Abstract
Helopeltis theivora Waterhouse is a predominant sucking pest in many tropic economic crops, such as tea, cocoa and coffee. Quantitative real-time PCR (qRT-PCR) is one of the most powerful tools to analyze the gene expression level and investigate the mechanism of insect physiology at transcriptional level. Gene expression studies utilizing qRT-PCR have been applied to numerous insects so far. However, no universal reference genes could be used for H. theivora. To obtain accurate and reliable normalized data in H. theivora, twelve candidate reference genes were examined under different tissues, developmental stages and sexes by using geNorm, NormFinder, BestKeeper, Delta Ct and RefFinder algorithms, respectively. The results revealed that the ideal reference genes differed across the treatments, and the consensus rankings generated from stability values provided by these programs suggested a combination of two genes for normalization. To be specific, RPS3A and Actin were the best suitable reference genes for tissues, RPL13A and GAPDH were suitable for developmental stages, EF1α and RPL13A were suitable for sexes, and RPL13A and RPS3A were suitable for all samples. This study represents the first systematic analysis of reference genes for qRT-PCR experiments in H. theivora, and the results can provide a credible normalization for qRT-PCR data, facilitating transcript profiling studies of functional genes in this insect.
Subject terms: Transcription, Physiology
Introduction
Quantitative real-time polymerase chain reaction (qRT-PCR) was developed and became been widely used in modern biology and biomedical sciences, due to the technical advantages of high sensitivity and repeatability, more rapidity and possibility to investigate several target genes simultaneously1,2. This method has been applied in many research directions, including amplification specificity and mRNA expression level detection, restriction fragment length polymorphism (RFLP) and single nucleotide polymorphism (SNP) analysis, etc3,4. However, the accuracy of quantitative analysis can be influenced by the quality and completeness of RNA and cDNA and amplification and experimental procedures among treatments5,6. Therefore, it is essential to minimize the biases by qRT-PCR normalization using internal reference genes7–9. Generally, internal reference genes are derived from housekeeping genes, which roles are maintaining the basic processes of cell cycle. So they are a class of genes expressed relatively stable in theory3,10,11. Nonetheless, the housekeeping genes expression levels is variable substantially in different treatments12,13. Hence, a particular and integrated research for each targeted species and experimental background is recommended for the validation of appropriate reference genes14,15.
The selection of a subset of reference genes that are constitutively expressed in various experiment conditions, including biotic and abiotic factors, is the first committed step for the accurately quantitative analysis of gene expression by qRT-PCR2,16. So plenty of publicly available online statistical tools, such as geNorm7, NormFinder17 and BestKeeper18, have been developed to identify a series of applicable housekeeping genes as internal controls for qRT-PCR normalization. At present, several reference genes have been identified under certain circumstance in Hemipteran insects, including ribosomal protein S9 (RPS9), elongation factor 1 alpha (EF1α) and TATA-box binding protein (TBP) for Aphis glycines19, EF1α, ribosomal protein L5 (RPL5) and RPS18 for Bactericera cockerelli20, β-tubulin1, β-tubulin2 and succinate dehydrogenase subunit A1 (SdhA1) for Ericerus pela21, 16S ribosomal RNA(16SRNA), RPS18 and RPL13 for Lipaphis erysimi (Kaltenbach)22, β-tubulin and EF1α for Bemisia tabaci (Gennadius)23.
Helopeltis theivora Waterhouse (Hemiptera: Miridae) is an important pest of many economic crops, including tea (Camellia sinensis L.), cashew (Anacardium occidentale L.), cocoa (Theobroma cocoa L.), coffee (Coffea spp.), pepper (Piper nigrum L.), cinchona (Cinchona spp.) and guava (Psidium guajava L.), to which it causes considerable damages24. Both adults and nymphs of H. theivora take in the plant juice from young buds, leaves, fruits and shoots with their piercing-sucking mouthparts25. As a consequence, drying up of shoots and fruits can arise from their sucking behavior, which causes more than 25% of crop losses26,27. In addition, this pest demonstrates high persistence of populations against stressful environment through strong ability of adaptation. However, the molecular biology of H. theivora, including the functional study of target genes and physiological mechanisms of its adaptability, is less clarified so far. Lack of suitable reference gene may be a major hurdle for the deeper studies in this insect. So the objective of this study was to assess and validate the most applicable reference genes for data normalization of qRT-PCR under various conditions in H. theivora. Twelve candidate genes from five functional classes were identified and cloned, including 3 structure-related genes (Actin, β-Tubulin1 and β-Tubulin2), 3 ribosomal genes (RPL13A, RPS3A and 18SRNA), 3 protein production genes (EF1α, eukaryotic initiation factor 4A (EIF4A) and ubiquitin-conjugating protein (UBQ)), 2 metabolism-related genes (glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and glucose 6 phosphate dehydrogenase (G6PDH)), and 1 transcription factor gene (TBP). The expression stability of these genes in sixteen samples (six from tissues, six from developmental stages and four from sexes) was performed by five Microsoft Excel-based programs (geNorm, NormFinder, BestKeeper, Delta Ct and RefFinder). The outcome of the study will not only benefit the future quantification of gene expression in H. theivora but will find translational application in other closely related Mirid.
Results
Primer evaluation and amplification efficiency of candidate reference genes
The specificity of all the primers was validated by single amplification band of expected size and melt curve in RT-PCR and qRT-PCR, respectively (Figs S1 and S2). The primer amplification efficiency and linear correlation coefficient (R2), which met the standard requirement of conventional qRT-PCR, were determined by the standard curve. The primer efficiency ranged from 90.1 to 96.9% and the R2 was above 0.998 (Table 1).
Table 1.
