Abstract
Background:
Disparate research sites using identical or near-identical magnetic resonance imaging (MRI) acquisition techniques often produce results that demonstrate significant variability regarding volumetric quantification of white matter hyperintensities (WMH) in the aging population. The sources of such variability have not previously been fully explored.
New Method:
3D FLAIR sequences from a group of randomly selected aged subjects were analyzed to identify sources-of-variability in post-acquisition processing that can be problematic when comparing WMH volumetric data across disparate sites. The methods developed focused on standardizing post-acquisition protocol processing methods to develop a protocol with less than 0.5% inter-rater variance.
Results:
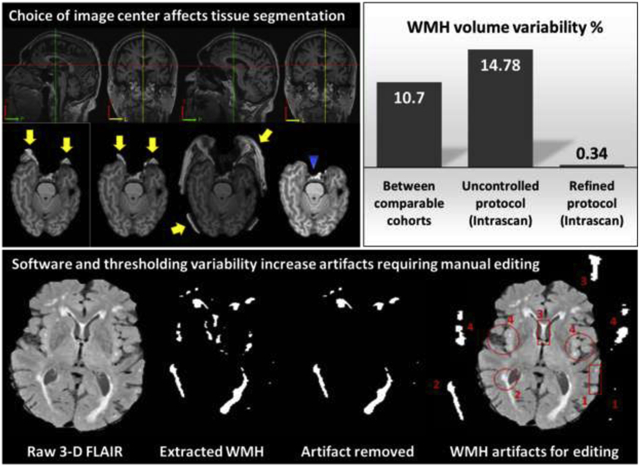
A series of experiments using standard MRI acquisition sequences explored postacquisition sources-of-variability in the quantification of WMH volumetric data. Sources-of-variability included: the choice of image center, software suite and version, thresholding selection, and manual editing procedures (when used). Controlling for the identified sources-of-variability led to a protocol with less than 0.5% variability between independent raters in post-acquisition WMH volumetric quantification.
Comparison with existing method(s):
Post-acquisition processing techniques can introduce an average variance approaching 15% in WMH volume quantification despite identical scan acquisitions. Understanding and controlling for such sources-of-variability can reduce postacquisition quantitative image processing variance to less than 0.5%.
Discussion:
Considerations of potential sources-of-variability in MRI volume quantification techniques and reduction in such variability is imperative to allow for reliable cross-site and crossstudy comparisons.
Keywords: cerebrovascular disease, white matter hyperintensity, volumetric analysis, sources of variability
Graphical Abstract
Significant variability in white matter hyperintensity quantification can occur as a result of variability in standardizing selection of the image center of gravity, software package, thresholding techniques, and manual editing procedures. Controlling for such variables can reduce the interscan post-acquisition processing variability to less than 0.5%.
1. INTRODUCTION
Neuroimaging is a critical tool for diagnosing neurodegenerative disease states (Abramson et al., 2015), such as vascular dementia and Alzheimer’s disease. The wide-spread availability, high spatial resolution, and variety of imaging-sequences afforded by magnetic resonance imaging (MRI) make it an ideal imaging modality for evaluation of cerebrovascular contributions to cognitive decline. Significant effort has gone into standardizing acquisition sequences for multisite studies such as the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (Vilar-Bergua et al., 2016), and the adoption of such consensus acquisition sequences beyond ADNI has allowed a greater degree of cross-study comparisons than afforded previously. Despite such standardization in acquisition protocols, post-acquisition processing techniques for subcortical white matter hyperintensity volume quantification (WMH-VQ) remain variable across studies and research sites. Few studies have examined the reliability and reproducibility of volumetric MRI postacquisition processing methods (De Guio et al., 2016).
The few studies addressing post-acquisition variability in MRI have focused exclusively on structural segmentation methods. Schnack and colleagues (2004) performed a multi-center MRI study focused on structural segmentation, where image processing was performed at a single site to reduce anticipated variability (Schnack et al., 2004). The study suggested that adding a thresholding calibration to the processing algorithm might allow more uniform segmentation across sites. However, this study did not assign multiple raters to verify their protocol nor did they validate the contention that a protocol including a standardized thresholding calibration would reduce cross-site or inter-rater variability. Ramirez and colleagues (2013) further addressed volumetric protocol reliability using three raters and two repeat scans (interval ~30 min – 50 days) for twenty subjects (Ramirez, Scott, & Black, 2013). However, the study did not examine variability between raters. They did comment on the issue of variance in the output volumes, which they attributed to brain structure changes during the long interval between the repeated scans, rather than inherent variability in post-acquisition processing. No such studies have as of yet focused on assessing the inter-rater reliability of WMH-VQ techniques.
Visual rating scales have been developed for assessing WMH burden. While visual rating scales are reasonable choices for clinical evaluation, given their ease of use in facilities lacking modern post-acquisition image processing facilities, they are limited by floor and ceiling effects and do not allow for the precise quantification necessary for detecting subtle changes in imaging characteristics over time (Pantoni et al., 2002). For this reason, semi-automated and automated techniques have been developed as more reliable and sensitive measures for WMH-VQ (Iorio et al., 2013). Despite the inherent benefits of automated post-acquisition WMH-VQ techniques, the mean values of WMH volume derived from distinct studies often demonstrate significant variability with mean volumes ranging from 0.5 – 11.2 cc3 (~5% of the average WMH-VQ across subjects), across otherwise comparable cohorts (Ambarki, Wahlin, Birgander, Eklund, & Malm, 2011; Carmichael et al., 2010; Promjunyakul et al., 2015; Ramirez et al., 2016; van den Heuvel et al., 2006; van der Flier et al., 2004; Wen & Sachdev, 2004; Wu et al., 2006). Frequently, such differences are assumed to be due to differential cohort characteristics. However, given the large number of competing protocols in widespread use, it is also possible that inherent sources-of-variability in post-acquisition image processing techniques contribute to such variability (Wu et al., 2006).
Despite advances in the field of quantitative neuroimaging, no universally agreed upon or standardized methodologies for WMH-VQ post-acquisition processing exist today, nor have the potential sources-of-variability in such protocols been systematically identified and addressed. In general, protocols for WMH-VQ use the same basic concepts regardless of differences in processing tools (software and algorithms), type of algorithm (semi or fully automated), or study design (cross-sectional or longitudinal) including: 1) image registration, 2) nonbrain tissue stripping, 3) intensity estimation and thresholding, and 4) manual editing (as deemed necessary), yet such differences may influence variability in WMH-VQ. As such, an understanding of the sources-of-variability inherent in WMH-VQ is critical for comparisons of findings across centers and for the integrity of multi-site studies that do not utilize a centralized processing site or a standardized, validated, multi-site post acquisition processing protocol. Furthermore, such understanding of WMH-VQ variability is essential for interpretation of longitudinal studies examining within-subject change, as the potential variability inherent in different quantification protocols (due to advances in software or other scientific/technologic factors), whether semi- or fully automated, can exceed the annual rate of change in WMH volumes for any given subject. The present study systematically analyzed potential sources-of-variability in WMH-VQ procedures that may potentially increase variability resulting in difficulty comparing cross-center data, limit the reliability of multi-center studies, and further preclude an accurate understanding of longitudinal within-subject WMH-VQ changes.
