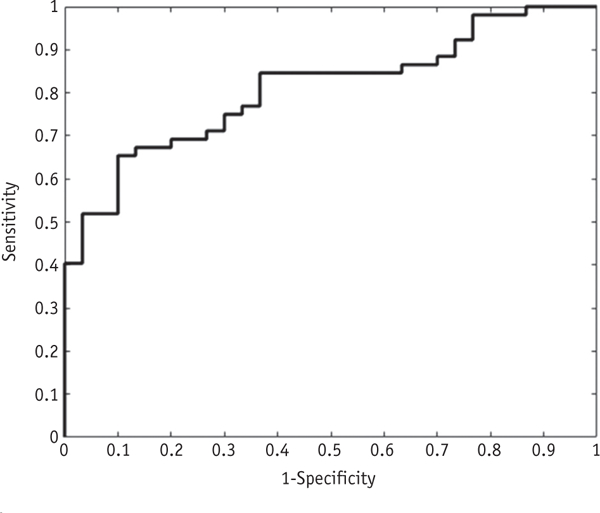
Receiver operating characteristic curve for IsoSVM model in leave-one-out cross-validation. Area under the curve (AUC) was 0.81, indicating excellent performance of the model on leave-one-out cross-validation.
On neuroradiology review, 22 of 82 cases (27%) were classified as indeterminate or mixed. For the remaining 60 cases, 55 (92%) were classified by radiologic interpretation as malignant progression, with a sensitivity of 97% and specificity of 19%.
Discussion
We have demonstrated that advanced machine learning, coupled with radiomic feature extraction using brain MRI, improved differentiation between true progression and treatment effect. In particular, our model substantially increased specificity as compared to the neuroradiologist’s interpretation, suggesting that machine learning methods can be used as an adjunct to traditional radiology review.
Improvement in the ability to distinguish true progression from treatment effect after SRS for brain metastases has important practical implications for patients and clinicians. For example, systemic therapies may be prematurely discontinued or salvage radiation therapy unnecessarily administered in cases of apparent progression without confirmatory pathology. Many positron emission tomography— and MRI-based methods have been proposed to diagnose treatment effect after SRS, but no standard approach has yet emerged (6, 12–17).
Integrated radiomic analysis may provide a new approach for improving the performance of conventional imaging of brain lesions. The results of the present study are consistent with other reports that have applied radio-mics for the diagnosis of growing brain metastases after SRS (18–20). For the most part, prior attempts have been limited by a very small number of lesions with pathologic confirmation (18, 19). In our dataset, nearly all lesions were verified by pathology; pathologic confirmation is critical to evaluating the true performance of these models, given the inaccuracy of conventional clinical judgment based on MRI or patient symptoms alone (12, 21). Some studies, for example, have examined the predictive ability of measuring mismatch in lesion appearance on T1 and T2 sequences (22, 23), but this approach failed to be predictive when assessed in an independent dataset (21).
Given the invasive nature of neurosurgery and the typically limited lifespan in this patient population, management often consists of observation until the patient becomes acutely or intractably symptomatic, and pathologic confirmation is, therefore, often lacking. In a recently published work, Zhang et al collected a larger series with pathologic confirmation to make predictions using an RUSBoost classifier with a reported AUC of 0.73 (20). Their method relied heavily on comparing differences in radiomic feature values at different imaging time points, termed “delta radiomics.” When they used radiomic features from a single time point, the AUC was lower at 0.65. In the present study, we curated a similarly large series of metastatic lesions with pathologic confirmation, and our methods resulted in a better AUC using only a single imaging time point. Future investigations are planned to determine the optimal number of time points to provide the best prognostic performance for our model.
