Abstract
Tachyplesin I, II and III are host defense peptides from horseshoe crab species with antimicrobial and anticancer activities. They have an amphipathic β-hairpin structure, are highly positively-charged and differ by only one or two amino acid residues. In this study, we compared the structure and activity of the three tachyplesin peptides alongside their backbone cyclized analogues. We assessed the peptide structures using nuclear magnetic resonance (NMR) spectroscopy, then compared the activity against bacteria (both in the planktonic and biofilm forms) and a panel of cancerous cells. The importance of peptide-lipid interactions was examined using surface plasmon resonance and fluorescence spectroscopy methodologies. Our studies showed that tachyplesin peptides and their cyclic analogues were most potent against Gram-negative bacteria and melanoma cell lines, and showed a preference for binding to negatively-charged lipid membranes. Backbone cyclization did not improve potency, but improved peptide stability in human serum and reduced toxicity toward human red blood cells. Peptide-lipid binding affinity, orientation within the membrane, and ability to disrupt lipid bilayers differed between the cyclized peptide and the parent counterpart. We show that tachyplesin peptides and cyclized analogues have similarly potent antimicrobial and anticancer properties, but that backbone cyclization improves their stability and therapeutic potential.
Keywords: tachyplesin, host defense peptide, anticancer, antimicrobial, antibiofilm, peptide-membrane interaction, structure-activity, model membranes, nuclear magnetic resonance solution structure
1. Introduction
The host defense peptides (HDPs) tachyplesin I, II and III (TI, TII and TIII) are active against a broad range of Gram-negative and Gram-positive bacteria and fungi [1,2,3,4] and possess anticancer properties [5,6,7,8,9,10,11,12,13,14,15]. Each analogue was isolated from a different species of horseshoe crab, but they share high sequence homology (Table 1). TI, TII and TIII possess 17 amino acid residues, two disulfide bonds, a C-terminal α-amidation, and their structure is organized in a β-hairpin [2,3,16]. Like other HDPs [17], TI, TII and TIII possess an amphipathic secondary structure (i.e., positively charged and hydrophobic amino acids segregate into distinct clusters), thought to be essential for their antimicrobial activity.
Table 1.
The sequence and physicochemical properties of tachyplesin I–III (TI-TIII) and their cyclic analogues (cTI-cTIII).
| Peptide | Sequence 1 | Mass (Da) 2 | RT (min) 3 | Charge 4 | |
|---|---|---|---|---|---|
| Calc. | Obs. | ||||
| TI | KWCFRVCYRGICYRRCR * | 2263.8 | 2263.5 | 18.04 | +7 |
| TII | RWCFRVCYRGICYRKCR * | 2263.8 | 2263.5 | 17.79 | +7 |
| TIII | KWCFRVCYRGICYRKCR * | 2235.8 | 2235.6 | 17.68 | +7 |
| cTI | KWCFRVCYRGICYRRCRG | 2303.8 | 2303.7 | 18.69 | +6 |
| cTII | RWCFRVCYRGICYRKCRG | 2303.8 | 2303.7 | 18.53 | +6 |
| cTIII | KWCFRVCYRGICYRKCRG | 2275.8 | 2275.5 | 18.44 | +6 |
1 Peptide sequences and amino acid residues differing from TI are in bold. * denotes C-terminal amidation. 2 Average mass calculated (Calc.) from the amino acid sequence and experimentally observed (Obs.) using synthetic peptide and determined from m/z 3+ in ESI-MS. 3 Retention time (RT) of peptides on an analytical RP-HPLC; chromatograms shown in Supplementary Figure S1a were obtained with a 2%/min gradient of 0–40% solvent B (90% acetonitrile; 0.05% trifluoroacetic acid (TFA) (v/v)) in solvent A (H2O, 0.05% TFA (v/v)) at a flow rate of 0.3 mL/min. 4 Charge of the peptides at pH 7.4.
Cationic amphipathic HDPs selectively target the anionic surfaces of microbes, rather than the neutral surface of host cells, and kill them by a mechanism that involves binding to and insertion into cell membranes. The initial binding is mediated by electrostatic attractions between the positively-charged residues of HDPs and the anionic microbial surface [18], and is followed by the insertion of hydrophobic residues into lipid membranes in a process that involves van-der-Waal’s interactions with the phospholipids [18,19].
Similar to bacterial cells, the surface of cancer cells is negatively-charged due to the increased expression and exposure of phospholipids containing the anionic phosphatidylserine (PS) headgroup [20,21,22,23]. In contrast, cell membranes of healthy mammalian cells are asymmetric and phospholipids containing PS-headgroups are found exclusively in the inner leaflet. Variations in the overall cell surface charge [24] in phospholipids with exposed [25,26] membrane fluidity and curvature [18,27], have been shown to modulate the selective toxicity of HDPs towards cancer cells. These differences regulate the affinity of peptides for cell membranes, the effective peptide-to-lipid ratio and the ability for HDP to kill cancerous cells or pathogens rather than healthy cells [28,29,30].
Several studies have investigated the activity, structure and mechanism of the action of TI, but few have examined the activity of TII and TIII. Early studies suggested that TI kills bacterial cells by a mechanism involving inner membrane permeabilization and the rapid efflux of K+ [31,32,33]. Later, TI was shown to translocate across lipid bilayers, cause a phospholipid flip-flop and form toroidal pores [33,34]. In cancerous cells, TI was reported to induce cell disruption and late apoptosis/necrosis [12,35]. Paredes-Gamero et al. proposed that the cancer cell death mechanism was dependent on the peptide dose: at high concentrations, the peptide-induced direct cell membrane disruption, and at lower concentrations, it activated intracellular cell death mechanisms [12].
Because of the expression of TI, TII and TIII in distinct species of horseshoe crab, we were specifically interested in comparing their structure, activity and mode-of-action. As these peptides have a potential application as anticancer and/or antimicrobial agents, we investigated whether backbone cyclization would increase the stability and maintain activity. Our studies show that TI, TII and TIII, and their cyclic analogues cTI, cTII and cTIII, have similar structures and activities against bacteria and cancerous cells. Backbone cyclization reduced the hemolytic activity and increased peptide stability while maintaining potent anticancer and antimicrobial activities. cTI and cTIII especially showed potential to be considered for the development of anticancer peptide-based drugs.
2. Results
2.1. Properties of Tachyplesin I–III and Their Cyclic Analogues
The amino acid sequences of TI, TII and TIII differ in positions 1 or 15, which can be a lysine or an arginine residue (Table 1). So far, TI is the most studied and the only analogue with reported structure calculations [36,37,38]. We were interested in comparing TI, TII and TIII and their backbone-cyclized analogues (cTI–III) to identify similarities and differences in their three-dimensional structure and stability; and to determine whether these characteristics affect the membrane interactions and biological activity of the peptides.
All peptides were synthesized using solid-phase peptide synthesis, oxidized and correctly folded, as suggested by the observed masses using electrospray ionization mass spectroscopy (ESI-MS; Supplementary Figure S1 and Table 1) and confirmed through clearly dispersed peaks in the amide region of their respective One-dimensional (1D) 1H NMR spectra [39]. The peptides were purified to >95%, as confirmed by analytical reverse-phase high-performance liquid chromatography (RP-HPLC; see chromatograms of pure peptides in Supplementary Figure S1a).
Despite minor differences, their overall hydrophobicity follows the trend cTI > cTII > cTIII > TI > TII > TIII (Table 1), as indicated by their retention time (RT) on analytical RP-HPLC (Supplementary Figure S1). The cyclic analogues appear to be overall more hydrophobic (less polar) than the parent peptides, which is consistent with the loss of the N-terminal charge and the C-terminal amidation resulting from cyclization.
2.2. Structure of Tachyplesins and Backbone-Cyclised Analogues
The three-dimensional (3D) structures of TII and TIII, and of the backbone-cyclized analogues cTI–cTIII were determined with solution NMR spectroscopy. All backbone resonances were fully assigned apart from the N-terminal amides of the two linear peptides, and the R1/G18 amides of cTII, reflective of some degree of flexibility in these regions. The secondary αH chemical shifts of the native peptides and the cyclic analogues were highly similar, indicating a negligible change in the backbone structure and the β-strands (W2-Y8; I11-R17) (Figure 1a,b).
Figure 1.
The NMR structures of TI and cTI (red), TII and cTII (cyan), TIII and cTIII (magenta). (a) Secondary αH chemical shift at 298 K determined from 1H NMR spectra. Shifts were calculated by subtraction of random coil 1H NMR shifts [41] from the experimental values. For tachyplesin I (TI), the shifts of the peptide synthesized in our lab were compared to shifts obtained from data banks: TI—PDB ID 2RTV [37], and TI—BMRB ID 1135 [38]. Positive shifts greater than 0.1 ppm suggest β-strands, indicated by grey arrows. (b) Overlay of the cyclic analogues (colored backbone) with their respective parent peptide (grey backbone). Hydrogen bonds between β-strands are indicated in blue. The structure of TI was obtained from the PDB (ID 1WO0). (c) Overlay of TI–TIII and cTI–cTIII. (d) Mobility of the residues (K or R) at position 1 and 15, which differ between the peptides, and of W2 of the native sequences and cyclic analogues, respectively.
The 3D solution structures of each peptide were calculated from distance restraints, ranging from 154 for the linear peptides, to 172–284 for the cyclic analogues, along with dihedral angle restraints totaling 34 to 36. The final family of structures for each of the peptides has good structural and energy statistics, as indicated by an overall MolProbity score of less than 1.6, shown in Table S1. Analysis of the structures by PROMOTIF [40] defines antiparallel β-strands being formed by residues W2-Y8 and I11-R17 in all but one of the peptides. The exception is TIII which has slightly shorter strands formed between residues C3-Y8 and I11-C16. All disulfide bonds are defined by PROMOTIF as adopting the short right-hand hook configuration. Structures of the tachyplesin peptides and the cyclized analogues differed primarily in the flexibility of the N- and C-termini of the parent peptides (Figure 1b–d). The reduction of the amino acid side-chain flexibility in the terminal regions due to backbone cyclization is emphasized in Figure 1d with the side chain of the residues K/R1 and K/R15, which differ between TI/cTI, TII/cTII and TIII/cTIII; the side chain of W2 is also shown, as this residue is used to monitor the peptide partitioning into lipid bilayers (see Section 2.5.2). No significant cation-π interactions between the R/K1 and W2 were noted for any of the tachyplesin peptides. Such an interaction might be revealed by NMR in the form of substantial deviations of the chemical shifts of arginine/lysine residues but none were observed (see Figure 1a). The lack of cation-π interactions is also supported by the NMR solution structures that reveal a high degree of flexibility in the termini of the linear peptides, and by the different orientations that these residues acquire in the cyclic peptides, as shown in Figure 1d.
The peptide structures were deposited with the Protein Data Bank (PDB) and the Biological Magnetic Resonance Data Bank (BMRB): TII—PDB ID: 6PI2, BMRB ID: 30617; TIII—PDB ID: 6PI3, BMRB ID: 30618; cTI—PDB ID: 6PIN; BMRB ID: 30619; cTII—PDB ID: 6PIO, BMRB ID: 30620; cTIII—PDB ID: 6PIP, BMRB ID: 30621.
2.3. Improved Stability and Reduced Hemolytic Activity of Cyclized Tachyplesin Peptides
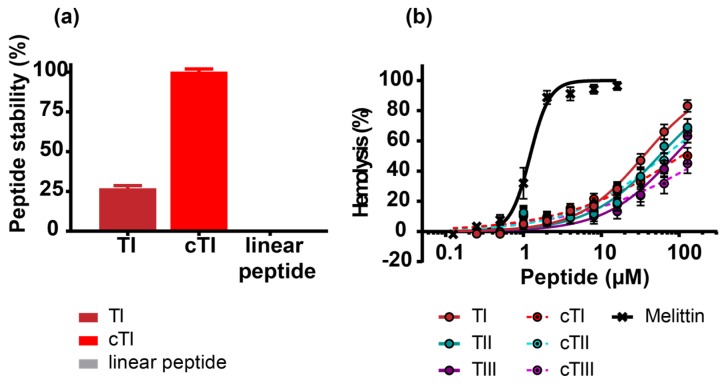
Cyclization has been shown to increase stability [42,43] and reduce the hemolytic activity of some peptides [44,45]. Comparison of resistance to human proteases showed that the cTI analogue did not degrade after treatment for 24 h in 25% (v/v) human serum, whereas only 25% of TI remained in the solution (see analytical RP-HPLC chromatograms in Supplementary Figure S2). A peptide (linear and without disulfide bonds) used as a control was fully degraded under the same conditions (Figure 2a). TI has previously been shown to be completely stable for 2 h in mouse or human serum [46].
