Abstract
Massive efforts to sequence cancer genomes have compiled an impressive catalogue of cancer mutations, revealing the recurrent exploitation of a handful of ‘hallmark cancer pathways’. However, unraveling how sets of mutated proteins in these and other pathways hijack pro-proliferative signaling networks and dictate therapeutic responsiveness remains challenging. Here, we show that cancer driver protein–protein interactions are enriched for additional cancer drivers, highlighting the power of physical interaction maps to explain known, as well as uncover new, disease-promoting pathway interrelationships. We hypothesize that by systematically mapping the protein–protein and genetic interactions in cancer—thereby creating Cancer Cell Maps—we will create resources against which to contextualize a patient’s mutations into perturbed pathways/complexes and thereby specify a matching targeted therapeutic cocktail.
Protein–protein and genetic interactions fill the gap between genotype and phenotype
The advent of next-generation sequencing technology has fueled a massive accumulation of genomic sequences over the past decade. In fact, any individual can now sequence their entire genome for only $200, gaining unprecedented insight into their ancestry and predisposition for certain diseases, particularly those with mono-genetic or Mendelian drivers. Furthermore, genomic sequencing of cancerous tissues is practically commonplace in many clinics, from whole genome [1] to targeted arrays [2], bestowing oncologists large quantities of data from which to, possibly, guide treatment decisions [3]. However, translating a list of patient-derived cancer mutations into patient-specific treatment decisions remains challenging, with only a few robust examples of success existing today (e.g. imatinib for the BCR–ABL fusion gene [4]).
Nevertheless, the analysis of genomic sequencing data has provided significant insight into our understanding of cancer biology. Numerous large-scale genomic studies, such as those from The Cancer Genome Atlas (TCGA) consortium, have demonstrated that cancer exploits a recurring set of ‘hallmark cancer pathways’—pan-cancer analyses estimate 90% of tumors have a driver alteration in at least one of ten hallmark signaling pathways [5••]. However, complexity persists, as each patient’s tumor possesses a heterogeneous mixture of mutations within these, as well as many other, pathways. Even two patients with the same tumor type (e.g. breast cancer) can possess strikingly different sets of mutations, with some patients possessing over 1000 mutations of unknown function, many of which may regulate these hallmark pathways in an unknown fashion. Consequently, and unsurprisingly, this mutational diversity between tumors drives differences in drug sensitivity, which are currently extremely difficult to reliably predict on a patient-by-patient basis.
The challenge of translating patient-derived mutational profiles into patient-tailored drug cocktails highlights a fundamental knowledge gap between genotype and phenotype in mammalian cell biology. In our view, navigating between genotype (e.g. mutations, copy number alterations, mRNA/protein expression) and phenotype (e.g. cell proliferation and drug sensitivity) requires maps depicting how each protein assembles into protein–protein interaction networks (by creating protein–protein interaction maps), and thus into particular pathways and protein complexes, and an understanding of how these interactions functionally compel cellular processes (by creating genetic interaction maps), such as cancer progression. In order to appropriately characterize disease-specific alterations, protein–protein and genetic interaction maps must be built by comparing ‘healthy’ (i.e. non-transformed) to diseased cells, contrasting mutant to wild-type networks, and evaluating differences across tissue types. The creation of ‘differential’ [6] cancer cell maps will provide a resource against which to contextualize new patient mutation data into known cancer pathways or propose synthetic lethal targeting strategies for pharmacological intervention (Figure 1).
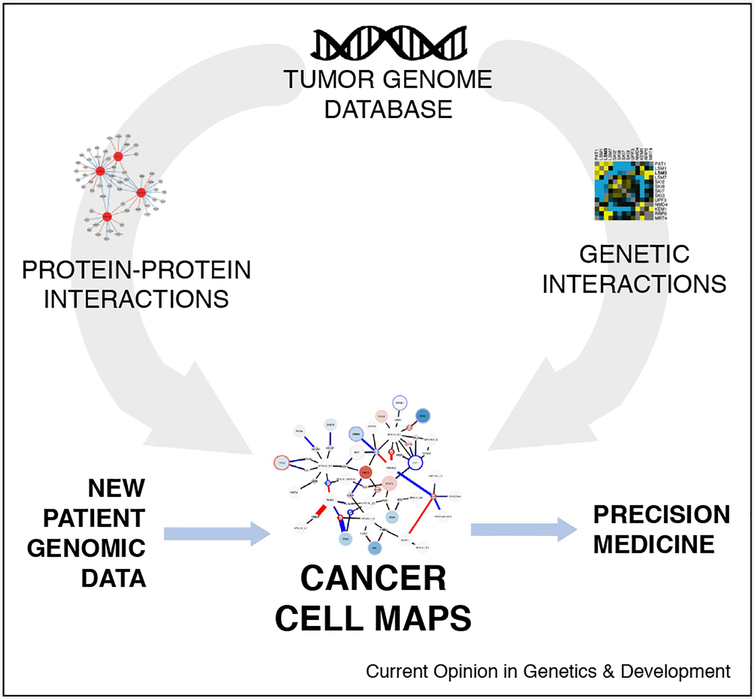
Figure 1.
