Abstract
Coronaviruses are enveloped, single-stranded RNA viruses that are distributed worldwide. They include transmissible gastroenteritis virus (TGEV), porcine epidemic diarrhea virus (PEDV), and the human coronaviruses severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV), many of which seriously endanger human health and well-being. Only alphacoronaviruses and betacoronaviruses harbor nonstructural protein 1 (nsp1), which performs multiple functions in inhibiting antiviral host responses. The role of the C terminus of betacoronavirus nsp1 in virulence has been characterized, but the location of the alphacoronavirus nsp1 region that is important for virulence remains unclear. Here, using TGEV nsp1 as a model to explore the function of this protein in alphacoronaviruses, we demonstrate that alphacoronavirus nsp1 inhibits host gene expression. Solving the crystal structure of full-length TGEV at 1.85-Å resolution and conducting several biochemical analyses, we observed that a specific motif (amino acids 91–95) of alphacoronavirus nsp1 is a conserved region that inhibits host protein synthesis. Using a reverse-genetics system based on CRISPR/Cas9 technology to construct a recombinant TGEV in which this specific nsp1 motif was altered, we found that this mutation does not affect virus replication in cell culture but significantly reduces TGEV pathogenicity in pigs. Taken together, our findings suggest that alphacoronavirus nsp1 is an essential virulence determinant, providing a potential paradigm for the development of a new attenuated vaccine based on modified nsp1.
Keywords: protein motif, pathogenesis, crystal structure, virulence factor, virus, coronavirus, host gene expression, immune evasion, nonstructural protein 1 (nsp1), transmissible gastroenteritis virus (TGEV)
Introduction
Coronaviruses (CoVs)2 belong to a large family of enveloped, single-stranded RNA viruses that are serious threats to public health and have caused considerable damage (1–5). Depending on serotype, CoVs can be generally divided into Alpha-, Beta-, Gamma-, and Deltacoronavirus (α-CoV, β-CoV, γ-CoV, and δ-CoV, respectively) genera (2). The α-CoVs include transmissible gastroenteritis virus (TGEV), porcine epidemic diarrhea virus (PEDV), human coronavirus 229E (HCoV-229E), human coronavirus NL63 (HCoV-NL63), and feline infectious peritonitis virus (FIPV) (1). The β-CoVs include murine hepatitis virus (MHV), SARS-CoV, and MERS-CoV (1).
CoV nonstructural protein 1 (nsp1) is the N-terminal component of the pp1a polyprotein. Only α-CoVs and β-CoVs encode nsp1, whereas γ-CoVs and δ-CoVs lack this protein (6). Nsp1 sequences identified using standard search tools, such as BLAST, are divergent among different CoVs. The sizes of α-CoV nsp1 and β-CoV nsp1 differ; the α-CoVs encode nsp1 proteins of ∼9 kDa, which are substantially smaller than the ∼20-kDa nsp1 proteins of β-CoVs. Although the sequence homologies among TGEV, PEDV, and SARS-CoV nsp1 proteins are low, the core structures share a relatively conserved domain (7–9). This high structural similarity may explain why CoV nsp1 has the conserved biological function of inhibiting host gene expression. However, the critical region of β-CoV nsp1 required to inhibit protein synthesis is different from that of α-CoV nsp1 (9–12). This difference could imply that the detailed mechanisms through which CoV nsp1 proteins inhibit host gene expression are unpredictable. To date, the ability of β-CoV nsp1 to inhibit host gene expression has been deeply studied. SARS-CoV nsp1 not only prevents mRNA translation but also promotes its degradation by binding to the 40S ribosomal subunit (13). Furthermore, MERS-CoV nsp1 selectively targets mRNA synthesized in the host cell nucleus for degradation and thus inhibits translation in host cells (14). TGEV and PEDV nsp1 proteins cannot bind the 40S ribosomal subunit to inhibit the translation of host mRNA; however, HCoV-229E and HCoV-NL63 nsp1 proteins might bind the 40S ribosomal subunit to affect host mRNA stability (9, 15, 16).
At present, the specific pathway by which α-CoV nsp1 inhibits host gene expression is unknown. However, the function of CoV nsp1 in viral immune system evasion has been well-characterized in cell culture. For example, SARS-CoV nsp1 suppresses interferon (IFN) expression and host antiviral signaling pathways in infected cells (10, 17). Furthermore, MHV nsp1 efficiently interferes with the type I IFN system (11), whereas PEDV nsp1 mediates cAMP-response element-binding protein (CREB)-binding protein (CBP) and NF-κB degradation to inhibit type II IFN responses (12, 18). Thus, nsp1 is considered a possible major virulence factor for CoVs. Moreover, the contribution of nsp1 to CoV pathogenesis has been directly demonstrated for MHV and SARS-CoV (11, 17). However, little direct evidence has indicated that α-CoV nsp1 is a virulence factor. Here, we present the crystal structure of full-length TGEV nsp1. Further structural and biochemical analyses indicated that a motif (amino acids 91–95) is important for the inhibition of host gene expression by α-CoV nsp1. In addition, we demonstrate that the loss of nsp1-induced inhibition of host protein synthesis does not affect the replication of TGEV but can significantly reduce its virulence in piglets. This research improves our understanding of why α-CoV nsp1 is necessary for virulence and may aid in the development of a new attenuated vaccine.
Results
Transient α-CoV nsp1 expression affects cellular gene expression
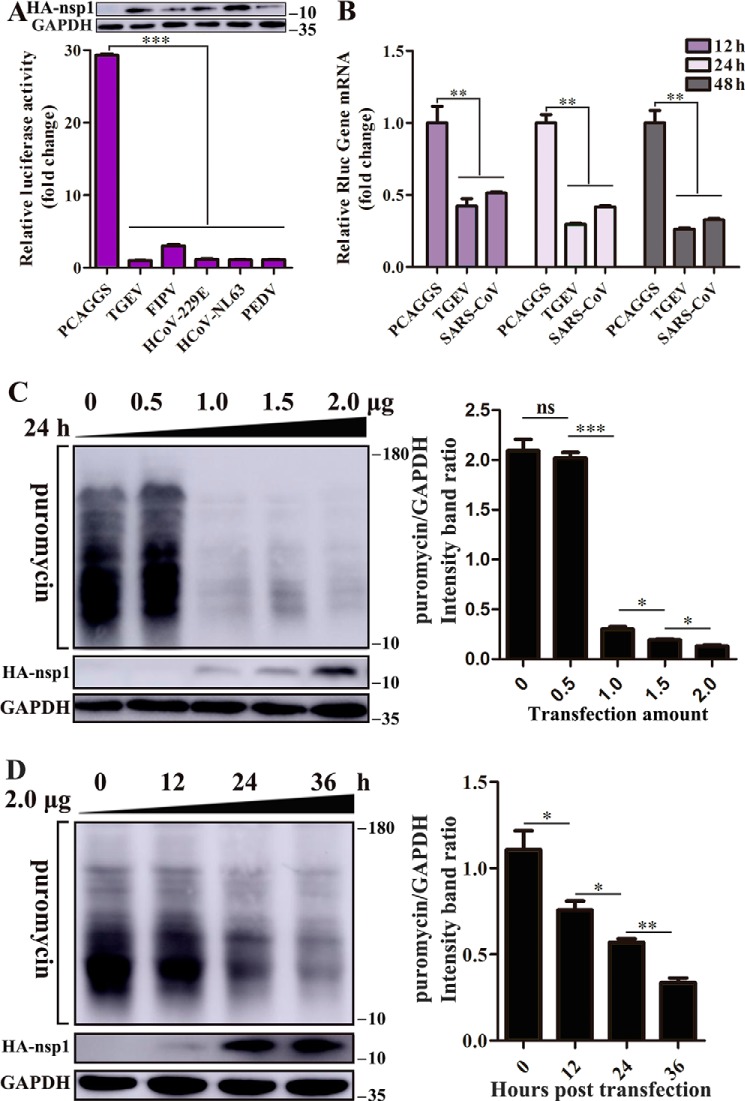
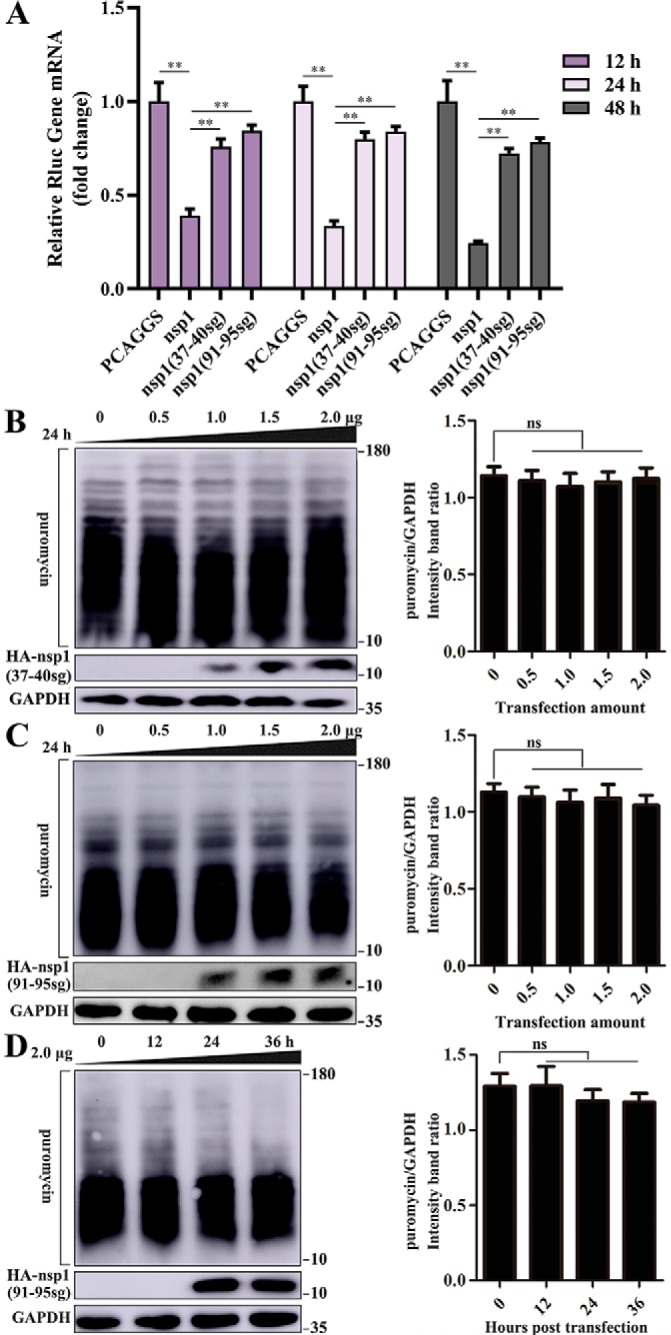
Using Renilla luciferase (Rluc) assays, we assessed several representative α-CoV nsp1 proteins for their ability to interfere with host-cell gene expression. Human embryonic kidney (HEK-293T) cells were cotransfected with plasmids expressing the nsp1 protein and pRL-SV40 plasmids expressing SV40 promoter–driven Rluc. The results showed that TGEV, FIPV, HCoV-229E, HCoV-NL63, and PEDV nsp1 significantly reduced luciferase reporter gene expression. Furthermore, subsequent Western blot analysis using equal amounts of intracellular proteins confirmed this finding (Fig. 1A).
