Abstract
Mammalian spermatozoa are infertile immediately after ejaculation and need to undergo a functional maturation process to acquire the competence to fertilize the female egg. During this process, called capacitation, the actin cytoskeleton dramatically changes its organization. First, actin fibers polymerize, forming a network over the anterior part of the sperm cells head, and then it rapidly depolymerizes and disappears during the exocytosis of the acrosome content (the acrosome reaction (AR)). Here, we developed a computational model representing the actin dynamics (AD) process on mature spermatozoa. In particular, we represented all the molecular events known to be involved in AD as a network of nodes linked by edges (the interactions). After the network enrichment, using an online resource (STRING), we carried out the statistical analysis on its topology, identifying the controllers of the system and validating them in an experiment of targeted versus random attack to the network. Interestingly, among them, we found that cyclin-dependent kinase (cyclin–CDK) complexes are acting as stronger controllers. This finding is of great interest since it suggests the key role that cyclin–CDK complexes could play in controlling AD during sperm capacitation, leading us to propose a new and interesting non-genomic role for these molecules.
Keywords: spermatozoa, cyclin–CDK complexes, actin dynamics, capacitation, acrosome reaction, biological network, computational model
1. Introduction
According to WHO, infertility is a “disease of the reproductive system defined by the failure to achieve a clinical pregnancy after 12 months or more of regular unprotected sexual intercourse” [1], and it affects around the 15−20% of couples. Among these, the male factor is responsible for approximately 50% of the cases (30% alone and 20% co-responsible with female factors [2]). There are many known causes associated with the appearance of male infertility, but in spite of the great effort carried out by researchers in recent years, 40−50% of the cases are due to still unknown causes (idiopathic male infertility [3]). This condition is characterized by a worsening of patient life conditions with medical and psychological negative consequences [4], an increase in the costs for both the couples and the National Health Services, and often by the necessity to undergo repeated cycles of medically assisted procreation (MAP). For these reasons, further research is still needed to decipher the exact molecular mechanisms related to the sperm function, in order to improve the current diagnosis tools and to develop specific treatments for men suffering from idiopathic infertility.
In this context, it is very important to consider that spermatozoa, achieve their fertilizing ability in the female oviduct, where they reside for a period varying from hours to days (depending on the species) bound to the epithelium, forming a sperm reservoir [5,6]. Here, they undergo a series of events collectively known as capacitation, a process involving different events, such as the cholesterol efflux from the sperm membrane [7,8], the increase of intracellular Ca2+ concentrations [9,10,11], the production of reactive oxygen species (ROS) [12], the activation of a bicarbonate/cAMP/PKA-dependent pathway and the subsequent proteins phosphorylation [13,14,15], and the cytoskeletal remodeling of actin fibers [16,17,18]. The latest is one of the keys events in driving the relocalization of membrane antigens involved in capacitation, in sperm-egg recognition and binding, and in the exocytosis of enzymes from the acrosome (the acrosome reaction (AR)), allowing the sperm cell to surpass the cumulus oocyte complex and the zona pellucida barriers to fertilize the egg.
Since the molecular mechanisms involved in control of actin dynamics (AD) during capacitation remains not completely understood, recently, our group carried out an in silico and in vitro study. Its results suggested that cyclin-dependent kinase (cyclin–CDK) complexes could be involved in control of this event, as demonstrated by the negative effects of a cyclin–CDK-specific inhibitor (Aminopurvalanol A) on actin polymerization with dramatic consequences on sperm capacitation and on in vitro fertilization outcome [19]. On this basis, here we developed a computational model devoted to describe the role of cyclin–CDK complexes in controlling the actin dynamic in mammalian mature spermatozoa. To this aim, in agreement with biological networks theory, we represented the molecules participating in the AD as nodes linked by directed edges (the interactions among them). Then, by the study of network topology, we inferred important information, useful to improve our knowledge on sperm physiology.
2. Results
2.1. Networks Creation, Analysis, and Visualization
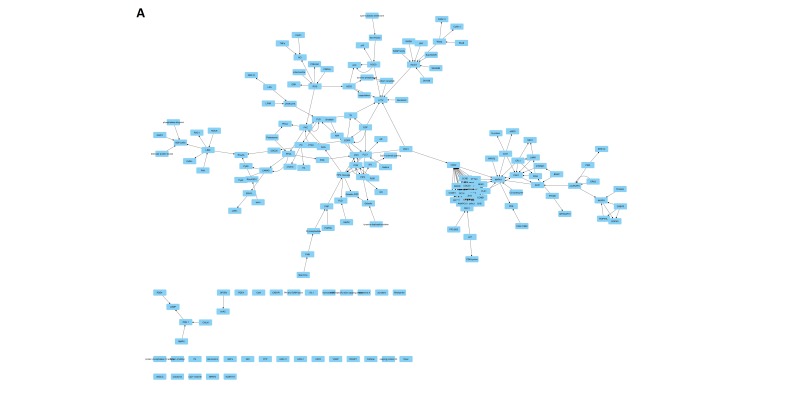
The experimental design, including all the steps for the creation and analysis of the network, is illustrated in Figure 1. As result of a manually-curated data analysis, we obtained a network (Data Base Network, DB Network) with 167 nodes and 274 links. After the STRING analysis of proteins involved in actin remodeling, we obtained a second network (STRING Network) with 49 nodes and 287 links. After merging the two networks, we obtained the final network (Merged Network, MN) constituted of 188 nodes and 558 links (Figure 2). The “in node degree” distribution and “out node degree” distribution follow an exponential law, characterized by a negative exponent (−1.183 and −1.314, respectively); the value of clustering coefficient is very low (0.127) and is uncorrelated with Node Degree (R2 = 0.003). The values of its main topological parameters are listed in Table 1 and in Supplementary Material S1.
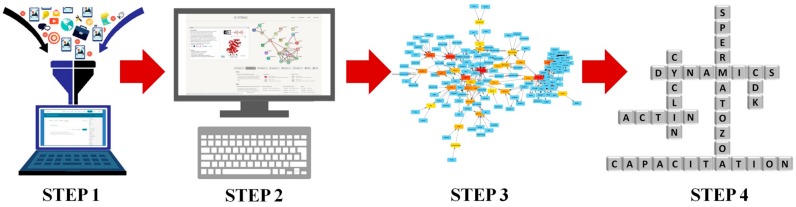
Figure 1.
Experimental design. The figure illustrate the four step followed during the development of the work. Step 1 represents the data collection from the literature; step 2 entails the data enrichment by using specific system biology tools; step 3 implies the creation, analysis, and visualization of the network; and step 4 represents the inferences about the mechanisms controlling actin dynamics (AD).