The cancer cell map for precision oncology. The analysis of large numbers of tumor genomes has revealed key mutations that drive cancer pathology. However, connecting the genotype of tumor mutational profiles to phenotypes of drug sensitivity is impeded by a fundamental lack of understanding into how protein–protein (physical) and genetic (functional) interactions are configured to drive cellular processes, such as proliferation and death. Seeded by this large-scale analysis of tumor genomes (top), what is needed is a systematic, context-diverse network mapping of the protein–protein interactions for mutated and wild-type oncogenes and tumor suppressors (left) as well as the identification of synthetic lethal genetic interactions (right). PPI and GI mapping, two highly complementary data sources, can then be assembled to form Cancer Cell Maps, providing a working scaffold of molecular interactions and the cell types/conditions under which they are active. New patient data can then be queried against this resource by mapping alterations to these hallmark cancer networks, which can inform precision medicine (blue arrows).
Physical interactors of cancer drivers are often cancer drivers, too
Protein–protein interactions (PPIs) often implicate a functional relationship between proteins. For example, PPIs typically denote proteins that work together to accomplish a specific cellular task or co-drive disease [7]. Often times, interactions can mediate crosstalk between distinct cellular pathways. For example, the physical interaction between PIK3CA and KRAS, discovered over two decades ago [8], reveals a poignant mechanism of crosstalk between the ERK and PI3K signaling pathways. This finding highlights the capability of protein–protein interactions to reveal how the cancer signaling network is configured, with implications for cancer therapy [9]. Furthermore, the disruption or strengthening of PPIs by mutations, which are found to frequently occur at the interface between other proteins or ligands, highlights the wide role these interactions may play in regulating signaling activity or downstream effectors [10]. In sum, proteins that physically interact with known disease genes are themselves, also, potential disease drivers.
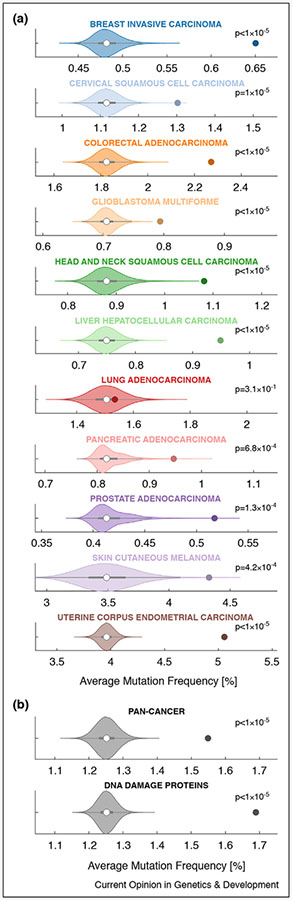
Given this line of thinking, one might expect protein interactors of major cancer drivers to be enriched for additional cancer drivers. To probe this hypothesis, we ask: are physical interactors of major cancer drivers also frequently mutated in cancer? To answer this question, we assembled the union of several large protein–protein interaction networks, including BioPlex [11], IRefIndex [12], Mentha [13], Human Interactome [14], and HPRD [15], and extract physical interactors (first neighbors) for each of the fifteen most commonly mutated proteins (copy number variations excluded) in each of eleven distinct cancer types [16]. Interestingly, we found the average mutation frequency of these interactors to be significantly higher than size-matched random controls for each cancer type, with the exception of lung adenocarcinoma (Figure 2a). We observed the same trend when collapsing across all eleven cancer types—’Pan-Cancer’ analysis (Figure 2b, top)—and also when assessing the interactomes of 12 canonical DNA damage proteins (Figure 2b, bottom). In sum, there appears to be a significant enrichment of frequently mutated cancer proteins that bind to other frequently mutated cancer proteins.
Figure 2.
Physical interactors of cancer drivers are often cancer drivers, too. (a) A human protein–protein interaction network was assembled from publicly available data (union of BioPlex, IRefIndex, Mentha, HumanInteractome, and HPRD networks, which amounted to 18 227 nodes and 333 490 edges) and probed for physical interactors (i.e. first neighbors) of the top 15 most frequently mutated proteins (copy number variations excluded) for each cancer type. This typically amounted to ~1500 interactors per cancer type. We then calculated the average mutation frequency across these cancer driver physical interactors (colored dots) for each cancer type (e.g. for breast invasive carcinoma, the average was ~0.65%) and compared them to the average mutation frequencies of 100 000 random size-matched permutations taken from the same dataset (violin histogram). We calculated an empirical p-value for each PPI set, defined as the fraction of random permutation sets of greater value. Interestingly, we find the average mutation frequency of the true cancer driver PPI sets for each cancer type, with the exception of lung adenocarcinoma, to be mutated significantly more often than expected by chance. (b) A pan-cancer analysis, performed by calculating the mutation frequency for each gene across all 11 cancer types in (a), also showed the PPIs of the top 15 most frequently mutated proteins to be mutated significantly more often than expected by chance (top; ‘Pan-Cancer’). In addition, we also assessed the average mutation frequency of known physical interactors of 12 canonical DNA damage proteins (CHEK1, CHEK2, RAD51, BRCA1, BRCA2, MLH1, MSH2, ATM, ATR, MDC1, PARP1, and FANCF), which we also found to be mutated significantly more often than expected by chance (mutation frequencies as calculated for ‘Pan-Cancer’ analysis). These results highlight the role of PPIs as potential drivers of cancer pathology and suggest an additional point of regulation for therapeutic intervention. Empty white dots denote the median of the randomly permuted sets, and the adjoining grey rectangles denote the interquartile range.