Figure 1.
α-CoV nsp1 inhibits protein synthesis. A, α-CoV nsp1 significantly inhibited the expression of reporter genes in HEK-293T cells. The expression of nsp1 and GAPDH was detected by Western blot analysis using an anti-HA antibody and an anti-GAPDH antibody, respectively (top). B, HEK-293T cells were cotransfected with pRL-SV40 and WT plasmids. At 12, 24, and 48 h post-transfection, the cells were lysed and subjected to real-time quantitative PCR analysis. TGEV nsp1 significantly inhibited the synthesis of Rluc mRNA; SARS nsp1 was used as the positive control. C, cells were transfected with different doses of the TGEV nsp1 plasmid (0–2.0 μg) for 24 h. The cells were pulsed with 3 μm puromycin for 1 h at 37 °C and then subjected to Western blot analysis (left). The grayscale values of the protein bands were analyzed by ImageJ (right). D, cells were pulsed with 3 μm puromycin for 1 h after 0, 12, 24, and 36 h of transfection and then subjected to Western blot analysis (left). The corresponding grayscale values of the protein bands were analyzed by ImageJ (right). The error bars show the S.D. of the results from three independent experiments. *, p < 0.05, significant; **, p < 0.01, highly significant; ***, p < 0.001, extremely significant.
Then, we used TGEV nsp1 as a model to study the detailed function of α-CoV nsp1. Using the SARS-CoV nsp1 protein as a positive control (13), we examined Rluc mRNA in HEK-293T cells using real-time quantitative PCR and found that TGEV nsp1 inhibited Rluc mRNA synthesis at different time points (Fig. 1B). To further explore whether TGEV nsp1 could broadly inhibit gene expression in HEK-293T cells, we examined host proteins using a ribopuromycylation assay. The broad-spectrum inhibitory activity of TGEV nsp1 on host protein synthesis depended on both concentration and time (Fig. 1, C and D).
Identification of the critical region of TGEV nsp1 required for inhibition of protein synthesis
Although the structure of a truncated TGEV nsp1 protein (Protein Data Bank (PDB) code 3ZBD) has been reported (8), we wondered whether the C terminus could affect the overall structure. We performed initial crystallization trials for the full-length TGEV nsp1 (residues Met-1 to Arg-109). Fortunately, we were able to determine the crystal structure of full-length TGEV nsp1 using the molecular replacement method and refined it to 1.85-Å resolution in space group P21. The details of the phasing and refinement steps are given in Table 1. Although the crystal resolution was very high, the specific conformation of the C terminus remained unclear. This was probably because the C-terminal loop was flexible, for which it is difficult to obtain accurate information. Structural characterization demonstrated that the structure of full-length TGEV nsp1 displayed eight β-sheets and two α-helices (Fig. 2A), and the full-length protein shared a common skeleton with the truncated protein. The similarity of the three-dimensional conformations indicated that the C terminus had little influence on the overall TGEV nsp1 structure.
Table 1.
Data collection and refinement statistics
The highest-resolution values are indicated in parentheses. Rmerge = ∑∑|Ih − 〈I〉|/∑∑Ih where Ih is the intensity measurement of reflection h and 〈I〉 is the average intensity from multiple observations. Rwork = ∑‖Fo| − |Fc‖/∑|Fo| where Fo and Fc are the observed and calculated structure factors, respectively. Rfree is equivalent to Rwork, but 5% of the measured reflections were excluded from the refinement and set aside for cross-validation.
| TGEV nsp1 | TGEV nsp1(91–95sg) | |
|---|---|---|
| Data collection | ||
| Space group | P21 | P21 |
| Cell parameters a, b, c (Å) | 36.16, 67.01, 90.10 | 33.44, 59.24, 57.17 |
| α, β, γ (°) | 90.00, 93.98, 90.00 | 90.00, 97.54, 90.00 |
| Wavelength | 1.0 | 0.97918 |
| Resolution range (Å) | 34.31-1.85 | 40.95-1.98 |
| Completeness (%) | 96.9 (96.9) | 97.6 (97.6) |
| Rmerge (last shell) | 0.056 (0.476) | 0.025 (0.403) |
| I/σ (last shell) | 35.12 (4.43) | 42.35 (9.42) |
| Redundancy (last shell) | 4.0 (3.9) | 3.9 (3.8) |
| Refinement | ||
| Resolution (Å) | 34.31-1.80 | 40.95-1.97 |
| Rwork/Rfree | 0.179/0.232 | 0.191/0.237 |
| No. of reflections | 35,753 | 15,284 |
| No. of protein atoms | 3,490 | 1,684 |
| No. of solvent atoms | 430 | 103 |
| No. of ions/ligands | 0 | 0 |
| Mean B factor (Å2) | 20.94 | 26.55 |
| r.m.s.d. | ||
| Bond length (Å) | 0.007 | 0.007 |
| Bond angle (°) | 0.864 | 0.784 |
| Ramachandran plot | ||
| Favored (%) | 98.34 | 97.58 |
| Allowed, outlier (%) | 1.66, 0.00 | 2.42, 0.00 |
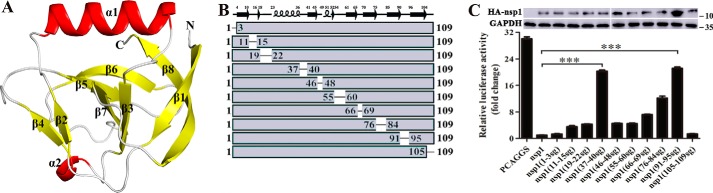
Figure 2.
Structure of full-length TGEV nsp1 and identification of the region that inhibits protein synthesis. A, crystal structure of TGEV nsp1. The structure of nsp1 is shown as a cartoon; α-helices are shown in red, and β-sheets are shown in yellow. B and C, based on the structure of TGEV nsp1, plasmids with mutated loop regions were constructed. The recombinant plasmids were as follows: nsp1(1–3sg), nsp1(11–15sg), nsp1(19–22sg), nsp1(37–40sg), nsp1(46–48sg), nsp1(55–60sg), nsp1(66–69sg), nsp1(76–84sg), nsp1(91–95sg), and nsp1(105–109sg). HEK-293T cells were transfected with the control plasmid (PCAGGS) or nsp1 plasmids. At 24 h, the cells were lysed and subjected to Rluc assays and Western blot analysis. The error bars show the S.D. of the results from three independent experiments. The asterisks indicate statistical significance calculated using Student's t test. ***, p < 0.001.