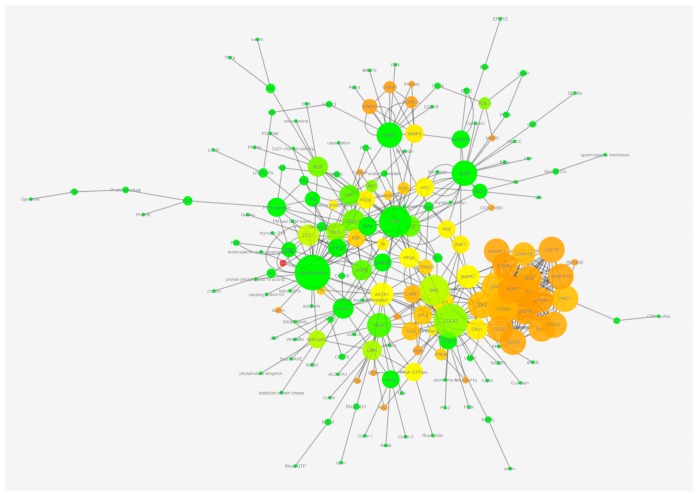
Figure 2.
Network of AD. The network represents the AD during the post-ejaculatory life of spermatozoa. The network was created with Cytoscape 3.1.2, and the node size is proportional to the number of links per node, with the color depending on the clustering coefficient (from green = 0 to red = 1).
Table 1.
Main Topological Parameters of ADCN_E network. List of the main topological parameters evaluated in the network with their values.
| Parameter | DB Network | STRING Network | Merged Network |
|---|---|---|---|
| N of nodes | 167 | 49 | 188 |
| N of edges | 274 | 287 | 558 |
| Clustering Coefficient | 0.037 | 0.341 | 0.127 |
| Diameter | 14 | 5 | 12 |
| Shortest Path Length | 8837 (31%) | 436 (19%) | 14832 (41%) |
| Characteristic Path Length | 5.760 | 1.505 | 5.246 |
| Averaged Number of Neighbors | 2.922 | 11.633 | 5.471 |
| In Degree | |||
| Exponent (γ) | −1.314 | −0.608 | −1.183 |
| Coefficient of Correlation (r) | 0.996 | 0.734 | 0.988 |
| Coefficient of Determination (R2) | 0.910 | 0.527 | 0.871 |
| Out Degree | |||
| Exponent (γ) | −1.708 | −0.675 | −1.314 |
| Coefficient of Correlation (r) | 0.989 | 0.935 | 0.991 |
| Coefficient of Determination (R2) | 0.885 | 0.735 | 0.879 |
| Node Degree vs. Cluster. Coeff. | |||
| Coefficient of Determination (R2) | 0.333 | 0.085 | 0.003 |
2.2. Identification of Controllers within the Network
To identify the Controller active trough the network, we identified the nodes that are both hubs (i.e., the nodes with higher connectivity) and bottlenecks (i.e., the nodes that are mainly involved in control of flow within the network).
The hubs were identified based on the following formula [20]:
| (1) |
where:
ND = node degree
μ = averaged node degree
σ = standard deviation of node degree
They represent about the 16% of nodes (31 on 188) and are listed in Table 2.
Table 2.
Network hubs. List of the highly connected nodes (hubs) with their corresponding number of edges.
| Node Name | Node Degree |
|---|---|
| AD | 36 |
| CDC42 | 34 |
| PKA | 30 |
| SRC | 28 |
| CDK1 | 24 |
| CCNB1 | 23 |
| CCNA2 | 22 |
| CDK2 | 22 |
| PLK1 | 22 |
| PRC1 | 21 |
| BUB1 | 20 |
| BUB1B | 20 |
| CCNB2 | 20 |
| MAD2L1 | 20 |
| CDC16 | 20 |
| CDC20 | 20 |
| CDC23 | 20 |
| PTTG1 | 20 |
| AKAP3 | 19 |
| ANAPC1 | 19 |
| CCNE1 | 19 |
| CKS1B | 19 |
| ANAPC10 | 19 |
| BUB3 | 19 |
| cAMP | 19 |
| NEK2 | 18 |
| [Ca2+]i | 15 |
| ACTA1 | 15 |
| CDKN1B | 15 |
| MAPK1 | 15 |
| RhoA | 13 |
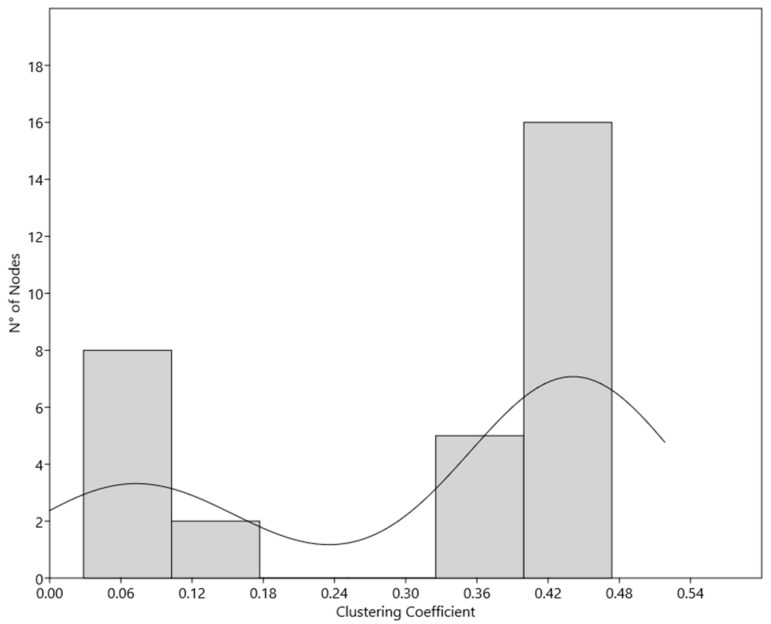
As expected, among the hubs, the node degree and the clustering coefficient did not correlate (r = −0.271, R2 = 0.114), while the Kernel density estiamtion (KDE) analysis (whose results are presented in Figure 3) shows the existence of two different subpopulations of hubs, characterized by different values of clustering coefficient (0−0.2 and 0.3−0.5, respectively), as shown in Table 3.
Figure 3.
KDE analysis. Histogram shows the subpopulations in hubs based on the Clustering Coefficient value.
Table 3.
