Abstract
ALDH16 is a novel family of the aldehyde dehydrogenase (ALDH) superfamily with unique structural characteristics that distinguish it from the other ALDH superfamily members. In addition to structural characteristics, there is an evolutionary-related grouping within the ALDH 16 genes. The ALDH16 isozymes in frog, lower animals, and bacteria possess a critical Cys302 residue in their active site, which is absent from ALDH16 in mammals and fish. Genomic analysis and plasma metabolomic studies have associated ALDH16A1 with the pathogenesis of gout in humans, although its actual involvement in this disease is poorly understood. Insight into the structure of ALDH16A1 is an important step in deciphering its function in gout. Herein, we report our efforts towards the structural characterization of Xenopus tropicalis ALDH16B1 (the homolog of human ALDH16A1) that was predicted to be catalytically-active. Recombinant ALDH16B1 was expressed in Sf9 cells and purified using affinity and size exclusion chromatography. Crystallization of ALDH16B1 was achieved by vapor diffusion. A data set was collected at 2.5 Å and preliminary crystallographic analysis showed that the frog ALDH16B1 crystals belong to the P 212121 space group with unit cell parameters a= 80.48 Å, b = 89.73 Å, c = 190.92 Å, α= β= γ= 90.00°. Structure determination is currently in progress.
Keywords: human ALDH16A1, frog ALDH16B1, catalytic residue, crystallization, aldehyde dehydrogenase
Introduction
The ALDH superfamily is a group of enzymes that catalyze the oxidation of endogenous and exogenous aldehydes using nicotinamide adenine dinucleotide (NAD) or nicotinamide adenine dinucleotide phosphate (NADP) as a cofactor. Through this enzymatic function, the human ALDH isoenzymes participate in biochemical pathways regulating alcohol detoxification, neuronal function, vitamin and amino acid metabolism and the synthesis of γ-aminobutyric acid and retinoic acid [1–3]. Abnormalities in the catalytic activity of ALDHs have been associated with severe disorders, including cancer, neurodegenerative conditions, cardiovascular diseases, diabetes, and stroke [4–7].
The members of the ALDH superfamily share a similar quaternary structure which comprises an NAD(P)+ binding domain, a catalytic domain that contains the critical cysteine (Cys) residue, and a smaller oligomerization domain. Although the most common oligomeric state of ALDHs is tetrameric, both dimeric and hexameric biological assemblies have also been reported [8]. Among the nineteen members of the ALDH superfamily, the ALDH16 family is the least characterized. The crystal structure of bacterial ALDH16 was recently reported [8]. Structurally, the members of this family contain an extra C-terminal domain, the role of which is poorly understood. While the average size of an ALDH monomer is about 500 amino acids, the members of ALDH16 family contain around 800 amino acids. Based on their evolutionary patterns, ALDH16 genes have been divided into four subfamilies, viz. 16A-mammals, 16B-amphibians, and lower animals 16C-bacteria, and 16D-fish [9]. The ALDH16A1 gene appears to be conserved in mammals (i.e., human, chimpanzee, and mouse) and is expressed as two spliced variants called the long and short forms [9]. ALDH enzymes with catalytic activity normally possess a critical cysteine residue (e.g., Cys302) in their active site. In the case of ALDH16, such a residue is found in frog, lower animals, and bacteria, but it is absent from mammals and fish. Due to this characteristic, human ALDH16A1 has been predicted to be enzymatically-inactive [9].