Figure 2.
The peptide stability in human serum and activity against human red blood cells (RBCs). (a) Stability of TI and cyclic analogue cTI in 25% (v/v) human serum for 24 h. The samples were analyzed by analytical RP-HPLC and the percentage of peptide remaining was determined from the area under the peptide peak in the chromatogram in comparison to peak area at time zero. A linear 18 amino acid peptide (KGGGGSGQLIDSMANSFV) was included as the positive control. (b) Hemolytic activity of tachyplesins and their cyclic analogues were tested up to 128 µM against human RBCs (0.25% (v/v), 1 h incubation at 37 °C). Melittin, a highly hemolytic peptide, was included as the positive control.
The percentage of hemolysis of human red blood cells (RBCs) followed the trend TI > TII > TIII > cTII > cTI > cTIII when compared at 128 µM, the highest concentration of peptide tested. TI was the most hemolytic of the native sequences. Backbone cyclization reduced the hemolytic activity of the cTI and cTIII compared to their parent counterparts, but led to no clear improvement for cTII. The C-amidated peptides TI–TIII lysed 66%, 56% and 41% of RBCs at 64 µM respectively (Figure 2b and Table 2). A similar trend but lower hemolytic activity had been reported for TI, TII, and TIII lacking C-terminal amidation at higher peptide concentrations [47], which is known to impact peptide activity [48,49].
Table 2.
Concentration of tachyplesin I–III (TI-TIII) and of their cyclic analogues (cTI-cTIII) required to induce 50% lysis in RBCs (HC50). a.
| Peptide | HC50 (µM) |
|---|---|
| TI | 34.9 ± 2.8 |
| TII | 55.4 ± 6.6 |
| TIII | 86.4 ± 12.2 |
| cTI | 106.9 ± 21.0 |
| cTII | 64.1 ± 9.4 |
| cTIII | >128 |
a Values were determined from a minimum of three independent replicates and depicted as mean ± SEM.
The positive control melittin, a hemolytic peptide from honeybee venom, induces 100% hemolysis in human RBCs at concentrations above 2 µM. By comparison, at this concentration, all tachyplesin peptides have a low hemolytic activity of around 10%.
2.4. Biological Activity of Tachyplesin I–III and of Their Cyclic Analogues
2.4.1. Activity against Bacteria
The Gram-negative Escherichia coli strains ATCC 25922 and DC2 CGSC 7139 and the Gram-positive Staphylococcus aureus strains ATCC 25923 and ATCC 6538 were used to determine the antimicrobial activity of the parent tachyplesin peptides and their cyclic analogues against bacteria with differences in the physical and chemical properties of their cell wall. E. coli ATCC 25922 (“smooth” LPS) and S. aureus ATCC 25923 are common control strains for antimicrobial susceptibility testing [50,51]. E. coli DC2 CGSC 7139 is hypersensitive to antibacterial agents and more permeable to dyes [52], while S. aureus ATCC 6538 is known to form biofilms [53]. Tachyplesin peptides and their cyclic analogues were tested against planktonic cultures of all strains and against S. aureus ATCC 6538 in the biofilm form for direct comparison of their antimicrobial activity.
Tachyplesin I–III were two-to-four times more potent than their cyclic analogues against all bacterial strains in their planktonic growth form (Table 3). Generally, the Gram-negative strains were more susceptible than the Gram-positive strains. Similar MICs have been reported previously for TI against E. coli ATCC 25922, [1] and TI and TII against S. aureus ATCC 25923 [2,3]. Tachyplesin peptides were most active against E. coli DC2 CGSC 7139 and E. coli ATCC 25922. The activity against these bacterial strains was reduced when the peptides were cyclized. The peptides were least active against S. aureus ATCC 6538 in the planktonic form, and no difference in activity was observed between tachyplesin peptides and cyclic analogues (Table 3).
Table 3.
The minimal inhibitory concentrations (MICs) of tachyplesin I–III (TI-TIII) and of their cyclic analogues (cTI-cTIII) against planktonic bacteria.
| MIC (µM) | ||||
|---|---|---|---|---|
| Peptide |
E. coli DC2 CGSC 7139 |
E. coli ATCC 25922 |
S. aureus ATCC 25923 |
S. aureus ATCC 6538 |
| TI | 0.5–1 | 0.0625–0.5 | 1–4 | 4–8 |
| TII | 0.5–1 | 0.125–0.5 | 1–4 | 4–8 |
| TIII | 0.25–0.5 | 0.0625–0.5 | 1–4 | 4–8 |
| cTI | 4 | 1 | 2–8 | 8 |
| cTII | 4–8 | 1–2 | 2–8 | 8 |
| cTIII | 4–8 | 1–2 | 2–8 | 8 |
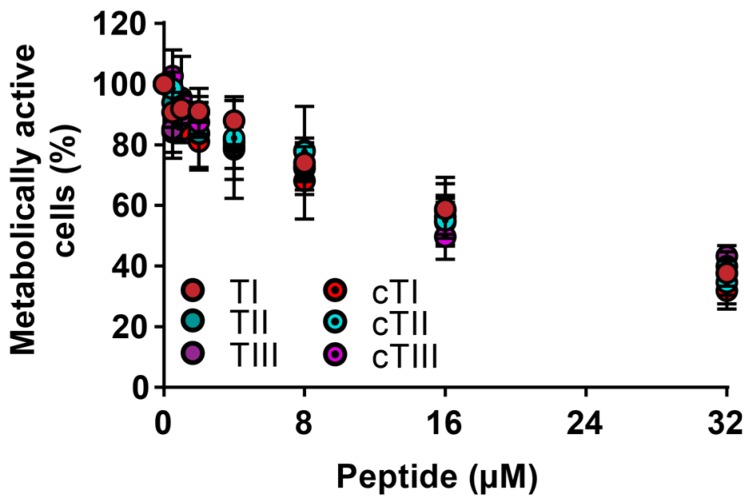
To determine whether the tachyplesin peptides could act against bacterial biofilms, the cells metabolic activity was measured for biofilms formed by S. aureus ATCC 6538 (Figure 3 and Table 4). All peptides exhibited similar activity against the bacteria in the biofilm form, with a 50% loss of cell metabolic activity observed at ~20 µM. However, approximately 40% of the biofilm was metabolically active at 32 µM, the highest concentration tested, suggesting a reduced activity against biofilm compared to the planktonic S. aureus ATCC 6538 bacteria.
Figure 3.
The activity of tachyplesin peptides and their cyclic analogues against an S. aureus ATCC 6538 biofilm. The response curve of biofilm treated with increasing the concentrations of peptide (n = 4, ± SEM).
Table 4.
Activity of tachyplesin I–III (TI–TIII) and cyclic analogues (cTI–cTIII) against S. aureus ATCC 6538 in the biofilm growth form.
| Peptide | CC50 (µM) 1 | Metabolically Active Cells (%) 2 |
|---|---|---|
| TI | 21.5 ± 2.2 | 37.7 ± 4.3 |
| TII | 23.1 ± 2.9 | 39.9 ± 2.9 |
| TIII | 24.2 ± 4.1 | 43.3 ± 4.1 |
| cTI | 17.8 ± 3.6 | 31.9 ± 6.0 |
| cTII | 19.4 ± 1.5 | 34.6 ± 7.1 |
| cTIII | 18.0 ± 1.4 | 37.5 ± 7.2 |
1 Peptide concentration required to induce the reduction of 50% of the metabolically active cell population (CC50) determined using one-site specific binding with the Hill slope. 2 Percentage of cells remaining metabolically active at 32 µM, the highest peptide concentration tested.
2.4.2. Activity against Cancerous Cells
TI, TII, TIII, and their cyclic analogues were tested against three melanoma (MM96L, HT144 and WM164) and one cervical cancer (HeLa) cell line (Supplementary Tables S2 and S3) to determine whether the peptides exhibit different cytotoxic activities. The aneuploid immortal keratinocyte cell line HaCaT was included as a non-cancerous control. Cytotoxicity toward cancer cell lines was compared by determining peptide concentrations required to achieve 50% of cell death (CC50) from dose-response curves (Table 5).
Table 5.
The cytotoxicity of tachyplesin I–III (TI-TIII) and cyclic analogues (cTI-cTIII) against cultured cells.
| CC50 (µM) 1 | Melanoma Selectivity 5 | |||||
|---|---|---|---|---|---|---|
| Peptide | MM96L 2 | HT144 2 | WM164 2 | HeLa 3 | HaCaT 4 | |
| TI | 1.5 ± 0.1 | 1.7 ± 0.2 | 2.5 ± 0.1 | 13.1 ± 1.2 | 11.6 ± 1.6 | 2–21 |
| TII | 1.6 ± 0.1 | 2.0 ± 0.1 | 1.6 ± 0.1 | 18.0 ± 3.9 | 3.7 ± 0.2 | 3–35 |
| TIII | 1.8 ± 0.1 | 2.0 ± 0.1 | 1.7 ± 0.1 | 21.7 ± 1.1 | 7.3 ± 0.5 | 5–48 |
| cTI | 1.3 ± 0.1 | 1.4 ± 0.1 | 2.7 ± 0.1 | 6.7 ± 0.6 | 7.9 ± 0.5 | 2–76 |
| cTII | 1.1 ± 0.1 | 0.8 ± 0.04 | 2.4 ± 0.3 | 7.2 ± 0.4 | 2.4 ± 0.3 | 3–58 |
| cTIII | 1.7 ± 0.1 | 0.9 ± 0.03 | 1.3 ± 0.1 | 9.3 ± 0.4 | 7.5 ± 0.3 | 3–98 |
1 The concentration necessary to kill 50% of cells was calculated from dose-response curves (n ≥ 3, ± SEM). 2 Melanoma cell lines: MM96L, HT144, WM164, 3 cervical cancer cell line: HeLa, 4 healthy epithelial control cell line: HaCaT (aneuploid immortal keratinocyte). Description and verification of each cell line are detailed in Supplementary Tables S2 and S3). 5 selectivity for melanoma cell lines was estimated through the activity-toxicity index (ATI). ATI = MHC/MCC50 (modified from Reference [46]), with MHC being the minimal concentration necessary to induce 10% (lower value) or 50% (higher value) cell death in human RBCs and MCC50 being the median of CC50 values of all melanoma cell lines. Values above 1 indicate a higher selectivity for the cancerous cells over RBCs.
The cytotoxic activities of the tachyplesin peptides and their cyclic analogues are dependent on the cell lines (Table 5). All peptides were most effective against the melanoma cell lines MM96L, HT144 and WM164 (cytotoxic activities ranged between CC50 0.8–2.7 µM). Compared to the control cell line HaCaT, the cytotoxic activities of the peptides against melanoma were significantly different (p < 0.05) with the exception of cTII against WM164. The cervical cancer cell line HeLa was more resistant towards the tachyplesin peptides compared to the melanoma cell lines and higher peptide concentrations were necessary to reach 50% cell death. Differences in cytotoxic activity against HeLa, compared to the control cell line HaCaT, can be observed for all peptides except TI and cTI. The cyclic analogues cTI–cTIII were approximately 2x more potent against HeLa than the parent peptides TI–TIII (p < 0.05).
The peptides have a similar selectivity for the melanoma cell lines at lower peptide concentrations which would induce ≤10% hemolysis in RBCs. At concentrations inducing ≤50% hemolysis in RBCs, the cyclic analogues cTI–cTIII were more selective. Overall, the most promising therapeutic range was observed for TIII, cTI and cTIII (Table 5).
2.5. Mechanistic Studies
Peptides with an amphipathic arrangement of charged and hydrophobic residues are known to act against bacterial and cancer cells via selective membrane targeting, penetration and/or lysis. To characterize how peptide-membrane interactions affect biological activity, we undertook detailed peptide-membrane binding studies that compare parent and cyclic tachyplesin peptides.
