Abstract
Cell dielectrophoretic responses have been extensively studied for biomarker expression, blood typing, sepsis, circulating tumor cell separations, and others. Surfactants are often added to the analytical buffer in electrokinetic cellular microfluidic systems to lower surface/interfacial tensions. In nonelectrokinetic systems, surfactants influence cell size, shape, and agglomeration; this has not been systematically documented in electrokinetic systems. In the present work, the impacts of the Triton X-100 surfactant on human red blood cells (RBCs) were explored via ultraviolet-visible spectroscopy (UV-Vis) and dielectrophoresis (DEP) to compare nonelectrokinetic and electrokinetic responses, respectively. The UV-Vis spectra of Triton X-100 treated RBCs were dramatically different from that of native RBCs. DEP responses of RBCs were compared to RBCs treated with low concentrations of Triton X-100 (0.07–0.17 mM) to ascertain surfactant effects on dielectric properties. A star-shaped electrode design was used to quantify RBC dielectric properties by fitting a single-shell oblate cell model to experimentally-derived DEP spectra. The presence of 0.07 and 0.11 mM of Triton X-100 shifted the RBC’s DEP spectra yielding lower crossover frequencies . The single-shell oblate model revealed that cell radius and membrane permittivity are the dominant influencers of DEP spectral shifts. The trends observed were similar for 0.11 mM and 0.07 mM Triton X-100 treated cells. However, a further increase of Triton X-100 to 0.17 mM caused cells to only exhibit negative DEP. The magnitude of the DEP force increased with Triton X-100 concentration. This work indicates that dynamic surfactant interactions with cell membranes alter cell dielectric responses and properties.
I. INTRODUCTION
Microfluidic platforms leverage micrometer-scale dimensions for molecular and cellular analysis enabling lower sample consumption, lower-costs, and shorter responses and quantification times. Advances in microfluidic platforms could transform patient access via point of care medical diagnostics, the need for which was recently articulated in the World Health Organization's List of Essential In Vitro Diagnostics and included blood hematocrit determination (#1),1 blood typing (#9),2 and malaria diagnosis (#18).3,4
Microfluidic platforms frequently employ electrokinetics due to simplicity and precise molecular control. Dielectrophoresis (DEP) is a powerful and flexible electrokinetic tool capable of discerning molecular expression on cell membranes for blood typing,2 monitoring sepsis,5,6 circulating tumor cell separations,7 and stem cell separations for therapeutic applications.8,9 DEP is field induced polarization and translational motion of polarizable particles or cells in nonuniform AC electric fields.10 Microelectrodes positioned in spatially nonuniform geometries delivering AC potentials have achieved a plethora of particle/cell separations,11 trappings,12 and other manipulations.13–15 Performing DEP within microfluidic platforms allows (sub)cellular electrical property characterizations including determination of permittivity and conductivity of membrane and cytoplasm. DEP frequency dependent responses have been used to obtain cell population distributions of surface biomarkers, cellular function, and biological mechanisms via studying cell's membrane barrier functionality and cytoplasm composition.16,17 However, variations confound reproducibility and comparisons between platforms, cells, or patients.18
Surfactants have been widely used in microfluidic platforms to achieve droplet generation and stabilization,19 lysed cells,20 reduce Joule heating,21 control electrolysis,22 split conglomerated cells/particles,23–26 and prevent biomolecule adsorption to aqueous, nonaqueous, or solid interfaces.26,27 Low concentrations (<1%) of nonionic surfactants are routinely added to cell-buffer solutions to increase accuracy and reproducibility within microfluidic platforms without disrupting protein content of cell membranes or cell viability.1,19–21,28 However, in nonelectrokinetic and nonmicrofluidic systems, surfactant molecules have been observed to interact with the cell membrane by disrupting the normal lipid bilayer architecture and reducing membrane surface tension.28–34 These interactions could alter DEP responses. The first demonstration of tuning DEP behavior with a surfactant was reported using an ionic surfactant, sodium dodecyl sulfate (SDS).26 Upon SDS treatment, the conductivity of cell surface significantly enhanced, making the cells exhibit a strong positive DEP force when the cells previously exhibited a negative DEP force. This result illustrates the importance of evaluating interactions between surfactants and cell membranes and simultaneously illustrates the need to explore the impacts of a nonionic surfactant on cell dielectric properties via DEP analysis.
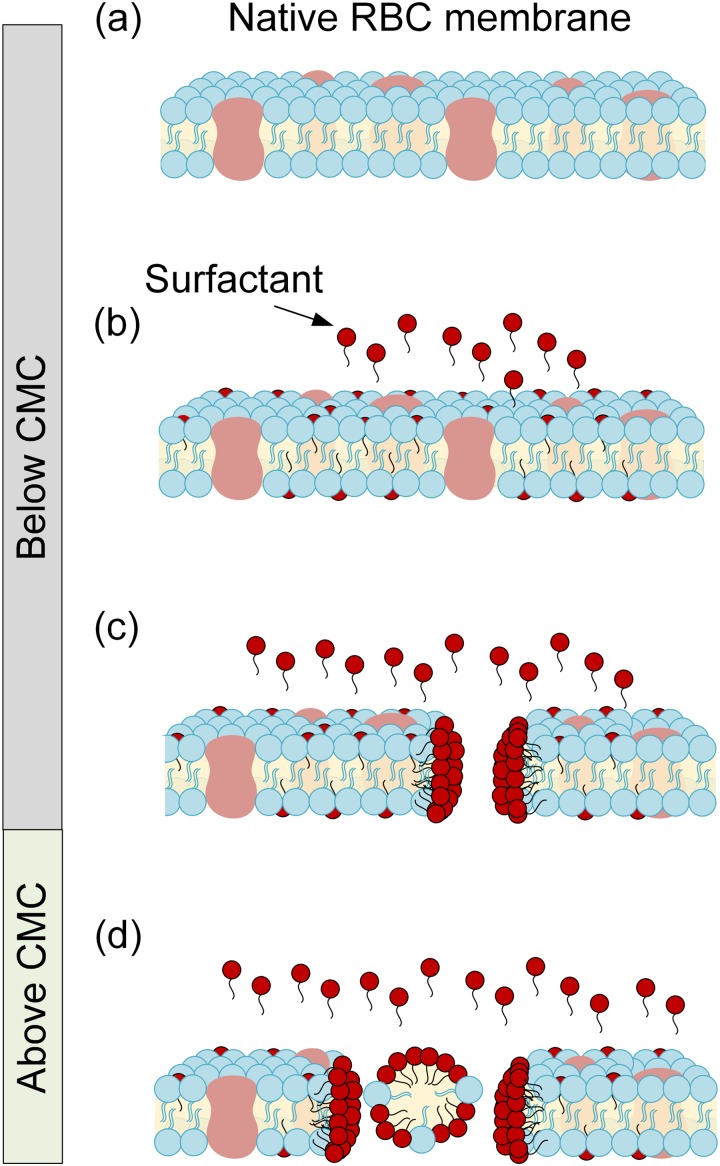
Red blood cells (RBCs) are excellent models to study surfactant interactions with cell components such as membranes and cytoplasm because RBCs have a simpler cell structure lacking a nucleus.35 In nonelectrokinetic and nonmicrofluidic systems, surfactant and cell membrane interactions are concentration dependent.28–34 Above the critical micelle concentration (CMC), surfactants solubilize cell membranes yielding hemolysis, while they protect cell membranes below the CMC.28–34 Figure 1 cartoons the interactions between the RBC membrane and surfactants at concentrations below and above the CMC. This biphasic behavior has been observed for both ionic and nonionic surfactants.28–34,36,37 However, microfluidic applications tend to utilize nonionic surfactants to minimize conductivity disruptions and protein conformational changes. Nonionic examples include poly[oxyethylene (n) nonylphenol],30 polyoxyethylene alkyl ethers,31 octyl-β-D glucopyranoside,32 and Triton.34 Two different mechanisms have been proposed to explain the osmotic resistance (e.g., reduced lysis) of RBCs below the CMC. At low concentrations, surfactant molecules insert into the cell membrane causing membrane cell enlargement by lowering the lateral surface tension. Thus, RBCs swell and increase the critical lytic volume.36,37 The second proposed mechanism is an increase in membrane permeability due to diffusion rate changes through the cell membrane.36,37 Effective membrane permeability is an important parameter in cell electrophysiology, since it ultimately influences membrane dielectric constant and permittivity.38 Thus, we hypothesize that surfactant molecules can intercalate within the RBC's membrane and potentially change both cell radius as well as membrane charge distribution and permittivity, both of which can be discerned from measuring DEP spectra.
FIG. 1.
Schematic of interaction between a red blood cell (RBC) membrane and surfactant molecules at concentrations below and above the critical micelle concentration (CMC). (a) Native RBC membrane schematic in the absence of surfactants. (b) Surfactant molecule intercalation within the RBC membrane, below CMC. (c) Pore formation within the RBC membrane as the surfactant concentration approaches the CMC. (d) Surfactant solubilization of the cell membrane yielding hemolysis above the CMC. The cartoon was adapted from a previous study.28
A. Dielectrophoresis theory and background
Dielectrophoresis utilizes AC signals to manipulate particles or cells within a medium. The dielectrophoretic force on a homogenous spherical particle (a rough first approximation to a cell) is given by10
| (1) |
where is the outer particle (or cell) radius, is the real part of the Clausius–Mossotti factor , and is the electric field gradient squared. The Clausius–Mossotti factor predicting positive and negative DEP responses can be defined as follows:10
| (2) |
where and are the complex permittivity of the particle and medium, respectively. Complex permittivity is dependent upon , where is the permittivity, is the conductivity, is angular frequency, and j is . If the particle's complex permittivity is greater than the complex permittivity of the medium (fCM > 0), the net force on the particle is toward maximum electric field strength regions moving the particle up the electric field gradient with a positive DEP (p-DEP) force. If complex permittivity of the particle is lower than the medium ( fCM < 0), the net force direction is toward the minimum electric field strength regions causing movement down the gradient with a negative DEP (n-DEP) force. The frequency at which the net force is zero and cell responses gradually shift from n-DEP to p-DEP or from p-DEP to n-DEP spectra is termed the crossover frequency .
