Abstract
Telomeric repeats in fungi of the subphylum Saccharomycotina exhibit great inter- and intra-species variability in length and sequence. Such variations challenged telomeric DNA-binding proteins that co-evolved to maintain their functions at telomeres. Here, we compare the extent of co-variations in telomeric repeats, encoded in the telomerase RNAs (TERs), and the repeat-binding proteins from 13 species belonging to the Yarrowia clade. We identified putative TER loci, analyzed their sequence and secondary structure conservation, and predicted functional elements. Moreover, in vivo complementation assays with mutant TERs showed the functional importance of four novel TER substructures. The TER-derived telomeric repeat unit of all species, except for one, is 10 bp long and can be represented as 5′-TTNNNNAGGG-3′, with repeat sequence variations occuring primarily outside the vertebrate telomeric motif 5′-TTAGGG-3′. All species possess a homologue of the Yarrowia lipolytica Tay1 protein, YlTay1p. In vitro, YlTay1p displays comparable DNA-binding affinity to all repeat variants, suggesting a conserved role among these species. Taken together, these results add significant insights into the co-evolution of TERs, telomeric repeats and telomere-binding proteins in yeasts.
Subject terms: Evolutionary biology, Telomeres
Introduction
Telomeres are dynamic and complex nucleoprotein structures located at the ends of linear chromosomes. Their principal function is to solve both the end-replication and the end-protection problems, while also contributing to the regulation of gene expression, chromosome movement and localization1–6. In most eukaryotic organisms, telomeres of nuclear chromosomes are composed of short DNA tandem repeats arranged in a double-stranded array and terminating in 3′ single-stranded (usually G-rich) overhang. The prevalent mechanism of telomere maintenance is based on a specialized reverse transcriptase called telomerase. The catalytic subunit of telomerase (TERT, Est2 in budding yeast) employs an RNA molecule (TER) as a scaffold, an anchor to the telomere and a template for the elongation of the 3′ overhang7. Telomeric repeats are bound by specific DNA-binding proteins, which, along with their interaction partners, form the shelterin complex6,8–11. These proteins protect telomeric DNA against recombination and undue DNA repair throughout most of the cell cycle, but relinquish telomeres to telomerase and DNA polymerases during S-phase12,13.
In order to ensure the dynamic switching between the inaccesible and extensible states of telomeres, when the 3′ overhang can be associated with telomerase and elongated, telomeric repeats have to satisfy specific criteria: telomeric proteins need to bind them strongly enough to secure the correct assembly of shelterin, but they also need to be able to transiently dissociate from telomeric DNA to allow its spatio-temporal accessibility to replication and transcription machineries14–16.
Given their essential roles in maintaining linear chromosomes, it is not surprising that the fundamental features of telomeres are conserved. On the other hand, there are several notable variations in how the conserved concepts are achieved. Perhaps the most dramatic deviation is represented by Drosophila and related species, where the short telomeric repeats are replaced by long complex retrotransposons17. Another example of variability in the mechanisms of telomere maintenance is represented by chromosomal termini of ascomycetous yeasts. Their telomeric sequences are rapidly evolving in both primary sequence and length18–20 and, with the exception of fission yeasts, their telomeric nucleoprotein complex differs significantly from the typical vertebrate shelterin. The double-stranded regions of telomeres in these species are bound by a variety of distantly related Myb-domain containing proteins21 and the CST complex regulates the maintenance of both G- and C-rich telomeric strands22. The reason for this rapidly divergent evolution of telomeres in yeasts is unknown. However, it is clear that the changes in sequences of telomeric repeats, derived from altered TER templates and degenerate template use, are accompanied by co-evolution of telomere-binding proteins, whose structures are adapting to the rapidly evolving cognate DNA21. In some phylogenetic branches, especially those comprising species that contain heterogenous telomeric repeats, this selection pressure resulted in the emergence of proteins exhibiting flexibility in DNA-binding specificity (such as Rap1 from Saccharomyces cerevisiae20 or Taz1 from Schizosacharomyces pombe23). Ascomycetous yeasts thus provide an ideal opportunity to study the co-evolution of TERs, telomeric repeats and the DNA-binding proteins that recognize them.
TERs from several yeast species, including S. cerevisiae, S. pombe and Kluyveromyces lactis, were thoroughly studied in the past, revealing the mechanism by which the sequence of the template domain of TER specifies the sequence of the telomeric repeats24–29. The extremely high degree of variability in length and sequence of TERs, even in closely related species, was later attributed to the fact that parts of TER may serve as a flexible scaffold for the assembly of protein complexes which bind only specific small parts of the RNA molecule30. The interactions between TER and a number of regulatory proteins were reported to modulate the activity, processivity and stability of the telomerase complex, affecting the overall telomere length and possibly also the sequence of telomeric repeats. For example, in S. cerevisiae, the variability of telomeric repeats is caused by low nucleotide incorporation processivity of telomerase29,31–34. Moreover, the important roles of specific RNA chaperones such as TCAB1 and assembly regulators including Lar7 and Pof8 in regulation of telomerase activity were described recently35–37. Comparative and functional analyses of yeast TERs also revealed several domains responsible for the assembly of the catalytic core of telomerase (pseudoknot, three-way junction (TWJ)), defining the telomeric repeat (template domain, template boundary element) and interaction with additional protein regulators (Ku-binding domain, Est1-binding domain, Sm site, Pop6, Pop7-binding/P3-like domain)18,38–44. In addition, several features of TER sequence facilitate spliceosomal cleavage, a special type of post-transcriptional processing discovered in S. pombe45. In this process, the TER precursor RNA is processed by the spliceosome, but undergoes only the first trans-esterification reaction of splicing, which releases the first exon as a mature TER45. The required sequence features differ among the yeast species where spliceosomal cleavage was later identified as a mode of TER processing, including the Schizosaccharomyces genus, Aspergillus and Neurospora crassa18,46–48.
Some functional domains are present in all TERs, although their sequence is not completely conserved, while others were described only in specific lineages (e.g. the snoRNA typical H/ACA box is conserved in vertebrate TERs, but not present in yeast TERs). In such cases, the presence or absence of a specific feature in TER often depends on the presence and activity of its interaction partner, so the co-evolution of TERs and their regulatory proteins takes place side by side with the co-evolution of telomeric repeats and telomere-binding proteins49,50. Hence, detailed analyses of conserved domains of TERs from phylogenetically distant species, such as those of filamentous fungi46,51, as well as the identification of novel features with potential to reveal previously unknown TER-protein interactions, are instrumental for better understanding of the evolution of yeast telomeric repeats and ultimately the entire telomere-protecting system.
Tracing back the actual evolutionary steps which led to the present variability of yeast telomeric repeats is difficult because most of these sequences are too divergent for simple alignment and the identification of conserved positions is complicated by their variable length. However, the telomeric repeat of Yarrowia lipolytica (5′-TTagtcAGGG-3′), which belongs to one of the basal phylogenetical lineages of Saccharomycotina, is relatively short, includes the canonical vertebrate motif 5′-TTAGGG-3′ and is bound both in vitro and in vivo by Tay1p52,53. Tay1 protein exhibits high affinity binding to both mammalian and Y. lipolytica telomeric repeats with stronger preference for the mammalian type54. Although it lacks a putative dimerization region, it contains two Myb domains exhibiting higher similarity to the Myb domains of human TRF1 and TRF2 than to Myb domains of other yeast telomere-binding proteins such as Taz1 or Rap153. Interestingly, Rap1p from several yeast species also possess 2 Myb domains, which are able to form a stable complex with the DNA55, but the overall structural and sequential similarity between Rap1p and Tay1p as well as their respective Myb domains is limited21. Homologues of Tay1p in other species are either absent or play roles at nontelomeric loci23,56,57 reflecting a major deviation of their telomeric repeats from the 5′-TTAGGG-3′ sequence. Y. lipolytica may therefore serve as a model of the ancestral tipping point, where the canonical type of telomere was converted to its divergent derivatives present in other ascomycetous yeasts.
To investigate this transition in more detail, we focused on analyses of TERs, telomeric repeats and telomere-binding proteins from 13 species belonging to the Yarrowia clade58, i.e. twelve species of the genus Yarrowia and Candida hispaniensis, which was used as an outgroup. These species exhibit a pronounced sequence divergence, with Y. galli and Y. deformans being the closest species to Y. lipolytica with 87–89% similarity in average between orthologs59. Y. alimentaria and Y. phangngensis appear to be the most divergent and dynamic species, as exemplified by the evolution of the lipase gene family60. The comparison of telomeric repeat sequences between individual species enabled us to identify conserved positions within a repeat required for the binding by Tay1p homologs that seem to retain their telomeric functions. We identified putative genomic loci for TER in all species and showed that a ∆ter mutant of Y. lipolytica undergoes rapid (yet reversible) shortening of telomeres. Comparative analysis of TER sequences resulted in the identification of conserved as well as novel structural elements. These elements were subjected to functional analysis to test their involvement in proper functioning of telomerase in vivo. Our results illustrate how investigation of telomeres in Yarrowia clade species may be instrumental in understanding the paths that eventually resulted in an unprecedented diversification of telomeres in yeasts.
