Abstract
Astrocytoma is an invasive carcinoma occurring in the nervous system and currently lacks effective treatment options. A deeper understanding of the mechanisms of tumorigenesis and tumor progression is needed in order to develop novel therapeutic strategies. Recent advances in in vitro culture systems have demonstrated that the use of three-dimensional (3D) culture models could be more relevant for this purpose as compared to monolayer or two-dimensional (2D) models due to their resemblance to in vivo cancer pathology. High-throughput techniques such as RNA sequencing, microarray analyses and cloning could provide useful insights into the relevance of these systems to the native tissue. Previous studies have reported RNA extraction protocols needed for such applications. We have modified these protocols to suit the isolation of total RNA from monolayer and hydrogel cultures of astrocytoma established using basement membrane matrix, GeltrexTM. We have used this method to demonstrate the differences in the expression of genes involved in autophagy, a process deregulated in many cancer types, in monolayer and hydrogel cultures using quantitative polymerase chain reaction (qPCR). This protocol can be adopted by the researchers who wish to understand the molecular basis of gene expression in hydrogel cultures of normal as well as cancer cell lines.
Keywords: RNA, Monolayer, 3D, Hydrogel, Glioblastoma, Cancer, TRIzol, Cell recovery solution, Astrocytoma
Background
Astrocytomas are central nervous system tumors of glial origin. Poor survival rates among patients within 5 years of diagnosis and almost inevitable relapse are the hallmarks of this life-threatening disease ( Krex et al., 2007 ). Our understanding of disease pathogenesis is limited, resulting in the lack of effective intervention strategies. In vitro culture systems could prove useful for studying cell-cell interactions and intracellular signaling events in astrocytoma, and provide platforms for testing potential therapeutic compounds ( Reardon et al., 2006 ; Baker and Chen, 2012). Three-dimensional (3D) in vitro culture platforms have especially gained popularity in recent years due to their biological relevance to in vivo tumors ( Li et al., 2007 ; Whiteside, 2008; Tibbitt and Anseth, 2009; Xu et al., 2014 ; Jogalekar and Serrano, 2018). 3D cultures established using various tissue engineering scaffolds (e.g., hydrogels) allow for multi-layered growth of cells in a fashion similar to that of native tissue, unlike two-dimensional (2D) cultures, making them better predictors of responses to drug candidates.
Hydrogel culture systems could be exploited in parallel with monolayer cultures to further our understanding of cell-cell and cell-matrix interactions with high-throughput applications that analyze cellular gene expression, such as RNA-sequencing, microarray analysis and cloning. To accomplish this, a pure, intact, high quality RNA needs to be isolated in parallel from separately cultured monolayer and hydrogel cultures so that both cultures are processed under identical conditions and are therefore comparable. Several protocols have been reported for extracting RNA from 3D cultures established using a variety of commonly used tissue engineering scaffolds such as Matrigel and collagen ( Lee et al., 2007 ; Mroue and Bissell, 2013). However, there is a paucity of information available with respect to RNA extraction from 3D cultures maintained in GeltrexTM, a relatively new basement membrane matrix derived from murine Engelbreth-Holm-Swarm tumors and manufactured by Invitrogen.
When we used existing protocols to carry out RNA isolation from multiple cell lines including astrocytoma, we found that our astrocytoma cell line (CCF-STTG1) was highly sensitive to the reagents that dissolve extracellular matrices used in 3D cultures and could not yield an intact RNA at the end of the procedure, although this procedure worked well with other mammalian and amphibian cell lines we used in our experiment. Therefore, we modified RNA isolation protocols available in the literature to extract total RNA from monolayer and hydrogel cultures of astrocytoma. We used this RNA to carry out gene expression analyses for key autophagy pathway genes, Beclin-1 (BECN1) and microtubule associated protein 1 light chain 3 beta (MAP1LC3B), using quantitative polymerase chain reaction (qPCR) ( Jogalekar et al., 2017 ). Our findings indicate a modest upregulation of these genes in hydrogel cultures of astrocytoma and are consistent with previous reports suggesting their upregulation in correlation with poor prognosis in glioma patients ( Pirtoli et al., 2009 ; Giatromanolaki et al., 2014 ).