B. Single-shell oblate DEP polarization model
The magnitude and direction of DEP forces depend highly on cellular shape and composition.39 Deviation from homogeneous cell sphericity significantly reduces accuracy of Eq. (1). RBCs are biconcave disks filled with cytoplasm and no nucleus. Thus, RBCs can be better described as single-shell oblate spheres with relatively conductive cytoplasm surrounded by thin dielectric membranes.40,41 Typically, the dielectric shell model, subjected to DEP forces, exhibits frequency dispersion. RBCs exhibit α- (sub-hertz to kilohertz), β- (kilohertz to megahertz range), and γ- (megahertz to gigahertz range) frequency dispersion. This work explores 300–700 kHz in the β-dispersion region whereby RBC membranes are conductive and polarized via Maxwell–Wagner interfacial polarizations.10 Charge distributions on membrane surfaces change due to dynamic polarization effects as the frequency changes. Treating RBCs as single-shell oblate spheroids with a long semimajor r radius and a semiminor c radius, where c is the cell half-length, results in a DEP force expression as42
| (3) |
where is the cell volume and the single-shell oblate spheroid is42
| (4) |
where is depolarization from outside the membrane to infinity, which reflects the effects of outer-membrane (shell) interactions and distortions to the local electrical field. For a single-shell oblate model, the complex permittivity of the cell is a function of the membrane complex permittivity and the cytoplasm complex permittivity can be determined as follows:40,42,43
| (5) |
where is the inner-membrane depolarization factor, and v is the ratio of outer-membrane volume to inner-membrane volume and can be calculated via , where d is the membrane thickness. Since a RBC’s thickness is three orders of magnitude less than its radius, the approximation is valid and can be calculated as follows:42,43
| (6) |
where for inner-membrane depolarization, and for outer-membrane depolarization. For a spherical cell, and , which means that Eq. (4) can be simplified to the expression presented in Eq. (2). Healthy human red blood cells have a semimajor radius r = 3.2 μm, , and .42,43 Reported parameters for RBC (blood type unspecified) membrane conductivity, cytoplasm conductivity, and media conductivity are <10−6 S/m, 0.31 ± 0.03 S/m, and 0.1 S/m, respectively. The membrane permittivity, cytoplasm permittivity, and media permittivity reported were 10.5 ± 0.45, 59 ± 6, and 80, respectively.43
C. Quantifying DEP spectra from cell velocities
The DEP force acting on cells can be determined by measuring cell velocities .44 If Brownian motion and buoyancy forces are neglected, the net DEP force is the sum of the Stokes drag force and the acceleration force : . For an oblate spheroidal cell, the DEP force can be described as45
| (7) |
where is acceleration, is the fluid's dynamic viscosity, and is the friction factor derived from the cell's size, shape, and surface roughness. For an oblate cell spheroid (with ) whose motion is perpendicular to the symmetry axis, was derived analytically to be46
| (8) |
One of the key elements of a DEP spectra is the crossover frequency, , since a cell's dielectric properties can be deduced from this number. Using the lower in combination with the minima and maxima of the DEP response spectra, membrane dielectric properties (permittivity, conductance, and capacitance) can be estimated.43 The is an easily discernible point from experiments that depends upon the cell dielectric properties relative to the suspending medium. Each cell has unique dielectrophoretic spectra as a function of frequency that reflects molecular structure and dielectric properties of both the cell's membrane and internal structure. A single-shell model has two interfaces: the inner interface between the cytoplasm and the membrane and the outer interface between the membrane and the surrounding medium. Multiple interfaces lead to the possibility of multiple fCO's. The lower is determined by cell size, shape, and the outer-membrane interface in the β-dispersion region, while the second, higher is influenced by the cell cytoplasm and inner interface. Prior research and compositional knowledge mean that RBC membranes can be assumed to be less conductive than the cytoplasm. Under these conditions, by setting in Eq. (4), we obtain the crossover frequency as follows:43
| (9) |
where and and are the specific capacitance and conductance of the cell membrane, respectively.43 Membrane capacitance is a measure of the membrane area that acts as a charge barrier and depends upon the membrane thickness, composition/dielectric properties, and morphological complexity. Membrane conductance, on the other hand, reflects the net transport of ionic species across the membrane through pores, ion channels, and defects under the influence of the applied field. As Eq. (9) demonstrates how enables monitoring of changes in cell membrane capacitance and conductance in response to external stimuli, in this case surfactants. Thus, measuring cell DEP behavior can infer cell dielectric properties, which can then be judiciously harnessed to compare, identify, separate, and study cell physiology. Due to prior knowledge of RBC , the frequency range utilized in this study was from 700 kHz to 300 kHz, whereby Maxwell–Wagner polarizations occur at the interface between the cell membrane and the suspending medium.10 Therefore, it becomes possible to probe surfactant interactions with the RBC membrane by quantifying and analyzing the DEP spectra of RBCs.
We test this hypothesis by investigating the effects of nonionic surfactant Triton X-100 on DEP responses of RBCs within a microfluidic device. Concentrations of Triton X-100 below the CMC (CMC = 0.22–0.24 mM) were added into isotonic RBC suspensions in phosphate buffer saline (PBS) subjected to nonuniform electric fields.47 The impacts of low surfactant concentrations on RBCs were systematically compared across traditional nonelectrokinetic cell characterization techniques (UV-Vis spectroscopy and optical microscopy) as well as via an electrokinetic technique, namely, DEP characterizations. We calculated cell dielectric properties from experimentally quantified cell velocities converted into DEP force spectra as a function of frequency. This work provides mechanistic understanding into the chemical interactions between a nonionic surfactant and RBC membranes inferred from DEP cell responses in a microfluidic platform.
II. MATERIALS AND METHODS
A. Chemicals and preparation
Potassium phosphate monobasic (ACS, ≥99%) and potassium phosphate dibasic (ACS, ≥99%) were purchased from EMD Millipore (Billerica, MA). Dextrose (anhydrous, ≥99.5%), and nonionic surfactant Triton X-100 (laboratory grade) were purchased from Sigma-Aldrich (St. Louis, MO). Sodium chloride (>99%, ACS) was purchased from Macron Chemicals (Center Valley, PA).
All RBC experiments were conducted in controlled isotonic PBS solution with a conductivity of 0.1 S/m. Phosphate Buffered Saline (PBS) stock solution was made from the following salts: 2.7 g of potassium phosphate monobasic, 3.5 g of potassium phosphate dibasic, and 1.2 g of sodium chloride were mixed in 99.9 mL of E-pure water (18.2 Ω cm, EMD Millipore Simplicity Ultrapure 185, Billerica, MA). To formulate PBS solution with the conductivity of 0.1 S/m, 1.2 g of dextrose and 64.3 μL of PBS stock solution were mixed in 24.9 mL of E-pure water, stored at 4 °C and used within one week.
B. Microfluidic device design and fabrication
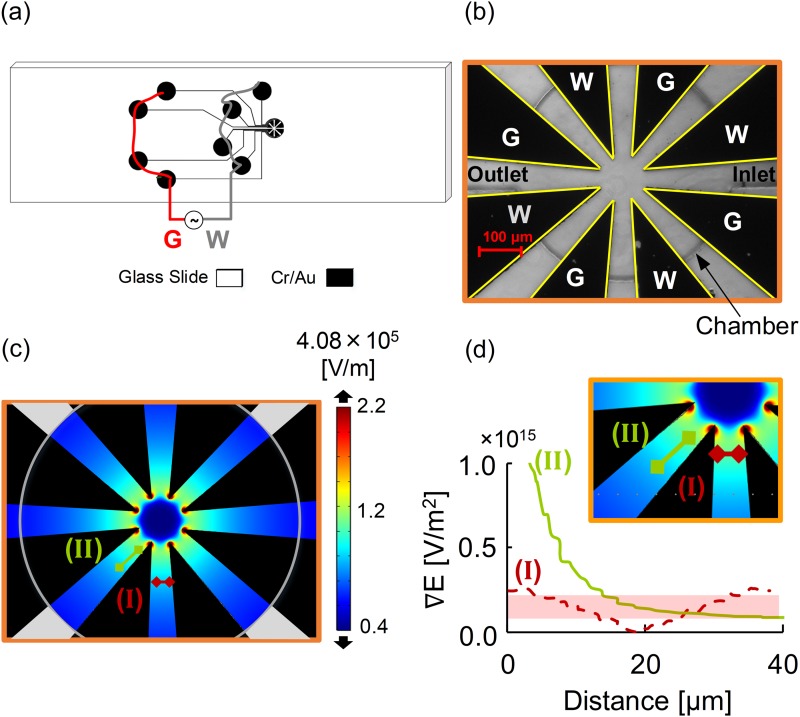
Figure 2 shows a star-shaped electrode array for the microfluidic device. The star-shaped electrode, as shown in Fig. 2, had four pairs of triangular electrodes converging symmetrically at a center point.48,49 The electrodes were fabricated with a thickness of 95 nm of Au above 10 nm Cr sputtered onto 25 × 76 × 1 mm glass microscope slides (AmScope.com). Photolithography, sputtering, and lift-off were employed to fabricate the microelectrodes on glass microscope slides following previously detailed procedures.48,49 Briefly, a master wafer was created by photolithography to obtain polydimethylsiloxane (PDMS) castings. Microelectrodes were then overlaid by the PDMS circular microfluidic chamber with a diameter of 590 μm and a depth of ∼70 μm. Disposable biopsy punches (Robbins Instruments, Chatham, NJ) were utilized to create inlet and outlet ports with diameters of 300 μm.
