Abstract
Progressive metastatic disease is a major cause of mortality for patients diagnosed with multiple types of solid tumors. One of the long-term goals of our laboratory is to identify molecular interactions that regulate metastasis, as a basis for developing agents that inhibit this process. Toward this goal, we recently demonstrated that intercellular adhesion molecule-2 (ICAM-2) converted neuroblastoma cells from a metastatic to a non-metastatic phenotype, a previously unknown function for ICAM-2. Interestingly, ICAM-2 suppressed metastatic but not tumorigenic potential in preclinical models, supporting a novel mechanism of regulating metastasis. We hypothesized that the effects of ICAM-2 on neuroblastoma cell phenotype depend on the interaction of ICAM-2 with the cytoskeletal linker protein α-actinin. The goal of the study presented here was to evaluate the impact of α-actinin binding to ICAM-2 on the phenotype of neuroblastoma tumor cells.
We used in silico approaches to examine the likelihood that the cytoplasmic domain of ICAM-2 binds directly to α-actinin. We then expressed variants of ICAM-2 with mutated α-actinin binding domains, and compared the impact of ICAM-2 and each variant on neuroblastoma cell adhesion, migration, anchorage-independent growth, co-precipitation with α-actinin, and production of localized and disseminated tumors in vivo.
The in vitro and in vivo characteristics of cells expressing ICAM-2 variants with modified α-actinin binding domains differed from cells expressing ICAM-2 wild type (WT) and also from cells that expressed no detectable ICAM-2. Like the WT protein, ICAM-2 variants inhibited cell adhesion, migration and colony growth in vitro. However, unlike the WT protein, ICAM-2 variants did not completely suppress development of disseminated neuroblastoma tumors in vivo. The data suggest the presence of α-actinin-dependent and α-actinin-independent mechanisms, and indicate that the interaction of ICAM-2 with α-actinin is critical to conferring an ICAM-2-mediated non-metastatic phenotype in neuroblastoma cells.
Keywords: ICAM-2, alpha-actinin, neuroblastoma, metastasis, in silico modeling, actin cytoskeleton, cell motility
Introduction
Intercellular adhesion molecule-2 (ICAM-2) is a transmembrane glycoprotein belonging to the immunoglobulin superfamily of cell adhesion molecules1,2. When expressed by vascular endothelial cells, the extracellular domain (ED) of ICAM-2 binds to β2-integrins on the surface of leukocytes as an initial step in immune responses3,4. When expressed by neovascular endothelium and leukocytes, the cytoplasmic domain (CD) of ICAM-2 binds to cytoskeletal linker proteins, but the functional significance of these interactions is not well understood. In tumor cells, we recently identified a unique role for ICAM-2 when we demonstrated that ICAM-2 conferred a non-metastatic phenotype in neuroblastoma (NB) cells5. SK-N-AS NB cells expressing ICAM-2 produced no tumors in an experimental metastasis model, while SK-N-AS cells expressing no detectable ICAM-2 produced disseminated tumors. Of note, the CD of ICAM-2 was essential to conferring a non-metastatic phenotype. The current study examines the hypothesis that the CD of ICAM-2 interacts with the cytoskeletal linker protein α-actinin and that this interaction is critical to the function of ICAM-2 in NB cells. We envision that identification of specific molecular interactions that regulate the metastatic process may provide a basis for development of anti-tumor agents that prevent or delay progressive metastatic disease in patients with solid tumors. We propose that the direct binding of the CD of ICAM-2 to α-actinin represents this type of regulatory interaction.
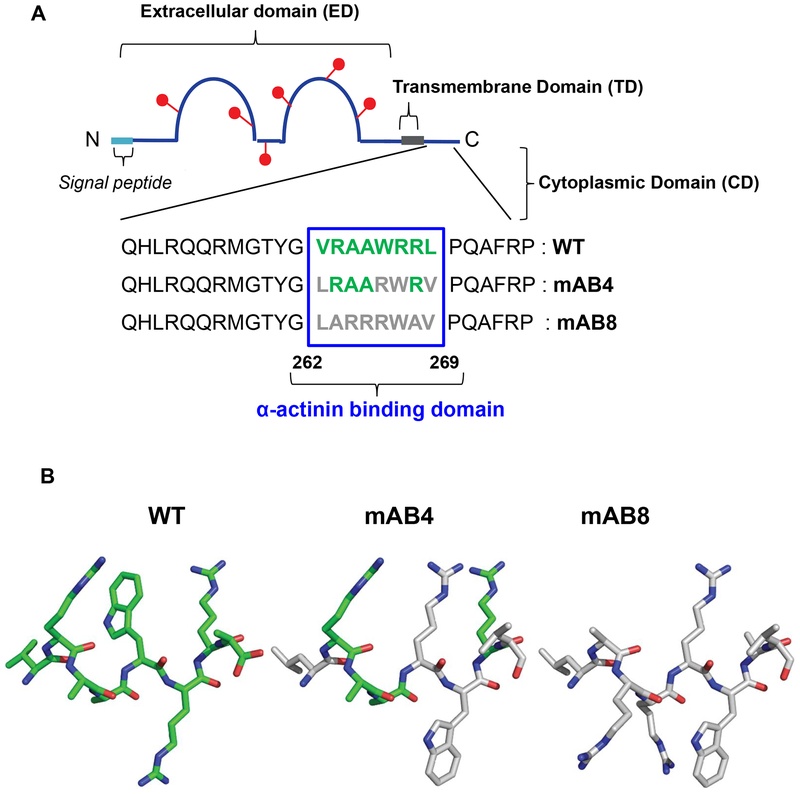
The sequence of ICAM-2 wild type (WT) expressed by normal cells6,7 and by NB cells is identical. The mature protein comprises a 202-amino acid N-terminus extracellular domain (ED) with two immunoglobulin-like loop structures, and 26-amino acid transmembrane (TD) and cytoplasmic domains (CD) (Figure 1). A 21-amino acid signal peptide routes the nascent protein through the endoplasmic reticulum, but is not present in the mature protein. Multiple proteins bind to the ED and to the CD of ICAM-28–13. These protein interactions are thought to mediate outside-in or inside-out signaling across the cell membrane, but the biological significance of this signaling remains to be elucidated. What is well established is that the actin network is a primary regulator of cell motility, and that actin cytoskeletal linker proteins that associate with ICAM-2 (ezrin, radixin, moesin, α-actinin)10,12 play roles in inducing or suppressing tumorigenic and/or metastatic properties of tumor cells14–17. Linker proteins are thought to “connect” actin to the cell membrane, by binding simultaneously to actin and to membrane-bound cell adhesion molecules. The hypothesis to be tested in this study is that ICAM-2 suppresses NB cell motility and therefore metastatic potential by binding to the actin cytoskeletal linker protein α-actinin. We used in silico modeling to predict the likelihood that the CD of ICAM-2 binds directly to α-actinin, and functional assays to compare the phenotypes of NB cells transfected to express ICAM-2 WT or variants with no detectable interaction with α-actinin (mAB4 and mAB8). These variants are not expressed in situ, but were expressed in transfected cells to determine whether variants that retain overall ICAM-2 WT structure but that do not associate with α-actinin conferred the same phenotype as the WT protein.
Figure 1. Schematic structure of ICAM-2 and ICAM-2 variants.
(A) Sequence of the cytoplasmic domains of endogenous human ICAM-2 wild type (WT) protein and variants mAB4 and mAB8. The extracellular domain (ED) of the protein contains two immunoglobulin-like domains with six N-glycosylation sites (red lollipops), a 26-amino acid transmembrane domain (TD) and a 26-amino acid cytoplasmic (CD) domain. The N-terminus of the nascent protein contains a 21-amino acid signal peptide (sp) that is not present in the 254 amino acid mature protein. (B) Structures of the 8-mer proposed α-actinin binding domains of ICAM-2 WT, mAB4 and mAB8 were generated using PDB IJ19 and PyMol43. The predicted structure of this domain of ICAM-2 WT is in green. The amino acid side chains of this domain of ICAM-2 mAB4 and mAB8 that have spatial orientations similar to those in the WT protein are also in green.
Results
We demonstrated previously that the CD of human ICAM-2, which contains a broadly defined α-actinin binding sequence12, was essential to conferring a non-metastatic phenotype in NB cells5. We now show a high likelihood that the CD of ICAM-2 binds to EF-hand domain of α-actinin and that this binding contributes to the phenotype of NB cells.