2. METHODS
2.1. Subjects
MRI acquisitions for 71 subjects (65 – 85 years old, spanning the cognitive continuum from normal through MCI to dementia) from the Sanders-Brown Center on Aging (University of Kentucky) research cohort were collected using a standard protocol. A random sample of scans from 21 participants were used for the discovery phase of the study with the remaining 50 participant scans used for validation. Details of the clinical characterization of this cohort has been published previously (Schmitt et al., 2012). This study was approved by the University of Kentucky Institutional Review Board under the protocols used to acquire the clinical data and MRI images.
2.2. MRI Acquisition
All MRI scans were acquired at the University of Kentucky, Magnetic Resonance Imaging and Spectroscopy Center using a Siemens 3T TIM-Trio MRI scanner (Siemens Healthcare, Erlangen, Germany). A 32-channel head coil was used to scan the subjects. Two acquisition sequences were executed for this study: 1) T1-weighted Magnetization-Prepared Rapid Acquisition Gradient Echo (3D MPRAGE), echo time (TE) 2.3 milliseconds, repetition time (TR) 2,530 milliseconds, inversion recovery time (IR) 1,100 milliseconds, flip angle 7°, 1×1×1 mm resolution full-brain coverage; 2) T2-weighted fluid-attenuated inversion recovery (FLAIR) image,. TE 388 milliseconds, TR 6,000 milliseconds, IR 2,200 milliseconds, 3D 1×1×1 mm. No gap between slices. All subjects included were scanned used identical imaging acquisition protocols, along with the same scanner and head coil.
2.3. Image Processing
MRI images were processed using an automated WMH-VQ method, described previously (Bahrani et al., 2017). Briefly, all MRI images were normalized for intensity. Two T1-weighted Magnetization-Prepared Rapid Acquisition Gradient Echo (MPRAGE) images were acquired and co-registered using statistical parametric map software (SPM8 or SPM12) (http://www.fil.ion.ucl.ac.uk/spm) and averaged. The averaged-MPRAGE were then registered to the single 3-D FLAIR image. Nonbrain tissue was stripped from the registered averaged- MPRAGE image using a brain extraction tool (FSL-BET), FSL-FMRIB software library (FSL v5.0.9) (http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/BET). Remaining scalp tissue was removed slice-by-slice manually, as needed, using the medical image processing analysis and visualization (MIPAV v7.4.0) application (http://mipav.cit.nih.gov). FLAIR images were generated from the binary mask of the stripped averaged-MPRAGE and were further segmented using the SPM unified regime. Five segmented images including, gray matter (GM), two white matter (WM) subsegments, cerebrospinal fluid (CSF), and the unclassified tissue (UT) masks were created in a native-space using an in-house segmentation template created from 145 images of healthy normal adult subjects, demographically similar to the subjects in this study (C. D. Smith et al., 2016). The two WM masks were generated for different WM classes that cannot be captured by one mask (tissue class) and were further summed to create a binary WM mask that was multiplied by the FLAIR. This step isolates all of the classified white matter voxels in the FLAIR image. The intensity distribution of these voxels was then fit with a Gaussian curve. The maximum and minimum threshold values were computed from the Gaussian distribution mean and standard deviation (SD). The threshold value was then applied to the stripped FLAIR images to obtain the final WMH-VQ mask.
2.4. Study Design
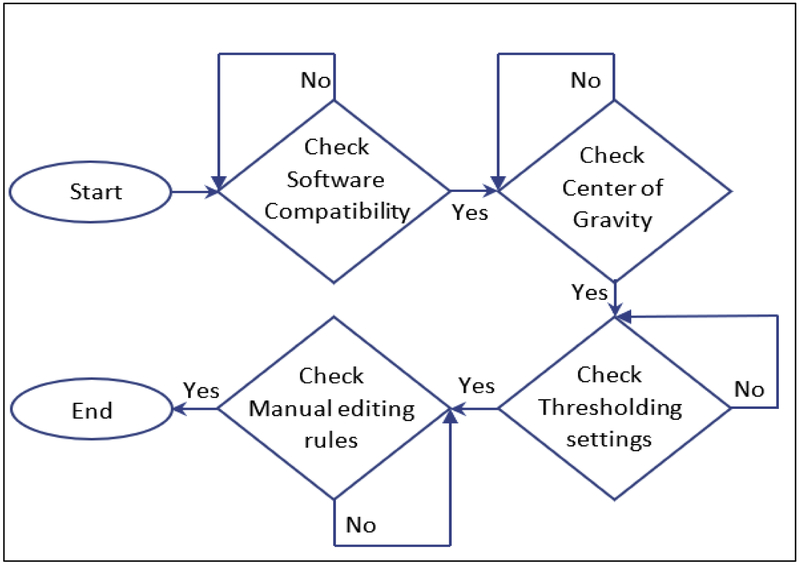
MRI images were used for both discovery and validation arms of the project as follows: twenty-one scans were used for analyzing the variability associated with software and system compatibility, choice of the center of gravity (CoG), threshold calculations, and manual editing procedures as part of the discovery dataset (Figure 1). An independent sample of 50 MRI images was used to anlayze the validation dataset after controlling for sources-of-variability identified in the discovery phase of the study.
Figure 1:
Flow chart summarizing the use of the discovery dataset (n=21) that examined distinct sources of variability inherent in white matter hyperintensity volumetric quantification (WMH-VQ) processing techniques.
2.5. Software Compatibility
The computers for this study are Linux operating systems and have the same software versions, MATLAB a2015b (MathWorks, Inc), MIPAV v7.4.0, and FSL 5.0.9. Two versions of SPM including SPM8 and SPM12 were used to examine variance inherent in specific software versions. For this experimental aim, we did not vary other software programs and so recognize that our findings may not generalize across all software systems and versions. Variability was assessed by comparing the WMH-VQ measurements from identical scans using both SPM8 and SPM12-based analyses.
