Abstract
Protein synthesis is one of the most energy demanding cellular processes. The ability to regulate protein synthesis is essential for cells under normal as well as stress conditions, such as nutrient deficiencies. One mechanism for protein synthesis suppression is the dimerization of ribosomes into hibernation complexes. In most cells, this process is promoted by the hibernating promoting factor (HPF) and in a small group of Gram‐negative bacteria (γ‐proteobacteria), the dimer formation is induced by a shorter version of HPF (HPFshort) and by an additional protein, the ribosome modulation factor. In most bacteria, the product of this process is the 100S ribosome complex. Recent advances in cryogenic electron microscopy methods resulted in an abundance of detailed structures of near atomic resolutions 100S complexes that allow for a better understanding of the dimerization process and the way it inhibits protein synthesis. As ribosomal dimerization is vital for cell survival, this process is an attractive target for the development of novel antimicrobial substances that might inhibit or stabilize the complex formation. As different dimerization processes exist among bacteria, including pathogens, this process may provide the basis for species‐specific design of antimicrobial agents. Here, we review in detail the various dimerization mechanisms and discuss how they affect the overall dimer structures of the bacterial ribosomes.
Keywords: 100S, hibernation, ribosome, single particle cryo‐EM
Abbreviations
- Anti‐SD
anti Shine–Dalgarno
- A‐tRNA
A‐site bound tRNA
- cryo‐EM
cryogenic electron microscopy
- CTD
C‐terminal domain
- E‐tRNA
tRNA located at the ribosome’s exit site
- HPFlong
long version of the hibernation promoting factor
- HPF short
short version of the hibernation promoting factor
- HPF
hibernation promoting factor
- LSU
Large subunit of the ribosome
- NTD
N‐terminal domain
- PBC
platform binding center
- PTC
peptidyl transferase center
- P‐tRNA
P‐site bound tRNA
- RMF
Ribosome modulation factor
- SD
Shine–Dalgarno
- SSU
small subunit of the ribosome
- tRNA
transfer RNA
Introduction
Ribosome hibernation is a cellular response to stress
Proteins synthesis, which is performed by the ribosome in all living cells, is one of the most energy demanding cellular processes 1, 2, 3. Ribosomes, which are the universal multicomponent ribonucleoprotein assemblies that translate the genetic code into proteins, are comprised of two structurally independent subunits of unequal sizes that associate for creating the functioning ribosome. Within the active ribosome, the small subunit (SSU) binds the mRNA and provides the decoding site. The large subunit (LSU) contains the ribosomal catalytic site, namely the peptidyl transferase center (PTC), in which peptide bonds are being formed, and the exit tunnel through which the nascent proteins emerge out of the ribosome (Fig. 1A). The ability to control the pace of protein synthesis is essential for cell survival during the exponential phase as well as under stress conditions 4. One of such translation‐suppressing mechanism is the dimerization of two ribosomes into hibernation complexes (100S in prokaryotes and 110S in eukaryotes). This process benefits from the ribosomal internal flexibility, which is needed for protein synthesis processivity 5, and has been observed in bacterial, plastids 6, 7, 8, 9, 10 and mammalian cells 11. In most cells, it is facilitated by a hibernating promoting factor (HPF) 10, 12, 13. The importance of the dimer formation for cell survival has been demonstrated in several systems. Among them, HPF knock‐out in Staphylococcus aureus causes ribosome breakdown upon entering the stationary phase that correlates with the onset of cell death and attenuated virulence 14, 15. Deletion of HPF in Lactococcus lactis resulted in decreased viability after resuscitation from starvation conditions 8. Similarly, HPF‐depleted Bacillus subtilis cells' ability to regrow from the stationary phase has been decreased 12. In addition, deletion of hpf in Listeria monocytogenes impairs the survival of the bacteria in a murine model of infection 7 and compromises its tolerance to aminoglycosides 16. Ribosome hibernation is a reversible process. It has been shown that in S. aureus, 100S complexes are disassembled by a GTPase HflX under heat stress and a hitherto unknown major dissociation factor 17, 18. A minor, yet clinically important group of Gram‐negative bacteria (γ‐proteobacteria) that includes Escherichia coli, Salmonella typhimurium, Yersinia pestis and Pseudomonas aeruginosa, carries a shorter form of HPF (HPFshort). In E. coli, HPFshort by itself does not induce dimer formation. Instead, it works in concert with the ribosome modulation factor (RMF) such that RMF (that is found only in species with HPFshort) induces dimerization of two ribosomes, and forms immature 90S complex. Then the following binding of HPF converts 90S ribosomes to a mature, inactive 100S complex 19, 20, 21, 22, 23, 24. γ‐Proteobacteria also carry an additional protein, YfiA, which binds and inactivates 70S ribosomes 25, 26. YfiA is an antagonist to HPFshort and RMF by preventing 100S formation 19, 21. Early cryo electron microscopy (cryo‐EM) studies yielded structures of ribosomal dimers at rather low resolution, sufficient only to demonstrate that the two 70S ribosomes are bound to each other via their SSU and that the dimer interface has some degree of flexibility 8, 27, 28, 29.
