Abstract
The trimethylamine methyltransferase MttB is the founding member of a widely distributed superfamily of microbial proteins. Genes encoding most members of the MttB superfamily lack the codon for pyrrolysine that distinguishes previously characterized trimethylamine methyltransferases, leaving the function(s) of most of the enzymes in this superfamily unknown. Here, investigating the MttB family member MtpB from the human intestinal isolate Eubacterium limosum ATCC 8486, an acetogen that excretes N-methyl proline during growth on proline betaine, we demonstrate that MtpB catalyzes anoxic demethylation of proline betaine. MtpB along with MtqC (a corrinoid protein) and MtqA (a methylcorrinoid:tetrahydrofolate methyltransferase) was much more abundant in E. limosum cells grown on proline betaine than on lactate. We observed that recombinant MtpB methylates Co(I)-MtqC in the presence of proline betaine and that other quaternary amines are much less preferred substrates. MtpB, MtqC, and MtqA catalyze tetrahydrofolate methylation with proline betaine, thereby forming a key intermediate in the Wood–Ljungdahl acetogenesis pathway. To our knowledge, MtpB methylation of Co(I)-MtqC for the subsequent methylation of tetrahydrofolate represents the first described anoxic mechanism of proline betaine demethylation. The activities of MtpB and associated proteins in acetogens or other anaerobes provide a possible mechanism for the production of N-methyl proline by the gut microbiome. MtpB's activity characterized here strengthens the hypothesis that much of the MttB superfamily comprises quaternary amine-dependent methyltransferases.
Keywords: bacterial metabolism, microbiology, energy metabolism, folate, microbiome, enzyme catalysis, acetogenesis, cobalamin, one-carbon metabolism, trimethylamine methyltransferase, cobalamin
Introduction
The MttB superfamily of proteins is widespread, with numerous examples found in both bacteria and archaea. MttB, the founding member of the superfamily, is a trimethylamine:corrinoid methyltransferase notable for possessing the genetically encoded amino acid pyrrolysine (1–4). To date, all verified trimethylamine:corrinoid methyltransferases have possessed pyrrolysine. The MttB family members encoded by bacterial and archaeal genes with a pyrrolysine codon form a single clade on the MttB superfamily phylogenetic tree. The remaining clades encompass the vast majority of family members and are encoded by genes lacking a pyrrolysine codon, leaving the function(s) of this diverse family in question (5).
MtgB is one of these “non-pyrrolysine” (non-Pyl)2 MttB family members found in the anaerobically respiring bacterium Desulfitobacterium hafniense. MtgB was shown to be a glycine betaine:cobalamin methyltransferase and is the first component of a corrinoid-dependent methyltransferase system (5). Such systems generally contain three components: a substrate-specific methyltransferase that demethylates the substrate to methylate a corrinoid-binding protein, which serves as the methyl donor for a second methyltransferase that subsequently methylates a cellular cofactor (6). In the case of D. hafniense glycine betaine metabolism, MtgB, MtgC, and MtgA are the glycine betaine-specific methyltransferase, the proposed cognate corrinoid-binding protein, and a methylcobalamin:tetrahydrofolate (THF) methyltransferase, respectively (5). Highly similar homologs of MtgB, MtgC, and MtgA are encoded in a polycistronic operon with a glycine betaine transporter in acetogenic Acetobacterium woodii (7).
The notable sequence diversity found in the MttB superfamily led Ticak et al. (5) to hypothesize that members of the family may have diversified their substrate range to utilize other quaternary amines. If true, demethylation of different quaternary amines could be important in environments inhabited by organisms harboring MttB superfamily members, such as the human gut. Quaternary amines of many types are abundant in certain foods. Metabolism of various quaternary amines by members of the gut microbiota has been implicated as having diverse impacts on human health (8–11).
In this work, it is demonstrated for the first time that an MttB family member is a proline betaine methyltransferase. Proline betaine (N,N-dimethyl l-proline or stachydrine) is a quaternary amine that functions as an osmo- and cryoprotectant in plants and microbes (12, 13). Proline betaine is present at high levels in citrus fruit (14) and is therefore a common component of human diets. N-methyl l-proline (hygric or hygrinic acid), a derivative of proline betaine lacking a methyl group, is also found in citrus fruit, but generally at 10-fold lower concentrations than proline betaine (14). Proline betaine and N-methyl l-proline have long been detected in human serum and urine (15, 16). Proline betaine has received recent interest for use as a biomarker of citrus intake (17, 18), whereas untargeted metabolomic studies also identify proline betaine as a biomarker of adherence to the Mediterranean diet (19) and proline betaine and N-methyl proline for adherence to the DASH diet (20). A number of beneficial health effects have been attributed to proline betaine (9), and urinary proline betaine is inversely correlated with blood pressure in adherents to healthy diets (10). In contrast, both proline betaine and N-methyl proline were negatively correlated with physical performance in a serum metabolomic profile of functionally limited older adults (11). Although many metabolomic studies do not specifically consider the effects of gut microbiota, a comparison of the colonic luminal metabolome of germ-free versus ex-germ-free mice found that N-methyl proline was only detected when gut microbiota were present (21). A possible source of microbially produced N-methyl proline would be the demethylation of proline betaine. However, to our knowledge, no pure (or mixed) culture of anaerobe(s) has been shown to demethylate proline betaine and excrete N-methyl proline. The only known microbial enzymatic mechanism of proline betaine demethylation proceeds via Rieske type demethylases, requiring oxygen as an intermediate step to further degradation of the compound (22). Therefore, the mechanism of microbial N-methyl proline production in the anoxic gut has remained an open question.
Here we show that Eubacterium limosum 8486, an acetogenic and butyrogenic human fecal isolate that possesses 42 mttB homologs (23, 24), consumes proline betaine and releases N-methyl proline during growth. MtpB (a non-Pyl MttB family member), together with a corrinoid protein and a corrinoid-dependent THF methyltransferase, were markedly more abundant in cells grown on proline betaine than lactate. MtpB and these same proteins together carried out a proline betaine:THF methyltransferase reaction. MtpB catalyzes the first step of THF methylation by methylating a cognate corrinoid protein with proline betaine. Identification of the proline betaine methyltransferase and the proline betaine:THF methyltransferase system expands the known substrate range of the MttB family and reveals a novel anoxic mechanism for microbial demethylation of proline betaine.
Results
E. limosum demethylates proline betaine during growth
E. limosum strains were previously shown to utilize quaternary amines, including glycine betaine and choline (25). To test whether E. limosum ATCC 8486 can grow on proline betaine, we synthesized l-proline betaine and N-methyl l-proline. Both compounds were verified by MS and TLC (Fig. S1). We found that E. limosum can indeed grow on proline betaine (Fig. 1, A and B). In the absence of proline betaine, little growth was observed in complex medium containing yeast extract and casamino acids. Growth was markedly stimulated in the presence of proline betaine. Yeast extract and casamino acids were not essential, and proline betaine-dependent growth also occurred in a defined medium (Fig. 1B). Cultures in either complex or defined media had similar doubling times of ∼7 h.
Figure 1.
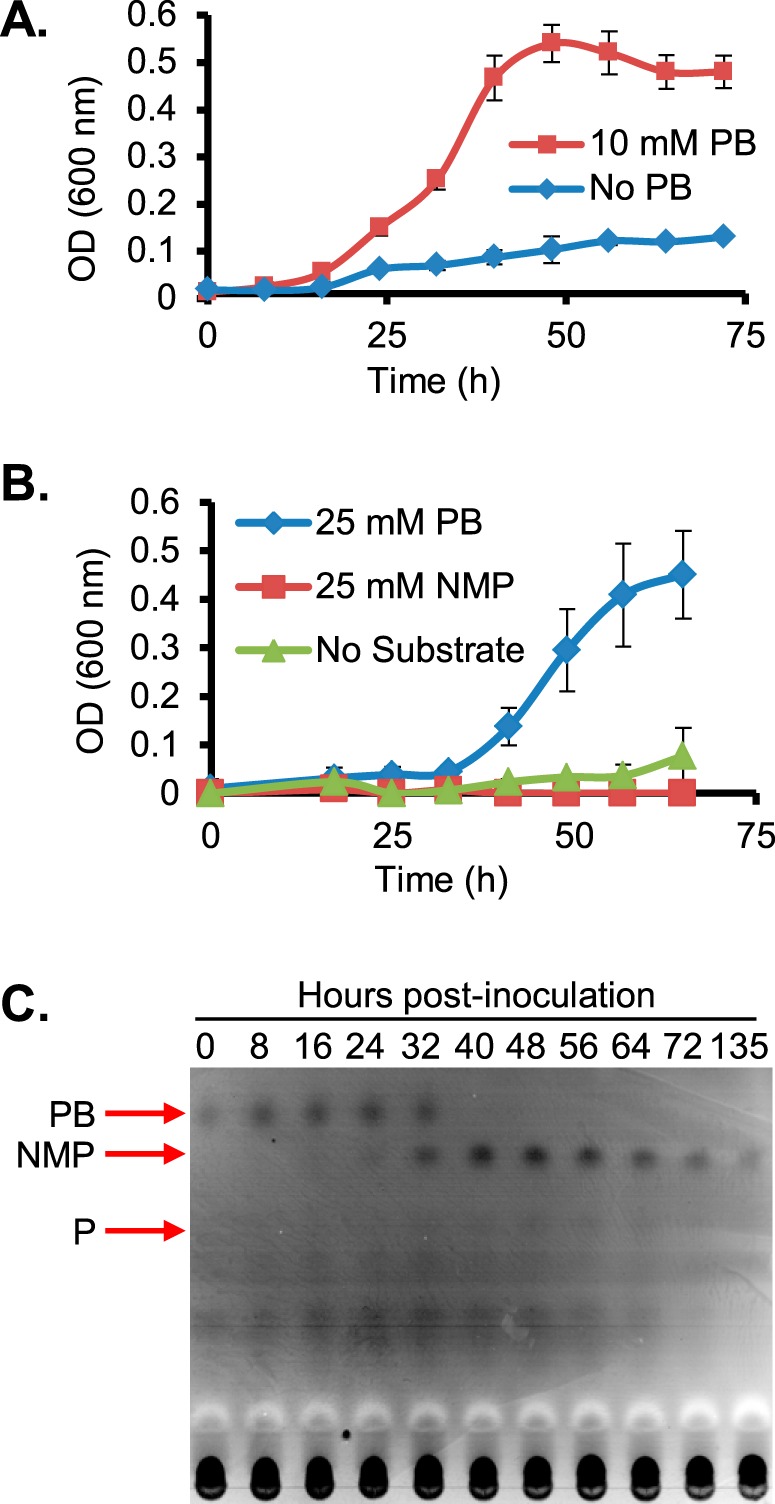
Growth of E. limosum is coupled to l-proline betaine demethylation. A, cells were grown in the presence and absence of 10 mm proline betaine in LS medium supplemented with 0.2% yeast extract and 0.2% casamino acids. B, cells were grown in defined LS medium lacking yeast extract or casamino acids in the presence of 25 mm l-proline betaine, 25 mm N-methyl l-proline, or no substrate. In both A and B, the error bars represent S.D. of the mean OD600 nm readings from three independent cultures. C, TLC of supernatants of E. limosum cultures with proline betaine shown in A demonstrates consumption of proline betaine and release of N-methyl proline during growth. The migration position of a proline standard is also indicated. Red arrows represent migration of chemical standards on the same plate. PB, proline betaine; NMP, N-methyl proline; P, proline.