Results
Identification and characterization of TER loci in the Yarrowia clade species
To identify the TER locus of Y. lipolytica, we first performed a BlastN search of its genome using the sequence of 1.5 telomeric repeat (5′-GTTAGTCAGGGTTAG-3′) as a query. Using this approach we identified a single intergenic locus (TER locus) on chromosome B that is actively transcribed. Although based on our RNA-seq analysis (see also below), the longest possible TER transcripts would be 1400–1500 nt long, ribo-depleted RNA-seq data showed that most reads mapped to this area end at ~950 nt downstream from the 5′ end of TER (Supplementary Fig. S1). The Y. lipolytica TER locus lies between two open reading frames encoding homologs of S. cerevisiae FRE2 (YALI0B13046g) and FRE3 (YALI0B13090g) genes (Fig. 1). In the reference genome of strain E150, the TER locus is flanked on one side by a retrotransposon that is absent in strain H222-S4 used for functional analyses. Similar search criteria as for Y. lipolytica were used to identify the putative TER loci in other species and the final candidates were chosen with respect to synteny within the TER locus containing region in Y. lipolytica. The synteny was retained in most of the species with the exceptions of Y. hollandica and Y. phangngensis, where the TER locus was transferred into another part of the genome due to two independent translocational events (Supplementary Fig. S2). In the case of C. hispaniensis, further changes in the genome structure altered the positions of both TER locus and neighbouring genes, resulting in the loss of synteny in this species. The coordinates of the TER in each species were determined with RNA-seq data (data not shown).
Figure 1.
Comparison of the sequences of telomeric repeats and template domains of TERs in the Yarrowia clade species. Bases in red indicate substitutions in the telomeric repeat and the template domain of TER, respectively, compared with Y. lipolytica TER. Sequences in grey boxes represent the vertebrate telomeric motif (Telomeric repeat column) or the template sequence required for its synthesis (Template domain in TER column).
Evolution of telomeric repeats in the species belonging to the Yarrowia clade
The sequences of telomeric repeats of 12 species belonging to the Yarrowia clade were determined by searching the ends of the scaffolds for short repeats resembling the telomeric sequence of Y. lipolytica or identifying these sequences in predicted TERs (see below) and deducing the exact sequence of the repeat from the conserved template region. In all 13 species (including Y. lipolytica) the telomeric repeat unit is composed of 10 nucleotides. Except for C. hispaniensis, these repeats are represented by a sequence 5′-TTNNNNAGGG-3′, where a canonical (vertebrate-type) telomeric repeat is interrupted by an insertion of four nucleotides (Fig. 1). This indicates that 5′-TT—AGGG-3′ sequences are less prone to substitutions than the spacer region, probably reflecting the binding preferences of telomere-binding proteins in the Yarrowia clade species (see below). More dramatic changes within the telomeric repeat occurred in C. hispaniensis, where four substitutions within the spacer region were accompanied by one change (G-to-A) in the canonical part of the repeat (Fig. 1). This reflects a relatively distant relationship of C. hispaniensis to the other species of this group.
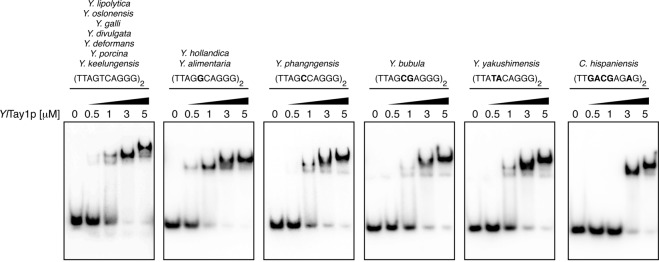
Next, we asked whether the changes in telomeric repeats are reflected by the amino acid sequences of Myb domains of Tay1, the major telomere-binding protein in Y. lipolytica. Comparison of the Myb domains of Tay1p homologs from the Yarrowia clade species revealed that in the 7 species containing the Y. lipolytica-type telomeric repeat (5′-TTAGTCAGGG-3′) the Myb domains are with one exception identical (Supplementary Fig. S3). In Y. porcina there is one (Myb1) and two (Myb2) substitutions whose effect on affinity of the protein to telomeres was not tested. Importantly, in three out of five species with a single or double substitutions in the spacer region of the repeat (Y. bubula, Y. hollandica and Y. phangngensis), both Myb domains are identical to those of YlTay1 protein. Interestingly, even in C. hispaniensis, where 5 out of 10 nucleotides within the repeat sequence are different when compared with Y. lipolytica, the Myb domains of Tay1p homologue experienced only 4 (Myb1) and 2 (Myb2) amino acid substitutions that may be associated with changes in their specificity toward the telomeric repeats. Yet, the conservation of Myb domains of Tay1p homologues in the species with a variant version of telomeric repeat implies that the protein either tolerates the changes in the spacer part of the telomeric sequence, or it was replaced by another DNA-binding protein. To address the first possibility, we analyzed the DNA-binding properties of purified YlTay1p using all variants of telomeric repeats found in the Yarrowia clade species as a substrate (Fig. 2). The electrophoretic-mobility shift assay (EMSA) experiments demonstrated that YlTay1p binds most of these sequences with affinity comparable to the natural telomeric repeats of Y. lipolytica. Slightly decreased affinity toward the probe was observed only in case of C. hispaniensis repeat. Altoghether, our data indicate that YlTay1p binds with comparable affinities to the telomeric sequences containing motif 5′-TT—AGGG-3′.
Figure 2.
Effect of the substitutions in telomeric repeats on the affinity of YlTay1p. EMSA experiments used purified YlTay1p at the indicated concentrations and end-labelled double-stranded DNA probes (with the shown sequences corresponding to the telomeric repeat of the listed species).
Deletion of the putative TER locus affects telomere length in Y. lipolytica
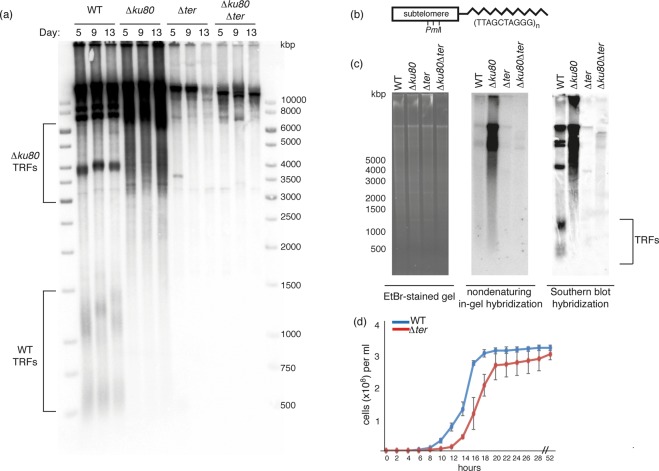
To assess the phenotype of Y. lipolytica cells lacking functional telomerase RNA, we constructed a strain (∆ter) with the entire TER locus replaced by a deletion cassette containing the URA3 selection marker. In parallel, we deleted the TER locus in a ∆ku80 strain lacking the functional YlKU80 gene61, whose product was shown to be involved in telomere maintenance in several model organisms62–65. The resulting four strains (WT, ∆ku80, ∆ter and ∆ku80∆ter) were subjected to the measurement of their telomere length using telomere restriction fragment (TRF) analysis. As reported earlier, the length of TRFs in the WT strain after digestion with PmlI varies between 500 and 1500 nt52 (Fig. 3a,b). The nature of longer fragments hybridizing to the telomeric probe is unclear. They may represent population of DNA fragments from chromosomal ends lacking PmlI site in the vicinity of telomeric tract and/or nontelomeric DNA fragments containing telomere-like repeats that are resistant to BAL-31 treatment52.
Figure 3.
Deletions of TER and/or YlKU80 genes affect telomere length, single-stranded telomeric overhang and growth in Y. lipolytica. (a) The strains with indicated genotypes were passaged 2 times (each passage took 4 days) and the length of telomeres was assessed by TRF analysis. (b) Scheme of a chromosomal end containing a subtelomere with multiple PmlI restriction sites (modified from Kinsky et al., 2010). (c) The length of telomeric overhangs was measured by in-gel hybridization under nondenaturing conditions using a C-rich telomeric probe (central panel) and compared with the hybridization signal obtained by standard Southern blot hybridization (right panel). Ethidium bromide stained gel served as a loading control (left panel). (d) The comparison of growth rates of wild-type (WT) and Δter strain. The growth curve of Δter strain starts 5 days (~30 generations) after the transformation of cells with the deletion cassette.
In contrast to the WT, the TRFs are absent in ∆ter strain and a complete loss of telomeric DNA was observed already after the first passage (~25 generations). The telomeres in the ∆ku80 strain are prolonged and heterogeneous, similar to the situation in mutants lacking active Ku heterodimer in C. albicans or the plant model Arabidopsis thaliana66,67. The double mutant ∆ku80∆ter exhibited loss of telomeres comparable to ∆ter, indicating that the telomeres in ∆ku80 strain are maintained by telomerase.
The length of the 3′ telomeric overhang of Y. lipolytica was measured via in-gel hybridization of non-denaturated TRFs to oligonucleotide probe (Fig. 3c). The weak signal obtained for the WT suggests that similarly to other yeast species (e.g. S. cerevisiae), the overhang is relatively short. The increased size and heterogeneity of TRFs in ∆ku80 mutant are accompanied by an increased length of 3′ overhang. In contrast, the strains lacking the TER locus exhibited a complete loss of telomeric single-stranded DNA, regardless of the absence or presence of a functional Ku70/80 complex.
The deletion of TER locus also resulted in slightly delayed (~3 hours compared with the WT) onset of the exponential growth phase in ∆ter strain, probably due to critical telomere shortening and senescence of a subpopulation of cells (Fig. 3d). The rapid recovery of the ∆ter cells from this crisis suggests that there is an effective back-up system activated in Y. lipolytica cells lacking functional telomerase. The double mutant ∆ku80∆ter exhibited a more pronounced growth defect (data not shown), probably caused by the lack of end-protection by Ku70/80.