Figure 1.
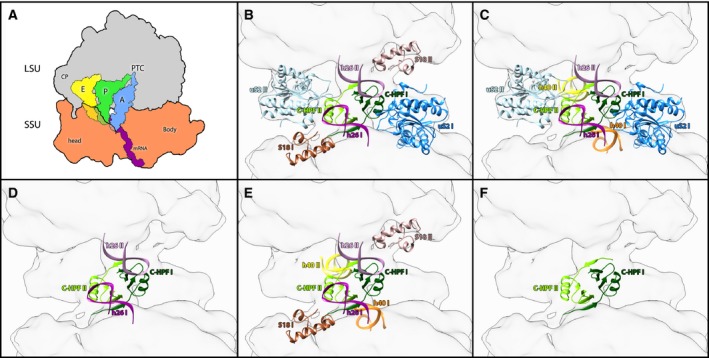
Species specificity of dimer interface of various bacterial systems. (A) Simplified scheme of the translating ribosome. The LSU in gray, SSU in orange, A‐site (A), P‐site (P) and E‐site (E) tRNA are colored in blue, green and yellow, respectively. The mRNA is marked in purple. The PTC, mRNA, central protuberance and the head and body domain in the SSU are also marked. (B–F) The specificity of dimer interface is demonstrated by highlighting the ribosomal components that form part of dimer interface of each species: Bacillus subtilis, Staphylococcus aureus 34 , S. aureus 33, L. lectococcus and T. thermpophilus, respectively. h26 is colored magenta‐sienna, h40 is colored orange‐yellow, C‐terminal HPF is colored green‐dark green, uS2 is colored blue‐light blue and s18 is colored brown‐rosy brown.
Results and Discussion
Structural studies decipher species‐specific ribosome dimerization
Recent advances in cryo‐EM methods resulted in an abundance of detailed structures of the 100S complexes at near atomic resolutions (3.0–5.9 Å for an individual 70S composing the dimer and 4.1–11 Å for the whole dimer) 30, 31, 32, 33, 34, 35, 36. These demonstrated that in all systems dimer formation benefits from the ribosomal inherent flexibility. They also revealed not only fundamental differences between dimers that include HPFshort and RMF and those that include only a long version of HPF (HPFlong) but also indicated both subtle and significant differences among the latter dimer type of the various bacterial species. Furthermore, the structures of the 100S hibernation complexes of B. subtilis 30, S. aureus 33, 34, L. lactis 32, and Thermus thermophilus 31 that carry HPFlong showed that this protein has two conserved functional domains [N‐terminal domain (NTD), and C‐terminal domain (CTD)] that are linked by a flexible linker. In all of the mentioned above structures, HPFlong‐NTD is bound at the space between the ‘head’ and ‘body’ (Fig. 1A) of the SSU, near the subunit interface and adopts α/β fold (β1‐α1‐β2‐β3‐β4‐α2). The binding of HPFlong‐NTD to the ribosome causes steric hindrance for transfer RNA (tRNA) and mRNA binding, thus inactivating the ribosome. HPFlong‐NTD is stabilized in its pocket by interacting with the 16S rRNA and various ribosomal proteins. Once bound, HPFlong‐NTD also overlaps with the binding sites of several antibiotics that bind the SSU (e.g. hygromycin B, tetracycline, edeine, kasugamycin, and pactamycin).
