Abstract
Protein methyltransferases mediate posttranslational modifications of both histone and nonhistone proteins. Whereas histone methylation is well-known to regulate gene expression, the biological significance of nonhistone methylation is poorly understood. Methyltransferase-like 21c (Mettl21c) is a newly classified nonhistone lysine methyltransferase whose in vivo function has remained elusive. Using a Mettl21cLacZ knockin mouse model, we show here that Mettl21c expression is absent during myogenesis and restricted to mature type I (slow) myofibers in the muscle. Using co-immunoprecipitation, MS, and methylation assays, we demonstrate that Mettl21c trimethylates heat shock protein 8 (Hspa8) at Lys-561 to enhance its stability. As such, Mettl21c knockout reduced Hspa8 trimethylation and protein levels in slow muscles, and Mettl21c overexpression in myoblasts increased Hspa8 trimethylation and protein levels. We further show that Mettl21c-mediated stabilization of Hspa8 enhances its function in chaperone-mediated autophagy, leading to degradation of client proteins such as the transcription factors myocyte enhancer factor 2A (Mef2A) and Mef2D. In contrast, Mettl21c knockout increased Mef2 protein levels in slow muscles. These results identify Hspa8 as a Mettl21c substrate and reveal that nonhistone methylation has a physiological function in protein stabilization.
Keywords: molecular chaperone, myogenesis, development, autophagy, skeletal muscle, chaperone-mediated autophagy (CMA), methyltransferase-like 21c (Mettl21c), myocyte enhancer factor 2 (Mef2), protein methylation, skeletal muscle, posttranslational modification, heat shock protein family A (Hsp70) member 8 (HSPA8), Hsc70
Introduction
The skeletal muscle provides body support, powers body movements, and regulates systemic energy homeostasis. Mammalian skeletal muscles are heterogeneous, composing two general types of muscle cells (myofibers): the slow-twitch (type I) and fast-twitch (type II) myofibers (1). The myocyte enhancer factor 2 (Mef2)3 family transcription factors are indispensable for muscle development and for the establishment of type-I myofibers (2–4). However, the level and activity of Mef2 proteins must be controlled in the adult during muscle maintenance, as aberrantly enhanced Mef2 activity is associated with myotonia and compromised mitochondria function (5, 6). Current understanding about the regulation of Mef2 is mainly on its transcriptional activity. Mef2 is activated by Ca2+/calmodulin kinases (7), calcium-activated serine/threonine phosphatase calcineurin (8), and mitogen-activated protein kinases, such as p38 and extracellular signal–regulated kinase 5 (9, 10). By contrast, the activity of Mef2 is repressed by class II histone deacetylases (11, 12). Recent studies have also pointed to a role of posttranscriptional regulation of Mef2 by microRNAs (6).
At the posttranslational level, Hspa8/Hsc70 plays a role in regulating Mef2 proteins through chaperone-mediated autophagy (CMA). In CMA, cytoplasmic client proteins with CMA-targeting motif are recognized and delivered by Hspa8/Hsc70 to lysosome through the lysosomal receptor, lysosomal-associated membrane protein 2A (Lamp2a) (13). Hspa8 has been shown to mediate the degradation of two major isoforms of the Mef2 proteins, Mef2A and Mef2D, in neuronal cells (14, 15). However, there is lack of a physiological context linking Hspa8 and Mef2 proteins in skeletal muscle.
Protein lysine methyltransferases (PKMTs) transfer a methyl group (CH3) from S-adenosyl-l-methionine (SAM) to lysine residuals of substrates (16). Methylation alters the hydrophobic and steric properties of a lysine residue, leading to changes in protein stability, protein-protein interactions, and protein-DNA interactions (17). Histone lysine methylation regulates chromatin structures and thus modulates gene expression (18), but the biological significance of nonhistone methylation is poorly understood (19). Recently, a group of 10 nonhistone protein lysine methyltransferases was characterized as methyltransferase family 16 (MTF16). Among this family METTL21A-D forms a subgroup based on phylogenetic analysis (20, 21). Intriguingly, METTL21C is identified as a pleiotropic gene for both skeletal muscle and bone through a genome-wide association study (22). Knockdown of Mettl21c impairs myogenesis of C2C12 myoblasts (22). A recent study identifies that Mettl21c is a type I myofiber–specific protein and functions to trimethylate valosin-containing protein (Vcp/p97), a protein required for autophagy (23, 24). Mettl21c−/− mice have normal muscle patterning, but dysregulated autophagy and muscle weakness (23). In human cells, METTL21C physically interacts with heat shock 70 kDa (HSPA) proteins (21), but it is unknown whether the HSPA proteins are bona fide substrates of METTL21C.
In this study, we used the Mettl21cLacZ knockin/knockout mouse model to track the developmental expression of Mettl21c. We found that Mettl21c was absent during myogenesis and was only expressed in mature type I myofibers. We then demonstrated that Mettl21c methylates and stabilizes Hspa8, thereby reducing the level of Hspa8 client proteins Mef2A and Mef2D. These data demonstrate an in vivo physiological function of Mettl21c and reveal the biological significance of nonhistone methylation in the skeletal muscle.
Results
Mettl21c is not expressed during myogenesis and only expressed in mature slow myofibers
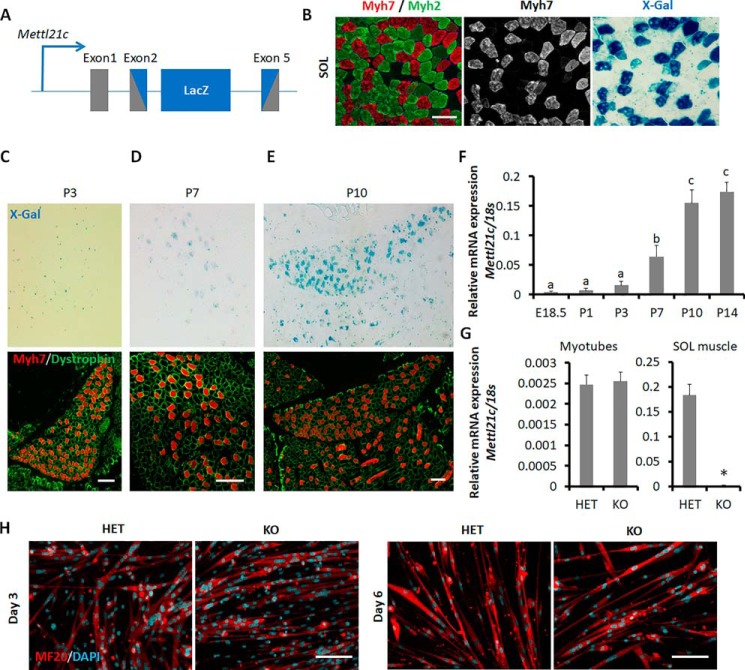
We tracked the expression of Mettl21c using Mettl21cLacZ/+ mice, which contain the β-gal reporter targeted to replace exon 2–5 of Mettl21c (Fig. 1A). Immunofluorescence staining of the slow (type I) myofiber–specific myosin heavy chain (Myh7) and X-Gal staining in serial sections of adult soleus (SOL) muscle indicate type I myofiber–specific expression of Mettl21c (Fig. 1B). We also isolated single myofibers from WT mice and detected the expression of Mettl21c specifically in type I myofibers expressing Myh7 in a separate study (25), which confirms X-Gal signal as a reporter of Mettl21c expression. We then investigated the developmental expression of Mettl21c. Whole-mount X-Gal staining showed no discernible positive signals in limb muscles of E18.5 embryos (data not shown). Additionally, there were no X-Gal signals in SOL muscle of postnatal day 3 (P3) mice, although Myh7+ (type I) myofibers were readily detectable at the same stage (Fig. 1C), indicating that the expression of Myh7 precedes that of Mettl21c and excluding a possible role of Mettl21c in type I myofiber specification. X-Gal signals were first shown in a group of Myh7+ myofibers in SOL muscles of P7 mice (Fig. 1D). The X-Gal signals became stronger and nearly all Myh7+ myofibers had positive X-Gal staining in SOL muscle of P10 mice (Fig. 1E). Consistent with the X-Gal staining patterns, mRNA levels of Mettl21c were low from E18.5 to P3, then up-regulated at P7, reaching the plateau levels at P10 in hind limb muscles (Fig. 1F). Hence, Mettl21c is expressed after the establishment of Myh7+ myofiber and the expression level is increased with the maturation of Myh7+ myofibers.