A whole genome study based on 457 Icelanders showed that a low-frequency singlenucleotide polymorphism (SNP) in one of ALDH16A1 exons is associated with high levels of urate and gout [10], an inflammatory disorder characterized by elevated blood levels of uric acid (hyperuricaemia) and severe joint pain [11]. Another study revealed maspardin, a protein encoded by spastic paraplegia 21 (SPG21) gene, to be involved in direct or indirect interactions with ALDH16A1 [12]. While the physical interaction between maspardin and ALDH16A1 has been confirmed by pull-down experiments, the role of ALDH16A1 in the pathogenesis of mast syndrome is yet to be explored. Human ALDH16A1 has also been predicted to physically interact with other proteins, such as deoxyribose-phosphate aldolase (DERA), enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase (EHHADH), glycine dehydrogenase (GLDC), ATP citrate lyase (ACLY), alanine-glyoxylate aminotransferase 2-like 2 (AGXT2L2), alanine-glyoxylate aminotransferase 2-like 1 (AGXT2L1), 4-aminobutyrate aminotransferase (ABAT), and hypoxanthine-guanidine phosphoribosyltransferase (HPRT1) [9, 13, 14]. Among all of the proteins predicted to interact with ALDH16A1, HPRT1 is the most relevant to gout due to its active role in uric acid metabolism.
To enhance our understanding of biological role of ALDH16A1 in physiology and pathology, we sought to crystallize members of the ALDH16 family. We were able to crystallize and collect X-ray diffraction data from frog ALDH16B1. Given that frog ALDH16B1 has a similar structural architecture to its human homolog, its three-dimensional structure is expected to provide a better understanding of the role of critical cysteine in the catalytic function of ALDH16, as well as critical structural insights that will help clarify the potential association of ALDH16A1 with gout.
Materials and Methods
ALDH16B1 expression and purification.
The open reading frame of the frog ALDH16B1 gene (encoding 829 amino acids) was sub-cloned into a baculovirus expression vector (pFB-LIC-Bse) by KpnI and EcoRI restriction digestion in-frame with a 6xHis-tag and a Tobacco Etch Virus (TEV) cleavage site at the amino terminus (Fig. 1a). The resultant plasmid was used for infection of Sf9 cells by the Tissue Culture Core at the University of Colorado according to standard procedures. The multiplicity of infection (MOI) was equal to one and expression was carried out over a 48h period. Cells were harvested, washed in PBS, and stored at −80°C for later use. Typically, the ALDH16B1 protein was purified from one-liter cell cultures. Cells were resuspended in 40 ml ice-cold lysis buffer (20 mM Tris, 500 mM NaCl, 35 mM imidazole, 1 mM TCEP, pH 7.4) supplemented with two tablets of mini-complete EDTA-free protease inhibitor tablets (Sigma-Aldrich). Lysis was induced by sonication on ice, applying two 10 sec medium-power pulses with a 50 sec interval time. Lysates were centrifuged at 31,000xg at 4°C for 1 h (Beckman Coulter Avanti J-E). Supernatants were passed through a 0.22 μm PVDF-based filter (Millipore), and the clarified lysates were loaded (1ml/min) into an FPLC system (Biorad NGC Quest 10) connected to a 5ml HisTrap FF column pre-equilibrated in lysis buffer at 4°C. Following an 800 ml wash, ALDH16B1 was eluted with imidazole added step-wise in lysis buffer reaching a final concentration of 500mM imidazole. The eluate protein was desalted in working buffer (20mM Tris, 150mM NaCl, 1mM TCEP, pH 7.4) by a dilution-concentration approach using an Amicon Ultra-15 concentrator (with regenerated cellulose) at a 10 kDa MW cutoff (Millipore). The recombinant protein was concentrated to 3.5 ml, centrifuged, and injected into the FPLC system connected to a HiLoad 16/600 Superdex 200pg gel filtration column (GE Healthcare) pre-equilibrated with working buffer and run at 0.5 ml/min at 4°C. Fractions that eluted under the major monodisperse peak were pooled and adjusted to have a final concentration of 0.36 mg/ml (Bradford assay). TEV protease was added for 2-3 h at 30°C according to manufacturer’s instructions (Genscript). The digest was centrifuged to remove precipitants and applied to the HisTrap FF column pre-equilibrated with working buffer (20mM Tris, 150mM NaC1, 1mM TCEP, pH 7.4). TEV protease was 6xHis-tagged. Therefore, the flow-through represented homogeneous cleaved ALDH16B1. The flow-through was concentrated and re-applied to the HiLoad 16/600 200pg column, equilibrated in working buffer. Fractions were pooled, concentrated, quantified (BCA method), aliquoted and stored at −80°C until required. Purity for all steps was verified by standard SDS-PAGE analysis.