These results highlight an opportunity to uncover new biological processes and molecular mechanisms implicated in cancer by probing these physical interactions. In this article, we briefly review two such examples, which possess a dual physical/functional relationship, and showcase themes gaining momentum in the scientific literature. First, we explore the physical and functional interface between inflammatory proteins and oncogenes. Second, we highlight recent insights into the interrelationships between DNA damage proteins and upstream proliferation-driving oncogenes. Lastly, we review a few recent large-scale genetic interaction studies that demonstrate the overlap between genetic (i.e. functional) and protein–protein (i.e. physical) interactomes, illustrating the synergy between these mapping approaches.
Physical interactions between inflammatory proteins and oncogenes
The relationship between inflammation and cancer is not a recent discovery. In fact, the first known observation spans back to 1863, when Rudolf Virchow noticed cancer at sites of chronic inflammation [17]. Inflammation is well appreciated to play a role in several aspects of tumor development, from oncogenesis to metastasis, and patients with inflammatory diseases such as hepatitis, colitis, and pancreatitis display a higher incidence of cancer [18]. Conversely, treatment with non-steroidal anti-inflammatory drugs (NSAIDs) has displayed anti-cancer activity for several cancers [19–21], as have anti-inflammatory physiological processes, such as getting regular exercise [22,23]. Although many important observations over the years have linked cancer with inflammatory processes, the direct physical regulation of cancer driver proteins by inflammatory proteins is only recently being investigated [24]. Our cancer driver PPI analysis above identified several inflammatory proteins—including STAT3, SMAD3, NFKBIA, STAT5B, and LRP1—that bind to top cancer drivers. To what extent do physical interactions between inflammatory molecules and cancer drivers mediate inflammation-induced cancer pathology? This remains an open question in the field and highlights exciting avenues for future research.
As an example, our analysis highlights a relationship between EGFR and STAT3 signaling. Specifically, EGFR is known to interact with STAT3 as well as the protein tyrosine phosphatase PTPRD, a commonly mutated [25,26] known negative regulator of STAT3 [27]. EGFR is known to activate and maintain STAT3 signaling, driving transcriptome-rewiring events that promote cancer progression and drug resistance [28]. In addition, the EGFR-STAT3 system leads to the production of Il-6, an inflammatory cytokine, a feedback process which is thought to underlie tumor initiation [29] and metastasis [30]. Inhibition strategies that co-ablate phosphorylated EGFR and STAT3 may be a potent anti-cancer therapy in some contexts [31]. This is but one example of several representing a dual physical and functional relationship between an inflammatory protein and a cancer driver; though, many more are expected to emerge in the future.
Physical interactions between DNA damage response proteins and oncogenes that drive proliferation
Another signaling interface of interest is between DNA damage response (DDR) proteins and oncogenes that drive proliferation, an interrelationship increasingly reported in the scientific literature though not fully understood. A deeper understanding of this interface could help identify patient-specific therapies, such as new combinations of PARP and kinase inhibitors targeting angiogenesis [32], PI3K/AKT [33], RAS/MAPK [34•], WEE-1 [35] and ATR/CHEK1 [36] pathways in order to overcome PARP inhibitor resistance. In order to identify known physical interactions between DDR proteins and pro-proliferative oncogenes, we searched our PPI network for physical interactors of 12 canonical DDR proteins (CHEK1, CHEK2, RAD51, BRCA1, BRCA2, MLH1, MSH2, ATM, ATR, MDC1, PARP1, and FANCF). Interestingly, we find these DDR gene inter-actors to be more frequently altered than expected by chance, calling attention to the central role DDR proteins and their interactors play in cancer progression (Figure 2b, bottom).
Of particular interest, we observed several interactions between DDR proteins and both the Ras/MAPK and PI3K/AKT signaling pathways, interrelationships that are gaining increasing attention in the scientific literature. For instance, a recent study highlighted the interplay between PARP and MAPK signaling [34•], finding strong synergy between dual MEK and PARP inhibition for RAS mutant cancers in vivo. This interplay is corroborated by several known physical interactions between PARP1 and various MAPK pathway members, including MAPK1, MAPK3, and MAPK13. An additional example is the interplay between BRCA1 and AKT signaling; specifically BRCA1 deficiency was found to drive AKT activation [37], which was involved in mediating tumorigenesis in mice likely by promoting chromosome instability [38]. Activated AKT in BRCA-deficient cells was found to impair CHEK1 nuclear localization and CHEK1 interaction with RAD51, leading to defects in homologous recombination [39]. Furthermore, AKT inhibition displayed efficacy against BRCA1-mutated mammary tumors in mice [40]. Publicly available PPI data indicate a physical interaction between AKT1 and several DDR proteins, including BRCA1, CHEK1, and MSH2 as well as between PIK3CA and ATR. These examples illustrate how PPIs can help explain known relationships between distinct pathways, or even uncover new potential pathway relationships. Considering these insights, further defining how interactions between DDR proteins and pro-proliferative oncogenes regulate cancer progression, growth suppression, and/or drug resistance is an exciting avenue for future research.