The three-dimensional (3D) structure of the α-actinin binding domain of ICAM-2 WT likely differs from those of variants mAB4 and mAB8.
We first modeled the α-actinin binding subdomain for ICAM-2 WT and variants mAB4 and mAB8, predicting that each variant would have minimal interaction with α-actinin (Figure 1A). The variants differed from ICAM-2 WT by having four (mAB4) or eight (mAB8) of the eight amino acids comprising the proposed α-actinin binding domain of ICAM-2 scrambled, to limit interaction with α-actinin. We then compared predicted structures for these forms of ICAM-2 in silico. Modeling was based on the known sequence and structure of the CD of murine ICAM-2, and accomplished by aligning amino acids 250–277 of the murine protein with amino acids 262–269 of human ICAM-2 WT, mAB4 and mAB8 (PDB 1J199). Resulting structures reflected a relative hydrophobic core (AAW) adjacent to hydrophilic amino acids (R) of ICAM-2 WT, compared to ICAM-2 mAB8 which contains a positively charged amino acid cluster (RRR) surrounded by hydrophobic amino acids (WA). ICAM-2 mAB4 contains a mixture of hydrophilic and hydrophobic (AAR) at the same positions. Structures demonstrated marked contrasts among the three constructs (Figure 1B). Readily apparent was divergent orientation of tryptophan (W) in the variant peptides, even though this amino acid was displaced by only one position in the variants. ICAM-2 WT appeared to contain “open” sites that might facilitate protein interactions. ICAM-2 mAB4 retained the side chain spatial configuration of four amino acids compared to ICAM-2 WT (similar orientations in green), while ICAM-2 mAB8 retained the spatial configuration of none of these side chains. These data predict that the structures and therefore the functions of the two variants differed from ICAM-2 WT, suggesting that the proposed constructs were suitable for comparing the function of ICAM-2 WT with variants that interacted inefficiently with α-actinin.
Modeling reveals a likely high-affinity interaction between the cytoplasmic domain of ICAM-2 and the EF-hand domain of α-actinin.
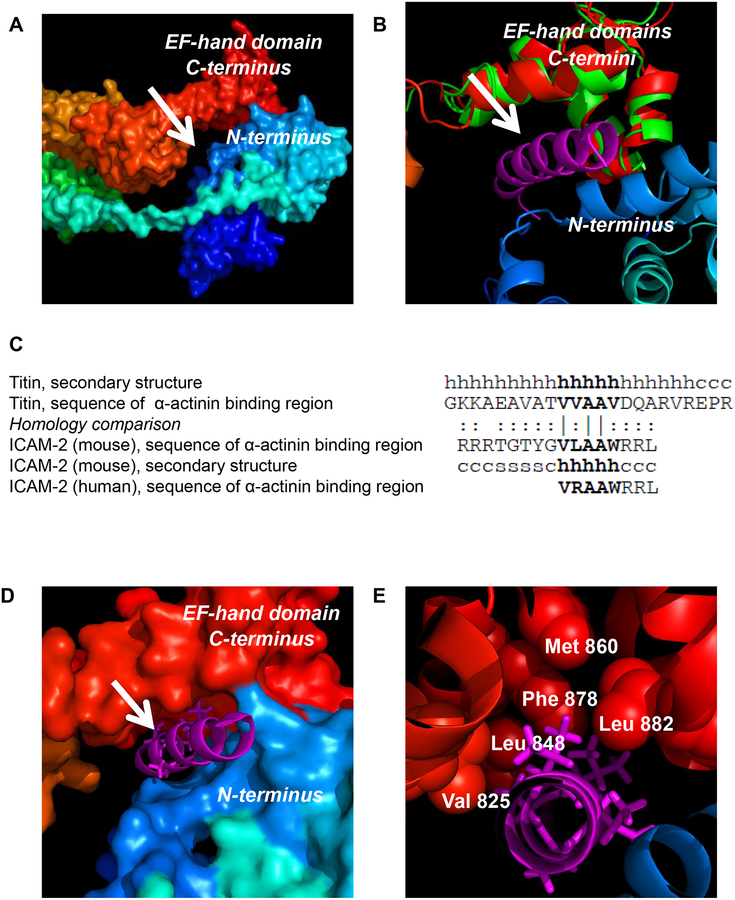
The region of α-actinin that binds to ICAM-2 has been only broadly defined12. To assess the likelihood that ICAM-2/α-actinin binding occurs in situ, it was necessary to identify a specific domain of α-actinin likely to interact with ICAM-2. We accomplished this using published structural data for full length chicken α-actinin (PDB 1SJJ18), the CD of murine ICAM-2 (PDB 1J199), and the co-structure of the EF-hand domain of human α-actinin with the α-actinin binding domain of rabbit titin (PDB 1H8B19). In a surface representation model of chicken α-actinin (PDB 1SJJ)(Figure 2A), the EF-hand domain in the C-terminus (red) and the actin binding domain (ABD) in the N-terminus (blue) are evident. Clearly visible is a potential binding cleft (arrow) formed by the EF-hand domain and ABD of the protein. Since chicken and human α-actinins are 87% identical18, we hypothesized that human α-actinin contains an analogous domain and that this domain is the site of interaction with human ICAM-2. Figure 2B shows the ribbon structure of the EF-hand domain of chicken α-actinin (red) and the ABD (blue). Aligned with the structure of chicken α-actinin is the EF-hand domain of human α-actinin (green). The structural alignment of these domains demonstrated an RMSD (root-mean-square deviation) of 1.47 Å, indicating a high-confidence alignment and a high likelihood that the 3D structure of this domain of human α-actinin is similar to the known structure of chicken α-actinin.
Figure 2. Modeling of ICAM-2 interaction with α-actinin.
(A) Surface representation of the full-length chicken α-actinin structure (PDB code 1SJJ) reveals a potential binding cavity (arrow) in the cleft between the actin binding domain (ABD, blue) and EF-hand (red) domains. The N-terminus to C-terminus is colored from blue (ABD) to red (EF-hand domain). (B) Structural alignment between full-length chicken α-actinin (red) and the EF-hand domain of human α-actinin (green)(PDB code 1H8B) demonstrated a high confidence alignment (RMSD of 1.47 Å). Structural alignment of the EF-hand domain of chicken α-actinin (red) with EF-hand domain of human α-actinin (residues 823–894, green) complexed to the α-actinin binding domain helix of rabbit titin (known structure, magenta) placed this helix in the putative binding pocket suggested by the structure in “A”. (C) The sequences of the α-actinin binding domains of rabbit titin, murine ICAM-2, and human ICAM-2 have high homology. The secondary structure of the rabbit and murine peptides are indicated as: “h” = helix, “s” = β-sheet and “c” = coil. Amino acid homology comparison is indicated as: “|” = identical; “:” = similar. (D) Modeling reveals a likely high-affinity interaction between the structurally conserved alpha-helix of titin (magenta) and the cytoplasmic domains of murine and human ICAM-2 (by structural and sequence homology) with the EF-hand domain of α-actinin (red). (E) Specific hydrophobic residues of the EF-hand domain of α-actinin and adjacent hydrophobic residues of a helical α-actinin binding structure suggest that ICAM-2 associates directly with α-actinin through hydrophobic interactions. The magenta stick structures represent the “VVAAV” component of the titin helix that is most conserved in ICAM-2.
Next, based on the known co-structure depicting the binding of rabbit titin with the EF-hand domain of human α-actinin19, we modeled the helical structure of titin (Figure 2B; arrow, residues 645–694) that interacts with α-actinin in its orientation relative to the EF-hand domain of human α-actinin. This procedure placed the helical α-actinin binding domain in the “binding pocket” of full-length α-actinin suggested by the structure in Figure 2A. Notably, the conserved sequence of the α-actinin binding domain of titin and the analogous domain of murine ICAM-2 (known structure) and human ICAM-2 (by sequence homology)(Figure 2C) suggested that the interaction of the CD of human ICAM-2 with α-actinin would be similar to the known binding of titin and α-actinin.
To further support this hypothesis, we aligned the surface structure of full-length chicken α-actinin with the α-actinin binding domain titin peptide (magenta) that is structurally conserved in murine ICAM-2 and human ICAM-2 (Figure 2D). This helical domain remained fitted into the proposed binding pocket of α-actinin. Specific residues of α-actinin (spheres, Figure 2E) adjacent to the helix comprising the α-actinin binding domain of titin include hydrophobic amino acids Val 825, Leu 848, Met 860, Phe 878, and Leu 882 of α-actinin. These residues comprise an available interactive domain for interaction with hydrophobic cores of α-actinin binding domains of titin and of murine and human ICAM-2.