FIG. 2.
(a) Macroscale schematic demonstrates the Cr/Au star-shaped electrode design in black on a glass slide. (b) Microscope (10×) image of the star-shaped microelectrodes within the circular chamber. (c) The electric field (V/m) distribution in the microfluidic chamber. Regions around electrode tips have the maximum field strength. (d) Plotted cutline of the electric field gradient approximately parallel to the electrode side is shown in the green line marked II, while the electric field gradient (V/m2) perpendicular to the working and ground electrode is shown in the dashed red line marked I. Locations of the green and red cut lines are shown in (c) and inset (d).
C. Blood experiments
Human blood samples from anonymous donors were obtained following Institutional Review Board (IRB)-approved protocols (M0540, [318164-12]) and stored at 4 °C, then tested on the day of donation. Blood was centrifuged at 132 rcf for 10 min. Supernatant plasma was removed, and the packed RBCs were washed twice with isotonic 0.9% wt./v NaCl solution via cycles of resuspension and centrifugal separation. For experiments, RBCs were mixed with 0.1 S/m PBS at 1.0% v/v. For the surfactant study, RBCs were also resuspended to 1.0% v/v while achieving final Triton X-100 concentrations of 0.00, 0.07, 0.11, 0.17, and 0.50 mM.
D. UV-Vis experiments
Ultra Violet Visible absorption spectroscopy is one of the simplest, label-free techniques yielding direct insight into size, shape, chemical composition, and optical properties of cells and/or proteins.50,51 UV-Vis absorbance readings and spectral profiles were obtained using a Genesis 10 UV scanning spectrophotometer (Thermo Fisher Scientific, Waltham, MA). UV-Vis was utilized as a nonelectrokinetic technique to study both hemolysis and RBC integrity.
1. Hemolysis
The extent of hemolysis over time was studied by exposing different concentrations of Triton X-100 to RBCs. Samples were kept in ambient conditions (25 °C) and were performed in triplicate within cuvettes (Spectrocell Inc., Oreland, PA). Negative controls were comprised of RBC suspensions in PBS in the absence of Triton X-100 (0.00 mM). Positive controls were 0.50 mM RBC/Triton X-100 whereby 100% lysis was expected.52,53 After 2 h, RBC surfactant and nonsurfactant suspensions were centrifuged at 132 rcf for 10 min. The supernatant was collected and used to measure free hemoglobin absorbance readings at 380, 415, and 450 nm. A PBS blank control was used to correct absorbance of free hemoglobin ,52,53
| (10) |
Hemolysis extent was determined by measuring UV-Vis absorbance of free hemoglobin (Hb) released from the RBCs.52,53 Changes to the Hb absorption spectrum could be discerned using the Soret band ,
| (11) |
Percent hemolysis was calculated relative to the 100% hemolysis positive control to compare across surfactant exposure conditions. Statistical analysis was conducted whereby * represents p-values < 0.05 and ** represents p-values < 0.01. Both * and ** were considered statistically significant differences.
2. RBC integrity and cell radii
RBC health and integrity were studied by exposing different concentrations of Triton X-100 (0.00, 0.07, 0.11, and 0.17 mM) to RBCs over time (0, 30, 60, and 120 min). Samples were diluted in 0.25% v/v RBC/PBS and kept in ambient conditions (25 °C) to obtain the spectra. UV-Vis spectrums were obtained from 250–750 nm in 1 nm increments.
For each solution condition, RBC radii (n ≅ 100) were concurrently measured with ImageJ® software (NIH, https://imagej.nih.gov/ij/) from a 2D microscopy image Zeiss Axiovert microscope (Zeiss, Oberkochen, Germany). RBC cell radii histograms were plotted to represent the cell size distribution of each cell population. Mean and standard deviation of the cell radius were also reported. Statistical analysis was conducted.
E. Data acquisition and analysis
RBC suspensions were introduced via ports to the dielectrophoretic microfluidic device mounted on the Zeiss Axiovert microscope. Low frequency alternating current electric fields were applied via a custom-built function generator (Microdevice Engineering Inc., Houghton, MI). To monitor impacts on RBC membranes, experiments were conducted at 5 Vpp potential (electric field density = 0.1 Vpp/μm) over a frequency range from 700 kHz to 300 kHz and a frequency sweep rate of 2400 Hz/s. The electric field density was calculated from the applied potential over the distance between opposing electrode tips (50 μm). Once the potential was applied, cell movement was recorded at 10× magnification at 1 fpm for 167 s.
As shown in Fig. 2, the star-shape device was designed to yield symmetric electric field nonuniformities to induce DEP motion. Cells in red, orange, and yellow regions in Fig. 2(c) experienced a greater DEP force than cells in green, teal, and blue regions. Thus, to make appropriate comparisons, cells were selected for analysis from similar electric field gradient regions to accurately and reproducibly quantify cell displacement. These data were used to calculate cell velocity and subsequently individual cell's DEP force spectra as a function of frequency. Cells initially located in the electric field strength regions between lines marked I and II in Figs. 2(c) and 2(d) (inset) were tracked forward in time. Figure 2(d) plots the electric field gradient along lines I and II. Cell velocities were only quantified for cells in the shaded region of interest.
Semiautomated and automated techniques were compared for analyzing cell displacement. The semiautomated technique with ImageJ® utilized recorded images to manually acquire cell displacement with time. Data were compiled into MATLAB to calculate the cell velocity and . The automated technique employed Tracker 4.11.0 software (https://physlets.org/tracker/) to directly obtain velocity profiles. Finally, Eq. (7) was utilized to calculate the dielectrophoretic force, which was plotted for samples without and with Triton X-100 concentrations of 0.07, 0.11, and 0.17 mM. RBC sizes (measured previously) were utilized to adjust the calculation.
III. RESULTS AND DISCUSSION
This study examined the interactions of low concentrations of Triton X-100 surfactant below the CMC with human RBCs using UV-Vis spectroscopy for cell integrity, optical microscopy for cell size, and dielectrophoretic techniques for membrane dielectric properties.
A. Hemolysis and cell size dependence on Triton X-100 concentrations
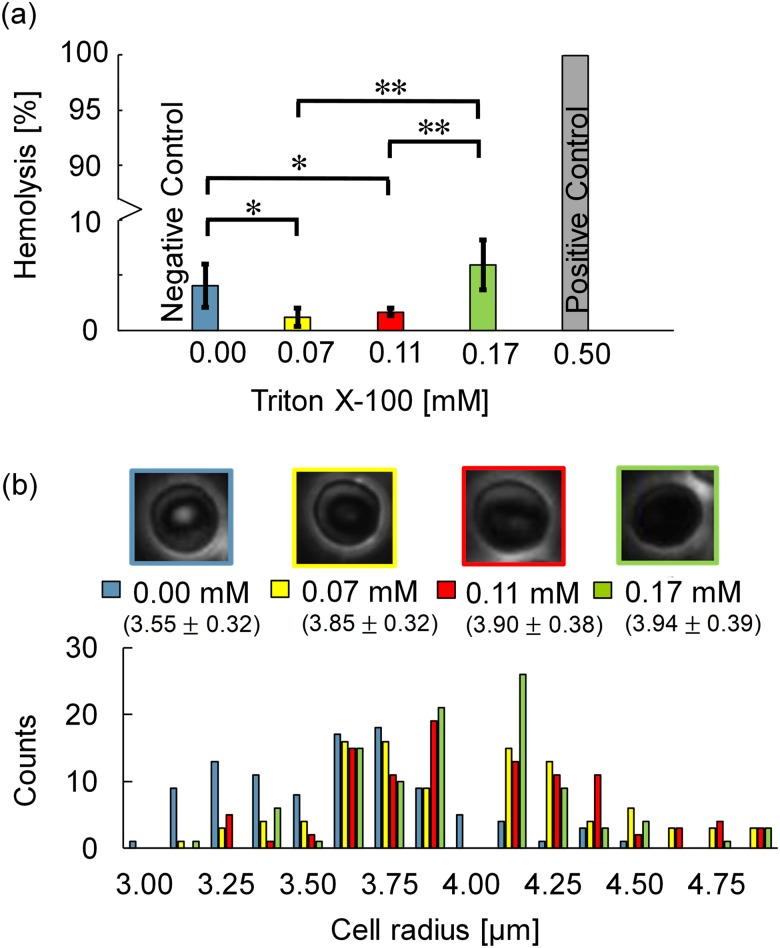
RBC integrity in the absence and presence of Triton X-100 was examined via traditional UV-Vis spectroscopy techniques.52,53 Figure 3(a) shows hemolysis percentages obtained via Eqs. (10) and (11). The positive control of 0.5 mM Triton X-100 was selected as more than twice the CMC = 0.22 mM and yielded 100% hemolysis.47 The negative control of RBCs in isotonic PBS (0.1 S/m) showed 4.05% hemolysis, which can be attributed to slight osmotic pressure changes and/or stress from centrifugation. Hemolysis percentages of 1.20% and 1.67% for 0.07 and 0.11 mM Triton X-100 treated RBCs, respectively, were notably lower than native RBCs (negative control, *p-value < 0.05). In 0.17 mM Triton X-100, hemolysis was 5.94%, which is considerably higher compared to 0.07 and 0.11 mM Triton X-100 (**p-value < 0.01). These results are consistent with previously reported biphasic phenomena,36,37 and further suggest that below the CMC, there is a narrow range of Triton X-100 concentrations whereby hemolysis is reduced from native conditions and the surfactant protects RBCs against rupture.