Primer amplification characteristics of candidate genes for qRT-PCR in H. theivora.
| Gene (Abbreviation) | Accession Number | Primer sequence | Product length (bp) | Tm (°C) | R 2 | E (%) |
|---|---|---|---|---|---|---|
| Actin | MN027587 | 5′ TACCGCAAGATTCCATACCCA 3' | 109 | 60 | 0.999 | 90.58 |
| 5′ AAGTCCTACGAGCTTCCCGAC 3' | ||||||
| β-Tubulin1 | MN027588 | 5′ GATGAAGGAAGTTGACGAGCA 3' | 237 | 57 | 0.999 | 90.10 |
| 5′ TCACCAGTATACCAATGGAGG 3' | ||||||
| β-Tubulin2 | MN027589 | 5′ TCAGGAAAAGGAGGCGGCAAA 3' | 365 | 61 | 0.999 | 90.99 |
| 5′ GGCGAGGGAACGACAGAGTAG 3' | ||||||
| RPL13A | MN027592 | 5′ ACACAAAATCGAAAGGGGAAA 3' | 153 | 59 | 1.000 | 93.76 |
| 5′ CGACCAAGATGACAGTAGGCA 3' | ||||||
| EF1α | MN027594 | 5′ TCGCTTTCGTTCCAATCTCTG 3' | 107 | 60 | 0.999 | 91.66 |
| 5′ GGCTTTGCCTTCTTTCCTTTC 3' | ||||||
| RPS3A | MN027598 | 5′ GCAGCATGGTGAAAAAGTGGC 3' | 151 | 60 | 0.999 | 91.21 |
| 5′ ACCTGGTTGTGTTGAGCGTAA 3' | ||||||
| GAPDH | MN027590 | 5′ CAATGACCCCTTTTTGGGCG 3' | 163 | 60 | 0.999 | 96.69 |
| 5′ CCATGGGATGGCTTTGGGAT 3' | ||||||
| 18SRNA | MN027591 | 5′ GCGATGTTGAACGTTGCTGT 3' | 110 | 59 | 0.999 | 96.32 |
| 5′ ACGCTTATGGCTCCGTCAAA 3' | ||||||
| G6PDH | MN027593 | 5′ CGTCTGCATATAGCGTCCGT 3' | 237 | 60 | 0.999 | 95.01 |
| 5′ GAGAACCACGTCGTCCACAT 3' | ||||||
| EIF4A | MN027595 | 5′ TGGTTCGAAACATGCCGAGA 3' | 184 | 60 | 0.999 | 93.64 |
| 5′ GCGTTGCTGAATAGCGGAAG 3' | ||||||
| TBP | MN027596 | 5′ CATCCCCAGCATCGGTACTC 3' | 296 | 60 | 0.999 | 93.79 |
| 5′ AAGGCGTCATCGGTGTCATT 3' | ||||||
| UBQ | MN027597 | 5′ GATTATCCGCTGAGCCCTCC 3' | 226 | 60 | 1.000 | 96.87 |
| 5′ CATTAGCGCCACTTTCGTCG 3' |
Expression profiles of candidate reference genes
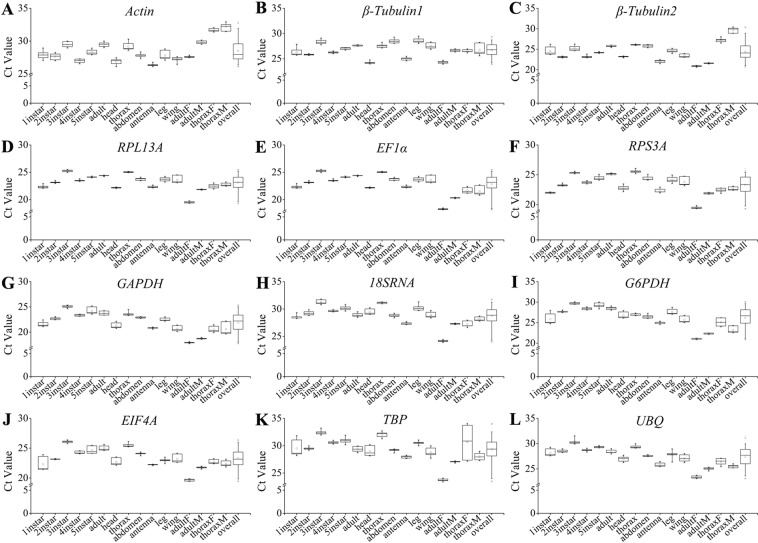
Expression levels of all candidate reference genes in sixteen samples (six tissues, six development stages and four sexes) were detected by qRT-PCR and summarized as Box-Whisker plots. The raw Ct values are available in Table S2. In all samples, GAPDH was found to be the most abundant with the lowest mean Ct value at 21.89 ± 1.98, whereas TBP transcript showed the lowest level with the highest mean Ct value at 29.33 ± 2.08 (Fig. 1G,K). Other candidate genes were also expressed at moderate levels, with mean Ct values at 28.52 ± 1.67, 26.69 ± 1.36, 24.41 ± 2.22, 23.13 ± 1.40, 22.87 ± 2.00, 23.33 ± 1.56, 28.78 ± 1.75, 26.23 ± 2.46, 23.30 ± 1.60, and 27.40 ± 1.83 in response to Actin, β-Tubulin1, β-Tubulin2, RPL13A, EF1α, RPS3A, 18SRNA, G6PDH, EIF4A and UBQ (Fig. 1). The largest and smallest dispersion of Ct values were identified for β-Tubulin1 gene at 4.44 cycles and Actin gene at 2.82 cycles in tissues (Fig. 1A,B), G6PDH gene at 3.88 cycles and UBQ gene at 2.23 cycles in developmental stages (Fig. 1I,L), β-Tubulin2 gene at 8.75 cycles and β-Tubulin1 gene at 2.47 cycles in sexes, and β-Tubulin2 gene at 8.75 cycles and β-Tubulin1 gene at 4.44 cycles in all samples, respectively (Fig. 1B,C).
Figure 1.
Average Ct values of twelve candidate reference genes at different tissues, development stages and sexes in H. theivora.