Notably, the helical structure critical to the binding of titin with human α-actinin19 and, independently, of murine ICAM-2 with human α-actinin appears to be a conserved aspect of these interactions. Of particular relevance, the original study describing the interaction between titin Z-repeats Zr1 and Zr7 of this helical domain and the EF-hand domain of α-actinin reported a nanomolar binding affinity for the interaction19. Together, published data, Figures 1 and 2, and the conserved sequence of the CDs of murine and human ICAM-2 suggest a high likelihood that the CD of human ICAM-2 interacts with high affinity to EF-hand domain of α-actinin, and that the structures of ICAM-2 mAB4 and mAB8 differ from ICAM-2 WT and alter the interaction of these variants with α-actinin.
To then examine the impact of ICAM-2/α-actinin interaction on NB cell phenotype, we first documented expression and cell membrane localization of ICAM-2 WT, mAB4 and mAB8 since alterations of cytoplasmic domains have been reported to affect the subcellular localization of mutated proteins20–22.
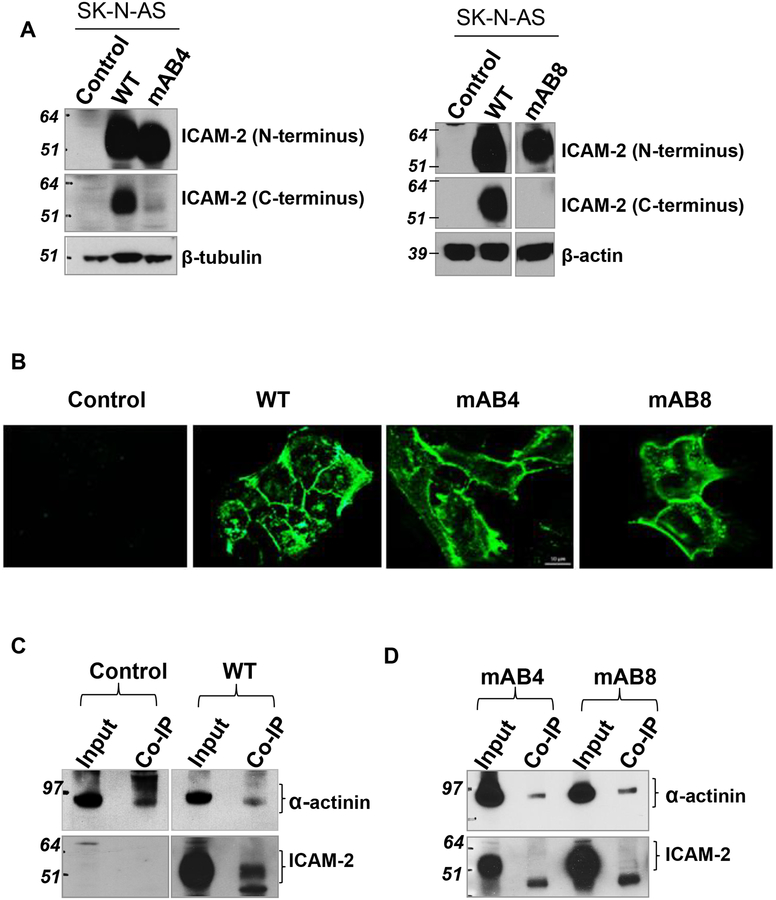
ICAM-2 localizes to cell membranes in transfected SK-N-AS NB cells.
The SK-N-AS cell line was derived from NB tumor cells in a bone marrow specimen obtained from a pediatric patient with Stage IV NB. This cell line is not MYCN-amplified and expresses no detectable endogenous ICAM-25. We transfected these cells to express ICAM-2 WT, mAB4 or mAB8. Immunoblots (Figure 3A) with antibodies to the extracellular N-terminus (AF244) or the intracellular C-terminus (sc-1512) of ICAM-2 confirmed its expression. Appropriately, only the antibody to the N-terminus of ICAM-2 detected ICAM-2 variants with scrambled CDs. Immunofluorescence (IF) analysis verified the cell membrane localization of ICAM-2 constructs (Figure 3B). Vector-transfected SK-N-AS cells (Control) showed no detectable ICAM-2 expression.
Figure 3. ICAM-2 WT and variants localized to cell membranes.
(A) Immunoblots confirmed that tranfected SK-N-AS cells expressed readily detectable levels of ICAM-2 WT, mAB4 or mAB8. Data in both membranes were generated in the same experiment, but immunoblotted individually. (B) Immunofluorescence staining demonstrated that ICAM-2 WT, mAB4 and mAB8 localized to cell membranes. Scale bar represents 10 μm for all panels. Details of procedures used are in the Methods. (C, D) ICAM-2 WT, but not mAB4 and mAB8, co-precipitated with α-actinin. Immunoprecipitations (IP) were performed using whole cell lysates and a mouse monoclonal antibody to α-actinin (MAB1682, Millipore-Fisher Scientific). The presence of all three types of ICAM-2 proteins in the “input” whole cell lysates (wcl) was confirmed prior to IP.
ICAM-2 WT co-precipitated w ith α-actinin.
We then used co-immunoprecipitation to address whether ICAM-2 with a partially (mAB4) or fully scrambled (mAB8) α-actinin binding domain associated with α-actinin. We immunoprecipitated (IP) α-actinin and immunoblotted (IB) for ICAM-2. IP/IB results for positive (WT) and negative (Control) controls (Figure 3C) demonstrated that ICAM-2 WT associated with α-actinin. In contrast, IP/IB showed no detectable association of ICAM-2 mAB4 or mAB8 with α-actinin (Figure 3D). The data indicate that rearrangement of amino acids 262–269 of the WT protein minimized ICAM-2/α-actinin interaction. We next evaluated the effects of ICAM-2 WT, mAB4 and mAB8 on cell phenotype.
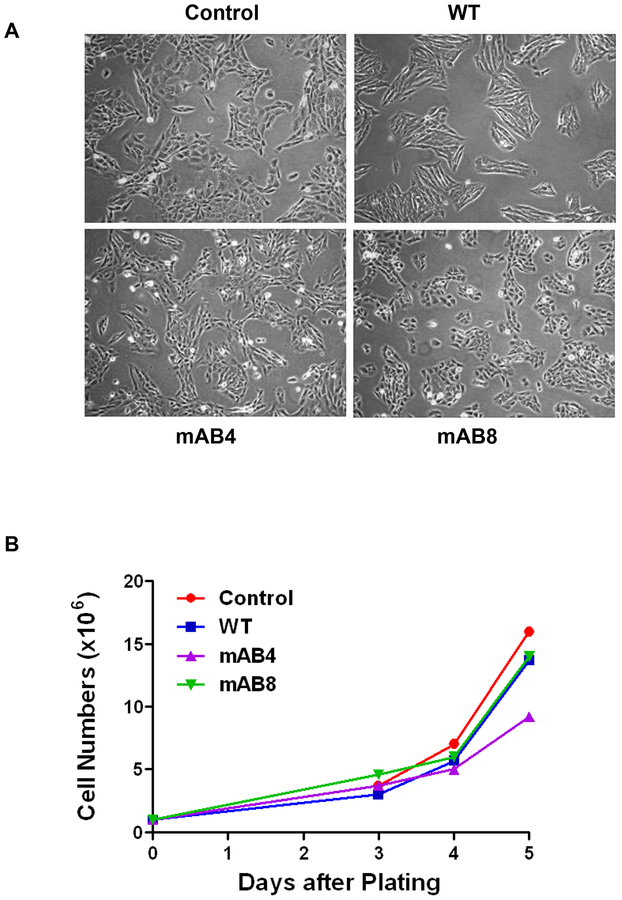
ICAM-2 expression did not affect cell morphology or doubling time.
Photomicrographs (Figure 4A) demonstrated that cell lines expressing ICAM-2 WT, mAB4 or mAB8 variants grew in vitro as attached monolayers, had similar nascent neuronal morphology, were similar in size when detached (assessed by Coulter Counter sizing, not shown), and had equivalent doubling times (30.4 ± 4.7 hours)(Figure 4B). All transfectants grew as tight cell groups. Groupings of ICAM-2 WT transfectants appeared more cohesive and angular than Control transfectants, as is sometimes characteristic of less aggressive phenotypes23.