FIG. 3.
(a) The impact of 0.00, 0.07, 0.11, 0.17, and 0.50 mM Triton X-100 on percent hemolysis. Hemolysis decreased by 2.9% upon adding 0.07 mM Triton X-100, then increased by 4.7% as the concentration of Triton X-100 increased from 0.07 to 0.17 mM. Significance of differences was determined by calculating p-values; * represents p-values < 0.05 and ** represents p-values < 0.01. (b) Microscopy images of RBCs, mean and histogram of RBC radii at 0.00, 0.07, 0.11, and 0.17 mM Triton X-100 concentrations. RBC radii increased with Triton X-100 concentrations, which are also noted in Table I.
Per proposed mechanism one, as surfactant molecules intercalate into cell membranes,28–34 they reduce strain within the membrane allowing cell volume to increase. This mechanism elevates the threshold for hemolysis, aka the critical lytic volume.36,37 When cell volume increases exceed the critical lytic volume, hemolysis occurs. Figure 3(b) shows cell radii distribution histograms, cell morphology, and mean radii for the RBC populations in the absence and presence of Triton X-100 below the CMC. Native RBCs typically have a biconcave shape with radii that range between 3.10 and 4.10 μm in isotonic physiological conditions.54 In the absence of Triton X-100, 75% RBCs were between 3.10 and 4.10 μm. In 0.07 mM and 0.11 mM Triton X-100, this shifted up such that 75% of the RBC population had a cell radius between 3.50 and 4.50 μm. As the Triton X-100 concentration increased to 0.17 mM, 85% had cell radii between 3.50 and 4.50 μm. Given the broader size distributions with both the 0.00 and 0.11 mM Triton X-100 treated RBC radii populations, a pairwise statistical t-test analysis yielded a *p-value < 0.05, which is classified as significant.55 Increasing radii with increasing Triton X-100 concentrations were attributed to the surfactant's intercalation into the RBC membranes thus elevating the critical lytic volumes.
As radii increased, images showed that RBCs lost their biconcave morphology consistent with prior observations.36,56 As shown in Fig. 3(b), most RBCs transformed from biconcave with bright circular interiors to stomatocytes with dark ring/slit-like profiles as Triton X-100 concentrations increased from 0.0 to 0.11 mM. At 0.17 mM Triton X-100 concentrations, most RBCs showed swollen spherocyte shapes whereby the ring/slit-like shadow disappeared as shown in Fig. 3(b). Both cell size and morphology influence the dielectrophoretic forces experienced by the cells. For DEP calculations, average radii were utilized, and the biconcave cells and stomatocytes in 0.0 mM (native), 0.07 mM, and 0.11 mM Triton X-100 were modeled with an oblate single-shell geometry. RBCs in 0.17 mM were spherocytes but did not display a crossover frequency, so the DEP model was not utilized. To increase accuracy, the DEP model incorporated in cell size and morphology as discussed in Sec. III E.
B. RBC integrity analysis via UV-Vis spectroscopy
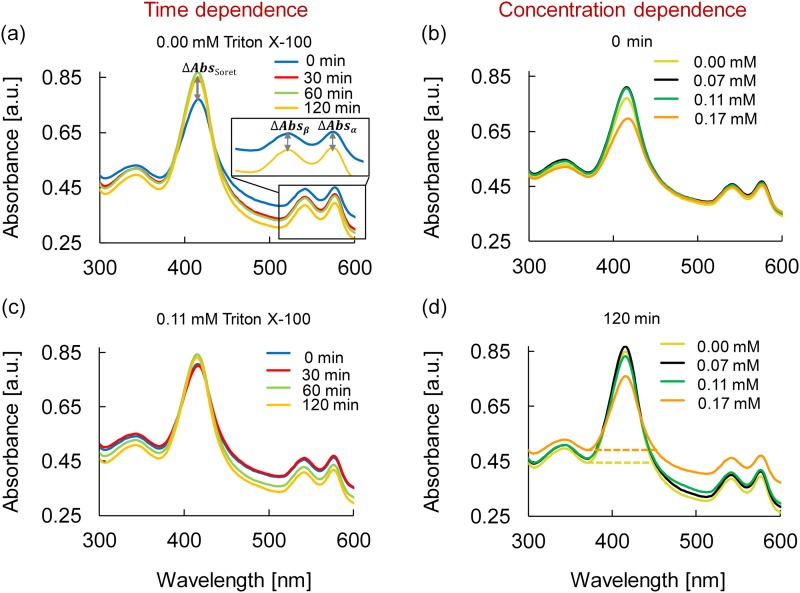
Beyond the standard hemolysis analysis, UV-Vis absorbance spectra can provide additional insights into the effects of low concentrations of Triton X-100 on RBC membranes and integrity by examining peaks from interior proteins such as hemoglobin. When a RBC membrane ruptures and the cell lyses, the interior contents of the cell are exposed to the surrounding media initiating secondary degradation reactions such as oxidation of hemoglobin.57,58 Figure 4 shows absorbance from 250 to 750 nm wavelengths for native and Triton X-100 treated RBC suspensions. Hemoglobin free in solution demonstrates characteristic absorption peaks of the Soret band at 415 nm and α and β bands at 576 and 541 nm, respectively (Fig. 4).59–67 Hemoglobin proteins are comprised of four heme groups; two within α chains and two within β chains.50,51 As shown previously, Soret bands are indicative of free hemoglobin absorbance and can be used to calculate the extent of hemolysis [Eqs. (10) and (11)] as well as hemoglobin monodispersity.52,53 Moreover, the peak shape, wavelength, and absorption maximum intensity of Soret and α/β bands can be utilized to monitor free hemoglobin, heme oxidation, orientation and protein denaturization.59–67 Upon Triton X-100 addition to the RBC suspensions, the wavelength position of the Soret, α, and β bands did not change over 120 min for any concentration [Figs. 4(a)–4(d)]. Thus, changes in the peak shape and absorbance intensity of the Soret, α, and β bands were used to ascertain Triton X-100 surfactant impacts over time [Fig. 4(a)].
FIG. 4.
UV-Vis absorbance spectra of 0.25 v/v% RBC/PBS from 250 to 750 nm. Time dependence was studied for (a) native RBCs (0.00 mM Triton X-100) and (c) 0.11 mM Triton X-100 treated RBCs at 0, 30, 60, and 120 min. Concentration dependence was studied for RBCs treated at 0.00, 0.07, 0.11, and 0.17 mM Triton X-100 at (b) 0 min and (d) 120 min. Changes in and peak absorption (ΔAbsα and ΔAbsβ) indicate and heme chain oxidation/protein degradation, while the Soret band (ΔAbsSoret) provides information on the extent of hemolysis.
Comparison of spectra in Figs. 4(a) and 4(c) illustrate native RBCs (0.00 mM Triton X-100) and 0.11 mM Triton X-100 treated RBCs, respectively, as a function of time. For native RBCs, the Soret peak intensity increased by 10.3% over 120 min, which is an indication of a slight increase in free hemoglobin as a result of baseline hemolysis (negative control).52,53,68 Furthermore, the intensity of the native α and β peaks at 576 and 541 nm decreased within 30 min, and after 120 min, the peaks decreased by 12.7% and 13.3%, respectively [Fig. 4(a)]. These peak decreases are indicative of baseline heme oxidation.62–64 Spectra for 0.11 mM in Fig. 4(c) illustrate an increase of 3.10% for the Soret peak after 120 min. Comparison between these Soret band increases of 10.3% and 3.10% for 0.00 mM (native) and 0.11 mM is consistent with previously reported biphasic phenomena36,37 whereby the presence of Triton X-100 reduced free hemoglobin by reducing hemolysis.
Heme changes can be inferred from α and β peaks. Changes in α and β peak intensity occurred for 0.11 mM Triton after 30 min but before 60 min with decreases of 10.3% and 9.96%, respectively, after 120 min. Thus, heme oxidation is hindered slightly in the presence of 0.11 mM Triton, consistent with the Soret band conclusion that RBC membranes remained intact for longer when Triton X-100 molecules were present. Similar heme UV-Vis spectral trends were obtained for 0.07 mM Triton X-100 treated RBCs (data not shown); the Soret band increased by 7.03%, while α and β peak intensities decreased by 12.0% and 12.8%. Trends differed slightly for 0.17 mM as discussed below.
Figures 4(b) and 4(d) show the concentration dependent impacts for 0.00, 0.07, 0.11, and 0.17 mM Triton X-100 treated RBCs at 0 and 120 min, respectively. At time 0, the spectra are nearly identical for Triton X-100 concentrations from 0.00 to 0.11 mM with less than 4.59% variation in the 415 nm Soret band [Fig. 4(b)]. However, the 0 min Soret band intensity decreased by 9.48% for 0.17 mM Triton X-100 compared to the native RBC sample. This result was puzzling and inconsistent with previous measures of hemolysis. However, previous work reports that a change in hemoglobin structure can cause a profound and abrupt decrease in the absorption maximum of the Soret band [Fig. 4(b)].69 The Soret peak shape broadened by ∼12 nm with 0.17 mM Triton X-100, which is an indicator of protein denaturation via a conformational change within the heme region.66,67 From the initial reading at time 0 to 120 min, the Soret peak intensity for 0.17 mM Triton X-100 increased by 9.03% indicative of an increase in free hemoglobin [compare Figs. 4(b) and 4(d)]. Furthermore, the intensities of both α and β peaks changed by only 3.12% and 3.07% over the 120 min [compare Figs. 4(b) and 4(d)], which is considered insignificant oxidization.