Figure 1.
Dynamic expression of Mettl21c during muscle development and myogenesis. A, genetic targeting of Mettl21c through insertion of LacZ into exons 2–5. The gray box indicates the exon of Mettl21c gene, and the blue box indicates LacZ gene. B, immunostaining of Myh7 (type I myofibers, red) and Myh2 (type IIa myofibers, green) and X-Gal staining of serial sections of SOL muscles isolated from Mettl21cLacZ/+ mice. Scale bar: 100 μm. C–E, immunostaining of Myh7 (red) and dystrophin (green), and X-Gal staining of serial sections of SOL muscles of P3 (C), P7 (D), and P10 (E) Mettl21cLacZ/+ mice. Scale bar: 100 μm. F, qPCR analysis of Mettl21c in different developmental stage. Error bars represent mean + S.D. of six mice with three technical repeats. Different letters indicate p < 0.05 (one-way ANOVA). G, qPCR analysis of Mettl21c in myocytes differentiated for 3 days or SOL muscles isolated from Mettl21cLacZ/+ (HET) or Mettl21cLacZ/LacZ (KO) mice. Error bars represent mean + S.D. of five independent biological experiments with three technical repeats. * indicates p < 0.05 (Student's t test). H, immunostaining of MF20 (myosin heavy chain, red) and DAPI (cyan) in differentiated myoblasts isolated from Mettl21cLacZ/+ mice at different days after differentiation induction. Scale bar: 50 μm.
To further confirm the absence of Mettl21c expression during myogenesis, we cultured primary myoblasts isolated from the Mettl21cLacZ/+ (HET) or Mettl21cLacZ/LacZ (KO) mice and induced the cells to differentiate. Quantitative PCR analyses showed that the mRNA levels of Mettl21c in the HET myotubes were identical to those of KO myotubes (both were nearly undetectable), indicative of the lack of Mettl21c expression in the HET myotubes (Fig. 1G). In contrast, the level of Mettl21c in the adult SOL muscles of HET was over 100 times higher than that in the KO muscles (Fig. 1G). In addition, there were no morphological differences between HET and KO myotubes (Fig. 1H), suggesting that Mettl21c is dispensable for myogenic differentiation. These results suggest that Mettl21c mainly functions in mature type I myofibers.
Mettl21c is indispensable for proper muscle function
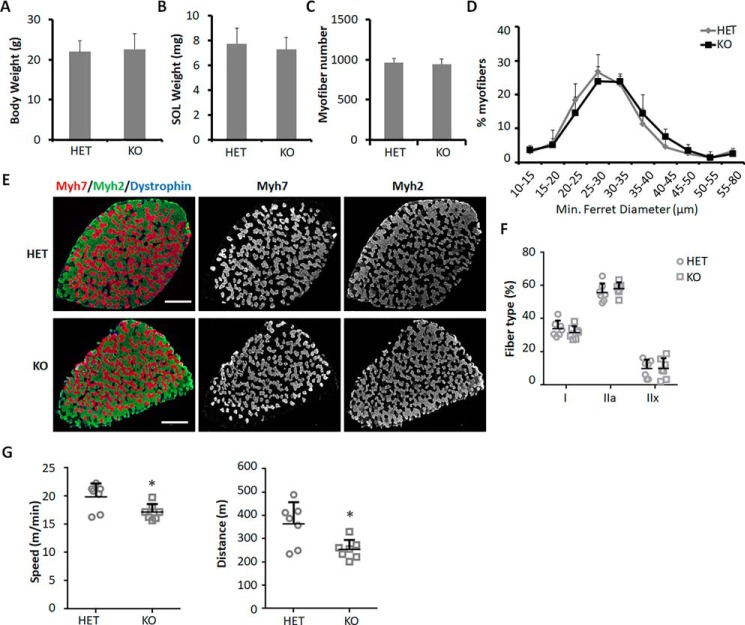
We next studied the function of Mettl21c in vivo using KO mice models. To exclude the potential influence of β-gal, we compared KO mice with Mettl21cLacZ/+ mice. KO mice were indistinguishable from HET and WT mice in gross morphology, body weight, and SOL muscle weight (Fig. 2, A and B). Total numbers and sizes of myofibers in SOL muscles were comparable between HET and KO mice (Fig. 2, C and D). Immunostaining of Myh isoforms showed that the percentage of type I myofibers in SOL muscle was comparable between HET and KO mice (Fig. 2, E and F), suggesting that Mettl21c KO does not affect slow myofiber development or maintenance. However, KO mice run shorter distance and had lower speed than HET mice (Fig. 2G), indicating that Mettl21c is necessary for proper functioning of skeletal muscles.
Figure 2.
Mettl21c is required for proper muscle function. A and B, relative body weight (A) and SOL muscle weight (B) of HET and KO mice. C, myofiber numbers in SOL muscles of HET and KO mice. D, myofiber size in SOL muscles of HET and KO mice. E, immunostaining of Myh7 (red) and Myh2 (green) in SOL muscles of HET or KO mice. Scale bar: 500 μm. F, quantification of different types of myofibers in SOL muscles of HET or KO mice. G, running speed and distance of HET and KO mice in treadmill test. Error bars represent mean + S.D. of seven mice. * indicates p < 0.05 (Student's t test).
Hspa8 is a substrate of Mettl21c
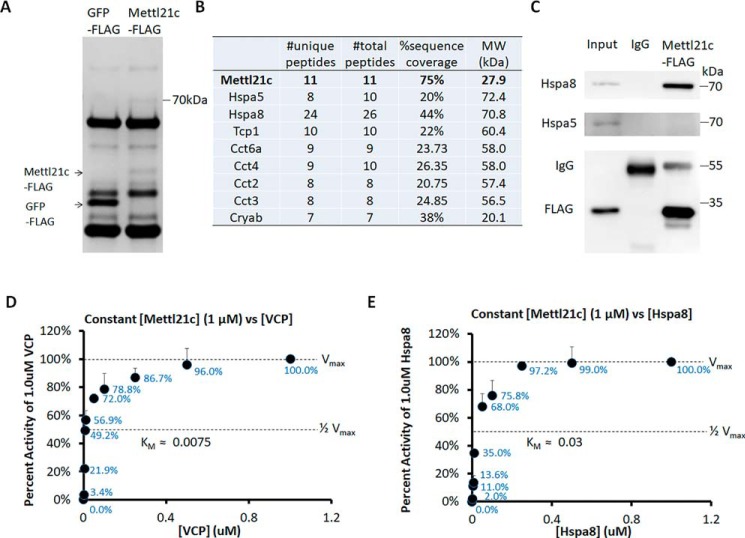
To understand the function of Mettl21c, we first attempted to identify Mettl21c-associated proteins. We used FLAG antibody to immunoprecipitate (IP) protein complexes from myocytes transduced with Mettl21c-FLAG or GFP-FLAG adenoviral vectors. SDS-PAGE analysis of the proteins obtained by IP revealed an ∼70 kDa protein co-purified with Mettl21c but not with GFP (Fig. 3A). Using affinity purification-MS (AP-MS), we identified several chaperone proteins specifically in Mettl21c-associated complexes, but not in GFP-associated complexes (Fig. 3B). Among these chaperone proteins, Hspa5 and Hspa8 have a molecular mass at ∼70 kDa. IP analysis confirmed that Mettl21c binds with Hspa8 (Fig. 3C), but not with Hspa5 in myocytes (Fig. 3C). These results indicate a physical association between Mettl21c and Hspa8.
Figure 3.
Mettl21c binds and methylates Hspa8. A, Coomassie Blue–stained SDS-PAGE of proteins pulled down by the FLAG antibody from myocytes transduced with Mettl21c-FLAG or GFP-FLAG adenoviral vectors. B, mass spectrometry analysis of Mettl21c binding proteins. C, detection of Hspa8 but not Hspa5 in Mettl21c-FLAG protein complex after immunoprecipitation from myocytes. The lysates of myocytes was set as a positive control to indicate the position of Hspa8 and Hspa5. D and E, In vitro methyltransferase activity of Vcp (D) and Hspa8 (E) as substrates of Mettl21c. Mettl21c was held constant at 1 μm in the presence of 13 μm [14C]SAM and increasing concentrations of either Vcp or Hspa8 as the substrate. The data are represented as a percentage of cpm (representing methyl groups transferred) at 1 μm substrate. Error bars represent mean + S.D. of three experiments performed in duplicate.