2.5.1. Peptide Binding to Model Membranes
The ability of TI to bind to model membranes has been previously shown and a preference for negatively-charged membranes was found [31,46]. We were interested in comparing the membrane-binding properties of the parent tachyplesins versus the cyclic analogues to determine whether differences in membrane binding could explain the relative biological activities (see Table 5). Given the similar potencies among the three tachyplesins, and among the three cyclic analogues, we compared the interaction of TI and cTI, as representative of parent and cyclic tachyplesin peptides, respectively, with model membranes using surface plasmon resonance (SPR). Phospholipids containing PC-headgroups are the most common in the outer leaflet of the mammalian plasma membrane [54,55]; thus, we prepared model membranes composed of the zwitterionic POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine), which forms fluid bilayers at 25 °C and mimics the overall fluidity and neutral surface of healthy eukaryotic cells [56]. Phospholipids containing the negatively-charged PS-headgroups are normally restricted to the inner leaflet in eukaryotic cell membranes but are exposed at the cell surface of cancerous cells [20,22,23]. Therefore, we used model membranes with 20% of POPS (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine), POPC/POPS (4:1 molar ratio), to represent the negatively-charged surface of cancer cells.
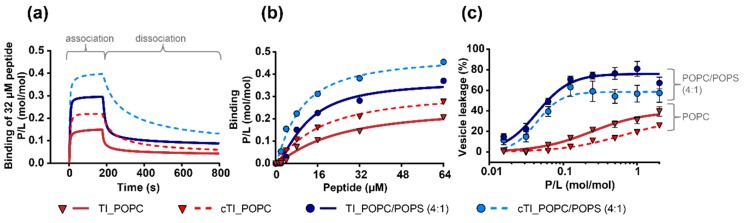
Both TI and cTI bound with higher affinity to negatively-charged POPC/POPS (4:1) than to zwitterionic POPC lipid bilayers. cTI had stronger affinity to both lipid systems than the parent peptide, as shown by a higher peptide-to-lipid (P/L) ratio during association, a slower dissociation rate (koff) from the lipids, and a higher amount of peptide remaining associated to the membrane at the end of dissociation (P/Loff) (Figure 4a,b and Table 6). Additionally, TI and cTI disrupted large unilamellar vesicles (LUVs) of POPC/POPS (4:1) with higher efficacy than LUVs of POPC, which agrees with their preference for negatively-charged over neutral membranes. However, TI disrupted LUVs of POPC and of POPC/POPS (4:1) more efficiently than the cyclic analogue cTI (Figure 4c, Table 6). Thus, the higher binding saturation and affinity of cTI (see P/Lmax and kinetic parameters in Table 6) to the membranes did not correlate with its ability to disrupt membranes. The higher efficacy of TI in disrupting membranes, compared to the cyclic analogue, might explain the higher activity of the parent TI–TIII for planktonic bacterial cells (see Table 3), compared to their cyclic analogues.
Figure 4.
Membrane binding and disruption induced by tachyplesin I (TI) and cyclic tachyplesin I (cTI). Model membranes composed of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and POPC/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine (POPS) (4:1) were compared. (a) Surface plasmon resonance sensorgrams obtained with 32 µM peptide injected over lipid bilayers deposited on an L1 chip surface for 180 s (association); dissociation was monitored for 600 s. Response units (RU) were converted into a peptide-to-lipid ratio (P/L (mol/mol)) to take into consideration the differences in lipid packing resulting in different amounts being deposited to cover the chip surface. (b) The dose-response curves show P/L obtained at the end of the association phase (t = 170 s) and plotted as a function of peptide concentration injected. (c) Percentage of vesicle leakage determined by fluorescence emission intensity of 5-carboxyfluorescein (λex = 490 nm, λem = 513 nm) leaking from LUVs at increasing concentrations of peptide. Lipid concentration used was 5 µM, and the peptide was tested up to 10 µM. Dose-response curves were fitted with one-site specific binding with Hill slope equation in GraphPad Prism.
Table 6.
The kinetic and affinity parameters from surface plasmon resonance analysis of the interaction of 32 µM tachyplesin I (TI) and cyclic tachyplesin I (cTI) with neutral (POPC) and negatively-charged model membranes (POPC/POPS (4:1)) and leakage induced by the same peptides and lipid systems.
| Peptide | Lipid System |
P/Lmax
(mol/mol) 1 |
KD
(µM) 1 |
koff (x 10−2 s−1) 2 |
P/Loff (mol/mol) 2 |
LCmax (%) 3 |
|---|---|---|---|---|---|---|
| TI | POPC | 0.26 ± 0.06 | 22.4 ± 10.9 | 1.50 ± 0.11 | 0.046 ± 0.001 | 39.5 ± 4.6 |
| cTI | 0.33 ± 0.07 | 16.7 ± 8.4 | 0.91 ± 0.03 | 0.065 ± 0.001 | 32.9 ± 9.4 | |
| TI | POPC/POPS (4:1) | 0.37 ± 0.04 | 11.8 ± 2.4 | 2.75 ± 0.22 | 0.096 ± 0.001 | 76.0 ± 2.8 |
| cTI | 0.48 ± 0.08 | 9.2 ± 3.4 | 0.70 ± 0.03 | 0.137 ± 0.002 | 58.5 ± 3.6 |
1 P/Lmax and KD were calculated from the dose-response curves (one-site specific binding with Hill slope equation, GraphPad Prism) in Figure 4b. The P/Lmax value represents the peptide-to-lipid ratio (mol/mol) when peptide-lipid binding reaches saturation, KD is the peptide concentration necessary to reach the half-maximal binding response. 2 koff is the dissociation constant and P/Loff is the peptide-lipid ratio at the end of association phase calculated from the sensorgrams obtained with 32 µM peptide in Figure 4a. koff and P/Loff were fitted in GraphPad Prism, assuming a Langmuir kinetic. 3 Percentage of leakage achieved when incubating 10 µM peptide with 5 µM LUVs. POPC is 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine; POPS is 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine.
To examine the ability of cTI to bind model membranes that mimic bacterial cell membranes, we prepared vesicles with an E. coli polar lipid extract composed of zwitterionic phosphatidylethanolamine (PE)-phospholipids, negatively-charged phosphatidylglycerol (PG)-phospholipids, and cardiolipin (CA) in the proportion 67:23.2:9.8 (wt/wt%). The SPR sensorgram and dose-response curves (Supplementary Figure S3) show that cTI has a high affinity for E. coli lipids. Comparison of the dose-response curves and fitted parameters show that the maximum amount of cTI bound to E. coli lipids (P/Lmax, Supplementary Table S4) is not as high as for the other tested negatively-charged membranes or for the zwitterionic POPC, but the P/Loff is higher than from the other tested membranes (see Supplementary Table S4), suggesting that a large amount of peptide remains bound to the bilayers that mimic bacterial membranes. To investigate whether cTI distinguishes the negatively-charged headgroups present in bacteria (i.e., PG) from those in cancer cells (i.e., PS), we compared the binding of cTI to POPC/POPG (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol; 4:1)) and to POPC/POPS (4:1). cTI has a slightly higher affinity for model membranes containing PS-phospholipids than to those containing PG-phospholipids, as shown by the higher maximum amount of peptide bound to POPC/POPS (4:1), and a slower dissociation rate (see P/Lmax, koff and P/Loff in Supplementary Figure S3).
2.5.2. Partitioning of Trp Residue into Model Membranes
The fluorescence emission of Trp is sensitive to the local environment. In an aqueous environment, the fluorescence emission spectrum of Trp has a maximum at ~350 nm (λex = 280 nm), in a hydrophobic environment the fluorescence emission spectrum is blue-shifted and an increase in the fluorescence quantum yield is usually observed [57,58,59]. Tachyplesin peptides have one Trp residue: the fluorescence emission properties of this residue can be used to examine the environment surrounding it and inform on the partitioning and orientation of the peptides bound to model lipid bilayers. In the current study, we followed the changes in Trp fluorescence emission spectra of TI–TIII and of the cyclic analogues cTI–cTIII upon titration with LUVs composed of zwitterionic (POPC) or of negatively-charged membranes with two distinct negatively-charged headgroups, i.e., PS and PG (Figure 5).
Figure 5.
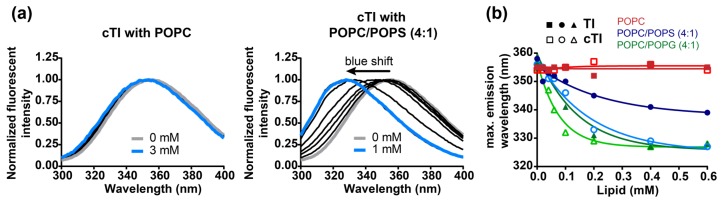
The partitioning of the 12.5 µM of tachyplesin I–III (TI-TIII) and cyclic analogues (cTI-cTIII) into lipid membranes. (a) Normalized fluorescence emission spectra of cTI in aqueous solution (grey) and upon titration with LUVs composed of POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine; up to 3 mM) or of POPC/POPS (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine; 4:1) up to 1 mM). Fluorescence emission spectra were obtained with excitation at 280 nm (b) Wavelength at which TI (solid symbol) and cTI (open symbol) have their maximum fluorescence emission in buffer and with increasing concentrations of LUVs composed of different lipid mixtures (POPC: red; POPC/POPS (4:1): blue; POPC/POPG (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol; 4:1): green).
TI and cTI bound to both zwitterionic POPC and negatively-charged POPC/POPS (4:1) bilayers, as shown with SPR (see Figure 4a,b); however, a blue shift in the Trp fluorescence emission spectra was only observed for model membranes containing negatively-charged phospholipids (Figure 5a, Table 7). Even at the highest concentration of 3 mM POPC LUVs, no change in the Trp fluorescence emission spectra was observed for any of the peptides. In contrast to POPC, a blue shift was detected when peptides were incubated with negatively-charged model membranes POPC/POPS (4:1) and POPC/POPG (4:1) at lipid concentrations as low as 0.1 mM (Figure 5).
Table 7.
Shift in the fluorescence emission maximum wavelength of tachyplesin I–III (TI-TIII) and cyclic analogues (cTI-cTIII) in buffer and in the presence of model membranes.
| POPC | POPC/POPS (4:1) | POPC/POPG (4:1) | |||
|---|---|---|---|---|---|
| Peptide | shift (nm) 1 | shift (nm) | 0.5 [L] (mM) 2 | shift (nm) | 0.5 [L] (mM) |
| TI | 2 | 22 | 0.19 | 30 | 0.09 |
| TII | 0 | 19 | 0.30 | 29 | 0.09 |
| TIII | −3 | 18 | 0.30 | 23 | 0.10 |
| cTI | 0 | 28 | 0.11 | 28 | 0.06 |
| cTII | 1 | 27 | 0.08 | 29 | 0.05 |
| cTIII | 3 | 27 | 0.12 | 28 | 0.08 |
1 blue shifts observed with 3 mM POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine); 1 mM POPC/POPS (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine; 4:1), or 1 mM POPC/POPG (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol; 4:1) LUVs. 2 lipid concentrations required to achieve half of the maximum blue shift observed (0.5 [L] in mM).
In the presence of negatively-charged membranes, the Trp fluorescence emission maxima of cTI–cTIII had a larger shift than the respective parent peptides TI–TIII (Table 7), as illustrated with TI and cTI (Figure 5b). The differences between parent and cyclic peptides were most pronounced with POPC/POPS (4:1) bilayers: TI–TIII required more than twice the amount of POPC/POPS (4:1) LUVs compared to cTI–cTIII (0.19–0.30 mM vs. 0.08–0.12 mM) to induce half of the maximal shifts of the Trp fluorescence emission spectra (Figure 5b, Table 7). Comparison among parent peptides shows similar spectral shifts for a given model membrane (see for instance Trp fluorescence emission shifts of TI–TIII when titrated with POPC/POPS); the same is true among cyclic analogues. These results suggest that the mutations R/K1 or R/K15 have a weak influence, whereas backbone cyclization between R/K1 and G18 impacts the insertion of the Trp residue into lipid membranes.
The Trp fluorescence emission spectra of all tachyplesin analogues displayed larger blue shifts in the presence of POPC/POPG (4:1) compared to POPC/POPS (4:1) LUVs, which suggests that the Trp residue partitions better and/or inserts further into PG-containing membranes than into PS-containing membranes (Table 7). Despite the blue shift in the fluorescence emission spectra of all the peptides when in the presence of negatively-charged membranes, no increase in quantum yield was detected (data not shown). This could be explained by fluorescence quenching induced by neighboring amino acid side chains, local carbonyl groups [58], or through photon re-absorption between Trp residues of neighbor peptides molecules due to their closer proximity once inserted into lipidic membranes.