Figure 4. Morphologies and growth rates of SK-N-AS cells transfected to express ICAM-2 WT, mAB4 or mAB8 are equivalent.
(A) All transfectants grew as attached monolayers and had nascent neuronal morphologies; all four transfectants grew as tight cell groups but groupings of cells transfected to express ICAM-2 WT appeared more cohesive and more angular than Control transfectants. (B) Cell proliferation assays showed that transfectancts had similar cell doubling times of 30.4 ± 4.7 hours.
ICAM-2 inhibited NB cell adhesion to extracellular matrix (ECM) proteins, independent of interaction with α-actinin.
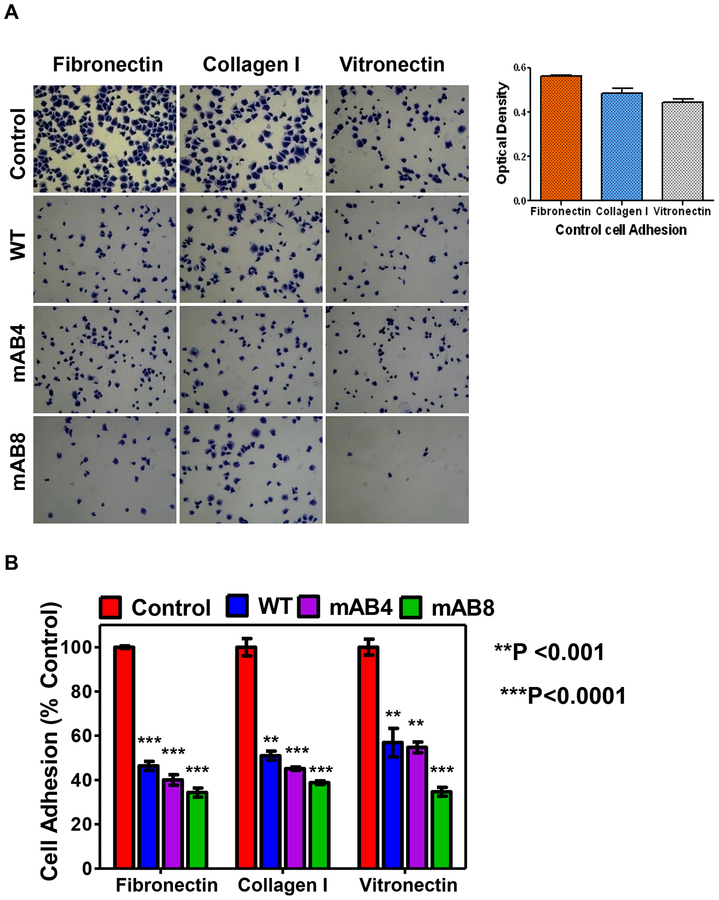
Adhesive and migratory properties are critical determinants of the metastatic potential of tumor cells23,24, with both processes involving a myriad of intracellular and extracellular proteins. Initial Matrigel invasion assays (Figure S1) indicated that extracellular matrix proteins (ECM) might participate in the phenotype conferred by ICAM-2. To identify specific ECM proteins involved, we performed adhesion assays with fibronectin, collagen I or vitronectin. Control transfectants adhered equally well to all three ECM proteins (Figure 5), and ICAM-2 WT, mAB4 and mAB8 inhibited adhesion to each of these proteins. Inhibition did not depend on ICAM-2/α-actinin interaction, as both variants produced results equivalent to the WT protein.
Figure 5. ICAM-2 WT and mAB variants inhibit NB cell adhesion to ECM proteins.
(A) NB cells adherent to each ECM protein were imaged after staining. The optical density for Control cells (used in calculations and presented as 100%) were for fibronectin=0.5630 ± 0.0032, for collagen I=0.4855 ± 0.019, and for vitronectin=0.4443 ± 0.0160 (mean ± SEM), as shown in right panel. (B) NB cell adhesion was quantitated by determination of the optical density of crystal violet-stained, adherent cells by standard methods44. Values for ICAM-2 transfectants were significantly different from Control transfectants (**P<0.001 or ***P<0.0001, as indicated).
ICAM-2 WT, mAB4 and mAB8 inhibited tum or cell motility in vitro and grow thin soft agar, in an α-actinin-dependent manner.
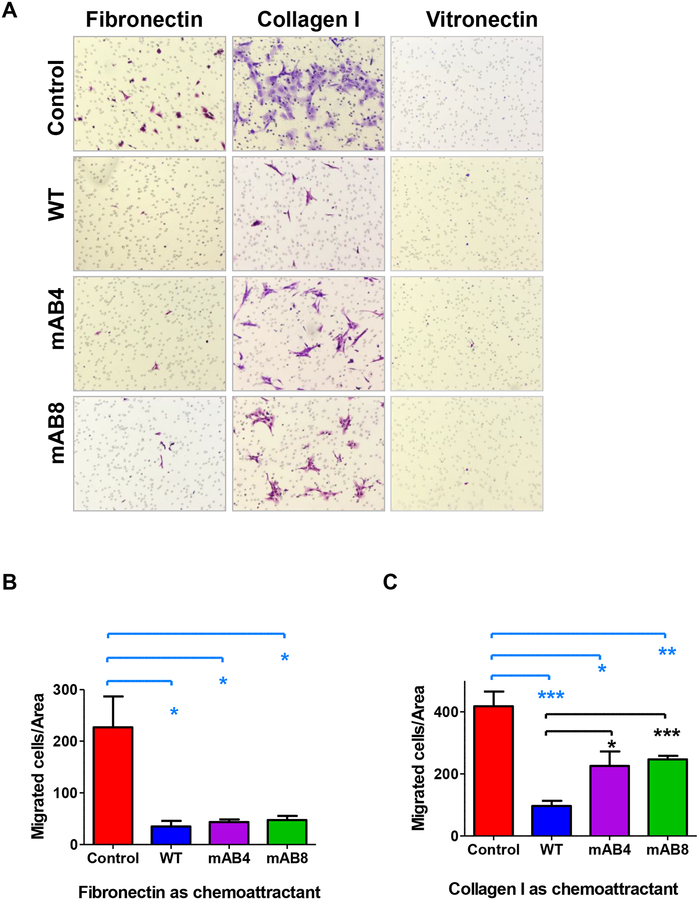
We then used Boyden migration assays to evaluate whether fibronectin, collagen I or vitronectin impacted migration of NB cells expressing ICAM-2. Collagen I was the strongest chemoattractant of the ECM proteins (Figure 6). Little migration was seen with vitronectin. All three ICAM-2 constructs inhibited migration mediated by fibronectin, indicating that this inhibition was independent of α-actinin. In contrast, ICAM-2 WT inihibited migration toward collagen I in an α-actinin-dependent manner. When collagen I was used as chemoattractant, ICAM-2 mAB4 and mAB8 conferred a phenotype intermediate to Control and ICAM-2 WT. Inclusion of antibody CBR IC2/23 (5 μg/mL), which inhibits extracellular binding of ICAM-2 to β2-integrin, had no effect on migration (data not shown). The data are consistent with the conclusion that ICAM-2/α-actinin interaction contributes to ICAM-2-mediated migratory properties of NB cells, in the presence of specific ECM proteins.
Figure 6. ICAM-2 WT and mAB variants inhibit NB cell motility in Boyden chamber migration assays.
(A) Transfected NB cells that migrated to the distal side of Boyden chamber membranes were imaged after an 18-hour incubation. Representative images were acquired with a Zeiss Axio Observer Z.1 microscope and Zen 2011 Blue software. (B) Migrated cell numbers were determined manually, and analyzed by t-test. ICAM-2 WT, mAB4 and mAB8 inhibited cell motility toward fibronectin compared to cells that expressed no detectable ICAM-2 (Control) (P<0.05*), in an α-actinin-independent manner. (C) ICAM-2 WT, mAB4 and mAB8 inhibited cell migration using collagen I as chemoattractant in an α-actinin-dependent manner (*P=0.0365; **P=0.0018; ***P<0.0001; *P=0.0155; ***P<0.0001).