2.6. Center of Gravity
The center of gravity (CoG) is linked to the nonbrain extracting process (Segonne et al., 2004). An accurate CoG enables a smooth stripping process with virtually no additional manual editing required. To allow assessment of potential variance that is associated with a differential selection of the CoG, two random CoGs were selected for each subject in addition to the systematic CoG. The systematic CoG was chosen by displaying; the registered averaged T1-weighted image using the Triplaner display module in MIPAV to locate the CoG visually (C1), using the cursor (estimating the brain center as one half the brain anterior-posterior, left-right, and inferior-superior distances). The second CoG (C2) was selected using the default CoG of the Triplaner display. The third Cog (C3) was randomly chosen manually by the post-acquisition analyst but its location was restricted to within a 0.5 cm diameter of C1. Variability was assessed by comparing the WMH-VQ measurements from identical scans using C1, C2, and C3 as the independent variables.
2.7. Threshold Calculation and WMH volume quantification
To extract the WMH volume, the WMH distribution must be defined (Anbeek, Vincken, van Osch, Bisschops, & van der Grond, 2004; Caligiuri et al., 2015). Variability in WMH-VQ are exacerbated when the minimum WMH intensity distribution overlaps with the normal appearing WM intensity distribution, leading either to over- or under-estimating WMH-VQ due to inconsistent thresholding. We used 10% of the maximum FLAIR WM voxel intensity as the minimum value to obtain the histogram distribution of the WM tissue. This lower limit is flexible and does not appear to contribute significant error in the fitting procedure. However, the upper threshold value is a critical factor for quantifying WMH volume. A two-Gaussian curve fit to the distribution (MATLAB curve-fitting tool) was used for computing the mean and SD. The mean and SD were applied to the thresholding equations to calculate the maximum and minimum thresholds. The thresholds were mean + 3 x SD for the lower bound and mean + 15 x SD for the upper bound (Bahrani et al., 2017). The upper bound eliminated extreme values occasionally seen as intensity artifacts in FLAIR images. All threshold values were expressed to the second decimal place. WMH mask artifacts were reduced using a Gaussian filter (1 × 1 × 1 mm). Total WMH volume was calculated from the final WMH mask.
We tested two parameters in our experiment to study their influence on the thresholding values and in turn on the WMH-VQ. First, we compared the mean and SD of the histogram distribution of the WM voxels extracted from the FLAIR image using voxel intensity and position on the Gaussian curve, versus voxels intensity and volume in mm3 rather than position on the Gaussian curve. Second, we tested the impact of the precision of the mean and SD on the calculation of the thresholding values. We choose the mean and the SD using the systematic algorithm described above carried to two significant digits (decimal places) as increasing the precision beyond this (i.e. adding additional significant digits (decimal places) did not further contribute to accuracy in the resultant WMH-VQ derived. This threshold was then compared to setting the same mean and SD threshold at a single or no significant digits (an integer).
2.8. Manual Editing
The sources-of-variability assessed above are all operator independent, but do not consider artifact removal which can be an additional source-of-variability that may require one or more manual editing steps. In order to define the variability associated with manual editing, two manual editing steps were included in the protocol to ensure that artifact did not confound the conclusions drawn regarding post-acquisition processing variability. Manual editing was performed on: 1) the whole brain mask after the nonbrain tissue extraction process and 2) the final WMH mask.
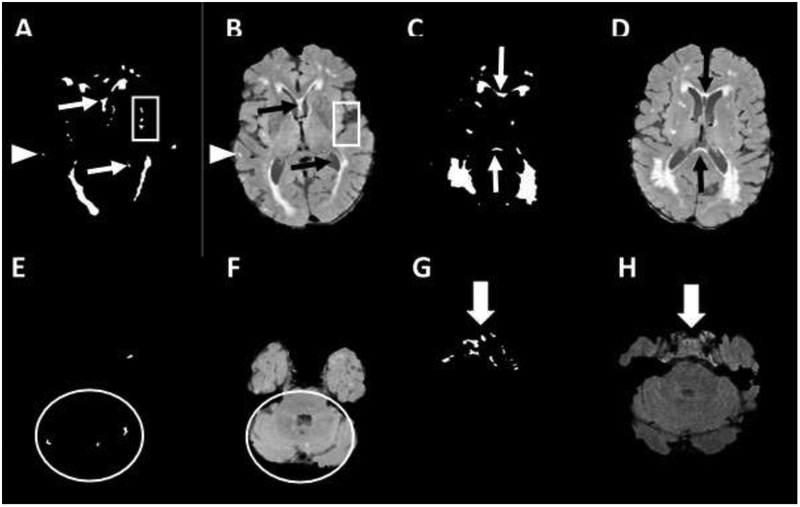
Manual editing was performed independently without standardization of procedures and again after developing a standard editing protocol to minimize operator-dependent error in these steps as follows. Extraneous voxels of T2 hyperintensity that are generated due to pulsation and flow artifacts were removed manually, guided by the original FLAIR image. A FLAIR image was displayed with a standard Gaussian-fit mean center and ten x SD grey scale window value side-by-side with the WMH mask. and the second image was kept with its original values, to allow maximal recognition of false positive and negative voxels. Figure 2 demonstrates the spectrum of false hyperintensity signals that were removed from the gray matter (GM), lateral sulcus and pineal gland, the voxels between and inside the ventricles, voxels in the cerebellum, and the voxels in the pons and lower brainstem. A synopsis of our manual protocol guidelines is presented in Table 1. Variability due to manual editing was assessed by comparing the WMH-VQ measurements from identically processed scans using both unstandardized and standardized manual editing protocols as the independent variables.
Figure 2:
Common hyperintensity signal artifacts in the white matter hyperintensity (WMH) mask include: Gray matter signals (GM), panels A, and B, (arrowhead); Lateral sulcus and pineal gland, panels A, and B, (rectangle); Voxels in between and inside the ventricles, panels A, B, C, and D, (narrow arrow); Voxels in cerebellum panels E and F (circle); Voxels in the pons and lower slices panels G and H (large arrow).
Table 1.