The two different hubs subpopulations, based on node clustering coefficient.
| Node Name | Clustering Coefficient | |
|---|---|---|
| Subpopulation 1 | CCNE1 | 0,4737 |
| ANAPC1 | 0,4708 | |
| CKS1B | 0,4708 | |
| ANAPC10 | 0,4708 | |
| CCNB2 | 0,4684 | |
| CDC16 | 0,4684 | |
| CDC23 | 0,4684 | |
| BUB3 | 0,4591 | |
| BUB1 | 0,4579 | |
| BUB1B | 0,4579 | |
| MAD2L1 | 0,4579 | |
| CDC20 | 0,4579 | |
| PTTG1 | 0,4474 | |
| NEK2 | 0,4444 | |
| CCNA2 | 0,4048 | |
| PLK1 | 0,4048 | |
| CDKN1B | 0,3952 | |
| CCNB1 | 0,3893 | |
| PRC1 | 0,3789 | |
| CDK2 | 0,3788 | |
| CDK1 | 0,3775 | |
| Subpopulation 2 | MAPK1 | 0,1429 |
| ACTA1 | 0,1346 | |
| SRC | 0,1000 | |
| CDC42 | 0,0860 | |
| [Ca2+]i | 0,0758 | |
| RhoA | 0,0641 | |
| PKA | 0,0498 | |
| AKAP3 | 0,0381 | |
| AD | 0,0331 | |
| cAMP | 0,0286 |
In the following step, we listed the nodes based on their values of bottleneck score and selected the first thirty-one nodes (the same number than hubs), in keeping with a previous study [20]. The list of the selected nodes is presented in Table 4.
Table 4.
Nodes and bottleneck score. The table shows the first thirty-one nodes with the bottleneck score for each node.
| Name | Bottleneck Score |
|---|---|
| PKA | 81 |
| CDC42 | 59 |
| AD | 57 |
| SRC | 40 |
| p-Tyr | 22 |
| cAMP | 16 |
| RhoA | 14 |
| PIP3 | 12 |
| LIMK | 11 |
| Wasp | 11 |
| Arp2/3 | 11 |
| ROS | 8 |
| AKAP3 | 8 |
| PP2A | 8 |
| CDK1 | 8 |
| PIP2 cleavage | 7 |
| PLK1 | 7 |
| PLD | 6 |
| RAF1 | 6 |
| CAMKII | 5 |
| [Ca2+]i | 5 |
| PI4P | 5 |
| sAC | 5 |
| AKAP4 | 5 |
| PIP2 | 4 |
| proAKAP4 | 4 |
| MAPK1 | 4 |
| RhoA GTPase | 4 |
| HCO3- | 4 |
| CAV1 | 3 |
| LPAR-LPA | 3 |
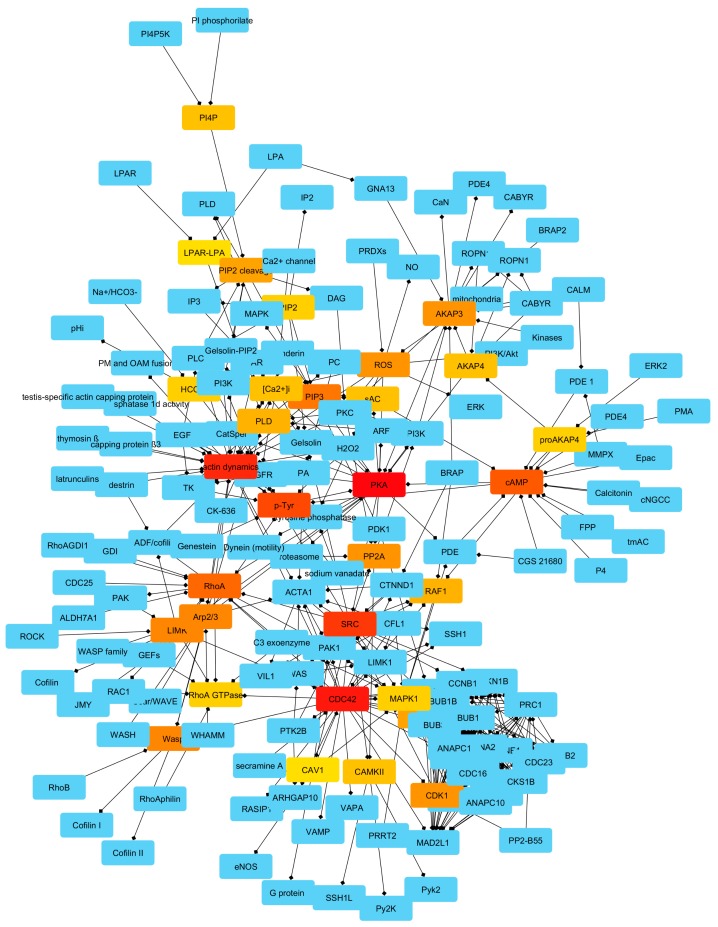
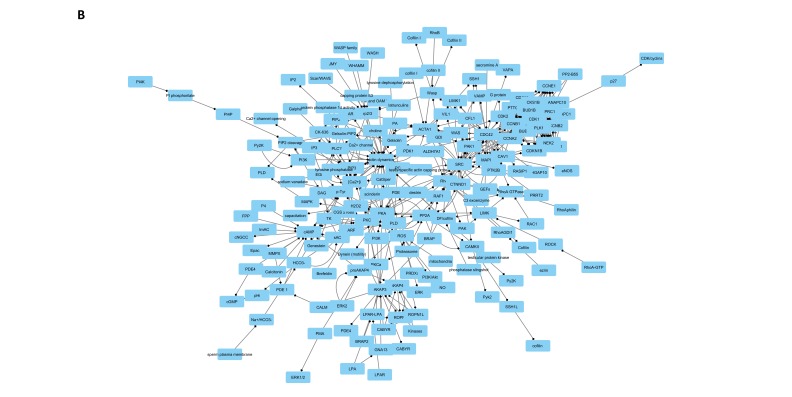
By using the CytoHubba plug-in, we computed the network of bottlenecks, which represents the back-bone of information flow control within MN (see Figure 4).
Figure 4.
Bottlenecks network active in control of information flow within MN. The node color is depending on their bottleneck score (form light yellow for lower values to dark red for higher ones).
Finally, as computed with an online open software (http://bioinformatics.psb.ugent.be/webtools/Venn, 06/12/2019), eleven nodes (see Figure 5) were either hubs and bottlenecks, listed in Table 5.
Figure 5.