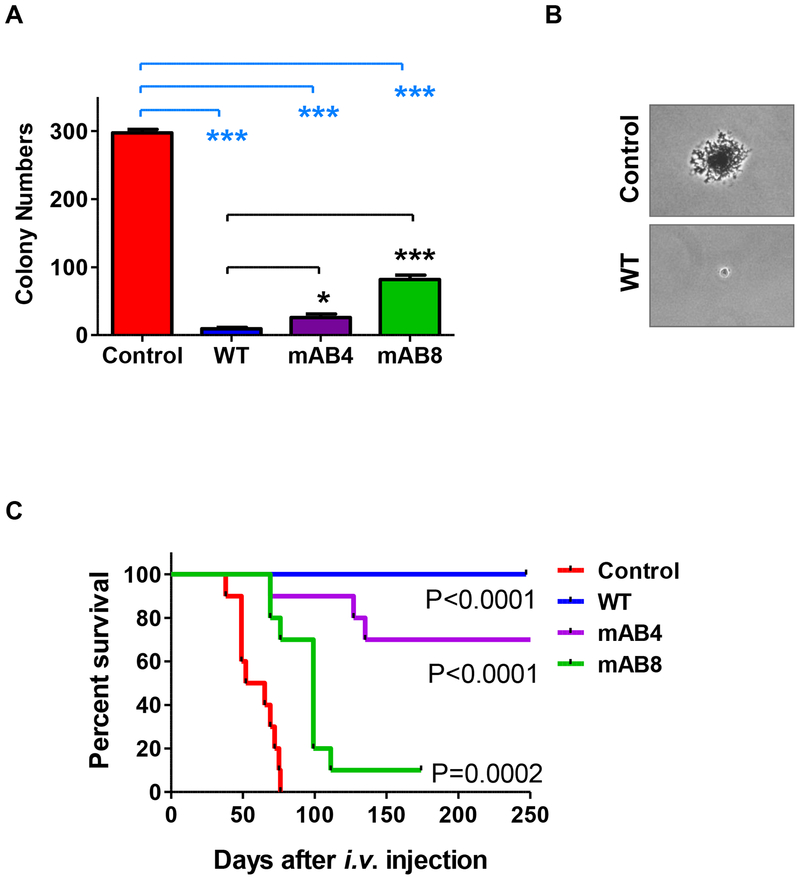
ICAM-2 WT also inhibited anchorage-independent growth. As previously published, SKN-AS cells expressing ICAM-2 WT formed smaller and fewer colonies in soft agar (9.3 ± 2.0) than control transfectants (297.2 ± 5.2 colonies) (P<0.0001) (Figures 7A,B)25. Cells expressing mAB4 and mAB8 variants also produced fewer colonies than Control cells (P<0.0001), but more than cells expressing ICAM-2 WT (P=0.00135 and P<0.0001). ICAM-2 variants conferred a partial phenotype that was, like migration, affected by the association of ICAM-2 with α-actinin. To confirm that results seen were not unique to a single NB cell line, we also performed anchorage-independent growth assays with Control and ICAM-2 WT transfectants of NB-1691 cells (Figure S2A). Unlike SK-N-AS cells NB-1691 cells have an amplified MYCN gene copy number, an unfavorable prognostic indicator clinically. As was seen with SK-N-AS cells, ICAM-2 inhibited the growth in soft agar of MYCN-amplified NB-1691 cells transfected to upregulate expression of ICAM-2; ICAM-2 was functional in MYCN-amplified (NB-1691) and -non-amplified (SK-N-AS) NB cells.
Figure 7. ICAM-2 inhibited colony growth in soft agar and development of disseminated tumors in vivo. ICAM-2 mAB8 delayed but did not inhibit development of disseminated tumors in an in vivo model of metastatic neuroblastoma.
(A) ICAM-2 WT, mAB4 and mAB8 suppressed anchorage-independent growth in vitro. Colonies of >20 cells were visualized 14–21 days after plating. Results were quantitated manually and analyzed using a two-tailed t-test and GraphPad Prism software (*P=0.0135 and ***P<0.0001). (B) Photomicrographs of representative colonies for Control and ICAM-2 WT transfectants were acquired at 400× magnification using a Dr-5 digital camera (Southern Microscope) mounted to a Nikon Eclipse TS100 inverted microscope and archived with TSView software (version 7.1.04, Tucsen Imaging Technology Co./Southern Microscope). (C) Kaplan-Meier survival plots of mice that received i.v. injections of SK-N-AS cell transfectants were analyzed by log-rank (Mantel-Cox) test using GraphPad Prism 5 software (Version 5.02) showed that mice receiving cells expressing mAB variants survived longer than mice receiving cells expressing no detectable ICAM-2, but not as long as mice receiving cells expressing ICAM-2 WT. *Data for control transfectants were previously published in BMC Cancer by BioMed Central25. Reprinted with permission.
Notably, the differences observed in anchorage-independent growth in vitro can reflect differences in tumorigenic or metastatic potential26–27. Therefore, we evaluated production of localized (tumorigenic potential) and disseminated (metastatic potential) tumors in vivo.
ICAM-2 did not affect development of subcutaneous tumors (tumorigenic potential) by NB cells.
We injected 5,000 or 50,000 or 500,000 Control or ICAM-WT transfectants subcutaneously (s.c.) into SCID mice and observed the mice for tumor formation. When 500,000 cells were implanted, the two transfectants produced tumors of equivalent size 45 ± 7 days following implantation (Figure S3). However, neither transfectant produced tumors within the >2-month duration of the experiment when 5,000 or 50,000 cells were implanted. We detected no impact of ICAM-2 on the tumorigenicity of SK-N-AS NB cells.
ICAM-2 WT suppressed development of disseminated tumors in vivo (metastatic potential of NB cells). ICAM-2 mAB4 and mAB8 delayed, but did not completely suppress development of disseminated tumors.
We demonstrated previously that ICAM-2 WT confers a non-metastatic phenotype in an in vivo model of disseminated NB. Kaplan-Meier survival plots demonstrated that mice receiving Control cells intravenously (i.v.) (Figure 7C) developed tumors at multiple anatomic sites, requiring euthanasia within 1–3 months25. Mice receiving ICAM-2 WT transfectants developed no detectable tumors and survived significantly longer (P<0.0001 compared to Control). Tofirst demonstrate that this function of ICAM-2 was not unique to a single cell line, we used the same model system to evaluate the function of ICAM-2 WT in NB-1691 cells. As was seen with SK-N-AS cells, upregulation of ICAM-2 suppressed development of disseminated tumors in SCID mice receiving i.v. injections of NB-1691 cell transfected to upregulate expression of ICAM-2 (P<0.02)(Figure S2C).
To then determine the extent to which ICAM-2/α-actinin interaction affected the in vivo phenotype of NB cells, we injected i.v. SK-N-AS cells transfected to express ICAM-2 mAB4 or mAB8, and observed the mice for tumor development. Expression of ICAM-2 variants delayed but did not prevent development of disseminated tumors in vivo (Figure 7C). Disseminated tumors developed in three mice receiving cells expressing the mAB4 variant and in nine mice receiving cells expressing the mAB8 variant. Survival data showed that mice receiving cells expressing ICAM-2 WT, mAB4 and mAB8 survived longer (P<0.0002) than mice receiving Control transfectants, and that mice receiving cells expressing ICAM-2 WT survived longer (P<0.0001) than mice receiving cells expressing ICAM-2 mAB4 or mAB8 variants. Mutation of the α-actinin binding domain of ICAM-2 attenuated the ability of ICAM-2 to suppress metastatic properties of NB cells. In vitro and in vivo data indicate that the α-actinin binding domain of ICAM-2 contributes significantly to the function of ICAM-2 in NB cells. Taken together, the data suggest that ICAM-2 functions as a suppressor of metastasis rather than of tumorigenesis in NB cells.