Comparative and functional analysis of TER sequences from the Yarrowia clade species
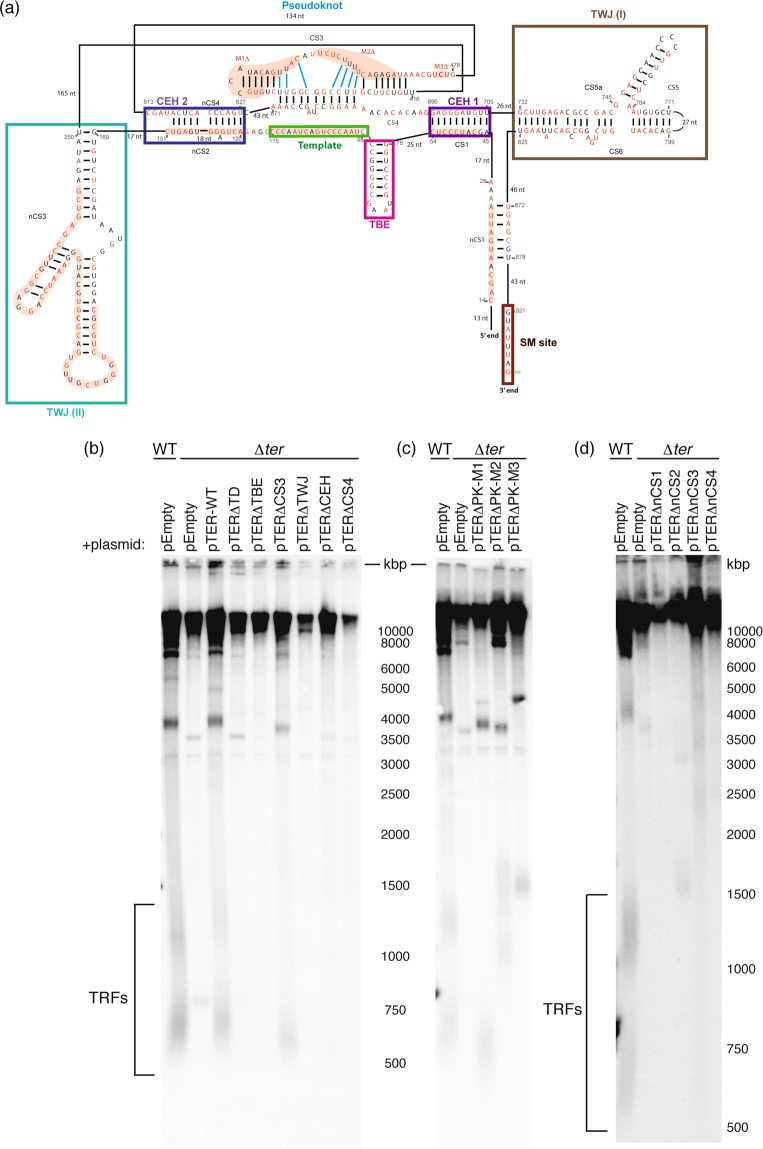
For the identification of putative functional elements involved in telomerase function, we aligned the nucleotide sequences of TER genes from 10 species using ClustalX68 (Supplementary Fig. S4; TERs of Y. phangngensis, Y. alimentaria and C. hispaniensis are too divergent, only allowing the alignment of regions corresponding to template domain, pseudoknot and the TWJ). Among these sequences, the level of conservation was high enough for generating a reliable alignment for 1–930 nt of Y. lipolytica TER, but beyond this point the sequences diverged too much and could not be aligned reliably. Furthermore, we identified an imperfect Sm site at position 920 nt, suggesting that longer transcripts could be processed to a shorter mature RNA with an Sm site close to its 3′ end, which is consistent with the results of Northern blot analysis (data not shown). For the purpose of covariation-based structure prediction we used the alignment that ends at the potential Sm site as an input for the RNAalifold69. Then we used the resulting RNAalifold-predicted helices as constraints for Mfold70 (Supplementary Fig. S5, Fig. 4a). Using this approach, we identified four conserved elements of the core structure of TER, known in yeasts and vertebrates: template, template-boundary element (TBE), a triple-helix containing pseudoknot (Supplementary Fig. S6), and a core-enclosing helix upstream of the template (CEH1). Interestingly, we identified another conserved helix downstream of the template, termed core-enclosing helix 2 (CEH2). Both CEH1 and CEH2 position the pseudoknot across the template providing further support for the role of the pseudoknot in regulating the template copying by telomerase. In addition to the template domain, we identified the conserved TWJ (Supplementary Fig. S7) and a novel element composed by conserved sequence nCS3, forming another TWJ with two stem-loops (TWJ (II)).
Figure 4.
Effect of the deletions of conserved and novel functional elements on the ability of TER to complement Δter mutation in Y. lipolytica. (a) A simplified scheme of Y. lipolytica TER with highlighted conserved domains and sequences subjected to functional analysis (for a more detailed structure of TER see Supplementary Fig. S5). (b–d) Wild-type (WT) and Δter strains were transformed with the plasmid constructs bearing the TER locus or its deletion variants (deletions of specific sequences are indicated), followed by the TRF analysis to test the ability of the plasmids to restore telomeric fragments in Δter mutant.
To perform functional analysis of the identified structural elements, we first constructed an episomal plasmid (pTER) bearing the entire intergenic sequence located between FRE2 and FRE3 genes of Y. lipolytica, carrying the TER locus and both 5′ and 3′ flanking sequences ensuring the presence of regions necessary for the regulation of transcription. Transformation of ∆ter strain by pTER resulted in the restoration of standard-length TRFs, indicating that the telomere defect due to TER deficiency is reversible and can be complemented by ectopic expression of TER (Fig. 4b). Subsequently, we used this system for testing the ability of deletion variants of TER lacking specific structural elements to restore the TRFs in the ∆ter strain. The deletion of template domain, template-boundary element, core-enclosing helix 1, TWJ and CS4 part of the pseudoknot caused a complete loss of the corresponding TER variants ability to restore TRFs. On the other hand, the variant lacking the CS3 sequence partially retained this ability. The restored TRFs in these cells were slightly shortened, but maintained throughout several (~120) cell divisions (Fig. 4b).
To characterize the elements of the pseudoknot structure in more detail, we analyzed three TER variants, lacking specific parts of the CS3 sequence (Fig. 4c). The deletion of motif 1 (M1), which includes a non-conserved stemloop (nucleotides 438–452), led to similar reduction of the TRF size as the deletion of the entire CS3 domain, suggesting this structure is crucial for the proper arrangement of CS3. Interestingly, the deletion of motif 2 (M2, nucleotides 455–468) resulted in prolonged TRFs (1000–1900 bp), implying that this mutation altered the spatial organisation of TER and increased the overall telomerase activity. The deletion variant lacking motif 3 (M3, nucleotides 484–486), which lies 8 nucleotides downstream from the 3′ end of CS3, also resulted in elongated TRFs (~1800 bp). Based on these data, we suggest that the pseudoknot might be involved in both positive and negative regulation of telomerase activity in Y. lipolytica.
Next, we performed the functional analysis of TER variants lacking the putative novel functional elements (nCS1–4). All four of them are located in the conserved part of TER (Supplementary Fig. S4, Fig. 4a) and their secondary structure is preserved in different species. The first element (nCS1) is located ~15 nt downstream from the 5′ end of the molecule, forms a stem structure and its deletion caused a loss of TRFs comparable to the ∆ter strain (Fig. 4d). In the structure prediction (Supplementary Fig. S5), the nCS2 element is paired with nCS4, forming a helix (CEH2) which stabilizes the core structure of TER. Deletion of either nCS2 or nCS4 was predicted to alter this structure and indeed abolished the ability of corresponding TER variant to restore standard TRFs in ∆ter strain (Fig. 4d). The deletion variant of TER lacking nCS3 also failed to restore normal TRFs pattern, suggesting that this structure plays an essential role in telomerase activity (Fig. 4d). Thus all four tested elements seem to be essential for proper telomerase function. Interestingly, some mutations in TER caused telomere elongation, indicating that their detailed analysis might be instrumental in shedding more light on telomerase regulation.
Sequences involved in the regulation of expression and stability of TER in Y. lipolytica
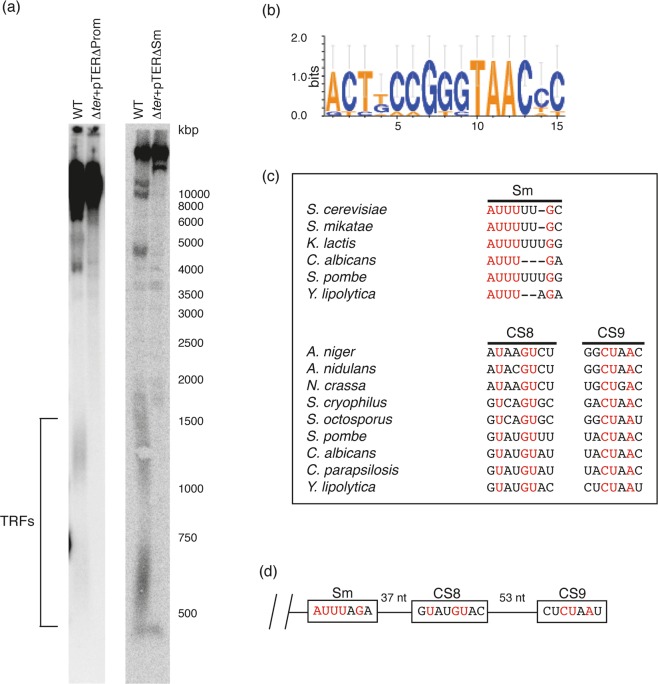
The comparative analysis of TERs from the Yarrowia clade species also revealed a conserved sequence lying ~40 nt upstream of the transcription start site, representing a putative TER promoter (Supplementary Fig. S4). This sequence includes the conserved motif 5′-TAAC-3′, which is present in all of the 10 aligned TERs (Fig. 5b). In agreement with the prediction that this sequence is functionally important, the plasmid construct bearing the complete TER sequence, yet lacking the putative promoter was unable to restore the TRFs in ∆ter strain (Fig. 5a).
Figure 5.