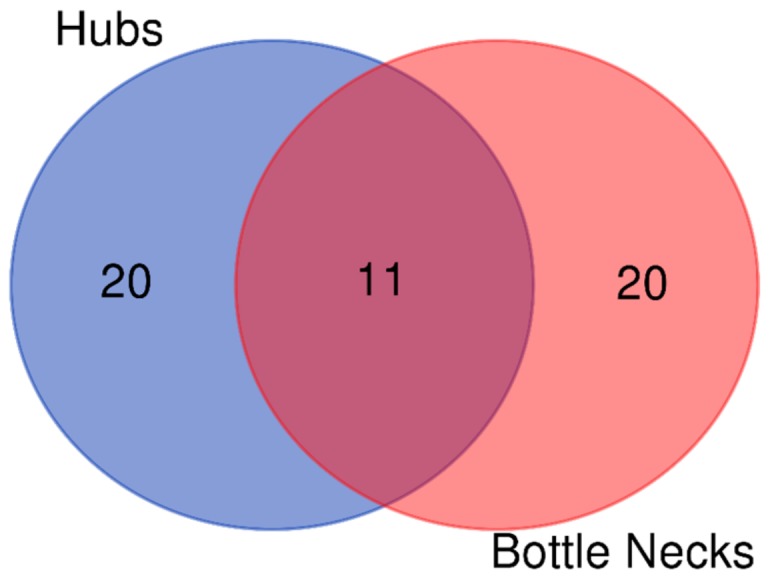
Venn diagram used to compute the nodes that are both hubs and Bottle Necks.
Table 5.
Role of MN controllers in mammalian sperm physiology, where known.
| Node Name | Node Degree | Bottleneck Score | Function (with Focus on Mammalian Spermatozoa Physiology) |
|---|---|---|---|
| AD | 36 | 82 | |
| PKA | 30 | 88 | Key effector of the bicarbonate-dependent cAMP/protein kinase A (PKA) pathway that leads to the control of p-Tyr of sperm proteins during capacitation. Its activation is correlated to a myriad of biochemical events. |
| CDC42 | 34 | 34 | Controller of cell cycle, controller of sperm AD. |
| SRC | 28 | 23 | A non-receptor tyrosine kinase protein that in humans is encoded by the SRC gene. This protein phosphorylates specific tyrosine residues in other tyrosine kinases. An elevated level of activity of c-Src tyrosine kinase is suggested to be linked to cancer progression by promoting other signals. Mutations in this gene could be involved in the malignant progression of colon cancer. |
| CCNA2 | 22 | 14 | Controller of cell cycle, controller of sperm AD. |
| cAMP | 19 | 13 | Second messenger of the bicarbonate-dependent cAMP/protein kinase A (PKA) pathway. |
| AKAP3 | 19 | 13 | It is expressed in spermatozoa and localized to the acrosomal region of the sperm head, as well as the length of the principal piece. It may function as a regulator of motility, capacitation, and the acrosome reaction (AR) |
| CDK1 | 24 | 6 | Controller of cell cycle, controller of sperm AD. |
| ACTA1 | 15 | 15 | Polymerizes and depolymerizes during capacitation. |
| BUB1 | 20 | 7 | It plays a key role in the establishment of the mitotic spindle checkpoint and chromosome congression. |
| [Ca2+]i | 15 | 12 | Second messenger involved in virtually all the biochemical event related to the capacitation. |
| RhoA | 13 | 12 | It interacts with proteins involved in capacitation and the AR, and RhoA signaling in sperm may be targeted by AKAPs. |
2.3. Targeted vs. Random Attack to MN
To test the reliability of our inference, we tested the effects of node removal from the networks with a computational experiment: we removed the controller (targeted removal) or different sets of 11 nodes randomly identified (function “=randbetween (min; max)” Excel 2003). In the first case, we obtained the network collapse, while in the second, we did not find evident effects in term of network stability (see Figure 6, for an exemplification).
Figure 6.
Diagram showing the effects of the removal from the network of controllers identified by our analysis (target attack), Panel A, compared with the effect of the removal of the same number of randomly selected nodes (random attack), Panel B. As it is evident, in the first case the network collapsed, in the second one no important changes in network topology are evident.
3. Discussion
It is well known that mammalian spermatozoa, once ejaculated, must undergo a complex process called capacitation to become fully fertile. During capacitation several biochemical changes affect the whole sperm physiology, leading to modifications in the chemical composition of the membrane and to a deep cytoskeleton reorganization [8,21,22]. This last event has been deeply studied by different groups and in different species, with the delivering of interesting molecular data that unfortunately still remain unorganized.
Here, we adopted a computational biology approach to gather the available data in order to reconstruct the active pathway responsible for the control of AD in spermatozoa. This strategy has been recently developed and is very promising in giving important data in several branches of biology, from cancer [23] to biochemical machinery [24] to germ cells physiology [20,25,26,27]. More in detail, we used the biological networks to carry out an operation of reverse engineering in four steps: first, we collected the data from the literature (Step 1); we enriched them by using a specific systems biology tool (STRING) (Step 2); we created, analyzed, and visualized the networks (Step 3); and, finally, we took the inference about AD control mechanisms (Step 4). By performing this kind of analysis, it is possible to compute some important topological parameters.
The first one to be discussed is the number of nodes (i.e., the number of molecules involved in AD pathway). Interestingly, these results are similar to prior ones found in several others pathways involved in control of important biological events, such as input (phototransduction) and output events (muscle contraction), information processing-(neurotransmitters release cycle) and energy metabolism-related pathways (ATP, glucose metabolism), endocrine control (insulin), cells cycle regulators (p53, pRb and c-Kit), and whole body coordination events (circadian clock) [28]. This leads us to speculate that this parameter could be influenced by the balance between the energetic costs of maintaining the integrity of each molecule and the need of stability within a system. Specifically, the metabolic investment to codify, synthesize, maintain, modulate, and, finally, catabolize each molecule could led to the evolution of transduction pathways characterized by the smallest possible number of molecules, in order to reduce the energy cost. At the same time, the need to ensure the highest stability for the system could favor the evolution of pathways composed by the largest number of molecules, since any fluctuation or failure of a single element will affect in a much more limited way the whole system the higher is the number of its components [28].