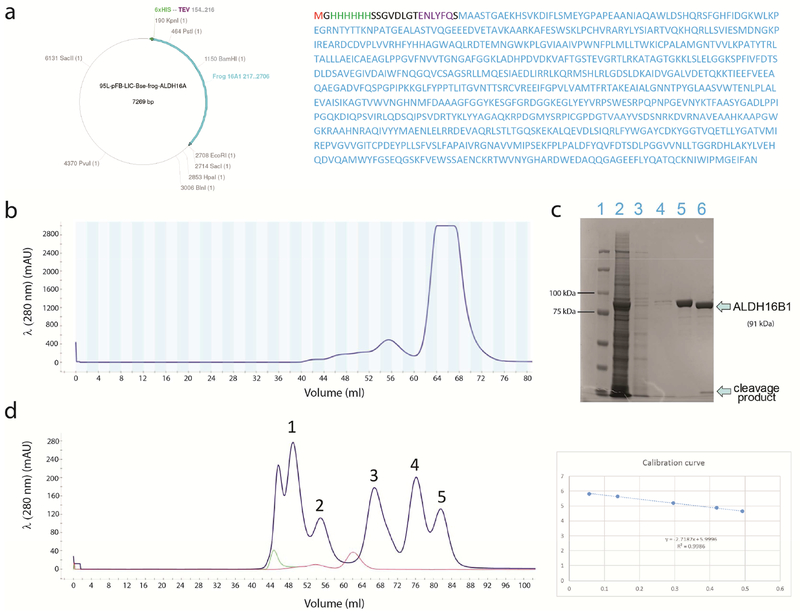
Figure 1. ALDH16B1 expression and purification.
a) ALDH16B1 expression plasmid and encoded protein. The 6-His tag and TEV cleavage sequence are shown in green and purple, respectively. The amino acid sequence of ALDH16B1, expressed by the plasmid, is shown in cyan. b) After buffer optimization, ALDH16B1 protein was eluted from Superdex-200 with a small percentage of aggregates. The aggregates are shown on the left site of the main peak. The main peak is homogeneous protein. c) SDS-page analysis was used to evaluate ALDH16B1 protein purity and TEV cleavage. From left to the right, the gel shows the (1) protein ladder, (2) cell lysate, (3) flow-through, (4) washes, (5) TEV-untreated ALDH16B1 and (6) TEV-treated ALDH16B1. d) The data from the analytical size exclusion chromatography revealed that the biological assembly of ALDH16B1 is homodimeric. From left to the right, the protein standard bands are: (1) thyroglobulin (669 kDa), (2) ferritin (440 kDa), (3) aldolase (158 kDa), (4) conalbumin (75 kDa), and (5) ovalbumin (44 kDa). Blue dextran (2000 kDa) (shown in light green) was used to determine the void volume (Vo) of the column. ALDH16B1 (shown in red) eluted between ferritin and aldolase. The calibration curve is shown on the right side of panel d.
ALDH16B1 crystallization.
Initial crystallization trials were carried out in 96-well microplates by mixing 100 nl ALDH16B1 protein solution (17.6 mg/ml) with 100 nl crystallization screening solutions. The crystallization screens were derived from Hampton Research (Index HT, PEG/Ion HT, SaltRx HT, Natrix HT, Crystal Screen HT and PEGRx HT) and Molecular Dimensions (JCSG-plus, Wizard 1,2 and Wizard 3,4). The procedure was carried out at the Yale School of Medicine Macromolecular X-ray Facility using a Mosquito (TTP LabTech) liquid handing robot. The plates were stored in a Rock Imager 1000 at 20°C and the drops were monitored on a regular basis. Low quality needle cluster crystals of ALDH16B1 manifested within 12 h and achieved their maximum size within 24 h. After several rounds of optimization trials in 96-well and 24-well plates, the ALDH16B1 protein crystals were optimized to single crystals. The crystals were flash frozen in the mother liquor that contained 30%-35% PEG 3350 as cryoprotectant.