The HPFlong‐CTD is bound to the interface between the two ribosomes comprising the dimer in all the structures mentioned above. One HPFlong‐CTD creates a homodimer with the HPFlong‐CTD of the second ribosome of the 100S complex. Mainly electrostatic interactions and domain swapping stabilize the HPFlong‐CTD homodimer. Interestingly, the first β strand of one HPFlong‐CTD crosses over and forms hydrophobic interactions with the last β strand of the other HPFlong‐CTD (Fig. 1F). HPFlong‐CTD homodimerization is the major contributor to the dimer formation as it is the single trait that is shared among all the 100S structures induced by HPFlong.
In most 100S structures, the dimer interface was found to be further stabilized by additional inter‐ribosomal interactions (Fig. 1B–F). In B. subtilis, HPFlong‐CTD of one ribosome further interacts with the N terminus of ribosomal protein bS18 of the second ribosome. In addition, the N‐terminal β‐hairpin and proximal region of the α2‐helix of protein uS2 establish a large interaction surface with the stem‐loop of helix h26 of the 16S rRNA 30. A similar interaction between uS2 and h26 was also observed in S. aureus 100S; however, in this structure bS18 does not seem to be involved in the dimer interface connections; instead, h40 interacts with HPFlong‐CTD 34. Additionally, in another S. aureus 100S structure, the HPFlong‐CTD does not interact with any other ribosomal components, whereas h26 of one ribosome interacts with its counter h26 instead of uS2 33. The finding that two independent studies on the same particle yielded different structure suggests that the 100S may adopt various conformations, thus further illustrating the flexibility of the dimer interface. On the other hand, it is plausible that this variation could result from the different experimental procedures employed for the creation of the dimer. In one study 34 the dimer was purified from a S. aureus strain carrying a high copy plasmid with the hpf gene, resulting in an increased amount of 100S ribosomes, whereas in the other study 33, a recombinant HPF was introduced to purified 70S ribosome at a 1 : 1 ratio. Similar to the first 100S structure mentioned above, in L. lactis the HPFlong‐CTD of one ribosome is interacting with h40 (and probably uS18); however, in this structure h26 is not interacting with uS2. Instead it interacts with the h26 of the opposite ribosome 32, like in the second 100S structure. In T. thermophilus, 100S formation is solely attributed to HPFlong‐CTD dimerization and not to any additional inter‐ribosome interactions 31. In most 100S dimer structures (excluding T. thermophilus), h26 forms a part of the dimer interface. In T. thermophilus, h26 is shorter compared to h26 in the other species (the differences are 30 nucleotides in S. aureus and B. subtilis, 29 in L. lactis and 23 in T. thermophilus), thus h26 of one ribosome is located too far from any other ribosomal component of the other ribosome in the dimer and therefore cannot form any contacts that may contribute to the dimer stabilization.
The linker that connects the two domains of HPFlong could not be detected in most of the available 100S structures. This finding has been attributed to the flexibility of this linker. The sequences of both NTD and CTD domains of HPFlong are rather conserved among different bacteria, whereas the sequence and composition of the linker is hardy conserved 32. It was postulated that the linker might traverse the RMF binding site 30 thus interfering with proper helix formation between Shine–Dalgarno (SD) and anti‐SD sequence during translation initiation, hence causing inhibition of translation.
Mechanisms for dimerization
Long version of HPF is a homodimer in solution 10, 14, 30, 31, 32, 33, 34. Therefore, it has been suggested that within the cell environment, HPFlong is present at a dimeric state and the long linker of the dimeric HPFlong enables the HPFlong‐NTD to interact with two independent 70S ribosomes, bringing them into close proximity (Fig. 2A). Then HPFlong‐CTD that is in a dimeric form binds to the other ribosomal component at the 30S subunit rims (depending on the type of 100S complex) to further stabilize the ribosomal dimers 30, 31, 32. Alternatively, one cannot exclude the possibility that in the cell, HPFlong is a monomer and upon binding to the ribosome, dimerization occurs via homodimerization of the HPFlong‐CTD.
Figure 2.