2.5.3. Insertion of Trp Residue into Model Membranes
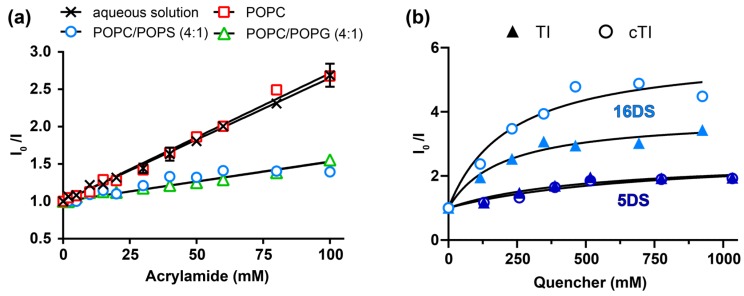
The in-depth location of the Trp residue of tachyplesin peptides bound to lipid membranes was investigated using fluorescence quenching methodologies. Acrylamide, an aqueous quencher unable to partition into lipid bilayers, was used to quench the fluorescence of Trp residues exposed to the aqueous solution. If the Trp residue inserts into the lipid bilayer, it becomes inaccessible to quenching by the aqueous-soluble acrylamide. In addition to acrylamide quenching, we used the lipidic quenchers 5- and 16-doxyl stearic acids (5DS and 16DS) to gain information on the in-depth location of TI and cTI within the membrane. The acids 5- and 16DS are fatty acids that insert into the lipid bilayer and possess the quencher moiety, nitroxide, located at carbon 5 and 16 of the fatty acid chain, respectively. The proximity of the nitroxide moiety to the Trp residue is required for quenching of its fluorescence emission; thus, comparison between the 5- and 16DS quenching efficacies gives information on the location of the Trp residue when the peptide is partitioned into membranes [60,61]. Changes in the Trp fluorescence emission of individual peptides were followed upon titration with each of the quenchers. The fluorescence emission intensity in the absence and presence of the quencher (I0/I) were plotted as a function of the quencher concentration and used to determine the Stern-Volmer constants (KSV) (Figure 6), which are proportional to the accessibility of the quencher to the fluorophore.
Figure 6.
The effect of aqueous (acrylamide) and lipidic (5DS and 16DS) quenching on the tryptophan fluorescence intensity to investigate peptide in-depth location within model membranes. (a) The Trp fluorescence emission of 12.5 µM cTI in aqueous solution or in the presence of LUVs (1 mM POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine), 1 mM POPC/POPS (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine; 4:1), 0.1 mM POPC/POPS (4:1) or 0.1 mM POPC/POPG (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol; 4:1)) in the absence (I0) and upon titration with increasing concentrations of acrylamide (I). Data are shown as Stern-Volmer plots (i.e., I0/I versus concentration of quencher). (b) Quenching of Trp fluorescence emission of 12.5 µM TI and cTI incorporated in 1 mM POPC/POPS (4:1) membranes by 5DS or 16DS.
A reduction in the slope (KSV) of the fitted data points in the presence of lipids, compared to the aqueous solution, indicates the reduced quenching efficacy by acrylamide. The percentage of Trp accessible to acrylamide was estimated by the ratio of KSV obtained in the presence and absence of LUVs, assuming that the Trp residue is fully exposed to acrylamide when the peptide is in an aqueous solution. Acrylamide quenched the Trp fluorescence emission of both TI and cTI with a similar efficacy in the buffer and in the presence of 1 mM POPC LUVs (Figure 6a, Table 8). These results show that when TI and cTI are bound to POPC, their Trp residue is accessible to acrylamide and likely to be exposed to the aqueous environment. In contrast, when in the presence of 1 mM POPC/POPS (4:1), the Trp fluorescence emission of TI and cTI was not efficiently quenched by acrylamide, suggesting that their Trp residue is not accessible to acrylamide. A similar result is expected for all the analogues given the lack of insertion of the Trp residue of TI–TIII and of cTI–cTIII when bound to POPC membranes and the large blue shift in the fluorescence emission spectra of the Trp residue of all the analogues when bound to POPC/POPS (4:1) membranes (see Table 7).
Table 8.
Fluorescence quenching of 12.5 µM tachyplesin I-III (TI-TIII) or cylic tachyplesin I-III (cTI-cTIII) by acrylamide.
| Acrylamide Accessibility (%) 1 | ||||
|---|---|---|---|---|
| 1 mM | 1 mM | 0.1 mM | 0.1 mM | |
| Peptide | POPC | POPC/POPS (4:1) | POPC/POPS (4:1) | POPC/POPG (4:1) |
| TI | 90.5 ± 4.3 | 16.2 ± 21.9 | 72.3 ± 3.9 | 54.2 ± 5.8 |
| TII | 76.2 ± 3.2 | 56.8 ± 6.9 | ||
| TIII | 82.1 ± 4.7 | 75.4 ± 4.6 | ||
| cTI | 103.8 ± 2.3 | 22.5 ± 14.3 | 32.3 ± 8.3 | 32.0 ± 4.1 |
| cTII | 61.4 ± 5.5 | 36.1 ± 7.7 | ||
| cTIII | 48.7 ± 6.5 | 46.2 ± 5.7 | ||
1 Tryptophan accessibility of tachyplesin I-III (TI–TIII) and cyclic tachyplesin (cTI–cTIII) in the presence of LUVs (1 mM POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine), 1 mM POPC/POPS (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine; 4:1), 0.1 mM POPC/POPS (4:1) or 0.1 mM POPC/POPG (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol; 4:1)) to the aqueous quencher acrylamide. The accessibility of the Trp residue to the quencher was calculated from the Stern-Volmer constants, KSV, which were derived from the linear fit of the data points (Equation (1)) of the peptide in buffer and peptide in the presence of lipids (see Figure 6a). Full exposition (100%) of the Trp residue to an aqueous environment was assumed in the buffer.
To identify potential differences between the six analogues, we monitored the quenching of Trp fluorescence emission by acrylamide in the presence of 0.1 mM POPC/POPS (4:1) or of 0.1 mM POPC/POPG (4:1) membranes. In the presence of 0.1 mM POPC/POPS (4:1) LUVs, acrylamide quenched the Trp fluorescence emission of TI–TIII with a higher efficacy than that of the cyclic analogues, suggesting that the Trp residue of cTI–cTIII is more protected from acrylamide. The Trp of the native tachyplesin peptides was less accessible for quenching in the presence of 0.1 mM POPC/POPG (4:1) LUVs compared to 0.1 mM POPC/POPS (4:1) LUVs through insertion into the membrane. The accessibility of the acrylamide to the Trp residues of the cyclic analogues remained approximately the same with both lipid systems (Table 8). These results suggest that when compared at the same lipid concentrations, more TI–TIII are inserted into POPC/POPG (4:1), than into POPC/POPS (4:1) membranes, whereas the cyclic analogues did not distinguish between the two negatively-charged lipid mixtures.
Comparison of the quenching efficacy by 5DS and 16DS tested with POPC/POPS (4:1) bilayers, showed that 16DS quenched the Trp fluorescence emission of TI and cTI with a higher efficacy than 5DS, which was particularly evident for the cyclic analogue (see Stern-Volmer plots and KSV values in Figure 6b, Table 9). The higher quenching efficacy induced by 16DS suggests that the Trp residue of TI and of cTI is located within the hydrophobic core of the POPC/POPS (4:1) bilayers. The negative deviation to the linearity (see Figure 6b and fb values in Table 9) suggests that the Trp residue of a fraction of peptide molecules is not accessible to the quenchers. This can be an indication of peptide molecules adopting different orientations within the membrane; nevertheless, the average location of the Trp residue of cTI and TI confirms that both peptides have their Trp residue deeply inserted and, in particular, the cyclic analogue. Overall, these results suggest that the Trp residue of cyclic analogues partitioned with a higher efficacy and/or were inserted deeper into anionic lipid bilayers, than parent tachyplesin peptides (Figure 7).
Table 9.
Fluorescence quenching of tachyplesin I (TI) and and cyclic tachyplesin I (cTI) partitioned into 1 mM POPC/POPS (4:1) LUVs by the quenchers 5DS and 16DS. 1.
| 5DS | 16DS | ||||
|---|---|---|---|---|---|
| Peptide | KSV (M−1) | fb | KSV (M−1) | fb | Z (Å) 2 |
| TI | 3.7 ± 1.3 | 0.63 ± 0.57 | 19.6 ± 4.3 | 0.74 ± 0.37 | 8.8 |
| cTI | 4.9 ± 1.8 | 0.61 ± 0.54 | 26.3 ± 7.1 | 0.83 ± 0.52 | 5.5 |
1 The KSV and fb (the fraction of light emitted by the peptide accessible to the quencher) were determined using the Lehrer equation (Equation (2)). 2 The calculated average distance, Z in Å, of the Trp residue from the bilayer center was determined using the parallax method (Equations (3) and (4)) [62].
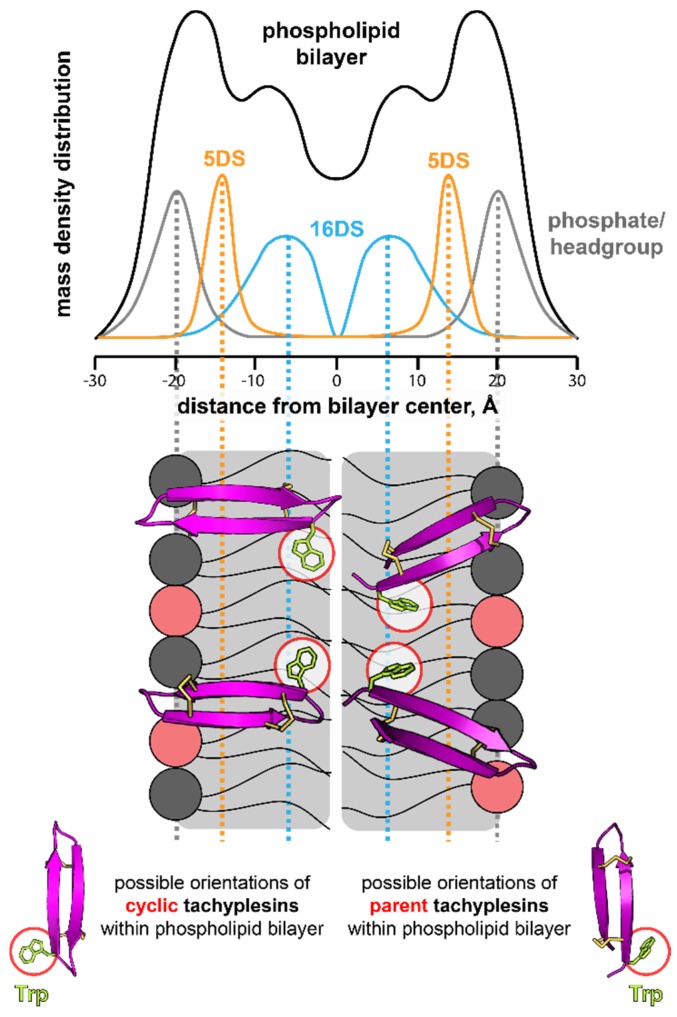
Figure 7.
Schematic representation of the proposed orientations of tachyplesin peptides and cyclic analogues, when inserted into negatively-charged membranes based on the tryptophan fluorescence quenching, results with 5- and 16DS. The phospholipids in grey represent PC headgroups and the phospholipids in red represent PS headgroups. The peptides have a backbone length of approximately 25 Å (measured in PyMol) and are able to span across a lipid monolayer. Other orientations are possible. The distribution of the 5DS and 16DS quencher within a phospholipid bilayer was illustrated based on previous publications [60,63].
3. Discussion
In this study we demonstrated the high structural homology of the three tachyplesin peptides TI, TII and TIII. The primary sequence of these peptides differs only between Lys or Arg residues at positions 1 or 15, and both of these positions are located close to the flexible peptide termini. The cyclic analogues cTI, cTII and cTIII share a high structural homology with each other and with their respective parent peptide, but the backbone cyclization reduced the range of motion of the amino acid side chains located in the region of the termini (see Figure 1d).