Manual editing protocol developed to systematically reduce sources of variation in the assessment of subcortical small vessel ischemic disease. Sources of variability and areas of analysis that require increased diligence and further development of standardized methodology are identified.
| Areas to systematically review for T2 artifact |
Rationale | Illustrations in Figure 1 |
|---|---|---|
| Common extraneous voxels | False intensities identified compared to original FLAIR image | Panels A and B |
| Cortical GM | Extends beyond anatomic boundaries of subcortical disease, but may be considered important for some studies | Panels A and B, (arrowhead) |
| Lateral sulcus/insular cortex and pineal gland | Signal artifact due to CSF boundary | Panels A and B, (rectangle) |
| Areas of contrast with GM and CSF between and inside the ventricles | Artifact due to CSF boundary and pulsation | Panels A, B, C, and D, (narrow arrow) |
| Cerebellum | Prone to infra-tentorial artifact and extensive CSF boundaries, but may be considered important for some studies | Panels E and F (circle) |
| Pons and lower slices | CSF pulsation from forth vertical may produce hyperintensity voxels in the pons. The extensive artifact in lower slices due to bone & CSF boundaries | Panels G and H (large arrow) |
| Pituitary gland & cavernous sinus | Extensive artifact due to bone & CSF boundaries | Panels G and H (large arrow) |
| Basal ganglia & thalamus | Deep GM artifacts due to homogeneous T2 signal need to be distinguished from true small vessel ischemic disease | Not shown |
2.9. Validation of a standard protocol to reduce variance
Controlling for selection of CoG, WM segmentation in SPM, curve fitting and threshold setting on the WM histogram, and manual editing produced a final protocol that was validated in an independent set of 50 MRI scans. We set the threshold for success at 0.5% variability as an acceptable limit of variability well within the range of anticipated within-subject annual longitudinal change. The current variability for WMH-VQ was calculated as a mean of the variability assessed across all parameters studied at −15%, based on the assumptions that inter-study, and intra-site inter-rater reliability would represent an average rather than cumulative (additive) effect on WMH-VQ assessments.
2.10. Statistical Analysis
Using the 21 discovery images, WMH volumes were calculated in a four-step process as follows. First, two raters assessed WMH volume under the protocol described in Section 2.3 above, one using SPM8 (OA) and one using SPM12 (AB). Variability was measured by the percent difference in the two raters’ ratings, as given below. Next, the software package was fixed (SPM12), and one of the two raters (AB) calculated WMH volume based on different CoG (as described in Section 2.4.2). Then, both software and CoG were fixed, and the rater (AB) calculated WMH volumes under different thresholding conditions, as described in Section 2.4.3. The distribution of the WM voxel intensity and position on the curve was visualized using histograms. Finally, software, CoG, and threshold were fixed, and manual editing was applied by both raters. The percentage difference (PD) for each set of ratings for each image, which was defined as the difference between the two sets of measurements divided by the average value of the two methods, for each source of variability (i.e., software compatibility, CoG, thresholding, and manual editing):
These summary PDs were used to quantify the approximate measurement error associated with each source of variability. The overall PD for each discovery image was calculated by taking the average of the four individual PDs. The WMH volumes obtained after implementing all four steps are referred to hereafter as “standardized” WMH.
Once the analyses based on the discovery data were completed, the two raters each calculated WMH volume for the set of 50 validation images based on the unstandardized and standardized protocols. Interrater agreement was assessed using the Pearson correlation coefficient and the Interrater Reliability (IRR). SigmaPlot 13 (Systat Software Inc., San Jose, California) was used for statistical data analysis.
Additionally, the permutation test (aka randomization test; MATLAB function https://www.mathworks.com/matlabcentral/fileexchange/63276-permutation-test) was applied to the 50 standardized WMH volumes to test whether mean WMH volume was different between raters (50,000 permutations). Finally, the Dice similarity test (using MATLAB) was utilized to find the similarity and dissimilarity of the WMH final masks before and after the manual editing between the two raters.
3. RESULTS
3.1. Subjects
The mean age of this cohort was 74.1 (± 8.0) years, the mean educational attainment was 16.9 (±3.3) years, and the mean WMH volume was 14.5 cc3 (± 23.0 cc3). In addition, 54% were female, 66% were hypertensive, 26% were diabetic, 10 were smokers, and 56% had hyperlipidemia. Finally, 30% of the cohort were cognitively normal, and the remaining 70% had a diagnosis of mild cognitive impairment at the time of the scan. There were no significant demographic or clinical differences between the discovery and validation data set participants in this study.
3.2. Software versions and compatibility
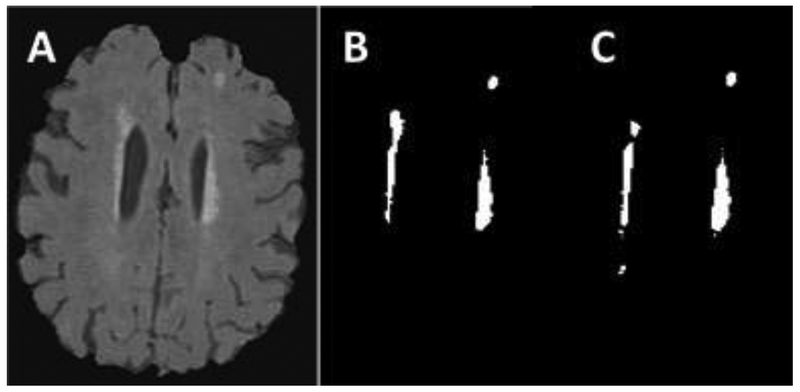
Different SPM software versions and software compatibility were found to be a significant source-of-variability. Analysis using SPM8 resulted in an overestimated WMH-VQ compared to analyses using SPM12, 36.44% before editing and 93.26% after editing (n = 21). Figure 3 shows the difference between the two processed WMH masks in contrast to the FLAIR image (Panel A). Panel B is the WMH mask resulting from the use of SPM8, while Panel C is the mask utilizing SPM12. These data demonstrate the importance of software version (even from the same source) in affecting variability in WMH-VQ.
Figure 3:
Example of a case where WMH masks differ based on SPM versions used. A: is the original T2 FLAIR image. B: WMH mask using MATLAB 2015 and SPM8. It shows an overestimate volume comparing to the FLAIR image and C which is the WMH mask that quantified using MATLAB 2015 and SPM12.
3.3. Selection of Center
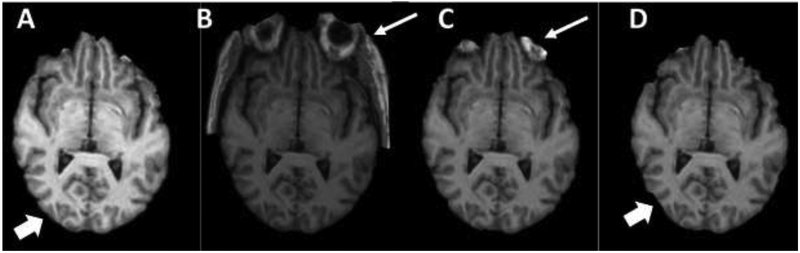
The use of different CoGs introduced a variability of approximately 11% in final WMH volumes. The percentage error in WMH-VQ (mean ± standard error of the mean (SE) in mm3) determined using C2, (28360 ± 7460), and C3, (33235 ± 8036), compared to C1, (33755 ± 7907), were 20.9% and 16.1%, respectively (n = 21). Figure 4 demonstrates the artifacts leading to increased WMH-VQ variability as a result of the choice of CoG.