We further performed in vitro methyltransferase assay to determine whether Hspa8 is methylated by Mettl21c. Various concentrations of recombinant Mettl21c and Hspa8 proteins were reacted in the presence of [14C]SAM, and the incorporation of methyl groups into substrates was measured as TCA-insoluble radioactivity. To establish the validity of the assay, we used recombinant Vcp as a positive control, which was recently shown to be methylated by Mettl21c (23). We found that Vcp was dose-dependently methylated by Mettl21c, with a Km at ∼0.0075 (Fig. 3D). We next held the Mettl21c at a constant concentration (1 μm) with varying concentration (0, 0.001, 0.005, 0.0075, 0.01, 0.05, 0.1, 0.25, 0.5, and 1.0 μm) of Hspa8, and found that Hspa8 was dose-dependently methylated by Mettl21c, with a Km at ∼0.03 (Fig. 3E). The Km of Mettl21c on Hspa8 was slightly higher than that on Vcp, indicating that Mettl21c-Hspa8 had a relatively lower reaction rate than that of Mettl21c-Vcp. These data provide compelling biochemical evidence that Hspa8 is a new substrate of Mettl21c.
Mettl21c methylates Hspa8 at Lys-561
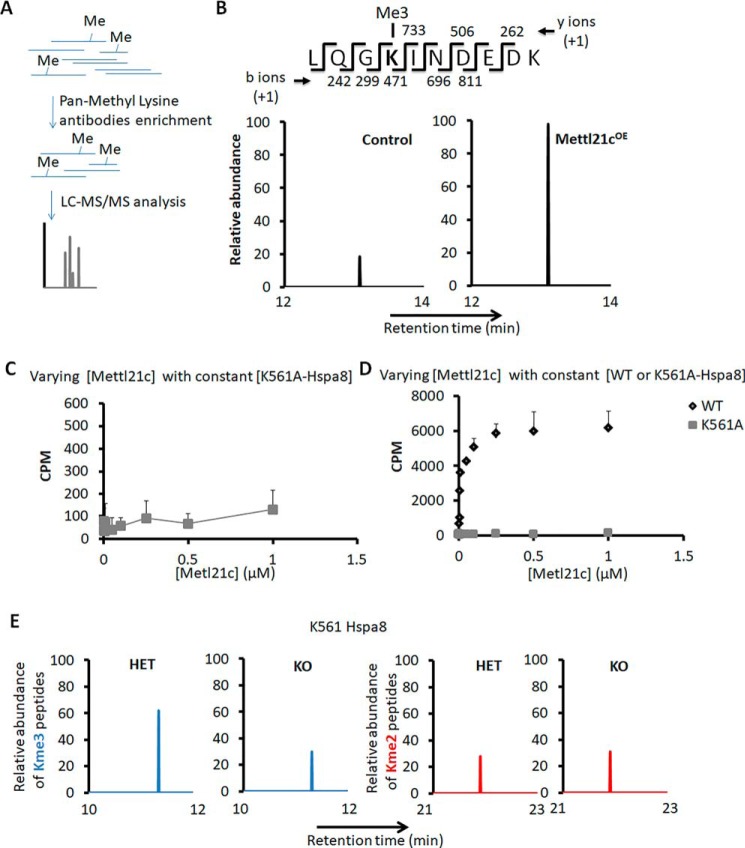
Then we utilized AP-MS to characterize methylated peptides enriched by IP using a pan-methyl lysine antibody in myocytes transduced with Mettl21c (Mettl21cOE) or GFP (as control) adenoviral vectors (Fig. 4A). This led to the identification of lysine (K) 561 in Hspa8 that was highly trimethylated in primary myoblasts transduced by Mettl21c-expressing adenovirus in relative to cells transduced by GFP control (Fig. 4B). Specifically, the abundance of the trimethylated Hspa8 peptide was 5-fold higher in Mettl21cOE myocytes than in control myocytes (Fig. 4B), indicating that Mettl21c is responsible for the Lys-561 trimethylation. To verify the methylation site, we mutated the Lys-561 to nonmethylatable alanine (A) and used the recombinant K561A-Hspa8 mutant as a substrate with increasing concentrations of Mettl21c in the methyltransferase assay. Whereas the WT Hspa8 was robustly methylated by Mettl21c, the K561A mutation abolished the dose-dependent incorporation of radioactivity and methylation by Mettl21c (Fig. 4, C and D), indicating that Lys-561 was the only site in Hspa8 that can be methylated by Mettl21c. We further utilized the same AP-MS assay to investigate methylated peptides in SOL muscles of HET and KO mice. We did not detect any Lys-561 monomethylation in Hspa8, and only detected either dimethylation or trimethylation of Lys-561 (Fig. 4E). Importantly, loss of Mettl21c decreased the abundance of trimethylated Hspa8 peptide by 51%, compensated by 11% increase in the abundance of dimethylated Hspa8 peptide (Fig. 4E). Our data demonstrate that Mettl21c trimethylates Hspa8 at Lys-561.
Figure 4.
Mettl21c methylates Hspa8 at Lys-561. A, flow chart of the analysis of methylated peptides. B, chromatogram view for the trimethylated Hspa8 peptide in myoblasts transduced with Mettl21c-GFP (Mettl21cOE) or GFP only (as control) adenoviral vectors. The MS/MS fragmentation pattern of Hspa8 peptide is highlighted. The b- and y-ions support trimethylation of Hspa8 at Lys-561. C and D, In vitro methyltransferase activity of Mettl21c and mutated Hspa8 (C) or WT Hspa8 (D). The substrate (K561A-Hspa8 or WT Hspa8) was held constant at 4 μm in the presence of 13 μm [14C]SAM and increasing concentrations of Mettl21c. The data are represented as cpm. Error bars represent mean + S.D. of three experiments performed in duplicate. E, chromatogram view for the dimethylated or trimethylated Hspa8 peptide in SOL muscles of HET or KO mice; MS/MS fragmentation pattern of Hspa8 peptide.
Lys-561 methylation stabilizes Hspa8
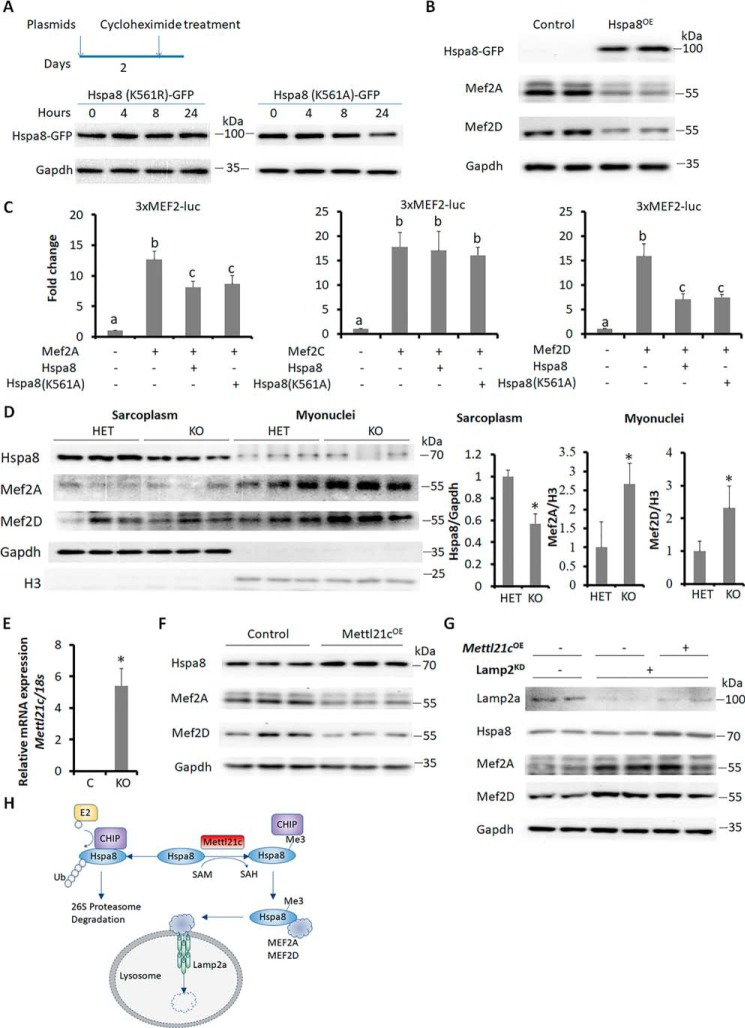
We next investigated the biological consequences of Lys-561 methylation. It has been reported that the methylation perturbs interaction between Hspa8 and an E3 ubiquitin ligase C terminus of Hsp70 interacting protein (CHIP), leading to decreased ubiquitination on Hspa8 (26). Thus, we hypothesize that the Lys-561 methylation prevents Hspa8 from degradation. We transfected 293A cells with K561R or K561A mutant pcDNA5–GFP-Hspa8 plasmids to mimic methylated or unmethylated Lys-561, and treated the 293A cells with a protein synthesis inhibitor cycloheximide (CHX) to observe the degradation of the two mutants (Fig. 5A). Although the protein level of K561A mutant was decreased within 8 h after CHX treatment, the protein level of K561R mutant was unchanged within 24 h after CHX treatment (Fig. 5A), indicating that Lys-561 methylation increased the stability of Hspa8.