In order to determine whether proline betaine is demethylated by E. limosum, we analyzed supernatants taken from the same cultures depicted in Fig. 1A. Supernatant samples were subjected to TLC followed by staining with bromocresol green. These experiments confirmed that proline betaine is consumed during growth (Fig. 1C). A spot co-migrating with authentic N-methyl proline began to accumulate during log phase and persisted in the supernatant into stationary phase (Fig. 1C). The area with an Rf corresponding to the putative N-methyl proline spot was extracted from an unstained lane loaded with supernatant from log phase. The extract was then subjected to MS, which confirmed the presence of an ion whose measured m/z corresponded to the theoretical m/z of N-methyl proline with an error of 3.8 ppm (Fig. S2). No compound with an Rf corresponding to authentic proline standard was observed during the experiment. Furthermore, we did not observe that N-methyl proline was used as a substrate for growth by E. limosum (Fig. 1B), suggesting that the organism may not be capable of completely demethylating proline betaine.
Proteomic analysis of E. limosum grown on proline betaine reveals a putative proline betaine:THF multicomponent methyltransferase system
To identify putative methyltransferases involved in proline betaine demethylation, the proteomes of E. limosum cells growing either on proline betaine or lactate were compared. We reasoned that growth on various substrates requires many of the same core enzymes of acetogenesis and butyrogenesis, but synthesis of proteins required for the assimilation of proline betaine would likely be regulated by the presence of the substrate. Such is the case for genes encoding components of a catabolic lactate dehydrogenase that are strongly up-regulated during growth on that substrate in Acetobacterium woodii (26). Two sets of independent cultures (n = 4) were grown to mid-log phase in defined medium containing either lactate or proline betaine. Cell pellets were lysed and subjected to trypsin digestion in preparation for subsequent detection of peptides by LC-MS/MS. Peptides were mapped against the proteins predicted to be encoded by the completely sequenced E. limosum ATCC 8486 genome (24) and quantified using exponentially modified protein abundance index (emPAI) scores, which allowed calculation of the molar percent abundance of each protein (27). Relative abundances of different proteins in cells grown on lactate or proline betaine were then compared. The significance of differences in relative abundance for a given protein between the two growth conditions was evaluated by Student's t test.
The E. limosum ATTC 8486 genome sequence curated at the National Center for Biotechnology (https://www.ncbi.nlm.nih.gov) encodes 4023 annotated proteins. We were able to identify 1727 proteins between the two data sets we obtained from proteomic analysis of E. limosum cells grown on either proline betaine or lactate, corresponding to 43% of the proteins annotated in the genome. Among these, many enzymes of the Wood–Ljungdahl pathway were abundant (Table 1) in cells grown on either lactate or proline betaine, although approximately half were significantly elevated (p < 0.05) in cells grown on proline betaine. As expected, components of the lactate dehydrogenase system were most abundant in lactate-grown cells and much less abundant, or not detectable, in proline betaine–grown cells (Table 2). Conversely, proteins homologous to those involved in multicomponent THF methyltransferase systems were very abundant in cells grown on proline betaine but less abundant or not detectable in cells grown on lactate. The identified methyltransferase components included a non-Pyl MttB family member (WP_38353400, designated MtpB), a homolog of a corrinoid-binding protein (WP_03832545, designated MtqC), and a homolog of a corrinoid:THF methyltransferase (WP_038351870, designated MtqA), which were 3.8 ± 0.67, 0.22 ± 0.13, and 1.5 ± 0.97% of the total detected protein, respectively (Table 3). MtqC and MtqA were 28- and 15-fold more abundant in cells grown on proline betaine relative those grown with lactate. MtpB was not detectable in cells grown on lactate, despite its high abundance in proline betaine–grown cells. A homolog of a corrinoid activation enzyme (WP_038351871, designated RamQ) was also detected in cells grown on proline betaine, although at much lower abundance than MtpB, MtqC, and MtqA. Homologs of such ATP-dependent activation proteins are found in both archaea (28) and bacteria (29, 30) and reduce inactive Co(II)-corrinoid proteins to the active Co(I) state.
Table 1.
Core proteins of the Wood–Ljungdahl pathway were detected in both growth conditions
| Name | Accession number | Possible function | Percentage of total detected protein in lactate-grown cells | Percentage of total detected protein in proline betaine–grown cells | -Fold change (proline betaine/lactate) | p value |
|---|---|---|---|---|---|---|
| mol % | mol % | -fold | ||||
| FdhA | WP_038354071.1 | Formate dehydrogenase subunit α | 0.028 ± 0.0077 | 0.050 ± 0.0052 | 1.8 | 0.0028 |
| Fhs1 | WP_038351869.1 | Formate-THF ligase | 0.16 ± 0.030 | 0.43 ± 0.085 | 2.8 | 0.00084 |
| FchA | WP_038351868.1 | Methenyl-THF cyclohydrolase | 0.022 ± 0.0064 | 0.028 ± 0.012 | 1.2 | 0.44 |
| FolD | WP_038351867.1 | Methylene-THF dehydrogenase | 0.12 ± 0.015 | 0.59 ± 0.40 | 4.9 | 0.058 |
| MetV | WP_038351866.1 | 5,10-Methylene-THF reductase | 1.4 ± 0.92 | 3.4 ± 0.50 | 2.4 | 0.0098 |
| MetF | WP_081571099.1 | 5,10-Methylene-THF reductase | 0.082 ± 0.0085 | 0.082 ± 0.021 | 1.0 | 0.95 |
| AcsE | WP_013381869.1 | Carbon monoxide dehydrogenase | 6.5 ± 1.7 | 3.9 ± 0.69 | 0.60 | 0.028 |
| AscC | WP_038352891.1 | Acetyl-CoA synthase subunit γ | 0.77 ± 0.39 | 1.6 ± 0.82 | 2.1 | 0.10 |
| AcsD | WP_038352892.1 | Acetyl-CoA synthase subunit δ | 2.4 ± 0.42 | 3.9 ± 0.70 | 1.6 | 0.012 |
| AcsA | WP_038352890.1 | Carbon-monoxide dehydrogenase catalytic subunit | 0.14 ± 0.012 | 1.0 ± 0.80 | 7.0 | 0.074 |
| AcsB | WP_038352888.1 | Bifunctional acetyl-CoA decarbonylase/synthase complex subunit α/β | 0.051 ± 0.013 | 0.43 ± 0.65 | 8.4 | 0.29 |
Table 2.
Proteins specifically involved in lactate metabolism are significantly more abundant during growth on lactate relative to growth on proline betaine
| Name | Accession number | Function | Percentage of total detected protein in proline betaine–grown cells | Percentage of total detected protein in lactate-grown cells | -Fold change (lactate/proline betaine) | p value |
|---|---|---|---|---|---|---|
| mol % | mol % | -fold | ||||
| LctA | WP_052237246.1 | Transcriptional regulator | 0.0013 ± 0.00062 | 0.0038 ± 0.0017 | 2.9 | 0.036 |
| LctB | WP_038352354.1 | Electron transfer flavoprotein subunit β | 0.00013 ± 0.00026 | 2.1 ± 0.96 | 16,000 | 0.0053 |
| LctC | WP_038352355.1 | Electron transfer flavoprotein subunit α | NDa | 2.4 ± 0.86 | 0.0013 | |
| LctD | WP_013380257.1 | Lactate dehydrogenase | ND | 1.9 ± 0.566 | 0.00061 | |
| LctE | WP_038352356.1 | Lactate permease | ND | 0.0029 ± 0.00082 | 0.00041 | |
| LctF | WP_038352908.1 | Lactate racemase | 0.0039 ± 0.00086 | 0.23 ± 0.11 | 59 | 0.0057 |
| PorA | WP_038354167.1 | Pyruvate:ferredoxin oxidoreductase | 0.16 ± 0.03 | 2.3 ± 1.2 | 14 | 0.01 |
a ND, not detected.