Selection of credible reference genes
geNorm analysis
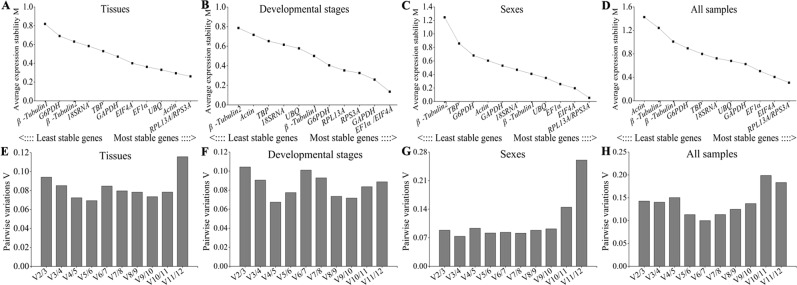
Two parameters of the M values and the pairwise variation V were calculated by geNorm software. The lower M values indicate that the gene was expressed more stably. The M values of twelve candidate genes were lower than 1.5 in different treatments, indicated that the expression levels of all genes were relatively stable. RPL13A and RPS3A were ranked as the most stable reference genes among in different tissues, sexes and all samples, which with the minimum M value 0.260, 0.054 and 0.308, respectively (Fig. 2A,C,D). However, EF1α and EIF4A were recommended as the most stable genes in different developmental stages, and the M value of the two genes were both at 0.135 (Fig. 2B). The role of pairwise variation V is determines the optimal number of reference genes for normalization. As shown in Fig. 2E–H, the pairwise variations V2/V3 were less than 0.15 in four different treatments, including tissues, developmental stages, sexes and all samples, suggesting that the optimal number of reference genes for normalization would be two. In addition, the least stable reference genes were β-Tubulin1 (M value = 0.819) in the case of tissues, β-Tubulin2 (M value = 0.786, 1.245) in different developmental stages and sexes, and Actin (M value = 1.425) in the case of all samples.
Figure 2.
Expression stability and optimal number for normalization of candidate reference genes analyzed by geNorm. (A–D) The stability M values of candidate genes in tissues, developmental stages, sexes and all sample, respectively. (E–H) The pairwise variations Vn/Vn+ 1 of candidate genes in four treatments mentioned above.
NormFinder analysis
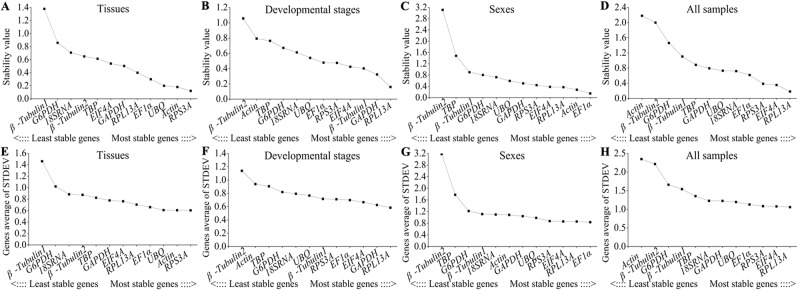
The optimal normalization gene among a set of candidate genes was identified by NormFinder software, providing a stability value of each genes that was negatively correlated with their stability. Based on the rank order assigned by NormFinder, RPL13A (stability value = 0.159, 0.321) was the best independent gene in different developmental stages and all samples, whereas RPS3A (stability value = 0.121) and EF1α (stability value = 0.150) were the optimal reference genes in different tissues and sexes, respectively (Fig. 3A–D). Similar to geNorm analysis, β-Tubulin1, β-Tubulin2 and Actin with maximum stability values (1.379, 1.057, 3.112 and 2.181) were found to be the least stable reference genes in four treatments (Fig. 3A–D).
Figure 3.
Stability analyses of candidate reference genes by NormFinder and Delta Ct algorithms. (A–D) The stability values of candidate genes calculated by NormFinder software in tissues, developmental stages, sexes and all sample, respectively. (E–H) The average STDEV values of candidate genes calculated by Delta Ct algorithms in four treatments mentioned above.
Delta Ct analysis
The Delta Ct algorithm is similar to geNorm, which also relies on the relative pairwise comparisons. The average standard deviation (SD) value of each gene was inversely proportional to the stability. The results showed that the most and least stable genes in four treatments recommended by Delta Ct were in accordance with the analysis by NormFinder (Fig. 3A–C,E,F). In all samples, RPL13A with the lowest SD value at 1.058 was ranked as the most stable gene, followed by EIF4A (1.076), RPS3A (1.085), EF1α (1.128), UBQ (1.194), GAPDH (1.224), 18SRNA (1.227), TBP (1.354), β-Tubulin1 (1.541), G6PDH (1.659), β-Tubulin2 (2.215) and Actin (2.348) (Fig. 3H).
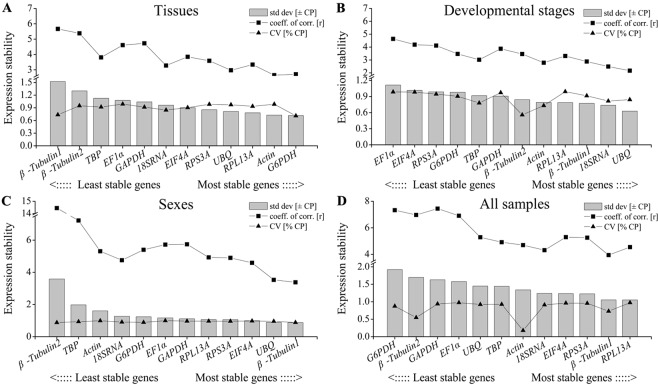
BestKeeper analysis
BestKeeper program is used to estimate three major parameters of each candidate gene from raw Ct values, including the SD value, coefficient of variation (CV) and correlation coefficient (r), and the more stable genes usually have the lower SD and CV values, and higher r value. The results showed that G6PDH (SD value = 0.716), UBQ (SD value = 0.628), β-Tubulin1 (SD value = 0.880) and RPL13A (SD value = 1.053) with the lowest SD values were recommended as the most stable reference genes in tissues, developmental stages, sexes and all samples, respectively (Fig. 4). Interestingly, G6PDH and β-Tubulin1 expression showed significant variation, which were ranked as least reference gene in all samples and tissues although they were the best genes in tissue and sex treatments, respectively (Fig. 4).
Figure 4.
Expression stability of candidate reference genes in four experimental groups calculated by Bestkeeper.