In addition, based on node degree distribution and its relationship with clustering coefficient, it was possible to classify the network as a scale free, in accordance with the Barabasi-Albert model. In this model, the node degree follows the general power law:
| (2) |
without being correlated with the clustering coefficient. This is a very intriguing finding because this topology confers to the network biologically relevant properties. First, as it has been extensively discussed, Barabasi-Albert networks are robust against random attacks and, at the same time, vulnerable to targeted attacks (see below for the experimental validation). It is possible to explain this, considering that most of the nodes are scarcely linked. Consequently, when their function will be affected by an external perturbation, the consequences would be negligible in terms of network stability. As it has been calculated (http://networksciencebook.com/chapter/1) in scale-free networks with γ < 3 the breakdown threshold converges to 1. Thus, it would be necessary to remove virtually all the nodes to break the network apart. Since the actin dynamic is a crucial event in sperm cells survival and acquisition of fertilizing ability, it is a useful pattern to guarantee that it will remain unaffected by eventual external and internal perturbations, therefore increasing the probability to fertilize.
On the other side, the presence of a low number of nodes (or molecules) able to exert a great control over the whole network allows it to save energy. Thus, the whole system can be modulated with high efficiency by controlling only a small number of molecules, reducing in this way the energetic cost and facilitating or accelerating the cell response. As already noted, incidentally, the identification of a small numbers of hubs could be helpful to control the biological systems with the purpose of developing diagnostic or therapeutic strategies for infertility or male contraception [28].
In addition, due to the small values of clustering coefficient it is possible to exclude the presence of hierarchical patterns, thus avoiding loops or clusters that could slow or interfere with the propagation of messages [28,29], with the exception of cyclin–CDK system (see below). Taken together, with the easy navigability of the network, demonstrated by the characteristic path length values of about 6.8, these features confer to the network a behavior both globally and locally efficient [30]. Indeed, the small number of passages through the network minimizes the loss of information due to the signal decrease and any local perturbation could influence the whole network in a very short time. Moreover, it increases the responsiveness of spermatozoa cytoskeleton organization to intracellular and extracellular stimuli.
Looking more in detail to the network clusterization, using the Cytocluster plug-in (IPCA algorithm for detection of dense subgraph in protein-protein networks), it has been possible to identify a dense structure involving the cyclin–CDK complexes. This is a noteworthy finding since it highlights a specific feature of the system. Indeed, likely as a safety strategy, the activity of these complexes is well modulated by the balancing of several entities interacting between each other, promoting the stability of the system and avoiding problems in it functioning.
Afterwards, by adopting a double step approach, we isolated the molecules exerting a greater control within the network. In particular, we identified the hubs, finding that it is possible to recognize two different hubs subpopulation, on the basis of their clusterization. This difference could to be in agreement with the hypothesis of Han and colleagues, which defined two different types of hubs ‘party’ hubs, which interact with most of their partners simultaneously, characterized by lower values of clustering coefficient, and ‘date’ hubs, which bind their different partners at different times or locations, more clustered [22]. In our network, the subpopulation 1 could account for date or local hubs and contains nodes related to the signal transduction and, mainly elements of cyclin–CDK system. The other one, could represent the party or general hubs list, and contains nodes, such as actin, AD and the universal messengers active in modulation of virtually all the pathways, such as Ca2+ of cAMP.
Then, we analyzed a second class of nodes that exert a strong control: that of bottlenecks, which are mainly involved in control of information flow within the network, and we computed their network. In this case, also, elements of the cyclin–CDK complexes seem to exert an important role.
Finally, we were able to identify 11 nodes that are both hubs and bottlenecks, as key controllers of AD network. It is very important to note two findings:
I: The process of actin cytoskeleton remodeling seems to be under the control of 11 molecules. It means that about 5% of nodes (11 on 188) are able to regulate such complex and important event.
II: On 11 controllers, 2 are directly related to actin (AD and ACTA1), and on the other nine, one third (3 on 9) are belonging to cyclin–CDK system (CCNA2, CDK1, CDC42).
To test the reliability of our hypothesis we performed an experiment by comparing the consequences in term of network integrity of the removal of the 11 putative controllers with that of several sets of 11 randomly selected nodes (see Figure 6). We found that the targeted removal of selected ones caused the collapse of the network, while the removal of 11 randomly selected nodes in the most of the cases did not have detectable consequences on network topology. In our opinion, these data are very interesting, because they clarify the signal cascade involved in control of actin dynamic and allow to confirm our data [19] re-interpret our data from Amaral and co-workers [31], that indicated the presence of several pathways previously neglected in the male gamete physiology. Concretely, they pointed out the presence of cell cycle-related proteins, in particular of cyclin–CDK) complexes and suggested that these proteins might be remnants of the spermatogenesis with no relevant function in mature spermatozoa. Indeed, although with controversy [32,33,34,35,36], it is well established that mature ejaculated spermatozoa do not have nuclear transcription activity, there is no protein synthesis from nuclear-encoded genes and they are not able to activate the cell cycle. Here, on the contrary, we could hypothesize that cyclin–CDK complexes are actively involved in control of actin function through capacitation. Until now it has been proposed that the bicarbonate present in great concentrations in the oviductal fluid (or in artificial systems), may induce the activation, via a sAC/cAMP/PKA-mediated pathway, of the lipid scrambling at the plasma membrane level. While gradually the plasma membrane acquires the ability to fuse with the outer acrosome membrane (OAM) thanks to the cholesterol extraction [7,37,38,39,40,41], the G-actin polymerizes forming a network of F-actin, which acts as a diaphragm between the two membranes avoiding their premature fusion. Once the capacitation is completely achieved and the physiological stimulus (zona-pellucida proteins) are met, the induced calcium peak causes the fast depolymerization of actin structure, and the AR can occur.
In this context, the model proposed could shed some light on the underling molecular mechanisms and could open new perspectives in the study of spermatozoa signaling. In particular, the data exposed suggest that in sperm cells cyclin–CDK complexes and their related molecules could be active, with a non-genomic effect, in controlling AD. On the other hand, we know that they remain active in several other cellular models as non-genomic effectors, as it has been well reviewed by some authors [42,43].
What still remains to elucidate is the possible role of actin cytoskeleton as an effector, rather than a passive target, in the signaling systems. In fact, it has been proposed that AD could have a role as a general coordinator of several important biological events. It is well-known that polymerized actin forms a continuous and dynamic connection between virtually all the cellular structures and that it presents an enormous surface area on which proteins and other molecules can dock [44]. This datum is strengthened by the finding that the plasma membrane surface area of a 20-μm-diameter generic cell is on the order of 700 μm2. By contrast, the total surface area of a typical concentration of 10 mg/mL F-actin is 47,000 μm2 [44] and that the diffusion along the cytoskeleton could be a reliable alternative for intracellular trafficking and signaling [45]. Our group has recently proposed that, together with other well-known molecules involved in intracellular signaling, such as Ca2+ or ATP, AD might provide a signal transduction route and acts as a macromolecular scaffold, probably contributing in this way to the spatial organization of those signaling pathways components participating in the spermatozoa functional maturation [29].