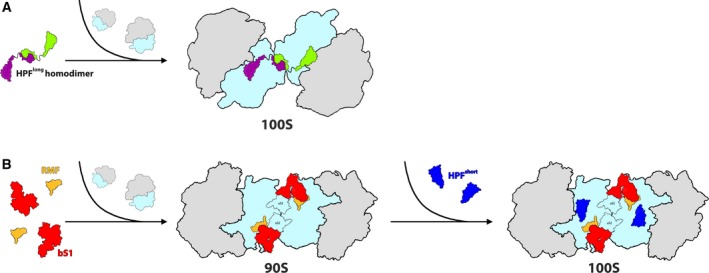
Overview of hibernation modes across bacteria. (A) 100S Ribosome formation is mediated by the binding of HPFlong‐NTD (colored in green and purple to differentiate between each HPF molecule which binds to a different ribosome) to the ribosomal SSU (colored in light blue). Dimerization of the two ribosomes is facilitated by homodimerization of two HPFlong‐CTDs. Secondary contacts may involve other ribosomal components, depending on the species. LSU is colored in gray. (B) Ribosome hibernation in Escherichia coli (and other γ‐proteobacteria). The initial binding of RMF and rProtein bS1 (colored in yellow and red, respectively) to the ribosome induces the formation of the immature 90S complex. Later HPFshort (colored in blue), binds at the catalytic site of the SSU (colored in light blue), resulting in the formation of the hibernating 100S dimer. LSU is colored in gray.
The presence of HPFlong does not necessarily result in ribosome dimer formation, as demonstrated by the structures of ribosomes from Mycobacterium smegmatis 37, 38 (which is frequently used as a model of Mycobacterium tuberculosis) and the spinach chloro‐ribosome 35, in complex with HPFlong. Both organisms carry HPFlong; but ribosomal dimers have not been reported in these species. In both structures, the presence of ribosomal proteins S1 (bS1 in M. smegmatis and bS1c in spinach) is suggested to be hindering the dimer formation by steric clashes. Despite being the largest of the ribosomal proteins (61 kDa), only parts of bS1 have been observed in crystal structures and in cryo‐EM reconstructions of various structures of the SSU, due to its weak interactions with the ribosome 39, 40, 41, 42, 43. In the chloroplasts, bS1c is tightly associated with the chloro‐ribosome and M. smegmatis has an rRNA extension, H54a, a unique feature of mycobacteria, which seems to bind to bS1 and stabilize it, as well as H54a itself. Sequence comparison of HPFlong‐CTD from M. smegmatis, M. tuberculosis, S. aureus, and L. lactis shows that one out of the five conserved residues proposed to be involved in the formation of 100S dimers is not conserved in Mycobacteria 32, 37. However, it is not clear that this is the reason that prevents 100S dimer formation in M. smegmatis. The structure of M. smegmatis in complex with HPFlong also suggests a small difference in the HPFlong‐NTD binding site since it does not clash with the binding site of tRNA located at the ribosome’s exit site (E‐tRNA) (unlike the previously described dimers), as indicated by the existence of a density for the E‐tRNA that has been observed in the M. smegmatis cryo‐EM maps.
As previously mentioned, γ‐proteobacteria carry the HPFshort version and along with RMF, they induce the 100S dimer formation. HPFshort is very similar to HPFlong‐NTD in terms of both sequence and structure, as demonstrated by the structure of E. coli 100S dimer 36. They also occupy the same binding pocket in all the structures mentioned above. However, in E. coli, HPFshort directly interacts with E‐site tRNA, similar to what has been observed in the M. smegmatis structure. Unlike HPFshort, RMF shares minor similarities to HPFlong‐CTD, which is bound at different locations on the ribosome. RMF interacts with both rRNA and proteins and it is located on the back of the small ribosomal subunit. RMF stabilizes a defined conformation of the 3' end of the 16S rRNA, which encompasses part of the anti‐SD sequence, similarly to the HPFlong linker. In the past, it was postulated that since neither HPF nor RMF is directly involved in the interactions of the two ribosomes, the dimer is formed due to conformational changes induced by the binding of these two proteins. This theory proved to be incorrect as no conformational changes, namely the swiveling of the SSU head, were detected. Instead, the cavity in which RMF is bound is capped by a large mass of additional density that was attributed to bS1. In the E. coli 100S structure, bS1 was found in an inactive, compacted conformation such that the C terminus of bS1 is folded back onto the 30S subunit, rather than extending into the solvent. The E. coli 100S is formed by two major bridges at the dimer interface where bS1 and uS2 from each 30S interact with uS4 and uS3, respectively, of the symmetry‐related SSU (Fig. 2B). Additionally, the C terminus of uS2 from one ribosome extends toward the mRNA entrance channel of its counterpart, thus effectively plugging the mRNA entrance channel, suggesting that such an interaction would not be possible on an already active translating ribosome to which mRNA is bound. The existence of two different mechanisms of ribosome dimerization (Fig. 2) and the conservation of protein uS2 as the main interaction partner support the idea that dimerization has been rescued in γ‐proteobacteria by the rmf gene upon loss of the CTD of HPFlong 32. Yet, it is still unclear why the presence of bS1 would promote dimer formation in γ‐proteobacteria, whereas in M. smegmatis and spinach that carry the HPF long version would hinder the dimerization. Perhaps the presence of the HPFlong‐CTD does not allow the two ribosomes to bind to each other in a manner that is similar to E. coli 100S.