To identify which region in TGEV nsp1 was essential for its inhibitory activity, we used methods that have been previously used to study important functional regions in PEDV nsp1 (9). Based on the crystal structure of TGEV nsp1, we designed the following series of plasmids in which random coils were replaced with flexible Ser-Gly-Ser-Gly (sg) linkers: nsp1(1–3sg), nsp1(11–15sg), nsp1(19–22sg), nsp1(37–40sg), nsp1(46–48sg), nsp1(52–60sg), nsp1(66–69sg), nsp1(76–84sg), nsp1(91–95sg), and nsp1(105–109sg) (Fig. 2, B and C). The Rluc assay showed that the loops comprising amino acids 37–40 and 91–95 were the most important regions for the ability of TGEV nsp1 to inhibit reporter gene expression (Fig. 2C). Subsequently, Western blot analysis indicated that nsp1(91–95sg) expression was significantly higher than nsp1 expression (Fig. 2C), again demonstrating that nsp1 could inhibit its own expression (9, 15). We examined Rluc mRNA expression in HEK-293T cells by real-time quantitative PCR and found that TGEV nsp1(91–95sg) and TGEV nsp1(37–40sg) did not inhibit Rluc mRNA synthesis at different time points (Fig. 3A). We conducted the ribopuromycylation assay, and the results showed that TGEV nsp1(37–40sg) significantly reduced the inhibition of host gene expression (Fig. 3B). Because the TGEV nsp1(37–40sg) protein was found to be insoluble in the Escherichia coli supernatant, the motif (amino acids 37–40) was not analyzed further. Based on the above experiments, we focused on TGEV nsp1(91–95sg). Finally, the ribopuromycylation assay demonstrated that the mutant protein no longer inhibited host protein synthesis in a dose- or time-dependent manner (Fig. 3, C and D). These data indicate that the motif comprising amino acids 91–95 is critical for TGEV nsp1–induced suppression of host gene expression.
Figure 3.
The motif comprising amino acids 91–95 is a critical region for TGEV nsp1-induced inhibition of host protein synthesis. A, HEK-293T cells were cotransfected with pRL-SV40 encoding the Rluc reporter gene downstream of the SV40 promoter and one of the following plasmids: PCAGGS, PCAGGS-TGEV-nsp1-HA, PCAGGS-TGEV-nsp1(37–40sg)-HA, and PCAGGS-TGEV-nsp1(91–95sg)-HA, which encode no gene, TGEV nsp1, TGEV nsp1(37–40sg), and TGEV nsp1(91–95sg), respectively. At 12, 24, and 48 h post-transfection, the cells were lysed and subjected to real-time quantitative PCR analysis. The values of TGEV nsp1, TGEV nsp1(37–40sg), and TGEV nsp1(91–95sg) were normalized to those of the untreated empty vector (PCAGGS) control, which were set to 1. B, HEK-293T cells were transfected with different doses of the TGEV nsp1(37–40sg) plasmid (0–2.0 μg) for 24 h. The cells were pulsed with 3 μm puromycin for 1 h at 37 °C and then subjected to Western blot analysis (left). The grayscale values of the protein bands were analyzed by ImageJ (right). C, cells were transfected with different doses of the TGEV nsp1(91–95sg) plasmid (0–2.0 μg) for 24 h. The cells were pulsed with 3 μm puromycin for 1 h at 37 °C and then subjected to Western blot analysis (left). The grayscale values of the protein bands were analyzed by ImageJ (right). D, the cells were pulsed with 3 μm puromycin for 1 h after transfection for 0, 12, 24, and 36 h (left) and then subjected to Western blot analysis (right). The grayscale values of the protein bands were analyzed by ImageJ. Data are represented as mean ± S.D., n = 3. **, p < 0.01; ns, not significant.
Structural similarity of WT and mutant TGEV nsp1
To explore whether a loss of protein activity by TGEV nsp1(91–95sg) was due to structural changes, we constructed a TGEV nsp1(91–95sg) plasmid. Expression and purification were performed as described for the full-length construct. This new construct yielded crystals under different crystallization conditions than those of the full-length protein, and native data were collected to 1.98 Å. The details of the phasing and refinement steps are given in Table 1. A comparison of surface electrostatics and shape between TGEV nsp1 and TGEV nsp1(91–95sg) led us to speculate that the sg linker mutation from amino acids 91 to 95 did not significantly change the overall structure of TGEV nsp1 (Fig. 4, A–F).
Figure 4.
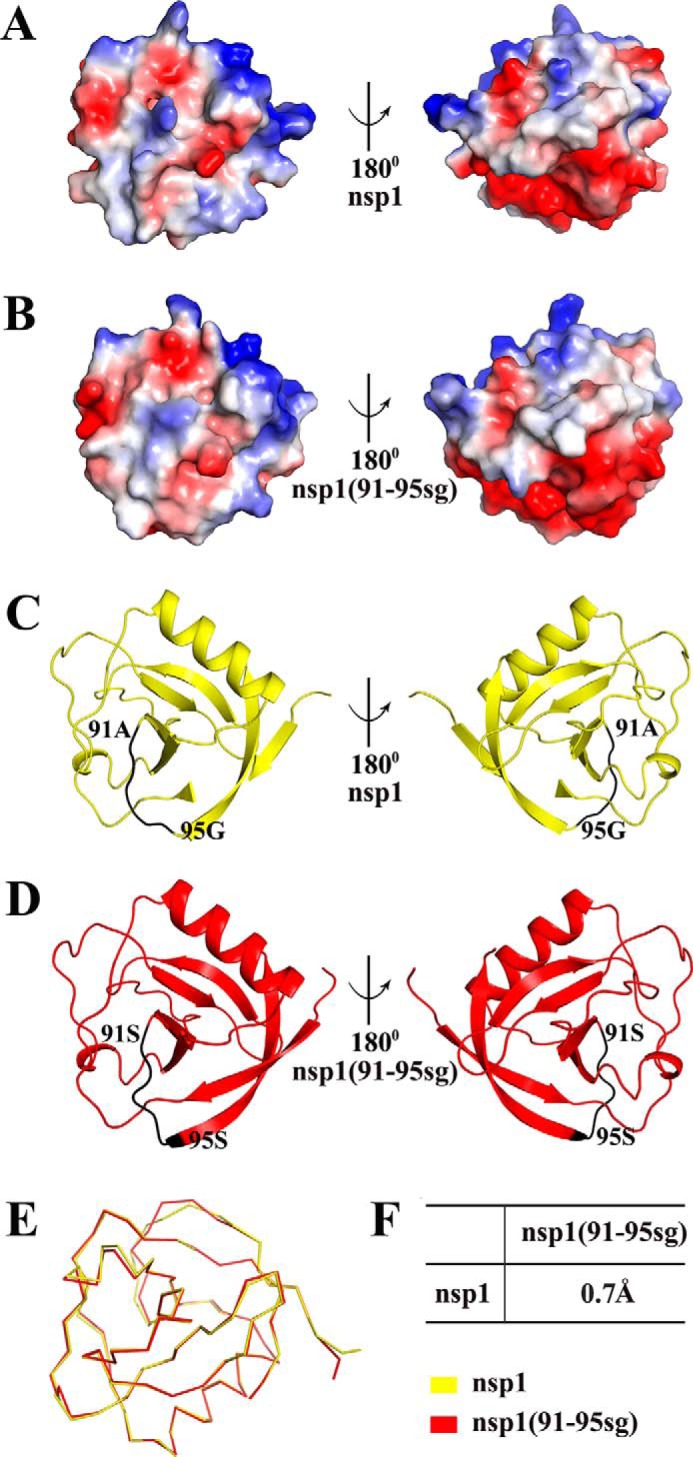
Structural comparisons of TGEV nsp1 and TGEV nsp1(91–95sg). A, views from opposite sides of the electrostatic surface of TGEV nsp1. B, views from opposite sides of the electrostatic surface of TGEV nsp1(91–95sg). Positive charges are shown in blue, and negative charges are shown in red. C, views from opposite sides of a cartoon model of TGEV nsp1 with the motif (amino acids 91–95) marked in black. D, views from opposite sides of a cartoon model of TGEV nsp1(91–95sg) with the motif (amino acids 91–95) marked in black. TGEV nsp1 is shown in yellow, and TGEV nsp1(91–95sg) is shown in red. E, ribbon diagrams of the two structures. F, r.m.s.d. values between TGEV nsp1 and TGEV nsp1(91–95sg). The r.m.s.d. values were calculated using PDBeFold.