Figure 5.
Lys-561 methylation stabilizes Hspa8. A, Western blot analysis of mutated Hspa8 after cycloheximide-induced inhibition of translation. B, Western blot analysis showing the effect of Hspa8 overexpression (Hspa8OE) on Mef2A and Mef2D proteins in myotubes. Primary myoblasts were induced to differentiate for 2 days and then were transduced with Hspa8OE or control adenoviral vectors for 3 more days. C, luciferase assays showing the effects of Hspa8 on the transcriptional activity of Mef2A, Mef2C, and Mef2D using the 3XMEF2-luc reporter. Error bars represent mean + S.D. of five independent biological experiments. * indicates p < 0.05 (Student's t test). D, Western blot analysis of indicated proteins in sarcoplasm and myonuclei proteins of SOL muscles from HET and KO mice. E, qPCR analysis of Mettl21c in differentiated myoblasts transduced with Mettl21c-GFP (Mettl21cOE) or GFP only (as control) adenoviral vectors. Primary myoblasts were induced to differentiate for 2 days and then were transduced with Mettl21cOE or control adenoviral vectors for 3 more days. Error bars represent mean + S.D. of five independent biological experiments. * indicates p < 0.05 (Student's t test). F, Western blot analysis of indicated proteins in control and Mettl21cOE myotubes. G, Western blot analysis of indicated proteins in control and Mettl21cOE myotubes after Lamp2 knockdown. Primary myoblasts were treated with lentivirus for 2 days and selected in puromycin (1 μg/ml) for 2 additional days. These primary myoblasts were then transduced with Mettl21cOE or control adenoviral vectors. Two days after adenoviral transduction, the myoblasts were induced to differentiate for 3 days. H, diagram summarizing the role of Mettl21c in type I myofibers. Mettl21c methylates Hspa8 at Lys-561 to interrupt its binding with an E3-ligase CHIP, and to prevent its ubiquitination and proteasome-dependent degradation. The surviving Hspa8 carries Mef2A and Mef2D to lysosome for degradation in chaperone-mediated autophagy pathway.
It has been reported that Hspa8 targets Mef2A and Mef2D to lysosome for degradation through CMA in neurons (14, 15). We therefore examined how Hspa8 affects Mef2 proteins in myotubes. We transduced myotubes by adenovirus expressing Hspa8-GFP (Hspa8OE) or GFP only (control), and found that Hspa8OE reduced the levels of Mef2A and Mef2D in myotubes (Fig. 5B). Consistently, Hspa8OE decreased the transcriptional activities of Mef2A and Mef2D, but not Mef2C (Fig. 5C). Notably, K561A-Hspa8 similarly inhibited the activity of Mef2A and Mef2D (Fig. 5C), indicating that K561 methylation was not required for the inhibition of Mef2A and Mef2D.
We further investigated the effect of Lys-561 trimethylation on Hspa8 stability. We detected the level of Hspa8 and its target proteins Mef2A and Mef2D in SOL muscles of Mettl21cKO mice. Mettl21cKO reduced the Lys-561 trimethylation and the level of Hspa8 in sarcoplasm and increased the protein levels of Mef2A and Mef2D in myonuclei (Figs. 4E and 5D). To examine the effect of Mettl21c-mediated Lys-561 trimethylation on Hspa8 level and function, differentiated myoblasts were transduced with Mettl21c (Mettl21cOE) or GFP (as control) adenoviral vectors. The expression of Mettl21c was over 2000-fold higher in Mettl21cOE myotubes than in control myotubes (Fig. 5E). Mettl21cOE increased the trimethylation and the level of Hspa8, concomitant with a decrease in the level of Mef2A and Mef2D (Figs. 4B and 5F). These results provide compelling evidence that Lys-561 methylation stabilizes Hspa8.
As Mettl21c trimethylates and maintains the level of Hspa8, which was shown to function as carrier to facilitate degradation of Mef2A and Mef2D through the CMA pathway, we hypothesize that the regulation of Mef2A and Mef2D by Mettl21c is CMA-dependent. To test this hypothesis, we selectively blocked CMA by silencing Lamp2a using lentiviral shRNA (27). Lamp2a shRNA efficiently reduced the level of Lamp2a, accompanied by an accumulation of Mef2A and Mef2D (Fig. 5G). Importantly, Lamp2a silencing prevented Mettl21cOE-mediated reduction of Mef2A and Mef2D (Fig. 5, F and G), indicating that the CMA pathway mediates the degradation of Mef2A and Mef2D in Mettl21cOE myotubes. These results together establish a model in which Mettl21c methylates Hspa8 to facilitate degradation of its client proteins such as Mef2A and Mef2D (Fig. 5H).
The expression of Mettl21c is absent during muscle regeneration
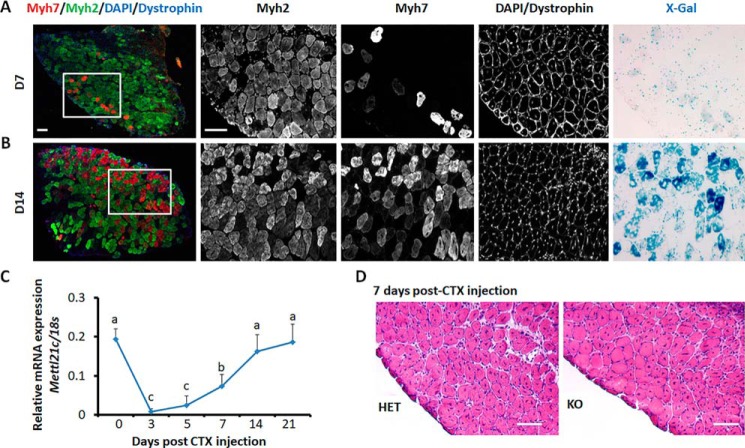
Although high levels of Mef2 proteins are detrimental to mature muscles (6), Mef2 transcription factors are required for muscle regeneration (3). As Mettl21c regulates the levels of Mef2A and Mef2D, we predict that Mettl21c expression should be down-regulated during muscle regeneration to allow accumulation of Mef2 proteins. We thus examined the expression of Mettl21c using the Mettl21cLaz/+ reporter mice during SOL muscle regeneration after cardiotoxin (CTX)-induced injuries. We performed immunofluorescence and X-Gal staining on sections of regenerated SOL muscles. At day 7 post CTX injection, only a small number of regenerating Myh7+ myofibers had weak X-Gal signals (Fig. 6A), indicating that Mettl21c is not expressed in newly regenerated type I myofibers. At day 14 post CTX injection, a substantial number of regenerated myofibers expressed Myh7, accompanied by increased levels of X-gal signal specifically in Myh7+ myofibers (Fig. 6B). In parallel, Mettl21c mRNA levels was sharply decreased by more than 90% within 3 days after CTX-induced muscle degeneration, and gradually increased in the following 2 weeks, reaching the maximal levels at day 14 post CTX injury (Fig. 6C). These results indicate that Mettl21c expression is down-regulated during muscle regeneration and suggests that Mettl21c is dispensable for muscle regeneration. We also investigated the regeneration of SOL muscles in HET and KO mice after CTX injury. Consistently, the regeneration of KO muscles was comparable to that of HET muscles at day 7 post CTX injury (Fig. 6D). These results ensure the notion that Mettl21c mainly functions in mature type I myofibers.