Table 3.
A non-Pyl MttB homolog and other components of a corrinoid-dependent methyltransferase system are significantly more abundant during growth on proline betaine relative to growth on lactate
| Name | Accession number | Function | Percentage of total detected protein in lactate-grown cells | Percentage of total detected protein in proline betaine–grown cells | -Fold change (proline betaine/lactate) | p value |
|---|---|---|---|---|---|---|
| mol % | mol % | -fold | ||||
| MtpB | WP_038353400.1 | Proline betaine:corrinoid methyltransferase | NDa | 3.8 ± 0.67 | <0.0001 | |
| MtqC | WP_038352545.1 | Cobalamin-binding protein | 0.0079 ± 0.0030 | 0.22 ± 0.13 | 28 | 0.017 |
| MtqA | WP_038351870.1 | Methyl-THF:corrinoid methyltransferase | 0.099 ± 0.033 | 1.5 ± 0.97 | 15 | 0.031 |
| RamQ | WP_038351871.1 | Corrinoid-activating enzyme | 0.0091 ± 0.0032 | 0.021 ± 0.0026 | 2.4 | 0.001 |
a ND, Not detected.
Genome context of the genes encoding MtpB, MtqC, and MtqA
The components of the putative proline betaine:THF methyltransferase systems are encoded in three separate parts of the chromosome (Fig. S3). MttB family member MtpB is encoded adjacent to a gene encoding a member of the betaine-choline-carnitine transporter (BCCT) family (Fig. S3A), whose documented substrates include glycine betaine, choline, γ-butyrobetaine, carnitine, and proline betaine (31). Also adjacent to the mtpB gene were genes encoding another non-Pyl MttB superfamily member and corrinoid protein homolog. MtqC is encoded by a gene adjacent to genes predicted to encode another corrinoid protein and transcriptional regulator (Fig. S3B). The proteins encoded adjacent to both mtqC and mtpB were detectable in the proteome of proline betaine–grown E. limosum, but at <0.005% molar abundance. It should be noted that the protocol employed for cellular digestion is primarily targeted at soluble proteins and generally underrepresents membrane-bound proteins in the proteome, such as a BCCT transporter or another type of proline betaine transporter. Finally, the genes encoding MtqA and RamQ were found next to genes encoding enzymes predicted to participate in the oxidation of methyl-THF (Fig. S3C; see also Table 1).
MtpB is a proline betaine:corrinoid methyltransferase
MtpB is encoded adjacent to a quaternary amine transporter homolog and is highly abundant in proline betaine–grown cells compared with lactate-grown cells. Additionally, as MtpB is a non-pyrrolysine MttB homolog, we hypothesized that this protein might act as a proline betaine:corrinoid methyltransferase. Therefore, the mtpB gene from E. limosum was cloned, and recombinant MtpB was isolated from E. coli (Fig. S4). As the glycine betaine methyltransferase MtgB can methylate free cob(I)alamin (5), we first tested MtpB for proline betaine:cob(I)alamin methyltransferase activity. However, MtpB methylation of cob(I)alamin as a substrate was undetectable. We next tested MtqC, the methylotrophic corrinoid protein most abundant in proline betaine–grown cells, as a substrate. Recombinant MtqC was isolated as an apoprotein from E. coli and reconstituted with hydroxocobalamin (Fig. S4). The recombinant MtqC holoprotein had a UV-visible absorbance spectrum characteristic of the catalytically inactive Co(II) state, as indicated by the peak at 475 nm (Fig. 2A). Although Ti(III)citrate is able to reduce free cobalamin, incubation of Co(II)-MtqC with MgATP and Ti(III)citrate resulted in little or no reduction to the Co(I) state (Fig. 2B). Reduction was greatly stimulated upon the addition of recombinant RamQ (Fig. 2B; see Fig. S4 for SDS-PAGE of RamQ), as observed by the disappearance of the 475-nm peak and appearance of the 386-nm peak indicative of the Co(I) corrinoid cofactor (Fig. 2, A and B). The addition of proline betaine to Co(I)-MtqC did not result in a change in the spectrum. However, upon the addition of MtpB, Co(I)-MtqC was methylated as demonstrated by disappearance of the 386-nm peak corresponding to Co(I)-MtqC and the appearance of a peak at 534 nm, corresponding to the formation of CH3-Co(III) MtqC (Fig. 3A). Sharp isosbestic points at 439 and 576 nm indicate that the conversion of Co(I)-MtqC to methyl-Co(III)-MtqC occurred without sizable appreciation of another corrinoid form, such as Co(II) MtqC, that might compromise rate determinations. Empirically determined Δϵ values allowed calculation of initial rates of Co(I)-MtqC disappearance and MtqC methylation (Fig. 3B). A series of MtqC methylation assays was performed to measure rate dependence on the concentration of proline betaine. Nonlinear regression was used to fit the Michaelis–Menten equation to the resulting data, yielding an apparent Km of 8 ± 2 mm proline betaine and an apparent Vmax of 3.1 μmol min−1 mg−1 MtpB, corresponding to a kcat of 170 ± 12 min−1 (Fig. 4). Other quaternary amines were evaluated as potential substrates for MtpB. No MtpB-dependent methylation of MtqC was observed with 100 mm choline. However, some quaternary amines were able to support limited MtpB-dependent methylation of MtqC. Initial rates of MtqC methylation (in μmol min−1 mg−1 MtpB) with 100 mm carnitine, phosphocholine, glycine betaine, or betonicine (trans-4-hydroxyproline betaine) were 0.00084, 0.032, 0.13, and 0.50, respectively (average of two reactions each). In summary, these results clearly show that MtpB can catalyze the methylation of MtqC, with proline betaine being the most active substrate tested.
Figure 2.
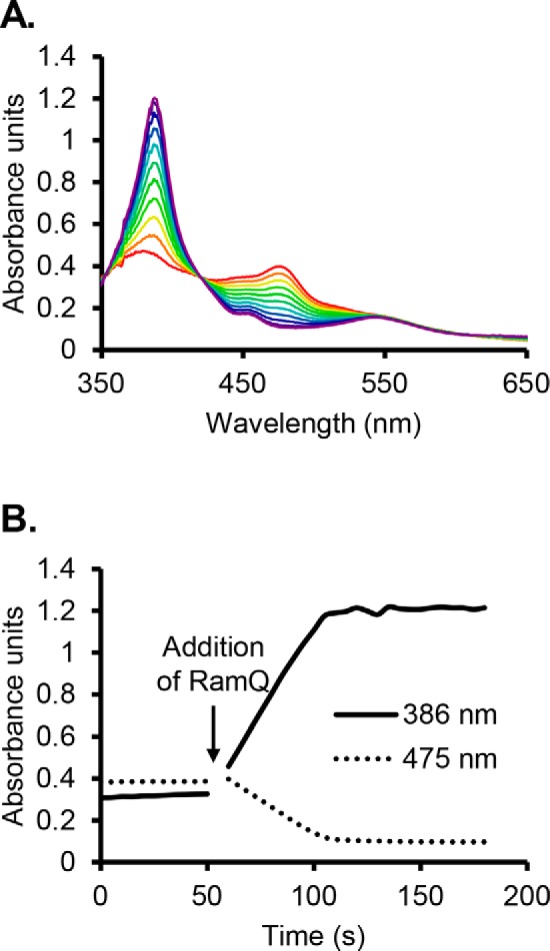
RamQ stimulates reduction of reconstituted MtqC from the Co(II) state to the Co(I) state in vitro. Reduction is observed with UV-visible spectroscopy. The Co(II) form of MtqC has a characteristic absorbance peak at 475 nm, whereas the Co(I) form has a signature absorbance peak at 386 nm. A, absorbance spectra were taken upon the addition of RamQ (red) and subsequently in 5-s intervals until equilibrium was reached at 110 s (purple). B, the addition of MtqC to reaction mix containing Ti(III) citrate and MgATP results in slow reduction of MtqC. MtqC reduction was dramatically enhanced by the addition of RamQ.
Figure 3.
