Abstract
The chaperonin GroEL and its co-chaperonin GroES form both GroEL–GroES bullet-shaped and GroEL–GroES2 football-shaped complexes. The residence time of protein substrates in the cavities of these complexes is about 10 and 1 s, respectively. There has been much controversy regarding which of these complexes is the main functional form. Here, we show using computational analysis that GroEL protein substrates have a bimodal distribution of folding times, which matches these residence times, thereby suggesting that both bullet-shaped and football-shaped complexes are functional. More generally, co-existing complexes with different stoichiometries are not mutually exclusive with respect to having a functional role and can complement each other.
Keywords: chaperonin, protein folding, GroEL, kinetics, bioinformatics, molecular machines, PSIPRED
Introduction
The Escherichia coli GroE system assists protein folding in vivo and in vitro by an ATP-dependent mechanism (1). It comprises GroEL, a complex of two back-to-back stacked rings, each made up of seven identical subunits, and its cofactor GroES, which is a single homoheptameric ring. Binding of GroES to GroEL forms a cage in which substrate proteins can fold in isolation from bulk solution (1). Early EM and biochemical studies showed that GroEL and GroES are able to form both GroEL–GroES2 symmetric football-shaped (2–5) and GroEL–GroES asymmetric bullet-shaped (6, 7) complexes. The crystal structure of the bullet complex was solved more than 2 decades ago (8), and, more recently, crystal structures of the football complex have also become available (9, 10). The existence of both types of complexes is, thus, not in any dispute. Moreover, both types of complexes were found to co-exist under certain conditions (11). The main controversy has concerned the identity of the predominant functional form of the GroE machine. Some studies show that, in the presence of substrate protein, the relative populations of the asymmetric and symmetric complexes shift in favor of the latter species (12, 13). By contrast, others have argued that the asymmetric complex is the functional form and that the symmetric species accumulates only in the presence of unfoldable protein substrates (14). Given that GroEL has become a paradigm for molecular machines, in general, and chaperonins, in particular, resolving the controversy regarding the identity of its main functional form is important and has been the subject of reviews (e.g. see Refs. 11 and 15). It is also of broad interest because other protein complexes with varying subunit stoichiometries are known to co-exist in vitro and in vivo (16). An example is the 20S core particle of the proteasome, which forms both 1:1 and 1:2 complexes with the 19S lid complex (17).
The debate regarding the functional roles of the bullet- and football-like complexes is of importance for understanding GroE's mechanism of action, in part, because it bears on the residence time of substrate proteins encapsulated in its cavity. In the case of the asymmetric complex, the residence time is estimated to be 10–15 s (18, 19), whereas, in the case of the symmetric complex, it is only about 1 s (13). Short residence times are consistent with the iterative annealing model (20) according to which the role of encapsulation is to subject the substrate protein to forced unfolding upon ATP and GroES-promoted conformational changes. According to the iterative annealing model, GroEL assists folding by unfolding misfolded proteins, thereby providing them with further opportunity to fold correctly inside or outside the cavity. The short residence times in the symmetric complex would, therefore, result in more iterations and higher folding yields. By contrast, long residence times are more consistent with a model according to which encapsulation favors folding owing to confinement and properties of the cavity walls and the cavity-confined water (1). Given that GroEL has evolved to facilitate the folding of a range of protein substrates (21, 22) with different properties (23), we consider here the possibility that symmetric and asymmetric complexes specialize in assisted folding of different protein substrate subsets and that both are, therefore, functional forms.
Results and discussion
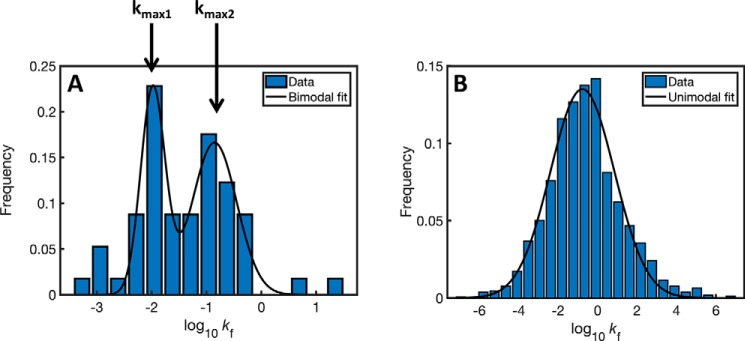
Assuming that slow folders might benefit from longer encapsulation times, we hypothesized that GroEL substrates have a bimodal distribution of folding rates (i.e. times) in accord with the different residence times in the two GroE complexes. We first compared the distribution of folding rates of the 57 obligatory GroEL substrates (21, 22) with that of all of the cytosolic proteins in E. coli. Given that many of the relevant three-dimensional structures are not available, the folding rate (kF) of each protein was calculated from its length and predicted secondary structure (24) as described before (25). It may be seen that the distribution of folding rates of the obligatory GroEL substrates is indeed found to be bimodal (Fig. 1A), whereas that of all of the ∼1600 cytosolic proteins is unimodal (Fig. 1B). The modality of these distributions was confirmed using the Bayesian information criterion (26). Strikingly, the ratio of the folding times (t = ln2/kF) corresponding to the two maxima of the bimodal distribution matches very closely to the ratio of experimentally determined residence times in the bullet and football forms (log(kmax1/kmax2) = log(t2/t1) = −1.95 + 0.82 = −1.13 (i.e. t2/t1 = 0.07), whereas the ratio of estimated residence times (13, 18, 19) is between (i.e. 0.1) and (i.e. 0.07)).