Figure 6.
Expression of Mettl21c during muscle regeneration. A and B, immunostaining of Myh7 (red), Myh2 (green), and dystrophin (blue), and X-Gal staining of serial sections of SOL muscles at day 7 (A) and day 14 (B) after CTX injection. Scale bar: 100 μm. C, qPCR analysis of Mettl21c at different days during SOL muscle regeneration. Error bars represent mean + S.D. of six mice with three technical repeats. Different letters indicates p < 0.05 (one-way ANOVA). D, H&E staining of SOL muscles from HET and KO mice 7 days after CTX injection. Scale bar: 100 μm.
Discussion
In this study we dissect the dynamic expression and in vivo function of a type I myofiber-specific methyltransferase Mettl21c. The expression of Mettl21c is absent during myogenesis, indicating that Mettl21c is not required for the development of skeletal muscle, inconsistent with the normal muscle morphology in KO mice. Despite the normal muscular morphology, KO mice have decreased motor activity. Wiederstein et al. (23) report a similar weak muscle phenotype and they additionally show that Mettl21c KO leads to dysregulated autophagy program potentially through Vcp.
We identified Hspa8 as a novel substrate of Mettl21c and pinpointed Lys-561 as the specific residue being trimethylated. Intriguingly, Jakobsson et al. (28) reported that human HSPA8, which shares similar primary structure with murine Hspa8, is trimethylated at Lys-561 by METTL21A, a paralog of METTL21C. A potential compensatory role of murine Mettl21a may explain why Mettl21c KO only partially blocks the trimethylation of Hspa8. Our AP-MS results indicate that Hspa8 is also dimethylated at Lys-561 in skeletal muscles. However, the Lys-561 dimethylation is not mediated by Mettl21c as deletion of Mettl21c leads to increased levels of dimethylated Hspa8. Although what dimethylates Hspa8 is unclear, SETD1A has been reported to dimethylate Lys-561 of Hspa1, a paralog of Hspa8 (29).
At the functional level, Lys-561 methylation of Hspa8 decreases its affinity for the E3 ligase, CHIP, and thus reduces the ubiquitination of Hspa8 (26). The latter event prevents ubiquitin-mediated proteasomal degradation of Hspa8 and stabilizes Hspa8. Consistently, we show that the mimetic of methylated Hspa8 has better stability than unmethylated Hspa8. In addition, we found that Mettl21cOE or Mettl21cKO increases or decreases the protein level of Hspa8, respectively.
Hspa8 plays several different roles in the autophagy program (30). For example, Hspa8 cooperates with BAG3, which mediates macro-autophagy and maintains mitochondria homeostasis (31, 32). Intriguingly, impaired Hspa8-BAG3–mediated autophagy results in muscle weakness (33). Hspa8 inhibitors, such as P140 and 2-phenylethynesulfonamide, have been shown to disrupt the binding of Hspa8 with the other chaperones and down-regulate autophagy program (30, 34, 35). Thus, the decreased level of Hspa8 may contribute to the impaired autophagy and weak muscle in the KO mice (23).
In addition, Hspa8 is a key regulator in the CMA pathway (36). It selectively recognizes KFERQ-like motif–containing proteins, including Mef2A and Mef2D, and shuttles them to lysosome for degradation. The degradation of Mef2A and Mef2D is blocked by interrupting their interactions with Hspa8 in neuronal cells (14, 15). Here, we identify a conserved role of Hspa8 in skeletal muscles by demonstrating that Hspa8OE reduced the protein levels of Mef2A and Mef2D. Thus, the expression of Mettl21c could harness the level of Mef2 proteins through Hspa8. The expression of Mettl21c is absent during myogenesis and muscle regeneration, allowing Mef2 transcription factors to accumulate and function during muscle establishment. Upon completion of muscle development or regeneration, Mettl21c is then increased to lower the level of Mef2 transcription factors. A proper level of Mef2 is key for normal muscle function, as a high level of Mef2A has been shown to compromise mitochondria function in the skeletal muscle (6). Collectively, our study establishes Mettl21c as a regulator of Hspa8 to maintain its level in mature type I myofibers.
Experimental procedures
Mice
The Mettl21cLacZ/+ mice (stock no. 025271) were from The Jackson Laboratory (23). Unless otherwise indicated, we used 2-month-old male mice for experiments. All procedures involving mice were approved by Purdue University's Animal Care and Use Committee under protocol no. 1112000440.
Primary myoblast isolation, culture, and differentiation
Primary myoblasts were isolated from 5-week-old WT female mice. The hind limb skeletal muscles were minced and digested in type I collagenase and dispase B mixture (Roche Applied Science). The digestion was stopped with Ham's F-10 medium containing 17% fetal bovine serum (FBS), and the cells were filtered from debris, centrifuged, and cultured in growth media (Ham's F-10 medium supplemented with 17% FBS, 4 ng ml−1 basic fibroblast growth factor and 1% penicillin-streptomycin) on uncoated dishes for 3 days when 5 ml growth media were added each day. Then the supernatants were collected, centrifuged, and trypsinized with 0.25% trypsin. After washing off the trypsin, primary myoblasts were seeded on collagen-coated dishes, and the growth medium was changed every 2 days. Myoblasts were induced to differentiate on Matrigel-coated dishes and cultured in differentiation media (DMEM supplemented with 2% horse serum and 1% penicillin-streptomycin). Differentiation media were replaced every day. Primary myoblasts were cultured in normal humidified tissue culture incubators with 5% CO2.
Immunostaining, X-gal staining, and image acquisition
For immunostaining of muscle samples, they were processed following the protocol described by Wang et al. (37). Generally, muscle slides were directly blocked with blocking buffer (5% goat serum, 2% BSA, 0.2% Triton X-100, and 0.1% sodium azide in PBS) for at least 30 min. Primary antibodies were diluted with blocking buffer. Myh2 (2F7 from Developmental Studies Hybridoma Bank (DSHB)) and Myh7 (BA-F8 from DSHB) antibodies were diluted at a ratio 1:300. Dystrophin (ab15277, Abcam) antibody was diluted at a ratio 1:1000. Blocking buffer was removed from the sections and diluted primary antibodies were added on sections overnight at 4 °C. After washing with PBS, the samples were incubated with respective secondary antibodies and DAPI for 45 min at room temperature. Myotubes were fixed with 4% paraformaldehyde, and then were blocked with blocking buffer for at least 30 min. Then the samples were incubated with primary antibodies (1:200 in blocking buffer) (MF20, DSHB) overnight. After washing with PBS, the samples were incubated with respective secondary antibodies and DAPI for 45 min at room temperature. Fluorescent images were captured using a Leica DM 6000B fluorescent microscope. For X-gal staining, tissue sections were fixed with 4% paraformaldehyde for 5 min. Then samples were washed with washing buffer (0.1 m phosphate buffer, pH 7.3, 2 mm MgCl2, and 0.02% Nonidet P-40) for at least three times. Then samples were stained with staining buffer (washing buffer containing 1 mg ml−1 X-Gal in dimethyl formamid, 2.12 mg ml−1 potassium ferrocyanide, and 1.64 mg ml−1 potassium ferricyanide) overnight at 37 °C. After staining, the staining buffer was poured off and replaced with washing buffer.
RNA extraction and real-time qPCR
Total RNA of muscles or myoblasts were extracted using TRIzol Reagent. RNA was treated with RNase-free DNase I to remove genomic DNA. The purity and concentration of total RNA were measured by NanoDrop 3000 (Thermo Fisher). Random primers and Moloney murine leukemia virus reverse transcriptase were used to convert RNA into cDNA. Real-time PCR was performed using Roche Light Cycler 480 PCR System with SYBR Green Master Mix. Forward and reverse primers for Mettl21c were 5′-AGGAGCTCAAGTCACAGCAACAGA and 5′-AGAGGCCAGCACGTAGTCATAACA, respectively. Ct value of 18S rRNA was used as internal control and 2−ΔΔCt method was used to analyze the relative mRNA expression of various genes.