Sequences at the 5′ and 3′ region of TER are involved in the regulation of expression and stability of TER in Y. lipolytica. (a) Wild-type (WT) and Δter strains were transformed with the plasmid constructs carrying TER lacking putative promoter (Prom) or Sm site (Sm), followed by the TRF analysis to test the ability of the plasmids to restore telomeric fragments in Δter mutant. (b) Sequence logo representing consensus sequence of TER promoter in Yarrowia clade species. (c) Comparison of the sequences of Sm site, CS8 and CS9 from Y. lipolytica TER with those of other yeast models. (d) Scheme representing the 3′ end of Y. lipolytica TER with indicated positions of putative conserved elements.
An Sm consensus sequence, which is bound by the Sm proteins and facilitates the assembly of the telomerase ribonucleoprotein complex, was found at the 3′ ends of TERs of filamentous fungi, budding and fission yeast18,38,41,45,46. Furthermore, consensus 5′ splicing site and branch point were found in Candida, Schizosaccharomyces and Aspergillus species, and shown in S. pombe to facilitate 3′ end-processing by a partial splicing reaction18,45,46. At the predicted 3′ end of Y. lipolytica TER, a consensus Sm site was found with sequence similar to the Sm site of C. albicans TER (Fig. 5c, Supplementary Fig. S5). The position of this sequence, consistent with the RNA-seq results (Supplementary Fig. S1), shows that detectable TER transcripts are about 950 nt long. The Yarrowia species TER sequences downstream of CS6 are too diverged for a reliable multiple sequences alignment, thus we could not obtain phylogenetic support for this prediction. However, the deletion variant of TER lacking this sequence was unable to complement the loss of TRFs in ∆ter strain (Fig. 5a), suggesting that it is a functional Sm site required for the proper processing of TER and the assembly of active telomerase in vivo. Importantly, homologs of all 7 human Sm proteins known to bind the Sm site and stabilize the TER molecule (SNRPB – YALI0D14102p, SNRPD1 – YALI0A04961p, SNRPD2 – YALI0F06644p, SNRPD3 – YALI0A19030p, SNRPE – YALI0D01155p, SNRPF – YALI0A05423p, SNRPG – YALI0F30426p) were also identified in the genome, suggesting the mechanism of TER stabilization is conserved in yeasts.
We also identified several candidate sequences for putative 5′ splice sites (CS8) and for branch point (CS9) in the Y. lipolytica TER. The best CS8 candidate downstream of the Sm site is nearly identical to CS8 from S. pombe and is followed by a degenerate CS9 motif (Fig. 5c,d). Candidate CS8 and CS9 sequences were also found in the other Yarrowia species TERs, but at different distances from CS6, suggesting that the 3‘ end of the mature TER transcripts vary significantly.
Transcriptome response to the loss of telomerase activity
Diverse telomeric repeats recruit not only diverse DNA-binding proteins, but indirectly influence also other proteins associated with telomere, including DNA repair factors, replication-assisting proteins and factors responding to cellular stress. The vertebrate-like telomeric repeat of Y. lipolytica, bound by different proteins, could thus trigger different stress response than other telomeric sequences found in budding and fission yeasts. To adress this possibility, we studied the transcriptomic response to telomere loss of ∆ter strain in this yeast. This analysis led to the identification of 111 differentially expressed genes (DEGs) annotated in the H222-S4 genome. Among these DEGs, only one has a homolog among the known telomere-related proteins. YALI0C17061g, up-regulated in the ∆ter strain, is a homolog of PARP1 and PARP3 proteins that are involved in poly(ADP-ribosylation) of TRF2, telomere length regulation and telomere integrity maintenance in human cells71–73. In addition, PARylation executed by PARPs mediates DNA damage response at DNA breaks and participates in oxidative stress response (OSR)74. Interestingly, other genes putatively involved in the OSR were also detected as over-expressed in ∆ter strain (Table 1). These involve YALI0C16621g, a predicted superoxide dismutase (SOD) gene, YALI0E02266g, containing a Cu-Zn binding SOD domain and YALI0B13200g, containing a leucine zipper domain of YAP family transcription factors that are involved in stress responses. Moreover, the overexpression in ∆ter was also detected for YALI0C17567g, a homolog of DDR48, a DNA-damage response factor linked to the OSR and DNA replication stress in S. cerevisiae75. An additional functional enrichment analysis has shown that the genes differentially expressed in ∆ter strain can be grouped in four major functional categories (Table 2). Previously, a number of environmental stress response genes have been found to be overexpressed in both S. cerevisiae and H. polymorpha TER deletion mutants. However, OSR has been observed in telomerase mutant of H. polymorpha (with telomeric repeat 5′-GGGTGGCG-3′ and undescribed telomere-binding proteins)76, mouse cells lacking telomerase77 and human cells lacking specific components of telomerase holoenzyme78, but not in S. cerevisiae79. In conclusion, these results suggest that telomerase loss in Y. lipolytica induces OSR, a feature it shares with some yeasts and higher eukaryotes.
Table 1.
Several genes overexpressed in ∆ter strain are possibly involved in the oxidative stress response.
| DEG | Putative function/functional domain | Fold change (∆ter/WT) |
|---|---|---|
| YALI0C17061g | Poly(ADP-ribose) polymerase | 2.07 |
| YALI0C16621g | mitochondrial superoxide dismutase | 7.62 |
| YALI0E02266g | Cu-Zn binding SOD domain | 2.19 |
| YALI0B13200g | YAP transcription factor leucine zipper domain | 2.98 |
| YALI0C17567g | DNA damage-responsive protein 48 | 2.82 |
Table 2.
A functional enrichment analysis has shown several pathways possibly induced in ∆ter strain.
| Functional enrichment category | Number of genes |
|---|---|
| Transporters/transmembrane proteins | 36 |
| Iron metabolism & dehydrogenases | 13 |
| Aldo/keto reductases and oxidases | 9 |
| Secondary metabolites production | 7 |
The categories are listed according to the number of included DEGs. Category “Transporters/transmembrane proteins” was reported as two separate enrichment groups by DAVID analysis. As the DAVID software does not provide the enrichment groups with labels, the names of the categories were chosen arbitrarily, to fit the contained functional terms.
Discussion
The species of the Yarrowia clade provide an excellent opportunity to assess co-evolution of the components of telomere maintenance system. Remarkably, while they are relatively closely related, they do exhibit a clear diversification of their telomeric repeats. Taking advantage of these characteristics we addressed two questions: (1) how are the changes in the primary sequence of telomeres reflected by the DNA-binding properties of telomere-binding proteins, and (2) what are the common structural features of the RNA component of telomerase that are required for its activity in vivo. Answering these questions is relevant not only for a distinct group of fungi, but is also important for gaining general insights into the evolution of eukaryotic telomeres.
The double-stranded region of Y. lipolytica telomeres is bound by Tay1 protein, which is in many ways different from the other yeast dsDNA-binding telomeric proteins. It possesses 2 Myb domains and binds the canonical mammalian 5′-TTAGGG-3′ repeats with higher affinity than the telomeric sequence of Y. lipolytica54. It is therefore possible that the insertion in the template domain of TER that changed the sequence of telomeric repeat in the common ancestor of the Yarrowia clade (and possibly all Saccharomycotina) was tolerated while Tay1p was still able to bind the telomeric repeat, providing it retained the core of the ancestral motif as in case of Y. lipolytica (5′-TTAGTCAGGG-3′). According to this scenario, the inner four nucleotides (5′-AGTC-3′) represent a part of the telomeric repeat whose alterations should not have a substantial effect on Tay1p binding. Importantly, all 13 species contain an ortholog of YlTay1p whose Myb domains underwent only minor (if any) amino acid substitutions (Supplementary Fig. S3), suggesting the crucial role of this protein in telomere maintenance might be conserved in the whole clade. Particularly interesting is C. hispaniensis, whose telomeric repeat encountered substitutions in all four positions of the spacer region plus G-to-A substitution in the 9th position of the repeat affecting the core 5′-TT—AGGG-3′ sequence (Fig. 1). YlTay1p exhibited a decreased affinity toward C. hispaniensis telomeric repeat (Fig. 2), supporting the importance of the intact core sequence for an optimal Tay1 binding. However, similarly to other species of the Yarrowia clade, C. hispaniensis seems to lack the homologs of SpTaz1 or ScRap1 proteins that exhibit flexible binding to telomeric repeat variants. It is thus likely that even though the affinity for an altered telomeric repeat is decreased, it is still sufficient for the Tay1 protein to fulfill the crucial telomeric functions. According to our hypothesis, more extensively altered telomeric repeats of other yeast species might not have been recognized by Tay1p homologs and as a result, evolutionary novel telomeric proteins such as Rap1 or Taz1 emerged21, while Tay1p homologs (such as SpTeb1) were either lost or retained to act as transcription factors recognizing TTAGGG-like repeats in regulatory sequences of their target genes23.