Data set collection and preliminary crystallographic analysis.
ALDH16B1 crystals were screened and a full data set at 2.8 Å was collected using a Rigaku Pilatus 200K Detector with a Rigaku 007 rotating copper anode X-ray generator (wavelength=1.5418 Å). Another data set was collected from the National Synchrotron Light Source II (NSLS-II) at 2.5 Å (wavelength=1.7438 Å). Diffraction data analyses for the home source and NSLS-II data sets were carried out using the HKL3000 [15] and XDS [16] program suites, respectively. The 2.5 Å data collection statistics are provided in Table 1. Molecular replacement was carried out using Phaser [17] and MOLREP [18] without obtaining a solution. The various models used for molecular replacement (e.g., PDB:1O01, PDB: 4QF6 and PDB: 4FR8) possessed sequence identities around 40% but only for the first ∼460 amino acids of ALDH16B1. For the C-terminal domain, the sequence identity between the models and our protein was around 27%.
Table 1.
Data collection statistics for ALDH16B1
| ALDH16B1 | |
|---|---|
| Space group | P 212121 |
| Cell dimensions | |
| a, b, c (Å) | 80.48 89.73 190.92 |
| α, β, γ, (°) | 90.00, 90.00, 90.00 |
| Resolution (Å) | 50.00 - 2.50 (2.56 - 2.50)* |
| Completeness (%) | 99.8 (98.0) |
| Total No. of reflections | 1270333 |
| No. of unique reflections | 92719 |
| Rmerge | 0.17 (1.48) |
| Rmeas | 0.18 (1.55) |
| I / σ | 11.8 (1.72) |
| CC (1/2) | 99.8 (70. 8) |
| Redundancy | 13.7 (12.8) |
| Matthews coefficient (Å3 Da−1) | 1.93 |
| % solvent | 36.38 |
| No. of molecules per ASU | 2 |
values in parentheses are for the highest resolution shell.
Results and Discussion
ALDH16B1 purification.
Initial purification trials of ALDH16B1 protein were carried out at pH 7.4 with 150 mM NaC1 and in the absence of a reducing agent. Under these conditions, the protein aggregated due to its multiple free cysteines. Aggregation could not be avoided, even after the addition of 5% glycerol. Stable ALDH16B1 protein was finally obtained in the presence of 1mM TCEP and 150 mM NaC1. After this optimization step, the bulk of ALDH16B1 protein eluted from the gel filtration column as a single monodisperse peak with a smaller shoulder peak representing aggregates and higher order multimers (Fig. 1b). ALDH16B1 protein purification and TEV cleavage were evaluated using SDS-PAGE (Fig. 1c). Analytical size exclusion chromatography has suggested the species under the major peak to be a dimer (Fig. 1d). Typically, 10mg purified ALDH16B1 could be recovered from a one-liter culture.
Crystallization and preliminary crystallographic analysis.