In conclusion, the model here stated clearly shows how cyclin–CDK complexes are active in sperm signaling and in control of AD, thus opening new perspectives in the potential design of diagnostic and therapeutic strategies that could help in the approaching of male infertility.
4. Materials and Methods
4.1. Data Collection, Network Creation, and General Analysis
As the data source, we used a validated and curated database [46], which describes the molecular events occurring during the capacitation-dependent actin remodeling process [46] updated with the recent literature from an accurate search in PubMed and Google Scholar.
The freely available and diffusible molecules, such as H2O, CO2, Pi, H+, and O2 were omitted, since they were considered not necessary and in some cases the record did not represent a single molecule but a complex event, due to the fact that most of the single molecular determinant of the phenomenon are still unknown. The quality control was realized as previously described [27]. Briefly, as a reference, two expert researchers on spermatozoa biology carried out an independent literature analysis on papers that referred to the sperm AD during capacitation, using the same key words, in curated scientific literature databases (PubMed, Scopus, Web of Science). Then, the databases were compared, and a third researcher verified the correctness of the record inserted and resolved eventual conflicts.
The database was realized in Microsoft Excel 2013 (Microsoft Corporation, Albuquerque, NM, USA) and contains the following fields:
Molecule involved in biochemical reaction (source): molecules participating in the interaction as source;
Interaction: kind of interaction between the molecules or the structures;
Molecule or anatomical structure involved in biochemical reaction (target): molecules participating to the interaction as target;
Alias: eventual aliases;
Role: physiological and/or pathological role of the molecule/reaction related to fertility;
Reference: article reporting the above mentioned data;
Notes: any further information that could be useful in the study.
Once the databases were created, we obtained the Actin Dynamic Control Network (ADCN).
Once the database was completed and the networks were realized, the main topological parameters (exposed in Table 6) were assessed by using the Network Analyzer plug-in for Cytoscape (open source bioinformatics software platform for visualizing molecular interaction networks and integrating with gene expression profiles and other state data).
Table 6.
Main topological parameters assessed. The twelve main topological parameters that have been examined are defined.
| Parameter | Definition |
|---|---|
| Connected Components | Number of networks in which any two vertices are connected to each other by links, and which is connected to no additional vertices in the network. |
| Number of nodes | Total number of molecules involved. |
| Number of edges | Total number of interactions found. |
| Clustering coefficient | Calculated as CI = 2nI/kI(kI–1), where nI is the number of links connecting the kI neighbors of node I to each other. It is a measure of how the nodes tend to form clusters. |
| Network diameter | The longest of all the calculated shortest paths in a network. |
| Shortest paths | The length of the shortest path between two nodes n and m is L (n, m). The shortest path length distribution gives the number of node pairs (n, m) with L(n,m) = k for k = 1,2,… |
| Characteristic path length | Expected distance between two connected nodes. |
| Averaged number of neighbors | Mean number of connections of each node. |
| Node degree | It is the number of interaction of each node. |
| Node degree distribution | It represents the probability that a selected node has k links. |
| γ | Exponent of node degree equation. |
| R2 | Coefficient of determination of node degree vs. number of nodes, on logarithmized data. |
4.2. Identification of Predicted Interactions with STRING
To identify and predict new molecular interactions of molecules included in we used STRING (Search Tool for the Retrieval of Interacting Genes/Proteins, https://string-db.org/ 06/12/2019) [47]. It is a database that includes known and predicted protein interactions. They could be either direct (physical) or indirect (functional) associations, and are derived from different sources: genomic context, high-throughput experiments, conserved co-expression, and previous knowledge. From the data obtained using STRING a new network was obtained, the Enriched Actin Dynamic Control Network (ADCN_E) by filtering the data for the Homo sapiens species and adopting a medium confidence score of 0.400.
4.3. Network Exhaustive Analysis and Visualization
All the networks were realized and analyzed using Cytoscape 3.1.2 [48]. The analysis was carried out considering them as directed and assessing the topological parameters listed and described in Table 4.
The identification of bottlenecks within the ADCN was carried out by using the Cytoscape plug-in CytoHubba. It implements the following algorithm for bottleneck calculation: let Ts be a shortest path tree rooted at node s. BN(v) = Σs∈V ps(v) where ps(v) = 1 if more than |V(Ts)|/ 4 paths from node s to other nodes in Ts meet at the vertex v; otherwise ps(v) = 0 [49]. This parameter is a measure of the node centrality expressed as the number of shortest paths in which the node is involved, and is related to the importance of nodes in controlling the information flux within directed networks [50].
We identify the bottleneck of network, and on their basis, we computed the relative network.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/17/4236/s1.
Author Contributions
Conceptualization, N.B.; Methodology, N.B., M.R.S., J.M.S., A.O., A.T., G.C.; Validation, B.B.; Formal Analysis, N.B.; Data Curation, M.R.S., J.M.S., A.O., A.T.; Writing – Original Draft Preparation, N.B., M.R.S., L.V.; Writing – Review & Editing, N.B., M.R.S., L.V.; Visualization, L.V.; Funding Acquisition, N.B., M.R.S., J.M.S., B.B.