TI–TIII and cTI–cTIII were active against representative Gram-positive and Gram-negative bacteria at low micromolar MICs (see Table 3). The antimicrobial activities of the three tachyplesin peptides were similar, as were the activities of the three cyclic analogues; however, differences were observed between the two groups. Overall, the parent tachyplesin peptides exhibited stronger antimicrobial activities than the cyclic analogues against all bacteria tested in the planktonic growth form. The Gram-negative E. coli strains were more sensitive to the peptides than the Gram-positive S. aureus strains. Against the S. aureus biofilm (see Figure 3 and Table 4), the tachyplesin peptides were >5-fold less active compared to the planktonic cells, likely due to the reduced accessibility of the peptides to the bacterial cell in biofilm form or the low ability of the peptides to penetrate the extracellular matrix of exopolysaccharides and access the cells [64]. The activities of the tachyplesin peptides against biofilms are comparable to other tested antimicrobial peptides [53]. The efficacy against the biofilms could be increased by using higher concentrations of the tachyplesin peptides or by using the peptide in combination with other antibiofilm or extracellular matrix disrupting agents [65]. Interestingly, TIII had been previously shown to prevent the formation of a Pseudomonas aeruginosa biofilm in a rat model of ureteral stent infection [66]; thus, even if the peptides cannot be used to completely disrupt the biofilm at the tested concentration, they could be useful for preventing biofilm formation on biomedical devices [67].
TI–TIII and cTI–cTIII peptides had a higher efficacy against the melanoma cell lines MM96L, HT144 and WM164 than to non-cancerous cells or the HeLa cervical cancer cell line (see Table 5). Interestingly, the cyclic tachyplesin analogues were more active against the cervical cancer cell line HeLa, than the parent tachyplesins. Differences in membrane composition between the different cells might explain the selectivity of the six analogues for melanoma over other cell lines [43], whereas a variation in the lipid-binding affinity and orientation within the membrane between parent versus cyclic analogues might explain more subtle differences observed against individual cell lines.
The experiments with model membranes confirmed the preference of all peptides for negatively-charged membranes (see Figure 4). TI, and its cyclized analogue cTI had a higher affinity for the negatively-charged POPC/POPS (4:1) than for neutral POPC bilayers, as shown in SPR experiments. cTI bond stronger and dissociated slower from POPC/POPS (4:1) and POPC bilayers than TI; however, TI disrupted both lipid bilayers with a higher efficacy than cTI. The ability of tachyplesin peptides to disrupt negatively-charged and neutral membranes with higher efficacy than their respective cyclic analogues, corroborates with their higher antimicrobial and hemolytic activities.
The fluorescence spectroscopy experiments show that the Trp residue of all six tachyplesin analogues partition and/or insert deeper into POPC/POPG (4:1), than into POPC/POPS (4:1) lipid bilayers (see Figure 5); in contrast, SPR studies show that cTI has a slightly higher affinity for POPC/POPS (4:1) than for POPC/POPG (4:1) membranes. This apparent contradiction emphasizes that specific phospholipid headgroups and membrane composition play a role in regulating and fine-tuning peptide-lipid binding interactions and the location of specific residues within the membrane.
The orientation of the peptide bound to membranes might also be affected by the phospholipid headgroup. When TI–TIII and cTI–cTIII were bound to neutral model membranes, their tryptophan residue did not insert into the bilayer. These results support an orientation in which both parent and cyclic tachyplesin peptides are parallel to the water-lipid interface, or with an orientation in which the β-turn (Y8-I11) inserts into the hydrophobic region. In the presence of negatively-charged membranes, the Trp residue of both parent and cyclic tachyplesin peptides partitioned into the membrane. However, the Trp residue in cyclic peptides cTI–cTIII seemed to be more deeply inserted into the bilayer and less exposed to the aqueous environment (see Figure 6, Table 8 and Table 9).
Previous studies detected minor alterations to the peptide backbone of TI: the bending of the termini when the peptide was in contact with lipid membranes [36,68]. Doherty et al. [69] suggested that large amplitude motions of TI in the plane of the lipid membrane are essential for translocation, pore formation and membrane disruption. Since backbone cyclization impacts the flexibility of the termini region, cyclic tachyplesin analogues could be more restricted in the conformational changes of their backbone upon interaction with the membrane. The lower flexibility in the termini region might explain the lower ability of the cyclic analogue cTI to disrupt lipid bilayers compared to the parent tachyplesin. Furthermore, backbone cyclization created a second turn (Y17-W2) at which the Trp residue is located (see Figure 1d). The cyclic tachyplesin peptides are likely to insert deeply into the lipid membrane with the second turn (Y17-W2) inserting close to the center of the bilayer (see Figure 6 and Table 9), whereas in the parent peptides, the Trp residue is slightly closer to the membrane interface. A shallower location of the Trp residue could result from the parent peptides adopting a tilted orientation within the membrane (see Figure 7). Differences in the orientation of the peptides when inserted into lipid membranes could explain the higher membrane-disruptive properties and hemolytic activity of TI, compared to cTI (see Figure 4c and Table 6).
Backbone cyclization was shown to increase stability to proteolytic degradation, reduce membrane disruption and decrease hemolysis (see Figure 2). Interestingly, cTII has a slightly higher hemolytic activity compared to cTI and cTIII. cTII has an Arg residue at position 1 and a Lys at position 15, whereas cTI and cTIII have Lys residues at position 1. Arg and Lys residues establish different interactions with the phospholipid headgroups. The side chain of Arg residues can establish hydrogen bonds (H-bond) with two phospholipid headgroups, whereas the side chain in Lys residues can only form one H-bond. In contrast to Lys, Arg residues can form an H-bond through their side chains while being involved in cation-π-interactions [70]. Taking into consideration the increased toxicity of cTII due to an Arg residue, a Lys residue at position 1 favors the selectivity towards cancer cells.
In conclusion, TI–TIII and their cyclized analogues cTI–cTIII have high structural homology. Backbone cyclization increased peptide stability against proteolytic degradation, reduced the flexibility of amino acid side chains located in the terminal regions (i.e., R/K1, W2), and hemolytic activity while maintaining potent anticancer and antimicrobial activities. Since high resistance to proteolysis and low toxicity for host cells are preferred properties for peptide-based drug templates, the cyclic analogues have a higher potential than the parent counterparts, despite a possible loss of potency depending on the target. The Arg at position 1 of the amino acid sequence of TII and cTII proved to be non-beneficial for reducing hemolytic activity and improving selectivity for cancerous cells. Thus, cTI and cTIII have a higher potential than cTII as peptide-based anticancer drug templates. We also found that melanoma cell lines are more susceptible to the treatment with the tachyplesin peptides than the cervical cancer cell line HeLa. Systematic changes of the amino acid sequence might be applied to further increase the selectivity and/or anticancer activity of cyclic tachyplesin analogues (i.e. cTI or cTIII) to target specific cancers.
4. Material and Methods
4.1. Peptide Synthesis, Folding and Purification
The synthesis, folding and purification of synthetic peptides were carried out as previously described [42,71]. Briefly, the peptides were synthesized using 9-fluorenylmethoxycarbonyl (Fmoc) solid-phase peptide synthesis (SPPS) on an automatic peptide synthesizer (Symphony, Protein Technologies Inc., Tucson, USA). Rink amide resin was used for the synthesis of parent tachyplesin peptides and 2-chlorotrityl (2-CTC) resin for the backbone cyclized analogues. The peptides were oxidized overnight in 0.1 M ammonium bicarbonate buffer at pH 8.5 and purified using reverse-phase HPLC (solvent A: H2O, 0.05% (v/v) trifluoroacetic acid (TFA), solvent B: 90% (v/v) acetonitrile, 0.05% (v/v) TFA) until the desired purity of >95%. The correct peptide mass was confirmed with ESI-MS, while native disulfide connectivity was inferred from the dispersion of peaks in the 1D NMR spectra using a Bruker Avance 600 MHz spectrometer (Billerica, USA). The peptide concentration was determined from the absorbance at 280 nm (ɛ280 = 8730 M−1.cm−1 as estimated extinction coefficient based on the contribution of Tyr and Trp residues, and disulfide bonds).
4.2. NMR Spectroscopy
For the structural analysis of TI–III and cTI–III, peptide (1 mg/mL) was dissolved in H2O/D2O (10:1, v/v) and the pH adjusted to pH 4–5. 1D 1H spectra, two-dimensional total correlated spectroscopy (TOCSY) and nuclear Overhauser effect spectroscopy (NOESY) were acquired with a Bruker Avance 600 MHz NMR spectrometer (Billerica, USA) at a temperature of 298 K. Additional spectra for 1H-13C HSQC and 1H-15N HSQC in H2O/D2O (10:1, v/v) and exclusive correlation spectra (E.COSY) in D2O were acquired. Spectra were referenced to an internal standard 2,2-dimethyl-2-silapentone-5-sulfonate (DSS) at 0 ppm. CYANA 3.97 was used to automatically calculate and refine structures based on distance restraints derived from the NOESY spectra [72], and torsion angles (ϕ and φ) generated using TALOS-N and Hα, Cα, Cβ, HN chemical shifts derived from NOESY, 1H-13C HSQC and 1H-15N HSQC spectra [73]. Several χ1 side-chain angle restraints were added based on E.COSY and NOESY data. A final set of structures was generated with CNS [74] using torsion angle dynamics, refinement and energy minimization in explicit solvent. Final structures were assessed for stereochemical quality using MolProbity [75].
4.3. Serum Stability
The serum stability assay was carried out as previously described [42] with some modifications. Briefly, tachyplesin variants were incubated in 25% (v/v) human serum diluted in phosphate-buffered saline (PBS) at a final concentration of 50 µM at 37 °C. Triplicates were collected at time 0 h and 24 h and the serum proteins were precipitated with acetonitrile (1:3 ratio) supplemented with 3% TFA. Samples were kept on ice for 10 min before centrifugation at 17,000× g for 10 min at 4 °C. The peptide containing supernatant of each sample was harvested and quantified using RP-HPLC (10 to 45% solvent B, 1%/min gradient). The percentage of peptide stability in human serum was calculated by comparing the area of the peptide peak obtained at 24 h to that at time 0 h. A linear peptide containing 18 amino acid residues (KGGGGSGQLIDSMANSFV) was included as a control susceptible to proteolytic degradation.
4.4. Hemolytic Studies
A small amount of blood was collected from three healthy human donors. The blood was immediately diluted in PBS and centrifuged 4–5 times for 1 min at 4000 rpm to wash and separate the human red blood cells (RBCs). RBCs suspension (0.25% (v/v) in PBS) was incubated with peptides with two-fold serial dilutions of the peptide (highest concentration tested was 128 µM, and the lowest was 0.25 µM) in a 96-well plate. Melittin, a membrane disruptive peptide, was used as control. The plates were incubated for 1 h at 37 °C. After incubation, the plates were centrifuged for 5 min at 1000 rpm to pellet any non-lysed RBCs. A total of 100 µL of the supernatant were transferred to a new 96-well plate [76]. The hemoglobin released into the supernatant from lysed cells was measured by absorbance at 415 nm using the Tecan infinite M1000Pro multiplate reader (Männedorf, Switzerland).
4.5. Cell Culture
Cells were grown in cell culture flasks and incubated in a humidified atmosphere (5% CO2, 37 °C). The cancer cell lines HeLa and the control cell line HaCaT were grown in a DMEM medium supplemented with 1% (v/v) penicillin/streptomycin and 10% (v/v) fetal bovine serum (FBS). The cancer cell lines MM96L, HT144 and WM164 were grown in a RPMI medium supplemented with 1% (v/v) penicillin/streptavidin, 10% (v/v) FBS, 20 mM l-glutamine, and 10 mM sodium pyruvate. Cell cultures were maintained by dilution upon reaching confluence, each 48–72 h. More information about the cell lines [77] is available in Tables S2 and S3.
4.6. Cytotoxicity Assays
Cells were seeded into 96-well flat-bottom plates at 5 × 103 cells/well and incubated overnight. The medium was removed and replaced with 90 µL serum-free medium, 10 µL of 10x concentrated peptide solutions in PBS were added. PBS was added as blank and 0.1% (v/v) Triton X-100 was used to establish 100% of cell death. After 2 h incubation at 37 °C, 10 µL of filtered 0.05% (w/v) resazurin solution was added to each well [70]. Resazurin is converted to the pink and fluorescent compound resorufin by viable cells [78]. After incubation overnight, the fluorescence intensity (λex = 565 nm and λem = 584 nm) was measured with the Tecan infinite M1000Pro multiplate reader (Männedorf, Switzerland).
The selectivity was calculated through the activity-toxicity index (ATI) [46]. ATI = MHC/MCC50, with MHC being the minimal concentration necessary to induce 10% or 50% cell death in human red blood cells and MCC50 the median of cytotoxic concentrations (CC50) of all tested melanoma cell lines.