Initial crystallization “hits” were obtained by four different conditions: i) 25% (w/v) PEG 1500, 100 mM MMT buffer pH 9.0 (D,L-Malic acid, MES and Tris at a molar ratio of 1:2:2, pH 9.0) (Molecular Dimensions, Maumee, OH), ii) 0.15 M D,L-Malic acid, 20% (w/v) PEG 3350 (Molecular Dimensions, Maumee, OH), iii) 0.2 M ammonium tartrate dibasic, 20% (w/v) PEG 3350 (Hampton Research, Aliso Viejo, CA ), and iv) 20% (w/v) PEG 10000, 100 mM HEPES/sodium hydroxide, pH 7.5 (Molecular Dimensions, Maumee, OH). In all cases, the crystal grew shaped as a very thin and small needle, and required optimization. A representative example is shown in Fig. 2a. Optimization efforts were initially focused on expanding around the two reagents (0.15 M DL-Malic acid, 20 % w/v PEG 3350) that displayed the most promising results. In these trials, we also varied final protein concentrations (9 mg/ml, 4.5 mg/ml, 2.25mg/ml and 1.25 mg/ml) and crystallization temperatures (20°C and 4°C). The crystallization temperature (20°C), final protein concentration (4.5 mg/ml), and crystallization conditions were then optimized to obtain single crystals. The optimization trials are illustrated in Fig. 2b. Good quality ALDH16B1 protein crystals were grown in a 24-well plate with 0.1 M Tris-HCl (pH 8.4-8.8), 10-15% PEG3350, 0.2M guanine and 0.02M urea. The presence of guanidine is critical for obtaining good diffraction quality crystals. Screening for identification of the best cryoprotectant was also carried out. Complete data sets were collected from both NSLS-II and home source (Yale School of Medicine Macromolecular X-ray Facility) at 2.5 Å and 2.8 Å, respectively (Fig. 3a, b). Further crystallographic analysis was focused on the data set collected from NSLS-II as this one yielded the highest resolution. The ALDH16B1 crystals (Table 1) belong to the primitive orthorhombic space group (P 212121) with unit cell parameters a= 80.48 Å, b = 89.73 Å, c = 190.92 Å, and α= β= γ= 90.00°. Overall completeness was determined to be 99.8%. The volume of the asymmetric unit was compatible with two ALDH16B1 molecules (Matthews coefficient, = 1.93 Å3 Da−1) with a solvent content estimated at 36%. Our efforts to solve the crystal structure by molecular replacement using homologous ALDH structures failed due to the low sequence identity between the target sequence and the searching models.
Figure 2. ALDH16B1 crystallization trials.
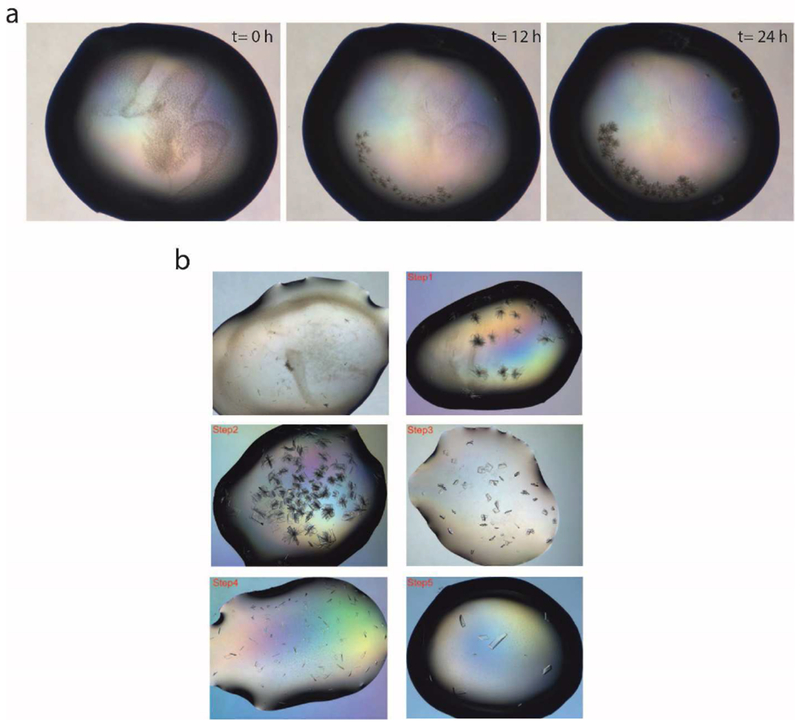
a) Representative example of the time-dependent growth of a needle-shaped cluster of ALDH16B1 crystals. b) Representative images showing effectiveness of optimization steps in obtaining single and good-diffracting ALDH16B1 crystals. ALDH16B1 crystal optimization involved pH, temperature, protein concentration, salts, PEGs and additives screening.