Protein extraction and Western blot analysis
Muscle samples and cultured myoblasts were washed with PBS and homogenized with radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS). Protein concentrations were determined using Pierce BCA Protein Assay Reagent (Pierce Biotechnology). Proteins (100 μg) were separated by 10% SDS-PAGE, electrotransferred onto PVDF membrane (EMD Millipore, Billerica, MA), and incubated with specific primary antibodies. Mef2A (B-4), Mef2D (H-11), GFP (B-2), and GAPDH (6C5) antibodies (1:1000 in 5% w/v nonfat dry milk) were from Santa Cruz Biotechnology, and Hspa8/Hsc70 (ab2788), NOQ/Myh7 (ab11083), and Lamp2a (ab18528) were from Abcam. Immunodetection was performed using ECL Western Blotting Substrate (Pierce Biotechnology) and detected with FluoChem R imaging system (ProteinSimple).
Myonuclei and sarcoplasm protein preparation
SOL muscles were homogenized in the cell lysis buffer (10 mm HEPES, pH 7.5, 10 mm KCl, 0.1 mm EDTA, 1 mm DTT, 0.5% Nonidet P-40, 0.5 mm PMSF and 1× protease inhibitor mixture) and allowed to swell on ice for 15–20 min with intermittent mixing. Tubes were vortexed to disrupt cell membranes and then centrifuged at 12,000 × g at 4 °C for 10 min. The supernatant is the sarcoplasm extract. The pelleted nuclei were washed twice with the cell lysis buffer and resuspended in the nuclear extraction buffer (20 mm HEPES, pH 7.5, 400 mm NaCl, 1 mm EDTA, 1 mm DTT, 1 mm PMSF and 1× protease inhibitor mixture) and incubated in ice for 30 min. Myonuclei extract was collected by centrifugation at 12,000 × g for 15 min at 4 °C.
Adenovirus generation
The adenoviruses with Hspa8, Mettl21c, or Mettl21c-FLAG were generated using the AdEasy system. Hspa8 ORF, Mettl21c ORF, and Mettl21c-FLAG ORF were cloned with primers 5′-AAGCGGCCGCATGTCTAAGGGACCTGCAGTTGG/5′-CGTCTAGATTAATCCACCTCTTCAATGG for Hspa8; 5′-AAGCGGCCGCATGGATCAGCATCTCCACATAG/5′-CGTCTAGATCACTCCCACTTTAATATCCC for Mettl21c; 5′-AAGCGGCCGCATGGATCAGCATCTCCACATAG/5′-CGTCTAGATCACTTGTCGTCATCGTCTTTGTAGTCCTCCCACTTTAATATCCC for Mettl21c-FLAG. These cloned DNA sequences were inserted into pAdTrack-CMV plasmid (the cloned Mettl21c-FLAG or Mettl21c ORF was also inserted into pcDNA3.1) and then were digested by PmeI and transfected the DH5a competent cell with pAdEasy-1. The steps were exactly following the methods described by Wang et al. (38).
Immunoprecipitation to pull down Mettl21c-binding complex
Vectors containing Mettl21c-FLAG or GFP-FLAG were transduced into primary myoblasts (70–80% confluent) by adenovirus. Two days post transduction, cells were induced to differentiation for 3 days and then were scraped with ice-cold PBS (from 10 × 100 mm plates for each plasmid) and were centrifuged. The cell pellet was completely resuspended with 1 ml lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 1% Nonidet P-40 with 1× protease inhibitor mixture), on ice, and sonicated with 1-s pulse and 5-s interval for 10 times. After 5 min on ice, cell lysate was centrifuged with 14,000 rpm at 4 °C, and then the supernatant (containing 5 mg proteins determined by the Pierce bicinchoninic acid assay) was transferred to 50 μl precleaned anti-FLAG magnetic beads slurry (M8823, Sigma). After incubation on a rotator for 3 h in a cold room (4 °C), the magnetic beads were washed three times with lysis buffer. Then, 500 μl ddH2O were added, up and down several times to remove salt or solvent remainder (twice). Bead captured proteins were eluted with 100 μl of 50 mm triethyl amine and 5 mm DTT on thermal shaker (99 °C, 5 min). The protein elution was centrifuged with CentriVap Concentrator (Labconco) to partially remove triethyl amine, and then was adjusted to 100 μl, pH 8.0 with 1% acetic acid, and 15 mm iodoacetamide and placed in dark for 1 h. Before desalting the protein elution with C18 ZipTips (NT3C18.96, Glygen), 1 μg trypsin was reacted with the 100 μl adjusted protein elution for 16 h at 37 °C. The following steps for mass spectrometric data acquisition and analysis were according to the descriptions by Wang et al. (38).
Mass spectrometry analysis of peptides with methylated lysine
Peptides with methylated lysine were enriched by immunoprecipitation using the PTMScan Pan-Methyl Lysine Kit (14809, Cell Signaling Technology) according to the manufacturer's instructions. In brief, myocytes or SOL muscle lysates were prepared using urea lysis buffer (20 mm HEPES, pH 8.0, 9 m urea, 2.5 mm sodium pyrophosphate). Protein concentrations were determined and adjusted to be the same. DTT (4.5 mm) and iodoacetamide (10 mm) were used to introduce reduction and alkylation of proteins. The urea was then diluted with 20 mm HEPES, pH 8.0, to a final concentration of ∼2 m. Before desalting the protein elution with C18 Sep-Paks, 1 μg trypsin was reacted with the 100 μl adjusted protein lysates for 16 h at 37 °C. The protein elution was frozen (−80 °C freezer) for 2 h and the frozen peptide solution was lyophilized for 2 days. Lyophilized peptides were resuspended using the IAP buffer provided in the kit. Then the IAP-peptides solution was mixed with antibody-bead slurry and incubated on a rotator for 2 h at 4 °C. The antibody-beads were washed three times with IAP buffer and washed three more times with ddH2O. Bead-captured peptides were eluted with 100 μl of 0.15% trifluoroacetic acid (TFA). Eluted peptides were purified with StageTips. The following steps for mass spectrometric data acquisition and analysis were according to the descriptions by Wang et al. (38).
Generation of mutated Hspa8
The pcDNA5-GFP-Hspa8 (K561A) and pcDNA5-GFP-Hspa8 (K561R) were generated using Q5 Site-Directed Mutagenesis Kit (E0554, New England Biolabs) according to the manufacturer's protocol. In brief, the pcDNA5-GFP-Hspa8 was amplified by PCR primers: Hspa8-A-f: GCGATTAACGATGAGGACAAAC or Hspa8-R-f: AGGATTAACGATGAGGACAAAC and Hspa8-Mu-r: GCCTTGAAGTTTCTCATCTTC. Note the plasmid concentrated should be less than 25 ng. PCR products were then reacted with KLD enzyme mix (provided in the kit). KLD mix was incubated with chemically competent cells. Plasmids were extracted from transfected cells and sequenced to confirm the correct mutation. To detect the degradation of mutated Hspa8, 293A cells were transfected with indicated plasmid. Two days after the transfection, 293A cells were treated with 100 μg ml−1 cycloheximide (C7698, Sigma) for up to 8 h to block protein translation.
Recombinant protein production and purification
pETDuet-1–derived plasmids incorporating His6-tagged proteins were transformed into the Escherichia coli expression strain BL21 (DE3) (Invitrogen). Cells were cultured in LB media with 0.1 mg/ml ampicillin at 37 °C in a shaking incubator at 220 rpm until the absorbance at 600 nm reached 0.8 OD. The culture was induced with 100 μm isopropyl β-d-thiogalactoside (Gold Biotechnology) and the temperature was lowered to 18 °C for 18 h. Cells were harvested by centrifugation at 7000 × g. Cell pellets were resuspended in lysis buffer (50 mm Tris, pH 7.5, 500 mm NaCl, 10% (w/v) glycerol, 30 mm imidazole, 3 mm 2-mercaptoethanol, 0.5% Triton X-100, 1 tablet cOmpleteTM EDTA-free Protease Inhibitor Mixture (Roche), and 2 mm AEBSF (Gold Biotechnology). Cells were lysed via probe sonication and underwent centrifugation at 100,000 × g for 1 h. The supernatant was rocked with nickel-nitrilotriacetic acid resin (Thermo Scientific) at 4 °C. After 1 h, resin was washed with buffer (50 mm Tris-HCl, pH 7.5, 500 mm NaCl, 10% glycerol, and 30 mm imidazole), followed by an additional wash with the addition of 0.5 m KCl. Recombinant proteins were removed from resin with elution buffer (50 mm Tris-HCl, pH 7.5, 500 mm NaCl, 300 mm imidazole). The eluted protein was concentrated in Amicon Ultra MWCO concentrators columns (Millipore) with appropriate molecular mass cutoffs. Proteins were aliquoted and stored at −80 °C and were thawed on ice prior to use in assays. Concentrations of each protein were determined via Bradford protein assay (Thermo Scientific).