In addition to RUSBoost, other groups have also used various incarnations of SVM algorithms in their classification models (18–20). In the current study, we used IsoSVM, a hybrid SVM model that applies a nonlinear dimensionality-reduction machine learning method for both feature selection and classification that has been previously applied to distinguish benign from malignant breast lesions (11). There is a need for standardization of approaches for radiomics-based classification problems, and the many disparate approaches highlight the critical need for further research in this area. Moreover, there is a lack of methodology for the systematic selection of the optimal gray levels and binning needed for MRI-based radiomics publications, and this is an area of active investigation. The choice of machine learning algorithm and tuning of algorithms for specific datasets is a critical and an unmet challenge. For example, Parmar et al systematically evaluated 12 different machine learning classifiers in lung cancer radiomics using computed tomography and found that the choice of machine learning algorithm had a dominant influence on performance variance (24). Our ability to achieve a high AUC in this work likely reflects a combination of factors, including the specific choice and tuning of our algorithm and a sizeable sample of cases with pathologic confirmation.
As with most of these studies, a major limitation of the present work is a relatively small sample size. This limits the performance of classifier algorithms that are highly dependent on data for training. Random noise in small datasets often can be mistakenly interpreted as meaningful (a problem known as overfitting); as a consequence, the model may not perform as well in independent datasets. Given the difficulty of collecting pathologically proven cases, an important next step will be to collaborate with partner institutions to create larger datasets for model training and independent validation. Public, large-scale imaging datasets, such as those of the Quantitative Imaging Network, would be invaluable, but to date no Quantitative Imaging Network for brain metastases has been established (25).
Another limitation of this study is our simplified labeling of mixed-pathology specimens as cases of true progression. Although this mirrors our clinical practice of treating mixed cases as true progression, an ideal model would account for the exact ratio of tumor to necrosis on pathology to optimize model training and more precisely measure model performance.
Compared with studies in which patients were scanned on a single scanner or in which SRS treatment was delivered by a single modality (eg, Gamma Knife), we describe results over a long period with varying scan acquisition parameters and where SRS was delivered on a variety of treatment machines with different fractionation schemes. Although normalization of MRI data has been proposed to standardize arbitrary differences in intensities of scans between patients and over time (26), the degree of importance and benefit of normalization for radiomic prediction models to classify cancer progression or treatment effect has not been well established. In particular, it is unclear whether intensity normalization over the entire magnetic resonance image is beneficial when the particular region of interest has a high average intensity. Indeed, of the brain radiomics studies discussed, 2 earlier studies performed intensity normalization, whereas the latest study did not (18–20). Given that we did not apply normalization or other preprocessing in the present study, our ability to generate a predictive model with excellent performance, despite substantial variability in treatment and image acquisition, likely increases the generalizability of our findings.
Linking radiomic features with underlying biologic characteristics can be extremely challenging (27). For example, NGTDM coarseness (2 of our top 5 ranked features) also has been used to distinguish head and neck tumors (less coarse) from normal tissues (more coarse) on positron emission tomography scan (28), but it is uncertain whether this may represent a common underlying biologic process across sites, imaging modalities, and clinical scenarios. Another highly ranked feature in this dataset was T1c GLRLM gray level nonuniformity. Similar nonuni-formity features were also top-performing features in a large, combined study of head and neck cancer and lung cancer radiomics (29); in that study, the authors found that these features, which measure intralesional heterogeneity, were predictive for both clinical prognosis and cell cycling gene expression levels, with increased heterogeneity correlating with greater proliferation. In our dataset, it appears that nonuniformity was higher in true progression cases (Table 2), concordant with the idea of using radiomic heterogeneity as a biomarker for proliferation. Kurtosis, our fourth-ranked feature, also has been linked with heterogeneity; in particular, higher mean values have predicted for local failure in head and neck cancers treated with radiation (30, 31).
Conclusions
Our results indicate that radiomics and machine learning methods hold great promise for discriminating between treatment effect and true progression in brain metastases treated with SRS. A predictive model built on radiomic features from an institutional cohort with pathologic validation of cancer progression performed excellently on cross-validation testing. Such work will prove invaluable for guiding the management of individual patients and assessing the outcomes of novel interventions.