Ribosome hibernation heterogeneity as a potential drug target
Structures of various HPFlong‐mediated 100S ribosomes determined by cryo‐EM show a high degree of conformational homogeneity, with the exception of T. thermophilus 100S that is staggered slightly differently (Fig 3A,B). The reason for this is probably the lack of inter‐ribosomal interactions additional to the HPFlong‐CTD homodimerization. In addition, head swiveling of the SSU, induced by HPFlong binding was observed only in one S. aureus study 34, benefiting from the inherent flexibility of the ribosomal SSU 44. In contrast, the HPFlong‐mediated 100S dimers have markedly different conformations, compared to the E. coli 100S dimer (mediated by HPFshort and RMF) (Fig. 3C,D). Whereas E. coli 100S complex is created by ‘back‐to‐back’ interactions of the SSUs of each ribosome, the other HPFlong‐mediated 100S complexes involve more ‘side‐to‐side’ (platform‐to‐platform) interactions of the SSU.
Figure 3.
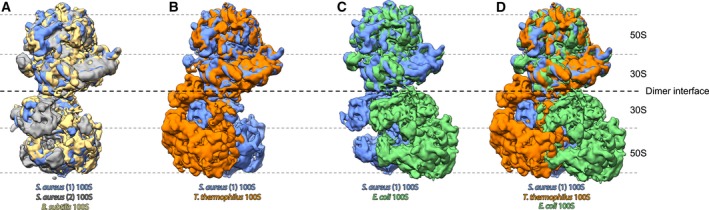
Conformations of 100S structures from different bacteria. Superposition of (A) Bacillus subtilis 100S (EMD‐3664, yellow) and two independently studied Staphylococcus aureus 100S (EMD‐3637, gray and EMD‐3638, blue) structures; (B) Thermus thermophilus 100S (colored orange) and S. aureus 100S (EMD‐3638, blue) structures; (C) Escherichia coli 100S (EMD‐0139, green) and S. aureus 100S (EMD‐3638, blue) structures; and (D) T. thermophilus 100S, E. coli 100S and S. aureus 100S (same coloring as A–C) structures. Both S. aureus 100S and B. subtilis 100S structures show a high degree of conformational conservation while T. thermophilus and E. coli 100S ribosomes are clearly staggered in a different conformation (thus, in B‐D, only one of the S. aureus 100S is shown as a representative of the second S. aureus 100S as well as B. subtilis 100S).
The HPFlong‐NTD by itself is sufficient for silencing the ribosomes as well as HPFshort and RMF that are in fact not directly involved in the dimer interface. These structural findings raise the question: what is the biological significance of the ribosomal 100S dimer? As mentioned above, in E. coli 100S, the arrangement of the dimer allows for one N terminus of uS2 to block the mRNA entrance channel of the other ribosome. In addition, 100S dimers are less susceptible to degradation by RNases 39, 41, 43, 44. Nevertheless, the reason for the 100S formation is not well understood yet. Nevertheless, several hypotheses have been raised. Since 100S formation does not significantly alter the large rRNA surface exposed to RNases, it was suggested 30 that 100S formation may interfere with a specific ribosome degradation pathway, rather than preventing nonspecific RNase action on ribosomes. In addition, bacterial translation and transcription are coupled and the binding sites (e.g., uS2) of RNA polymerase on the 70S ribosome overlap with the 70S dimerization interface 40, 45, thus decreasing the translation activity 31, 34. Finally, proteins uS2, uS7, and uS11, along with rRNA helices 26 and 40, are part of the mRNA ‘platform binding center’ (PBC) 5, 17. This platform has been proposed to be a common site dedicated to mRNAs binding prior to the actual translation initiation to increase control over translation 5. uS2 was shown to possess a high level of flexibility and is involved in SSU dimerization in crystals 5. It takes active part in the dimer formation (as well as h26 and h40 in some of the 100S complexes reported above). Franken et al 32 propose that the interface of the dimer blocks the PBC by acting as a general, rather than a specific, inhibitor of mRNA translation initiation. Whether or not the 100S dimer formation does prevent protein synthesis in all or in some of the manners suggested above, it is clear that the hibernation complex manages this process by multiple mechanisms.