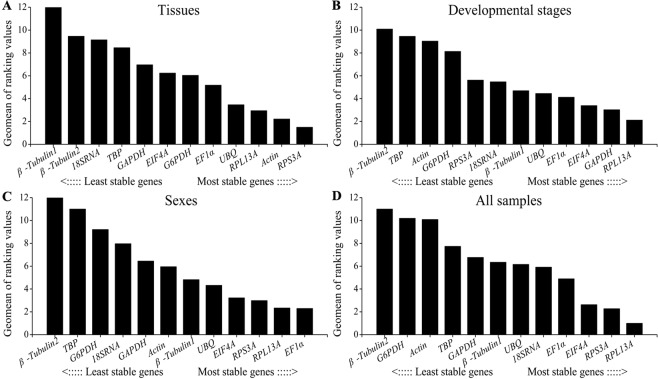
Comprehensive ranking of RefFinder
RefFinder is a comprehensive web-based program that integrates all four above- mentioned software tools to rank the stability of candidate reference genes based on the Geomean of ranking values. The comprehensive rankings of candidate genes in four treatments were showed in Fig. 5. In tissue, developmental stage and sexes samples, RPS3A, RPL13A and EF1α were the most desired reference genes and followed with Actin, GAPDH and RPL13A, respectively (Fig. 5A–C). In all samples, the overall ranking followed by RPL13A, RPS3A, EIF4A, EF1α, 18SRNA, UBQ, β-Tubulin1, GAPDH, TBP, Actin, G6PDH and β-Tubulin2 (Fig. 5D). In addition, the stability rankings of all candidate genes by five analysis software were showed in Table 2.
Figure 5.
Comprehensive stability of candidate reference genes in four experimental groups determined by RefFinder.
Table 2.
Expression stability ranking of the twelve candidate reference genes.
| Experimental conditions | Methods | Stability ranking | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | ||
| Tissues | geNorm | RPL13A/RPS3A | Actin | UBQ | EF1α | EIF4A | GAPDH | TBP | 18SRNA | β-Tubulin2 | G6PDH | β-Tubulin1 | |
| NormFinder | RPS3A | Actin | UBQ | EF1α | RPL13A | GAPDH | EIF4A | TBP | β-Tubulin2 | 18SRNA | G6PDH | β-Tubulin1 | |
| Delta Ct | RPS3A | Actin | UBQ | EF1α | RPL13A | EIF4A | GAPDH | TBP | β-Tubulin2 | 18SRNA | G6PDH | β-Tubulin1 | |
| Bestkeeper | G6PDH | Actin | RPL13A | UBQ | RPS3A | EIF4A | 18SRNA | GAPDH | EF1α | TBP | β-Tubulin2 | β-Tubulin1 | |
| RefFinder | RPS3A | Actin | RPL13A | UBQ | EF1α | G6PDH | EIF4A | GAPDH | TBP | 18SRNA | β-Tubulin2 | β-Tubulin1 | |
| Developmental stages | geNorm | EF1α/EIF4A | GAPDH | RPS3A | RPL13A | G6PDH | β-Tubulin1 | UBQ | 18SRNA | TBP | Actin | β-Tubulin2 | |
| NormFinder | RPL13A | GAPDH | β-Tubulin1 | EIF4A | RPS3A | EF1α | UBQ | 18SRNA | G6PDH | TBP | Actin | β-Tubulin2 | |
| Delta Ct | RPL13A | GAPDH | EIF4A | EF1α | RPS3A | β-Tubulin1 | UBQ | 18SRNA | G6PDH | TBP | Actin | β-Tubulin2 | |
| Bestkeeper | UBQ | 18SRNA | β-Tubulin1 | RPL13A | Actin | β-Tubulin2 | GAPDH | TBP | G6PDH | RPS3A | EIF4A | EF1α | |
| RefFinder | RPL13A | GAPDH | EIF4A | EF1α | UBQ | β-Tubulin1 | 18SRNA | RPS3A | G6PDH | Actin | TBP | β-Tubulin2 | |
| Sexes | geNorm | RPL13A/RPS3A | EIF4A | EF1α | UBQ | β-Tubulin1 | 18SRNA | GAPDH | Actin | G6PDH | TBP | β-Tubulin2 | |
| NormFinder | EF1α | Actin | RPL13A | EIF4A | RPS3A | GAPDH | UBQ | 18SRNA | G6PDH | β-Tubulin1 | TBP | β-Tubulin2 | |
| Delta Ct | EF1α | RPL13A | EIF4A | RPS3A | UBQ | GAPDH | Actin | 18SRNA | β-Tubulin1 | G6PDH | TBP | β-Tubulin2 | |
| Bestkeeper | β-Tubulin1 | UBQ | EIF4A | RPS3A | RPL13A | GAPDH | EF1α | G6PDH | 18SRNA | Actin | TBP | β-Tubulin2 | |
| RefFinder | EF1α | RPL13A | RPS3A | EIF4A | UBQ | β-Tubulin1 | Actin | GAPDH | 18SRNA | G6PDH | TBP | β-Tubulin2 | |
| All samples | geNorm | RPL13A/RPS3A | EIF4A | EF1α | GAPDH | UBQ | 18SRNA | TBP | G6PDH | β-Tubulin1 | β-Tubulin2 | Actin | |
| NormFinder | RPL13A | EIF4A | RPS3A | EF1α | 18SRNA | UBQ | GAPDH | TBP | β-Tubulin1 | G6PDH | β-Tubulin2 | Actin | |
| Delta Ct | RPL13A | EIF4A | RPS3A | EF1α | UBQ | GAPDH | 18SRNA | TBP | β-Tubulin1 | G6PDH | β-Tubulin2 | Actin | |
| Bestkeeper | RPL13A | β-Tubulin1 | RPS3A | EIF4A | 18SRNA | Actin | TBP | UBQ | EF1α | GAPDH | β-Tubulin2 | G6PDH | |
| RefFinder | RPL13A | RPS3A | EIF4A | EF1α | 18SRNA | UBQ | β-Tubulin1 | GAPDH | TBP | Actin | G6PDH | β-Tubulin2 | |
Validation of recommended candidate reference genes
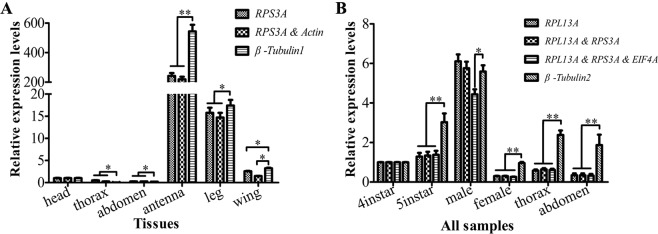
To validate the suitability of recommended reference genes, the mRNA expression of target gene OBP3 in six tissues and all samples (including two tissues, two developmental stages and two sexes) was quantified by the best and worst candidate genes (or gene combinations). Among six tissues, the normalized expression results of OBP3 were similar to those determined using RPS3A (the best candidate gene) and the combination of RPS3A/Actin (the best two), respectively. However, the expression levels of OBP3 normalized using β-Tubulin1 were significantly higher than using RPS3A or combination of RPS3A/Actin in three tissues (antenna, leg and wing) and lower in two tissues (thorax and abdomen) (Fig. 6A). Similarly, the expression of OBP3 quantified by β-Tubulin2 (the worst gene) were significantly greater than by the combination of RPL13A/RPS3A (the best two genes) or RPL13A/RPS3A/EIF4A (the best three genes) in all samples except male adult (Fig. 6B).