Figure 4.
Several examples of cases that highlight the effect of the center of gravity (CoG) on bone extraction method using BET-FSL tools. Panel A: demonstrates optimal bone extraction with almost clean brain tissue. Panel B and C show non-brain tissue remaining (narrow arrows) due to choosing an alternate CoG. Panel D demonstrates a loss of a portion of GM due to the non-tissue extraction process as a result of choosing an alternate CoG.
3.4. Thresholding
Fitting the histogram distribution of the WM intensities to the Gaussian curve was also shown to contribute to interrater reliability variance in WMH volume before and after manual editing. The percentage variance of fitting the WM histogram distribution of the WM voxel intensities and volume, mean ± SE (39427 ± 8299), versus the WM voxel intensities and position, (39759 ± 8237) on the Gaussian curve was found to be 2.5% (n=21). Thresholding the FLAIR mask to compute the WMH volume was also shown to be a significant source-of-variability. The percentage error between the thresholding values carried to either none or one significant digits, versus the maximal selection of two significant digits was −19.9% and 10.2%, respectively. This percentage error is maximally evident whenever the distribution is not corrected for the natural left-handed skew deviation inherent in community-based samples such as ours and the many others that have been studied to date.
3.5. Manual editing
All steps in the WMH-VQ protocol represent automated processes that can be standardized to reduce variability. While the protocol is fully automated, artifacts can create erroneous volume estimates, and so manual editing may be desired in order to remove artifacts when present. Variability due to non-systematic manual editing was 1.7% (rater-I, 28503 ± 8683 and rater-II, 28394 ± 8667) compared to systematic manual editing. Using this systematic manual editing protocol, the variability in WMH-VQ was reduced to 0.34% overall.
3.6. Validation of a standardized protocol
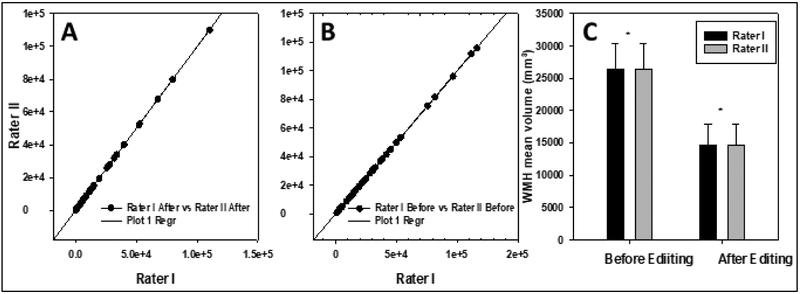
In order to investigate whether controlling for these sources of variability could result in a protocol with a minimal acceptable variability (defined as < 0.5% WMH-VQ) could be developed, we studied the performance characteristic of standardized protocol using identical acquisitions, with post-processing performed by independent raters using independent workstations, Inter-rater analysis, using Spearman correlations and linear regression models for WMH masks before and after editing (Figure 5), demonstrated r2 values = 0.999, with SE = 118.7 and 68.1 respectively, and p < 0.001 for the 50 scans used in the validation study. WMH volume variance in the refined protocol was 0.23% before manual editing (all processes automated) and this increased only slightly to 0.34% after manual editing once all sources-of-variability were addressed in a systematic fashion. The permutation test showed the observed mean difference in WMH volume before manual editing was 12.37, and P-value = 0.998; the observed mean difference was 0.97 and P-value = 0.999 after editing, which again shows a good concordance between the raters. As well, the Dice similarity test confirmed that result with 0.99 (dissimilarity: 0.009) before editing and 0.98 (dissimilarity: 0.018) after manual editing.
Figure 5.
Regression curve for WMH volumes before and after editing (Panel A and B, respectively, (n = 50)). Panel C, the mean value of WMH volume for both raters before and after editing (n = 50). R2 = 0.999, Standard error estimation before editing 118.7 and after editing 68.1.( p < 0.001).
4. DISCUSSION
This study demonstrates that even automated post-acquisition WMH-VQ techniques have several inherent sources-of-variability that can lead to discrepant results between raters and centers using different post-acquisition protocols. The importance of this finding should not be understated. The data generated and the conclusions drawn from different raters and centers, even when using standardized data acquisition and source images such as those acquired in ADNI or other large multi-center collaboratives, can be quite discrepant if post-acquisition protocols have not been refined to address such sources-of-variability.
The present data further demonstrate that systematically identifying and addressing potential sources-of-variability inherent in post-acquisition WMH-VQ techniques can result in a dramatic reduction in intra-scan variability from ~15% to less than 0.5%. Sources-of-variability identified in the present study, and methods to overcome these confounds, include the selection of CoG, thresholding effects, software versions, and manual editing procedures (as included in the protocol). Specific discussion focused on each identified source-of-variability and methods developed to reduce such variability are presented below.
The present data demonstrate the importance of software compatibility for any longitudinal, multi-center study lacking: 1) a central uniform post-acquisition processing center, 2) central processing centers that undergo software upgrades between acquisitions and processing of images, or 3) for between-study comparisons using different post-acquisition processing regimens. SPM is based on the use of MATLAB scripts. Updating one of these software packages without updating the other produced significant variability in intra-scan WMH-VQ. As software versions are constantly evolving, it is necessary to re-evaluate potential sources-of-variability introduced with each new software version employed both within and across sites. As such, one should also consider the issue of variability introduced when combining legacy data with recently acquired data if software versions are upgraded (as they are likely to be) over time. While such upgrades are important for enabling technological progress in WMH-VQ measurements, unless legacy scan data are reprocessed with the same software, drawing conclusions regarding longitudinal datasets from post-acquisition data derived from protocols using different software versions may be problematic. The present data demonstrate that considerations of increased variability in such samples could be at least partially responsible for changes in longitudinal trajectories or analyses examining historical or birth cohort effects.
Another source of variability lies in the selection of the CoG, which can affect non-brain tissue extraction. Nonbrain tissue extraction is essential for optimal brain segmentation (Xue et al., 2007). The BET stripping tool is a common brain extraction tool that is easy to both use and to script (Despotovic, Goossens, & Philips, 2015; Shattuck, Prasad, Mirza, Narr, & Toga, 2009). In order to obtain an accurate non-tissue extraction result with BET, the CoG should be consistently and uniformly assigned across protocols (Boesen et al., 2004; M S. Atkins, 2002). The closer the CoG is to the center of the brain (tissue to be analyzed), the less non-brain tissue artifact will be seen (see Figure 4). Random estimation of the CoG or variability in such estimation that differs by protocol could increase the sources-of-variability due to inclusion of residual of nonbrain tissue. This problem may be solved by either performing manual editing, increasing the number of BET iterations (S. M. Smith et al., 2007), or editing the CoG manually to ensure uniformity. The selection of three distinct CoGs isolated as independent variables, allowed us to examine the variability associated with such selection independent of other procedures. While many automated protocols select identical CoGs, the exact CoG selected often differs by protocol, and many protocols do not take into account differences in brain center coordinates that may vary from subject to subject due to subject positioning in the scanner. Certain CoG selections can increase artifacts related to excess inclusion of nonbrain tissue. Standardized selction of CoG, necessary to develop uniform protocols across diparate raters, centers, and studies will require the development of consensus best-practices in the field of post-acquisition processing.