As the hibernation pathway is crucial for cell survival, it represents an attractive target for the development of novel antimicrobial agents. As different dimerization processes exist among different bacteria, including pathogens, this process may provide the basis for the species‐specific design of antimicrobial agents. Such potential inhibitors could interfere with HPF dimerization either by perturbing the function or of the critical flexible linker, or by competing with the HPF binding to the ribosomes. In addition, the stimulation of 100S disassembly factor activity may be used. Thus, bacterial growth inhibition by targeting 100S assembly or disassembly opens a new path for developing novel antibacterial therapy.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
All authors participated in the interpretation of the results and in writing the article.
Acknowledgements
We thank the entire Yonath group for their interest and experimental support; MNY was supported by the National Institutes of Health grant GM121359. AY was supported by European Research Council Grants 322581 (NOVRIB) and the Kimmelman Center for Macromolecular Assemblies. AY holds the Martin S. and Helen Kimmel Professorial Chair at the Weizmann Institute of Science.
References
- 1. Buttgereit F & Brand MD (1995) A hierarchy of ATP‐consuming processes in mammalian cells. Biochem J 312, 163–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Russell JB & Cook GM (1995) Energetics of bacterial growth: balance of anabolic and catabolic reactions. Microbiol Rev 59, 48–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Szaflarski W & Nierhaus KH (2007) Question 7: optimized energy consumption for protein synthesis. Orig Life Evol Biosph 37, 423–428. [DOI] [PubMed] [Google Scholar]
- 4. Starosta AL, Lassak J, Jung K & Wilson DN (2014) The bacterial translation stress response. FEMS Microbiol Rev 38, 1172–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bashan A & Yonath A (2008) The linkage between ribosomal crystallography, metal ions, heteropolytungstates and functional flexibility. J Mol Struct 890, 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yoshida H & Wada A (2014) The 100S ribosome: ribosomal hibernation induced by stress. Wiley Interdiscip Rev RNA 5, 723–732. [DOI] [PubMed] [Google Scholar]
- 7. Kline BC, McKay SL, Tang WW & Portnoy DA (2015) The Listeria monocytogenes hibernation‐promoting factor is required for the formation of 100S ribosomes, optimal fitness, and pathogenesis. J Bacteriol 197, 581–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Puri P, Eckhardt TH, Franken LE, Fusetti F, Stuart MC, Boekema EJ, Kuipers OP, Kok J & Poolman B (2014) Lactococcus lactis YfiA is necessary and sufficient for ribosome dimerization. Mol Microbiol 91, 394–407. [DOI] [PubMed] [Google Scholar]
- 9. Tagami K, Nanamiya H, Kazo Y, Maehashi M, Suzuki S, Namba E, Hoshiya M, Hanai R, Tozawa Y & Morimoto T (2012) Expression of a small (p) ppGpp synthetase, YwaC, in the (p) ppGpp0 mutant of Bacillus subtilis triggers YvyD‐dependent dimerization of ribosome. Microbiologyopen 1, 115–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ueta M, Wada C, Daifuku T, Sako Y, Bessho Y, Kitamura A, Ohniwa RL, Morikawa K, Yoshida H & Kato T (2013) Conservation of two distinct types of 100S ribosome in bacteria. Genes Cells 18, 554–574. [DOI] [PubMed] [Google Scholar]
- 11. Krokowski D, Gaccioli F, Majumder M, Mullins MR, Yuan CL, Papadopoulou B, Merrick WC, Komar AA, Taylor DJ & Hatzoglou M (2011) Characterization of hibernating ribosomes in mammalian cells. Cell Cycle 10, 2691–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Akanuma G, Kazo Y, Tagami K, Hiraoka H, Yano K, Suzuki S, Hanai R, Nanamiya H, Kato‐Yamada Y & Kawamura F (2016) Ribosome dimerization is essential for the efficient regrowth of Bacillus subtilis . Microbiology 162, 448–458. [DOI] [PubMed] [Google Scholar]