Genetic interaction mapping often reveals functional protein–protein interactions
Uncovering functional PPIs can be challenging, as many physical interactions, when perturbed, do not enact a phenotypic change. Genetic interaction (GI) mapping, pioneered in the early 2000s, is a powerful technique to systematically reveal functional relationships between genes, which often indicate the presence of a physical interaction. GI mapping involves the pairwise perturbation of genes (e.g. knockout, knockdown or overexpression) in order to elucidate how one gene modulates the phenotype of the other. Typically, cell viability is used as the phenotypic readout, where GIs that increase cellular fitness are said to be ‘positive’ and GIs that decrease cellular fitness are said to be ‘negative’. GI mapping is often used to uncover new functions of genes [41,42], enabling a hierarchical organization of gene products into functional complexes and pathways, and to identify synthetic–lethal interactions with relevance to cancer combination therapy. As alluded to, the presence of a functional (i.e. genetic) interaction often indicates a physical interaction, and vice versa, highlighting the synergy between combined GI and PPI mapping initiatives.
The majority of GI studies were performed in the budding yeast Saccharomyces cerevisiae [43,44]. More recently, however, GI mapping has entered mammalian cell contexts. In human cells, GI screens have already tackled diverse topics, including chromatin regulation [45,46], ricin susceptibility [47], drug target interactions [48], and functioning of cancer oncogenes/tumor suppressors [49–51], among others. In one of the largest GI screens ever conducted in human cells, Horlbeck et al. assessed 222 784 gene pairs using a pooled dual-sgRNA CRISPR interference (dCas9-KRAB) loss-of-function lentiviral vector screen in K526 and Jurkat cells, two human immune cell lines [52••]. Interestingly, gene pairs with highly correlated GI fitness profiles were enriched for known physical interactions from the STRING physical interaction database. They further uncovered new mechanism, discovering TMEM261, a previously poorly characterized gene, as a critical regulator of oxidative phosphorylation similarly to core mitochondrial complex I proteins, to which TMEM261 is known to physically associate [53].
Computational analyses of GI datasets have also harnessed much insight, most of which further highlight the intersection between GIs and PPIs. Pan et al. [54] utilized data from Project Achilles [55], a compendium of lentivirus-based pooled shRNA and CRISPR/Cas9 genome-wide across cell lines, to cluster GI fitness profiles across cell lines (N = 342) [56•]. They found ~40% of complexes from the CORUM [57] protein complex data base to possess significantly correlated GI profiles. In addition, correlated GI profiles were also used to annotate previously unknown protein complexes. In another study, Rauscher et al. developed a computational framework (‘MINGLE’) to integrate 85 CRISPR/Cas9 screens with mutation, copy number, and mRNA expression data from 60 different cancer cell lines [58••]. This study used a random effects statistical model to identify significant relationships between normalized CRISPR scores and genetic alterations across cell lines (17 545 significant GIs). Again, genes with similar interaction profiles were enriched for co-complex membership. This study revealed PRKCSH, GANAB, and UGP2 as novel positive regulators of the Wnt/β-catenin pathway by searching for genes possessing a negative genetic interaction score with RNF43, a known negative Wnt pathway regulator. Interestingly, PRKCSH and GANAB also physically interact to form the glucosidase II complex.
These studies highlight the cohesion between GI and PPI mapping approaches—where the presence of one interaction type typically implicates the other—and depicts the synergy between the two approaches in systematically uncovering new biology with relevance to cancer and other diseases.
Building cancer cell maps for precision medicine
As mentioned above, genomic sequencing of cancerous tissues has led to an impressive catalogue of disease-driving mutations. Although many cancers harbor alterations in well-known cancer drivers, such as TP53 or EGFR, a tumor may additionally possess anywhere from 10 to over 1000 rare somatic genetic alterations that remain uncharacterized. We and others [59, 60, 61••] hypothesize that while these mutations may appear rare when viewed independently, they likely converge on a smaller number of protein complexes, signaling cascades, and transcriptional regulatory circuits. By recapitulating this at network-level, we anticipate significant pathway-based signals to emerge, signals likely imperceptible by considering individual mutations in isolation. Towards building a network-level view, we believe it critical to compare networks from ‘healthy’/non-transformed cells to diseased cells, to study both mutant and wild-type protein interactomes, and to compare across distinct cancer types. The unbiased and systematic collection of network-level data in this way, utilizing both physical (e.g. large-scale AP-MS) and genetic (e.g. large-scale CRISPR screens) approaches, is needed to create Cancer Cell Maps that delineate the disease-specific molecular wiring of the cell and how it differs between cancer types. To probe these questions and hypotheses, we [61••] and others [62,63] have pioneered various cell mapping initiatives to delineate the molecular interactions that drive cancer. Once created and validated, we envision Cancer Cell Maps will be integrated into the oncologist’s toolkit, against which a patient’s specific mutations can be queried in order to identify the pathways and protein complexes that are perturbed, enabling the rational and precise selection of appropriate targeted therapies (Figure 1).