In vitro methyltransferase reaction
Methyltransferase reactions were performed in 50-μl volumes for 1 h at 37 °C in methyltransferase reaction buffer (50 mm Tris, pH 7.5, 50 mm KCl, 5 mm MgCl2, 1 mm ATP), 13 μm [14C]SAM (2 μCi), 1 μm methyltransferase enzyme, and varying concentrations of substrate. The reactions were stopped by precipitating proteins with 50-μl 10% (v/v) TCA at 4° for 1 h. The reactions were spotted onto glass fiber filters (Whatman, 45 um) and the acid-insoluble material was retained during vacuum filtration. The reaction tubes were rinsed with an additional 50 μl 10% (v/v) TCA and applied to the filters. The filters were then washed with 1 ml of 10% (v/v) TCA, followed by 1 ml 100% ethanol, and left to dry for 10 min at room temperature. The dried filters were placed in scintillation vials with 10 ml Bio-Safe IITM biodegradable scintillation mixture (Research Products International) and radioactivity was measured by scintillation counting.
Luciferase assay
Transient transfections were performed with Lipofectamine2000 (Thermo) according to manufacturer's instructions. HEK293 cells were grown in DMEM supplemented with 10% FBS. Briefly, 100 ng of reporter 3XMEF2-luc, 10 ng Renilla, and 300 ng of each other plasmids (pCGN-MEF2A, pcDNA3.1-MEF2C-HA, pCGN-MEF2D, pcDNA5-GFP-Hspa8 (K561A), and pcDNA5-GFP-Hspa8) was mixed with 2 μl of Lipofectamine2000 and added to 293 cells in 24-well plates. After 48 h, cells were harvested and analyzed with the Dual-Luciferase Reporter Assay System (Promega). The total amount of DNA added in each transfection was kept constant by referring to the Renilla signal. The 3XMEF2-luc (Addgene plasmid 32967), pCGN-MEF2A (Addgene plasmid 32958), and pCGN-MEF2D (Addgene plasmid 32963) were gifts from Ron Prywes; the pcDNA3.1-MEF2C-HA was a gift from Andrew Lassar (Addgene plasmid 32515); and the pcDNA5-HSPA8 was a gift from Harm Kampinga (Addgene plasmid 19460).
Lentivirus generation
To deplete endogenous Lamp2a, shRNAs target 5′-GACTGCAGTGCAGATGAAG, 5′-CTGCAATCTGATTGATTA, and 5′-TAAACACTGCTTGACCACC, corresponding to the exon 8 of Lamp2a, were chosen according to the description by Massey et al. (27). The hairpin (sense-loop-antisense) for these sequences was cloned in the Plko.1 plasmid. Lentiviral particles containing Plko.1-Lamp2a-shRNAs or Plko.1-Scramble (a gift from David Sabatini, Addgene plasmid 1864) (39) were produced according to the protocol described by Wang et al. (40).
Statistical analysis
The data were presented with mean and S.D. p values were calculated using unpaired two-tailed Student's t test for two groups' comparison and one-way ANOVA for multiple groups' comparison. p values < 0.05 were considered to be statistically significant.
Author contributions
C. W. and S. K. conceptualization; C. W., J. A., A. C. R., J. C., H. E. H., Y. N., F. Y., and S. K. investigation; C. W. methodology; C. W. and S. K. writing-original draft; C. W. and S. K. writing-review and editing; C. A. H., W. A. T., and S. K. resources; S. K. supervision; S. K. funding acquisition.
Acknowledgments
We thank Jessie Ellis for technical assistance, Jun Wu and Mary Larimore for mouse colony maintenance, and members of the Kuang laboratory for valuable comments.
This work was supported by the National Institutes of Health Grant R01AR071649 and the National Institute of Food and Agriculture Grant NC-1184. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- Mef2
- myocyte enhancer factor 2
- CMA
- chaperone-mediated autophagy
- SOL
- soleus
- P
- postnatal day
- HET
- heterozygous Mettl21cLacZ/+
- IP
- immunoprecipitate
- AP
- affinity purification
- CHIP
- C terminus of Hsp70 interacting protein
- CHX
- cycloheximide
- CTX
- cardiotoxin
- DSHB
- Developmental Studies Hybridoma Bank
- ANOVA
- analysis of variance.
References
- 1. Schiaffino S., and Reggiani C. (2011) Fiber types in mammalian skeletal muscles. Physiol. Rev. 91, 1447–1531 10.1152/physrev.00031.2010 [DOI] [PubMed] [Google Scholar]
- 2. Potthoff M. J., Wu H., Arnold M. A., Shelton J. M., Backs J., McAnally J., Richardson J. A., Bassel-Duby R., and Olson E. N. (2007) Histone deacetylase degradation and MEF2 activation promote the formation of slow-twitch myofibers. J. Clin. Invest. 117, 2459–2467 10.1172/JCI31960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu N., Nelson B. R., Bezprozvannaya S., Shelton J. M., Richardson J. A., Bassel-Duby R., and Olson E. N. (2014) Requirement of MEF2A, C, and D for skeletal muscle regeneration. Proc. Natl. Acad. Sci. U.S.A. 111, 4109–4114 10.1073/pnas.1401732111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Potthoff M. J., Arnold M. A., McAnally J., Richardson J. A., Bassel-Duby R., and Olson E. N. (2007) Regulation of skeletal muscle sarcomere integrity and postnatal muscle function by Mef2c. Mol. Cell. Biol. 27, 8143–8151 10.1128/MCB.01187-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wu H., and Olson E. N. (2002) Activation of the MEF2 transcription factor in skeletal muscles from myotonic mice. J. Clin. Invest. 109, 1327–1333 10.1172/JCI0215417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wüst S., Dröse S., Heidler J., Wittig I., Klockner I., Franko A., Bonke E., Günther S., Gärtner U., Boettger T., and Braun T. (2018) Metabolic maturation during muscle stem cell differentiation is achieved by miR-1/133a-mediated inhibition of the Dlk1-Dio3 mega gene cluster. Cell Metab. 27, 1026–1039.e6 10.1016/j.cmet.2018.02.022 [DOI] [PubMed] [Google Scholar]
- 7. Passier R., Zeng H., Frey N., Naya F. J., Nicol R. L., McKinsey T. A., Overbeek P., Richardson J. A., Grant S. R., and Olson E. N. (2000) CaM kinase signaling induces cardiac hypertrophy and activates the MEF2 transcription factor in vivo. J. Clin. Invest. 105, 1395–1406 10.1172/JCI8551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu H., Rothermel B., Kanatous S., Rosenberg P., Naya F. J., Shelton J. M., Hutcheson K. A., DiMaio J. M., Olson E. N., Bassel-Duby R., and Williams R. S. (2001) Activation of MEF2 by muscle activity is mediated through a calcineurin-dependent pathway. EMBO J. 20, 6414–6423 10.1093/emboj/20.22.6414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ornatsky O. I., Cox D. M., Tangirala P., Andreucci J. J., Quinn Z. A., Wrana J. L., Prywes R., Yu Y. T., and McDermott J. C. (1999) Post-translational control of the MEF2A transcriptional regulatory protein. Nucleic Acids Res. 27, 2646–2654 10.1093/nar/27.13.2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang C. C., Ornatsky O. I., McDermott J. C., Cruz T. F., and Prody C. A. (1998) Interaction of myocyte enhancer factor 2 (MEF2) with a mitogen-activated protein kinase, ERK5/BMK1. Nucleic Acids Res. 26, 4771–4777 10.1093/nar/26.20.4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McKinsey T. A., Zhang C. L., Lu J., and Olson E. N. (2000) Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408, 106–111 10.1038/35040593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chang S., McKinsey T. A., Zhang C. L., Richardson J. A., Hill J. A., and Olson E. N. (2004) Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol. Cell. Biol. 24, 8467–8476 10.1128/MCB.24.19.8467-8476.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eskelinen E. L., and Saftig P. (2009) Autophagy: A lysosomal degradation pathway with a central role in health and disease. Biochim. Biophys. Acta 1793, 664–673 10.1016/j.bbamcr.2008.07.014 [DOI] [PubMed] [Google Scholar]