The rapid loss of TRFs in the Y. lipolytica strain lacking putative TER locus confirmed that this sequence is indeed transcribed into functional telomerase RNA (Fig. 3a). The complete loss of TRFs was observed in very early generations (less than 50 cell divisions), unlike what has been observed in S. cerevisiae ∆tlc1 or S. pombe ∆ter1 mutants, where even after more than 100 generations, short TRFs can be detected24,26. This phenotype is reminiscent of the situation in Y. lipolytica ∆est2 cells lacking the catalytic subunit of telomerase52 and suggests that telomerase dysfunction results in very rapid telomere shortening, accompanied by the activation of a back-up system which allows a subpopulation of cells to overcome the early growth crisis and escape senescence. Since both ∆ter and ∆ku80∆ter strains are able to overcome the crisis, the back-up system does not require the activity of Ku70/80 heterodimer. The complete loss of detectable TRFs in both strains also suggests that this process does not include telomerase-independent amplification of telomeric DNA. In contrast to the ∆ku80∆ter strain of Y. lipolytica, S. cerevisiae cells lacking both TLC1 and YKU80 genes are not viable, suggesting the mechanism of chromosome end maintenance in ∆ter mutant of Y. lipolytica is different from that of ∆tlc1 mutants of S. cerevisiae62. On the other hand, ∆ku80 mutants of K. lactis expressing a modified allele of TER (ter1-4LBsr) and thus introducing altered repeats into its telomeric tract are viable, which is in line with our observations65. In agreement with previous studies on ∆est2 strain52, it is possible that in a Y. lipolytica ∆ter strain, chromosomal ends are maintained via amplification of subtelomeric repeats by homologous recombination. In ∆ku80 strain, TRFs appear to be prolonged and heterogenous (Fig. 3a), similarly to C. albicans ∆ku70/∆ku70 strain66, suggesting that Ku70/80 heterodimer is a negative regulator of telomere length in Y. lipolytica. In line with this hypothesis is the observation that ∆ku80∆ter strain exhibits the same telomeric phenotype as the single ∆ter strain (Fig. 3a).
The telomeric 3′ overhang is prolonged in the ∆ku80 strain of Y. lipolytica (Fig. 3c), which is most probably due to increased nucleolytic degradation of telomeric C-strand62,63. This phenotype is also reminiscent of S. cerevisiae strains lacking functional Ku70/80 heterodimer, whose ssDNA overhangs are prolonged throughout the cell cycle and occupied by an increased amount of Cdc13p molecules80. In ∆ter and ∆ku80∆ter strains, the loss of TRFs is accompanied by a complete loss of telomeric ssDNA, suggesting the alternative mechanism of chromosome-end maintenance does not involve preserving the 3′ overhang composed of telomeric repeats.
To further investigate how the divergence of telomeric repeats of the Yarrowia clade species co-evolved with the corresponding RNA components of telomerase, we performed a bioinformatic and functional analysis of TERs from all 13 species. The results underlined the overall diversity of these structures observed in different sets of organisms18,41,81 (Supplementary Fig. S8). Even the 10 species, whose TERs are similar enough for sequence alignment, exhibited major differences in its length and sequence. As expected, the most conserved of the entire TER is the template domain, where we were able to identify the conserved motif 5′-TAACCC-3′ (which presumably serves as the template for synthesis of one vertebrate-type telomeric repeat) in each of the 10 aligned TERs. Strong conservation can also be observed in the regions representing the TWJ and the CS4 subunit of pseudoknot. Interestingly, the sequence of CS3 is only partially conserved with several variants of the central part and it is not essential for the telomerase function. The TRFs observed after transformation of ∆ter strain with plasmid bearing TER variant lacking CS3 are shorter than the wild-type TRFs, but they are maintained throughout several cell divisions, suggesting this deletion variant of TER is able to partially associate with catalytic subunit of telomerase (Fig. 4b).
There are also several features typical for the conventional yeast TERs missing in the TER of Y. lipolytica (and also TERs from other species of the Yarrowia clade). The absence of Est1-binding arm is in agreement with the fact that there is no clear homolog of the ScEST1 gene in the genome of Y. lipolytica and other Yarrowia species. These data imply that the connection between telomeric DNA and telomerase is mediated by a yet unknown and possibly unique set of regulatory proteins in these species. The 5′ arm with the binding site for Ku heterodimer, typical for Saccharomyces species and Candida glabrata82 is also absent in Yarrowia clade TERs, although Ku70/80 is a key regulator of telomere maintenance, suggesting the association of Ku70/80 with telomerase is indirect and might involve other interacting partners.
Another feature, essential for telomerase activity in S. cerevisiae is the P3-like domain. In TLC1 RNA, this domain provides the binding site for Pop6 and Pop7 proteins, facilitating the assembly of the telomerase Est1-Cdc13 recruitment module43,83. In Y. lipolytica TER, we were not able to identify a homologous domain by a simple sequence alignment, however, the nCS1 element, which is essential for telomerase activity, shares several similarities with this structure. Specifically, the secondary structure prediction shows that similarly to S. cerevisiae P3-like domain, nCS1 also forms a bulged stem structure with conserved A-U and G-C pairs at the base of the stem (Supplementary Fig. S5)43. This indicates that nCS1 element might be involved in the assembly of Y. lipolytica telomerase in a similar way as the P3-like domain of TLC1 RNA. Another candidate for the P3-like domain in Y. lipolytica TER is the TWJ (II), which lies in a similar position as the P3-like domain of S. cerevisiae TLC1 RNA. However, its role in telomerase assembly is yet to be tested experimentally. On the other hand, the protein composition of telomerase recruitment module in Y. lipolytica is probably different from that of S. cerevisiae, since there is no clear ortholog of ScPOP6 or ScPOP7 gene present in its genome (YALI0E29007g is the ortholog of ScPOP1).
We also assessed the transcriptome response to the lack of TER using the RNA-seq analysis. Surprisingly, the analysis did not reveal a change in expression of a distinct group of telomere-related genes such as TAY1, possibly reflecting the essential, non-telomeric roles of Tay1p53 (Table 2). However, several genes possibly involved in OSR were found to be up-regulated in the ∆ter strain (Table 1). This may point to a more conserved role of telomere damage triggering the OSR—in human cells mediated by p53 and p21 pathway84. Although the homologs of p53 and p21 are not present in the Y. lipolytica genome and many other genes involved in the OSR are not affected, it would be interesting to further elucidate the variability of factors mediating the OSR in species with different telomere composition.
Conclusion
Our study, aimed at comparative analysis of the Yarrowia clade species, in essence caught evolution of telomeres in action. We have shown that changes in telomeric repeats are constrained by the DNA-binding properties of the Tay1 protein, which tolerates substitutions within the region outside the core 5′-TTAGGG-3′ motif. It is possible that C. hispaniensis, a species with a more diverged sequence of the telomeric repeat is just at the tipping point of the phylogenetic tree where Tay1-like telomeric proteins were replaced by more flexible telomere binding factors allowing further diversification of the telomeric sequences. Additionally, our work demonstrated that comparative studies of TERs such as the one reported here are still worthwhile as they can lead to identification of novel conserved structural motifs whose functions are important for telomerase activity in vivo.
Materials and Methods
Yeast strains
In this study, Y. lipolytica strain H222-S4 (MATA, ura3-302 SUC2) (Barth and Gaillardin, 1996) was used as the wild type. Wild type strain and H222-SW6 strain (MATA, ura3-302 SUC2 ∆ku80) lacking YlKU80 gene (∆ku80)61 were kindly provided by Gerold Barth (Technische Universität Dresden, Dresden, Germany). Both strains lacking the TER locus (∆ter, ∆ku80∆ter) were derived from H222-S4 and H222-SW6 strains, respectively, using the strategy described below.
Cultivation and media
For each experiment, yeast cells were cultivated in YPD [1% (w/v) yeast extract, 2% (w/v) Bacto Peptone, 2% (w/v) glucose] or SD [0.17% (w/v) yeast nitrogen base, 0.5% (w/v) ammonium sulfate, 2% (w/v) glucose] medium at 28 °C with constant aeration. Cultures used for construction of growth curves started at 105 cells/ml in YPD medium, 5 µl aliquotes were collected every 2 hours and cells were counted in a Bürker chamber. Each experiment was carried out in triplicate, the total amounts of cells were counted separately for the three independent cultures and the indicated numbers were calculated as means. The error bars represent the standard deviation.
Tay1 protein purification and electrophoretic mobility shift assays (EMSA)
Recombinant Tay1 protein was purified as described previously53. The 10 µl EMSA reactions contained 20 mM HEPES-NaOH (pH 7.3), 100 mM NaCl, purified Tay1p at indicated concentrations and 16 nM dsDNA probe, prepared as follows: the G-rich single-stranded oligonucleotides (sequences are listed in Supplementary Table S1) were labelled at 5′ end by T4 polynucleotide kinase (Thermo Scientific) using [γ-32P]ATP (Hartmann Analytic), mixed with 3-fold molar excess of the unlabelled complementary oligonucleotide, heated for 10 min at 100 °C and slowly cooled down at room temperature to allow efficient formation of the double-stranded probes. The reaction mixtures were incubated for 10 min at room temperature, afterwards the glycerol was added to the final concentration of 4.5% (v/v) and the samples were loaded on 6% polyacrylamide gel in 0.5x TBE buffer [40 mM Tris-HCl (pH 8.3), 45 mM boric acid, 1 mM EDTA]. The electrophoresis was performed in cold 0.5x TBE buffer at 20 mA per gel for 20 min. The gels were fixed with 10% (v/v) methanol, 10% (v/v) acetic acid for 10 min, dried and exposed to a phosphor screen. Signal was detected using Personal Molecular Imager FX (BioRad). The contrast of images was adjusted using Adobe Photoshop 12.0.
Construction of the ∆ter and ∆ku80∆ter strains
The entire TER locus including ~1200 bp long upstream and downstream flanking regions was amplified (for the list of oligonucleotides see Supplementary Table S1) and cloned into pDrive cloning vector (QIAGEN). The resulting plasmid was digested with restriction enzymes NdeI and BglII to remove the transcribed part of the locus and gel-purified. The URA3 gene was PCR-amplified using primers containing the restriction sites for NdeI and BglII and ligated with the linearized plasmid. The full-length disruption cassette was PCR-amplified and used for transformation of H222-S4 and H222-SW6 cells. Transformants were selected after 3 days (~20 generations) of incubation at 28 °C on SD media lacking uracil and the disruption of the TER locus was verified by PCR and sequencing.