Funding
This research was funded by EU MSCA-ITN Horizon 2020, Project REP-BIOTECH 675526 (MSR) and Rep-Eat-H2020-MSCA-COFUND 713714 (JSM).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Barratt C.L.R., Björndahl L., De Jonge C.J., Lamb D.J., Osorio Martini F., McLachlan R., Oates R.D., van der Poel S., St John B., Sigman M., et al. The diagnosis of male infertility: An analysis of the evidence to support the development of global WHO guidance—Challenges and future research opportunities. Hum. Reprod. Update. 2017;23:660–680. doi: 10.1093/humupd/dmx021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Winters B.R., Walsh T.J. The epidemiology of male infertility. Urol. Clin. North Am. 2014;41:195–204. doi: 10.1016/j.ucl.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Ring J.D., Lwin A.A., Köhler T.S. Current medical management of endocrine-related male infertility. Asian J. Androl. 2016;18:357–363. doi: 10.4103/1008-682X.179252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inhorn M.C., Patrizio P. Infertility around the globe: New thinking on gender, reproductive technologies and global movements in the 21st century. Hum. Reprod. Update. 2015;21:411–426. doi: 10.1093/humupd/dmv016. [DOI] [PubMed] [Google Scholar]
- 5.Suarez S.S. Mammalian sperm interactions with the female reproductive tract. Cell Tissue Res. 2016;363:185–194. doi: 10.1007/s00441-015-2244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sostaric E., Dieleman S.J., van de Lest C.H.A., Colenbrander B., Vos P.L.A.M., Garcia-Gil N., Gadella B.M. Sperm binding properties and secretory activity of the bovine oviduct immediately before and after ovulation. Mol. Reprod. Dev. 2008;75:60–74. doi: 10.1002/mrd.20766. [DOI] [PubMed] [Google Scholar]
- 7.Leahy T., Gadella B.M. New insights into the regulation of cholesterol efflux from the sperm membrane. Asian J. Androl. 2000;17:561–567. doi: 10.4103/1008-682X.153309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ickowicz D., Finkelstein M., Breitbart H. Mechanism of sperm capacitation and the acrosome reaction: Role of protein kinases. Asian J. Androl. 2012;14:816–821. doi: 10.1038/aja.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ren D., Xia J., Aoki F., Sakai S., Kohmoto K., Arnoult C., Cardullo R., Lemos J., Florman H., Arnoult C., et al. Calcium signaling through CatSper channels in mammalian fertilization. Physiol. (Bethesda) 2010;25:165–175. doi: 10.1152/physiol.00049.2009. [DOI] [PubMed] [Google Scholar]
- 10.Navarrete F.A.1., García-Vázquez F.A., Alvau A., Escoffier J., Krapf D., Sánchez-Cárdenas C., Salicioni A.M., Darszon A.V.P. Biphasic role of calcium in mouse sperm capacitation signaling pathways. J. Cell. Physiol. 2015;230:1758–1769. doi: 10.1002/jcp.24873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunter R.H.F., Petersen H.H., Greve T. Ovarian follicular fluid, progesterone and Ca2+ ion influences on sperm release from the Fallopian tube reservoir. Mol. Reprod. Dev. 1999;54:283–291. doi: 10.1002/(SICI)1098-2795(199911)54:3<283::AID-MRD9>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 12.Aitken R.J., Baker M.A., Nixon B. Are sperm capacitation and apoptosis the opposite ends of a continuum driven by oxidative stress? Asian J. Androl. 2015;17:633–639. doi: 10.4103/1008-682X.153850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romarowski A., Battistone M.A., La Spina F.A., Puga Molina L., Del C., Luque G.M., Vitale A.M., Cuasnicu P.S., Visconti P.E., Krapf D., et al. PKA-dependent phosphorylation of LIMK1 and Cofilin is essential for mouse sperm acrosomal exocytosis. Dev. Biol. 2015;405:237–249. doi: 10.1016/j.ydbio.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Signorelli J., Diaz E.S., Morales P. Kinases, phosphatases and proteases during sperm capacitation. Cell Tissue Res. 2012;349:765–782. doi: 10.1007/s00441-012-1370-3. [DOI] [PubMed] [Google Scholar]
- 15.Cohen G., Rubinstein S., Gur Y., Breitbart H. Crosstalk between protein kinase A and C regulates phospholipase D and F-actin formation during sperm capacitation. Dev. Biol. 2004;267:230–241. doi: 10.1016/j.ydbio.2003.10.034. [DOI] [PubMed] [Google Scholar]
- 16.Correa L.M., Thomas A., Meyers S.A. The macaque sperm actin cytoskeleton reorganizes in response to osmotic stress and contributes to morphological defects and decreased motility. Biol. Reprod. 2007;77:942–953. doi: 10.1095/biolreprod.107.060533. [DOI] [PubMed] [Google Scholar]
- 17.Brener E., Rubinstein S., Cohen G., Shternall K., Rivlin J., Breitbart H. Remodeling of the Actin Cytoskeleton During Mammalian Sperm Capacitation and Acrosome Reaction. Biol. Reprod. 2002;68:837–845. doi: 10.1095/biolreprod.102.009233. [DOI] [PubMed] [Google Scholar]
- 18.Delgado-Buenrostro N.L., Mújica A., Chiquete-Felix N., Déciga-Alcaraz A., Medina-Reyes E.I., Uribe-Carvajal S., Chirino Y.I. Role of Wasp and the small GTPases RhoA, RhoB, and Cdc42 during capacitation and acrosome reaction in spermatozoa of English guinea pigs. Mol. Reprod. Dev. 2016;83:927–937. doi: 10.1002/mrd.22657. [DOI] [PubMed] [Google Scholar]
- 19.Bernabò N., Valbonetti L., Greco L., Capacchietti G., Ramal Sanchez M., Palestini P., Botto L., Mattioli M., Barboni B. Aminopurvalanol A, a Potent, Selective, and Cell Permeable Inhibitor of Cyclins/Cdk Complexes, Causes the Reduction of in Vitro Fertilizing Ability of Boar Spermatozoa, by Negatively Affecting the Capacitation-Dependent Actin Polymerization. Front. Physiol. 2017;8:1097. doi: 10.3389/fphys.2017.01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ordinelli A., Bernabò N., Orsini M., Mattioli M., Barboni B. Putative human sperm Interactome: A networks study. BMC Syst. Biol. 2018;12:52. doi: 10.1186/s12918-018-0578-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breitbart H., Rotman T., Rubinstein S., Etkovitz N. Role and regulation of PI3K in sperm capacitation and the acrosome reaction. Mol. Cell. Endocrinol. 2010;314:234–238. doi: 10.1016/j.mce.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 22.Breitbart H., Finkelstein M. Biochemical and Biophysical Research Communications Actin cytoskeleton and sperm function. Biochem. Biophys. Res. Commun. 2018;506:372–377. doi: 10.1016/j.bbrc.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 23.Nishi H., Tyagi M., Teng S., Shoemaker B.A., Hashimoto K., Alexov E., Wuchty S., Panchenko A.R. Cancer Missense Mutations Alter Binding Properties of Proteins and Their Interaction Networks. PLoS ONE. 2013;8:e66273. doi: 10.1371/journal.pone.0066273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharan R., Ideker T. Modeling cellular machinery through biological network comparison. Nat. Biotechnol. 2006;24:427–433. doi: 10.1038/nbt1196. [DOI] [PubMed] [Google Scholar]