Figure 1.
Distributions of folding rates of GroEL substrates and all cytosolic E. coli proteins. A, the distribution of folding rates of GroEL substrates is bimodal with maximum values at log(kF) of −1.95 and −0.82. Bimodality was confirmed using the Bayesian information criterion (BIC); BIC = nln(RSS/n) + kln(n), where n in the number of data points, RSS is the residual sum of squares between the model and the data, and k is the number of parameters in the model. The two models were compared using ΔBICi,j = BICi − BICj, where i and j represent the number of modes in the model. Here, we found ΔBIC1,2 = 16, indicating BIC2 < BIC1 and, therefore, that the bimodal distribution describes the data significantly better (lower BIC score is better). B, distribution of the folding rates of all E. coli cytosolic proteins is unimodal, with a maximum value of log(kF) = −0.72. Here, ΔBIC1,2 = −22, thereby indicating that the unimodal distribution describes the data significantly better.
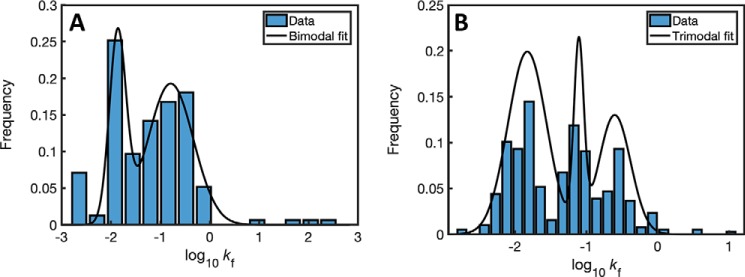
Next, we analyzed the distributions of folding rates of homologs of the 57 obligatory GroEL substrates in mollicutes (27), a class of bacteria that includes both organisms with a chaperonin system and (the only known) organisms without a chaperonin system. The distribution of folding rates of the 155 homologs of GroEL substrates in mollicutes with a chaperonin system was also found to be bimodal (Fig. 2A). By contrast, the distribution of folding rates of the 387 homologs of GroEL substrates in mollicutes without a chaperonin system was found to be multimodal (Fig. 2B). These results support our proposal that bimodality in the distribution of folding rates is a property of GroEL substrates that is linked to the mechanism of their GroE dependence.
Figure 2.
Distributions of folding rates of homologs of the GroEL substrates in mollicutes with and without a chaperonin system. A, distribution of folding rates of 155 homologs of GroEL substrates in mollicutes with a chaperonin system. For this class, ΔBIC1,2 = 15, indicating that the bimodal distribution describes the data better than a unimodal distribution, as found for the GroEL substrates in E. coli. B, distribution of folding rates of 387 homologs of GroEL substrates in mollicutes without a chaperonin system. For this class, ΔBIC1,2 = 6 and ΔBIC2,3 = 28, indicating that a trimodal distribution describes the data significantly better than unimodal or bimodal distributions.
In summary, there is compelling biochemical evidence that GroE footballs are functional. It is not clear, however, why inter-ring negative cooperativity (28), which is responsible for the presence of the bullet form, has evolved if the only functional form is the football. Our data suggest that both forms are functional with respect to substrates with different folding rates. It is also possible, however, that both forms are functional under different conditions. It has been shown, for example, that formation of footballs is favored under high ATP concentrations, but the range of ATP concentrations in E. coli cells varies widely and was reported to be 1.55 ± 1.22 mm (29). Consequently, the asymmetric species may also be the main functional form at low ATP concentrations. In general, co-existing complexes can complement each other and need not be mutually exclusive in having a functional role.