- 13. Ueta M, Wada C & Wada A (2010) Formation of 100S ribosomes in Staphylococcus aureus by the hibernation promoting factor homolog SaHPF. Genes Cells. 15, 43–58. [DOI] [PubMed] [Google Scholar]
- 14. Basu A & Yap M‐NF (2016) Ribosome hibernation factor promotes Staphylococcal survival and differentially represses translation. Nucleic Acids Res 44, 4881–4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Basu A, Shields KE, Eickhoff CS, Hoft DF & Yap MN (2018) Thermal and nutritional regulation of ribosome hibernation in Staphylococcus aureus . J Bacteriol 200, e00426–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McKay SL & Portnoy DA (2015) Ribosome hibernation facilitates tolerance of stationary‐phase bacteria to aminoglycosides. Antimicrob Agents Chemother 59, 6992–6999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Basu A & Yap M‐NF (2017) Disassembly of the Staphylococcus aureus hibernating 100S ribosome by an evolutionarily conserved GTPase. Proc Natl Acad Sci 114, E8165–E8173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gohara DW & Yap M‐NF (2018) Survival of the drowsiest: the hibernating 100S ribosome in bacterial stress management. Curr Genet 64, 753–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maki Y, Yoshida H & Wada A (2000) Two proteins, YfiA and YhbH, associated with resting ribosomes in stationary phase Escherichia coli . Genes Cells 5, 965–974. [DOI] [PubMed] [Google Scholar]
- 20. Ueta M, Ohniwa RL, Yoshida H, Maki Y, Wada C & Wada A (2008) Role of HPF (hibernation promoting factor) in translational activity in Escherichia coli . J Biochem 143, 425–433. [DOI] [PubMed] [Google Scholar]
- 21. Ueta M, Yoshida H, Wada C, Baba T, Mori H & Wada A (2005) Ribosome binding proteins YhbH and YfiA have opposite functions during 100S formation in the stationary phase of Escherichia coli . Genes Cells 10, 1103–1112. [DOI] [PubMed] [Google Scholar]
- 22. Wada A, Igarashi K, Yoshimura S, Aimoto S & Ishihama A (1995) Ribosome modulation factor: stationary growth phase‐specific inhibitor of ribosome functions from Escherichia coli . Biochem Biophys Res Commun 214, 410–417. [DOI] [PubMed] [Google Scholar]
- 23. Yamagishi M, Matsushima H, Wada A, Sakagami M, Fujita N & Ishihama A (1993) Regulation of the Escherichia coli rmf gene encoding the ribosome modulation factor: growth phase‐and growth rate‐dependent control. EMBO J 12, 625–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wada A, Yamazaki Y, Fujita N & Ishihama A (1990) Structure and probable genetic location of a" ribosome modulation factor" associated with 100S ribosomes in stationary‐phase Escherichia coli cells. Proc Natl Acad Sci 87, 2657–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Agafonov DE, Kolb VA, Nazimov IV & Spirin AS (1999) A protein residing at the subunit interface of the bacterial ribosome. Proc Natl Acad Sci 96, 12345–12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vila‐Sanjurjo A, Schuwirth B‐S, Hau CW & Cate JH (2004) Structural basis for the control of translation initiation during stress. Nat Struct Mol Biol 11, 1054. [DOI] [PubMed] [Google Scholar]
- 27. Kato T, Yoshida H, Miyata T, Maki Y, Wada A & Namba K (2010) Structure of the 100S ribosome in the hibernation stage revealed by electron cryomicroscopy. Structure 18, 719–724. [DOI] [PubMed] [Google Scholar]
- 28. Ortiz JO, Brandt F, Matias VR, Sennels L, Rappsilber J, Scheres SH, Eibauer M, Hartl FU & Baumeister W (2010) Structure of hibernating ribosomes studied by cryoelectron tomography in vitro and in situ . J Cell Biol 190, 613–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yoshida H, Maki Y, Kato H, Fujisawa H, Izutsu K, Wada C & Wada A (2002) The ribosome modulation factor (RMF) binding site on the 100S ribosome of Escherichia coli . J Biochem 132, 983–989. [DOI] [PubMed] [Google Scholar]