- 25.Saponaro I., Bernabò N., Todisco G., Lucidi P., Berardinelli P. Role of Actin in Spermatozoa Function Through Biological Network Theory. Volume 9783642232. Springer; Berlin/Heidelberg, Germany: 2012. [Google Scholar]
- 26.Bernabò N., Greco L., Ordinelli A., Mattioli M., Barboni B. Capacitation-Related Lipid Remodeling of Mammalian Spermatozoa Membrane Determines the Final Fate of Male Gametes: A Computational Biology Study. OMICS. 2015;19:712–721. doi: 10.1089/omi.2015.0114. [DOI] [PubMed] [Google Scholar]
- 27.Bernabò N., Agostino R.D., Ordinelli A., Mattioli M. Systems Biology in Reproductive Medicine The maturation of murine spermatozoa membranes within the epididymis, a computational biology perspective. Syst. Biol. Reprod. Med. 2016;62:299–308. doi: 10.1080/19396368.2016.1205679. [DOI] [PubMed] [Google Scholar]
- 28.Bernabò N., Mattioli M., Barboni B. Signal transduction in the activation of spermatozoa compared to other signalling pathways: A biological networks study. Int. J. Data Min. Bioinform. 2015;12:59. doi: 10.1504/IJDMB.2015.068953. [DOI] [PubMed] [Google Scholar]
- 29.Bernabò N., Berardinelli P., Mauro A., Russo V., Lucidi P., Mattioli M., Barboni B. The role of actin in capacitation-related signaling: An in silico and in vitro study. BMC Syst. Biol. 2011;5:47. doi: 10.1186/1752-0509-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Latora V., Marchiori M. Efficient Behavior of Small-World Networks. Phys. Rev. Lett. 2001;87:198701. doi: 10.1103/PhysRevLett.87.198701. [DOI] [PubMed] [Google Scholar]
- 31.Amaral A., Castillo J., Ramalho-Santos J., Oliva R. The combined human sperm proteome: Cellular pathways and implications for basic and clinical science. Hum. Reprod. Update. 2014;20:40–62. doi: 10.1093/humupd/dmt046. [DOI] [PubMed] [Google Scholar]
- 32.Miller D., Brinkworth M., Iles D. Paternal DNA packaging in spermatozoa: More than the sum of its parts? DNA, histones, protamines and epigenetics. Reproduction. 2010;139:287–301. doi: 10.1530/REP-09-0281. [DOI] [PubMed] [Google Scholar]
- 33.Amaral A., Ramalho-Santos J. The Male Gamete Is Not a Somatic Cell—The Possible Meaning of Varying Sperm RNA Levels. Antioxid. Redox Signal. 2013;18:179–185. doi: 10.1089/ars.2012.4715. [DOI] [PubMed] [Google Scholar]
- 34.Gur Y., Breitbart H. Protein synthesis in sperm: Dialog between mitochondria and cytoplasm. Mol. Cell. Endocrinol. 2008;282:45–55. doi: 10.1016/j.mce.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 35.Gur Y., Breitbart H. Mammalian sperm translate nuclear-encoded proteins by mitochondrial-type ribosomes. Genes Dev. 2006;20:411–416. doi: 10.1101/gad.367606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baker M.A. The ’omics revolution and our understanding of sperm cell biology. Asian J. Androl. 2011;13:6–10. doi: 10.1038/aja.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gadella B.M., Harrison R.A.P. The capacitating agent bicarbonate induces protein kinase A-dependent changes in phospholipid transbilayer behavior in the sperm plasma membrane. Development. 2000;2420:2407–2420. doi: 10.1242/dev.127.11.2407. [DOI] [PubMed] [Google Scholar]
- 38.Botto L., Bernabò N., Palestini P., Barboni B. Bicarbonate induces membrane reorganization and CBR1 and TRPV1 endocannabinoid receptor migration in lipid microdomains in capacitating boar spermatozoa. J. Membr. Biol. 2010;238:33–41. doi: 10.1007/s00232-010-9316-8. [DOI] [PubMed] [Google Scholar]
- 39.Barboni B., Bernabò N., Palestini P., Botto L., Pistilli M.G., Charini M., Tettamanti E., Battista N., Maccarrone M., Mattioli M. Type-1 Cannabinoid receptors reduce membrane fluidity of capacitated boar sperm by impairing their activation by bicarbonate. PLoS ONE. 2011;6:e23038. doi: 10.1371/journal.pone.0023038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brouwers J.F., Boerke A., Silva P.F.N., Garcia-Gil N., van Gestel R.A., Helms J.B., van de Lest C.H.A., Gadella B.M. Mass spectrometric detection of cholesterol oxidation in bovine sperm. Biol. Reprod. 2011;85:128–136. doi: 10.1095/biolreprod.111.091207. [DOI] [PubMed] [Google Scholar]
- 41.Gadella B.M., Luna C. Cell biology and functional dynamics of the mammalian sperm surface. Theriogenology. 2014;81:74–84. doi: 10.1016/j.theriogenology.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 42.Lim S., Kaldis P. Cdks, cyclins and CKIs: Roles beyond cell cycle regulation. Development. 2013;140:3079–3093. doi: 10.1242/dev.091744. [DOI] [PubMed] [Google Scholar]
- 43.Hydbring P., Malumbres M., Sicinski P. Non-canonical functions of cell cycle cyclins and cyclin-dependent kinases. Nat. Rev. Mol. Cell Biol. 2016;17:280–292. doi: 10.1038/nrm.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Janmey P.A. The Cytoskeleton and Cell Signaling: Component Localization and Mechanical Coupling. Physiol. Rev. 1998;78:763–781. doi: 10.1152/physrev.1998.78.3.763. [DOI] [PubMed] [Google Scholar]
- 45.Shafrir Y., ben-Avraham D., Forgacs G. Trafficking and signaling through the cytoskeleton: A specific mechanism. J. Cell Sci. 2000;113:2747–2757. doi: 10.1242/jcs.113.15.2747. [DOI] [PubMed] [Google Scholar]
- 46.Bernabò N., Ordinelli A., Ramal Sanchez M., Mattioli M., Barboni B. Networks Models of Actin Dynamics during Spermatozoa Postejaculatory Life: A Comparison among Human-Made and Text Mining-Based Models. Biomed Res. Int. 2016;2016 doi: 10.1155/2016/9795409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P., et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chin C.H., Chen S.H., Wu H.H., Ho C.W., Ko M.T., Lin C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014;8:S11. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pržulj N., Wigle D.A., Jurisica I. Functional topology in a network of protein interactions. 2004, 20, 340–348. Bioinformatics. 2004;20:340–348. doi: 10.1093/bioinformatics/btg415. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.