Materials and methods
The sequences of E. coli (K12 strain, organism ID 83333) proteins were extracted from Uniprot. The number of cytosolic proteins, which were defined as such if they did not include the keyword “membrane” in their Uniport accession, was found to be 1579. The list of GroEL homologs from mollicutes was taken from Ref. 27. PSIPRED (24) was used to predict the secondary structure of all of the proteins in this study. Folding rates were calculated using the equation, log10(kF) = 10.7 − 16.6(leff0.1 − 1), where leff is the effective protein length (25). The experimentally determined (30) folding rate of the GroEL substrate PepQ, for example, is 0.039 min−1 (log10(kF) = −3.23), in good agreement with the calculated rate of 0.026 min−1 (log10(kF) = −3.36).
Author contributions
L. S. B. data curation; L. S. B. and A. H. formal analysis; L. S. B. investigation; L. S. B. methodology; A. H. conceptualization; A. H. supervision; A. H. funding acquisition; A. H. writing-original draft.
Acknowledgments
We thank Drs. R. Gruber and E. M. Meiering for helpful comments on this paper.
Footnotes
This work was supported by United States–Israel Binational Science Foundation Grant 2015170 and the Minerva Foundation with funding from the Federal German Ministry for Education and Research and the Kimmelman Center for Biomolecular Structure and Assembly. The authors declare that they have no conflicts of interest with the contents of this article.
References
- 1. Hayer-Hartl M., Bracher A., and Hartl F. U. (2016) The GroEL-GroES chaperonin machine: a nano-cage for protein folding. Trends Biochem. Sci. 41, 62–76 10.1016/j.tibs.2015.07.009 [DOI] [PubMed] [Google Scholar]
- 2. Llorca O., Marco S., Carrascosa J. L., and Valpuesta J. M. (1994) The formation of symmetrical GroEL-GroES complexes in the presence of ATP. FEBS Lett. 345, 181–186 10.1016/0014-5793(94)00432-3 [DOI] [PubMed] [Google Scholar]
- 3. Schmidt M., Rutkat K., Rachel R., Pfeifer G., Jaenicke R., Viitanen P., Lorimer G., and Buchner J. (1994) Symmetric complexes of GroE chaperonins as part of the functional cycle. Science 265, 656–659 10.1126/science.7913554 [DOI] [PubMed] [Google Scholar]
- 4. Azem A., Kessel M., and Goloubinoff P. (1994) Characterization of a functional GroEL14(GroES7)2 chaperonin hetero-oligomer. Science 265, 653–656 10.1126/science.7913553 [DOI] [PubMed] [Google Scholar]
- 5. Harris J. R., Plückthun A., and Zahn R. (1994) Transmission electron microscopy of GroEL, GroES, and the symmetrical GroEL/ES complex. J. Struct. Biol. 112, 216–230 10.1006/jsbi.1994.1022 [DOI] [PubMed] [Google Scholar]
- 6. Ishii N., Taguchi H., Yoshida M., Yoshimura H., and Nagayama K. (1991) Image analysis by electron microscopy of two-dimensional crystals developed on a mercury surface of chaperonin from Thermus thermophilus. J. Biochem. 110, 905–908 [PubMed] [Google Scholar]
- 7. Saibil H. R., Zheng D., Roseman A. M., Hunter A. S., Watson G. M., Chen S., auf der Mauer A., O'Hara B. P., Wood S. P., Mann N. H., Barnett L. K., and Ellis R. J. (1993) ATP induces large quaternary rearrangements in a cage-like chaperonin structure. Curr. Biol. 3, 265–273 10.1016/0960-9822(93)90176-O [DOI] [PubMed] [Google Scholar]
- 8. Xu Z., Horwich A. L., and Sigler P. B. (1997) The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature 388, 741–750 10.1038/41944 [DOI] [PubMed] [Google Scholar]
- 9. Fei X., Ye X., LaRonde N. A., and Lorimer G. H. (2014) Formation and structures of GroEL:GroES2 chaperonin footballs, the protein-folding functional form. Proc. Natl. Acad. Sci. U.S.A. 111, 12775–12780 10.1073/pnas.1412922111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Koike-Takeshita A., Arakawa T., Taguchi H., and Shimamura T. (2014) Crystal structure of a symmetric football-shaped GroEL:GroES2-ATP14 complex determined at 3.8 Å reveals rearrangement between two GroEL rings. J. Mol. Biol. 426, 3634–3641 10.1016/j.jmb.2014.08.017 [DOI] [PubMed] [Google Scholar]