Figure 3. ALDH16B1 crystal diffraction.
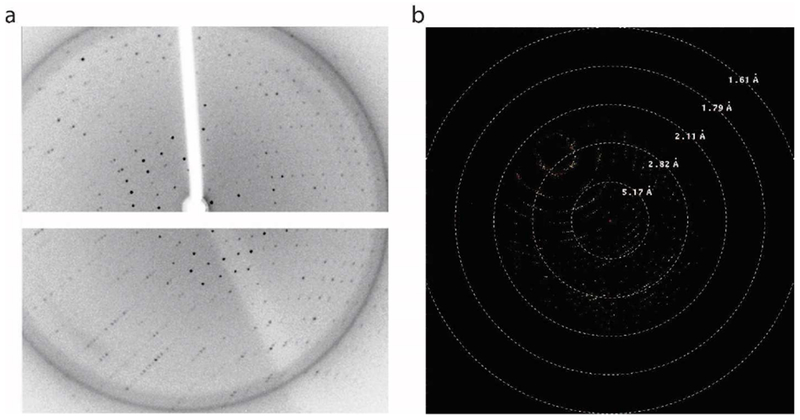
a) Diffraction data obtained from the home source at Yale School of Medicine Macromolecular X-ray Facility. A complete data set was collected at 2.8 Å (wavelength=1.5418 Å). b) A complete data set was also collected from the National Synchrotron Light Source II (NSLS-II) at 2.5 Å (wavelength=1.7438 Å).
Conclusions
We have developed a protocol for the expression and purification of frog ALDH16B1 from Sf9 cells that produce purified protein in milligram quantities. We have identified the optimal conditions that yield sizeable, single, and good-diffracting ALDH16B1 crystals. Determination of the crystal structure of ALDH16B1 has been unsuccessful using molecular replacement with other ALDH structures in the protein database. These molecular replacement techniques included a variety of strategies, such as removing regions of the model with low sequence identity to ALDH16B1, creating and using smaller search models that demonstrate acceptable sequence identity to the various domains of ALDH16B1, utilizing theoretical models, and last (but not least) testing polyalanine models. Low sequence identity between the available models and the C-terminal domain of ALDH16B1 may have contributed to the lack of success using molecular replacement. Alternate methods to solve the structure are underway and the results are expected to provide valuable insights into the biological functions of this structurally-unique macromolecule.
Supplementary Material
Highlights.
structural studies of frog ALDH16B1, a homolog of human ALDH16A1
overexpression and purification of frog ALDH16B1 in mg quantities
ALDH16B1 crystallization
size exclusion and crystallographic data support a homodimeric structure for ALDH16B1
Acknowledgments
This work was supported in parts by National Institutes of Health (NIH) Grants AR064137, and AA02205.