Discussion
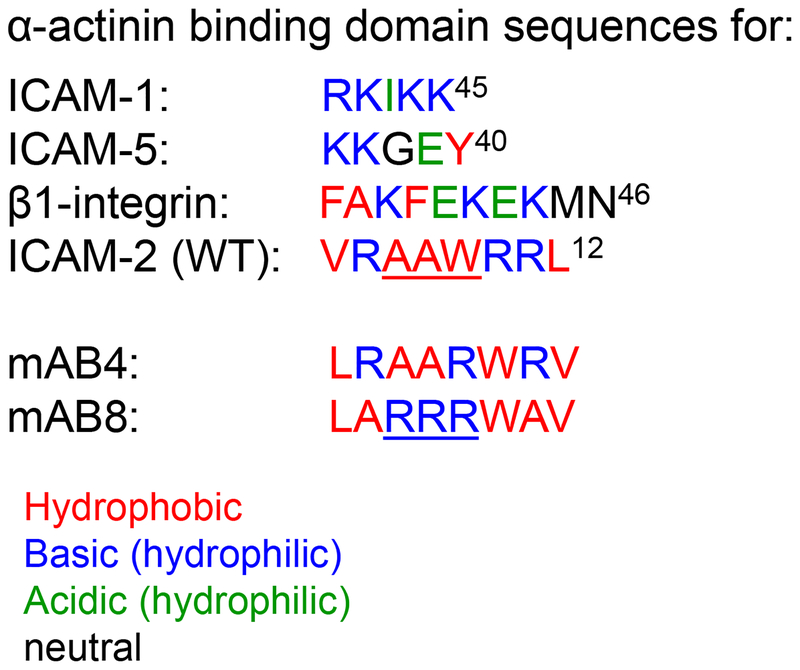
Development of metastatic disease is associated with a high mortality for patients diagnosed with solid tumors. The long-term goal of our laboratory is to identify molecular interactions critical to the metastatic process, with an end goal of designing therapeutic agents that prevent metastatic progression. Our earlier work5,25 demonstrated that ICAM-2 completely suppressed development of disseminated NB tumors in an experimental metastasis model, and that the CD of ICAM-2 was essential for this function. The current study determined that direct binding of ICAM-2 to α-actinin is essential to suppress the metastatic potential of these tumor cells. Together, in silico, in vitro and in vivo data indicate a high likelihood that the intracellular C-terminus of ICAM-2 interacts directly with the EF-hand domain of α-actinin to regulate tumor cell migration mediated by collagen I, anchorage-independent growth, and development of disseminated tumors in vivo. These studies are the first to model the interaction of α-actinin with a membrane-bound CAM. They predict that ICAM-2 interacts with α-actinin predominantly through hydrophobic residues (Figure 2E), in contrast to the hydrophilic or neutral interactions reported for α-actinin with ICAM-1, ICAM-5 or β1-integrin (Figure 8). The data also suggest a second, α-actinin-independent mechanism by which ICAM-2 inhibits adhesion to or migration mediated by specific ECM proteins. Neither mechanism for ICAM-2 has been reported previously.
Figure 8. Disparate α-actinin binding sequences in proteins belonging to two families of cell adhesion molecules: ICAM-145, ICAM-212, ICAM-540 and β1-integrin46.
Residues are indicated as hydrophobic (red); basic hydrophilic (blue); acidic hydrophilic (green), and neutral (black).
The most well-studied functions for ICAM-2 are immune-related: ICAM-2 enhanced the immune response of transfected colon carcinoma cells28; induced an anti-tumor immune response in pancreatic carcinogenesis29; and contributed to cytolysis of human pancreatic cancer cells by human γδ-T cells30. ICAM-2 also rendered transfected Jurkat T cells less sensitive to the cytotoxicity of staurosporine and anti-Fas antibody31; but did not change the sensitivity of NB cells to doxorubicin, vincristine, or etoposide (KJY, unpublished data). We propose that ICAM-2 has a function in NB cells that is distinct from functions previously described for this protein. Unpublished data from our laboratory indicate that ICAM-2 may have functions similar to those in NB in other types of solid tumors, including pancreatic and breast cancer cells.
While the data strongly suggest α-actinin-dependent and -independent mechanisms, some limited (undetectable) interaction may occur between the ICAM-2 variants and α-actinin to suppress adhesion to fibronectin and collagen I and to confer partial phenotypes in growth in soft agar and migration assays and in disseminated tumor production in vivo. The data are also consistent with multiple mechanistic possibilities. Firstly, intracellular ICAM-2/α-actinin binding may induce conformational changes in the extracellular domain of the same ICAM-2 molecule to inhibit or facilitate cellular interactions with ECM proteins, a type of inside-out signaling as has been reported for β1- and β2-integrin and ICAM-132–34. Secondly, effects of ICAM-2 on NB cell adhesion and migration be more indirect: Although immunoblots show no effect of ICAM-2 on level of expression of α5-, α2- or β1-integrin proteins in NB cells (data not shown), ICAM-2 may affect the function of these components of binding partners of fibronectin [α5β1] and collagen I [α2β1] (Figures 5 and 6). Thirdly, unpublished RNA microarray data (not shown) demonstrate that ICAM-2 upregulated expression of at least three phosphatases (~3- to 7-fold) that dephosphorylate Focal Adhesion Kinase, Src, and Rac and Rho GTPases, each of which plays a critical role in tumor cell motility. Less likely is the possibility that ICAM-2 induces tumor cell differentiation or de-differentiation, as RT-PCR data (Figure S4) and immunoblot data (not shown) reveal no differences in expression of EMT (E-cadherin, Occludin, Vimentin, Snail 1) or sternness (c-Myc) markers. Morphology (Figure 4) and tumorigenicity data (Figure S3) are consistent with this observation. The data indicate that ICAM-2 inhibits adhesive and migratory properties of tumor cells by two distinct mechansims, but is unlikely to affect NB cell differentiation or tumorigenicity. Further, ICAM-2/α-actinin binding is essential to the function of ICAM-2 in NB cells.
Materials and Methods
Cell lines
The parent human neuroblastoma cell line SK-N-AS (American Type Culture Collection, Manassas, VA) and transfectants were maintained in Dulbecco’s Modified Eagle Medium (Hyclone, Fisher Scientific, Savannah, GA) with 10% FBS (Atlanta Biologicals, Lawrenceville, GA) and 2 mM L-glutamine (Fisher Scientific, Savannah, GA, USA) at 37°C, 10% CO2.
Molecular models of ICAM-2/α-actinin binding
Modeling studies were based on published structural data (Protein Data Base [PDB1ZXQ and PDB1J19 for ICAM-2; PDB1HCI, PDB1SJJ, and PDB1H8B for α-actinin]). Molecular Dynamics simulations using AMBER 99 force field distribution was used to assess the flexibility35 of energy minimized structures. Overall structural quality was evaluated with PROCHECK36, Verify3Dimensional37 and MolProbity servers. Structures were evaluated by Ramachandran analysis, rotamer analysis, bond length, angle geometry, and clash analysis. Structures having Molprobity scores of 50% or better were deemed of sufficient quality for docking studies, as templates for protein modeling (Global Range Molecular Matching)38, and for evaluating compensatory surface changes.
ICAM-2 variants
The plasmid encoding human ICAM-2 and transfectants were generated as published5. Overlapping PCR using Pfu polymerase (QuickChange mutagenesis kit; Agilent Tech, Wilmington, DE, USA) and pIRES.ICAM-2 was performed to scramble 4/8 or 8/8 amino acids comprising the proposed α-actinin binding site of ICAM-2 WT (VRAAWRRL)(Figure 1). In variant mAB4, VRAAWRRL was replaced with LRAARWRV; in variant mAB8 VRAAWRRL was replaced with LARRRWAV. The sequence of plasmids encoding variants of ICAM-2 was confirmed by Sanger sequencing (St. Jude Hartwell Center, Memphis, TN, USA). In the Figures, transfected cells are referred to by the form of ICAM-2 expressed: WT, mAB4 or mAB8. Cells transfected with empty vector, SK-N-ASpIRESneo2, are labeled in the Figures as “Control”.
Cell doubling time
Cells (1×106) were plated in 75 cm2 flasks, and cell numbers determined by hemocytometer. Doubling times were calculated using www.doubling-time.com/compute.php39.
Anchorage-independent growth
Immunoprecipitations
Co-immunoprecipitations (IP) were done by standard methods as described previously5,25. The antibody for IP of α-actinin was MAB1682 (Fisher Scientific). Antibodies for IB of ICAM-2, α-actinin, and actin were AF244 (R&D Systems, Minneapolis, MN, USA) and sc-1512 (Santa Cruz Biotech, Santa Cruz, CA, USA), sc-7454R (Santa Cruz), 4968 (Cell Signaling Technology, Danvers, MA, USA), and A5316 (Sigma, St. Louis, MO, USA), respectively. AF244 recognizes the extracellular N-terminus of ICAM-2. Sc-1512 recognizes the intracellular C-terminus of ICAM-2.