Supplementary Material
Acknowledgments
This work was supported by the Nicholl Family Foundation, the National Institutes of Health (grant nos. 5P30CA06973 [IRAT], 1R01CA190299, and U01CA140204), and the NVIDIA Corporation (donation of Tesla K40 GPU card used for this research). The funding sources had no involvement in the design, collection, analysis, or interpretation of data or in the writing of the report or decision to submit the article for publication.
M. A. J. reports National Institutes of Health grants (nos. 5P30CA06973 [IRAT], 1R01CA190299, and U01CA140204) and nonfinancial support from NVIDIA Corporation during the conduct of the study. L. K. reports grants from the Nicholl Family Foundation during the conduct of the study and grants and personal fees from Novocure and Accuray, outside the submitted work. K. R. reports grants from Elekta AB, personal fees from AstraZeneca, personal fees from Medtronic, and grants from Accuray, outside the submitted work.
Footnotes
Conflict of Interest: M. A. J. and V. P. have patents US patents, nos. 20170112459 and 20160171695, issued relevant to the machine learning algorithms applied in this work.
Supplementary material for this article can be found at https://doi.org/10.1016/j.ijrobp.2018.05.041.
References
- 1.Nieder C, Grosu AL, Gaspar LE. Stereotactic radiosurgery (SRS) for brain metastases: A systematic review. Radiat Oncol 2014;9:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soliman H, Das S, Larson DA, et al. Stereotactic radiosurgery (SRS) in the modern management of patients with brain metastases. Onco-target 2016;7:12318–12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsao M, Xu W, Sahgal A. A meta-analysis evaluating stereotactic radiosurgery, whole-brain radiotherapy, or both for patients presenting with a limited number of brain metastases. Cancer 2012;118:2486–2493. [DOI] [PubMed] [Google Scholar]
- 4.Sahgal A, Aoyama H, Kocher M, et al. Phase 3 trials of stereotactic radiosurgery with or without whole-brain radiation therapy for 1 to 4 brain metastases: individual patient data meta-analysis. Int J Radiat Oncol Biol Phys 2015;91:710–717. [DOI] [PubMed] [Google Scholar]
- 5.Patel TR, McHugh BJ, Bi WL, et al. A comprehensive review of MR imaging changes following radiosurgery to 500 brain metastases. AJNR Am J Neuroradiol 2011;32:1885–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandsma D, Stalpers L, Taal W, et al. Clinical features, mechanisms, and management of pseudoprogression in malignant gliomas. Lancet Oncol 2008;9:453–461. [DOI] [PubMed] [Google Scholar]
- 7.Kohutek ZA, Yamada Y, Chan TA, et al. Long-term risk of radio-necrosis and imaging changes after stereotactic radiosurgery for brain metastases. J Neuro-Oncol 2015;125:149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images are more than pictures, they are data. Radiology 2016;278:563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parekh V, Jacobs MA. Radiomics: A new application from established techniques. Expert Rev Precis Med Drug Dev 2016;1:207–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parekh VS, Macura KJ, Harvey S, et al. Multiparametric deep learning tissue signatures for a radiological biomarker of breast cancer: Preliminary results. Available at: https://arxiv.org/abs/1802.08200. Accessed February 10, 2018. [DOI] [PMC free article] [PubMed]
- 11.Parekh VS, Jacobs MA. Integrated radiomic framework for breast cancer and tumor biology using advanced machine learning and multiparametric MRI. NPJ Breast Cancer 2017;3:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruzevick J, Kleinberg L, Rigamonti D. Imaging changes following stereotactic radiosurgery for metastatic intracranial tumors: Differentiating pseudoprogression from tumor progression and its effect on clinical practice. Neurosurg Rev 2014;37:193–201; [discussion 201]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiggenraad R, Bos P, Verbeek-de Kanter A, et al. Pseudo-progression after stereotactic radiotherapy of brain metastases: Lesion analysis using MRI cine-loops. J Neuro-Oncol 2014;119:437–443. [DOI] [PubMed] [Google Scholar]