Abbreviations
- ALDH
aldehyde dehydrogenase
- NADP
nicotinamide adenine dinucleotide phosphate
- SNP
single-nucleotide polymorphism
- TCEP
tris(2-carboxyethyl)phosphine
- MOI
multiplicity of infection
- MW
molecular weight
- TEV
Tobacco Etch Virus
- BCA
bicinchoninic acid
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
References
- [1].Seitz HK, Becker P, Alcohol metabolism and cancer risk, Alcohol Res Health, 30 (2007) 38–41, 44–37. [PMC free article] [PubMed] [Google Scholar]
- [2].Kim JI, Ganesan S, Luo SX, Wu YW, Park E, Huang EJ, Chen L, Ding JB, Aldehyde dehydrogenase 1a1 mediates a GABA synthesis pathway in midbrain dopaminergic neurons, Science, 350 (2015)102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Yoshida A, Rzhetsky A, Hsu LC, Chang C, Human aldehyde dehydrogenase gene family, Eur J Biochem, 251 (1998) 549–557. [DOI] [PubMed] [Google Scholar]
- [4].Chen CH, Ferreira JC, Gross ER, Mochly-Rosen D, Targeting aldehyde dehydrogenase 2: new therapeutic opportunities, Physiol Rev, 94 (2014) 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hellsten R, Johansson M, Dahlman A, Sterner O, Bjartell A, Galiellalactone inhibits stem cell-like ALDH-positive prostate cancer cells, PLoS One, 6 (2011) e22118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Munzel T, Daiber A, The potential of aldehyde dehydrogenase 2 as a therapeutic target in cardiovascular disease, Expert Opin Ther Targets, 22 (2018) 217–231. [DOI] [PubMed] [Google Scholar]
- [7].Pan G, Munukutla S, Kar A, Gardinier J, Thandavarayan RA, Palaniyandi SS, Type-2 diabetic aldehyde dehydrogenase 2 mutant mice (ALDH 2*2) exhibiting heart failure with preserved ejection fraction phenotype can be determined by exercise stress echocardiography, PLoS One, 13 (2018) e0195796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Liu LK, Tanner JJ, Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer, J Mol Biol, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Vasiliou V, Sandoval M, Backos DS, Jackson BC, Chen Y, Reigan P, Lanaspa MA, Johnson RJ, Koppaka V, Thompson DC, ALDH16A1 is a novel non-catalytic enzyme that may be involved in the etiology of gout via protein-protein interactions with HPRT1, Chem Biol Interact, 202 (2013) 22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sulem P, Gudbjartsson DF, Walters GB, Helgadottir HT, Helgason A, Gudjonsson SA, Zanon C, Besenbacher S, Bjornsdottir G, Magnusson OT, Magnusson G, Hjartarson E, Saemundsdottir J, Gylfason A, Jonasdottir A, Holm H, Karason A, Rafnar T, Stefansson H, Andreassen OA, Pedersen JH, Pack AI, de Visser MC, Kiemeney LA, Geirsson AJ, Eyjolfsson GI, Olafsson I, Kong A, Masson G, Jonsson H, Thorsteinsdottir U, Jonsdottir I, Stefansson K, Identification of low-frequency variants associated with gout and serum uric acid levels, Nat Genet, 43 (2011) 1127–1130. [DOI] [PubMed] [Google Scholar]
- [11].Ragab G, Elshahaly M, Bardin T, Gout: An old disease in new perspective - A review, J Adv Res, 8 (2017) 495–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hanna MC, Blackstone C, Interaction of the SPG21 protein ACP33/maspardin with the aldehyde dehydrogenase ALDH16A1, Neurogenetics, 10 (2009) 217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Charkoftaki G, Chen Y, Han M, Sandoval M, Yu X, Zhao H, Orlicky DJ, Thompson DC, Vasiliou V, Transcriptomic analysis and plasma metabolomics in Aldh16a1-null mice reveals a potential role of ALDH16A1 in renal function, Chem Biol Interact, 276 (2017) 15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M, BioGRID: a general repository for interaction datasets, Nucleic Acids Res, 34 (2006) D535–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Minor W, Cymborowski M, Otwinowski Z, Chruszcz M, HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes, Acta Crystallogr D Biol Crystallogr, 62 (2006) 859–866. [DOI] [PubMed] [Google Scholar]
- [16].Kabsch W, XDS, Acta Crystallogr D Biol Crystallogr, 66 (2010) 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ, Phaser crystallographic software, J Appl Crystallogr, 40 (2007) 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Vagin A, Teplyakov A, An approach to multi-copy search in molecular replacement, Acta Crystallogr D Biol Crystallogr, 56 (2000) 1622–1624. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.