Materials and Reagents
0.8 μm 150 ml filter unit (Thermo Scientific Nalgene, catalog number: 125-0080)
NuncTM Cell Culture Treated Flasks with Filter Caps (NuncTM, catalog number: 136196)
Falcon® Cell Scrapers, Sterile (Corning®, catalog number: 353085)
Falcon® 50 ml High Clarity PP Centrifuge Tubes (Corning®, catalog number: 352098)
RNase-free Microfuge Tubes (1.5 ml) (InvitrogenTM, catalog number: AM12400)
5cc BD Luer-LokTM Disposable Syringe (BD, catalog number: 309603)
21 G 1 needle (BD, catalog number: 305166)
10 ml serological pipette, sterile (USA Scientific, catalog number: 1071-0810)
FisherbrandTM SureOneTM Aerosol Barrier Pipette Tips (FisherbrandTM SureOneTM, catalog numbers: 02-707-404 [1,000 μl], 02-707-430 [200 μl], 02-707-432 [20 μl])
CCF-STTG1 cell line (ATCC, catalog number: CRL-1718)
RPMI-1640 Medium (ATCC, catalog number: 30-2001)
Fetal Bovine Serum (FBS) (ATCC, catalog number: 30-2020)
LDEV-Free Reduced Growth Factor Basement Membrane Matrix, GeltrexTM (Invitrogen, catalog number: A1413202)
70% Ethanol
RNaseZapTM RNase Decontamination Solution (AmbionTM, catalog number: AM9780)
TRIzolTM Plus RNA Purification Kit (AmbionTM, catalog number: 12183555)
Cell Recovery Solution (Corning, catalog number: 354253)
Phosphate buffered saline (PBS) (Sigma-Aldrich, catalog number: P3813)
Chloroform (Sigma-Aldrich, catalog number: C2432)
Nuclease-Free Water (not DEPC-Treated) (AmbionTM, catalog number: AM9937)
DNA-free DNA Removal Kit (AmbionTM, catalog number: AM1906)
RNA 6000 Nano Kit (Agilent, catalog number: 5067-1511)
Ethyl alcohol, Pure (Sigma-Aldrich, catalog number: E7023)
RPMI-1640 complete medium (see Recipes)
Sterile 1x PBS (see Recipes)
Equipment
GilsonTM PIPETMAN ClassicTM Pipettes (Gilson, catalog numbers: F123600 [P20], F123601 [P200], F123602 [P1000], F144802 [P10], F144801 [P2])
2100 bioanalyzer instrument (Agilent, catalog number: G2939BA)
Drummond Pipet-Aid, Plain, 110V (Drummond, catalog number: DP-110)
Biosafety cabinet (Nuaire, catalog number: NU-425-600)
4 °C centrifuge (Beckman Coulter, catalog number: AllegraTM 21R)
Centrifuge rotors
Water bath at 37 °C (Sheldon Manufacturing Inc., catalog number: 1211)
AutoFlow Water Jacket CO2 Incubator (Nuaire, catalog number: NU-4750)
-20 °C freezer
Software
2100 Expert (Agilent)
Photoshop CS6 (Adobe)
Procedure
-
Cell culture
Establish 2D and 3D cultures separately, of CCF-STTG1 cells using RPMI-1640 complete medium. For 3D culture, coat T25 flasks with 100 µl per cm2 GeltrexTM and allow it to solidify in a 37°C incubator for 30 min. Plate CCF-STTG1 cells on top of the solidified GeltrexTM at a density of 5 x 104 cells per cm2. For monolayer or 2D culture, plate equal number of cells directly in T25 flasks (do not coat these flasks with matrix prior to the addition of cells). Incubate the flasks at 37 °C and 5% CO2.
Aspirate the old media and add fresh media to culture flasks every other day. Monitor the cell growth periodically.
-
RNA isolation
Note: Make sure that all the supplies used for the RNA isolation such as centrifuge tubes, pipette tips, plastic pipettes and microcentrifuge tubes are sterile, and DNase- and RNase-free.
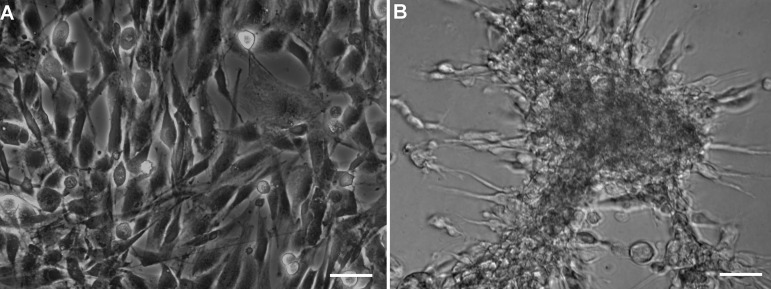
Begin RNA isolation when the cells in 2D and 3D cultures are ~80-90% confluent (Figure 1). This usually takes about a week, although it may vary with the cell and culture type. Process both culture flasks on the same day in parallel.