- 30. Beckert B, Abdelshahid M, Schäfer H, Steinchen W, Arenz S, Berninghausen O, Beckmann R, Bange G, Turgay K & Wilson DN (2017) Structure of the Bacillus subtilis hibernating 100S ribosome reveals the basis for 70S dimerization. EMBO J 36, 2061–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Flygaard RK, Boegholm N, Yusupov M & Jenner LB (2018) Cryo‐EM structure of the hibernating Thermus thermophilus 100S ribosome reveals a protein‐mediated dimerization mechanism. Nat Commun 9, 4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Franken LE, Oostergetel GT, Pijning T, Puri P, Arkhipova V, Boekema EJ, Poolman B & Guskov A (2017) A general mechanism of ribosome dimerization revealed by single‐particle cryo‐electron microscopy. Nat Commun 8, 722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Khusainov I, Vicens Q, Ayupov R, Usachev K, Myasnikov A, Simonetti A, Validov S, Kieffer B, Yusupova G & Yusupov M (2017) Structures and dynamics of hibernating ribosomes from Staphylococcus aureus mediated by intermolecular interactions of HPF. EMBO J 36, 2073–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Matzov D, Aibara S, Basu A, Zimmerman E, Bashan A, Yap M‐NF, Amunts A & Yonath AE (2017) The cryo‐EM structure of hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus . Nat Commun 8, 723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Boerema AP, Aibara S, Paul B, Tobiasson V, Kimanius D, Forsberg BO, Wallden K, Lindahl E & Amunts A (2018) Structure of the chloroplast ribosome with chl‐RRF and hibernation‐promoting factor. Nat Plant 4, 212. [DOI] [PubMed] [Google Scholar]
- 36. Beckert B, Turk M, Czech A, Berninghausen O, Beckmann R, Ignatova Z, Plitzko JM & Wilson DN (2018) Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1. Nature Microbiol 3, 1115‐1121. [DOI] [PubMed] [Google Scholar]
- 37. Mishra S, Ahmed T, Tyagi A, Shi J & Bhushan S (2018) Structures of Mycobacterium smegmatis 70S ribosomes in complex with HPF, tmRNA, and P‐tRNA. Sci Rep 8, 13587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li Y, Sharma MR, Koripella RK, Yang Y, Kaushal PS, Lin Q, Wade JT, Gray TA, Derbyshire KM, Agrawal RK & et al (2018) Zinc depletion induces ribosome hibernation in mycobacteria. Proc Natl Acad Sci USA 115, 8191–8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Byrgazov K, Grishkovskaya I, Arenz S, Coudevylle N, Temmel H, Wilson DN, Djinovic‐Carugo K & Moll I (2014) Structural basis for the interaction of protein S1 with the Escherichia coli ribosome. Nucleic Acids Res 43, 661–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Demo G , Rasouly A , Vasilyev N , Svetlov V , Loveland AB , Diaz‐Avalos R , Grigorieff N , Nudler E & Korostelev AA (2017) Structure of RNA polymerase bound to ribosomal 30S subunit. eLife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sengupta J, Agrawal RK & Frank J (2001) Visualization of protein S1 within the 30S ribosomal subunit and its interaction with messenger RNA. Proc Natl Acad Sci 98, 11991–11996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I & Franceschi F (2000) Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell 102, 615–623. [DOI] [PubMed] [Google Scholar]
- 43. Wimberly BT, Brodersen DE, Clemons WM Jr, Morgan‐Warren RJ, Carter AP, Vonrhein C, Hartsch T & Ramakrishnan V (2000) Structure of the 30S ribosomal subunit. Nature 407, 327. [DOI] [PubMed] [Google Scholar]
- 44. Zhang W, Dunkle JA & Cate JH (2009) Structures of the ribosome in intermediate states of ratcheting. Science 325, 1014–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kohler R, Mooney R, Mills D, Landick R & Cramer P (2017) Architecture of a transcribing‐translating expressome. Science 356, 194–197. [DOI] [PMC free article] [PubMed] [Google Scholar]