- 14.Sundgren PC. MR spectroscopy in radiation injury. AJNR Am J Neuroradiol 2009;30:1469–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cicone F, Minniti G, Romano A, et al. Accuracy of F-DOPA PET and perfusion-MRI for differentiating radionecrotic from progressive brain metastases after radiosurgery. Eur J Nucl Med Mol Imaging 2015;42: 103–111. [DOI] [PubMed] [Google Scholar]
- 16.Galldiks N, Stoffels G, Filss CP, et al. Role ofO-(2-(18)F-fluoroethyl)- L-tyrosine PET for differentiation of local recurrent brain metastasis from radiation necrosis. J Nucl Med 2012;53:1367–1374. [DOI] [PubMed] [Google Scholar]
- 17.Galldiks N, Langen KJ, Pope WB. From the clinician’s point of view - What is the status quo of positron emission tomography in patients with brain tumors? Neuro Oncol 2015;17:1434–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larroza A, Moratal D, Paredes-Sanchez A, et al. Support vector machine classification of brain metastasis and radiation necrosis based on texture analysis in MRI. J Magn Reson Imaging 2015;42:1362–1368. [DOI] [PubMed] [Google Scholar]
- 19.Tiwari P, Prasanna P, Wolansky L, et al. Computer-extracted texture features to distinguish cerebral radionecrosis from recurrent brain tumors on multiparametric MRI: A feasibility study. AJNR Am J Neuroradiol 2016;37:2231–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Z, Yang J, Ho A, et al. A predictive model for distinguishing radiation necrosis from tumour progression after gamma knife radio-surgery based on radiomic features from MR images. Eur Radiol 2018;28:2255–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stockham AL, Tievsky AL, Koyfman SA, et al. Conventional MRI does not reliably distinguish radiation necrosis from tumor recurrence after stereotactic radiosurgery. J Neuro-Oncol 2012;109:149–158. [DOI] [PubMed] [Google Scholar]
- 22.Kano H, Kondziolka D, Lobato-Polo J, et al. T1/T2 matching to differentiate tumor growth from radiation effects after stereotactic radiosurgery. Neurosurgery 2010;66:486–491; [discussion 491–492]. [DOI] [PubMed] [Google Scholar]
- 23.Dequesada IM, Quisling RG, Yachnis A, et al. Can standard magnetic resonance imaging reliably distinguish recurrent tumor from radiation necrosis after radiosurgery for brain metastases? A radiographic-pathological study. Neurosurgery 2008;63:898–903; [discussion 904]. [DOI] [PubMed] [Google Scholar]
- 24.Parmar C, Grossmann P, Bussink J, et al. Machine learning methods for quantitative radiomic biomarkers. Sci Rep-UK 2015;5:13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalpathy-Cramer J, Freymann JB, Kirby JS, et al. Quantitative imaging network: Data sharing and competitive algorithmvalidation leveraging the cancer imaging archive. Transl Oncol 2014;7:147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shinohara RT, Sweeney EM, Goldsmith J, et al. Statistical normalization techniques for magnetic resonance imaging. Neuroimage Clin 2014;6:9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou M, Scott J, Chaudhury B, et al. Radiomics in brain tumor: Image assessment, quantitative feature descriptors, and machine-learning approaches. AJNR Am J Neuroradiol 2018;39:208–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu H, Caldwell C, Mah K, et al. Coregistered FDG PET/CT-based textural characterization of head and neck cancer for radiation treat-ment planning. IEEE Trans Med Imaging 2009;28:374–383. [DOI] [PubMed] [Google Scholar]
- 29.Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Just N Improving tumour heterogeneity MRI assessment with histograms. BrJCancer 2014;111:2205–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.King AD, Chow KK, Yu KH, et al. Head and neck squamous cell carcinoma: Diagnostic performance of diffusion-weighted MR imaging for the prediction of treatment response. Radiology 2013;266: 531–538. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