Figure 6.
Validation of the gene stability measures. Expression levels of a target gene, OBP3, in six tissues (A) and all samples (including two tissues, two developmental stages, and two sexes) (B) were tested by different normalization reference genes. Bars represent the means and standard deviations of three biological replicates. Single asterisk indicates significant differences among different treatment within OBP3 expression by normalized by reference gene or combination (P < 0.05, Duncan’s test), and double asterisks show extremely significant differences in the expression of OBP3 (P < 0.01, Duncan’s test).
Discussion
qRT-PCR is one kind of conventional method that can quantify the expression of nucleic acidin genetic researches. Evaluate appropriate reference genes should be the key link to get rid of differences and obtain the exact qRT-PCR results15. However, it is difficult to apply a universal appropriate reference gene for different experimental samples. Terribly, the true consequences of target gene’s expression would be concealed if using the bad reference genes28,29. Hence, in order to ensure stable expression of reference genes in a given species, all candidate genes should be assessed under a certain treatments6,30,31. In our study, the stability of twelve candidate genes was identified by five algorithms under multiple tissue, developmental stage and sex treatments in H. theivora. So far, no suitable reference genes are available for this insect or the genus of Helopeltis.
Our results confirmed that the stability of reference genes was variable dramatically under different treatments. For example, Actin was stably expressed under the tissue-specific conditions in H. theivora; but its expression varies significantly among the rest of samples (Table 2). This is similar to the results from Mustard aphid, L. erysimi (Hemiptera: Aphididae), in which the expression of Actin was stable under artificial diet treatments, but variable across different developmental stages22. Meanwhile, the results of different softwares can vary based on different algorithms, which are consistent with the previous results of other insects32,33. For instance, under developmental stage samples in H. theivora, EF1α and EIF4A were recommended as internal standard by geNorm, RPL13A was rated as the best gene by both NormFinder and Delta Ct, whereas UBQ was top choice by BestKeeper (Table 2). Therefore, the combined use of these algorithms can ensure the results are more credible. To this end, RefFinder was used to calculate the integrated ranking of stability for all candidate reference genes.
In recent years, increasing researches have indicated that a single reference gene used for the normalization in qRT-PCR may influence the accuracy of expression level6,15. Analysis of relative gene expression utilizing multiple reference genes showed up as a more reliable alternative34–36. Generally, geNorm is used to determine the optimal number of reference genes6,7. By focusing our findings, according to analysis of the minimum number V by geNorm, all treatments including tissues, developmental stages, sexes and all samples were suitable for using two reference genes.
In this study, we discovered two types of candidate genes (proteins associated with ribosome and protein production) showed relative stability in most samples in H. theivora, which were widely used for qRT-PCR in several species or treatments. Eukaryotic ribosome, consisting of four rRNAs and more than eighty ribosomal proteins, is one of the most evolutionarily conserved organelles37. Ribosomal proteins participates in the process of ribosome synthesis, transcription and cell development regulation, cell differentiation and rRNA processing, etc38. Previous study showed that some ribosome associated genes have been used as stable internal reference for quantitative analysis. Shu et al. (2018) evaluated eight candidate reference genes for relative quantification in Spodoptera litura, and the results indicated that RPL13A and RPL7A were the best genes in larva and fat body samples, respectively. RPL13A was also considered to be stable for tissues, developmental stages and sexes in Aphidius gifuensis and for low-temperature treatments in Thitarodes armoricanus39,40. Other ribosomal proteins have also been identified as internal control, such as RPS18 for the gene expression associated with seasonal variation and division of labor in Apis mellifera41, RPS11 for heat stress conditions in Chilo suppressalis (Walker)42, RP49 for developmental stages in A. mellifera43, RPL32 and RPS15 for tissue samples in H. armigera12, RPL7A and RP18 for most tissues in Lethrus apterus11, and so on. Similarly, our results demonstrated that RPL13A and RPS3A were consistently stably expressed throughout a majority of experimental samples in H. theivora.
The conserved nuclear gene EF1α, a G-protein that binds and recruits aa-tRNAs to the A-site of the ribosome, has been valuable as a higher-level phylogenetic marker in insects and it has also been widely used for stable reference gene44,45. For example, EF1α was the relatively stable gene for developmental stages and photoperiods in Harmonia axyridis33, for cuticle in S. litura15, for developmental stages in Coleomegilla maculate32, and for RNAi treatments in Coccinella septempunctata9. In the present study, consistent with above results, EF1α was ranked as the most stable gene in sex samples of H. theivora. Interestingly, those structure and metabolism related proteins (Actin, Tubulin, GAPDH and G6PDH) were ranked as less stable genes in this study, though these genes have been commonly used as internal controls in many expression researches13,46–48.
In summary, this is the first report of reference genes expression in H. theivora. The expression profiles of twelve candidate reference genes under three treatments (different tissues, developmental stages and sexes) were investigated by five algorithms, geNorm, NormFinder, BestKeeper, Delta Ct and RefFinder. A pair of stable reference genes was particularly recommended for each experimental condition as follows: RPS3A and Actin for tissues, RPL13A and GAPDH for developmental stages, EF1α and RPL13A in sex samples, and RPL13A and RPS3A across all samples (three treatments). Our results demonstrate that there is no a universal reference gene can suitable for all experimental conditions. In addition, this study represents the critical first step to normalize qRT-PCR data for the functional genomics research in H. theivora.