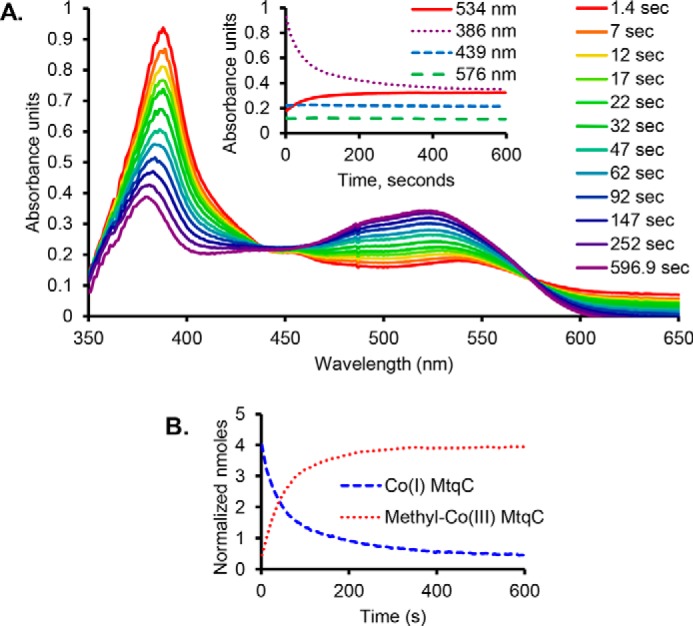
MtpB methylates Co(I) MtqC in vitro with proline betaine. The course of the reaction is observed with UV-visible spectroscopy. The Co(I) form of MtqC has an absorbance peak at 386 nm, and the methyl-Co(III) form of MtqC has an absorbance peak at 534 nm. A, Co(I) MtqC is methylated upon the addition of MtpB to a reaction containing previously reduced MtqC and proline betaine. The inset shows reaction progress. B, the signature peaks for Co(I) MtqC and CH3-Co(III) MtqC were used to calculate the loss of the superreduced form of MtqC and the formation of the methylated form. Empirically determined Δϵ from the Co(I) state to the CH3-Co(III) state was used for this plot, which is normalized to set total MtqC in both states to 4.5 nmol, the amount initially added to the reaction.
Figure 4.
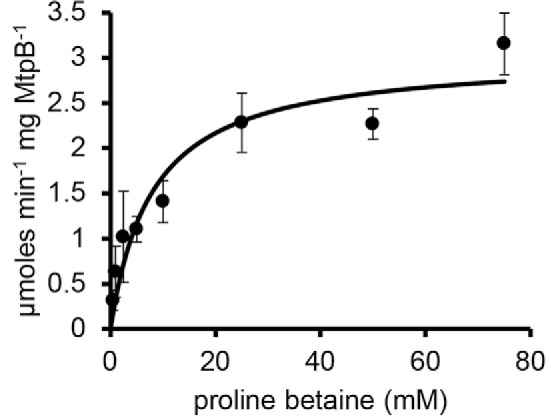
Michaelis–Menten kinetic analysis of MtpB with increasing proline betaine. Each point is the mean initial rate of three reactions at a given concentration. Error bars, S.D.
The glycine betaine methyltransferase has only trace activity with proline betaine
Several transporters have been noted to utilize either glycine betaine or proline betaine as substrates, so we considered it possible that the previously described glycine betaine methyltransferase MtgB (5) might use both betaines as substrates. We tested the ability of a range of proline betaine concentrations to support cob(I)alamin methylation by MtgB from D. hafniense. At 10, 50, or 100 mm proline betaine, cob(I)alamin methylation was undetectable (<0.7 nmol min−1 mg−1), and only trace activity (∼2 nmol min−1 mg−1) was detectable when the concentration was raised to 250 mm proline betaine. In contrast, our preparations of MtgB catalyze cob(I)alamin methylation with glycine betaine with an apparent Vmax of 0.7 μmol min−1 mg−1 and an apparent Km of 1.2 mm.
In vitro reconstitution of the proline betaine:THF methyltransferase reaction
The methylation of THF forms a key intermediate in the Wood–Ljungdahl pathway, and catabolism of methylated substrates proceeds via CH3-THF to support the growth of acetogens like E. limosum (32, 33). MtqA's homology to known corrinoid:THF methyltransferases, such as the THF methyltransferase domain of cobalamin-dependent methionine synthase (34), as well as the abundance of MtqA during growth of E. limosum on proline betaine and the genomic context of the mtqA gene led us to hypothesize that MtqA might enable the transfer of methyl groups from proline betaine to THF in the presence of MtpB and MtqC. Therefore, recombinant MtqA was produced from E. coli (Fig. S4).
Reaction mixtures containing MtpB, MtqC, and MtqA along with THF were preincubated in the presence of Ti(III)citrate, MgATP, and RamQ to allow activation of MtqC to the Co(I) state. Reactions were initiated by the addition of proline betaine. Methyl-THF production was monitored by reverse-phase HPLC and quantified using a standard curve prepared with CH3-THF. Equimolar amounts of MtpB and MtqA, in the presence of a 10-fold molar ratio of MtqC, methylated THF at a rate of 1.49 ± 0.045 μmol of CH3-THF min−1 mg−1 total protein in the assay, or 206.8 ± 6.3 mol of CH3-THF min−1 mol−1 MtqA (Fig. 5). No CH3-THF formation was detectable in control reactions lacking MtpB, MtqC, MtqA, or proline betaine (Fig. 5). Significantly, MtpB methylates MtqC, but the two together do not methylate THF except upon the addition of MtqA. This strongly indicates that MtqA demethylates MtqC to methylate THF during the proline betaine:THF methyltransferase reaction.
Figure 5.
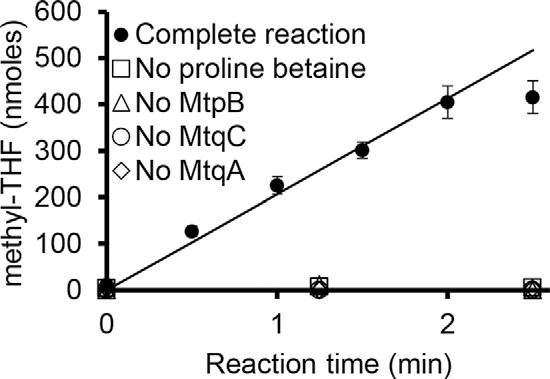
The addition of MtqA to MtpB and MtqC allows methylation of THF with proline betaine. Methyl-THF formation was monitored by HPLC analysis of time points taken from three independent reactions containing MtqC, RamQ, MtpB, MtqA, Ti(III)citrate, MgATP, and THF (complete) in 200 μl. Controls were performed in duplicate, as indicated, lacking either MtpB, MtqC, MtqA, or proline betaine. Error bars, S.D. Least-squares regression was performed on the linear portion of the reaction, from 0 to 2 min.
Discussion
Here, we demonstrate that E. limosum, an obligate anaerobe isolated from human feces, grows on proline betaine and releases N-methyl proline during growth. We further describe the functional diversification of the MttB superfamily to include a proline betaine:corrinoid methyltransferase. The MttB family member MtpB initiates the methylation of THF via interaction with the corrinoid protein MtqC and MtqA to form a multicomponent methyltransferase system with the novel function of methylation of THF with proline betaine. This system allows entry of a single methyl group of proline betaine into the central acetogenic metabolism with the subsequent excretion of N-methyl proline by the organism (Fig. 6). To our knowledge, this is the first time an anoxic mechanism of proline betaine demethylation has been described.
Figure 6.
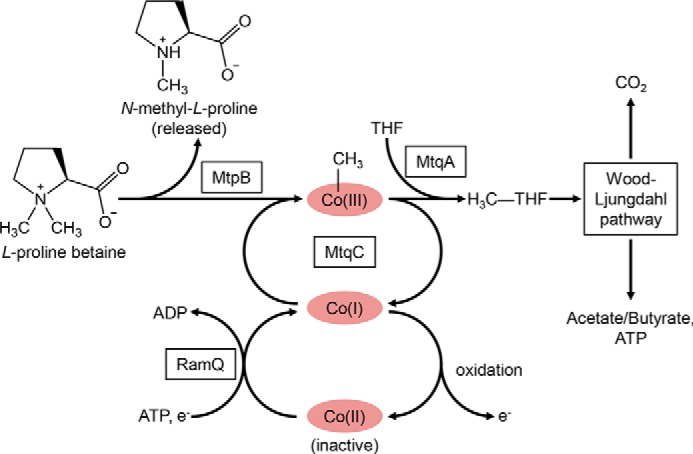
Model for proline betaine metabolism in E. limosum. The methyl group from proline betaine is transferred to Co(I) MtqC by MtpB, forming methyl-Co(III) MtqC. N-Methyl proline is produced for release into the culture supernatant, whereas the methyl group is subsequently transferred to THF by MtqA. The resulting methyl-THF enters the Wood–Ljungdahl pathway, in which a portion is oxidized to CO2 and the remainder is converted to cell carbon or acetate/butyrate. Co(I) MtqC can be adventitiously oxidized to catalytically inactive Co(II), which is reduced to active Co(I) by the ATP and reducing power–dependent activity of RamQ.
Bacteria, such as Sinorhizobium meliloti, have long been known to transport proline betaine, either for use as a compatible solute under conditions of high osmolarity or as a source of nitrogen, carbon, and energy under conditions of low osmotic stress (35). Genes for proline betaine degradation were first identified on the sym plasmid responsible for catabolism of several quaternary amines (36). The gene responsible for the initial demethylation of proline betaine was found to encode a Rieske iron-sulfur oxygenase-like protein (37). The proline betaine oxygenase activity and other predicted properties of the metalloprotein product were subsequently confirmed (38). A second oxidative demethylation follows, leading to the formation of proline with further degradation via glutamate (22). For several reasons, it seems unlikely that organisms harboring this pathway would be responsible for the observed production of N-methyl proline by members of the gut microbiota. Genes that may encode the oxygenase-requiring pathway have been found in the genomes of at least 100 organisms (22). Some of these are opportunistic human pathogens, but none are typical symbionts found in the gastrointestinal tract. Whereas oxygen concentrations are highly limiting in the intestine (39), it is possible that sufficient oxygen is found in some parts of the gastrointestinal tract to allow oxidative demethylation of proline betaine. However, in this circumstance, it would be expected that the second oxygenase in the pathway would demethylate N-methyl proline, followed by subsequent catabolism of the liberated proline (22).