Immunofluorescence (IF) staining
IF staining was performed using standard methods12,40 to visualize the distribution of ICAM-2 in situ. Plated cells were washed and fixed with cold methanol. Nonspecific binding was blocked 10% goat serum (Sigma). The antibody for ICAM-2 was CBR IC2/2 (Santa Cruz), diluted 1:200 in 1% goat serum. The secondary antibody was Alexa Fluor® goat anti-mouse 488 (Life Tech, Grand Island, NY, USA), diluted 1:500 in PBS. Image acquisition was accomplished with a Zeiss Axio Observer Z. 1 microscope with Apotome2 structured illumination slider and Zen 2011 Blue software (Carl Zeiss Microscopy, Thornwood, NY, USA). Optimal exposure time was determined with unstained controls, and pixel saturation was avoided. Images were initially acquired in the Zeiss.czi proprietary format, each channel having a grayscale dynamic range of 14-bits, and then pseudocolored. Files were exported to 8-bit TIFF for contrast/brightness adjustment. Scale bars were added by Zen Blue.
Morphology
Photomicrographs were acquired at 200× magnification using a Dr-5 digital camera (Southern Microscope, Haw River, NC, USA) mounted to a Nikon Eclipse TS100 inverted microscope Nikon Instruments Inc., Melville, NY, USA) and archived with TSView software (version 7.1.04, Tucsen Imaging Technology Co./Southern Microscope).
Boyden chamber migration
Neuroblastoma cells were tryspsinized and suspended in serum-free DMEM at 3 × 105 cells/mL. 100 μL of cell suspension added to the upper chamber of 6.5 mm Transwell® permeable supports (8 μm pore size). If included, CBR IC2/2 antibody was added to the cell suspension at 5 μg/mL. Lower chambers contained serum-free DMEM with 10 μg/mL of human collagen I (BD Biosciences), plasma fibronectin (BD Biosciences), or plasma vitronectin (Promega). After an 18h incubation, cells adherent to the membrane were fixed with 4% formaldehyde and stained with 0.02% crystal violet. Cells remaining on the apical side of the membrane were removed with cotton-tipped applicators, and photomicrographs of migratory cells attached to basal side of membrane were acquired and quantified manually. Image acquisition was with a Zeiss Axio Observer Z.1 microscope and Zen 2011 Blue software.
Adhesion
96 well plates (Corning) were coated for 2 hours at room temperature with 10 μg/mL human collagen I (BD Biosciences), plasma fibronectin (BD Biosciences), or plasma vitronectin (Promega). After 2 hours, ECM protein solutions were aspirated, wells washed twice with PBS, and cells incubated with 2% BSA for 30 min at 37°C to limit non-specific binding. Wells were then washed with serum-free DMEM, and each well plated with 100 μL of NB cells suspended in serum-free DMEM at 3 × 105 cells/mL. Plates were centrifuged at 10 × g for 1 minute and incubated at 37°C, 10% CO2 for 20 minutes. Non-adherent cells were removed with PBS and gentle agitation. Adherent cells were fixed with 4% formaldehyde, stained with 0.02% crystal violet, and photomicrographs acquired. Absorbances were recorded with a VersaMax microplate reader at 590nm (Molecular Devices).
Mouse model of metastatic neuroblastoma
This method has been reported in detail5,25. Briefly, female Es1e/SCID mice (6–8 weeks old), obtained from Dr. Phil Potter41, were housed in AAALAC accredited vivariums at St. Jude Children’s Research Hospital Animal Resources Center (Memphis, TN) or the University of Alabama at Birmingham (Birmingham, AL). Protocols were approved by the St. Jude or UAB Animal Care and Use Committee, respectively. Following i.v. injection of tumor cells, mice (10/group) were monitored daily and euthanized immediately if distress or discomfort was evident. Day of euthanasia was recorded as the day of death. Following s.c. injections, mice (3/group) were euthanized when tumors reached a volume of 1,000–1,500mm3, calculated by standard methods42.
Statistical analyses
Statistical analyses were performed and graphs generated using GraphPad Prism software 5 (GraphPad, San Diego, CA, USA). Each type of assay was done a minimum of three times, with a minimum of three replicates per cell line. Kaplan Meier survival data were analyzed using a log-rank (Mantel-Haenszel).
Supplementary Material
Acknowledgements
We are indebted to Dr. Mary Danks for her support. We thank Dr. Phil Potter for allowing us to use the mouse strain developed in his laboratory. We are also grateful to Joanna Remack and Rebecca Bush for outstanding technical assistance. This work was supported by the University of Alabama at Birmingham-Comprehensive Cancer Center- YSB-New Faculty Development Award (YSB-NFDA: P30 CA013148).
Abbreviations
- ICAM-2
Intercellular adhesion molecule-2
- NB
neuroblastoma
- ED
extracellular domain
- sp
signal peptide
- TD
transmembrane domain
- CD
cytoplasmic domain
- CAM
cell adhesion molecules
- IP
immunoprecipitation
- WT
wild type
- IB
immunoblot
- 3D
3-dimensional
- mAB4
ICAM-2 with partially scrambled α-actinin binding domain
- mAB8
ICAM-2 with fully scrambled α-actinin binding domain
- RMSD
root-mean-square deviation
- s.c.
subcutaneous
- i.v.
intravenous
Footnotes
Conflict of interest.
The authors declare no conflict of interest.
References
- 1.Casasnovas JM, Springer TA, Liu JH, Harrison SC, Wang JH. Crystal structure of ICAM-2 reveals a distinctive integrin recognition surface. Nature 1997; 387: 312–315. [DOI] [PubMed] [Google Scholar]
- 2.Nortamo P, Li R, Renkonen R, Timonen T, Prieto J, Patarroyo M et al. The expression of human intercellular adhesion molecule-2 is refractory to inflammatory cytokines. Eur J Immunol 1991; 21: 2629–2632. [DOI] [PubMed] [Google Scholar]
- 3.de Fougerolles AR, Stacker SA, Schwarting R, Springer TA. Characterization of ICAM-2 and evidence for a third counter-receptor for LFA-1. J Exp Med 1991; 174: 253–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kotovuori A, Pessa-Morikawa T, Kotovuori P, Nortamo P, Gahmberg CG. ICAM-2 and a peptide from its binding domain are efficient activators of leukocyte adhesion and integrin affinity. J Immunol 1999; 162: 6613–6620. [PubMed] [Google Scholar]
- 5.Yoon KJ, Phelps DA, Bush RA, Remack JS, Billups CA, Khoury JD. ICAM-2 expression mediates a membrane-actin link, confers a nonmetastatic phenotype and reflects favorable tumor stage or histology in neuroblastoma. PLoS One 2008; 3: e3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Staunton DE, Dustin ML, Springer TA. Functional cloning of ICAM-2, a cell adhesion ligand for LFA-1 homologous to ICAM-1. Nature 1989; 339: 61–64. [DOI] [PubMed] [Google Scholar]
- 7.Huang MT, Mason JC, Birdsey GM, Amsellem V, Gerwin N, Haskard DO et al. Endothelial intercellular adhesion molecule (ICAM)-2 regulates angiogenesis. Blood 2005; 106: 1636–1643. [DOI] [PubMed] [Google Scholar]
- 8.Diacovo TG, deFougerolles AR, Bainton DF, Springer TA. A functional integrin ligand on the surface of platelets: intercellular adhesion molecule-2. Journal of Clinical Investigation 1994; 94: 1243–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamada K, Shimizu T, Yonemura S, Tsukita S, Hakoshima T. Structural basis of adhesion-molecule recognition by ERM proteins revealed by the crystal structure of the radixin-ICAM-2 complex. EMBO J 2003; 22: 502–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yonemura S, Hirao M, Doi Y, Takahashi N, Kondo T, Tsukita S et al. Ezrin/radixin/moesin (ERM) proteins bind to a positively charged amino acid cluster in the juxta-membrane cytoplasmic domain of CD44, CD43, and ICAM-2. J Cell Biol 1998; 140: 885–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heiska L, Alfthan K, Gronholm M, Vilja P, Vaheri A, Carpen O. Association of ezrin with intercellular adhesion molecule-1 and −2 (ICAM-1 and iCaM-2). Regulation by phosphatidylinositol 4, 5-bisphosphate. J Biol Chem 1998; 273: 21893–21900. [DOI] [PubMed] [Google Scholar]