Materials and Methods
Insect rearing
H. theivora adults were collected from cocoa pod in the field of Spice and Beverage Research Institute, Chinese Academy of Tropical Agricultural Sciences, and reared on green beans and 10% sucrose solution in laboratory. A laboratory colony was established and maintained at 26 ± 1 °C, 75 ± 5% relative humidity and 14:10 light: dark cycle. Sample collection begins with the second generation insect.
Sample collection
Individuals at six developmental stages were collected, including 1st instar nymphs (30 individuals), 2nd instar nymphs (20 individuals), 3rd instar nymphs (15 individuals), 4th instar nymphs (10 individuals), 5th instar nymphs (5 individuals) and adults (whole body, 3 individuals). The six tissues of adults (10∼15 individuals), including antenna, head, thorax, abdomen, leg and wing, were dissected by scalpel and washed in cold phosphate buffered saline. The four sex samples were also collected and dissected, including adult (adultF and adultM, 3 individuals) and thorax (thoraxF and thoraxM, 10 individuals) of both female and male. All samples were placed in 1.5 ml centrifuge tubes (50∼100 mg each tube), flash frozen in liquid nitrogen and kept in −80 °C successively. Each treatment contained three biological replications.
Total RNA isolation and cDNA synthesis
Total RNA from different tissues, developmental stages and sexes were extracted by TRIzol Reagent (Invitrogen, USA) following the manufacturer’s protocol. Each sample was disrupted and homogenized with 1 ml TRIzol reagent. Depending on the pellet size, moderate amounts of RNase-free water was added to dissolve the precipitate. The concentration and purity of total RNA were measured by fluorescence microplate reader (BioTek, USA). The isolated RNA (1 µg per samples) was reversely transcribed to first-strand cDNA using PrimeScript RT reagent Kit with gDNA Eraser (TaKaRa, China) following the manufacturer’s instructions. The products were stored at −20 °C.
Real-time PCR (RT-PCR) and quantitative real-time PCR (qRT-PCR)
Twelve candidate reference genes were identified and cloned based on the transcriptome data of H. theivora. The primers and sequence data of twelve genes were listed in Table 1 and S1. RT-PCR amplifications were performed by denaturing at 94 °C for 3 min, followed by 35 cycles of 94 °C for 30 s, 60 °C for 30 s and 72 °C for 1 min, with a final extension at 72 °C for 10 min. The amplification products were detected by 1% agarose gel electrophoresis and extracted by E.Z.N.A.TM Gel Extraction Kit (Omega, USA). The DNA fragments were ligated with pMD-19T and transformed into Escherichia coli DH5α (TaKaRa, China). Plasmid were extracted by E.Z.N.A.TM Plasmid Miniprep Kit II (Omega, USA) and used as the templates for standard curve of candidate genes.
qRT-PCR reactions were performed with three technical replicates on BioRad CFX96 Real-Time PCR detection system using 2× TB Green Premix Ex Taq (TaKaRa, China). Amplification conditions consisted of an initial denaturation at 95 °C for 3 min, followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s. After reaction, a melting curve analysis from 65 °C to 95 °C was applied to all reactions to ensure consistency and specificity of the amplified product. A series of 10-fold dilution of plasmids were used to create the five-point standard curves using the linear regression model18. The regression equation was carried out to calculate the efficiency (E) and correlation coefficient (R2) of each primer pair. The efficiencies (E) of corresponding primers were estimated according to the equation:
Data mining and statistical analysis
The cycle threshold values (Ct values) from qRT-PCR were collected and the stability of candidate reference genes were ranked by using Microsoft Excel based software tools (or algorithm), including geNorm, NormFinder, BestKeeper, Delta Ct and RefFinder. The relative quantities converted from the raw Ct values (the highest relative quantity of gene was set to 1) were used as input data for geNorm and NormFinder. geNorm calculates the expression stability value (M) and pair wise variation (V). Gene expression is considered stable when the M value is below 1.5, and the lowest M values are produced by genes with the most stable expression. Besides, the value of Vn/Vn+1 was used for determine the optimal normalization reference gene number and geNorm Vn/Vn+1 < 0.15 could be the standard for better normalization7. NormFinder provides a stability value for each gene, which is a direct measure of the estimated both intra- and inter- group expression variation17. BestKeeper use raw Ct values and PCR efficiency to determine the optimal reference genes18. Delta Ct algorithm evaluated the stability rank order of reference genes by comparing the relative expression of pairwise genes within each sample49. Finally, the web-based program RefFinder (http://www.leonxie.com/referencegene.php), which is a comprehensive platform integrating above four algorithms, provided an overall ranking of the stability of candidate reference genes50.
Odorant binding protein-3 (OBP3, Accession number: MN062611) gene was used to evaluate the validity of selected reference genes under different experimental conditions. qRT-PCR amplification of OBP3 was obtained from the transcriptome data and performed with primers: Forward (CAGCGGAGTCGAAGATACCC) and Reverse (TGGCCTTCGCTAGTTTTGGT). The relative expression levels of OBP3 were determined according to the Ct values based on the 2−ΔΔCt method51. All the treatments were performed in three biological and technical replicates, respectively. One-way ANOVA was used to compare the effects of treatments. Statistical analyses were performed using SPSS 20.0 (SPSS, Inc., USA).
Supplementary information
Acknowledgements
This work was supported by grants from Natural Science Foundation of Hainan province (Grant No. 317272) and Central Public-interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences (Grant No. 1630142018007).
Author Contributions
Z.W., Q.M. and A.L. conceived and designed the research. Z.W., Q.M. and X.Z. performed the experiments. S.S., S.G. and Y.G. contributed regents and analytical tools. Z.W. and Q.M. analyzed data and constructed tables and figures. Z.W. wrote the manuscript. Q.M. and A.L. revised the draft. All authors reviewed and approved the final version.
Data Availability
All data generated or analyzed during this study are included in this published article and its Supplementary Information files.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zheng Wang and Qianqian Meng contributed equally.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-49479-1.