In contrast, the three-component proline betaine:THF methyltransferase system, composed of MtpB, MtqC, and MtqA, does not require oxygen to function. Release of abundant N-methyl proline by gut microbes that possess this type of enzyme system may provide a ready explanation for the anoxic production of N-methyl proline in the human intestine. Acetogens gain relatively little energy from methylotrophic catabolism (26), further ensuring a relatively high specific flux of proline betaine through the demethylation system. Excretion of a singly demethylated product, such as N-methyl proline, is not unusual for acetogens, and E. limosum was previously shown to demethylate glycine betaine and choline, excreting N,N-dimethylglycine and N,N-dimethylethanolamine, respectively (25). The excretion of singly demethylated products of quaternary amines raises the issue of how the substrate and product are moved across the membrane, given the low amount of energy conserved during acetogenesis. In this regard, it is tempting to speculate that a transporter, such as a BCCT family member, could carry out antiport of the quaternary amine substrate and demethylated product, in a manner analogous to BCCT family member CaiT, which simultaneously imports carnitine while exporting the reduction product, γ-butyrobetaine (31). However, given that many BCCT family members and other transporters carry out ion gradient driven symport of substrates, substrate transport (and product export) remains an important topic for future work on quaternary amine utilization by acetogens.
E. limosum is not likely to be the only Eubacterium species to utilize a close MtpB homolog to demethylate proline betaine. BLAST searches of microbial genomes curated at the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov) revealed close homologs (>93% identity) of MtpB encoded in the genomes of Eubacterium callanderi and Eubacterium maltosivorans, the latter of which originates from human feces (40). Acetogens may also not be the only type of organisms in the intestinal microbiota to undertake limited demethylation of proline betaine, as several types of anaerobic microbial catabolism are known to utilize similar corrinoid-dependent methyltransferase systems, such as respiration of nitrites or sulfites (5) and methanogenesis (4). These may utilize more distant homologs of MtpB.
Structural and genomic context can provide important clues to the activities of bacterial gene products of uncertain function (41). Here we show that, for certain microbes, protein abundance gleaned from label-free quantitative proteomics can also provide an important clue leading to the demonstration of novel catabolic functions within protein families. This is especially true of anaerobes, such as acetogens or methanogens. These organisms gain relatively little energy from their respective pathways (42, 43) and as a result use relatively large amounts of the growth substrate for catabolism, often using enzymes with low specific activities. The E. limosum proteomes we obtained illustrate the utility of a proteomic approach to identifying candidate enzymes in an unresolved catabolic pathway. Lactate– or proline betaine–grown cells contained the constituents of lactate dehydrogenase or the proline betaine:THF methyltransferase system, respectively, at ∼6–7% molar abundance; but each system was markedly less abundant in cells grown on the other substrate. It should be noted that E. limosum lactate dehydrogenase is highly similar to the biochemically characterized A. woodii lactate dehydrogenase (44), and the up-regulation of the encoding genes on lactate versus other substrates had been previously documented in A. woodii (26), which further validates the utility of a proteomic approach in identifying catabolic enzymes.
Approximately half of the identified enzymes belonging to the Wood–Ljungdahl pathway were significantly elevated (p < 0.05) in proline betaine–grown cells compared with those grown on lactate, whereas several others also appeared elevated but missed the p value significance cutoff (Table 1). An increased reliance on the Wood–Ljungdahl pathway of acetogenesis when E. limosum is grown on proline betaine relative to lactate would be reasonable, based on the probable pathways for acetogenesis from these two substrates. The Wood–Ljungdahl pathway comprises two distinct metabolic branches that together operate to produce acetyl-CoA from two one-carbon units (42, 45). During autotrophic growth, the enzymes composing the methyl-branch reduce CO2 to methyl-THF, whereas the carbonyl-branch reduces CO2 to CO. The two single-carbon units are then combined with CoA to produce acetyl-CoA via acetyl-CoA synthase. During methylotrophic acetogenesis, the same enzymes composing the methyl branch operate in the reverse direction, as methyl-THF is generated directly from the substrate (46). Methyl-THF must then be oxidized to generate the reducing power to make acetyl-CoA from methyl-THF and CO2. In contrast, during acetogenic lactate metabolism, two-thirds of the acetyl-CoA made derives from the operation of the catabolic lactate dehydrogenase and pyruvate:ferredoxin oxidoreductase, and only one-third of the acetyl-CoA derives from the Wood–Ljungdahl pathway (44). Therefore, carbon flux through the Wood–Ljungdahl pathway is much higher for cells grown on proline betaine, as production of each acetyl-CoA that is precursor to both catabolic end products and cellular material requires reducing power and carbon from the proline betaine methyl group.
MtpB joins MtgB, the glycine betaine methyltransferase, as a member of the MttB superfamily with a demonstrated ability to demethylate a quaternary amine, providing further support for the hypothesis that the great majority of this protein family, which lacks pyrrolysine, has specificity for N-demethylation of quaternary amines (5). Proline betaine and glycine betaine share some structural similarity, differing only by the two methylene groups joining the α-carbon and the quaternary amine of proline betaine. Some membrane receptors and transporters function effectively with both compounds (47, 48). Therefore, it could be said that that acquisition of proline betaine as a substrate by the MttB superfamily is not surprising. In this regard, MtpB has clearly undergone evolution toward specificity for the proline betaine substrate, as other quaternary amines, including glycine betaine, are much poorer substrates for the methylation of MtqC. Further work will be required to determine whether the apparent Km of 8 mm we observed for MtpB methylation of MtqC with proline betaine reflects an adaptation to the catabolic use of this commonly used cellular osmolyte or if this is an artifact of assaying a subreaction of the overall proline betaine:THF methyltransferase reaction catalyzed in concert with MtqA. In any case, it is becoming clear that the MttB superfamily has members that have evolved to utilize different substrates. As another example, we have recently described a non-Pyl MttB homolog with the ability to demethylate l-carnitine, further illustrating the specificity of multiple MttB superfamily members for distinct quaternary amines. Interestingly, in the proteome of carnitine-grown cells, MtpB was 2000-fold less abundant than in cells grown on proline betaine.3
Recent discoveries have made it clear that quaternary amine metabolism by the human gut microbiota can be useful in understanding the origins of different components of the human metabolome (19, 21) and, most importantly, has a significant impact on human health (8). This has led to emphasis on understanding how different members of the microbiota contribute to metabolism of quaternary amines and their derivatives. Such efforts often involve understanding the metabolic potential of the intestinal community via metagenomic sequencing. Mis-annotation of protein function is a fundamental issue inherent to computational annotation of sequence-based databases (49). Such has been the case with members of the MttB superfamily, whose functional diversification to different recognized quaternary amines is only now becoming clear. Given the importance of microbiotal quaternary amine metabolism to human health, functional characterization of members of the MttB superfamily and how different members recognize various quaternary amine substrates will be an important avenue for future experimentation.
Experimental procedures
Growth of E. limosum
E. limosum ATCC 8486 was obtained from the American Type Culture Collection and was cultured anaerobically at 37 °C. The LS medium used in growth experiments contained 1 g/liter NH4Cl, 0.1 g/liter MgSO4·7H2O, 0.05 g/liter CaCl2·2H2O, 2.5 g/liter NaCl, 0.1 mg/liter biotin, 0.2 mg/liter folic acid, 0.3 mg/liter pyridoxine-HCl, 0.2 mg/liter thiamine-HCl, 0.1 mg/liter riboflavin, 0.2 mg/liter nicotinic acid, 0.2 mg/liter d-pantothenate hemicalcium salt, 0.1 mg/liter hydroxocobalamin-HCl, 0.1 mg/liter p-aminobenzoic acid, and 0.1 mg/liter lipoic acid. Pfennig and Lippert's trace elements solution (50) was added at 3 ml/liter, and cultures were made anaerobic by flush/evacuation with 100% N2. 0.35 g/liter K2HPO4, 0.28 g/liter KH2PO4, 1.8 g/liter NaHCO3, 0.48 g/liter Na2S·9H2O, and 0.47 g/liter cysteine-HCl were added after autoclaving from sterile anoxic stock solutions. The medium was supplemented with yeast extract (3 g/liter) and casamino acids (3 g/liter) where indicated.