- 12.Heiska L, Kantor C, Parr T, Critchley DR, Vilja P, Gahmberg CG et al. Binding of the cytoplasmic domain of intercellular adhesion molecule-2 (ICAM-2) to alpha-actinin. J Biol Chem 1996; 271: 26214–26219. [DOI] [PubMed] [Google Scholar]
- 13.Geijtenbeek TB, Krooshoop DJ, Bleijs DA, van Vliet SJ, van Duijnhoven GC, Grabovsky V et al. DC-SIGN-ICAM-2 interaction mediates dendritic cell trafficking. Nat Immunol 2000; 1: 353–357. [DOI] [PubMed] [Google Scholar]
- 14.Honda K, Yamada T, Endo R, Ino Y, Gotoh M, Tsuda H et al. Actinin-4, a novel actin-bundling protein associated with cell motility and cancer invasion. J Cell Biol 1998; 140: 1383–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Honda K, Yamada T, Hayashida Y, Idogawa M, Sato S, Hasegawa F et al. Actinin-4 increases cell motility and promotes lymph node metastasis of colorectal cancer. Gastroenterology 2005; 128: 51–62. [DOI] [PubMed] [Google Scholar]
- 16.Sun CX, Robb VA, Gutmann DH. Protein 4.1 tumor suppressors: getting a FERM grip on growth regulation. J Cell Science 2002; 115: 3991–4000. [DOI] [PubMed] [Google Scholar]
- 17.Otey CA, Carpen O. Alpha-actinin revisited: a fresh look at an old player. Cell Motil Cytoskeleton 2004; 58: 104–111. [DOI] [PubMed] [Google Scholar]
- 18.Liu J, Taylor DW, Taylor KA. A 3-D reconstruction of smooth muscle alpha-actinin by CryoEm reveals two different conformations at the actin-binding region. J Mol Biol 2004; 338: 115–125. [DOI] [PubMed] [Google Scholar]
- 19.Atkinson RA, Joseph C, Kelly G, Muskett FW, Frenkiel TA, Nietlispach D et al. Ca2+- independent binding of an EF-hand domain to a novel motif in the alpha-actinin-titin complex. Nat Struct Biol 2001; 8: 853–857. [DOI] [PubMed] [Google Scholar]
- 20.DeLisser HM, Chilkotowsky J, Yan HC, Daise ML, Buck CA, Albelda SM. Deletions in the cytoplasmic domain of platelet-endothelial cell adhesion molecule-1 (PECAM-1, CD31) result in changes in ligand binding properties. J Cell Biol 1994; 124: 195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishiya T, DeFranco AL. Ligand-regulated chimeric receptor approach reveals distinctive subcellular localization and signaling properties of the Toll-like receptors. J Biol Chem 2004; 279: 19008–19017. [DOI] [PubMed] [Google Scholar]
- 22.Pasqualini R, Hemler ME. Contrasting roles for integrin beta 1 and beta 5 cytoplasmic domains in subcellular localization, cell proliferation, and cell migration. J Cell Biol 1994; 125: 447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer 2003; 3: 362–374. [DOI] [PubMed] [Google Scholar]
- 24.Eccles SA, Box C, Court W. Cell migration/invasion assays and their application in cancer drug discovery. Biotechnology annual review 2005; 11: 391–421. [DOI] [PubMed] [Google Scholar]
- 25.Feduska JM, Garcia PL, Brennan SB, Bu S, Council LN, Yoon KJ. N-glycosylation of ICAM-2 is required for ICAM-2-mediated complete suppression of metastatic potential of SK-N-AS neuroblastoma cells. BMC Cancer 2013; 13: 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cifone MA, Fidler IJ. Correlation of patterns of anchorage-independent growth with in vivo behavior of cells from a murine fibrosarcoma. Proc Natl Acad Sci U S A 1980; 77: 1039–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mori S, Chang JT, Andrechek ER, Matsumura N, Baba T, Yao G et al. Anchorage-independent cell growth signature identifies tumors with metastatic potential. Oncogene 2009; 28: 2796–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melero I, Gabari I, Corbi AL, Relloso M, Mazzolini G, Schmitz V et al. An anti-ICAM-2 (CD102) monoclonal antibody induces immune-mediated regressions of transplanted ICAM-2-negative colon carcinomas. Cancer Res 2002; 62: 3167–3174. [PubMed] [Google Scholar]
- 29.Hiraoka N, Yamazaki-Itoh R, Ino Y, Mizuguchi Y, Yamada T, Hirohashi S et al. CXCL17 and ICAM2 are associated with a potential anti-tumor immune response in early intraepithelial stages of human pancreatic carcinogenesis. Gastroenterology 2011; 140: 310–321. [DOI] [PubMed] [Google Scholar]
- 30.Liu Z, Guo B, Lopez RD. Expression of intercellular adhesion molecule (ICAM)-1 or ICAM-2 is critical in determining sensitivity of pancreatic cancer cells to cytolysis by human gammadelta-T cells: implications in the design of gammadelta-T-cell-based immunotherapies for pancreatic cancer. Journal of gastroenterology and hepatology 2009; 24: 900–911. [DOI] [PubMed] [Google Scholar]
- 31.Perez OD, Kinoshita S, Hitoshi Y, Payan DG, Kitamura T, Nolan GP et al. Activation of the PKB/AKT pathway by ICAM-2. Immunity 2002; 16: 51–65. [DOI] [PubMed] [Google Scholar]
- 32.Hogg N, Stewart MP, Scarth SL, Newton R, Shaw JM, Law SK et al. A novel leukocyte adhesion deficiency caused by expressed but nonfunctional beta2 integrins Mac-1 and LFA-1. The Journal of Clinical Investigation 1999; 103: 97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hynes RO. Integrins: versatility, modulation, and signaling in cell adhesion. Cell 1992; 69: 11–25. [DOI] [PubMed] [Google Scholar]
- 34.Yang Y, Jun CD, Liu JH, Zhang R, Joachimiak A, Springer TA et al. Structural basis for dimerization of ICAM-1 on the cell surface. Molecular cell 2004; 14: 269–276. [DOI] [PubMed] [Google Scholar]
- 35.Hariharan R, Pillai MR. Structure-function relationship of inhibitory Smads: Structural flexibility contributes to functional divergence. Proteins 2008; 71: 1853–1862. [DOI] [PubMed] [Google Scholar]
- 36.Lovell SC, Davis IW, Arendall WB 3rd, de Bakker PI, Word JM, Prisant MG et al. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins 2003; 50: 437–450. [DOI] [PubMed] [Google Scholar]
- 37.Eisenberg D, Luthy R, Bowie JU. VERIFY3D: assessment of protein models with three-dimensional profiles. Methods in enzymology 1997; 277: 396–404. [DOI] [PubMed] [Google Scholar]
- 38.Tovchigrechko A, Vakser IA. Development and testing of an automated approach to protein docking. Proteins 2005; 60: 296–301. [DOI] [PubMed] [Google Scholar]
- 39.Mohamet L, Lea ML, Ward CM. Abrogation of E-cadherin-mediated cellular aggregation allows proliferation of pluripotent mouse embryonic stem cells in shake flask bioreactors. PLoS One 2010; 5: e12921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nyman-Huttunen H, Tian L, Ning L, Gahmberg CG. alpha-actinin-dependent cytoskeletal anchorage is important for ICAM-5-mediated neuritic outgrowth. Journal of Cell Science 2006; 119: 3057–3066. [DOI] [PubMed] [Google Scholar]
- 41.Morton CL, lacono L, Hyatt JL, Taylor KR, Cheshire PJ, Houghton PJ et al. Activation and antitumor activity of CPT-11 in plasma esterase-deficient mice. Cancer chemotherapy and pharmacology 2005; 56: 629–636. [DOI] [PubMed] [Google Scholar]
- 42.Garcia PL, Council LN, Christein JD, Arnoletti JP, Heslin MJ et al. Development and histopathological characterization of tumorgraft models of pancreatic ductal adenocarcinoma. PLoS One 2013; 8(10): e78183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schrodinger LLC (2010). The PyMOL Molecular Graphics System, Version 1.3r1.
- 44.Humphries MJ. Cell-substrate adhesion assays. Curr Protoc Cell Biol 2001; Chapter 9: Unit 9 1. [DOI] [PubMed] [Google Scholar]
- 45.Carpen O, Pallai P, Staunton DE, Springer TA. Association of intercellular adhesion molecule-1 (ICAM-1) with actin-containing cytoskeleton and alpha-actinin. J Cell Biol 1992; 118: 1223–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Otey CA, Vasquez GB, Burridge K, Erickson BW. Mapping of the alpha-actinin binding site within the beta 1 integrin cytoplasmic domain. J Biol Chem 1993; 268: 21193–21197. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.