Treat all surfaces with ethanol followed by RNase ZAP prior to UV-including both centrifuge rotors, pipettes, and bench surfaces outside the tissue culture room.
Turn on the centrifuge so it cools to 4 °C.
Turn on and set the water bath at 37 °C.
UV-sterilize the tissue culture hood and the required materials for 30 min before beginning the RNA extraction procedure. Also, have cold 1x PBS ready (for use with cell recovery solution).
-
Without removing the media, detach cells from the bottom of the 2D and 3D culture flasks using cell scrapers and transfer them to 50 ml centrifuge tubes. Centrifuge for 5 min at 200 × g at 4 °C. Trypsin was not used in this procedure since it alters gene expression (Chaudhry, 2008).
Note: Cells can be lysed with TRIzol® reagent in a culture dish or flask directly if the dishes are made of materials that are compatible with TRIzol® reagent. Please check the product details and their compatibility before doing so.
Decant the supernatant from the tube containing 3D cells. Please make sure to leave equal volume of media in the other tube containing monolayer cell pellet, since these tubes will be processed in parallel under identical conditions.
-
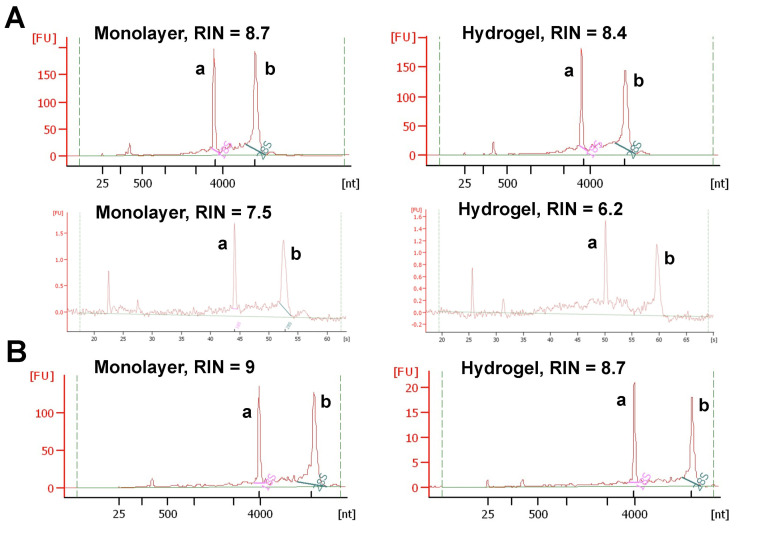
Separate the astrocytoma cells from the basement membrane matrix with one of the two ways–using TRIzol or cell recovery solution. The method of choice is determined by analyzing the sensitivity of cell lines of interest to both matrix-dissolving reagents (by trial and error method). We found that the cell recovery solution worked well with some cell lines (e.g., HCC70 cell line, Figure 2B), while others were found to be sensitive and died upon exposure (e.g., CCF-STTG1 cell line, Figure 2A). Therefore, we decided to use TRIzol for these cell lines. Since 2D and 3D cultures must be processed identically, 2D or monolayer cultures were also exposed to equal volumes of TRIzol or cell recovery solution although they do not contain matrix.
-
Using TRIzol reagent:
Add TRIzol® reagent to both tubes so that the sample volume is 10% of the total volume and incubate the samples for 5 min. For example, if the sample volume is 1 ml, then 9 ml of TRIzol should be added.
Lyse cells by passing them through 21 G x 1" needle and 5cc syringe at least 5-6 times.
After cell lysis, add 0.2 ml of chloroform per ml of TRIzol® reagent, invert the tubes to mix the contents for 15 s followed by the incubation for 2-3 min at room temperature.
Centrifuge at 15,000 × g for 15 min at 4 °C.