TRF analysis
Total genomic DNA (gDNA) was isolated as described by Barth and Gaillardin85. 3 µg of gDNA were digested using 6 U of PmlI overnight. The fragments were separated in 1% (w/v) agarose gel for 16 hours at 1.6 V/cm and stained with 0.5 µg/ml ethidium bromide solution for 20 minutes (stained gel served as a loading control). The gel was then incubated for 40 minutes in denaturation solution (1.5 M NaCl, 0.5 M NaOH), 30 minutes in neutralization solution (1.5 M NaCl, 0.5 M Tris, pH 7.4) and 30 minutes in 20x SSC (3 M NaCl, 0.3 M Na-citrate, pH 7.0). The DNA was then transferred to Immobilon NY+ membrane (EMD Millipore) with a VacuGene XL blotter (GE Healthcare) in 20x SSC and fixed by incubating the membrane at 80 °C for 1 hour. The membrane was pre-hybridized for 30 minutes at 65 °C in Church buffer [0.25 M sodium phosphate buffer (pH 7.2), 1 mM EDTA] and hybridized at 65 °C overnight in the same buffer containing 50 ng of denaturated telomere-specific probe (YlTEL probe 1; Supplementary Table S1) labelled with [α-32P] dCTP (Prime-a-Gene® Labelling System, Promega). The membrane was washed once with wash buffer I [0.15 M NaCl, 15 mM Na-citrate, 0.1% (w/v) SDS] for 20 minutes at room temperature and once with wash buffer II [7.5 mM NaCl, 0.75 mM Na-citrate, 0.1% (w/v) SDS] for 1 hour at 50 °C. The signal was detected by Personal Molecular Imager FX (BioRad). The contrast of images was adjusted using Adobe Photoshop 12.0.
In-gel hybridization
1.5 μg of gDNA digested with PmlI was separated in 0.7% (w/v) agarose gel for 16 hours at 60 V. The gel was placed on a double-layer of Whatman paper and mounted on a Gel Dryer (model 583, Bio-Rad). Drying was carried out for 15–20 minutes at room temperature. The dried gel was sealed in a plastic bag and hybridized with 50 ng of oligonucleotide probe (YlTEL probe 2; Supplementary Table S1) end-labelled with [γ-32P] ATP in the hybridization buffer (5x SSC, 2.5x Denhardt’s solution, 50 mM pyrophosphate, 1 mM Na2PO4, 0.4 mM ATP, 20 µg/ml salmon sperm DNA) overnight at 37 °C. After the removal of excess hybridization buffer, the gel was washed 3–4 times for 30 minutes in 0.25x SSC at room temperature with agitation, sealed in a bag and exposed to a phosphor screen. Afterwards, the gel was subjected to a denaturing Southern blot in order to visualize the total telomeric DNA. First, the gel was incubated for 10 minutes in 0.25 M HCl, then for 45 minutes in denaturation solution and 5 minutes in 0.4 M NaOH. The DNA was transferred to a Hybond-XL nylon membrane (GE Healthcare). After the transfer, the membrane was pre-hybridized for 1 hour in Church buffer and hybridized overnight at 50 °C with 50 ng of YlTEL probe 2. The membrane was washed for 20 minutes at room temperature in 2x SSC and exposed to a phosphor screen. The contrast of images was adjusted using Adobe Photoshop 12.0.
RNA-seq analysis of ∆ter mutant
Three independent cultures of both wild type and ∆ter yeast cells were cultivated in 5 ml liquid YPD medium until the exponential phase (OD600 = 0.6–0.8). Afterwards, total RNA was isolated using Direct-zolTM RNA miniprep kit (Zymo Research) and the quality of the RNA was determined by Agilent 2100 Bioanalyzer (Agilent) using RNA 6000 Nano Assay (Agilent). The RNA integrity number (RIN) of all samples was assessed to be 9.6–9.8. For every sample, the library of oriented reads with single reads of 75 bp (TruSeq Stranded mRNA, Illumina) was prepared from 1 µg of isolated RNA and the libraries were analyzed by NextSeq550 apparatus (Illumina). The six obtained libraries were named WT1 to WT3 and TER1 to TER3. The sequenced libraries were first cleaned with Trimmomatic tool (v. 0.38)86 to clip sequencing adapter fragments from reads and to remove low quality regions (Trimmomatic parameter values: ILLUMINACLIP: TruSeq3-SE.fa:2:15:5 LEADING: 5 TRAILING: 5 SLIDINGWINDOW: 5:20 MINLEN: 50). Then, the filtered reads were mapped against H222-S4 reference genome87 using HiSat2 aligner tool (v. 2.1.0)88 with the following options:–max-intronlen 4000. To report the number of reads associated with each coding gene we used featureCounts tools (v. 1.6.2)89 with the option largestOverlap. The reads for the RNA-seq have been deposited at the EMBL-ENA and are publicly available under the accession number PRJEB29941 (https://www.ebi.ac.uk/ena/data/view/PRJEB29941). Differential gene expression analysis was performed under the R environment (v. 3.3.3) using the BioConductor package DESeq2 (v. 1.20.0)90,91. A preliminary investigation of the read count matrix revealed important biases in two libraries (TER1 and WT1) leading to inconsistent variance estimations for a large number of genes. Thus, for this analysis, we discarded libraries TER1 and WT1. We considered two criteria to define differentially expressed genes (DEGs): (1) The DESeq2 adjusted p-value was < = 0.001; and (2) Expression variation was at least two-fold between the wild-type and deletion strain. As noted earlier76, some of the DEGs may reflect the activation of URA3-related metabolic pathways in the ter1::URA3 mutants. Thus, we performed a comparison of the 126 identified DEGs to another DEG dataset from a strain generated by URA3 integration (Y. lipolytica mhb1::URA392). This analysis yielded 15 shared DEGs, which were excluded from the set and only the remaining 111 DEGs were further analyzed (Supplementary Table S2). To detect telomere-related genes, the DEGs were searched against the SwissProt database (Release 2018_06) with blastp algorithm from BLAST+ 2.2.3193,94, E-value threshold <0.0001 and telomere-related proteins were extracted from the list of significant hits based on a set of manually selected GO terms related to telomere biology (Supplementary Table S3). Furthermore, the QuickGO tool (https://www.ebi.ac.uk/QuickGO/) was used to identify DEGs associated with the GO term GO:0006979: response to oxidative stress. An additional DEG functional clustering analysis was performed with the tool DAVID, v. 6.8 (https://david.ncifcrf.gov/95,96). A function “Functional annotation clustering” was used to identify the functional enrichment clusters with the defined defaults as a source of functional annotation and the list of DEGs submitted as a gene list. Y. lipolytica CLIB122 (E150) genome was selected automatically as a background.
Genome sequencing
For comparative genome analysis, 13 strains of the Yarrowia clade were used. Their genomes were sequenced, assembled and annotated at the Micalis Institute, and deposited at the EMBL-ENA (Supplementary Table S4). Different strategies of sequencing and assembly were used. Y. alimentaria CBS 10151, Y. galli CBS 9722, Y. phangngensis CBS 10407, Y. yakushimensis CBS 10253, and C. hispaniensis CBS 9996 were sequenced using Roche 454 technology. Single reads were retrieved from a shotgun library and paired-end reads from a 8-kb library for a total coverage ranging from 16.7X (Y. alimentaria) to 39.46X (C. hispaniensis). Celera assembler version 6.1 (version 5.3 for Y. galli) was used. Additional Illumina HiSeq data were used to correct 454 sequencing errors occurring especially at homopolymers for strains CBS 10151, CBS 10253, and CBS 9996. Y. bubula CBS 12934, Y. deformans CBS 2071, Y. divulgata CBS 11013, Y. hollandica CBS 4855, Y. keelungensis CBS 11062, Y. oslonensis CBS 10146, and Y. porcina CBS 12935 were sequenced using Illumina Solexa technology with a HiSeq2000 system. Both shotgun and 8-kb paired-end libraries were sequenced in paired-end (2 × 100 bp). Raw reads were trimmed with Trimmomatic v.0.3286 and Cutadapt v.1.8.397. Assemblies were generated using SOAPdenovo2 v.2.0498 with optimal kmer estimated with kmergenie v.1.6799. Gap closing was performed using GapCloser v1.1298.
Phylogeny
A set of 97 protein sequences was chosen among the 912 proteins used for a previously published phylogenetic tree drawn for 6 species60. The following criteria were used: genes do not possess any introns and have a known function in Y. lipolytica, genes are singleton in all species of the Yarrowia clade, alignment of the protein sequences subsequently cleaned with Gblocks100 represents at least 70% of the initial alignment, the resulting alignment is longer than 100 amino acids. The 97 alignments were then concatenated, leading to a 41496-residue alignment. Phylogenetic trees were constructed by maximum likelihood, with PhyML101 and a JTT substitution model corrected for heterogeneity between sites by a Γ-law distribution, with four different categories of evolution rates. The proportion of invariable sites and the α-parameter of the Γ-law distribution were optimized according to the data. A bootstrap value was calculated from 100 replicates.
Supplementary information
Acknowledgements
We thank members of our laboratories and dr. Peter Baumann (Johannes Gutenberg Universität Mainz, Germany) for fruitful discussions. This work was supported by grants from the Slovak Grant Agencies APVV (APVV-15-0022 (L.T.), APVV-14-0253 (J.N.)) and VEGA (1/0052/16 (L.T.) and 1/0027/19 (J.N.)), a grant from the Canadian Institutes of Health Research (FDN 154315) and the Canadian Research Chair on telomere biology (R.J.W.) and a grant by the United States-Israel Binational Science Foundation (grant number 2013344) (Y.T.). Genome sequencing and RNA-seq analysis for species of the Yarrowia clade were funded by the projects CALIN (Carburants Alternatifs et Systèmes d’Injection, grant N° 25331 awarded to Jean-Marc Nicaud), YALIP (INRA AIP-Bioressources 2011 (C.N.)), and CAER (DGAC, convention 2012 93 0805 awarded to Jean-Marc Nicaud).