Synthesis of l-proline betaine and N-methyl l-proline
The synthesis of l-proline betaine (stachydrine hydrochloride) was carried out by following the description of Cornforth and Henry (51). We add a few notes that lead to an increased yield and some further characterization. The yield is increased by re-extraction of the AgI with methanol and by treating the mother liquors with acetone. The total yield of crystalline material, mp 222–223 °C (lit. 222 °C), is 37–41%. It is worth mentioning that Cornforth and Henry do not mention the work of Karrer and Widmer (52), who used the same method to prepare stachydrine from N-methyl proline. IR(Nujol): 1783, 1704, 1481, 1417, 1403, 1251, 1233, 1190, 1039, 1026, 997, 952, 912, 801, and 728 cm−1. 1H NMR(D2O), 700 MHz: δ 2.17 (m, 2H, CH2), 2.28 (m, 1H, CH2), 2.48 (s, 1H, CH2), 3.10 (s, 3H, CH3-N), 3.29 (s, 3H, CH3-N), 3.54 (q, 1H, CH2-N), 3.71 (m, 1H, CH2-N), 4.08 (t, 1H, N-CH-COO). N-methyl-l-proline (hygric or hygrinic acid) was made by following the procedure of Lin et al. (53) (see also Aurelio et al. (54)). We modified the procedure by using only one-tenth the amount of PdC. We obtained crystals from ethanol, whose melting point and IR spectrum agreed with those reported. As with stachydrine, the yield is increased by treating the mother liquors with acetone. We obtained satisfactory exact masses for both compounds (Fig. S1).
Analysis of proline betaine and N-methyl proline in culture supernatants
TLC was performed as described previously (5). Standards included the above synthesized l-proline betaine and N-methyl l-proline and commercially available proline (Thermo Fisher Scientific). Cell-free supernatants (10 μl in total) were spotted onto Silica Gel 60 plates (Merck-Millipore) in 2.5-μl aliquots, which were dried before the addition of the next aliquot. Amines were detected with bromocresol green staining. For accurate mass analysis, unstained TLC spots were cut from the plate using stained lanes from the same plate as a reference. Compounds were extracted from cut segments and analyzed using a Bruker MaXis ESI Q-Tof instrument (Bruker, Billerica, MA). Full details are found in the supporting Methods.
Proteomic analysis of E. limosum
Two sets of four replicate cultures (10 ml each) were grown on defined LS medium; one set was grown with 50 mm l-proline betaine, and the second set was grown on 50 mm dl-lactate. Each culture was anaerobically harvested in mid-log phase (OD600 nm ≈ 0.45). Proteins were extracted and precipitated from cell lysate with TCA and subjected to trypsin digestion. Prior to MS/MS, samples were subjected to two-dimensional LC separation. MS/MS data were acquired with an Orbitrap Fusion mass spectrometer. Full details of sample preparation, LC, and MS/MS can be found in the supporting Methods.
Processed sequence information from the MS/MS data were searched using Mascot Daemon by Matrix Science version 2.5.1 (Boston, MA) and searched against databases of the protein sets encoded in genomes of E. limosum ATCC 8486 (24) and E. limosum KIST612 (55) maintained at the National Center for Biotechnology Information. Label-free quantitation was performed using the spectral count approach. Scaffold (Proteomic Software, Inc., Portland, OR) was used for data analysis and calculation of emPAI values to estimate the mol % of each identified protein within the total set of identified proteins (27). Student's t test was performed using Scaffold to evaluate whether the difference for certain proteins between growth conditions is significant (p < 0.05). Full details of data processing are found in the supporting Methods.
Cloning, expression, and purification of recombinant proteins
All primer pairs used for gene amplification and cloning are listed in Table S1. The genes encoding MtpB, MtqC, MtqA, and RamQ were amplified from E. limosum genomic DNA and inserted into pSpeed using the polymerase incomplete primer extension (PIPE) method (56). The pSpeed-based mtqC expression vector was modified by the removal of the N-terminal His6 tag and the insertion of a C-terminal Strep II tag using the PIPE method.
MtpB, MtqC, and MtqA were produced in E. coli BL21 (DE3). RamQ was produced anaerobically in E. coli SG13009. Recombinant His6-tagged MtpB, MtqA, and RamQ were purified using HisTrap FF crude columns (GE Healthcare). For MtpB and RamQ, lysis and purification steps were performed in an anoxic chamber (Coy Laboratories, Inc., Grass Lake, MI). MtpB was subject to further purification using two Superose 12 10/300 GL columns (GE Life Sciences) connected in tandem. N-terminally Strep-II–tagged MtqC apoprotein was purified with a Strep-Tactin XT high-capacity column (5-ml total volume, IBA Lifesciences, Gottingen, Germany) and reconstituted with cobalamin using a protocol adapted from Schilhabel et al. (30). Unincorporated cobalamin and other impurities were removed from reconstituted MtqC holoprotein using Superose 12 (GE Healthcare). MtqC, MtqA, and RamQ were stored in anaerobic vials at −80 °C until use. MtpB lost activity when stored at −80 °C, even in 10 or 50% glycerol as cryoprotectant. Activity was maximally retained for at least 2 weeks when stored in an anaerobic vial on ice in a 4 °C cold room as described in the supporting Methods. MtgB was recombinantly produced as described previously (5).
Reduction and corrinoid methylation assays
Reactions with MtqC were monitored in stoppered quartz sub-micro cuvettes (Starna Cells, Inc.) with a 1-cm pathlength masked to allow monitoring of either 50- or 100-μl reaction volumes. Reaction mixtures were assembled in an anaerobic chamber (Coy Laboratories, Inc.) with an atmosphere of 2% H2 in N2. Cuvettes were stoppered and removed from the chamber, with subsequent anaerobic additions made by Hamilton syringe. Reactions took place in red light at 37 °C, and UV-visible spectra were monitored using an HP 8453 diode-array biochemical analysis spectrophotometer. MtqC holoprotein (45 μm) was first reduced to the Co(I) state by incubation with RamQ (2.5 μm) in an anaerobic reaction mix containing 2 mm Ti(III) citrate, 2.5 mm MgCl2, 2.5 mm ATP, and substrate in 22 mm potassium phosphate buffer, pH 7.2. Nearly complete reduction to Co(I) was confirmed by the appearance of the characteristic peak at 386 nm, which did not increase with additional RamQ. Methylation was initiated by the addition of recombinant MtpB, and methylation was observed by recording spectra every second. Initial rates were determined for a range of substrate concentrations, and nonlinear regression was performed in JMP Pro 12 (SAS) to determine kinetic parameters. Molar extinction coefficients for the change from Co(I) MtqC to CH3-Co(III) MtqC were empirically determined. Reaction mixtures containing 30 μm MtqC, 1 mm Ti(III) citrate, 2.5 mm MgCl2, 2.5 mm ATP, and 100 mm proline betaine were incubated with 1 μm RamQ until the UV-visible spectra remained unchanged. Under these conditions, the addition of subsequent RamQ did not result in further spectral change. Methylation was initiated by the addition of 1 μm MtpB. Subsequent additions of MtpB did not result in spectral change. Δϵ534 nm (4144 cm·liter·mol−1) and Δϵ386 nm (−16,210 cm·liter·mol−1) were determined by subtracting the absorbance of fully reduced MtqC from the absorbance of fully methylated MtqC from three independent reactions. MtgB-dependent methylation of cob(I)alamin was assayed as described previously (5), except that 0.6–1.2 mg of MtgB were added to the reaction to detect trace activity with the indicated proline betaine concentrations.
Methylation of THF by MtpB, MtqC, and MtqA
Reactions were carried out in dim red light in an anaerobic chamber (Coy) with an atmosphere of 2% H2 in N2. Reaction mixtures containing 5 mm THF (Sigma-Aldrich), 5 μm MtpB, 50 μm MtqC, 5 μm MtqA, 2 μm RamQ, 1 mm MgCl2, 1 mm ATP, and 2 mm Ti(III) citrate in 50 mm potassium phosphate, pH 7.2, were preincubated for 5 min at 37 °C to allow MtqC reduction to the Co(I) state. Reactions were then initiated by the addition of 50 mm proline betaine, and 30-μl aliquots were removed every 30 s and stopped with 6 μl of saturated TCA. The reaction time points were then stored in stoppered, anaerobic HPLC vials in darkness at −80 °C. Thawed aliquots were analyzed via reverse-phase chromatography using a 250 × 4.6-mm Varian Microsorb MV-100 C18 column (Agilent, Santa Clara, CA) on a Dionex UltiMate 3000 HPLC system (Thermo Fisher Scientific). Samples were eluted with 7% (v/v) acetonitrile in 30 mm potassium phosphate buffer, pH 3.0, at 0.5 ml/min. THF (typical elution time 15.5 min) and CH3-THF (typical elution time 20 min) peaks were detected at 272 nm. Peak integration was performed in Chromeleon 6.8 (Dionex), and 5-methyltetrahydrofolic acid was quantified based on a standard curve prepared from commercial CH3-THF (Sigma-Aldrich). The initial rate of THF methylation was determined by least-squares linear regression in JMP Pro 12 (SAS).
Data availability
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD013806 and DOI 10.6019/PXD013806 (for lactate-grown cells) and PXD013962 and DOI 10.6019/PXD013962 (for proline betaine–grown cells).
Author contributions
J. W. P., L. Z., and J. A. K. data curation; J. W. P., E. J. B., L. Z., and J. A. K. formal analysis; J. W. P. validation; J. W. P., L. Z., and J. A. K. investigation; J. W. P. visualization; J. W. P., E. J. B., and L. Z. methodology; J. W. P. and J. A. K. writing-original draft; J. W. P., E. J. B., L. Z., and J. A. K. writing-review and editing; E. J. B. and L. Z. resources; E. J. B. and J. A. K. supervision; L. Z. software; J. A. K. conceptualization; J. A. K. funding acquisition; J. A. K. project administration.