References
- 1.Prunier FP, Chouteau M, Whibley A, Joron M, Llaurens V. Selection of valid reference genes for reverse transcription quantitative PCR analysis in Heliconius numata (Lepidoptera: Nymphalidae) J. Insect Sci. 2016;16:50. doi: 10.1093/jisesa/iew034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang L, et al. Selection of reference genes for qRT-PCR and expression analysis of high-altitude-related genes in grassland caterpillars (Lepidoptera: Erebidae: Gynaephora) along an altitude gradient. Ecol. Evol. 2017;7:9054–9065. doi: 10.1002/ece3.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Płachetka-Bożek A, Augustyniak M. Evaluation of candidate reference genes for quantitative gene expression analysis in Spodoptera exigua after long-time exposure to cadmium. Sci. Rep. 2017;7:8338. doi: 10.1038/s41598-017-08630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nguyen DQ, Eamens AL, Grof CPL. Reference gene identification for reliable normalisation of quantitative RT-PCR data in Setaria viridis. Plant Methods. 2018;14:24. doi: 10.1186/s13007-018-0293-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bustin SA, Benes V, Nolan T, Pfaffl MW. Quantitative real-time RT-PCR– a perspective. J. Mol. Endocrinol. 2005;34:597–601. doi: 10.1677/jme.1.01755. [DOI] [PubMed] [Google Scholar]
- 6.Yang XW, Pan HP, Yuan L, Zhou XG. Reference gene selection for RT-qPCR analysis in Harmonia axyridis, a global invasive lady beetle. Sci. Rep. 2018;8:2689. doi: 10.1038/s41598-018-20612-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vandesompele Jo, De Preter Katleen, Pattyn Filip, Poppe Bruce, Van Roy Nadine, De Paepe Anne, Speleman Frank. Genome Biology. 2002;3(7):research0034.1. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan HP, Yang XW, Siegfried BD, Zhou XG. A comprehensive selection of reference genes for RT-qPCR analysis in a predatory lady beetle, Hippodamia convergens (Coleoptera: Coccinellidae) PLoS ONE. 2015;10:e0125868. doi: 10.1371/journal.pone.0125868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang CX, et al. Selection of reference genes for RT-qPCR analysis in Coccinella septempunctata to assess un-intended effects of RNAi transgenic plants. Front. Plant Sci. 2016;7:1835. doi: 10.3389/fpls.2016.01672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen H, et al. Reference genes selection for quantitative gene expression studies in Pinus massoniana L. Trees-Struct. Func. 2016;30:685–696. doi: 10.1007/s00468-015-1311-3. [DOI] [Google Scholar]
- 11.Nagy NA, et al. Evaluation of potential reference genes for real-time qPCR analysis in a biparental beetle, Lethrus apterus (Coleoptera: Geotrupidae) PeerJ. 2017;5:e4047. doi: 10.7717/peerj.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang SD, et al. Identification and validation of reference genes for normalization of gene expression analysis using qRT-PCR in Helicoverpa armigera (Lepidoptera: Noctuidae) Gene. 2015;555:393–402. doi: 10.1016/j.gene.2014.11.038. [DOI] [PubMed] [Google Scholar]
- 13.Shi CH, et al. Evaluation of Housekeeping Genes for Quantitative Real-Time PCR Analysis of Bradysia odoriphaga (Diptera: Sciaridae) Int. J. Mol. Sci. 2016;17:1034. doi: 10.3390/ijms17071034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hruz T, et al. RefGenes: identification of reliable and condition specific reference genes for RT-qPCR data normalization. BMC Genomics. 2011;12:156. doi: 10.1186/1471-2164-12-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shu BS, et al. Evaluation of reference genes for real-time quantitative PCR analysis in larvae of Spodoptera litura exposed to azadirachtin stress conditions. Front. Physiol. 2018;9:372. doi: 10.3389/fphys.2018.00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saha P, Blumwald E. Assessing reference genes for accurate transcript normalization using quantitative real-time PCR in pearl millet [Pennisetum glaucum (L.) R. Br] PLoS ONE. 2014;9:e106308. doi: 10.1371/journal.pone.0106308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 18.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004;26:509–515. doi: 10.1023/B:BILE.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 19.Bansal R, Mamidala P, Mian MR, Mittapalli O, Michel AP. Validation of reference genes for gene expression studies in Aphis glycines (Hemiptera: Aphididae) J. Econ. Entomol. 2012;105:1432–1438. doi: 10.1603/EC12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ibanez F, Tamborindeguy C. Selection of reference genes for expression analysis in the potato psyllid, Bactericera cockerelli. Insect Mol. Biol. 2016;25:227–238. doi: 10.1111/imb.12219. [DOI] [PubMed] [Google Scholar]
- 21.Yu SH, et al. Identification and evaluation of reference genes in the Chinese white wax scale insect Ericerus pela. Springerplus. 2016;5:791. doi: 10.1186/s40064-016-2548-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koramutla MK, Aminedi R, Bhattacharya R. Comprehensive evaluation of candidate reference genes for qRT-PCR studies of gene expression in mustard aphid, Lipaphis erysimi (Kalt) Sci. Rep. 2016;6:25883. doi: 10.1038/srep25883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai TM, Lü ZC, Liu WX, Wan FH. Selection and validation of reference genes for qRT-PCR analysis during biological invasions: The thermal adaptability of Bemisia tabaci MED. PLoS ONE. 2017;12:e0173821. doi: 10.1371/journal.pone.0173821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roy S, Muraleedharan N, Mukhapadhyay A, Handique G. The tea mosquito bug, Helopeltis theivora Waterhouse (Heteroptera: Miridae): its status, biology, ecology and management in tea plantations. Int. J. Pest Manage. 2015;61:179–197. doi: 10.1080/09670874.2015.1030002. [DOI] [Google Scholar]
- 25.Prabhakaran P, Radhakrishnan B, Srikumar KK, Suresh Kumar B. Efficacy of certain common ferns against red spider mite Oligonychus coffeae and tea mosquito bug Helopeltis theivora infesting tea. Plant Protect. Sci. 2017;53:232–242. doi: 10.17221/23/2015-PPS. [DOI] [Google Scholar]
- 26.Bhau BS, Mech J, Borthakur S, Bhuyan M, Bhattacharyya PR. Morphological and genetic diversity studies among populations of tea mosquito bug, Helopeltis theivora from Assam, India. Mol. Biol. Rep. 2014;41:7845–7856. doi: 10.1007/s11033-014-3678-4. [DOI] [PubMed] [Google Scholar]