In summary, UV-Vis spectral analysis of hemoglobin served as an indirect indicator of RBC membrane integrity because relative Soret, α, and β peak absorptions changed once hemoglobin was released from the cell into the surrounding solution. Subsequent heme oxidation/protein degradation of the heme chains begin upon hemoglobin exposure to the PBS. In general, hemolysis increased with time and decreased with Triton X-100 concentrations.
C. Qualitative dielectrophoretic behavior
RBC membrane charge properties were further explored by correlating cell size and dielectrophoretic (DEP) responses of RBCs because these properties directly correlated with membrane polarization and integrity. The AC frequency range at which cells transition their movement from negative DEP to positive DEP was determined by applying a 0.1 Vpp/μm signal with a frequency sweeping from 700 to 300 kHz between four pairs of converging triangular and planar Cr/Au electrodes (Fig. 2). The frequency sweep was conducted from high to low frequency because the DEP forces cells experience while undergoing p-DEP at higher frequencies are larger in magnitude than those trapped at electric field minima while experiencing n-DEP [Eqs. (1) and (7)]. Previous studies revealed that p-DEP to n-DEP frequency sweep transitions yielded more reproducible and accurate results than static frequency tests.16
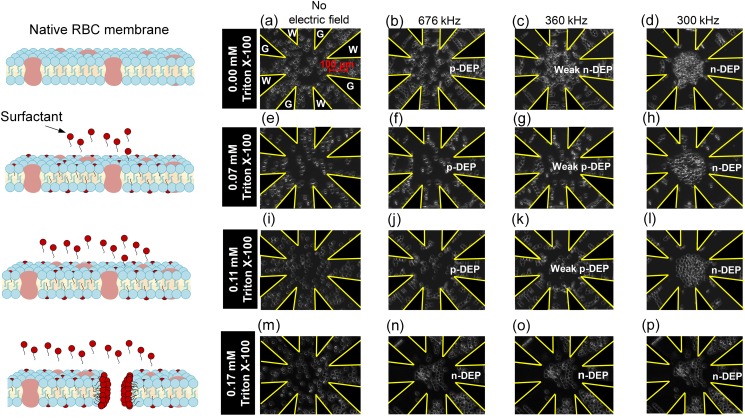
The p-DEP to n-DEP transition was explored as a function of Triton X-100 concentrations below and approaching the CMC (∼0.22 mM) since isolated and dispersed surfactant molecules show minimal lysing effects,28–34 while surfactant molecules assembled into micelles impact RBC integrity.36,37,47 All DEP experiments were conducted after RBCs were at steady state, whereby contact with Triton X-100 concentrations occurred for 2 h or less. Figure 5 shows images of RBC locations in the microfluidic chamber before, during, and after applying the frequency sweep. Prior to applying the potential, RBCs were uniformly distributed in the chamber [Figs. 5(a), 5(e), 5(i), and 5(m)]. Upon applying the sweeping frequency signal, RBCs were pulled toward regions with high electric field density, indicative of p-DEP, then transitioned toward lower electric field density regions indicative of n-DEP. For native (0.00 mM) RBCs, p-DEP occurred from 700 kHz down to 492 kHz [Figs. 5(a) and 5(b)]. DEP forces diminished to zero at the crossover frequency, [between Figs. 5(b) and 5(c)], and then DEP forces gradually increased to push the RBCs away from the electrodes toward the electric field minima at the chamber center from 472 kHz down to 300 kHz [Figs. 5(c) and 5(d)]. At the low Triton X-100 concentration of 0.07 mM, the p-DEP response was strong, but the crossover was a lethargic transition between 359 and 381 kHz [Figs. 5(e)–5(h)] before achieving n-DEP at 300 kHz. As the surfactant concentration increased to 0.11 mM [Figs. 5(i)–5(l)], a few cells exhibited n-DEP throughout the frequency sweep, while the majority of RBCs exhibited a lethargic between 365 and 385 kHz and again demonstrated n-DEP at 300 kHz. Triton X-100 at 0.17 mM caused cells to uniformly exhibit n-DEP from 700 down to 300 kHz [Figs. 5(m)–5(p)]. The literature indicates that 0.17 mM Triton X-100 is the lowest concentration to sufficiently permeabilize the RBC membrane prior to complete cell lysis at 0.22 mM (CMC).70 Such permeabilization would effect DEP polarizations. Furthermore, UV-Vis spectral results (Fig. 4) indicated Triton X-100 impacted hemolysis as well as hemoglobin structure at 0.17 mM.
FIG. 5.
Video microscopy images illustrating the time sequence of 1.0 v/v% red blood cells in the 0.1 S/m isotonic PBS medium at 0.00 (first row), 0.07 (second row), 0.11 (third row), and 0.17 mM (fourth row) Triton X-100 concentration at 0.1 Vpp/μm from 700 kHz to 300 kHz within star-shaped microfluidic devices. Images are shown at 0 s [(a), (e), (i), and (m)] corresponding to no field, 10 s corresponding to 676 kHz [(b), (f), (j), and (n)], 142 s and 360 kHz [(c), (g), (k), and (o)], and 167 s and 300 kHz [(d), (h), (l), and (p)]. Images illustrate that the p-DEP to n-DEP crossover frequency shifts to lower frequencies with increasing Triton X-100 concentrations. Images also reveal changes in RBC radii and shape as previously quantified in Fig. 3.
RBC size and morphology was also apparent in Figs. 5(d), 5(h), 5(l), and 5(p) whereby the smaller native RBCs tightly packed for n-DEP and comprised a larger portion of the chamber for 0.11 mM Triton X-100. In addition, RBC uniformity was altered for 0.17 mM Triton X-100.
D. Quantitative dielectrophoretic spectra
To better understand surfactant and RBC membranes interactions, DEP spectra were quantified to approximate the DEP force vs frequency via both semiautomated and automated analysis techniques as described previously. Agreement between the two analyses was excellent with consistent, but a small bias of <10 kHz in the crossover frequency prediction. Thus, only the automated technique is reported here.
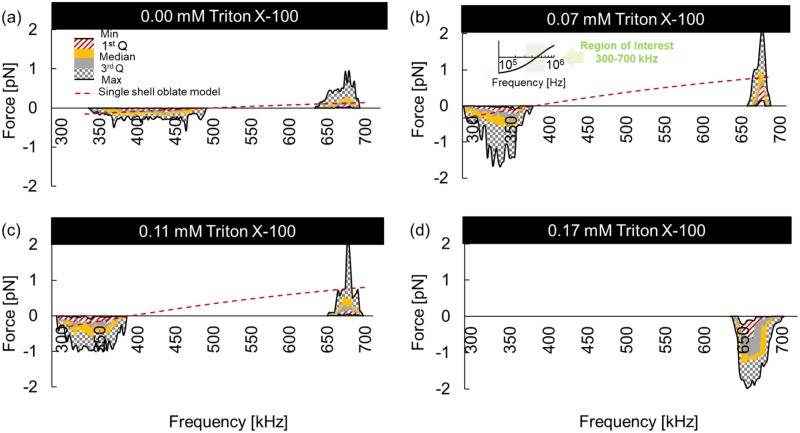
Figure 6 shows the DEP force profile inferred from the cell velocities via Eq. (7) of native and Triton X-100 treated RBC suspensions via a box-and-whisker-plot-inspired representation. As the key illustrates in Fig. 6, the red to gray patterned regions show the first, second, and third interquartile ranges, with boundary lines illustrating the min, median, and max. The min and max lines (or whiskers) bounding the patterned area show the full range of the DEP force. This representation is a concise tool to illustrate the motion of every cell in the microdevice chamber's region of interest.
FIG. 6.
Dielectrophoretic force as a function of frequency from 300 to 700 kHz of 0.00 mM (native) (a), 0.07 mM (b), 0.11 mM (c), and 0.17 mM (d) Triton X-100 treated RBCs. DEP force profiles were calculated from cell velocities and represented via a box-and-whisker-inspired representation whereby the colored regions show the interquartile ranges, while the min and max lines for the whiskers show the full force range (patterned area) for all cells in the test region. The median force is the boundary between yellow and gray.
The crossover from p-DEP to n-DEP behavior occurs at successively lower frequencies as the Triton X-100 concentration increases. Native cells yielded an of 482 ± 10 kHz, while the 0.07 mM treated RBCs yielded 370 ± 11 kHz and 0.11 mM yielded a similar value of 375 ± 10 kHz. Also apparent is the increase in the magnitude of the maximum positive DEP force, , obtained for Triton X-100 treated samples. Native RBCs experienced an average positive of +0.25 pN and an average negative of −0.15 pN. Treatment at 0.07 mM had the average positive and negative increased to +0.75 and −0.50 pN, respectively. However, Triton X-100 treatment at 0.11 mM only increased DEP forces to +0.65 and −0.50 pN. No was observed for 0.17 mM since n-DEP occurred for all frequencies tested and yielded an average DEP force of −1.17 pN. These results suggest that low concentrations of surfactant impacted the strength of RBC membrane polarizations and those polarizations vary with available surfactants. Several possible membrane polarization mechanisms are capable of shifting the dielectrophoretic spectra and are discussed below.