Author Contributions
F.Č., E.B. and M.S. performed the experiments; C.N. and H.D. obtained and analyzed genomic sequences; H.D., C.N. and K.J. performed the bioinformatic analysis of RNA-seq data; B.K., A.K., F.Č. and Y.T. performed the comparative analysis of TERs primary sequences and secondary structures; Y.T. depicted secondary structure of the whole TER as well as conserved sequences, F.Č. wrote the first draft of the paper; Y.T., C.N., R.J.W., J.N. and Ľ.T. supervised the research; Ľ.T. conceived the study, identified the candidate TER locus in Y. lipolytica, prepared first drafts of the figures and coordinated the research; F.Č., K.J., H.D., B.K., A.K., E.B., M.S., R.J.W., J.N., Y.T., C.N. and Ľ.T. edited and approved the manuscript.
Data Availability
All data generated or analysed during this study are included in this published article (and its Supplementary Information Files).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yehuda Tzfati, Email: tzfati@mail.huji.ac.il.
Cécile Neuvéglise, Email: cecile.neuveglise@inra.fr.
Ľubomír Tomáška, Email: lubomir.tomaska@uniba.sk.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-49628-6.
References
- 1.Zakian VA. Telomeres: beginning to understand the end. Science. 1995;270:1601–1607. doi: 10.1126/science.270.5242.1601. [DOI] [PubMed] [Google Scholar]
- 2.McEachern MJ, Krauskopf A, Blackburn EH. Telomeres and their control. Annu. Rev. Genet. 2000;34:331–358. doi: 10.1146/annurev.genet.34.1.331. [DOI] [PubMed] [Google Scholar]
- 3.de Lange T. How telomeres solve the end-protection problem. Science. 2009;326:948–952. doi: 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wellinger RJ, Zakian VA. Everything you ever wanted to know about Saccharomyces cerevisiae telomeres: beginning to end. Genetics. 2012;191:1073–1105. doi: 10.1534/genetics.111.137851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu RA, Upton HE, Vogan JM, Collins K. Telomerase mechanism of telomere synthesis. Annu. Rev. Biochem. 2017;86:439–460. doi: 10.1146/annurev-biochem-061516-045019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Lange T. Shelterin-mediated telomere protection. Annu. Rev. Genet. 2018;52:223–247. doi: 10.1146/annurev-genet-032918-021921. [DOI] [PubMed] [Google Scholar]
- 7.Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 8.Erdel F, et al. Telomere recognition and assembly mechanism of mammalian shelterin. Cell Rep. 2017;18:41–53. doi: 10.1016/j.celrep.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Lange T. How shelterin solves the telomere end-protection problem. Cold Spring Harb. Symp. Quant. Biol. 2010;75:167–177. doi: 10.1101/sqb.2010.75.017. [DOI] [PubMed] [Google Scholar]
- 10.Armstrong CA, Tomita K. Fundamental mechanisms of telomerase action in yeasts and mammals: understanding telomeres and elomerase in cancer cells. Open Biol. 2017;7:160338. doi: 10.1098/rsob.160338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linger BR, Price CM. Conservation of telomere protein complexes: shuffling through evolution. Crit. Rev. Biochem. Mol. Biol. 2009;44:434–446. doi: 10.3109/10409230903307329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li S, et al. Cdk1-dependent phosphorylation of Cdc13 coordinates telomere elongation during cell-cycle progression. Cell. 2009;136:50–61. doi: 10.1016/j.cell.2008.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jun HI, Liu J, Jeong H, Kim JK, Qiao F. Tpz1 controls a telomerase-nonextendible telomeric state and coordinates switching to an extendible state via Ccq1. Genes & Dev. 2013;27:1917–1931. doi: 10.1101/gad.219485.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greider CW. Regulating telomere length from the inside out: the replication fork model. Genes Dev. 2016;30:1483–1491. doi: 10.1101/gad.280578.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee YW, Arora R, Wischnewski H, Azzalin CM. TRF1 participates in chromosome end protection by averting TRF2-dependent telomeric R loops. Nat. Struct. Mol. Biol. 2018;25:147–153. doi: 10.1038/s41594-017-0021-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohki R, Ishikawa F. Telomere-bound TRF1 and TRF2 stall the replication fork at telomeric repeats. Nucleic Acids Res. 2004;32:1627–1637. doi: 10.1093/nar/gkh309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kordyukova M, Olovnikov I, Kalmykova A. Transposon control mechanisms in telomere biology. Curr. Opin. Genet. Dev. 2018;49:56–62. doi: 10.1016/j.gde.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Gunisova S, et al. Identification and comparative analysis of telomerase RNAs from Candida species reveal conservation of functional elements. RNA. 2009;15:546–59. doi: 10.1261/rna.1194009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lue NF. Plasticity of telomere maintenance mechanisms in yeast. Trends Biochem. Sci. 2010;35:8–17. doi: 10.1016/j.tibs.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steinberg-Neifach O, Lue NF. Telomere DNA recognition in Saccharomycotina yeast: Potential lessons for the co-evolution of ssDNA and dsDNA-binding proteins and their target sites. Front. Genet. 2015;6:1–10. doi: 10.3389/fgene.2015.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cervenak F, et al. Double-stranded telomeric DNA binding proteins: Diversity matters. Cell cycle. 2017;16:1568–1577. doi: 10.1080/15384101.2017.1356511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lue NF. Evolving linear chromosomes and telomeres: A C-strand-centric view. Trends. Biochem. Sci. 2018;43:314–326. doi: 10.1016/j.tibs.2018.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sepsiova R, et al. Evolution of telomeres in Schizosaccharomyces pombe and its possible relationship to the diversification of telomere binding proteins. PLoS One. 2016;11:e0154225. doi: 10.1371/journal.pone.0154225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singer MS, Gottschling DE. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 25.Tzfati Y, Fulton TB, Roy J, Blackburn EH. Template boundary in a yeast telomerase specified by RNA structure. Nature. 2000;288:863–867. doi: 10.1126/science.288.5467.863. [DOI] [PubMed] [Google Scholar]
- 26.Leonardi J, Box JA, Bunch JT, Baumann P. TER1, the RNA subunit of fission yeast telomerase. Nat. Struct. Mol. Biol. 2008;15:26–33. doi: 10.1038/nsmb1343. [DOI] [PubMed] [Google Scholar]
- 27.Webb CJ, Zakian VA. Identification and characterization of Schizosaccharomyces pombe TER1 telomerase RNA. Nat. Struct. Mol. Biol. 2007;15:26–33. doi: 10.1038/nsmb1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kabaha MM, Zhitomirsky B, Schwartz I, Tzfati Y. The 5′ arm of Kluyveromyces lactis telomerase RNA is critical for telomerase function. Mol. Cell. Biol. 2008;28:1875–1882. doi: 10.1128/MCB.01683-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forstemann K, Zaug AJ, Cech TR, Lingner J. Yeast telomerase is specialized for C/A-rich RNA templates. Nucleic Acids Res. 2003;31:1646–1655. doi: 10.1093/nar/gkg261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zappulla DC, Cech TR. Yeast telomerase RNA: a flexible scaffold for protein subunits. Proc. Natl. Acad. Sci. USA. 2004;101:10024–10029. doi: 10.1073/pnas.0403641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ritchie KB, Mallory JC, Petes TD. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell Biol. 1999;19:6065–6075. doi: 10.1128/MCB.19.9.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou J, Hidaka K, Futcher B. The Est1 subunit of telomerase binds the TLC1 telomerase RNA. Mol. Cell Biol. 2000;20:1947–1955. doi: 10.1128/MCB.20.6.1947-1955.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stellwagen AE, Haimberger ZW, Veatch JR, Gottschling DE. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosomal ends. Genes Dev. 2003;17:2384–2395. doi: 10.1101/gad.1125903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang M, Americ M, Lingner J. Telomerase repeat addition processivity is increased at critically short telomeres in Tel1-dependent manner in Saccharomyces cerevisiae. Genes Dev. 2007;21:2485–2494. doi: 10.1101/gad.1588807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen L, et al. An activity switch in human telomerase based on RNA conformation and shaped by TCAB1. Cell. 2018;174:218–230. doi: 10.1016/j.cell.2018.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Collopy LC, et al. LARP7 family proteins have conserved function in telomerase assembly. Nat. Commun. 2018;9:557. doi: 10.1038/s41467-017-02296-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mennie AK, Moser BA, Nakamura TM. LARP7-like protein Pof8 regulates telomerase assembly and poly(A)+TERRA expression in fission yeast. Nat. Commun. 2018;9:586. doi: 10.1038/s41467-018-02874-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tzfati Y, Knight Z, Roy J, Blackburn EH. A novel pseudoknot element is essential for the action of a yeast telomerase. Genes Dev. 2003;17:1779–1788. doi: 10.1101/gad.1099403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shefer K, et al. A triple helix within a pseudoknot is a conserved and essential element of telomerase RNA. Mol. Cell Biol. 2007;27:2130–2143. doi: 10.1128/MCB.01826-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bown Y, et al. A critical three-way junction is conserved in budding yeast and vertebrate telomerase RNAs. Nucleic Acids Res. 2007;35:6280–6289. doi: 10.1093/nar/gkm713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dandjinou AT, et al. A phylogenetically based secondary structure of yeast telomerase RNA. Curr. Biol. 2004;14:1148–1158. doi: 10.1016/j.cub.2004.05.054. [DOI] [PubMed] [Google Scholar]
- 42.Laterreur N, Eschbach SH, Lafontaine DA, Wellinger RJ. A new telomerase RNA element that is crucial for telomere elongation. Nucleic Acids Res. 2013;41:7713–7724. doi: 10.1093/nar/gkt514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lemieux B, et al. Active yeast telomerase shares subunits with ribonucleoproteins RNase P and RNase MRP. Cell. 2016;165:1171–1181. doi: 10.1016/j.cell.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Webb CJ, Zakian VA. Telomerase RNA is more than a DNA template. RNA Biol. 2016;13:683–689. doi: 10.1080/15476286.2016.1191725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Box JA, Bunch JT, Tang W, Baumann P. Spliceosomal cleavage generates the 3′ end of telomerase RNA. Nature. 2008;456:910–914. doi: 10.1038/nature07584. [DOI] [PubMed] [Google Scholar]