The selection of an appropriate threshold is critical for specifying the volume of WMH to include in the mask. If the threshold is set too high, it will reduce the sensitivity of WMH detection, while setting the threshold too low can increase the presence of WMH artifacts that may necessitate the inclusion of burdensome manual editing processes. The highest sensitivity to thresholding value effects exists for subjects with large WMH volumes and is less important for those with low levels of such imaging findings. The present analysis found that two independent Gaussian curves provided the most consistent principal fit to the mean of the hyperintensity distribution. Even though the histogram distribution of WM using intensity and voxel position vs. voxel volume showed a relatively small variance < 3%, it still remained one of the sources-of-variability in excess of the acceptable threshold set in our study aims.
Manual editing may be necessary for accurate WMH-VQ assessment, as the WMH mask will likely contain at least some FLAIR artifact. The decision to include a manual editing step(s) may be dependent on the protocol specifics that either limit or increase artifact representation in the WMH-VQ assessment. The present data demonstrate that WMH-VQ can be overestimated by as much as 42% using an automated process without manual editing. While such overestimates due to artifact may exhibit regression to the mean when analyzing large samples, they prohibit accurate assessments of the true WMH-VQ and further prevent accurate analyses when working with smaller samples or when considering within-subject change in WMH-VQ. While machine learning techniques are being developed to address editing procedures systematically (Ahmed et al., 2019; Bzdok, 2017; Doyle, Mehta, & Brammer, 2015; Mateos-Perez et al., 2018), manual editing may still be required for many studies depending on the sample size and the nature of the hypothesis being tested (Bzdok, 2017). It is important to also note that machine learning techniques often require the “ground truth” in the training set (Ahmed et al., 2019; Bzdok, 2017; Doyle et al., 2015; Mateos-Perez et al., 2018). Therefore, obtaining an accurate “ground truth” was a main purpose of the present study. Given these considerations, manual editing remains a common necessity for WMH-VQ protocols until improved automated machine learning techniques are introduced into the field (Cuadrado-Godia et al., 2018).
While introducing human bias with manual editing procedures, the present data demonstrate that the development of standard rules for manual editing can significantly reduce intra-scan variability in the final WMH masks and WMH-VQ results, despite such procedures. Specific editing rules that proved useful for reducing inter-rater variability included: 1) removal of T2 hyperintensity artifacts in CSF/GM junctions, especially those involving the septum pellucidum; 2) removal of all T2 hyperintensities below the level of the midbrain, including the cerebellum, as this area is highly prone to significant pulsation and other artifacts; 3) removal of T2 signal hyperintensities in the cortical GM; and 4) editing of the supratentorial deep GM structures (including the basal ganglia and thalamus) that require special attention as these structures are in end-arterial zones that are both subject to high levels of small vessel ischemic disease and are also prone to significant artifact. (Hegde, Mohan, Lath, & Lim, 2011; Lim, 2009) Irrespective of the specific rules for manual editing standardization that are applied to a given protocol, it is clear that specifying such procedures and standardizing them across raters, sites, and studies would help reduce the variability in WMH-VQ seen within and across disparate studies.
While the present findings and method developed focus on a cross-sectional analysis, the reduction in sources-of-variability suggested in the present methods are critically important for any studies assessing longitudinal change in WMH-VQ. As change in WMH-VQ is estimated at ~5%/year, any protocol that introduces a greater degree of variability in cross-sectional findings is likely to generate inaccurate longitudinal results. Our analyses of both the findings reported in the literature and those described within our study suggest that current variability demonstrated in WMH-VQ assessment is 10-15%, a figure that is simply unacceptable. As study protocols and software versions are constantly being modified for improvement overtime, re-grounding legacy data and longitudinal data collection based on the principles described is critical for scientific discovery in the field of WMH-VQ. This new method of addressing post-acquisition sources-of-variability overcomes this limitation and may prove to be even more useful if integrated with other acquisition methods to reduce variability, e.g. longitudinal data is acquired with the same imaging sequence and protocol on the same scanner.
Study limitations include our focus on a largely Caucasian, highly-educated, aged, study population that may limit the generalizability of our findings to other populations. Minority and underserved populations are at greater risk for cerebrovascular disease and WMH accumulation and are an important focus of future studies. In addition, caution should be used in interpreting these data in regards to disease processes that may affect younger populations, such as those with multiple sclerosis, as such subjects were not studied in our experimental design. Further limitations include the specific software programs that were analyzed and a statistical threshold-based analysis approach; it is possible that the present considerations studied may not be applicable to all software programs and version upgrades. In addition, we did not fully explore how a region of interest (ROI) analyses would be impacted by the use of standardized methodologies, although it is assumed that such analyses would benefit from the standardized approach presented. Further work in this area is clearly indicated. Despite such caveats, the present data suggest that careful attention to what may seem to be simple changes in software version (incidental upgrades) or selection of post-acquisition analysis parameters ( selection of CoG and thresholding limits), and standardization of operator-dependent steps (manual editing) may improve cross-site, cross-study and longitudinal WMH-VQ assessments in order to advance the field.
Future directions include analyzing the potential sources-of-variability in WMH-VQ across-sites to better identify which variables are most important for establishing cross-center reliability. A further focus on sources-of-variability that exist within subjects in longitudinal studies also need to be pursued before we can use within subject change in WMH-VQ as a reliable outcome measure for imaging findings related to vascular cognitive impairment or vascular dementia. Data from the present study are also being used currently as the “ground truth” in our collaborative development to advance artificial intelligence machine learning approaches to WMH-VQ.
The final validation study attempted to determine if addressing all the sources-of-variability identified in the study in composite would lead to a protocol with overall reduced WMH-VQ variability that we considered acceptable (defined as variability < 0.5%). The field is in need of protocol development adequate to study within subject WMH-VQ change accurately, as average WMH-VQ change is approximately 5% the total WMH-VQ measurement. This goal was achieved demonstrating a post-acquisition WMH-VQ variance well under our target of < 0.5%. The standardized protocol used in this study may not be ideal for many researchers, depending on their needs and the practical implementation of the data derived. However, the lessons learned in addressing potential sources-of-variability in WMH-VQ assessment techniques can be applied universally to help limit methodologic variability.