Supplementary Material
Acknowledgments
We express our appreciation to Katherine Huening for development of the protocol for production of recombinant Ram proteins and to Duncan Kountz for developing protocols and clones used in the production of MtqC and MtqA, preparing samples of cells grown on lactate for proteomic analysis, and development of the LS medium. We also thank Yu Cao for assistance in performing mass spectrometric analysis of proline betaine and N-methyl proline and Dr. Ruisheng Jiang for performing 1H NMR on proline betaine synthesized for this study. The Fusion Orbitrap instrument was supported by National Institutes of Health Grant S10 OD018056.
This research was supported by National Institute of Health Grant 1R01DK109345. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supporting Methods and Figs. S1–S4.
The MS proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifiers PXD013806 and PXD013962.
D. J. Kountz, L. Zhang, and J. A. Krzycki, submitted for publication.
- non-Pyl
- non-pyrrolysine
- OD
- optical density
- THF
- tetrahydrofolate
- TLC
- thin layer chromatography
- emPAI
- exponentially modified protein abundance index
- BCCT
- betaine-choline-carnitine transporter
- PIPE
- polymerase incomplete primer extension.
References
- 1. Ferguson D. J. Jr., and Krzycki J. A. (1997) Reconstitution of trimethylamine-dependent coenzyme M methylation with the trimethylamine corrinoid protein and the isozymes of methyltransferase II from Methanosarcina barkeri. J. Bacteriol. 179, 846–852 10.1128/jb.179.3.846-852.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Paul L., Ferguson D. J. Jr., and Krzycki J. A. (2000) The trimethylamine methyltransferase gene and multiple dimethylamine methyltransferase genes of Methanosarcina barkeri contain in-frame and read-through amber codons. J. Bacteriol. 182, 2520–2529 10.1128/JB.182.9.2520-2529.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Soares J. A., Zhang L., Pitsch R. L., Kleinholz N. M., Jones R. B., Wolff J. J., Amster J., Green-Church K. B., and Krzycki J. A. (2005) The residue mass of l-pyrrolysine in three distinct methylamine methyltransferases. J. Biol. Chem. 280, 36962–36969 10.1074/jbc.M506402200 [DOI] [PubMed] [Google Scholar]
- 4. Krzycki J. A. (2004) Function of genetically encoded pyrrolysine in corrinoid-dependent methylamine methyltransferases. Curr. Opin. Chem. Biol. 8, 484–491 10.1016/j.cbpa.2004.08.012 [DOI] [PubMed] [Google Scholar]
- 5. Ticak T., Kountz D. J., Girosky K. E., Krzycki J. A., and Ferguson D. J. (2014) A nonpyrrolysine member of the widely distributed trimethylamine methyltransferase family is a glycine betaine methyltransferase. Proc. Natl. Acad. Sci. U.S.A. 111, E4668–E4676 10.1073/pnas.1409642111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Matthews R. G. (2009) Cobalamin- and corrinoid-dependent enzymes. Met. Ions Life Sci. 6, 53–114 10.1039/BK9781847559159-00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lechtenfeld M., Heine J., Sameith J., Kremp F., and Müller V. (2018) Glycine betaine metabolism in the acetogenic bacterium Acetobacterium woodii. Environ. Microbiol. 20, 4512–4525 10.1111/1462-2920.14389 [DOI] [PubMed] [Google Scholar]
- 8. Tang W. H. W., Bäckhed F., Landmesser U., and Hazen S. L. (2019) Intestinal microbiota in cardiovascular health and disease: JACC state-of-the-art review. J. Am. Coll. Cardiol. 73, 2089–2105 10.1016/j.jacc.2019.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang R. H., Liu Z. K., Yang D. S., Zhang X. J., Sun H. D., and Xiao W. L. (2018) Phytochemistry and pharmacology of the genus Leonurus: the herb to benefit the mothers and more. Phytochemistry 147, 167–183 10.1016/j.phytochem.2017.12.016 [DOI] [PubMed] [Google Scholar]
- 10. Loo R. L., Zou X., Appel L. J., Nicholson J. K., and Holmes E. (2018) Characterization of metabolic responses to healthy diets and association with blood pressure: application to the Optimal Macronutrient Intake Trial for Heart Health (OmniHeart), a randomized controlled study. Am. J. Clin. Nutr. 107, 323–334 10.1093/ajcn/nqx072 [DOI] [PubMed] [Google Scholar]
- 11. Lustgarten M. S., Price L. L., Chalé A., and Fielding R. A. (2014) Metabolites related to gut bacterial metabolism, peroxisome proliferator-activated receptor-α activation, and insulin sensitivity are associated with physical function in functionally-limited older adults. Aging Cell 13, 918–925 10.1111/acel.12251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Trinchant J. C., Boscari A., Spennato G., Van de Sype G., and Le Rudulier D. (2004) Proline betaine accumulation and metabolism in alfalfa plants under sodium chloride stress: exploring its compartmentalization in nodules. Plant Physiol. 135, 1583–1594 10.1104/pp.103.037556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bashir A., Hoffmann T., Kempf B., Xie X., Smits S. H. J., and Bremer E. (2014) Plant-derived compatible solutes proline betaine and betonicine confer enhanced osmotic and temperature stress tolerance to Bacillus subtilis. Microbiology 160, 2283–2294 10.1099/mic.0.079665-0 [DOI] [PubMed] [Google Scholar]
- 14. Servillo L., Giovane A., Balestrieri M. L., Bata-Csere A., Cautela D., and Castaldo D. (2011) Betaines in fruits of citrus genus plants. J. Agric. Food Chem. 59, 9410–9416 10.1021/jf2014815 [DOI] [PubMed] [Google Scholar]
- 15. Chambers S. T., and Kunin C. M. (1987) Isolation of glycine betaine and proline betaine from human-urine: assessment of their role as osmoprotective agents for bacteria and the kidney. J. Clin. Invest. 79, 731–737 10.1172/JCI112878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lever M., Sizeland P. C. B., Bason L. M., Hayman C. M., and Chambers S. T. (1994) Glycine betaine and proline betaine in human blood and urine. Biochim. Biophys. Acta 1200, 259–264 10.1016/0304-4165(94)90165-1 [DOI] [PubMed] [Google Scholar]
- 17. Atkinson W., Downer P., Lever M., Chambers S. T., and George P. M. (2007) Effects of orange juice and proline betaine on glycine betaine and homocysteine in healthy male subjects. Eur. J. Nutr. 46, 446–452 10.1007/s00394-007-0684-5 [DOI] [PubMed] [Google Scholar]
- 18. Lang R., Lang T., Bader M., Beusch A., Schlagbauer V., and Hofmann T. (2017) High-throughput quantitation of proline betaine in foods and suitability as a valid biomarker for citrus consumption. J. Agric. Food Chem. 65, 1613–1619 10.1021/acs.jafc.6b05824 [DOI] [PubMed] [Google Scholar]
- 19. Almanza-Aguilera E., Urpi-Sarda M., Llorach R., Vázquez-Fresno R., Garcia-Aloy M., Carmona F., Sanchez A., Madrid-Gambin F., Estruch R., Corella D., and Andres-Lacueva C. (2017) Microbial metabolites are associated with a high adherence to a Mediterranean dietary pattern using a H-1-NMR-based untargeted metabolomics approach. J. Nutr. Biochem. 48, 36–43 10.1016/j.jnutbio.2017.06.001 [DOI] [PubMed] [Google Scholar]
- 20. Rebholz C. M., Lichtenstein A. H., Zheng Z., Appel L. J., and Coresh J. (2018) Serum untargeted metabolomic profile of the dietary approaches to stop hypertension (DASH) dietary pattern. Am. J. Clin. Nutr. 108, 243–255 10.1093/ajcn/nqy099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Matsumoto M., Kibe R., Ooga T., Aiba Y., Kurihara S., Sawaki E., Koga Y., and Benno Y. (2012) Impact of intestinal microbiota on intestinal luminal metabolome. Sci. Rep. 2, 233 10.1038/srep00233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kumar R., Zhao S. W., Vetting M. W., Wood B. M., Sakai A., Cho K., Solbiati J., Almo S. C., Sweedler J. V., Jacobson M. P., Gerlt J. A., and Cronan J. E. (2014) Prediction and biochemical demonstration of a catabolic pathway for the osmoprotectant proline betaine. Mbio 5, e00933–13 10.1128/mBio.00933-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Song Y., and Cho B.-K. (2015) Draft genome sequence of chemolithoautotrophic acetogenic butanol-producing Eubacterium limosum ATCC 8486. Genome Annouc. 3, e01564–14 10.1128/genomeA.01564-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Song Y., Shin J., Jeong Y., Jin S., Lee J. K., Kim D. R., Kim S. C., Cho S., and Cho B. K. (2017) Determination of the genome and primary transcriptome of syngas fermenting Eubacterium limosum ATCC 8486. Sci. Rep. 7, 13694 10.1038/s41598-017-14123-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Muller E., Fahlbusch K., Walther R., and Gottschalk G. (1981) Formation of N,N-dimethylglycine, acetic-acid, and butyric-acid from betaine by Eubacterium limosum. Appl. Environ. Microbiol. 42, 439–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schoelmerich M. C., Katsyv A., Sung W., Mijic V., Wiechmann A., Kottenhahn P., Baker J., Minton N. P., and Müller V. (2018) Regulation of lactate metabolism in the acetogenic bacterium Acetobacterium woodii. Environ. Microbiol. 20, 4587–4595 10.1111/1462-2920.14412 [DOI] [PubMed] [Google Scholar]