Acknowledgements
We would like to acknowledge members of the Cancer Cell Map Initiative (CCMI), including, but not limited to, Dr. Danielle Swaney, Dr. Jisoo Park, and Dr. Fan Zheng, for providing their expertise and for lending support to strengthen our mapping initiatives.
Funding
This work was supported by the National Institutes of Health U54 CA209891 (NJK and TI), U01 MH115747 (NJK), T32 EB009383 (ZZCN), and F32 CA239333 (MB). Additional support was received from the Martha and Bruce Atwater Breast Cancer Research Fund via UCSF Helen Diller Family Comprehensive Cancer Center (MK) and UCSF Prostate Cancer Program Research Pilot Funding (MK).
Footnotes
Conflict of interest statement
T.I. is co-founder of Data4Cure, Inc., is on the Scientific Advisory Board, and has an equity interest. T.I. is on the Scientific Advisory Board of Ideaya BioSciences, Inc., has an equity interest, and receives income for sponsored research funding. The terms of these arrangements have been reviewed and approved by the University of California San Diego in accordance with its conflict of interest policies.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Nakagawa H, Wardell CP, Furuta M, Taniguchi H, Fujimoto A: Cancer whole-genome sequencing: present and future. Oncogene 2015, 34:5943–5950 10.1038/onc.2015.90. [DOI] [PubMed] [Google Scholar]
- 2.Kline CN, Joseph NM, Grenert JP, van Ziffle J, Talevich E, Onodera C, Aboian M, Cha S, Raleigh DR, Braunstein S et al. : Targeted next-generation sequencing of pediatric neuro-oncology patients improves diagnosis, identifies pathogenic germline mutations, and directs targeted therapy. Neuro Oncol 2017, 19:699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hyman DM, Taylor BS, Baselga J: Implementing genome-driven oncology. Cell 2017, 168:584–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Savage DG, Antman KH: Imatinib mesylate—a new oral targeted therapy. N Engl J Med 2002, 346:683–693 10.1056/nejmra013339. [DOI] [PubMed] [Google Scholar]
- 5••. Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, Dimitriadoy S, Liu DL, Kantheti HS, Saghafinia S et al. : Oncogenic signaling pathways in the cancer genome atlas. Cell 2018, 173:321–337.e10.In this article, authors performed a genomic pan-cancer analysis, identifying ten signaling pathways that are altered at least once in 89% of tumors. These pathways are: cell cycle, Hippo, Myc, Notch, Nrf2, PI3K, RTK/Ras, TGFβ, p53, and β-Catenin/Wnt. They integrate genomic analyses with a pathway-centric perspective, providing comprehensive lists of alterations in these pathways. In addition, they assess patterns of alteration co-occurrence and mutual exclusivity, identifying pathways that potentially cross-talk. Furthermore, they find that 57% of tumors possess at least one alteration that is targetable by currently available drugs.
- 6.Ideker T, Krogan NJ: Differential network biology. Mol Syst Biol 2012, 8. 10.1038/msb.2011.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goh K-I, Goh K-I, Cusick ME, Valle D, Childs B, Vidal M, Barabasi A-L: The human disease network. Proc Natl Acad Sci U S A 2007, 104:8685–8690 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodriguez-Viciana P, Warne PH, Dhand R, Vanhaesebroeck B, Gout I, Fry MJ, Waterfield MD, Downward J: Phosphatidylinositol-3-OH kinase direct target of Ras. Nature 1994, 370:527–532. [DOI] [PubMed] [Google Scholar]
- 9.Di Nicolantonio F, Arena S, Tabernero J, Grosso S, Molinari F, Macarulla T, Russo M, Cancelliere C, Zecchin D, Mazzucchelli L et al. : Deregulation of the PI3K and KRAS signaling pathways in human cancer cells determines their response to everolimus. J Clin Invest 2010, 120:2858–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buljan M, Blattmann P, Aebersold R, Boutros M: Systematic characterization of pan-cancer mutation clusters. Mol Syst Biol 2018, 14:e7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huttlin EL, Bruckner RJ, Paulo JA, Cannon JR, Ting L, Baltier K, Colby G, Gebreab F, Gygi MP, Parzen H et al. : Architecture of the human interactome defines protein communities and disease networks. Nature 2017, 545:505–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Razick S, Magklaras G, Donaldson IM: iRefIndex: a consolidated protein interaction database with provenance. BMC Bioinformatics 2008, 9:405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calderone A, Castagnoli L, Cesareni G: Mentha: a resource for browsing integrated protein-interaction networks. Nat Methods 2013, 10:690–691 10.1038/nmeth.2561. [DOI] [PubMed] [Google Scholar]
- 14.Luck K, Sheynkman GM, Zhang I, Vidal M: Proteome-scale human interactomics. Trends Biochem Sci 2017, 42:342–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peri S, Navarro JD, Amanchy R, Kristiansen TZ, Jonnalagadda CK, Surendranath V, Niranjan V, Muthusamy B, Gandhi TKB, Gronborg M et al. : Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res 2003, 13:2363–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, Shen R, Taylor AM, Cherniack AD, Thorsson V, Akbani R, Bowlby R, Wong CK, Wiznerowicz M, Sanchez-Vega F, Robertson AG, Schneider BG, Lawrence MS, Noushmehr H, Malta TM, Cancer Genome Atlas Network, Stuart JM, Benz CC, Laird PW: Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell 2018, 173:291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balkwill F, Mantovani A: Inflammation and cancer: back to Virchow? Lancet 2001, 357:539–545. [DOI] [PubMed] [Google Scholar]