4.7. Antimicrobial Studies
S. aureus ATCC 25923, S. aureus ATCC 6538, E. coli ATCC 25922, and E. coli DC2 CGSC 7139 were grown in Mueller Hinton Broth (Sigma Aldrich, St. Luis, USA). Bacterial cultures in the exponential growth phase were diluted to an OD600nm of 0.001 and seeded into 96-well plates. Peptides at different concentrations, starting at 64 µM and with two-fold serial dilutions (i.e., 64, 32, 16, 8, 4, 2, 1, 0.5, 0.25, 0.125 and 0.0625 µM), were incubated with cells [43]; 0.05% (v/v) resazurin was added to the cultures the next day and the conversion to resorufin was measured after 1 h of incubation at 37 °C using a 565 nm excitation and 584 nm emission wavelength, as above.
4.8. Biofilm Studies
S. aureus ATCC 6538 (1 × 106 cfu/mL) was cultured in Tryptic Soy Broth, containing 0.25% (w/v) glucose and incubated in 96-well microtiter flat-bottomed polystyrene plates for 24 h at 37 °C. Preformed biofilms were then washed with Mueller Hinton Broth to remove non-adherent cells. Two-fold serial dilutions of each peptide (highest concentration tested was 32 µM and lowest was 0.25 µM) were added to the biofilms for 4 h. Untreated 24 h preformed biofilms were used as a control. The metabolic activity of biofilm-embedded cells was determined using a resazurin reduction fluorometric assay as previously described [53].
4.9. Lipid Vesicle Preparation
Mixtures with synthetic lipids (POPC, POPS, POPG, Avanti Polar Lipids) or E. coli lipid extract (Avanti Polar Lipids, Alabaster. USA) were extruded in HEPES buffer (10 mM HEPES, 150 mM NaCl, pH 7.4) to produce lipid vesicles, as previously described [43,79]. LUVs (Ø ≤ 100 nm) were used in fluorescence spectroscopy assays and small unilamellar vesicles (SUVs, Ø ≤ 50 nm) for SPR.
4.10. Fluorescence Spectroscopy Assays
Fluorescence emission spectra (300–400 nm, excitation at 280 nm, slits 3/3 mm) of 12.5 µM peptide and l-Trp in HEPES buffer (in quartz cuvettes, path length of 0.5 cm) were scanned upon titration with LUVs composed of various lipid compositions (up to 3 mM POPC, and up to 1.5 mM POPC/POPS (4:1) or POPC/POPG (4:1)) [80] using a FluoroMax-4 spectrofluorometer (Horiba, Kyoto, Japan). Integrated areas of the fluorescence emission spectra were corrected for fluorophore dilution and light dispersion due to titration with LUVs suspension; the blank was discounted.
4.11. Quenching of Tryptophan Fluorescence
The membrane in-depth location of the Trp residue within the peptides was followed using Acrylamide (Sigma Aldrich, St. Luis, USA) and 5- and 16-DS (Sigma Aldrich, St. Luis, USA). Quenching induced by acrylamide was monitored with 12.5 µM peptide in a HEPES buffer, in the presence of 1 mM of POPC, 0.1 mM POPC/POPS (4:1), 1 mM POPC/POPS (4:1), or 0.1 mM POPC/POPG (4:1) titrated with increasing concentration of acrylamide [81]. The fluorescence emission spectra were determined with an excitation wavelength of 290 nm to reduce the quencher/fluorophore light absorption ratio. The fluorescence emission spectra area was corrected for the inner filter effect [82] due to increased absorbance of acrylamide. Data points were analyzed using the Stern-Volmer representation (Equation (1)), the KSV is determined from the slope.
| (1) |
A total of 12.5 µM of TI, or of cTI, in HEPES buffer and 1 mM POPC/POPS (4:1) were titrated with increasing concentration of 5DS, or of 16DS. The fluorescence emission spectra area was corrected for the inner filter effect [82] due to increased absorbance of 5-,16DS. The effective concentration of 5DS and 16DS in the lipid bilayer was calculated: partition coefficients of 5DS and 16DS into fluid membranes are 89,000 and 9730, respectively [59,81]. The data points had a negative deviation to linearity and the KSV was determined by fitting the data with the Lehrer equation (Equation (2)) [59,81].
| (2) |
I0 = fluorescent intensity in buffer
I = fluorescent intensity in presence of quencher
KSV = Stern-Volmer constant
[Q] = quencher concentration
fB = fraction of light accessible to the quencher =
I0,B = fluorescent intensity of the accessible population of the quencher when [Q] = 0
The average distance of the Trp residue from the bilayer center (Å) was determined using the parallax method [62] following the equation,
| (3) |
| (4) |
in which the z1F is the distance between the Trp residue and the quencher moiety in 5DS, L21 is the distance between the quencher groups in 5 and 16DS (5 Å), F1 is the fluorescence of 12.5 µM of TI, or of cTI, in the presence of 1 mM POPC/POPS (4:1) and 0.4 mM 5DS, F2 is the fluorescence of 12.5 µM of TI, or of cTI, in the presence of 1 mM POPC/POPS (4:1) and 0.4 mM 16DS. C is the molar fraction of quencher within the total lipid concentration per unit area (assuming the surface of lipid to be 70 Å2) [62]. zcF is the distance between the Trp residue and the center of the bilayer, Lc1 is the distance between the center of the bilayers and the quencher group of 5DS (15 Å).
4.12. Surface Plasmon Resonance (SPR)
SPR was used to investigate the affinity and binding kinetics of peptides to membranes of different compositions. The experiments were conducted with an L1 biosensor chip (GE Healthcare) at 25 °C using a BIAcore 3000 instrument (GE Healthcare, Chicago, USA) [83,84]. The HEPES buffer was used for sample preparation and as a running buffer. Lipid bilayers were immobilized onto L1 chip by injection of SUVs at a flow rate of 2 µL/min. Peptide samples with two-fold serial dilutions (the highest concentration tested was 64 µM and the lowest was 1 µM) were injected over the lipid bilayer at a flow rate of 5 µL/min. Association of the peptides onto the lipid bilayer was followed for 180 s and the dissociation from the lipid for 600 s. The BIAeval software was used to analyze the sensorgrams. The response units (RU) were normalized to the peptide-to-lipid ratio (P/L); the P/L obtained at a fixed time point at the end of the association curve (at 170 s), at which the response has reached a plateau and the binding is close to equilibrium, was used to compare the affinity of the peptides to the different lipid systems [84].
4.13. Vesicle Leakage Assay
LUVs (Ø ≤ 100 nm) were prepared with the HEPES buffer containing 40 mM of the fluorescent 5-carboxyfluorescein (CF, Sigma Aldrich, St. Luis, USA). LUVs filled with CF at self-quenching concentrations were separated from the non-encapsulated dye on a Sephadex G-50 column equilibrated with HEPES buffer [85]. The concentration of CF-LUVs was determined through a calibration curve prepared from the original lipid mixture using Stewart’s assay (absorbance at 485 nm) [86]. Peptides were incubated (25 °C, 20 min) at two-fold dilutions (starting at 10 µM) with LUVs (5 µM) in the HEPES buffer in a 96-well flat-bottom black optiplates (Perkin Elmer, Waltham, USA). Vesicles and peptides were incubated for 20 min in dark and the release of CF was measured in a Tecan infinite M1000Pro multiplate reader using 490 nm as the excitation and 513 nm as emission wavelengths.
4.14. Statistical Analysis
Values (mean or fit ± SEM) were analyzed in GraphPad Prism 7 to test for significant differences in the cytotoxic activity of the peptides. The multiple t-test function with the Holm–Sidak method was applied when indicated. P values below 0.05 were considered to be significant.
Acknowledgments
Many thanks to QUEDDI, the Matt Cooper lab and the Helmut Schaider lab for the cell lines HaCaT, HT144 and WM164 respectively. The authors thank Olivier Cheneval and Joachim Weidmann (IMB, UQ) for their assistance with peptide synthesis.
Abbreviations
| 1D | One-dimensional |
| 2-CTC | 2-chlorotrityl resin |
| 3D | Three-dimensional |
| ACN | Acetonitrile |
| ATI | Activity/toxicity index |
| ATTC | American Type Culture Collection |
| BMRB | Biological magnetic resonance data bank |
| CC | Cytotoxic concentration |
| CF | 5-carboxyfluorescein |
| cTI | Cyclic Tachyplesin I |
| cTII | Cyclic Tachyplesin II |
| cTIII | Cyclic Tachyplesin III |
| cTI–cTIII | Cyclic Tachyplesin I, II and III |
| DS | Doxyl-stearic acid |
| E.COSY | Exclusive correlation spectra |
| ESI-MS | Electrospray ionization mass spectroscopy |
| FBS | Fetal bovine serum |
| Fmoc | 9-fluorenylmethoxycarbonyl |
| HC50 | Hemolytic concentration necessary for 50% lysis of RBCs |
| HDP | Host defense peptide |
| KSV | Stern – Volmer constant |
| LPS | Lipopolysaccharide |
| LUV | Large unilamellar vesicle |
| MCC | Minimal cytotoxic concentration |
| MHC | Minimal hemolytic concentration |
| MIC | Minimal inhibitory concentration |
| NMR | Nuclear magnetic resonance |
| NOESY | Nuclear Overhauser effect spectroscopy |
| PBS | Phosphate buffered saline |
| PDB | Protein data bank |
| PC | Phosphatidylcholine |
| PS | Phosphatidylserine |
| PG | Phosphatidylglycerol |
| P/L | Peptide-to-lipid ratio |
| POPC | 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine |
| POPS | 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine |
| POPG | 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) |
| RBCs | Red blood cells |
| RP-HPLC | Reverse-phase high-performance liquid chromatography |
| RT | Retention time |
| RU | Response unit |
| SEM | Standard error of mean |
| SPPS | Solid phase peptide synthesis |
| SPR | Surface plasmon resonance |
| SUV | Small unilamellar vesicle |
| TI | Tachyplesin I |
| TII | Tachyplesin II |
| TIII | Tachyplesin III |
| TI–TIII | Tachyplesin I, II and III |
| TFA | Trifluoroacetic acid |
| TOCSY | Total correlated spectroscopy |
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/17/4184/s1.
Author Contributions
F.V. conceptualized this study with supervision and input from S.T.H. and N.L. F.V., S.A.D., and P.J.H. conducted experimental work: F.V. folded and purified the peptides, conducted toxicity assays, model membranes, surface plasmon resonance and fluorescence spectroscopy assays; S.A.D. conducted the biofilm assay and helped F.V. with the antimicrobial assays; P.J.H. performed NMR spectroscopy and structural analysis of the peptides; Y.-H.H. established the serum stability assay. F.V. analyzed and interpreted data with assistance from S.T.H. and expert contributions from all the co-authors. S.T.H., N.L., P.J.H., D.J.C., Y.-H.H. and A.S.V. revised and edited the manuscript. S.T.H. and D.J.C. provided the resources and acquired the funding.