The motif comprising amino acids 91–95 is important for the regulation of host translation by α-CoV nsp1
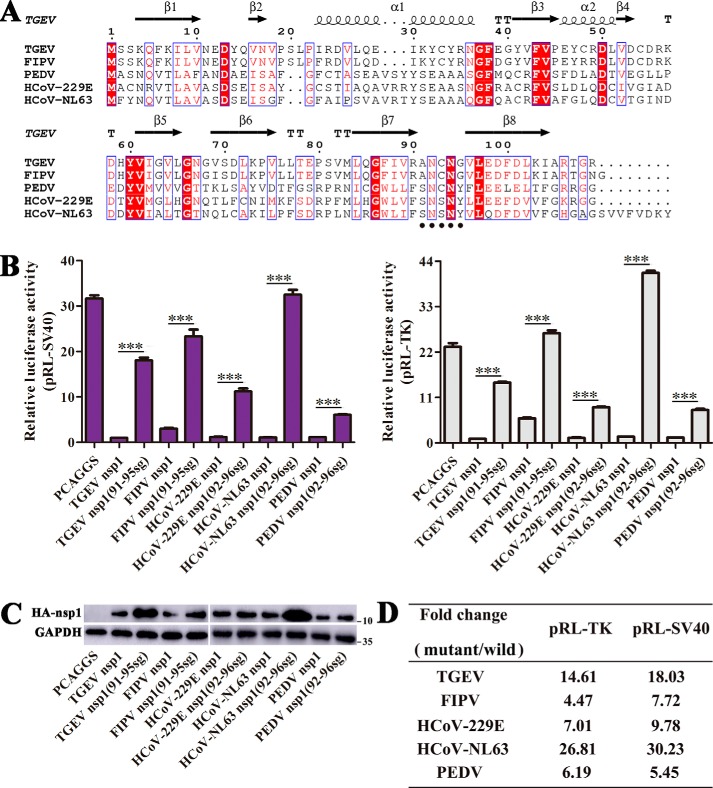
We next determined whether this motif (amino acids 91–95) plays an important role in inhibiting host protein synthesis in the context of other α-CoV nsp1 proteins. To explore the corresponding motif in α-CoV nsp1, we selected the previously mentioned representative sequences and aligned them using ClustalW2 software. The α-CoV nsp1 amino acid sequence alignment showed that the motif comprising amino acids 91–95 was a relatively conserved domain (Fig. 5A). Then, we constructed mutant nsp1 proteins from FIPV, HCoV-229E, HCoV-NL63, and PEDV to explore whether the α-CoV nsp1 motif comprising amino acids 91–95 was an important region for the suppression of host gene expression. Luciferase assays indicated that the important function of this motif (amino acids 91–95) in inhibiting host gene expression not only was exhibited by TGEV nsp1 but also could be extended to nsp1 proteins from other α-CoVs (Fig. 5B). Furthermore, Western blot analysis revealed that the expression of the mutant proteins was higher than that of the WT proteins, especially for TGEV nsp1 and HCoV-NL63 nsp1 (Fig. 5C), consistent with the results of the luciferase assays. In addition, we compared the Rluc values between the mutant and the WT nsp1 proteins (Fig. 5D). These data showed that the relevant motif (amino acids 91–95) might be responsible for inhibiting host gene expression, albeit to different degrees, among the α-CoV nsp1 proteins. Consistent with our previous results (9), when we mutated the motif (amino acids 91–95) in PEDV nsp1, we again observed partial recovery of the ability to suppress Rluc expression. Taken together, these results indicated that the motif comprising amino acids 91–95 was an important conserved region for the regulation of host translation by α-CoV nsp1.
Figure 5.
The motif comprising amino acids 91–95 is an important region for α-CoV nsp1-induced inhibition of gene expression. A, the following sequences from GenBank were used to create the sequence alignment (given as the abbreviation and GenBank accession number): TGEV, HQ462571.1; FIPV, ADL71484.1; PEDV, AJP67455.1; HCoV-229E, CAA49377.1; and HCoV-NL63, AFV53147.1. The residue numbers with reference to TGEV nsp1 are labeled on top of the panel. The regions comprising amino acids 91–95 are marked by small black dots in the α-CoV nsp1 proteins. Residues conserved in most sequences are shown in red and are boxed with a white background. The sequences were aligned with ClustalW2, and the figure was prepared with ESPript3.0. B and C, host protein synthesis inhibited by “sg” replacement mutants of α-CoV nsp1. HEK-293T cells were cotransfected with pRL-SV40 or pRL-TK and one of the following plasmids: PCAGGS (control), PCAGGS-TGEV-nsp1, PCAGGS-TGEV-nsp1(91–95sg), PCAGGS-FIPV-nsp1, PCAGGS-FIPV-nsp1(91–95sg), PCAGGS-HCoV-229E-nsp1, PCAGGS-HCoV-229E-nsp1(92–96sg), PCAGGS-HCoV-NL63-nsp1, PCAGGS-HCoV-NL63-nsp1(92–96sg), PCAGGS-PEDV-nsp1, and PCAGGS-PEDV-nsp1(92–96sg), which encode no protein, TGEV nsp1, TGEV nsp1(91–95sg), FIPV nsp1, FIPV nsp1(91–95sg), HCoV-229E nsp1, HCoV-229E nsp1(92–96sg), HCoV-NL63 nsp1, or HCoV-NL63 nsp1(92–96sg), respectively. At 24 h post-transfection, the cells were lysed and subjected to Rluc assays and Western blot analysis. Data are represented as mean ± S.D., n = 3. ***, p < 0.001. D, the -fold change in Rluc activity in the mutant protein relative to the corresponding WT protein is shown.
Construction and recovery of a TGEV mutant virus
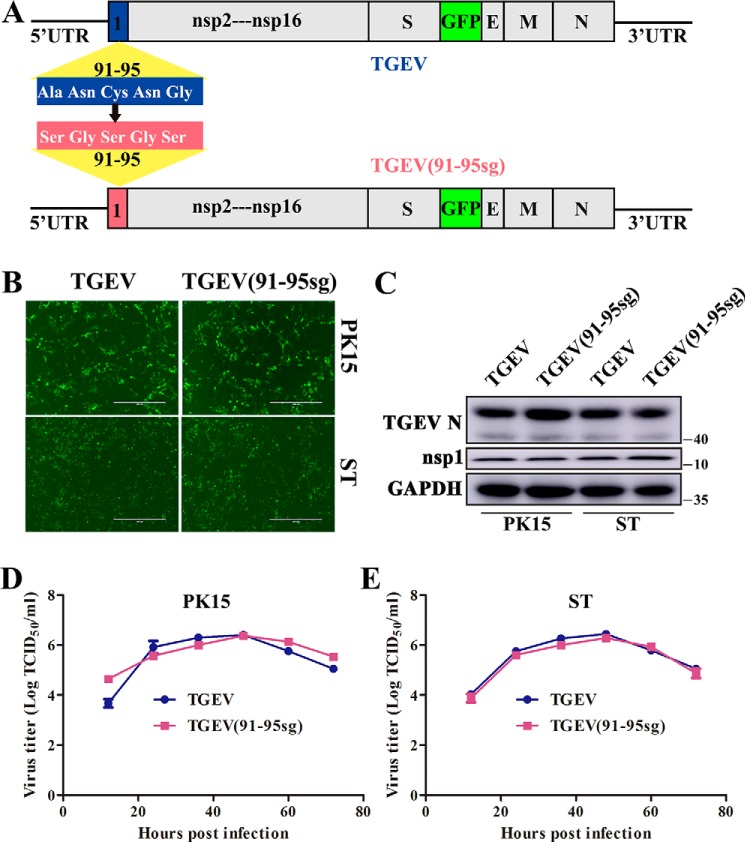
Although α-CoV nsp1 expression is known to suppress host gene expression, its biological functions in viral replication have been largely unexplored. To evaluate the role of TGEV nsp1 in inhibiting host gene expression during viral replication, we used our reverse-genetics system to construct a recombinant TGEV encoding a mutant nsp1 protein in which ORF3 was replaced by green fluorescent protein (GFP) (19). Based on the previously described results, we replaced the important motif (amino acids 91–95) in the nsp1-coding sequence. In the resulting mutant virus, the replicase gene start codon, translational reading frame, and residues required for proteolytic release of nsp1 from the replicase polyprotein were maintained (Fig. 6A). The mutant virus was successfully amplified by transfecting corresponding bacterial artificial chromosome (BAC) plasmids into HEK-293T cells. Then, we collected the amplified virus 24 h after transfection to infect TGEV-susceptible cells. Characteristic green fluorescence consistent with the parental virus phenotype was observed 36 h after infection (Fig. 6B). The recombinant virus, which was subsequently named TGEV(91–95sg), was verified by sequencing the corresponding replaced regions. To assess the stability of TGEV(91–95sg), we analyzed the nsp1-coding region by RT-PCR sequencing after seven passages in PK15 cells, and no nucleotide changes were detected. Furthermore, Western blot analysis confirmed that the virus successfully infected PK15 and swine testicle (ST) cells (Fig. 6C).
Figure 6.
Effects of the recombinant virus at the cellular level. A, DNA-based reverse-genetics BAC system for TGEV and TGEV(91–95sg). The relative positions of the mutated genes used for molecular cloning are indicated. B and C, the recombinant virus was successfully used to infect susceptible PK15 and ST cells. TGEV-BAC and TGEV(91–95sg)-BAC were transfected into HEK-293T cells with Lipofectamine 3000, and then the virus was inoculated into PK15 and ST cells 24 h later. Green fluorescence was observed after 36 h. The expression of TGEV N and TGEV nsp1 proteins in cells was also detected by Western blot analysis using anti-TGEV N and anti-TGEV nsp1 antibodies, respectively. GAPDH served as the loading control. D and E, multistep growth curves for TGEV and TGEV(91–95sg) in PK15 and ST cells at a multiplicity of infection of 0.1. Original magnification × 50 (scale bars 400 μm). The virus titers at different time points, as indicated, were determined by an end-point dilution assay. Data are represented as mean ± S.D., n = 3.