- 14. Yang Q., She H., Gearing M., Colla E., Lee M., Shacka J. J., and Mao Z. (2009) Regulation of neuronal survival factor MEF2D by chaperone-mediated autophagy. Science 323, 124–127 10.1126/science.1166088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang L., Sun Y., Fei M., Tan C., Wu J., Zheng J., Tang J., Sun W., Lv Z., Bao J., Xu Q., and Yu H. (2014) Disruption of chaperone-mediated autophagy-dependent degradation of MEF2A by oxidative stress-induced lysosome destabilization. Autophagy 10, 1015–1035 10.4161/auto.28477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lanouette S., Mongeon V., Figeys D., and Couture J. F. (2014) The functional diversity of protein lysine methylation. Mol. Syst. Biol. 10, 724 10.1002/msb.134974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hamamoto R., Saloura V., and Nakamura Y. (2015) Critical roles of non-histone protein lysine methylation in human tumorigenesis. Nat. Rev. Cancer 15, 110–124 10.1038/nrc3884 [DOI] [PubMed] [Google Scholar]
- 18. Black J. C., Van Rechem C., and Whetstine J. R. (2012) Histone lysine methylation dynamics: Establishment, regulation, and biological impact. Mol. Cell 48, 491–507 10.1016/j.molcel.2012.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Biggar K. K., and Li S. S. (2015) Non-histone protein methylation as a regulator of cellular signalling and function. Nat. Rev. Mol. Cell Biol. 16, 5–17 10.1038/nrm3915, 10.1038/nrn3875 [DOI] [PubMed] [Google Scholar]
- 20. Falnes P. Ø., Jakobsson M. E., Davydova E., Ho A., and Małecki J. (2016) Protein lysine methylation by seven-β-strand methyltransferases. Biochem. J. 473, 1995–2009 10.1042/BCJ20160117 [DOI] [PubMed] [Google Scholar]
- 21. Cloutier P., Lavallee-Adam M., Faubert D., Blanchette M., and Coulombe B. (2013) A newly uncovered group of distantly related lysine methyltransferases preferentially interact with molecular chaperones to regulate their activity. PloS Genet. 9, e1003210 10.1371/journal.pgen.1003210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang J., Hsu Y. H., Mo C., Abreu E., Kiel D. P., Bonewald L. F., Brotto M., and Karasik D. (2014) METTL21C is a potential pleiotropic gene for osteoporosis and sarcopenia acting through the modulation of the NF-κB signaling pathway. J. Bone Miner. Res. 29, 1531–1540 10.1002/jbmr.2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wiederstein J. L., Nolte H., Günther S., Piller T., Baraldo M., Kostin S., Bloch W., Schindler N., Sandri M., Blaauw B., Braun T., Hölper S., and Krüger M. (2018) Skeletal muscle-specific methyltransferase METTL21C trimethylates p97 and regulates autophagy-associated protein breakdown. Cell Rep. 23, 1342–1356 10.1016/j.celrep.2018.03.136 [DOI] [PubMed] [Google Scholar]
- 24. Ju J. S., Fuentealba R. A., Miller S. E., Jackson E., Piwnica-Worms D., Baloh R. H., and Weihl C. C. (2009) Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. J. Cell Biol. 187, 875–888 10.1083/jcb.200908115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang C., Zhang B., Ratliff A. C., Arrington J., Chen J., Xiong Y., Yue F., Nie Y., Hu K., Jin W., Tao W. A., Hrycyna C. A., Sun X., and Kuang S. (2019) Methyltransferase-like 21e inhibits 26S proteasome activity to facilitate hypertrophy of type IIb myofibers. FASEB J. 33, 9672–9684 10.1096/fj.201900582R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang H., Amick J., Chakravarti R., Santarriaga S., Schlanger S., McGlone C., Dare M., Nix J. C., Scaglione K. M., Stuehr D. J., Misra S., and Page R. C. (2015) A bipartite interaction between Hsp70 and CHIP regulates ubiquitination of chaperoned client proteins. Structure 23, 472–482 10.1016/j.str.2015.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Massey A. C., Kaushik S., Sovak G., Kiffin R., and Cuervo A. M. (2006) Consequences of the selective blockage of chaperone-mediated autophagy. Proc. Natl. Acad. Sci. U.S.A. 103, 5805–5810 10.1073/pnas.0507436103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jakobsson M. E., Moen A., Bousset L., Egge-Jacobsen W., Kernstock S., Melki R., and Falnes P. Ø. (2013) Identification and characterization of a novel human methyltransferase modulating Hsp70 protein function through lysine methylation. J. Biol. Chem. 288, 27752–27763 10.1074/jbc.M113.483248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cho H. S., Shimazu T., Toyokawa G., Daigo Y., Maehara Y., Hayami S., Ito A., Masuda K., Ikawa N., Field H. I., Tsuchiya E., Ohnuma S., Ponder B. A., Yoshida M., Nakamura Y., and Hamamoto R. (2012) Enhanced HSP70 lysine methylation promotes proliferation of cancer cells through activation of Aurora kinase B. Nat. Commun. 3, 1072 10.1038/ncomms2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stricher F., Macri C., Ruff M., and Muller S. (2013) HSPA8/HSC70 chaperone protein: Structure, function, and chemical targeting. Autophagy 9, 1937–1954 10.4161/auto.26448 [DOI] [PubMed] [Google Scholar]
- 31. Gamerdinger M., Hajieva P., Kaya A. M., Wolfrum U., Hartl F. U., and Behl C. (2009) Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 28, 889–901 10.1038/emboj.2009.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tahrir F. G., Knezevic T., Gupta M. K., Gordon J., Cheung J. Y., Feldman A. M., and Khalili K. (2017) Evidence for the role of BAG3 in mitochondrial quality control in cardiomyocytes. J. Cell. Physiol. 232, 797–805 10.1002/jcp.25476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Arndt V., Dick N., Tawo R., Dreiseidler M., Wenzel D., Hesse M., Fürst D. O., Saftig P., Saint R., Fleischmann B. K., Hoch M., and Höhfeld J. (2010) Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr. Biol. 20, 143–148 10.1016/j.cub.2009.11.022 [DOI] [PubMed] [Google Scholar]
- 34. Page N., Gros F., Schall N., Décossas M., Bagnard D., Briand J. P., and Muller S. (2011) HSC70 blockade by the therapeutic peptide P140 affects autophagic processes and endogenous MHCII presentation in murine lupus. Ann. Rheum. Dis. 70, 837–843 10.1136/ard.2010.139832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Leu J. I., Pimkina J., Frank A., Murphy M. E., and George D. L. (2009) A small molecule inhibitor of inducible heat shock protein 70. Mol. Cell 36, 15–27 10.1016/j.molcel.2009.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cuervo A. M., and Wong E. (2014) Chaperone-mediated autophagy: Roles in disease and aging. Cell Res. 24, 92–104 10.1038/cr.2013.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang C., Yue F., and Kuang S. (2017) Muscle histology characterization using H&E staining and muscle fiber type classification using immunofluorescence staining. Bio Protoc. 7, e2279 10.21769/BioProtoc.2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang C., Wang M., Arrington J., Shan T., Yue F., Nie Y., Tao W. A., and Kuang S. (2017) Ascl2 inhibits myogenesis by antagonizing the transcriptional activity of myogenic regulatory factors. Development 144, 235–247 10.1242/dev.138099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sarbassov D. D., Guertin D. A., Ali S. M., and Sabatini D. M. (2005) Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 10.1126/science.1106148 [DOI] [PubMed] [Google Scholar]
- 40. Wang C., Liu W., Liu Z., Chen L., Liu X., and Kuang S. (2015) Hypoxia inhibits myogenic differentiation through p53 protein-dependent induction of bhlhe40 protein. J. Biol. Chem. 290, 29707–29716 10.1074/jbc.M115.688671 [DOI] [PMC free article] [PubMed] [Google Scholar]