Funding
This project was funded by a National Health Medical Research Council (NHMRC) project grant (APP1084965). F.V. was supported by the UQ Research Scholarship, S.T.H. is an Australian Research Council (ARC) Future Fellow (FT150100398), D.J.C. is an ARC Australian Laureate Fellow (FL150100146). Marie Skłodowska-Curie Research and Innovation Staff Exchange grant (RISE; call: H2020-MSCA-RISE-2014, grant agreement 644167) funded secondments of S.A.D. and of A.S.V. to the University of Queensland. The Translational Research Institute is supported by a grant from the Australian Government.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Edwards I.A., Elliott A.G., Kavanagh A.M., Zuegg J., Blaskovich M.A.T., Cooper M.A. Contribution of Amphipathicity and Hydrophobicity to the Antimicrobial Activity and Cytotoxicity of β-Hairpin Peptides. ACS Infect. Dis. 2016;2:442–450. doi: 10.1021/acsinfecdis.6b00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miyata T., Tokunaga F., Yoneya T., Yoshikawa K., Iwanaga S., Niwa M., Takao T., Shimonishi Y. Antimicrobial peptides, isolated from horseshoe crab hemocytes, tachyplesin II, and polyphemusins I and II: chemical structures and biological activity. J. Biochem. 1989;106:663–668. doi: 10.1093/oxfordjournals.jbchem.a122913. [DOI] [PubMed] [Google Scholar]
- 3.Nakamura T., Furunaka H., Miyata T., Tokunaga F., Muta T., Iwanaga S., Niwa M., Takao T., Shimonishi Y. Tachyplesin, a class of antimicrobial peptide from the hemocytes of the horseshoe crab (Tachypleus tridentatus). Isolation and chemical structure. J. Biol. Chem. 1988;263:16709–16713. [PubMed] [Google Scholar]
- 4.Ohta M., Ito H., Masuda K., Tanaka S., Arakawa Y., Wacharotayankun R., Kato N. Mechanisms of antibacterial action of tachyplesins and polyphemusins, a group of antimicrobial peptides isolated from horseshoe crab hemocytes. Antimicrob. Agents Chemother. 1992;36:1460–1465. doi: 10.1128/AAC.36.7.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buri M.V., Torquato H.F.V., Barros C.C., Ide J.S., Miranda A., Paredes-Gamero E.J. Comparison of Cytotoxic Activity in Leukemic Lineages Reveals Important Features of beta-Hairpin Antimicrobial Peptides. J. Cell. Biochem. 2017;118:1764–1773. doi: 10.1002/jcb.25844. [DOI] [PubMed] [Google Scholar]
- 6.Chen J., Xu X.M., Underhill C.B., Yang S., Wang L., Chen Y., Hong S., Creswell K., Zhang L. Tachyplesin activates the classic complement pathway to kill tumor cells. Cancer Res. 2005;65:4614–4622. doi: 10.1158/0008-5472.CAN-04-2253. [DOI] [PubMed] [Google Scholar]
- 7.Ding H., Jin G., Zhang L., Dai J., Dang J., Han Y. Effects of tachyplesin I on human U251 glioma stem cells. Mol. Med. Rep. 2015;11:2953–2958. doi: 10.3892/mmr.2014.3021. [DOI] [PubMed] [Google Scholar]
- 8.Kuzmin D.V., Emel’yanova A.A., Kalashnikova M.B., Panteleev P.V., Ovchinnikova T.V. In Vitro Study of Antitumor Effect of Antimicrobial Peptide Tachyplesin I in Combination with Cisplatin. Bull. Exp. Biol. Med. 2018;165:220–224. doi: 10.1007/s10517-018-4134-6. [DOI] [PubMed] [Google Scholar]
- 9.Li Q.F., Ou Yang G.L., Li C.Y., Hong S.G. Effects of tachyplesin on the morphology and ultrastructure of human gastric carcinoma cell line BGC-823. World J. Gastroenterol. 2000;6:676–680. doi: 10.3748/wjg.v6.i5.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X., Dai J., Tang Y., Li L., Jin G. Quantitative Proteomic Profiling of Tachyplesin I Targets in U251 Gliomaspheres. Mar. Drugs. 2017;15:20. doi: 10.3390/md15010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ouyang G.L., Li Q.F., Peng X.X., Liu Q.R., Hong S.G. Effects of tachyplesin on proliferation and differentiation of human hepatocellular carcinoma SMMC-7721 cells. World J. Gastroenterol. 2002;8:1053–1058. doi: 10.3748/wjg.v8.i6.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paredes-Gamero E.J., Martins M.N., Cappabianco F.A., Ide J.S., Miranda A. Characterization of dual effects induced by antimicrobial peptides: regulated cell death or membrane disruption. Biochim. Biophys. Acta. 2012;1820:1062–1072. doi: 10.1016/j.bbagen.2012.02.015. [DOI] [PubMed] [Google Scholar]
- 13.Rothan H.A., Ambikabothy J., Ramasamy T.S., Rashid N.N., Yusof R. A Preliminary Study in Search of Potential Peptide Candidates for a Combinational Therapy with Cancer Chemotherapy Drug. Int. J. Pept. Res. Ther. 2017;25:115–122. doi: 10.1007/s10989-017-9646-9. [DOI] [Google Scholar]
- 14.Shi S.L., Wang Y.Y., Liang Y., Li Q.F. Effects of tachyplesin and n-sodium butyrate on proliferation and gene expression of human gastric adenocarcinoma cell line BGC-823. World J. Gastroenterol. 2006;12:1694–1698. doi: 10.3748/wjg.v12.i11.1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H.T., Wu J., Zhang H.F., Zhu Q.F. Efflux of potassium ion is an important reason of HL-60 cells apoptosis induced by tachyplesin. Acta Pharmacol. Sin. 2006;27:1367–1374. doi: 10.1111/j.1745-7254.2006.00377.x. [DOI] [PubMed] [Google Scholar]
- 16.Muta T., Fujimoto T., Nakajima H., Iwanaga S. Tachyplesins Isolated from Hemocytes of Southeast Asian Horseshoe Crabs (Carcinoscorpius rotundicauda and Tachypleus gigas): Identification of a New Tachyplesin, Tachyplesin III, and a Processing Intermediate of Its Precursor1. J. Biochem. 1990;108:261–266. doi: 10.1093/oxfordjournals.jbchem.a123191. [DOI] [PubMed] [Google Scholar]
- 17.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 18.Teixeira V., Feio M.J., Bastos M. Role of lipids in the interaction of antimicrobial peptides with membranes. Prog. Lipid Res. 2012;51:149–177. doi: 10.1016/j.plipres.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 19.Yeaman M.R., Yount N.Y. Mechanisms of Antimicrobial Peptide Action and Resistance. Pharmacol. Rev. 2003;55:27–55. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- 20.Utsugi T., Schroit A.J., Connor J., Bucana C.D., Fidler I.J. Elevated expression of phosphatidylserine in the outer membrane leaflet of human tumor cells and recognition by activated human blood monocytes. Cancer Res. 1991;51:3062–3066. [PubMed] [Google Scholar]
- 21.Ran S., Downes A., Thorpe P.E. Increased Exposure of Anionic Phospholipids on the Surface of Tumor Blood Vessels. Cancer Res. 2002;62:6132. [PubMed] [Google Scholar]
- 22.Zwaal R.F., Comfurius P., Bevers E.M. Surface exposure of phosphatidylserine in pathological cells. Cell. Mol. Life Sci. 2005;62:971–988. doi: 10.1007/s00018-005-4527-3. [DOI] [PubMed] [Google Scholar]
- 23.Riedl S., Rinner B., Asslaber M., Schaider H., Walzer S., Novak A., Lohner K., Zweytick D. In search of a novel target - phosphatidylserine exposed by non-apoptotic tumor cells and metastases of malignancies with poor treatment efficacy. Biochim. Biophys. Acta. 2011;1808:2638–2645. doi: 10.1016/j.bbamem.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwasaki T., Ishibashi J., Tanaka H., Sato M., Asaoka A., Taylor D., Yamakawa M. Selective cancer cell cytotoxicity of enantiomeric 9-mer peptides derived from beetle defensins depends on negatively charged phosphatidylserine on the cell surface. Peptides. 2009;30:660–668. doi: 10.1016/j.peptides.2008.12.019. [DOI] [PubMed] [Google Scholar]
- 25.Leite Natália B., Aufderhorst-Roberts A., Palma Mario S., Connell Simon D., Neto João R., Beales Paul A. PE and PS Lipids Synergistically Enhance Membrane Poration by a Peptide with Anticancer Properties. Biophys. J. 2015;109:936–947. doi: 10.1016/j.bpj.2015.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuzaki K., Sugishita K., Fujii N., Miyajima K. Molecular Basis for Membrane Selectivity of an Antimicrobial Peptide, Magainin 2. Biochemistry. 1995;34:3423–3429. doi: 10.1021/bi00010a034. [DOI] [PubMed] [Google Scholar]
- 27.Hoskin D.W., Ramamoorthy A. Studies on anticancer activities of antimicrobial peptides. Biochim. Biophys. Acta. 2008;1778:357–375. doi: 10.1016/j.bbamem.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris F., Dennison S.R., Singh J., Phoenix D.A. On the selectivity and efficacy of defense peptides with respect to cancer cells. Med. Res. Rev. 2013;33:190–234. doi: 10.1002/med.20252. [DOI] [PubMed] [Google Scholar]
- 29.Gaspar D., Veiga A.S., Castanho M.A. From antimicrobial to anticancer peptides. A review. Front. Microbiol. 2013;4:294. doi: 10.3389/fmicb.2013.00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuzaki K. Why and how are peptide–lipid interactions utilized for self-defense? Magainins and tachyplesins as archetypes. Biochim. Biophys. Acta. 1999;1462:1–10. doi: 10.1016/S0005-2736(99)00197-2. [DOI] [PubMed] [Google Scholar]
- 31.Matsuzaki K., Fukui M., Fujii N., Miyajima K. Interactions of an antimicrobial peptide, tachyplesin I, with lipid membranes. Biochim. Biophys. Acta. 1991;1070:259–264. doi: 10.1016/0005-2736(91)90173-6. [DOI] [PubMed] [Google Scholar]
- 32.Iwanaga S., Muta T., Shigenaga T., Seki N., Kawano K., Katsu T., Kawabata S. Structure-function relationships of tachyplesins and their analogues. Ciba Found. Symp. 1994;186:160–174. doi: 10.1002/9780470514658.ch10. [DOI] [PubMed] [Google Scholar]
- 33.Imura Y., Nishida M., Ogawa Y., Takakura Y., Matsuzaki K. Action mechanism of tachyplesin I and effects of PEGylation. Biochim. Biophys. Acta. 2007;1768:1160–1169. doi: 10.1016/j.bbamem.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 34.Matsuzaki K., Yoneyama S., Fujii N., Miyajima K., Yamada K., Kirino Y., Anzai K. Membrane permeabilization mechanisms of a cyclic antimicrobial peptide, tachyplesin I, and its linear analog. Biochemistry. 1997;36:9799–9806. doi: 10.1021/bi970588v. [DOI] [PubMed] [Google Scholar]
- 35.Kuzmin D.V., Emelianova A.A., Kalashnikova M.B., Panteleev P.V., Balandin S.V., Serebrovskaya E.O., Belogurova-Ovchinnikova O.Y., Ovchinnikova T.V. Comparative in vitro study on cytotoxicity of recombinant beta-hairpin peptides. Chem. Biol. Drug Des. 2017;91:294–303. doi: 10.1111/cbdd.13081. [DOI] [PubMed] [Google Scholar]
- 36.Laederach A., Andreotti A.H., Fulton D.B. Solution and micelle-bound structures of tachyplesin I and its active aromatic linear derivatives. Biochemistry. 2002;41:12359–12368. doi: 10.1021/bi026185z. [DOI] [PubMed] [Google Scholar]
- 37.Kushibiki T., Kamiya M., Aizawa T., Kumaki Y., Kikukawa T., Mizuguchi M., Demura M., Kawabata S., Kawano K. Interaction between tachyplesin I, an antimicrobial peptide derived from horseshoe crab, and lipopolysaccharide. Biochim. Biophys. Acta. 2014;1844:527–534. doi: 10.1016/j.bbapap.2013.12.017. [DOI] [PubMed] [Google Scholar]
- 38.Kawano K., Yoneya T., Miyata T., Yoshikawa K., Tokunaga F., Terada Y., Iwanaga S. Antimicrobial peptide, tachyplesin I, isolated from hemocytes of the horseshoe crab (Tachypleus tridentatus). NMR determination of the beta-sheet structure. J. Biol. Chem. 1990;265:15365–15367. [PubMed] [Google Scholar]
- 39.Mielke S.P., Krishnan V.V. Characterization of protein secondary structure from NMR chemical shifts. Prog. Nucl. Magn. Reson. Spectrosc. 2009;54:141–165. doi: 10.1016/j.pnmrs.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hutchinson E.G., Thornton J.M. PROMOTIF--a program to identify and analyze structural motifs in proteins. Protein Sci. 1996;5:212–220. doi: 10.1002/pro.5560050204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wishart D.S., Bigam C.G., Yao J., Abildgaard F., Dyson H.J., Oldfield E., Markley J.L., Sykes B.D. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 42.Chan L.Y., Zhang V.M., Huang Y.H., Waters N.C., Bansal P.S., Craik D.J., Daly N.L. Cyclization of the antimicrobial peptide gomesin with native chemical ligation: influences on stability and bioactivity. Chembiochem. 2013;14:617–624. doi: 10.1002/cbic.201300034. [DOI] [PubMed] [Google Scholar]