5. Conclusions:
The present study sought to systematically identify sources-of-variability in WMH-VQ techniques that can create challenges for both within-site and between-site data comparisons and conclusions. This exercise allowed the development of a standardized protocol, minimizing potential sources of bias and variability in the determination of WMH-VQ measurements in our study sample. While the developed protocol was found to be optimal for use in the present dataset for the detection of subcortical white matter disease, many other protocols exist in the field and may have unique attributes that make them optimal for specific study purposes. Such protocols should, in light of the present data, systematically evaluate the sources-of-variability inherent in their methodologies to move the field of post-acquisition processing of WMH-VQ into a more rigorous and standardized arena where data may be more reliably compared across studies and sites. In addition, data on WMH-VQ that may represent a more reliable “ground truth” is critical for the development and training of machine learning algorithms that may allow future artificial intelligence approaches to WMH-VQ assessment.
These data strongly support the notion that consensus “best-practices” should be developed in the field to aid such discovery. Only through such initiatives can we hope to advance our understanding of the risks, diagnosis, study outcome measures, and treatment modality considerations that might mitigate the impact of small vessel ischemic disease on the population today.
Highlights.
Current protocols for WMH volumetric quantification have substantial variability.
Selection of image center, software, threshold, and manual editing introduce variability.
Methods to address these sources-of-variability can be developed and are essential for reliable interpretation of data.
Standardizing techniques can reduce intra-scan variability to less than 0.5%.
Acknowledgments:
This study was funded by NIH P30 AG028383, UH2 NS100606, R01 NR014189, and R01 AG042419
Abbreviations:
- WMH
white matter hyperintensities
- WMH-VQ
WMH volumetric quantification
- FSL-BET
functional MRI software library-brain extraction tool
- MIPAV
medical image processing analysis and visualization
- SPM
statistical parametric map
- CoG
center of gravity
- ADNI
Alzheimer’s Disease Neuroimaging Initiative
- MRI
magnetic resonance imaging
- TE
echo time
- TR
repetition time
- IR
inversion recovery
- FLAIR
T2-weighted fluid-attenuated inversion recovery
- MPRAGE
T1-weighted magnetization-prepared rapid acquisition gradient echo
- SE
standard error
- SD
standard deviation
- GM
gray matter
- WM
white matter
- CSF
cerebral spinal fluid
- UT
the unclassified tissue
Footnotes
Publisher's Disclaimer: This is a PDF file of an article that has undergone enhancements after acceptance, such as the addition of a cover page and metadata, and formatting for readability, but it is not yet the definitive version of record. This version will undergo additional copyediting, typesetting and review before it is published in its final form, but we are providing this version to give early visibility of the article. Please note that, during the production process, errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: The authors have nothing to disclose
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Abramson RG, Burton KR, Yu JP, Scalzetti EM, Yankeelov TE, Rosenkrantz AB, … Subramaniam RM (2015). Methods and challenges in quantitative imaging biomarker development. Acad Radiol, 22(1), 25–32. doi: 10.1016/j.acra.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed MR, Zhang Y, Feng Z, Lo B, Inan OT, & Liao H (2019). Neuroimaging and Machine Learning for Dementia Diagnosis: Recent Advancements and Future Prospects. IEEE Rev Biomed Eng, 12, 19–33. doi: 10.1109/RBME.2018.2886237 [DOI] [PubMed] [Google Scholar]
- Ambarki K, Wahlin A, Birgander R, Eklund A, & Malm J (2011). MR imaging of brain volumes: evaluation of a fully automatic software. AJNR Am J Neuroradiol, 32(2), 408–412. doi: 10.3174/ajnr.A2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anbeek P, Vincken KL, van Osch MJ, Bisschops RH, & van der Grond J (2004). Probabilistic segmentation of white matter lesions in MR imaging. Neuroimage, 21(3), 1037–1044. doi: 10.1016/j.neuroimage.2003.10.012 [DOI] [PubMed] [Google Scholar]
- Bahrani AA, Powell DK, Yu G, Johnson ES, Jicha GA, & Smith CD (2017). White Matter Hyperintensity Associations with Cerebral Blood Flow in Elderly Subjects Stratified by Cerebrovascular Risk. J Stroke Cerebrovasc Dis, 26(4), 779–786. doi: 10.1016/j.jstrokecerebrovasdis.2016.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boesen K, Rehm K, Schaper K, Stoltzner S, Woods R, Luders E, & Rottenberg D (2004). Quantitative comparison of four brain extraction algorithms. Neuroimage, 22(3), 1255–1261. doi: 10.1016/j.neuroimage.2004.03.010 [DOI] [PubMed] [Google Scholar]
- Bzdok D (2017). Classical Statistics and Statistical Learning in Imaging Neuroscience. Front Neurosci, 11, 543. doi: 10.3389/fnins.2017.00543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caligiuri ME, Perrotta P, Augimeri A, Rocca F, Quattrone A, & Cherubini A (2015). Automatic Detection of White Matter Hyperintensities in Healthy Aging and Pathology Using Magnetic Resonance Imaging: A Review. Neuroinformatics, 13(3), 261–276. doi: 10.1007/s12021-015-9260-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael O, Schwarz C, Drucker D, Fletcher E, Harvey D, Beckett L, … Alzheimer's Disease Neuroimaging, I. (2010). Longitudinal changes in white matter disease and cognition in the first year of the Alzheimer disease neuroimaging initiative. Arch Neurol, 67(11), 1370–1378. doi: 10.1001/archneurol.2010.284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadrado-Godia E, Dwivedi P, Sharma S, Ois Santiago A, Roquer Gonzalez J, Balcells M, … Suri JS (2018). Cerebral Small Vessel Disease: A Review Focusing on Pathophysiology, Biomarkers, and Machine Learning Strategies. J Stroke, 20(3), 302–320. doi: 10.5853/jos.2017.02922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Guio F, Jouvent E, Biessels GJ, Black SE, Brayne C, Chen C, … Chabriat H. (2016). Reproducibility and variability of quantitative magnetic resonance imaging markers in cerebral small vessel disease. J Cereb Blood Flow Metab, 36(8), 1319–1337. doi: 10.1177/0271678X16647396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despotovic I, Goossens B, & Philips W (2015). MRI segmentation of the human brain: challenges, methods, and applications. Comput Math Methods Med, 2015, 450341. doi: 10.1155/2015/450341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle OM, Mehta MA, & Brammer MJ (2015). The role of machine learning in neuroimaging for drug discovery and development. Psychopharmacology (Berl), 232(21–22), 4179–4189. doi: 10.1007/s00213-015-3968-0 [DOI] [PubMed] [Google Scholar]