- 27. Ishihama Y., Oda Y., Tabata T., Sato T., Nagasu T., Rappsilber J., and Mann M. (2005) Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell Proteomics 4, 1265–1272 10.1074/mcp.M500061-MCP200 [DOI] [PubMed] [Google Scholar]
- 28. Ferguson T., Soares J. A., Lienard T., Gottschalk G., and Krzycki J. A. (2009) RamA, a protein required for reductive activation of corrinoid-dependent methylamine methyltransferase reactions in methanogenic Archaea. J. Biol. Chem. 284, 2285–2295 10.1074/jbc.M807392200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hennig S. E., Jeoung J. H., Goetzl S., and Dobbek H. (2012) Redox-dependent complex formation by an ATP-dependent activator of the corrinoid/iron-sulfur protein. Proc. Natl. Acad. Sci. U.S.A. 109, 5235–5240 10.1073/pnas.1117126109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schilhabel A., Studenik S., Vödisch M., Kreher S., Schlott B., Pierik A. Y., and Diekert G. (2009) The ether-cleaving methyltransferase system of the strict anaerobe Acetobacterium dehalogenans: analysis and expression of the encoding genes. J. Bacteriol. 191, 588–599 10.1128/JB.01104-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ziegler C., Bremer E., and Krämer R. (2010) The BCCT family of carriers: from physiology to crystal structure. Mol. Microbiol. 78, 13–34 10.1111/j.1365-2958.2010.07332.x [DOI] [PubMed] [Google Scholar]
- 32. Drake H. L., Gössner A. S., and Daniel S. L. (2008) Old acetogens, new light. Ann. N.Y. Acad. Sci. 1125, 100–128 10.1196/annals.1419.016 [DOI] [PubMed] [Google Scholar]
- 33. Jeong J., Bertsch J., Hess V., Choi S., Choi I. G., Chang I. S., and Müller V. (2015) Energy conservation model based on genomic and experimental analyses of a carbon monoxide-utilizing, butyrate-forming acetogen, Eubacterium limosum KIST612. Appl. Environ. Microbiol. 81, 4782–4790 10.1128/AEM.00675-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Evans J. C., Huddler D. P., Hilgers M. T., Romanchuk G., Matthews R. G., and Ludwig M. L. (2004) Structures of the N-terminal modules imply large domain motions during catalysis by methionine synthase. Proc. Natl. Acad. Sci. U.S.A. 101, 3729–3736 10.1073/pnas.0308082100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gloux K., and Le Rudulier D. (1989) Transport and catabolism of proline betaine in salt-stress Rhizobioum meliloti. Arch. Microbiol. 151, 143–148 10.1007/BF00414429 [DOI] [Google Scholar]
- 36. Goldmann A., Boivin C., Fleury V., Message B., Lecoeur L., Maille M., and Tepfer D. (1991) Betaine use by rhizosphere bacteria: genes essential for trigonelline, stachydrine, and carnitine catabolism in Rhizobium meliloti are located on pSym in the symbiotic region. Mol. Plant. Microbe Interact. 4, 571–578 10.1094/MPMI-4-571 [DOI] [PubMed] [Google Scholar]
- 37. Burnet M. W., Goldmann A., Message B., Drong R., El Amrani A., Loreau O., Slightom J., and Tepfer D. (2000) The stachydrine catabolism region in Sinorhizobium meliloti encodes a multi-enzyme complex similar to the xenobiotic degrading systems in other bacteria. Gene 244, 151–161 10.1016/S0378-1119(99)00554-5 [DOI] [PubMed] [Google Scholar]
- 38. Daughtry K. D., Xiao Y., Stoner-Ma D., Cho E., Orville A. M., Liu P., and Allen K. N. (2012) Quaternary ammonium oxidative demethylation: X-ray crystallographic, resonance Raman, and UV-visible spectroscopic analysis of a Rieske-type demethylase. J. Am. Chem. Soc. 134, 2823–2834 10.1021/ja2111898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zheng L., Kelly C. J., and Colgan S. P. (2015) Physiologic hypoxia and oxygen homeostasis in the healthy intestine. A review in the theme: cellular responses to hypoxia. Am. J. Physiol. Cell Physiol. 309, C350–C360 10.1152/ajpcell.00191.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Feng Y., Stams A. J. M., Sanchez-Andrea I., and de Vos W. M. (2018) Eubacterium maltosivorans sp. nov., a novel human intestinal acetogenic and butyrogenic bacterium with a versatile metabolism. Int. J. Syst. Evol. Microbiol. 10.1099/ijsem.0.003028 [DOI] [PubMed] [Google Scholar]
- 41. Zhao S., Kumar R., Sakai A., Vetting M. W., Wood B. M., Brown S., Bonanno J. B., Hillerich B. S., Seidel R. D., Babbitt P. C., Almo S. C., Sweedler J. V., Gerlt J. A., Cronan J. E., and Jacobson M. P. (2013) Discovery of new enzymes and metabolic pathways by using structure and genome context. Nature 502, 698–702 10.1038/nature12576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schuchmann K., and Müller V. (2014) Autotrophy at the thermodynamic limit of life: a model for energy conservation in acetogenic bacteria. Nat. Rev. Microbiol. 12, 809–821 10.1038/nrmicro3365 [DOI] [PubMed] [Google Scholar]
- 43. Thauer R. K., Kaster A. K., Seedorf H., Buckel W., and Hedderich R. (2008) Methanogenic archaea: ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 6, 579–591 10.1038/nrmicro1931 [DOI] [PubMed] [Google Scholar]
- 44. Weghoff M. C., Bertsch J., and Müller V. (2015) A novel mode of lactate metabolism in strictly anaerobic bacteria. Environ. Microbiol. 17, 670–677 10.1111/1462-2920.12493 [DOI] [PubMed] [Google Scholar]
- 45. Ragsdale S. W., and Pierce E. (2008) Acetogenesis and the Wood-Ljungdahl pathway of CO2 fixation. Biochim. Biophys. Acta 1784, 1873–1898 10.1016/j.bbapap.2008.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kremp F., Poehlein A., Daniel R., and Müller V. (2018) Methanol metabolism in the acetogenic bacterium Acetobacterium woodii. Environ. Microbiol. 20, 4369–4384 10.1111/1462-2920.14356 [DOI] [PubMed] [Google Scholar]
- 47. Boscari A., Mandon K., Dupont L., Poggi M. C., and Le Rudulier D. (2002) BetS is a major glycine betaine/proline betaine transporter required for early osmotic adjustment in Sinorhizobium meliloti. J. Bacteriol. 184, 2654–2663 10.1128/JB.184.10.2654-2663.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Webb B. A., Karl Compton K., Castañeda Saldaña R., Arapov T. D., Keith Ray W., Helm R. F., and Scharf B. E. (2017) Sinorhizobium meliloti chemotaxis to quaternary ammonium compounds is mediated by the chemoreceptor McpX. Mol. Microbiol. 103, 333–346 10.1111/mmi.13561 [DOI] [PubMed] [Google Scholar]
- 49. Schnoes A. M., Brown S. D., Dodevski I., and Babbitt P. C. (2009) Annotation error in public databases: misannotation of molecular function in enzyme superfamilies. PLoS Comp. Biol. 5, e1000605 10.1371/journal.pcbi.1000605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pfennig N., and Lippert K. D. (1966) Uber das Vitamin B12-Bedurfnis phototropher Schwefelbakterien. Arch. Mikrobiol. 55, 245–256 [Google Scholar]
- 51. Cornforth J. W., and Henry A. J. (1952) The isolation of l-stachydrine from the fruit of Capparis tomentosa. J. Chem. Soc. 1952, 601–603 [Google Scholar]
- 52. Karrer P., and Widmer R. (1925) Die konfiguration des nicotins: optisch active hygrinsäure. Helv. Chim. Acta 8, 364–368 10.1002/hlca.19250080157 [DOI] [Google Scholar]
- 53. Lin N.-H., He Y., and Wittenberger S. J. (May 14, 1996) Method of preparing enantiomerically-pure 3-methyl-5-(1-alkyl-2(s)-pyrrolidinyl)isoxazoles. United States Patent US5516912A
- 54. Aurelio L., Box J. S., Brownlee R. T., Hughes A. B., and Sleebs M. M. (2003) An efficient synthesis of N-methyl amino acids by way of intermediate 5-oxazolidinones. J. Org. Chem. 68, 2652–2667 10.1021/jo026722l [DOI] [PubMed] [Google Scholar]
- 55. Roh H., Ko H. J., Kim D., Choi D. G., Park S., Kim S., Chang I. S., and Choi I. G. (2011) Complete genome sequence of a carbon monoxide-utilizing acetogen, Eubacterium limosum KIST612. J. Bacteriol. 193, 307–308 10.1128/JB.01217-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Klock H. E., Koesema E. J., Knuth M. W., and Lesley S. A. (2008) Combining the polymerase incomplete primer extension method for cloning and mutagenesis with microscreening to accelerate structural genomics efforts. Proteins 71, 982–994 10.1002/prot.21786 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD013806 and DOI 10.6019/PXD013806 (for lactate-grown cells) and PXD013962 and DOI 10.6019/PXD013962 (for proline betaine–grown cells).