- 27.Roy S, Prasad AK. Sex-based variation in insecticide susceptibility and tolerance related biochemical parameters in tea mosquito bug Helopeltis theivora. Phytoparasitica. 2018;46:405–410. doi: 10.1007/s12600-018-0670-x. [DOI] [Google Scholar]
- 28.VanGuilder HD, Vrana KE, Freeman WM. Twenty-five years of quantitative PCR for gene expression analysis. BioTechniques. 2008;44:619–626. doi: 10.2144/000112776. [DOI] [PubMed] [Google Scholar]
- 29.Dzaki N, Ramli KN, Azlan A, Ishak IH, Azzam G. Evaluation of reference genes at different developmental stages for quantitative real-time PCR in Aedes aegypti. Sci. Rep. 2017;7:43618. doi: 10.1038/srep43618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tellin O, et al. Housekeeping genes as internal standards: use and limits. J. Biotechnol. 1999;75:291–295. doi: 10.1016/S0168-1656(99)00163-7. [DOI] [PubMed] [Google Scholar]
- 31.Zhu X, et al. Selection and evaluation of reference genes for expression analysis using qRT-PCR in the beet armyworm Spodoptera exigua (Hübner) (Lepidoptera: Noctuidae) PLoS ONE. 2014;9:e84730. doi: 10.1371/journal.pone.0084730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang CX, et al. Selection of reference genes for RT-qPCR analysis in a predatory biological control agent, Coleomegilla maculata (Coleoptera: Coccinellidae) Sci. Rep. 2015;5:18201. doi: 10.1038/srep18201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qu C, et al. Selection and evaluation of reference genes for expression analysis using quantitative real-time PCR in the Asian Ladybird Harmonia axyridis (Coleoptera: Coccinellidae) PLoS ONE. 2018;13:e0192521. doi: 10.1371/journal.pone.0192521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morales MA, et al. Selection of reference genes for expression studies of xenobiotic adaptation in Tetranychus urticae. Int. J. Biol. Sci. 2016;12:1129–1139. doi: 10.7150/ijbs.16319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fu W, et al. Exploring valid reference genes for quantitative real-time PCR analysis in Plutella xylostella (Lepidoptera: Plutellidae) Int. J. Biol. Sci. 2014;9:792–802. doi: 10.7150/ijbs.5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan HP, et al. Assessment of potential risks of dietary RNAi to a soil micro-arthropod, Sinella curviseta Brook (Collembola: Entomobryidae) Front. Plant Sci. 2016;7:1028. doi: 10.3389/fpls.2016.01028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dolezal JM, Dash AP, Prochownik EV. Diagnostic and prognostic implications of ribosomal protein transcript expression patterns in human cancers. BMC Cancer. 2018;18:275. doi: 10.1186/s12885-018-4178-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xue S, Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Bio. 2012;13:355–369. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu G, et al. Evaluation of reference genes for reverse transcription quantitative PCR studies of physiological responses in the ghost moth, Thitarodes armoricanus (Lepidoptera, Hepialidae) PLoS ONE. 2016;11:e0159060. doi: 10.1371/journal.pone.0159060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao XK, et al. Identification and validation of reference genes for gene expression analysis in Aphidius gifuensis (Hymenoptera: Aphidiidae) PLoS ONE. 2017;12:e0188477. doi: 10.1371/journal.pone.0188477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moon KH, Lee SH, Kim YH. Validation of quantitative real-time PCR reference genes for the determination of seasonal and labor-specific gene expression profiles in the head of western honey bee, Apis mellifera. PLoS ONE. 2018;13:e0200369. doi: 10.1371/journal.pone.0200369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu J, Lu MX, Cui YD, Du YZ. Selection and evaluation of reference genes for expression analysis using qRT-PCR in Chilo suppressalis (Lepidoptera: Pyralidae) J. Econ. Entomol. 2017;110:683–691. doi: 10.1093/jee/tow297. [DOI] [PubMed] [Google Scholar]
- 43.Lourenço AP, Mackert A, Cristino ADS, Simões ZLP. Validation of reference genes for gene expression studies in the honey bee, Apis mellifera, by quantitative real-time RT-PCR. Apidologie. 2008;39:372–385. doi: 10.1051/apido:2008015. [DOI] [Google Scholar]
- 44.Martoni F, Bulman SR, Pitman A, Armstrong KF. Elongation factor-1α accurately reconstructs relationships amongst psyllid families (Hemiptera: Psylloidea), with possible diagnostic implications. J. Econ. Entomol. 2017;110:2618–2622. doi: 10.1093/jee/tox261. [DOI] [PubMed] [Google Scholar]
- 45.Mateyak MK, et al. Demonstration of translation elongation factor 3 activity from a non-fungal species, Phytophthora infestans. PLoS ONE. 2018;13:e0190524. doi: 10.1371/journal.pone.0190524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Boer ME, et al. Reference genes for QRT-PCR tested under various stress conditions in Folsomia candida and Orchesella cincta (Insecta, Collembola) BMC Mol. Biol. 2009;10:54. doi: 10.1186/1471-2199-10-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Z, et al. Azadirachtin-induced apoptosis involves lysosomal membrane permeabilization and cathepsin L release in Spodoptera frugiperda Sf9 cells. Int. J. Biochem. Cell B. 2015;64:126–135. doi: 10.1016/j.biocel.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 48.Van Hiel MB, et al. Identification and validation of housekeeping genes in brains of the desert locust Schistocerca gregaria under different developmental conditions. BMC Mol. Biol. 2009;10:56. doi: 10.1186/1471-2199-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Silver N, Best S, Jiang J, Thein SL. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006;7:33. doi: 10.1186/1471-2199-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xie F, Xiao P, Chen D, Xu L, Zhang B. miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012;80:75–84. doi: 10.1007/s11103-012-9885-2. [DOI] [PubMed] [Google Scholar]
- 51.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C (T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its Supplementary Information files.