- 18.Coussens LM, Werb Z: Inflammation and cancer. Nature 2002, 420:860–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kune GA, Kune S, Watson LF: Colorectal cancer risk, chronic illnesses, operations and medications: case control results from the Melbourne Colorectal Cancer Study. 1988. Int J Epidemiol 2007, 36:951–957. [DOI] [PubMed] [Google Scholar]
- 20.Wang D: Prostaglandins and cancer. Gut 2006, 55:115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friis S, Riis AH, Erichsen R, Baron JA, Sørensen HT: Low-dose aspirin or nonsteroidal anti-inflammatory drug use and colorectal cancer risk. Ann Intern Med 2015, 163:347. [DOI] [PubMed] [Google Scholar]
- 22.Hojman P, Gehl J, Christensen JF, Pedersen BK: Molecular mechanisms linking exercise to cancer prevention and treatment. Cell Metab 2018, 27:10–21. [DOI] [PubMed] [Google Scholar]
- 23.Benatti FB, Pedersen BK: Exercise as an anti-inflammatory therapy for rheumatic diseases—myokine regulation. Nat Rev Rheumatol 2014, 11:86–97. [DOI] [PubMed] [Google Scholar]
- 24.Dibra D, Mishra L, Li S: Molecular mechanisms of oncogene-induced inflammation and inflammation-sustained oncogene activation in gastrointestinal tumors: an under-appreciated symbiotic relationship. Biochim Biophys Acta 2014, 1846:152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szaumkessel M, Wojciechowska S, Janiszewska J, Zemke N, Byzia E, Kiwerska K, Kostrzewska-Poczekaj M, Ustaszewski A, Jarmuz-Szymczak M, Grenman R et al. : Recurrent epigenetic silencing of the PTPRD tumor suppressor in laryngeal squamous cell carcinoma. Tumour Biol 2017, 39 1010428317691427. [DOI] [PubMed] [Google Scholar]
- 26.Walia V, Prickett TD, Kim J-S, Gartner JJ, Lin JC, Zhou M, Rosenberg SA, Elble RC, Solomon DA, Waldman T, Samuels Y: Mutational and functional analysis of the tumor-suppressor PTPRD in human melanoma. Hum Mutat 2014, 35:1301–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Veeriah S, Brennan C, Meng S, Singh B, Fagin JA, Solit DB, Paty PB, Rohle D, Vivanco I, Chmielecki J et al. : The tyrosine phosphatase PTPRD is a tumor suppressor that is frequently inactivated and mutated in glioblastoma and other human cancers. Proc Natl Acad Sci U S A 2009, 106:9435–9440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ung N, Putoczki TL, Stylli SS, Ng I, Mariadason JM, Chan TA, Zhu H-J, Luwor RB: Anti-EGFR therapeutic efficacy correlates directly with inhibition of STAT3 activity. Cancer Biol Ther 2014, 15:623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu J, Liu W, Williams JP, Ratner N: EGFR-Stat3 signalling in nerve glial cells modifies neurofibroma initiation. Oncogene 2017, 36:1669–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim G, Ouzounova M, Quraishi AA, Davis A, Tawakkol N, Clouthier SG, Malik F, Paulson AK, D’Angelo RC, Korkaya S et al. : SOCS3-mediated regulation of inflammatory cytokines in PTEN and p53 inactivated triple negative breast cancer model. Oncogene 2015, 34:671–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang C, Xu H, Zhou Z, Tian Y, Cao X, Cheng G, Liu Q: Blocking of the EGFR-STAT3 signaling pathway through afatinib treatment inhibited the intrahepatic cholangiocarcinoma. Exp Ther Med 2018, 15:4995–5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu JF, Barry WT, Birrer M, Lee J-M, Buckanovich RJ, Fleming GF, Rimel B, Buss MK, Nattam S, Hurteau J et al. : Combination cediranib and olaparib versus olaparib alone for women with recurrent platinum-sensitive ovarian cancer: a randomised phase 2 study. Lancet Oncol 2014, 15:1207–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Juvekar A, Burga LN, Hu H, Lunsford EP, Ibrahim YH, Balmañà J, Rajendran A, Papa A, Spencer K, Lyssiotis CA et al. : Combining a PI3K inhibitor with a PARP inhibitor provides an effective therapy for BRCA1-related breast cancer. Cancer Discov 2012, 2:1048–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34•. Sun C, Fang Y, Yin J, Chen J, Ju Z, Zhang D, Chen X, Vellano CP, Jeong KJ, Ng PK-S et al. : Rational combination therapy with PARP and MEK inhibitors capitalizes on therapeutic liabilities in RAS mutant cancers. Sci Transl Med 2017, 9 eaal5148.This article finds synergy when combining PARP and MEK inhibitors to treat tumors with alterations in the MAPK pathway, namely mutations in Ras. This relationship was found to be independent of BRCA1/2 or p53 mutation status, suggesting generalizable synergistic activity. Interestingly, synergy was most apparent when decreased expression of homologous recombination components was observed in addition to elevated PARP1 protein levels. Mechanistically, MEK pathway effects were mediated through FOXO3a, as enforced expression of FOXO3a was able to recapitulate MEK inhibitory effects. This article motivates the need for further clinical investigation into the effectiveness of dual PARP and MEK inhibition for Ras mutant cancers.