Growth properties of TGEV and TGEV(91–95sg)
Following the successful recovery of TGEV(91–95sg), we next evaluated the role of nsp1 in inhibiting host gene expression during viral replication. The growth properties of TGEV(91–95sg) in various cell lines were measured by performing a multistep growth assay with a viral multiplicity of infection of 0.1. The results showed that both viruses reached a plateau at ∼48 h postinfection in PK15 and ST cells. Furthermore, the viral growth and peak titers of TGEV(91–95sg) in susceptible cells were indistinguishable from those of the WT virus, indicating that the nsp1 gene was not critical for viral growth in cell culture (Fig. 6, D and E). To confirm this conclusion, we also tested the nsp1 expression level during viral infection and found that it was indistinguishable between the mutant and WT viruses (Fig. 6C).
Pathogenicity of TGEV and TGEV(91–95sg)
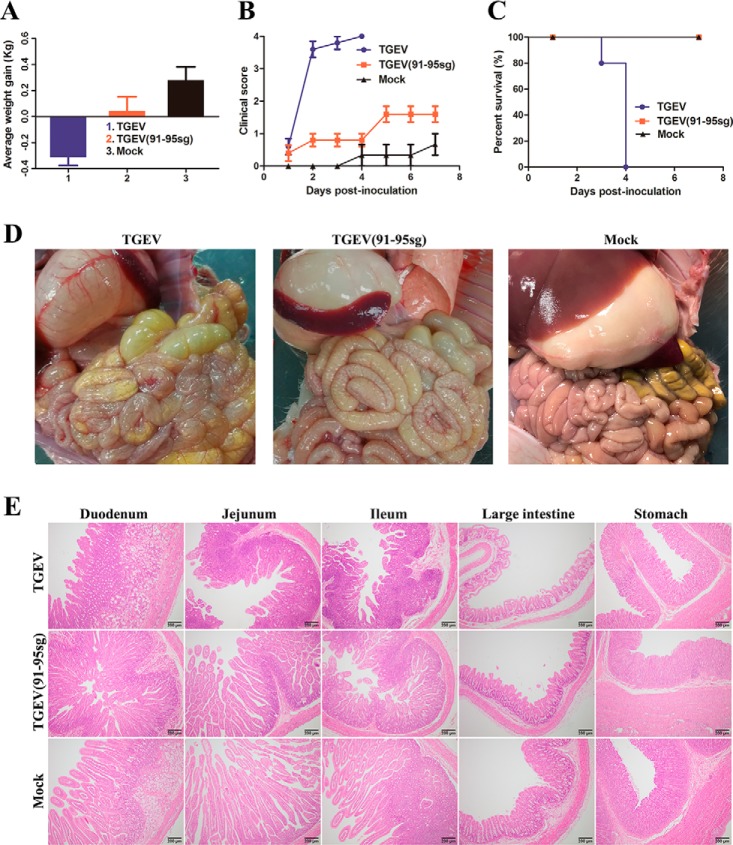
To test the pathogenicity of TGEV(91–95sg), an animal experiment was carried out. Piglets that had not been breastfed at birth were randomly divided into two groups with five piglets in each group; in addition, a mock-infected control group was formed that contained three piglets. The piglets were orally inoculated at a dose of 1 × 106 50% tissue culture infective dose (TCID50) with the respective chimeric virus or mock-infected with Dulbecco's modified Eagle's medium (DMEM). The animal experiments showed that replacement of the selected motif (amino acids 91–95) reduced the pathogenic properties of TGEV. Furthermore, in the TGEV group, all five piglets exhibited obvious dehydration and weight loss. Severe diarrhea began at 48 h postinfection, and all piglets died within 96 h, indicating the acquisition of lethal characteristics (Fig. 7C). Interestingly, the clinical signs were significantly less severe in the TGEV(91–95sg) group than in the TGEV group (Fig. 7, A–C). To further assess the attenuation of TGEV(91–95sg), we performed post-mortem examinations and hematoxylin and eosin (H&E) staining. In the TGEV group, the stomach was obviously inflated and had undigested flocculate. Moreover, the gastric fundus mucosa was congested, flushed, and bleeding. The stomach wall and small intestinal wall became thin, which was especially true for the jejunum, and animals presented with intestinal dilation with an abundance of yellow liquid. In the TGEV(91–95sg) group, the stomach was slightly inflated, and the gastric fundus mucosa was not bleeding. The small intestinal wall was slightly thinner than that in the mock group. Overall, the damage to the small intestine and stomach in the TGEV group was significantly worse than that in the TGEV(91–95sg) group (Fig. 7D). Moreover, H&E staining revealed extensive fine damage to different segments of the intestine, characterized by the fragmentation and shedding of intestinal villi. The results showed that the TGEV(91–95sg) group had only mild lesions in both the intestine and stomach compared with the TGEV group (Fig. 7E).
Figure 7.
Pathogenicity analysis of TGEV and TGEV(91–95sg). A, piglets were inoculated with the viruses. The average weight gain of the piglets was recorded on the day of death or euthanasia as the final time point. In the TGEV group, one piglet died by the 3rd day, and the other piglets died by the 4th day. In the TGEV(91–95sg) and mock groups, the piglets were euthanized on the 8th day. Data are represented as mean ± S.D., n = 3–5. B, clinical mental state scores of the piglets in the different groups. The following criteria were used for evaluation: 0, normal; 1, mild lethargy (slow to move; head down); 2, moderate lethargy (stands but tends to lie down); 3, heavier lethargy (lies down; occasionally stands); and 4, severe lethargy (recumbent; moribund). Data are represented as mean ± S.D., n = 3–5. C, survival rates of the piglets in each group. Survival curves for the piglets infected with the recovered viruses in each group are shown. D, gross lesions in piglets inoculated with the recombinant viruses. The intestinal lesions were examined on the day of death or after euthanasia at the final time points. The necropsy images show transparent intestines observed in piglets inoculated with TGEV and TGEV(91–95sg) but not in mock-inoculated piglets. E, histopathological examination of the intestines from the recombinant TGEV-infected piglets. Different segments, including the duodenum, jejunum, ileum, large intestine, and stomach, were taken from each group and then processed for H&E staining. Representative images are shown. Original magnification × 100 (scale bars 200 μm).
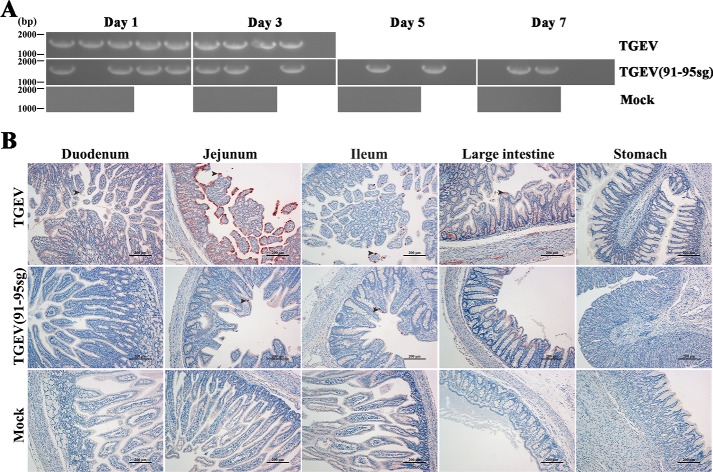
Next, we investigated how nsp1 contributed to the virulence of TGEV by analyzing the viral infection efficiency as reflected by virus shedding (Fig. 8A) and viral tissue distribution (Fig. 8B). Fecal samples were obtained with fecal swabs, and virus shedding was tested by regular PCR. The shedding of these groups was measured on days 1–7 and was terminated because of the quick deaths of the infected piglets. At 7 days postinoculation, all surviving piglets were euthanized to reduce the stress of the other piglets. The results showed that the challenged piglets had viruses in their feces. In contrast, no detectable virus shedding was found in the mock-infected pigs. Furthermore, we performed sequencing of the corresponding replaced regions to confirm the stability of the infective virus. Immunohistochemistry showed that the virus was present in susceptible intestinal tissues, consistent with the clinical results. Collectively, these data demonstrate that TGEV(91–95sg) is strongly attenuated in vivo, and we conclude that the nsp1 gene is necessary for virulence.
Figure 8.
Virus shedding in feces and immunohistochemistry of the intestine and stomach. A, daily virus shedding in feces from the different groups was measured by RT-PCR detection of viral genomes in fecal swabs. The length of the target fragment is ∼1800 bp. B, immunohistochemistry of different tissue sections, including the stomach, duodenum, jejunum, ileum, and large intestine, from the piglets. The black arrowheads point to the virus in the tissue. Original magnification × 100 (scale bars 200 μm).
Discussion
Many viral proteins suppress host gene expression and modify host cell environments to promote virus-specific translation (20, 21). As a result of such processes, these proteins are often considered important virulence factors that contribute to the design of live attenuated vaccines. For example, nonstructural protein 1 (NS1) of the influenza virus plays a crucial role in replication and pathogenesis by suppressing apoptosis-associated specklike protein ubiquitination; thus, vaccination with a live attenuated H5N1 influenza vaccine lacking NS1 is safe (22–24). The virion host shutoff (vhs) protein is the key regulator of the induced early host shutoff response, and herpes simplex viruses with vhs deleted have been proposed as live attenuated vaccines (25, 26). Human metapneumovirus protein M2-1 phosphorylation plays important regulatory roles in RNA synthesis, replication, and pathogenesis, and inhibition of M2-1 phosphorylation may serve as a novel approach for development of live attenuated vaccines (27).