- 46.Kuprys PV, et al. Identification of telomerase RNAs from filamentous fungi reveals conservation with vertebrates and yeasts. PLoS One. 2013;8:e58661. doi: 10.1371/journal.pone.0058661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kannan R, Helston RM, Dannebaum RO, Baumann P. Diverse mechanisms for spliceosome-mediated 3′ end processing of telomerase RNA. Nat. Commun. 2015;6:6104. doi: 10.1038/ncomms7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qi X, et al. Prevalent and distinct spliceosomal 3′-endprocessing mechanisms for fungal telomerase RNA. Nat. Commun. 2015;6:6105. doi: 10.1038/ncomms7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen JL, Greider CW. Telomerase RNA structure and function: implications for dyskeratosis congenita. Trends Biochem. Sci. 2004;29:183–192. doi: 10.1016/j.tibs.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 50.Theimer CA, Feigon J. Structure and function of telomerase RNA. Curr. Opin. Struct. Biol. 2006;16:307–318. doi: 10.1016/j.sbi.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 51.Qi X, et al. The common ancestral core of vertebrate and fungal telomerase RNAs. Nucleic Acids Res. 2013;41:450–462. doi: 10.1093/nar/gks980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kinsky S, Mihalikova A, Kramara J, Nosek J, Tomaska L. Lack of catalytic subunit of telomerase leads to growth defects accompanied by structural changes at the chromosomal ends in Yarrowia lipolytica. Curr. Genet. 2010;56:413–425. doi: 10.1007/s00294-010-0310-6. [DOI] [PubMed] [Google Scholar]
- 53.Kramara J, et al. Tay1 protein: a novel telomere-binding factor from Yarrowia lipolytica. J. Biol. Chem. 2010;285:38078–38092. doi: 10.1074/jbc.M110.127605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Visacka K, et al. Synergism of the two Myb domains of Tay1 protein results in high affinity binding to telomeres. J. Biol. Chem. 2012;287:32206–32215. doi: 10.1074/jbc.M112.385591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Konig P, Giraldo R, Chapman L, Rhodes D. The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA. Cell. 1996;85:125–136. doi: 10.1016/S0092-8674(00)81088-0. [DOI] [PubMed] [Google Scholar]
- 56.Valente LP, et al. Myb-domain protein Teb1 controls histone levels and centromere assembly in fission yeast. EMBO J. 2013;32:450–460. doi: 10.1038/emboj.2012.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vassetzky N, Gaden F, Brun C, Gasser SM, Gilson E. Taz1p and Teb1p, two telobox proteins in Schizosaccharomyces pombe, recognize different telomere-related DNA sequences. Nucleic Acids Res. 1999;27:4687–4694. doi: 10.1093/nar/27.24.4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Michely S, Gaillardin C, Nicaud JM, Neuveglise C. Comparative physiology of oleaginous species from the Yarrowia clade. PLoS One. 2013;8:e63356. doi: 10.1371/journal.pone.0063356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gaillardin, C., Mekouar, M. & Neuveglise, C. Comparative genomics of Yarrowia lipolytica in Yarrowia lipolytica (ed. Barth, G.) 1–30 (Springer, 2013).
- 60.Meunchan M, et al. Comprehensive analysis of a yeast lipase family in the Yarrowia clade. PLoS One. 2015;10:e0143096. doi: 10.1371/journal.pone.0143096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kretzschmar A, et al. Increased homologous integration frequency in Yarrowia lipolytica strains defective in non-homologous end-joining. Curr. Genet. 2013;59:63–72. doi: 10.1007/s00294-013-0389-7. [DOI] [PubMed] [Google Scholar]
- 62.Gravel S, Larrivee M, Labrecque P, Wellinger RJ. Yeast Ku as a regulator of chromosomal end structure. Science. 1998;280:741–744. doi: 10.1126/science.280.5364.741. [DOI] [PubMed] [Google Scholar]
- 63.Polotnianka RM, Li J, Lustig AJ. The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr. Biol. 1998;8:813–835. doi: 10.1016/S0960-9822(98)70325-2. [DOI] [PubMed] [Google Scholar]
- 64.d’Adda di Fagagna F, et al. Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr. Biol. 2001;11:1192–1196. doi: 10.1016/S0960-9822(01)00328-1. [DOI] [PubMed] [Google Scholar]
- 65.Carter SD, Iyer S, Xu J, McEachern MJ, Astrom SU. The role of non-homologous end-joining components in telomere metabolism in Kluyveromyces lactis. Genetics. 2007;175:1035–1045. doi: 10.1534/genetics.106.067447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chico L, Ciudad T, Hsu M, Lue NF, Larriba G. The Candida albicans Ku70 modulates telomere length and structure by regulating both telomerase and recombination. PLoS One. 2011;6:e23732. doi: 10.1371/journal.pone.0023732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gallego ME, Jalut N, White CI. Telomerase dependence of telomere lengthening in Ku80 mutant Arabidopsis. Plant Cell. 2003;15:752–789. doi: 10.1105/tpc.008623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 69.Bernhart SH, Hofacker IL, Will S, Gruber AR, Stadler PF. RNAalifold: improved consensus structure prediction for RNA alignments. BMC Bioinformatics. 2008;9:474. doi: 10.1186/1471-2105-9-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- 71.Gomez M, et al. PARP1 is a TRF2-associated poly(ADP-ribose)polymerase and protects eroded telomeres. Mol. Biol. Cell. 2006;17:1686–1696. doi: 10.1091/mbc.e05-07-0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Beneke S, et al. Rapid regulation of telomere length is mediated by poly(ADP-ribose)polymerase-1. Nucleic Acids Res. 2008;36:6309–6317. doi: 10.1093/nar/gkn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boehler C, et al. Poly(ADP-ribose)polymerase 3 (PARP3), a newcomer in cellular response to DNA damage and mitotic progression. Proc. Natl. Acad. Sci. USA. 2011;108:2783–2788. doi: 10.1073/pnas.1016574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hegedus C, Virag L. Inputs and outputs of poly(ADP-ribosyl)ation: relevance to oxidative stress. Redox Biol. 2014;2:978–982. doi: 10.1016/j.redox.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tkach JM, et al. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat. Cell Biol. 2012;14:966–976. doi: 10.1038/ncb2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Beletsky AV, et al. The genome-wide transciption response to telomerase deficiency in the thermotolerant yeast Hansenula polymorpha DL-1. BMC genomics. 2017;18:492. doi: 10.1186/s12864-017-3889-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sahin E, et al. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 2011;470:359–365. doi: 10.1038/nature09787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ibanez-Cabellos JS, et al. Acute telomerase components depletion triggers oxidative stress as an early event previous to telomeric shortening. Redox Biol. 2018;14:398–408. doi: 10.1016/j.redox.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nautiyal S, DeRisi JL, Blackburn EH. The genome-wide expression response to telomerase deletion in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2002;99:9316–9321. doi: 10.1073/pnas.142162499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fisher TS, Taggart AKP, Zakian VA. Cell cycle-dependent regulation of yeast telomerase by Ku. Nat. Struct. Mol. Biol. 2004;11:1198–1205. doi: 10.1038/nsmb854. [DOI] [PubMed] [Google Scholar]
- 81.Waldl M, et al. TERribly difficult: searching for telomerase RNAs in Saccharomycetes. Genes (Basel) 2018;9:E372. doi: 10.3390/genes9080372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kachouri-Lafond R, et al. Large telomerase RNA, telomere length heterogeneity and escape from senescence in Candida glabrata. FEBS Lett. 2009;583:3605–3610. doi: 10.1016/j.febslet.2009.10.034. [DOI] [PubMed] [Google Scholar]
- 83.Laterreur N, et al. The yeast telomerase module for telomere recruitment requires a specific RNA architecture. RNA. 2018;24:1067–1079. doi: 10.1261/rna.066696.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Westin ER, et al. The p53/p21(WAF/CIP) pathway mediates oxidative stress and senescence in dyskeratosis congenita cells with telomerase insufficiency. Antioxid. Redox Signal. 2011;14:985–997. doi: 10.1089/ars.2010.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Barth, G. & Gaillardin, C. Yarrowia lipolytica in Nonconventional yeasts in biotechnology (ed. Wolf, K) 318–338 (Springer, 1996).
- 86.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Devillers H, Neuveglise C. Genome sequence of the oleaginous yeast Yarrowia lipolytica H222. Microbiol. Resour. Announc. 2019;8:e01547–18. doi: 10.1128/MRA.01547-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liao Y, Smyth GK, Shi W. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 90.R Core Team. R: A language and enviroment for statistical computing. R Foundation for statistical computing, http://www.R-project.org/ (2014).
- 91.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bakkaiova J, et al. The strictly aerobic yeast Yarrowia lipolytica tolerates loss of a mitochondrial DNA-packaging protein. Eukaryot. Cell. 2014;13:1143–1157. doi: 10.1128/EC.00092-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 94.Camacho C, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huang DW, Shermanm BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 96.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Martin Marcel. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17(1):10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 98.Luo R, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chikhi R, Medvedev P. Informed and automated k-mer size selection for genome assembly. Bioinformatics. 2014;30:31–37. doi: 10.1093/bioinformatics/btt310. [DOI] [PubMed] [Google Scholar]
- 100.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 101.Guindon S, Gascuel O. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article (and its Supplementary Information Files).