- 35.Karnak D, Engelke CG, Parsels LA, Kausar T, Wei D, Robertson JR, Marsh KB, Davis MA, Zhao L, Maybaum J, Lawrence TS, Morgan MA: Combined inhibition of Wee1 and PARP1/2 for radiosensitization in pancreatic cancer. Clin Cancer Res 2014, 20:5085–5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim H, George E, Ragland RL, Rafail S, Zhang R, Krepler C, Morgan MA, Herlyn M, Brown EJ, Simpkins F: Targeting the ATR/CHK1 axis with PARP inhibition results in tumor regression in BRCA-mutant ovarian cancer models. Clin Cancer Res 2017, 23:3097–3108 10.1158/1078-0432.ccr-16-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiang T, Ohashi A, Huang Y, Pandita TK, Ludwig T, Powell SN, Yang Q: Negative regulation of AKT activation by BRCA1. Cancer Res 2008, 68:10040–10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guirouilh-Barbat JK, Wilhelm T, Lopez BS: AKT1/BRCA1 in the control of homologous recombination and genetic stability: the missing link between hereditary and sporadic breast cancers. Oncotarget 2010, 1:691–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jia Y, Song W, Zhang F, Yan J, Yang Q: Akt1 inhibits homologous recombination in Brca1-deficient cells by blocking the Chk1-Rad51 pathway. Oncogene 2013, 32:1943–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baek HJ, Kim SE, Kim JK, Shin DH, Kim TH, Kim KG, Deng C-X, Kim SS: Inhibition of AKT suppresses the initiation and progression of BRCA1-associated mammary tumors. Int J Biol Sci 2018, 14:1769–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M et al. : Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 2007, 446:806–810. [DOI] [PubMed] [Google Scholar]
- 42.Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, Weissman JS, Krogan NJ: Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell 2005, 123:507–519. [DOI] [PubMed] [Google Scholar]
- 43.Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JLY, Toufighi K, Mostafavi S et al. : The genetic landscape of a cell. Science 2010, 327:425–431 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tong AHY: Global mapping of the yeast genetic interaction network. Science 2004, 303:808–813 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 45.Roguev A, Talbot D, Negri GL, Shales M, Cagney G, Bandyopadhyay S, Panning B, Krogan NJ: Quantitative genetic-interaction mapping in mammalian cells. Nat Methods 2013, 10:432–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Du D, Roguev A, Gordon DE, Chen M, Chen S-H, Shales M, Shen JP, Ideker T, Mali P, Qi LS, Krogan NJ: Genetic interaction mapping in mammalian cells using CRISPR interference. Nat Methods 2017, 14:577–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bassik MC, Kampmann M, Lebbink RJ, Wang S, Hein MY, Poser I, Weibezahn J, Horlbeck MA, Chen S, Mann M et al. : A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell 2013, 152:909–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han K, Jeng EE, Hess GT, Morgens DW, Li A, Bassik MC: Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat Biotechnol 2017, 35:463–474 10.1038/nbt.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong ASL, Choi GCG, Cui CH, Pregernig G, Milani P, Adam M, Perli SD, Kazer SW, Gaillard A, Hermann M et al. : Multiplexed barcoded CRISPR-Cas9 screening enabled by CombiGEM. Proc Natl Acad Sci U S A 2016, 113:2544–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen JP, Zhao D, Sasik R, Luebeck J, Birmingham A, Bojorquez-Gomez A, Licon K, Klepper K, Pekin D, Beckett AN et al. : Combinatorial CRISPR–Cas9 screens for de novo mapping of genetic interactions. Nat Methods 2017, 14:573–576 10.1038/nmeth.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosenbluh J, Mercer J, Shrestha Y, Oliver R, Tamayo P, Doench JG, Tirosh I, Piccioni F, Hartenian E, Horn H et al. : Genetic and proteomic interrogation of lower confidence candidate genes reveals signaling networks in β-catenin-active cancers. Cell Syst 2016, 3:302–316.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52••. Horlbeck MA, Xu A, Wang M, Bennett NK, Park CY, Bogdanoff D, Adamson B, Chow ED, Kampmann M, Peterson TR et al. : Mapping the genetic landscape of human cells. Cell 2018, 174:953–967.e22.This article depicts one of the largest genetic interaction screens completed in human cells. They used a pooled dual-sgRNA CRISPR interference (dCas9-KRAB) loss-of-function lentiviral screen in K526 and Jurkat cells to perturb 222 784 gene pairs. The authors report this as being a fourfold increase over prior genetic interaction screens. Highly correlated GI pairs are found to be enriched for physical interactions and they discover TMEM261, a previously poorly characterized gene, as a critical regulator of oxidative phosphorylation.