- 43.Troeira Henriques S., Lawrence N., Chaousis S., Ravipati A.S., Cheneval O., Benfield A.H., Elliott A.G., Kavanagh A.M., Cooper M.A., Chan L.Y., et al. Redesigned Spider Peptide with Improved Antimicrobial and Anticancer Properties. ACS Chem. Biol. 2017;12:2324–2334. doi: 10.1021/acschembio.7b00459. [DOI] [PubMed] [Google Scholar]
- 44.Kondejewski L.H., Farmer S.W., Wishart D.S., Kay C.M., Hancock R.E., Hodges R.S. Modulation of structure and antibacterial and hemolytic activity by ring size in cyclic gramicidin S analogs. J. Biol. Chem. 1996;271:25261–25268. doi: 10.1074/jbc.271.41.25261. [DOI] [PubMed] [Google Scholar]
- 45.Tam J.P., Lu Y.A., Yang J.L. Marked increase in membranolytic selectivity of novel cyclic tachyplesins constrained with an antiparallel two-beta strand cystine knot framework. Biochem. Biophys. Res. Commun. 2000;267:783–790. doi: 10.1006/bbrc.1999.2035. [DOI] [PubMed] [Google Scholar]
- 46.Edwards I.A., Elliott A.G., Kavanagh A.M., Blaskovich M.A.T., Cooper M.A. Structure-Activity and -Toxicity Relationships of the Antimicrobial Peptide Tachyplesin-1. ACS Infect. Dis. 2017;3:917–926. doi: 10.1021/acsinfecdis.7b00123. [DOI] [PubMed] [Google Scholar]
- 47.Marggraf M.B., Panteleev P.V., Emelianova A.A., Sorokin M.I., Bolosov I.A., Buzdin A.A., Kuzmin D.V., Ovchinnikova T.V. Cytotoxic Potential of the Novel Horseshoe Crab Peptide Polyphemusin III. Mar. Drugs. 2018;16:466. doi: 10.3390/md16120466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hilchie A.L., Hoskin D.W., Power Coombs M.R. Anticancer Activities of Natural and Synthetic Peptides. In: Matsuzaki K., editor. Antimicrobial Peptides: Basics for Clinical Application. Springer Singapore; Singapore: 2019. pp. 131–147. [DOI] [PubMed] [Google Scholar]
- 49.Kuzmin D.V., Emelianova A.A., Kalashnikova M.B., Panteleev P.V., Ovchinnikova T.V. Effect of N- and C-Terminal Modifications on Cytotoxic Properties of Antimicrobial Peptide Tachyplesin I. Bull. Exp. Biol. Med. 2017;162:754–757. doi: 10.1007/s10517-017-3705-2. [DOI] [PubMed] [Google Scholar]
- 50.Rivera M., Bertasso A., McCaffrey C., Georgopapadakou N.H. Porins and lipopolysaccharide of Escherichia coli ATCC 25922 and isogenic rough mutants. FEMS Microbiol. Lett. 1993;108:183–187. doi: 10.1111/j.1574-6968.1993.tb06096.x. [DOI] [PubMed] [Google Scholar]
- 51.Treangen T.J., Maybank R.A., Enke S., Friss M.B., Diviak L.F., Karaolis D.K., Koren S., Ondov B., Phillippy A.M., Bergman N.H., et al. Complete Genome Sequence of the Quality Control Strain Staphylococcus aureus subsp. aureus ATCC 25923. Genome Announc. 2014;2:e01110-14. doi: 10.1128/genomeA.01110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clark D. Novel antibiotic hypersensitive mutants of Escherichia coli genetic mapping and chemical characterization. FEMS Microbiol. Lett. 1984;21:189–195. doi: 10.1111/j.1574-6968.1984.tb00209.x. [DOI] [Google Scholar]
- 53.Pinto S.N., Dias S.A., Cruz A.F., Mil-Homens D., Fernandes F., Valle J., Andreu D., Prieto M., Castanho M., Coutinho A., et al. The mechanism of action of pepR, a viral-derived peptide, against Staphylococcus aureus biofilms. J. Antimicrob. Chemother. 2019:dkz223. doi: 10.1093/jac/dkz223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watson H. Biological membranes. Essays Biochem. 2015;59:43–69. doi: 10.1042/bse0590043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ingolfsson H.I., Melo M.N., van Eerden F.J., Arnarez C., Lopez C.A., Wassenaar T.A., Periole X., de Vries A.H., Tieleman D.P., Marrink S.J. Lipid organization of the plasma membrane. J. Am. Chem. Soc. 2014;136:14554–14559. doi: 10.1021/ja507832e. [DOI] [PubMed] [Google Scholar]
- 56.Van Meer G., Voelker D.R., Feigenson G.W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eftink M.R. Fluorescence Quenching Reactions. In: Dewey T.G., editor. Biophysical and Biochemical Aspects of Fluorescence Spectroscopy. Springer US; Boston, MA, USA: 1991. pp. 1–41. [Google Scholar]
- 58.Ghisaidoobe A.B., Chung S.J. Intrinsic tryptophan fluorescence in the detection and analysis of proteins: a focus on Forster resonance energy transfer techniques. Int. J. Mol. Sci. 2014;15:22518–22538. doi: 10.3390/ijms151222518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Santos N.C., Prieto M., Castanho M.A.R.B. Quantifying molecular partition into model systems of biomembranes: an emphasis on optical spectroscopic methods. Biochim. Biophys. Acta. 2003;1612:123–135. doi: 10.1016/S0005-2736(03)00112-3. [DOI] [PubMed] [Google Scholar]
- 60.Fernandes M.X., García de la Torre J., Castanho M.A.R.B. Joint determination by Brownian dynamics and fluorescence quenching of the in-depth location profile of biomolecules in membranes. Anal. Biochem. 2002;307:1–12. doi: 10.1016/S0003-2697(02)00024-6. [DOI] [PubMed] [Google Scholar]
- 61.Lakowicz J.R. Topics in Fluorescence Spectroscopy Principles. Springer US; Boston, MA, USA: 2002. [Google Scholar]
- 62.Chattopadhyay A., London E. Parallax method for direct measurement of membrane penetration depth utilizing fluorescence quenching by spin-labeled phospholipids. Biochemistry. 1987;26:39–45. doi: 10.1021/bi00375a006. [DOI] [PubMed] [Google Scholar]
- 63.Kyrychenko A., Ladokhin A.S. Molecular Dynamics Simulations of Depth Distribution of Spin-Labeled Phospholipids within Lipid Bilayer. J. Phys. Chem. B. 2013;117:5875–5885. doi: 10.1021/jp4026706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Batoni G., Maisetta G., Esin S. Antimicrobial peptides and their interaction with biofilms of medically relevant bacteria. Biochim. Biophys. Acta. 2016;1858:1044–1060. doi: 10.1016/j.bbamem.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 65.Grassi L., Maisetta G., Esin S., Batoni G. Combination Strategies to Enhance the Efficacy of Antimicrobial Peptides against Bacterial Biofilms. Front. Microbiol. 2017;8:2409. doi: 10.3389/fmicb.2017.02409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Minardi D., Ghiselli R., Cirioni O., Giacometti A., Kamysz W., Orlando F., Silvestri C., Parri G., Kamysz E., Scalise G., et al. The antimicrobial peptide Tachyplesin III coated alone and in combination with intraperitoneal piperacillin-tazobactam prevents ureteral stent Pseudomonas infection in a rat subcutaneous pouch model. Peptides. 2007;28:2293–2298. doi: 10.1016/j.peptides.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 67.Yu K., Lo J.C., Yan M., Yang X., Brooks D.E., Hancock R.E., Lange D., Kizhakkedathu J.N. Anti-adhesive antimicrobial peptide coating prevents catheter associated infection in a mouse urinary infection model. Biomaterials. 2017;116:69–81. doi: 10.1016/j.biomaterials.2016.11.047. [DOI] [PubMed] [Google Scholar]
- 68.Oishi O., Yamashita S., Nishimoto E., Lee S., Sugihara G., Ohno M. Conformations and orientations of aromatic amino acid residues of tachyplesin I in phospholipid membranes. Biochemistry. 1997;36:4352–4359. doi: 10.1021/bi962171f. [DOI] [PubMed] [Google Scholar]
- 69.Doherty T., Waring A.J., Hong M. Dynamic structure of disulfide-removed linear analogs of tachyplesin-I in the lipid bilayer from solid-state NMR. Biochemistry. 2008;47:1105–1116. doi: 10.1021/bi701390t. [DOI] [PubMed] [Google Scholar]
- 70.Torcato I.M., Huang Y.H., Franquelim H.G., Gaspar D., Craik D.J., Castanho M.A., Troeira Henriques S. Design and characterization of novel antimicrobial peptides, R-BP100 and RW-BP100, with activity against Gram-negative and Gram-positive bacteria. Biochim. Biophys. Acta. 2013;1828:944–955. doi: 10.1016/j.bbamem.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 71.Cheneval O., Schroeder C.I., Durek T., Walsh P., Huang Y.H., Liras S., Price D.A., Craik D.J. Fmoc-based synthesis of disulfide-rich cyclic peptides. J. Org. Chem. 2014;79:5538–5544. doi: 10.1021/jo500699m. [DOI] [PubMed] [Google Scholar]
- 72.Güntert P. Automated NMR Structure Calculation with CYANA. In: Downing A.K., editor. Protein NMR Techniques. Humana Press; Totowa, NJ, USA: 2004. pp. 353–378. [DOI] [PubMed] [Google Scholar]
- 73.Shen Y., Bax A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR. 2013;56:227–241. doi: 10.1007/s10858-013-9741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brunger A.T. Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2007;2:2728. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 75.Chen V.B., Arendall W.B., 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang Y.H., Colgrave M.L., Clark R.J., Kotze A.C., Craik D.J. Lysine-scanning mutagenesis reveals an amendable face of the cyclotide kalata B1 for the optimization of nematocidal activity. J. Biol. Chem. 2010;285:10797–10805. doi: 10.1074/jbc.M109.089854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang Y., Liu Y., Zheng C., Shen C. Investigation of Cross-Contamination and Misidentification of 278 Widely Used Tumor Cell Lines. PLoS ONE. 2017;12:e0170384. doi: 10.1371/journal.pone.0170384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Riss T.L., Moravec R.A., Niles A.L., Duellman S., Benink H.A., Worzella T.J., Minor L. Cell Viability Assays. In: Sittampalam G.S., Coussens N.P., Brimacombe K., Grossman A., Arkin M., Auld D., Austin C., Baell J., Bejcek B., Chung T.D.Y., et al., editors. Assay Guidance Manual. Eli Lilly & Company and the National Center for Advancing Translational Sciences; Bethesda, MD, USA: 2016. [Google Scholar]
- 79.Mayer L.D., Hope M.J., Cullis P.R. Vesicles of variable sizes produced by a rapid extrusion procedure. BBA-Biomembranes. 1986;858:161–168. doi: 10.1016/0005-2736(86)90302-0. [DOI] [PubMed] [Google Scholar]
- 80.Henriques S.T., Pattenden L.K., Aguilar M.-I., Castanho M.A.R.B. The Toxicity of Prion Protein Fragment PrP(106−126) is Not Mediated by Membrane Permeabilization as Shown by a M112W Substitution. Biochemistry. 2009;48:4198–4208. doi: 10.1021/bi900009d. [DOI] [PubMed] [Google Scholar]
- 81.Henriques S.T., Castanho M.A.R.B. Environmental factors that enhance the action of the cell penetrating peptide pep-1: A spectroscopic study using lipidic vesicles. Biochim. Biophys. Acta. 2005;1669:75–86. doi: 10.1016/j.bbamem.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 82.Caputo G.A., London E. Using a novel dual fluorescence quenching assay for measurement of tryptophan depth within lipid bilayers to determine hydrophobic alpha-helix locations within membranes. Biochemistry. 2003;42:3265–3274. doi: 10.1021/bi026696l. [DOI] [PubMed] [Google Scholar]
- 83.Henriques S.T., Huang Y.H., Castanho M.A., Bagatolli L.A., Sonza S., Tachedjian G., Daly N.L., Craik D.J. Phosphatidylethanolamine binding is a conserved feature of cyclotide-membrane interactions. J. Biol. Chem. 2012;287:33629–33643. doi: 10.1074/jbc.M112.372011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Henriques S.T., Huang Y.-H., Rosengren K.J., Franquelim H.G., Carvalho F.A., Johnson A., Sonza S., Tachedjian G., Castanho M.A.R.B., Daly N.L., et al. Decoding the membrane activity of the cyclotide kalata B1: the importance of phosphatidylethanolamine phospholipids and lipid organization on hemolytic and anti- HIV activities. J. Biol. Chem. 2011;286:24231. doi: 10.1074/jbc.M111.253393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang Y.-H., Colgrave M.L., Daly N.L., Keleshian A., Martinac B., Craik D.J. Biological Activity of the Prototypic Cyclotide Kalata B1 Is Modulated by the Formation of Multimeric Pores. J. Biol. Chem. 2009;284:20699–20707. doi: 10.1074/jbc.M109.003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stewart J.C. Colorimetric determination of phospholipids with ammonium ferrothiocyanate. Anal. Biochem. 1980;104:10–14. doi: 10.1016/0003-2697(80)90269-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.