E. Single-shell oblate dielectrophoretic force model
MATLAB was used to model dielectrophoretic spectra by adjusting parameters in Eq. (9) to best approximate the experimentally obtained spectra in the presence and absence of Triton X-100. The single-shell oblate model was used to examine the relative contributions of RBC parameters including radius, membrane permittivity and conductivity , cytoplasm permittivity and conductivity , and medium permittivity and conductivity . Medium conductivity strongly impacts the cell behavior in the β-dispersion region; however, in our DEP force model, medium conductivity was kept constant to match the controlled experimental condition. For high frequency AC systems, medium conditions including pH, ionic concentration and ionic strength are assumed relatively stable over time due to negligible electrochemical reactions.48,49 Also, in the β-dispersion region, cytoplasm permittivity and conductivity have been shown to have no significant influence on crossover frequency .42 Thus, , , and were held constant in the model at 0.10 S/m, 0.31 ± 0.03 S/m, and 59.0 ± 6, while parametric sweeps were conducted for r, and . Membrane conductivity was explored over 2 orders of magnitude greater than those of native RBC membranes (10−6 S/m) without significantly shifting ; furthermore, when was reduced to 10−4 S/m, shifted down only by 10 kHz, which does not account for the 110 kHz shift observed experimentally. Thus, was fixed at 10−6 S/m for subsequent parametric sweeps exploring the impact of cell radius and membrane permittivity. To determine the cell membrane permittivity, a trial and error methodology was utilized. MATLAB code was written to incrementally increase the membrane permittivity parameter until the experimentally measured crossover frequency and the model-derived crossover frequency were well fitted with less than 0.5 kHz maximum different.
Maxwell-Wagner interfacial polarizations govern cell responses in the β-dispersion region whereby the electric field interacts with ions in the medium causing them to move and align around the cell. Cell size is critical to interfacial polarization; the larger the cell is, the longer the time scale for ions to move around, align, and induce cell polarization. Equations (7) and (9) explain how cell radius impacts DEP force magnitude and , respectively. For this analysis, cell size was accounted for in the radii and quantified from data in Fig. 3(b).
Cell radii for 0.00 and 0.07 mM Triton X-100 are shown in Table I. Average radii were utilized as the starting parameters for model fits to the experimental DEP spectra in Fig. 6. As the cell radius increased from 3.55 to 3.85 μm upon increasing Triton X-100 from 0.00 to 0.07 mM, the computed crossover frequency decreased from 482 to 444 kHz. However, does not match the experimentally observed fCO = 370 kHz. Thus, cell enlargement is not sufficient to account for the decrease in in the presence of 0.07 mM Triton X-100. Similarly, for 0.11 mM Triton X-100 treated RBCs, the model required radii values (5.2 μm) outside the range of experimentally observed cell radii, essentially requiring a 7.3% cell volume increase, which is not feasible without cell lysis.
TABLE I.
Optimized parameters for a single-shell oblate DEP polarization model for 0.00, 0.07, 0.11, and 0.17 mM treated RBCs in 0.1 S/m isotonic PBS. The experimentally measured crossover frequency is shown as fCO, while the model-derived crossover frequency is . Bold parameters were varied (normal font parameters were fixed) in the model to achieve the best agreement with experimental data.
| Triton X-100 (mM) | fCO (kHz) | r (μm)a | ɛmem | Cmem (mF/m2) | |
|---|---|---|---|---|---|
| 0.00 | 482 ± 10 | 3.55 ± 0.32 | |||
| Optimizing cell size and ɛmem on fCOb | 3.55 | 10.5 | 482 | 11.51 | |
| 0.07 | 370 ± 11 | 3.85 ± 0.32 | |||
| Optimizing cell size on fCO | 3.85 | 10.7 | 444 | ||
| Optimizing ɛmem on fCO | 3.55 | 13.4 | 370 | ||
| Optimizing cell size and ɛmem on fCOb | 3.85 | 12.7 | 370 | 14.05 | |
| 0.11 | 375 ± 10 | 3.90 ± 0.38 | |||
| Optimizing cell size on fCO | 3.9 | 10.7 | 432 | ||
| Optimizing ɛmem on fCO | 3.55 | 13.3 | 375 | ||
| Optimizing cell size and ɛmem on fCOb | 3.9 | 12.3 | 375 | 13.61 | |
| 0.17 | N/Ac | 3.94 ± 0.39 | N/A | N/A | N/A |
Statistical analysis between the cell radius of 0.00 and 0.17 mM Triton X-100 treated RBC populations revealed the p-value < 0.05, which indicates a significant difference.
Parameters utilized in model DEP spectra fits in Fig. 6.
Not applicable.
In addition to cell size effects, the movement and alignment of ions around the cell are influenced by the content and properties of the cell membrane. Thus, as cell surfaces change it affects polarization in distinctive ways and cause cells to display discretely different DEP spectra. One possible mechanism is that membrane permittivity solely causes interfacial polarization changes in the presence of Triton X-100. As the membrane permittivity was increased from 10.5 to 13.4, decreased from 482 to 370 kHz to match the experimentally observed of 370 kHz in the presence of 0.07 mM Triton X-100. Similarly, for 0.11 mM, a membrane permittivity value of 13.3 could achieve fCO = 375 kHz. These results suggest that the presence of Triton X-100 makes RBC membranes less permissive.
Although the contribution of membrane permittivity is a promising explanation for the observed shift in DEP spectra, cell size can also cause DEP spectral variations. Thus, the optimal mechanism likely includes both cell radius and membrane permittivity contributions to DEP spectral changes. The average cell radius was utilized as a starting parameter in the single-shell ablate model. In the presence of 0.07 mM Triton X-100, as the cell radius was increased from 3.55 to the experimentally measured 3.85 μm, the predicted changed from 482 to 444 kHz (a decrease of 8%). Thus, membrane permittivity would need to increase from 10.7 to 12.7 to achieve a drop from 444 to the experimentally observed 370 kHz (a 16% further decrease). This translates to a specific membrane capacitance increase from 11.51 mF/m2 for native to 14.05 mF/m2 for 0.07 mM Triton X-100 treated RBCs. Similar observations were obtained for 0.11 mM as shown in Table I whereby Cmem decreased to 13.61 mF/m2. Cell membrane capacitance depends on the cell dielectric properties, size, shape, and cell surface morphology associated with membrane blebbing or microvilli.71 These characteristics of membrane folding or smoothing change how charges distribute between the membrane surface and the medium, which shift as a function of frequency and thus contribute to a cell's DEP frequency spectra.72 Results support the hypothesis that the DEP force acting on a cell in the β-dispersion region was strongly influenced by both the cell radius and membrane permittivity, which ultimately reflects in the cell membrane capacitance values reported in Table I.
The cell membrane capacitance results are consistent with an effective increase in the total surface charge storing capacity. Ions from the surrounding medium accumulate on the outer cell membrane surface, while the internal counterions accumulate on the inner cell membrane surface. The accumulated ions repeatedly reverse their alignment with respect to each other during each half-cycle of the applied AC signal. At 0.07 and 0.11 mM Triton X-100, as the RBC diameter increased the ion transport path length increased around the cell. The combination of this effect with the increase in surface area for charge accumulation ultimately increased the time scale for charge displacement. This time scale becomes apparent in the crossover frequencies observed. In addition, at 0.07 and 0.11 mM Triton X-100 concentration, the polar head group (hydrophilic region) of the surfactant molecules binds with the phospholipid polar head in the outer surface of the cell membrane while the nonpolar tail group (hydrophobic region) intercalates deeper into the cell membrane (Fig. 1). The newly introduced surfactants interact differently with medium ions in the present of an AC potential. These alterations in distribution of polar groups and/or charges across the cell membrane distinctively impact the cell membrane permittivity as shown in Table I.
It is also valuable to note that as the concentration of Triton X-100 increased from 0.07 to 0.11 mM, the experimentally measured cell radius as well as the model-derived membrane permittivity did not change significantly. The 0.07 mM and 0.11 mM concentrations differ in the availability of surfactant molecules. However, this does not translate into structural changes within the RBC membrane, which is apparent from the hemolysis data in Fig. 3(a), the UV absorbance data in Fig. 4 as well as in the crossover frequencies qualitative and quantitative data in Figs. 5 and 6. Extension of the mechanism discussed in the previous studies,28–34 suggests that the RBC membrane is able to intercalate increasingly more surfactant molecules into the phospholipid bilayer until a saturation threshold is reached. Increasing numbers of isolated surfactant intercalations into the membrane do not appear to substantially change the membrane's structural and dielectric properties. From 0.07 to 0.11 mM Triton X-100 concentration, membrane saturation was not reached. However, as the surfactant concentration increased to 0.17 mM, which is closer to the CMC, surfactant-surfactant interactions become more prevalent, saturating the membrane and interfering with membrane functionality. This is apparent in hemolysis (Fig. 3), structure and size (Fig. 4), and dielectric data (Figs. 5 and 6).
The RBC parameters shown in Table I are the best fits for the single-shell oblate DEP model to experimentally measured DEP spectra for different Triton X-100 concentrations. The optimized model fit is also shown as the red dashed curve in Figs. 6(a)–6(d). Using the parameters in Table I, the DEP force spectra agreed in sign and order of magnitude with experimental observations within p-DEP and n-DEP regions. Since the 300 to 700 kHz range is narrow, the model fit appears slightly linear as shown in the inset. One limitation with the cell velocity analysis approach to ascertain the DEP force is that the cells move rapidly to an equilibrium position, especially with strong DEP responses. Once in that equilibrium position, additional cell movement is not detected, although the DEP force on the cell remains high. This is apparent when comparing Figs. 6 and 5 where a strong negative DEP response is observed, but no additional cell velocities are measured below ∼650 kHz. Thus, in these steady state periods, deviations between the experimental data and the DEP polarization model are not real, but a manifestation of the measurement approach.