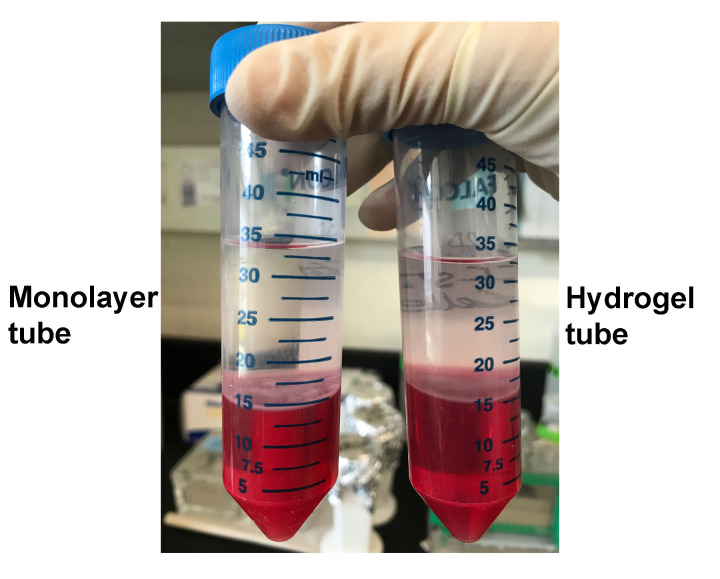
Transfer colorless upper phase to the RNase-free tube. The volume of colorless upper phase varies depending upon the volumes of sample and TRIzol/chloroform added in previous steps. For example, in our case, the volume of matrix containing cells was 2.8 ml, so we added 2.8 x 9 ml = 25.2 ml of TRIzol plus 0.2 x 25.2 ml = 5.04 ml of chloroform. So the total volume of the solution was ~33 ml. Out of that, the volume of colorless upper phase came out to be ~50%, i.e., 18 ml (Figure 3).
Add 70% ethanol (equal volume) to a final concentration of 35%. Use absolute or molecular biology grade alcohol to make 70% ethanol to reduce the chances of contamination.
Mix well by inverting the tube to disperse precipitate.
-
Using cell recovery solution:
Wash cells thrice with cold 1x PBS and add equal amounts of cell recovery solution (227µl per cm2) directly to T25 flasks.
Scrape the cells using cell scrapers and transfer to a cold 50 ml centrifuge tube.
-
Incubate on ice till GeltrexTM is dissolved (up to 1 h, varies for each cell line). Mix the suspension by inverting the tubes every 15 min.
Note: Please pay attention to exposure times for cell recovery solution used to dissolve GeltrexTM. Some cell lines may be more sensitive to this reagent than others. Optimize treatment times for your cell line before conducting a major experiment.
When the cells settle down at the bottom of the tube, centrifuge the mixture at 200 × g at 4 °C for 5 min.
Wash cells in cold 1x PBS twice.
-
Proceed with RNA extraction using the manufacturer’s (Ambion) protocol for isolating total RNA from the animal cells.
Use DNA-freeTM kit to remove DNA contamination and store pure RNA at -80 °C until use.
-
Check the quality of extracted RNA integrity with Agilent bioanalyzer (Figure 2).
Note: The samples with RNA integrity number (RIN) > 8 were considered pure and high quality, and used for experiments. If RNA concentration in the sample is less than 25 ng/μl, Agilent software may not show RIN values (Mueller et al., 2004). In that case, it is essential to determine RNA concentration using another method like Nanodrop. It is also important to confirm RNA integrity by visually inspecting electropherograms provided by Agilent Software even though RIN values are not displayed.
Figure 1. Phase contrast images showing 80-90% confluent CCF-STTG1 cells in monolayer and hydrogel cultures.
CCF-STTG1 cells were allowed to grow in A) Monolayer and B) Hydrogel cultures until they reached 80-90% confluency before beginning RNA extraction procedure.
Figure 2. Electropherograms showing the quality of total RNA extracted from monolayer and hydrogel cultures.
The total RNA was extracted from monolayer and hydrogel cultures of astrocytoma cell line, CCF-STTG1. One microliter of total RNA per sample was analyzed using Agilent Bioanalyzer. A. RNA isolated from CCF-STTG1 cells using TRIzol (Top panel) and cell recovery solution (Bottom panel). B. RNA extracted from HCC70 (breast cancer cell line) cells used as control to show that cell recovery solution works well for HCC70 cells, but not for CCF-STTG1 cells. a: 18s peak, b: 28s peak.
Figure 3. Colorless upper phase separated after centrifugation step.
Followed by incubation with TRIzol and chloroform, the monolayer and hydrogel samples of CCF-STTG1 were centrifuged for 15 min at 15,000 × g at 4 °C to obtain colorless upper phase which contains RNA.