This phenomenon is also widely found in CoVs. Envelope proteins affect virulence, and an attenuated virus with envelope gene deletion is a promising vaccine candidate (28–34). Although many viral proteins can induce host translational shutoff, nsp1 was the first CoV gene 1 protein recognized to play an important role in inhibiting host gene expression to regulate viral replication. In β-CoVs, deletion of the functional region of nsp1 will be critical for developing attenuated vaccines (11, 17, 35). However, the role of nsp1 in α-CoV pathogenesis is unknown. In our study, we replaced the important motif in the TGEV nsp1-coding sequence using our reverse genetics technology. Then, we successfully recovered the mutant virus. Subsequently, the viral growth and peak titers of the mutant virus in susceptible cells were found to be indistinguishable from those of the WT virus, indicating that the nsp1 gene was not a critical factor driving viral growth in cell culture (Fig. 6, D and E). To confirm this conclusion, we also tested the nsp1 expression level during viral infection and found that it was indistinguishable between the mutant and WT viruses (Fig. 6C). Furthermore, the animal experiments revealed that the determinant region within TGEV nsp1 was critical for viral virulence based on gain-of-function studies. Taken together, the results of this study demonstrate, for the first time, that loss of α-CoV nsp1–induced inhibition of host gene expression does not affect viral replication but significantly reduces virulence.
Among the four CoV genera, only α-CoV and β-CoV encode nsp1. Previous studies have indicated that the mechanisms by which nsp1 suppresses host gene expression may differ among CoV species (13–15, 36). The mechanisms of nsp1-induced host gene suppression have been well-characterized for β-CoVs. Nsp1 proteins of bat CoVs have been shown to bind viral RNA and to suppress host translation (37). SARS-CoV nsp1 uses a two-pronged strategy to inhibit host gene expression by first interacting with the 40S ribosomal subunit and then inactivating translation (13). MERS-CoV nsp1 selectively targets transcribed mRNA to inhibit host gene expression (14). MHV nsp1 interferes efficiently with the type I IFN system to enhance virulence (11). Nonetheless, structural analysis indicates that CoV nsp1s have a common origin (7–9) and that the nsp1 proteins of α-CoVs and β-CoVs share a biological function to inhibit host gene expression. Furthermore, previous studies have revealed that the C terminus of MHV and SARS-CoV nsp1 is an important functional region for inhibiting host protein synthesis (10, 11). However, the conserved important domain underlying the α-CoV nsp1–induced inhibition of host gene expression is unclear. Although we have reported that the loops at amino acid positions 67–71, 78–85, and 103–110 generate a stable functional region (9), this functional domain is not present in the TGEV nsp1 protein (12). To identify the common functional domain in α-CoV nsp1, we selected TGEV nsp1 as a model. Although the structure of a truncated TGEV nsp1 (PDB code 3ZBD) has been reported (8), we wondered whether the C terminus could affect the overall structure. Subsequently, we determined the structure of full-length TGEV nsp1. Structural characterization revealed that full-length TGEV nsp1 shared a common skeleton with truncated nsp1. These similar conformations suggested that the C terminus had no influence on the overall structure of TGEV nsp1. Although the 37–40 mutant protein significantly reduced the inhibition of host gene expression, the mutant virus with nsp1(37–40sg) was not successful. Although a previous study has also reported that 5′-UTR of coronavirus can effect viral replication (38), we are not sure whether the rescue failure of this mutant virus is due to the loss of nsp1 activity or the effect of 5′-UTR. Through structural analysis and biochemical experiments, we verified that the motif comprising amino acids 91–95 was necessary for the inhibition of host gene expression by nsp1. Interestingly, this motif could be expanded to other representative α-CoV nsp1 proteins (Fig. 5). In our study, we found that the motif comprising amino acids 91–95 is a conserved region in α-CoVs, which is conducive to understanding the mechanism of α-CoV nsp1 inhibition of host gene expression.
In summary, using a reverse-genetics platform, we obtained the first evidence that TGEV nsp1 does not affect replication ability but significantly reduces pathogenicity. We also found that the motif comprising amino acids 91–95 in α-CoV nsp1 is a common functional region involved in the inhibition of host gene expression. Overall, our results add detail to the molecular mechanism of α-CoV virulence, which might contribute to the design of a novel attenuated α-CoV vaccine based on nsp1 modifications.
Experimental procedures
Cells and viruses
HEK-293T and ST cells (ATCC) were maintained at 37 °C with 5% CO2 in Gibco DMEM (high glucose, 4.5 g/liter) containing 10% fetal bovine serum. PK15 cells (ATCC) were maintained in Gibco DMEM (low glucose, 1 g/liter) supplemented with 10% fetal bovine serum. The pBAC-TGEV-GFP plasmid containing a full-length infectious cDNA clone, which was derived from the highly virulent TGEV strain WH-1 (GenBankTM accession number HQ462571), has been described previously (19). To acquire the TGEV recombinant virus, the plasmid was first transfected into HEK-293T cells, and then the virus was recovered to infect virus-susceptible cells.
Plasmid construction
For expression in E. coli, the full-length gene sequence of TGEV nsp1 (GenBank accession number HQ462571.1) was cloned into a pET-42b(+) vector with an N-terminal His6 tag via PCR amplification. The forward and reverse primers contained the NdeI and XhoI restriction sites, respectively. The mutant TGEV nsp1 sequence was inserted into pET-42b using the same method.
For expression in eukaryotic cells, WT nsp1 flanked with an N-terminal hemagglutinin (HA) tag was cloned into the PCAGGS vector using the EcoRI and XhoI restriction sites. To determine the functional region of TGEV nsp1, we used a method reported previously (9). The following plasmids in which a random coil was replaced with a flexible sg linker were engineered : nsp1(1–3sg), nsp1(11–15sg), nsp1(19–22sg), nsp1(37–40sg), nsp1(46–48sg), nsp1(55–60sg), nsp1(66–69sg), nsp1(76–84sg), nsp1(91–95sg), and nsp1(105–109sg). All of the recombinant expression plasmids were sequenced, and no unexpected mutations occurred.
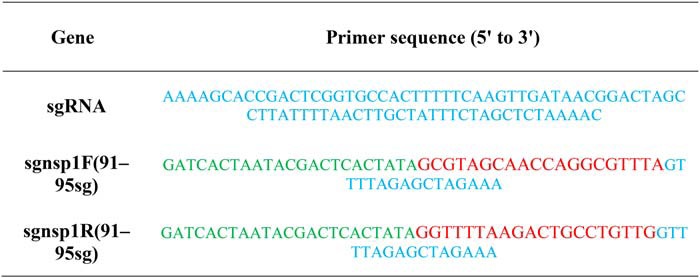
To construct the recombinant virus containing mutant nsp1, a method previously reported by our laboratory was used (19). The sequences of the relevant primers are shown in Table 2. Each corresponding mutant virus was efficiently constructed using CRISPR/Cas9 technology. Briefly, two specific restriction sites encompassing the TGEV nsp1 sequence were selected. Then, we synthesized two types of single-stranded DNA forward primers (sgnsp1F(91–95sg) and sgnsp1R(91–95sg)) and a constant reverse primer (sgRNA) corresponding to the two restriction sites. After performing annealing PCR using the forward and reverse primers, the purified short DNA fragments from the PCR products were transcribed using T7 RNA polymerase. The transcribed products corresponding to the two sites were incubated with the nuclease Cas9 to digest the pTGEV-GFP BAC plasmid in vitro, and the digestion yielded a linearized BAC and an ∼1.8-kb DNA fragment that included the TGEV nsp1 sequence. Specifically, pTGEV-GFP BAC was digested in a 50-μl reaction mixture with 5 μg of pTGEV-GFP BAC, 5 μl of Cas9 (New England Biolabs), 10 μg of sgRNA, and 5 μl of Nuclease Reaction Buffer (New England Biolabs) at 37 °C overnight.
Table 2.
Sequences of the primers used for CRISPR/Cas9
sgRNA is a common bottom oligo for transcription template DNA. The T7 promoter sequence is presented in green, and the guide sequence for the targeting of transcription template DNA is presented in red. The blue sequence regions of sgnsp1F(91–95sg)/sgnsp1R(91–95sg) are the regions that overlap with the sgRNA sequence.
Protein expression and purification
For protein expression, the recombinant plasmid was transformed into E. coli Trans BL21 (DE3) cells, which were grown at 37 °C in lysogeny broth (LB) medium containing 50 μg/ml kanamycin until the optical density at 600 nm (OD600) reached 0.6–0.8. Then, 1 mm isopropyl β-d-thiogalactopyranoside was added to induce cell growth for 8 h at 27 °C. Purification was performed using a method reported previously (9). The target protein was further purified by a Superdex 200 (GE Healthcare) column with elution buffer (20 mm Tris-HCl and 200 mm NaCl, pH 7.4). For crystallization, the purified protein was concentrated to ∼10 mg/ml.