- 11. Noshiro D., and Ando T. (2018) Substrate protein dependence of GroEL-GroES interaction cycle revealed by high-speed atomic force microscopy imaging. Philos. Trans. R. Soc. Lond. B Biol. Sci. 373, 20170180 10.1098/rstb.2017.0180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ye X., and Lorimer G. H. (2013) Substrate protein switches GroE chaperonins from asymmetric to symmetric cycling by catalyzing nucleotide exchange. Proc. Natl. Acad. Sci. U.S.A. 110, E4289–E4297 10.1073/pnas.1317702110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang D., Ye X., and Lorimer G. H. (2013) Symmetric GroEL:GroES2 complexes are the protein-folding functional form of the chaperonin nanomachine. Proc. Natl. Acad. Sci. U.S.A. 110, E4298–E4305 10.1073/pnas.1318862110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haldar S., Gupta A. J., Yan X., Miličić G., Hartl F. U., and Hayer-Hartl M. (2015) Chaperonin-assisted protein folding: relative population of asymmetric and symmetric GroEL:GroES complexes. J. Mol. Biol. 427, 2244–2255 10.1016/j.jmb.2015.04.009 [DOI] [PubMed] [Google Scholar]
- 15. Taguchi H. (2015) Reaction cycle of chaperonin GroEL via symmetric “football” intermediate. J. Mol. Biol. 427, 2912–2918 10.1016/j.jmb.2015.04.007 [DOI] [PubMed] [Google Scholar]
- 16. Chorev D. S., Ben-Nissan G., and Sharon M. (2015) Exposing the subunit diversity and modularity of protein complexes by structural mass spectrometry approaches. Proteomics 15, 2777–2791 10.1002/pmic.201400517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bard J. A. M., Goodall E. A., Greene E. R., Jonsson E., Dong K. C., and Martin A. (2018) Structure and function of the 26S proteasome. Annu. Rev. Biochem. 87, 697–724 10.1146/annurev-biochem-062917-011931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Weissman J. S., Rye H. S., Fenton W. A., Beechem J. M., and Horwich A. L. (1996) Characterization of the active intermediate of a GroEL-GroES-mediated protein folding reaction. Cell 84, 481–490 10.1016/S0092-8674(00)81293-3 [DOI] [PubMed] [Google Scholar]
- 19. Horwich A. L. (2011) Protein folding in the cell: an inside story. Nat. Med. 17, 1211–1216 10.1038/nm.2468 [DOI] [PubMed] [Google Scholar]
- 20. Todd M. J., Lorimer G. H., and Thirumalai D. (1996) Chaperonin-facilitated protein folding: optimization of rate and yield by an iterative annealing mechanism. Proc. Natl. Acad. Sci. U.S.A. 93, 4030–4035 10.1073/pnas.93.9.4030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kerner M. J., Naylor D. J., Ishihama Y., Maier T., Chang H. C., Stines A. P., Georgopoulos C., Frishman D., Hayer-Hartl M., Mann M., and Hartl F. U. (2005) Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell 122, 209–220 10.1016/j.cell.2005.05.028 [DOI] [PubMed] [Google Scholar]
- 22. Fujiwara K., Ishihama Y., Nakahigashi K., Soga T., and Taguchi H. (2010) A systematic survey of in vivo obligate chaperonin-dependent substrates. EMBO J. 29, 1552–1564 10.1038/emboj.2010.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Azia A., Unger R., and Horovitz A. (2012) What distinguishes GroEL substrates from other Escherichia coli proteins? FEBS J. 279, 543–550 10.1111/j.1742-4658.2011.08458.x [DOI] [PubMed] [Google Scholar]
- 24. Jones D. T. (1999) Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202 10.1006/jmbi.1999.3091 [DOI] [PubMed] [Google Scholar]
- 25. Ivankov D. N., and Finkelstein A. V. (2004) Prediction of protein folding rates from the amino acid sequence-predicted secondary structure. Proc. Natl. Acad. Sci. U.S.A. 101, 8942–8944 10.1073/pnas.0402659101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Priestly M. B. (1981) Spectral Analysis and Time Series, p. 375, Academic Press, New York [Google Scholar]
- 27. Schwarz D., Adato O., Horovitz A., and Unger R. (2018) Comparative genomic analysis of mollicutes with and without a chaperonin system. PLoS One 13, e0192619 10.1371/journal.pone.0192619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yifrach O., and Horovitz A. (1995) Nested cooperativity in the ATPase activity of the oligomeric chaperonin GroEL. Biochemistry 34, 5303–5308 10.1021/bi00016a001 [DOI] [PubMed] [Google Scholar]
- 29. Yaginuma H., Kawai S., Tabata K. V., Tomiyama K., Kakizuka A., Komatsuzaki T., Noji H., and Imamura H. (2014) Diversity in ATP concentrations in a single bacterial cell population revealed by quantitative single-cell imaging. Sci. Rep. 4, 6522 10.1038/srep06522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Weaver J., Jiang M., Roth A., Puchalla J., Zhang J., and Rye H. S. (2017) GroEL actively stimulates folding of the endogenous substrate protein PepQ. Nat. Commun. 8, 15934 10.1038/ncomms15934 [DOI] [PMC free article] [PubMed] [Google Scholar]