A DEP force of −1.17 pN for 0.17 mM Triton X-100 was inferred from the initial results between 700 and 640 kHz. As nonionic Triton X-100 surfactant molecules become more prevalent within the cell membrane structure,28 it follows that the cell membrane's propensity to polarize would be altered. There is evidence supporting this. HeLa cell membranes were reported to become permeabilized after 20 min of exposure to 0.17 mM Triton X-100.70 At 0.17 mM Triton X-100, more RBCs experienced irreversible damage from the surfactant as shown in Fig. 5 whereby the cells lost integrity and collapsed. The increase in overall hemolysis and cell radii indirectly suggest pore formation within the RBC membranes in the presence of 0.17 mM Triton X-100. Changes in the membrane permeability have a cascading effect by not only changing the membrane polarization but also altering internal cytoplasm conductivity and permittivity. Here, since the DEP tests were completed in the β-dispersion region, the DEP responses were not able to provide insight into changes in the RBC cytoplasm.
In summary, as shown in Fig. 6, the increased cell size and permittivity of Triton X-100 treated RBCs increased the exerted and decrease . This trend was also apparent in Table I showing membrane capacitance changes of 11.51, 14.05, and 13.61 mF/m2 for 0.00 mM, 0.07 mM, and 0.11 mM Triton X-100 treated cells, respectively. The strong media/membrane polarization dependency in the presence of Triton X-100 suggests that interactions between cells and surfactants alter dielectrophoretic polarizations and should be independently tested and controlled for in electrokinetic applications.
IV. CONCLUSIONS
Surfactants are regularly added to cell-buffer solutions to reduce cell sticking to surfaces and improve reproducibility between experimental runs. However, surfactant interactions with cell membranes and influences on the cell dielectric behaviors have only been explored in electrokinetic systems in an isolated study. In this work, surfactant interactions with cell membranes were explored in both nonelectrokinetic and electrokinetic systems. RBCs were treated with low concentrations of Triton X-100 (0.07–0.17 mM) below the critical micelle concentration because at and above the CMC, hemolysis is prevalent.
Traditional cell characterization techniques (UV-Vis spectroscopy and optical microscopy) were reproduced as comparison controls to previously reported nonelectrokinetic measures of RBC integrity and viability. Increasing Triton X-100 (0.00–0.17 mM) increased the RBC cell size and progressively altered cell membrane integrity as deduced from Soret and α and β chain peaks in the UV-Vis spectra.
Dielectrophoretic responses of RBCs pretreated with the same four Triton X-100 concentrations were then measured over the β-dispersion frequency region from 300 to 700 kHz where cell polarizations are membrane-dominated. To gain a better insight into the underlying mechanisms, a single-shell oblate model was utilized to fit dielectric parameters to experimentally quantified cell velocity and DEP force data. It is valuable to note that the DEP force magnitude was increased by 25% with the various Triton X-100 concentrations. The parameters most influential in shifting the crossover frequency from 482 ± 10 kHz at 0.00 mM down to 370 ± 11 kHz at 0.07 mM were cell radii and cell membrane permittivity. When comparing 0.07 mM and 0.11 mM, hemolysis, qualitative DEP spectra, and quantitative were not significantly different, which indicated that surfactant molecules likely interacted with the RBC membrane in isolation. However, as the Triton X-100 concentration increased to 0.17 mM, surfactant/surfactant interactions increased and were much more disruptive to membrane functionality as evidenced by RBC size and morphology, hemolysis, hemoglobin structural changes, and most prominently a major shift in RBC dielectric properties. At 0.17 mM Triton X-100, the complex permittivity of the RBCs remained lower than the surrounding medium effectively meaning that the RBCs lost the ability to switch from negative DEP to positive DEP.
This work shows that surfactant molecules traditionally added to mediate microfluidic device wall properties also dynamically interact with cell membranes and alter cell polarizability and electrokinetic responses. Since electrokinetic microfluidic systems are gaining commercial traction for portable medical diagnostic applications, it is important that the effects of the secondary chemical interactions between cells and surfactants be further explored. This work sets a foundation for measuring these phenomena and demonstrates that surfactant/membrane interactions should be separately attributed to cellular response data in order to accurately catalog cell properties and engineer reliable microfluidic electrokinetic devices.
ACKNOWLEDGMENTS
The authors gratefully acknowledge graduate teaching assistance support from Michigan Technological University and Portage Health Foundation Graduate Assistantship for Sanaz Habibi. Funding support for Hector Moncada-Hernandez came from the National Science Foundation (NSF) (No. IIP 1417187) and the State of Michigan's Technology Talent Transfer Network (T3N).
The authors are concurrently affiliated with MicroDevice Engineering Inc. (microdengineering.com), a spinoff from Michigan Tech. The work contained herein is not directly commercializable and thus no conflict of interest exists in making it public.
REFERENCES
- 1.Lee H. Y., Barber C., Rogers J. A., and Minerick A. R., Electrophoresis 36(7–8), 978–985 (2015). 10.1002/elps.201400466 [DOI] [PubMed] [Google Scholar]
- 2.Srivastava S. K., Daggolu P. R., Burgess S. C., and Minerick A. R., Electrophoresis 29(24), 5033–5046 (2008). 10.1002/elps.200800166 [DOI] [PubMed] [Google Scholar]
- 3.Gascoyne P., Satayavivad J., and Ruchirawat M., Acta Trop. 89(3), 357–369 (2004). 10.1016/j.actatropica.2003.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization, World Health Organization Model List of Essential In Vitro Diagnostics, 1st ed. (World Health Organization, Geneva, 2018).
- 5.Prieto J. L., Su H.-W., Hou H. W., Vera M. P., Levy B. D., Baron R. M., Han J., and Voldman J., Lab Chip 16(22), 4333–4340 (2016). 10.1039/C6LC00940A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuczenski R. S., Chang H.-C., and Revzin A., Biomicrofluidics 5(3), 032005 (2011). 10.1063/1.3608135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henslee E. A., Sano M. B., Rojas A. D., Schmelz E. M., and Davalos R. V., Electrophoresis 32(18), 2523–2529 (2011). 10.1002/elps.201100081 [DOI] [PubMed] [Google Scholar]
- 8.Adams T. N., Turner P. A., Janorkar A. V., Zhao F., and Minerick A. R., Biomicrofluidics 8(5), 054109 (2014). 10.1063/1.4895756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adams T. N. G., Jiang A. Y. L., Vyas P. D., and Flanagan L. A., Methods 133, 91–103 (2018). 10.1016/j.ymeth.2017.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pethig R. R., Dielectrophoresis: Theory, Methodology and Biological Applications (John Wiley & Sons, 2017). [Google Scholar]
- 11.Lapizco-Encinas B. H., Simmons B. A., Cummings E. B., and Fintschenko Y., Anal. Chem. 76(6), 1571–1579 (2004). 10.1021/ac034804j [DOI] [PubMed] [Google Scholar]
- 12.Green N. G., Morgan H., and Milner J. J., J. Biochem. Biophys. Methods 35(2), 89–102 (1997). 10.1016/S0165-022X(97)00033-X [DOI] [PubMed] [Google Scholar]
- 13.Mernier G., Piacentini N., Braschler T., Demierre N., and Renaud P., Lab Chip 10(16), 2077–2082 (2010). 10.1039/c000977f [DOI] [PubMed] [Google Scholar]
- 14.Suehiro J., Shutou M., Hatano T., and Hara M., Sens. Actuators B 96(1), 144–151 (2003). 10.1016/S0925-4005(03)00517-3 [DOI] [Google Scholar]
- 15.Lagally E. T., Lee S. H., and Soh H. T., Lab Chip 5(10), 1053–1058 (2005). 10.1039/b505915a [DOI] [PubMed] [Google Scholar]
- 16.Adams T. N. G., Leonard K. M., and Minerick A. R., Biomicrofluidics 7(6), 64114 (2013). 10.1063/1.4833095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J., Huang Y., Wang X.-B., Becker F. F., and Gascoyne P. R. C., Biophys. J. 78(5), 2680–2689 (2000). 10.1016/S0006-3495(00)76812-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mark D., Haeberle S., Roth G., Von Stetten F., and Zengerle R., Microfluidics Based Microsystems (Springer, 2010), pp. 305–376. [Google Scholar]