Crystallization, data collection, and structure determination
TGEV nsp1 and TGEV nsp1(91–95sg) were crystallized via the sitting-drop vapor-diffusion method at 20 °C. Optimal crystals were obtained under the following conditions through screening and optimization: 0.5 m nickel chloride and 4% PEG3350 for TGEV nsp1 and 0.2 m ammonium phosphate dibasic and 23% PEG3350 for TGEV nsp1(91–95sg). Single crystals were first washed with 5, 10, 15 and 30% ethylene glycol (v/v) as a cryoprotectant and then flash frozen in liquid nitrogen. All data collection steps were performed on beamline BL17U at the Shanghai Synchrotron Radiation Facility (SSRF) using a MAR 225 CCD detector (MAR Research). All of the diffraction images were integrated, merged, and scaled using HKL-3000 software (39). The structures were solved by molecular replacement with PHASER (40) using the truncated structure of TGEV nsp1 (PDB code 3ZBD) as a starting model. Manual model building was performed using Coot (41), and the structures were refined with Phenix (42). The refinement statistics are shown in Table 1. All of the structural figures were drawn using PyMOL (43). The root mean square deviation (r.m.s.d.) was analyzed using PDBeFold (https://www.ebi.ac.uk/msd-srv/ssm/).3 The amino acid sequences of the CoV nsp1 proteins were aligned using the ClustalW2 software program (44) and visualized with the ESPript 3 server (http://espript.ibcp.fr/ESPript/ESPript/)3 (46).
Reporter assay, ribopuromycylation assay, and Western blot analysis
The functional plasmid and reporter gene plasmids were transfected into HEK-293T cells using Lipofectamine 3000 (Thermo Fisher) according to the manufacturer's instructions. At 24 h post-transfection, cell lysates were prepared and subjected to an Rluc reporter activity assay (Promega). A ribopuromycylation assay was performed as described previously (9, 45). Briefly, cultured HEK-293T cells were transfected with different doses of the different plasmids. At various time points post-transfection, the cells were pulse-labeled with 3 μm puromycin and then incubated for an additional hour at 37 °C with 5% CO2.
To determine the success of plasmid transfection or virus infection in the cells, cell samples were collected for treatment. For treatment, the cells were first gently washed twice with precooled PBS and then centrifuged at 4000 rpm for 5 min. The supernatant was discarded, radioimmune precipitation assay lysis buffer (Beyotime) was added, and the samples were incubated with rotation at 4 °C for 20 min. The extracts were prepared in SDS-PAGE sample buffer. Protein expression was analyzed via Western blotting. The proteins were visualized using an anti-HA antibody (Ab; Signalway Antibody (SAB)), an anti-puromycin Ab (Millipore), an anti-TGEV N Ab (provided by the laboratory), and an anti-TGEV nsp1 Ab (provided by the laboratory). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression was detected with an anti-GAPDH monoclonal Ab (mAb; Proteintech) to confirm equal protein loading. The corresponding grayscale value of each protein band was analyzed with ImageJ.
Quantitative analysis
Quantitative analysis of mRNA was performed as described previously (9). Briefly, RNA was extracted from the tissue or cell samples, and a reverse transcription kit (Toyobo) was used to quantitatively reverse transcribe 100 ng of RNA in a 20-μl system. The reverse transcription program was performed according to the instructions of the reverse transcription kit (Toyobo). The target mRNA and the endogenous control RNA (18S rRNA) were amplified with a TaqMan One-Step Real-time PCR Master Mix Reagent kit (Bio-Rad). Following the manufacturer's instructions, PCR was performed on an ABI PRISM 7000 real-time thermocycler (Applied Biosystems, Foster City, CA). The amount of target mRNA was normalized to the amount of endogenous 18S rRNA.
Protein structure accession numbers and statistical analysis
The coordinates and structural characteristics of TGEV nsp1 and TGEV nsp1(91–95sg) were submitted to the Research Collaboratory for Structural Bioinformatics (RCSB) under PDB accession numbers 6IVC and 6IVD, respectively. Unpaired Student's t test and one-way analysis of variance with Bonferroni correction (Prism version 6.01, GraphPad Software) were used to analyze differences between groups. * indicates a significant difference (p < 0.05), ** indicates a highly significant difference (p < 0.01), and *** indicates an extremely significant difference (p < 0.001).
Animal experiments
Thirteen 1-day-old piglets from a TGEV-free sow were randomly divided into three groups and fed fresh liquid milk diluted in warm water every 3 h. All piglets were confirmed to be free of TGEV, PEDV, porcine deltacoronavirus, and rotavirus by RT-PCR analysis of piglet feces before viral challenge. The piglet weights were measured and recorded at the beginning of the challenge. The piglet challenge group was inoculated orally with chimeric viral particles at a dose of 1 × 106 TCID50 or mock-inoculated with 500 μl of DMEM. At 7 days postinoculation, all surviving piglets were euthanized to reduce the stress of the other piglets. Before necropsy, the weight of every piglet was recorded. At necropsy, four sections of the intestine (duodenum, jejunum, ileum, and large intestine) and the stomach were collected, fixed in 10% formalin for histopathological examination, and stained with H&E.
Ethics statement
The animal experiments were performed according to protocols approved by the Scientific Ethics Committee of Huazhong Agricultural University (permit number HZAUSW-2018-009). Animal care and maintenance procedures were in compliance with the recommendations by the Regulations for the Administration of Affairs Concerning Experimental Animals provided by the Ministry of Science and Technology of China.
Author contributions
Z. S. data curation; Z. S. software; Z. S. and G. W. formal analysis; Z. S. validation; Z. S. visualization; Z. S. writing-original draft; Z. S., G. W., Y. Y., and J. S. project administration; G. W. methodology; L. F., S. X., Z. F. F., and G. P. resources; F. L. and G. P. writing-review and editing; G. P. supervision; G. P. funding acquisition.
Acknowledgment
We thank the staff at the Shanghai Synchrotron Radiation Facility (SSRF) BL17U1 beamline for assistance with the X-ray data collection.
This work was supported by National Natural Science Foundation of China Grants 31873020 and 31722056, National Key Research and Development Plan of China Grant 2018YFD0500102, and Huazhong Agricultural University Scientific and Technological Self-Innovation Foundation Program 2662017PY028. The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (codes 6IVC and 6IVD) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- CoV
- coronavirus
- HCoV
- human coronavirus
- FIPV
- feline infectious peritonitis virus
- MHV
- murine hepatitis virus
- TGEV
- transmissible gastroenteritis virus
- PEDV
- porcine epidemic diarrhea virus
- SARS-CoV
- severe acute respiratory syndrome coronavirus
- MERS-CoV
- Middle East respiratory syndrome coronavirus
- nsp1
- nonstructural protein 1
- IFN
- interferon
- Rluc
- Renilla luciferase
- PDB
- Protein Data Bank
- sg
- Ser-Gly-Ser-Gly
- BAC
- bacterial artificial chromosome
- ST
- swine testicle
- TCID50
- 50% tissue culture infective dose
- DMEM
- Dulbecco's modified Eagle's medium
- r.m.s.d.
- root mean square deviation
- H&E
- hematoxylin and eosin
- NS1
- nonstructural protein 1
- vhs
- virion host shutoff
- HA
- hemagglutinin
- sgRNA
- single guide RNA
- Ab
- antibody
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase.