- Hegde AN, Mohan S, Lath N, & Lim CC (2011). Differential diagnosis for bilateral abnormalities of the basal ganglia and thalamus. Radiographics, 31(1), 5–30. doi: 10.1148/rg.311105041 [DOI] [PubMed] [Google Scholar]
- Iorio M, Spalletta G, Chiapponi C, Luccichenti G, Cacciari C, Orfei MD, … Piras F. (2013). White matter hyperintensities segmentation: a new semi-automated method. Front Aging Neurosci, 5, 76. doi: 10.3389/fnagi.2013.00076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CC (2009). Magnetic resonance imaging findings in bilateral basal ganglia lesions. Ann Acad Med Singapore, 38(9), 795–798. [PubMed] [Google Scholar]
- Atkins MS, K. S., Law B, Orchard J, Rosenbaum W (2002, 9 May 2002). Difficulties of T1 brain MRI segmentation techniques. Paper presented at the Medical Imaging 2002, San Diego, California, United States. [Google Scholar]
- Mateos-Perez JM, Dadar M, Lacalle-Aurioles M, Iturria-Medina Y, Zeighami Y, & Evans AC (2018). Structural neuroimaging as clinical predictor: A review of machine learning applications. Neuroimage Clin, 20, 506–522. doi: 10.1016/j.nicl.2018.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantoni L, Simoni M, Pracucci G, Schmidt R, Barkhof F, & Inzitari D (2002). Visual rating scales for age-related white matter changes (leukoaraiosis): can the heterogeneity be reduced? Stroke, 33(12), 2827–2833. [DOI] [PubMed] [Google Scholar]
- Promjunyakul N, Lahna D, Kaye JA, Dodge HH, Erten-Lyons D, Rooney WD, & Silbert LC (2015). Characterizing the white matter hyperintensity penumbra with cerebral blood flow measures. Neuroimage Clin, 8, 224–229. doi: 10.1016/j.nicl.2015.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez J, McNeely AA, Scott CJM, Masellis M, Black SE, & Alzheimer's Disease Neuroimaging I (2016). White matter hyperintensity burden in elderly cohort studies: The Sunnybrook Dementia Study, Alzheimer's Disease Neuroimaging Initiative, and Three-City Study. Alzheimers Dement, 12(2), 203–210. doi: 10.1016/j.jalz.2015.06.1886 [DOI] [PubMed] [Google Scholar]
- Ramirez J, Scott CJ, & Black SE (2013). A short-term scan-rescan reliability test measuring brain tissue and subcortical hyperintensity volumetrics obtained using the lesion explorer structural MRI processing pipeline. Brain Topogr, 26(1), 35–38. doi: 10.1007/s10548-012-0228-z [DOI] [PubMed] [Google Scholar]
- Schmitt FA, Nelson PT, Abner E, Scheff S, Jicha GA, Smith C, … Kryscio RJ (2012). University of Kentucky Sanders-Brown healthy brain aging volunteers: donor characteristics, procedures and neuropathology. Curr Alzheimer Res, 9(6), 724–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnack HG, van Haren NE, Hulshoff Pol HE, Picchioni M, Weisbrod M, Sauer H, … Kahn RS (2004). Reliability of brain volumes from multicenter MRI acquisition: a calibration study. Hum Brain Mapp, 22(4), 312–320. doi: 10.1002/hbm.20040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segonne F, Dale AM, Busa E, Glessner M, Salat D, Hahn HK, & Fischl B (2004). A hybrid approach to the skull stripping problem in MRI. Neuroimage, 22(3), 1060–1075. doi: 10.1016/j.neuroimage.2004.03.032 [DOI] [PubMed] [Google Scholar]
- Shattuck DW, Prasad G, Mirza M, Narr KL, & Toga AW (2009). Online resource for validation of brain segmentation methods. Neuroimage, 45(2), 431–439. doi: 10.1016/j.neuroimage.2008.10.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CD, Johnson ES, Van Eldik LJ, Jicha GA, Schmitt FA, Nelson PT, … Wellnitz CV (2016). Peripheral (deep) but not periventricular MRI white matter hyperintensities are increased in clinical vascular dementia compared to Alzheimer's disease. Brain Behav, 6(3), e00438. doi: 10.1002/brb3.438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Rao A, De Stefano N, Jenkinson M, Schott JM, Matthews PM, & Fox NC (2007). Longitudinal and cross-sectional analysis of atrophy in Alzheimer's disease: cross-validation of BSI, SIENA and SIENAX. Neuroimage, 36(4), 1200–1206. doi: 10.1016/j.neuroimage.2007.04.035 [DOI] [PubMed] [Google Scholar]
- van den Heuvel DM, ten Dam VH, de Craen AJ, Admiraal-Behloul F, van Es AC, Palm WM, … Group PS (2006). Measuring longitudinal white matter changes: comparison of a visual rating scale with a volumetric measurement. AJNR Am J Neuroradiol, 27(4), 875–878. [PMC free article] [PubMed] [Google Scholar]
- van der Flier WM, Middelkoop HA, Weverling-Rijnsburger AW, Admiraal-Behloul F, Spilt A, Bollen EL, … van Buchem MA (2004). Interaction of medial temporal lobe atrophy and white matter hyperintensities in AD. Neurology, 62(10), 1862–1864. [DOI] [PubMed] [Google Scholar]
- Vilar-Bergua A, Riba-Llena I, Nafria C, Bustamante A, Llombart V, Delgado P, & Montaner J (2016). Blood and CSF biomarkers in brain subcortical ischemic vascular disease: Involved pathways and clinical applicability. J Cereb Blood Flow Metab, 36(1), 55–71. doi: 10.1038/jcbfm.2015.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, & Sachdev P (2004). The topography of white matter hyperintensities on brain MRI in healthy 60- to 64-year-old individuals. Neuroimage, 22(1), 144–154. doi: 10.1016/j.neuroimage.2003.12.027 [DOI] [PubMed] [Google Scholar]
- Wu M, Rosano C, Butters M, Whyte E, Nable M, Crooks R, … Aizenstein HJ (2006). A fully automated method for quantifying and localizing white matter hyperintensities on MR images. Psychiatry Res, 148(2–3), 133–142. doi: 10.1016/j.pscychresns.2006.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue H, Srinivasan L, Jiang S, Rutherford M, Edwards AD, Rueckert D, & Hajnal JV (2007). Automatic segmentation and reconstruction of the cortex from neonatal MRI. Neuroimage, 38(3), 461–477. doi: 10.1016/j.neuroimage.2007.07.030 [DOI] [PubMed] [Google Scholar]