Data analysis
Determine the RNA quality using Agilent bioanalyzer 2100 and RNA 6000 Nano Kit.
Load RNA samples on the chip using the manufacturer’s (Agilent) instructions and read the chip using 2100 Bioanalyzer Expert Software (Figure 2).
Recipes
-
RPMI-1640 complete medium
RPMI 1640 basal medium
10% FBS
-
Mix both ingredients and filter sterilize the medium through a 0.8 μm filter unit
Note: We do not use antibiotics for cell culture in our laboratory but you can add them to the medium at the appropriate concentration (usually 1% of the total volume) depending upon your requirements.
Store at 4 °C and use within a month
-
-
Sterile 1x PBS
Dissolve the powdered contents from one pouch of PBS to 1,000 ml of distilled water to get 1x PBS, pH 7.4
Autoclave the solution and store at room temperature
Acknowledgments
We are grateful for the funding support provided by the New Mexico State University Manasse Chair Endowment to E. E. Serrano, and the National Institutes of Health (P50GM68762) grant. We thank V. B. Knight for providing technical assistance for this work. This protocol was modified from previously published RNA extraction methods ( Lee et al., 2007 ; Mroue and Bissell, 2013).
Competing interests
The authors declare no competing interests.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Baker B. M. and Chen C. S.(2012). Deconstructing the third dimension: how 3D culture microenvironments alter cellular cues. J Cell Sci 13): 3015-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chaudhry A. M.(2008). Induction of gene expression alterations by culture medium from trypsinized cells. J Biol Sci 8(1): 81-87. [Google Scholar]
- 3. Giatromanolaki A., Sivridis E., Mitrakas A., Kalamida D., Zois C. E., Haider S., Piperidou C., Pappa A., Gatter K. C., Harris A. L. and Koukourakis M. I.(2014). Autophagy and lysosomal related protein expression patterns in human glioblastoma. Cancer Biol Ther 15(11): 1468-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jogalekar M. P., Cooper L. G. and Serrano E. E.(2017). Hydrogel environment supports cell culture expansion of a grade IV astrocytoma. Neurochem Res 42(9): 2610-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jogalekar M. P. and Serrano E. E.(2018). Morphometric analysis of a triple negative breast cancer cell line in hydrogel and monolayer culture environments. PeerJ 6: e4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Krex D., Klink B., Hartmann C., von Deimling A., Pietsch T., Simon M., Sabel M., Steinbach J. P., Heese O., Reifenberger G., Weller M., Schackert G. and German Glioma N.(2007). Long-term survival with glioblastoma multiforme. Brain 10): 2596-2606. [DOI] [PubMed] [Google Scholar]
- 7. Lee G. Y., Kenny P. A., Lee E. H. and Bissell M. J.(2007). Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods 4(4): 359-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li H., Fan X. and Houghton J.(2007). Tumor microenvironment: the role of the tumor stroma in cancer. J Cell Biochem 101(4): 805-815. [DOI] [PubMed] [Google Scholar]
- 9. Mroue R. and Bissell M. J.(2013). Three-dimensional cultures of mouse mammary epithelial cells. Methods Mol Biol 945: 221-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mueller O., Lightfoot S. and Schroeder A.(2004). RNA integrity number(RIN)–standardization of RNA quality control. Agilent application note: 5989-1165EN. [Google Scholar]
- 11. Pirtoli L., Cevenini G., Tini P., Vannini M., Oliveri G., Marsili S., Mourmouras V., Rubino G. and Miracco C.(2009). The prognostic role of Beclin 1 protein expression in high-grade gliomas. Autophagy 5(7): 930-936. [DOI] [PubMed] [Google Scholar]
- 12. Reardon D. A., Rich J. N., Friedman H. S. and Bigner D. D.(2006). Recent advances in the treatment of malignant astrocytoma. J Clin Oncol 24(8): 1253-1265. [DOI] [PubMed] [Google Scholar]
- 13. Tibbitt M. W. and Anseth K. S.(2009). Hydrogels as extracellular matrix mimics for 3D cell culture. Biotechnol Bioeng 103(4): 655-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Whiteside T. L.(2008). The tumor microenvironment and its role in promoting tumor growth. Oncogene 27(45): 5904-5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xu X., Sabanayagam C. R., Harrington D. A., Farach-Carson M. C. and Jia X.(2014). A hydrogel-based tumor model for the evaluation of nanoparticle-based cancer therapeutics. Biomaterials 35(10): 3319-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]