References
- 1. Weiss S. R., and Navas-Martin S. (2005) Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 69, 635–664 10.1128/MMBR.69.4.635-664.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Weiss S. R., and Leibowitz J. L. (2011) Coronavirus pathogenesis. Adv. Virus. Res. 81, 85–164 10.1016/B978-0-12-385885-6.00009-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jung K., and Saif L. J. (2015) Porcine epidemic diarrhea virus infection: etiology, epidemiology, pathogenesis and immunoprophylaxis. Vet. J. 204, 134–143 10.1016/j.tvjl.2015.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhou P., Fan H., Lan T., Yang X. L., Shi W. F., Zhang W., Zhu Y., Zhang Y. W., Xie Q. M., Mani S., Zheng X. S., Li B., Li J. M., Guo H., Pei G. Q., et al. (2018) Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 556, 255–258 10.1038/s41586-018-0010-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fehr A. R., and Perlman S. (2015) Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 1282, 1–23 10.1007/978-1-4939-2438-7_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Woo P. C., Huang Y., Lau S. K., and Yuen K. Y. (2010) Coronavirus genomics and bioinformatics analysis. Viruses 2, 1804–1820 10.3390/v2081803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Almeida M. S., Johnson M. A., and Wüthrich K. (2006) NMR assignment of the SARS-CoV protein nsp1. J. Biomol. NMR 36, 46 10.1007/s10858-006-9018-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jansson A. M. (2013) Structure of alphacoronavirus transmissible gastroenteritis virus nsp1 has implications for coronavirus nsp1 function and evolution. J. Virol. 87, 2949–2955 10.1128/JVI.03163-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shen Z., Ye G., Deng F., Wang G., Cui M., Fang L., Xiao S., Fu Z. F., and Peng G. (2018) Structural basis for the inhibition of host gene expression by porcine epidemic diarrhea virus nsp1. J. Virol. 92, e01896 10.1128/JVI.01896-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Narayanan K., Huang C., Lokugamage K., Kamitani W., Ikegami T., Tseng C. T., and Makino S. (2008) Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 82, 4471–4479 10.1128/JVI.02472-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Züst R., Cervantes-Barragán L., Kuri T., Blakqori G., Weber F., Ludewig B., and Thiel V. (2007) Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog. 3, e109 10.1371/journal.ppat.0030109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang Q., Ma J., and Yoo D. (2017) Inhibition of NF-κB activity by the porcine epidemic diarrhea virus nonstructural protein 1 for innate immune evasion. Virology 510, 111–126 10.1016/j.virol.2017.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kamitani W., Huang C., Narayanan K., Lokugamage K. G., and Makino S. (2009) A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat. Struct. Mol. Biol. 16, 1134–1140 10.1038/nsmb.1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lokugamage K. G., Narayanan K., Nakagawa K., Terasaki K., Ramirez S. I., Tseng C. T., and Makino S. (2015) Middle East respiratory syndrome coronavirus nsp1 inhibits host gene expression by selectively targeting mRNAs transcribed in the nucleus while sparing mRNAs of cytoplasmic origin. J. Virol. 89, 10970–10981 10.1128/JVI.01352-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang C., Lokugamage K. G., Rozovics J. M., Narayanan K., Semler B. L., and Makino S. (2011) Alphacoronavirus transmissible gastroenteritis virus nsp1 protein suppresses protein translation in mammalian cells and in cell-free HeLa cell extracts but not in rabbit reticulocyte lysate. J. Virol. 85, 638–643 10.1128/JVI.01806-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang Y., Shi H., Rigolet P., Wu N., Zhu L., Xi X. G., Vabret A., Wang X., and Wang T. (2010) Nsp1 proteins of group I and SARS coronaviruses share structural and functional similarities. Infect. Genet. Evol. 10, 919–924 10.1016/j.meegid.2010.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wathelet M. G., Orr M., Frieman M. B., and Baric R. S. (2007) Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J. Virol. 81, 11620–11633 10.1128/JVI.00702-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang Q., Shi K., and Yoo D. (2016) Suppression of type I interferon production by porcine epidemic diarrhea virus and degradation of CREB-binding protein by nsp1. Virology 489, 252–268 10.1016/j.virol.2015.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang G., Liang R., Liu Z., Shen Z., Shi J., Shi Y., Deng F., Xiao S., Fu Z. F., and Peng G. (2019) The N-terminal domain of spike protein is not the enteric tropism determinant for transmissible gastroenteritis virus in piglets. Viruses 11, E313 10.3390/v11040313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lloyd R. E. (2006) Translational control by viral proteinases. Virus Res. 119, 76–88 10.1016/j.virusres.2005.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schneider R. J., and Mohr I. (2003) Translation initiation and viral tricks. Trends Biochem. Sci. 28, 130–136 10.1016/S0968-0004(03)00029-X [DOI] [PubMed] [Google Scholar]
- 22. Patnaik S., Basu D., Southall N., Dehdashti S., Wan K. K., Zheng W., Ferrer M., Taylor M., Engel D. A., and Marugan J. J. (2019) Identification, design and synthesis of novel pyrazolopyridine influenza virus nonstructural protein 1 antagonists. Bioorg. Med. Chem. Lett. 29, 1113–1119 10.1016/j.bmcl.2019.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Park H. S., Liu G., Thulasi Raman S. N., Landreth S. L., Liu Q., and Zhou Y. (2018) NS1 protein of 2009 pandemic influenza a virus inhibits porcine NLRP3 Inflammasome-mediated interleukin-1β production by suppressing ASC ubiquitination. J. Virol. 92, e00022–18 10.1128/JVI.00022-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nicolodi C., Groiss F., Kiselev O., Wolschek M., Seipelt J., and Muster T. (2019) Safety and immunogenicity of a replication-deficient H5N1 influenza virus vaccine lacking NS1. Vaccine 37, 3722–3729 10.1016/j.vaccine.2019.05.013 [DOI] [PubMed] [Google Scholar]
- 25. Liu Y. F., Tsai P. Y., Chulakasian S., Lin F. Y., and Hsu W. L. (2016) The pseudorabies virus vhs protein cleaves RNA containing an IRES sequence. FEBS J. 283, 899–911 10.1111/febs.13642 [DOI] [PubMed] [Google Scholar]
- 26. Pasieka T. J., Lu B., Crosby S. D., Wylie K. M., Morrison L. A., Alexander D. E., Menachery V. D., and Leib D. A. (2008) Herpes simplex virus virion host shutoff attenuates establishment of the antiviral state. J. Virol. 82, 5527–5535 10.1128/JVI.02047-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cai H., Zhang Y., Lu M., Liang X., Jennings R., Niewiesk S., and Li J. (2016) Phosphorylation of human metapneumovirus M2-1 protein upregulates viral replication and pathogenesis. J. Virol. 90, 7323–7338 10.1128/JVI.00755-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jimenez-Guardeño J. M., Nieto-Torres J. L., DeDiego M. L., Regla-Nava J. A., Fernandez-Delgado R., Castaño-Rodriguez C., and Enjuanes L. (2014) The PDZ-binding motif of severe acute respiratory syndrome coronavirus envelope protein is a determinant of viral pathogenesis. PLoS Pathog. 10, e1004320 10.1371/journal.ppat.1004320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. DeDiego M. L., Nieto-Torres J. L., Jimenez-Guardeño J. M., Regla-Nava J. A., Castaño-Rodriguez C., Fernandez-Delgado R., Usera F., and Enjuanes L. (2014) Coronavirus virulence genes with main focus on SARS-CoV envelope gene. Virus. Res. 194, 124–137 10.1016/j.virusres.2014.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Su S., Wong G., Shi W., Liu J., Lai A. C. K., Zhou J., Liu W., Bi Y., and Gao G. F. (2016) Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 24, 490–502 10.1016/j.tim.2016.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lamirande E. W., DeDiego M. L., Roberts A., Jackson J. P., Alvarez E., Sheahan T., Shieh W. J., Zaki S. R., Baric R., Enjuanes L., and Subbarao K. (2008) A live attenuated severe acute respiratory syndrome coronavirus is immunogenic and efficacious in golden Syrian hamsters. J. Virol. 82, 7721–7724 10.1128/JVI.00304-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Netland J., DeDiego M. L., Zhao J., Fett C., Álvarez E., Nieto-Torres J. L., Enjuanes L., and Perlman S. (2010) Immunization with an attenuated severe acute respiratory syndrome coronavirus deleted in E protein protects against lethal respiratory disease. Virology 399, 120–128 10.1016/j.virol.2010.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fett C., DeDiego M. L., Regla-Nava J. A., Enjuanes L., and Perlman S. (2013) Complete protection against severe acute respiratory syndrome coronavirus-mediated lethal respiratory disease in aged mice by immunization with a mouse-adapted virus lacking E protein. J. Virol. 87, 6551–6559 10.1128/JVI.00087-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Graham R. L., Donaldson E. F., and Baric R. S. (2013) A decade after SARS: strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 11, 836–848 10.1038/nrmicro3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lei L., Ying S., Baojun L., Yi Y., Xiang H., Wenli S., Zounan S., Deyin G., Qingyu Z., Jingmei L., and Guohui C. (2013) Attenuation of mouse hepatitis virus by deletion of the LLRKxGxKG region of Nsp1. PLoS One 8, e61166 10.1371/journal.pone.0061166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tohya Y., Narayanan K., Kamitani W., Huang C., Lokugamage K., and Makino S. (2009) Suppression of host gene expression by nsp1 proteins of group 2 bat coronaviruses. J. Virol. 83, 5282–5288 10.1128/JVI.02485-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gustin K. M., Guan B. J., Dziduszko A., and Brian D. A. (2009) Bovine coronavirus nonstructural protein 1 (p28) is an RNA binding protein that binds terminal genomic cis-replication elements. J. Virol. 83, 6087–6097 10.1128/JVI.00160-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guan B. J., Su Y. P., Wu H. Y., and Brian D. A. (2012) Genetic evidence of a long-range RNA-RNA interaction between the genomic 5′ untranslated region and the nonstructural protein 1 coding region in murine and bovine coronaviruses. J. Virol. 86, 4631–4643 10.1128/JVI.06265-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Minor W., Cymborowski M., Otwinowski Z., and Chruszcz M. (2006) HKL-3000: the integration of data reduction and structure solution-from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 10.1107/S0907444906019949 [DOI] [PubMed] [Google Scholar]
- 40. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 42. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., and Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 10.1107/S0907444902016657 [DOI] [PubMed] [Google Scholar]
- 43. Grell L., Parkin C., Slatest L., and Craig P. A. (2006) EZ-Viz, a tool for simplifying molecular viewing in PyMOL. Biochem. Mol. Biol. Educ. 34, 402–407 10.1002/bmb.2006.494034062672 [DOI] [PubMed] [Google Scholar]
- 44. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., and Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 45. Schmidt E. K., Clavarino G., Ceppi M., and Pierre P. (2009) SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods 6, 275–277 10.1038/nmeth.1314 [DOI] [PubMed] [Google Scholar]
- 46. Robert X., and Gouet P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 10.1093/nar/gku316 [DOI] [PMC free article] [PubMed] [Google Scholar]