- 19.Mazutis L., Gilbert J., Ung W. L., Weitz D. A., Griffiths A. D., and Heyman J. A., Nat. Protoc. 8(5), 870–891 (2013). 10.1038/nprot.2013.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ocvirk G., Salimi-Moosavi H., Szarka R. J., Arriaga E. A., Andersson P. E., Smith R., Dovichi N. J., and Harrison D. J., Proc. IEEE 92(1), 115–125 (2004). 10.1109/JPROC.2003.820551 [DOI] [Google Scholar]
- 21.Davalos R. V., McGraw G. J., Wallow T. I., Morales A. M., Krafcik K. L., Fintschenko Y., Cummings E. B., and Simmons B. A., Anal. Bioanal. Chem. 390(3), 847–855 (2008). 10.1007/s00216-007-1426-5 [DOI] [PubMed] [Google Scholar]
- 22.Lee H. Y., Barber C., and Minerick A. R., Electrophoresis 35(12–13), 1782–1789 (2014). 10.1002/elps.201400013 [DOI] [PubMed] [Google Scholar]
- 23.Khoshmanesh K., Zhang C., Tovar-Lopez F. J., Nahavandi S., Baratchi S., Kalantar-zadeh K., and Mitchell A., Electrophoresis 30(21), 3707–3717 (2009). 10.1002/elps.200900079 [DOI] [PubMed] [Google Scholar]
- 24.Rosales-Cruzaley E., Cota-Elizondo P. A., Sanchez D., and Lapizco-Encinas B. H., Bioprocess Biosyst. Eng. 36(10), 1353–1362 (2013). 10.1007/s00449-012-0838-6 [DOI] [PubMed] [Google Scholar]
- 25.Tang S. Y., Zhang W., Yi P., Baratchi S., Kalantar-zadeh K., and Khoshmanesh K., Electrophoresis 34(9–10), 1407–1414 (2013). 10.1002/elps.201200659 [DOI] [PubMed] [Google Scholar]
- 26.Tang S.-Y., Zhang W., Baratchi S., Nasabi M., Kalantar-zadeh K., and Khoshmanesh K., Anal. Chem. 85(13), 6364–6371 (2013). 10.1021/ac400741v [DOI] [PubMed] [Google Scholar]
- 27.Zhou J., Ellis A. V., and Voelcker N. H., Electrophoresis 31(1), 2–16 (2010). 10.1002/elps.200900475 [DOI] [PubMed] [Google Scholar]
- 28.Parsi K., Phlebology 30(5), 306–315 (2015). 10.1177/0268355514534648 [DOI] [PubMed] [Google Scholar]
- 29.Dufour S., Deleu M., Nott K., Wathelet B., Thonart P., and Paquot M., Biochim. Biophys. Acta 1726(1), 87–95 (2005). 10.1016/j.bbagen.2005.06.015 [DOI] [PubMed] [Google Scholar]
- 30.Galembeck E., Alonso A., and Meirelles N. C., Chem. Biol. Interact. 113(2), 91–103 (1998). 10.1016/S0009-2797(98)00006-4 [DOI] [PubMed] [Google Scholar]
- 31.Prete P. S., Gomes K., Malheiros S. V., Meirelles N. C., and de Paula E., Biophys. Chem. 97(1), 45–54 (2002). 10.1016/S0301-4622(02)00042-X [DOI] [PubMed] [Google Scholar]
- 32.Sblano C., Micelli S., Notarachille G., and Meleleo D., Open Biol. J. 5, 1–5 (2012). 10.2174/1874196701205010001 [DOI] [Google Scholar]
- 33.Schreier S., Malheiros S. V. P., and de Paula E., Biochim. Biophys. Acta Biomembr. 1508(1), 210–234 (2000). 10.1016/S0304-4157(00)00012-5 [DOI] [PubMed] [Google Scholar]
- 34.Trägner D. and Csordas A., Biochem. J. 244(3), 605–609 (1987). 10.1042/bj2440605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sackmann E., “Biological membranes architecture and function,” in Handbook of Biological Physics, Vol. 1 (Elsevier, 1995), pp. 1–63. 10.1016/s1383-8121(06)80018-7 [DOI] [Google Scholar]
- 36.Isomaa B., Hägerstrand H., and Paatero G., Biochim. Biophys. Acta Biomembr. 899(1), 93–103 (1987). 10.1016/0005-2736(87)90243-4 [DOI] [PubMed] [Google Scholar]
- 37.Sheetz M. P. and Singer S. J., Proc. Natl. Acad. Sci. U.S.A. 71(11), 4457–4461 (1974). 10.1073/pnas.71.11.4457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dilger J. P., McLaughlin S., McIntosh T. J., and Simon S. A., Science 206(4423), 1196–1198 (1979). 10.1126/science.228394 [DOI] [PubMed] [Google Scholar]
- 39.Pethig R. and Kell D. B., Phys. Med. Biol. 32(8), 933 (1987). 10.1088/0031-9155/32/8/001 [DOI] [PubMed] [Google Scholar]
- 40.Kakutani T., Shibatani S., and Sugai M., Bioelectrochem. Bioenerg. 31(2), 131–145 (1993). 10.1016/0302-4598(93)80002-C [DOI] [Google Scholar]
- 41.Irimajiri A., Hanai T., and Inouye A., J. Theor. Biol. 78(2), 251–269 (1979). 10.1016/0022-5193(79)90268-6 [DOI] [PubMed] [Google Scholar]
- 42.Gagnon Z., Gordon J., Sengupta S., and Chang H. C., Electrophoresis 29(11), 2272–2279 (2008). 10.1002/elps.200700604 [DOI] [PubMed] [Google Scholar]
- 43.Gascoyne P., Pethig R., Satayavivad J., Becker F. F., and Ruchirawat M., Biochim. Biophys. Acta Biomembr. 1323(2), 240–252 (1997). 10.1016/S0005-2736(96)00191-5 [DOI] [PubMed] [Google Scholar]
- 44.Morgan H. and Green N. G., AC Electrokinetics (Research Studies Press, 2003). [Google Scholar]
- 45.Su Y. H., Tsegaye M., Varhue W., Liao K. T., Abebe L. S., Smith J. A., Guerrant R. L., and Swami N. S., Analyst 139(1), 66–73 (2014). 10.1039/C3AN01810E [DOI] [PubMed] [Google Scholar]
- 46.Cruz J. M. and García-Diego F. J., J. Phys. D Appl. Phys. 31(14), 1745 (1998). 10.1088/0022-3727/31/14/020 [DOI] [Google Scholar]
- 47.Tan C. H., Huang Z. J., and Huang X. G., Anal. Biochem. 401(1), 144–147 (2010). 10.1016/j.ab.2010.02.021 [DOI] [PubMed] [Google Scholar]
- 48.An R., Massa K., Wipf D. O., and Minerick A. R., Biomicrofluidics 8(6), 064126 (2014). 10.1063/1.4904059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.An R., Wipf D. O., and Minerick A. R., Biomicrofluidics 8(2), 021803 (2014). 10.1063/1.4867557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koch A. L., J. Theor. Biol. 18(1), 133–156 (1968). 10.1016/0022-5193(68)90174-4 [DOI] [PubMed] [Google Scholar]
- 51.Alupoaei C. E. and García-Rubio L. H., Chem. Eng. Commun. 192(2), 198–218 (2005). 10.1080/00986440590473326 [DOI] [Google Scholar]
- 52.Henkelman S., Rakhorst G., Blanton J., and van Oeveren W., Mater. Sci. Eng. C 29(5), 1650–1654 (2009). 10.1016/j.msec.2009.01.002 [DOI] [Google Scholar]
- 53.Xiong G. M., Yuan S., Wang J. K., Do A. T., Tan N. S., Yeo K. S., and Choong C., Acta Biomater. 23, 240–249 (2015). 10.1016/j.actbio.2015.05.003 [DOI] [PubMed] [Google Scholar]
- 54.Turgeon M. L., Clinical Hematology: Theory and Procedures (Lippincott Williams & Wilkins, 2005). [Google Scholar]
- 55.Bhat P. G., Flanagan D. R., and Donovan M. D., J. Pharm. Sci. 85(6), 624–630 (1996). 10.1021/js950381s [DOI] [PubMed] [Google Scholar]
- 56.Khairy K., Foo J., and Howard J., Cell. Mol. Bioeng. 1(2), 173 (2008). 10.1007/s12195-008-0019-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wagener F. A., Volk H.-D., Willis D., Abraham N. G., Soares M. P., Adema G. J., and Figdor C. G., Pharmacol. Rev. 55(3), 551–571 (2003). 10.1124/pr.55.3.5 [DOI] [PubMed] [Google Scholar]
- 58.Jeney V., Balla G., and Balla J., Front. Physiol. 5, 379 (2014). 10.3389/fphys.2014.00379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Andrasko J., J. Forensic Sci. 42(4), 601–607 (1997). 10.1520/JFS14171J [DOI] [PubMed] [Google Scholar]
- 60.Hanson E. K. and Ballantyne J., PLoS One 5(9), e12830 (2010). 10.1371/journal.pone.0012830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wojdyla M., Raj S., and Petrov D., J. Biomed. Opt. 17(9), 097006 (2012). 10.1117/1.JBO.17.9.097006 [DOI] [PubMed] [Google Scholar]
- 62.Strader M. B., Kassa T., Meng F., Wood F. B., Hirsch R. E., Friedman J. M., and Alayash A. I., Redox Biol. 8, 363–374 (2016). 10.1016/j.redox.2016.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meng F., Kassa T., Strader M. B., Soman J., Olson J. S., and Alayash A. I., J. Biol. Chem. 294(11), 4145–4159 (2019). 10.1074/jbc.RA118.006452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Banerjee M., Pramanik M., Bhattacharya D., Lahiry M., Basu S., and Chakrabarti A., J. Biosci. 36(5), 809–816 (2011). 10.1007/s12038-011-9163-5 [DOI] [PubMed] [Google Scholar]
- 65.Meng F. and Alayash A. I., Anal. Biochem. 521, 11–19 (2017). 10.1016/j.ab.2017.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun W., Li X., Wang Y., Zhao R., and Jiao K., Electrochim. Acta 54(17), 4141–4148 (2009). 10.1016/j.electacta.2009.02.055 [DOI] [Google Scholar]
- 67.Rusling J. F. and Nassar A. E. F., J. Am. Chem. Soc. 115(25), 11891–11897 (1993). 10.1021/ja00078a030 [DOI] [Google Scholar]
- 68.Garabagiu S., Mater. Res. Bull. 46(12), 2474–2477 (2011). 10.1016/j.materresbull.2011.08.032 [DOI] [Google Scholar]
- 69.Anderson A. B. and Robertson C. R., Biophys. J. 68(5), 2091–2097 (1995). 10.1016/S0006-3495(95)80388-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koley D. and Bard A. J., Proc. Natl. Acad. Sci. U.S.A. 107(39), 16783–16787 (2010). 10.1073/pnas.1011614107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Irimajiri A., Asami K., Ichinowatari T., and Kinoshita Y., Biochim. Biophys. Acta Biomembr. 896(2), 214–223 (1987). 10.1016/0005-2736(87)90182-9 [DOI] [PubMed] [Google Scholar]
- 72.Asami K., J. Non Cryst. Solids 305(1–3), 268–277 (2002). 10.1016/S0022-3093(02)01110-9 [DOI] [Google Scholar]