- 53.Stroud DA, Surgenor EE, Formosa LE, Reljic B, Frazier AE, Dibley MG, Osellame LD, Stait T, Beilharz TH, Thorburn DR, Salim A, Ryan MT: Accessory subunits are integral for assembly and function of human mitochondrial complex I. Nature 2016, 538:123–126. [DOI] [PubMed] [Google Scholar]
- 54.Beltrao P, Cagney G, Krogan NJ: Quantitative genetic interactions reveal biological modularity. Cell 2010, 141:739–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cowley GS, Weir BA, Vazquez F, Tamayo P, Scott JA, Rusin S, East-Seletsky A, Ali LD, Gerath WF, Pantel SE et al. : Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci Data 2014, 1:140035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56•. Pan J, Meyers RM, Michel BC, Mashtalir N, Sizemore AE, Wells JN, Cassel SH, Vazquez F, Weir BA, Hahn WC et al. : Interrogation of mammalian protein complex structure, function, and membership using genome-scale fitness screens. Cell Syst 2018, 6:555–568.e7.This article utilized a computational approach to analyze genetic interaction (GI) data from Project Achilles [56]. By clustering genes according to correlations in GI profiles, they found ~40% of CORUM complexes to possess significant GI correlations, which formed connected functional modules. They used this approach to resolve novel functional modules within complexes lacking structural resolution, such as the SWI/SNF complex. By overlaying functional connectivity data with protein–protein interaction data from hu.MAP, they identified novel protein complexes with no known literature annotation. Specifically, they identified C16orf59 and C14orf80 as forming a heterodimeric complex with delta and epsilon tubulins, implicating them in regulating cell division and motility.
- 57.Giurgiu M, Reinhard J, Brauner B, Dunger-Kaltenbach I, Fobo G, Frishman G, Montrone C, Ruepp A: CORUM: the comprehensive resource of mammalian protein complexes-2019. Nucleic Acids Res 2019, 47:D559–D563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58••. Rauscher B, Heigwer F, Henkel L, Hielscher T, Voloshanenko O, Boutros M: Toward an integrated map of genetic interactions in cancer cells. Mol Syst Biol 2018, 14:e7656.This article discusses the creation of a novel computational framework, called MINGLE, to integrate data from CRISPR screens performed across 60 cancer cell lines with mutation, copy number, and mRNA expression data from COSMIC and CCLE. They developed a two-step normalization process to compare across distinct genetic interaction (GI) maps, generating a finalized list of 17 545 predicted gene–gene interactions (FDR < 20%). Clustering genes by GI profiles identified network modules with similar functional characteristics. Furthermore, they identified novel positive regulators of the Wnt/β-Catenin signaling pathway. Importantly, this work creates computational standards for how to integrate, and maximize the use of, the growing number of large-scale CRISPR screens.
- 59.Califano A, Butte AJ, Friend S, Ideker T, Schadt E: Leveraging models of cell regulation and GWAS data in integrative network-based association studies. Nat Genet 2012, 44:841–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ideker T, Dutkowski J, Hood L: Boosting signal-to-noise in complex biology: prior knowledge is power. Cell 2011, 144:860–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61••. Krogan NJ, Lippman S, Agard DA, Ashworth A, Ideker T: The cancer cell map initiative: defining the hallmark networks of cancer. Mol Cell 2015, 58:690–698.In this article, we describe the Cancer Cell Map Initiative (CCMI), an effort driven by multiple investigators at UCSF and UC San Diego, aimed at systematically detailing the complex interactions among frequently hijacked cancer genes and how they differ between diseased and healthy states. CCMI is motivated by genomic observations that cancer frequently exploits a recurring set of hallmark cancer pathways and networks. The effort seeks to contextualize mutations, seemingly rare when viewed independently, into a smaller number of protein complexes, signaling cascades, and transcriptional circuits. By integrating protein-protein and genetic interaction mapping approaches, as well as predictive network modeling, and structural work on newly identified pathway connections, the long-term goal of the project is to create Cancer Cell Maps against which a patient’s mutations can be queried in order to predict the most effective therapeutic regimen.
- 62.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A et al. : Proteomics. Tissue-based map of the human proteome. Science 2015, 347 1260419. [DOI] [PubMed] [Google Scholar]
- 63.Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, Gill S, Harrington WF, Pantel S, Krill-Burger JM et al. : Defining a cancer dependency map. Cell 2017, 170:564–576. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]