Abstract
Karyotypic data from Australian native freshwater fishes are scarce, having been described from relatively few species. Golden perch (Macquaria ambigua) and Murray cod (Maccullochella peelii) are two large-bodied freshwater fish species native to Australia with significant indigenous, cultural, recreational and commercial value. The arid landscape over much of these fishes’ range, coupled with the boom and bust hydrology of their habitat, means that these species have potential to provide useful evolutionary insights, such as karyotypes and sex chromosome evolution in vertebrates. Here we applied standard and molecular cytogenetic techniques to characterise karyotypes for golden perch and Murray cod. Both species have a diploid chromosome number 2n = 48 and a male heterogametic sex chromosome system (XX/XY). While the karyotype of golden perch is composed exclusively of acrocentric chromosomes, the karyotype of Murray cod consists of two submetacentric and 46 subtelocentric/acrocentric chromosomes. We have identified variable accumulation of repetitive sequences (AAT)10 and (CGG)10 along with diverse methylation patterns, especially on the sex chromosomes in both species. Our study provides a baseline for future cytogenetic analyses of other Australian freshwater fishes, especially species from the family Percichthyidae, to better understand their genome and sex chromosome evolution.
Keywords: sex determination, florescence in situ hybridisation (FISH), comparative genomic hybridisation (CGH), karyogram, DNA methylation
1. Introduction
Morphological characteristics of chromosomes, like the size and number (including ploidy) may vary within taxonomic groups such as families, genera and even species [1]. Moreover, functional differences (e.g., accumulation of repetitive sequences, location of a certain gene or DNA methylation) often occur among species as well as among populations or among individuals of the same species [2,3,4]. Consequently, cytogenetic studies provide a useful tool for establishing phylogenetic and evolutionary relationships among species. Karyotype analysis can also facilitate the discovery of heteromorphic or heterochromatic sex chromosomes.
Identification of sex chromosomes is important in dioecious species as it provides information on the evolution of sex determination and insights into the effect of environment in driving sex ratios of populations through upregulation and downregulation of genes in gonadal development [5,6]. The importance of understanding the sex chromosome systems in fish is heightened by the existence of diverse genetic and epigenetic features such as genetic sex determination (GSD), environmental sex determination (ESD), mixed GSD–ESD and rapid sex reversal within this large group of animals [7].
Fish comprise more than fifty percent of extant vertebrate species [8], with around 32,500 species, including more than 15,000 freshwater species [9]. Freshwaters are thus hyperdiverse, considering this habitat constitutes less than 0.3% [10] of available global water. Freshwater fish are thus an important component of global biodiversity and evolutionary studies.
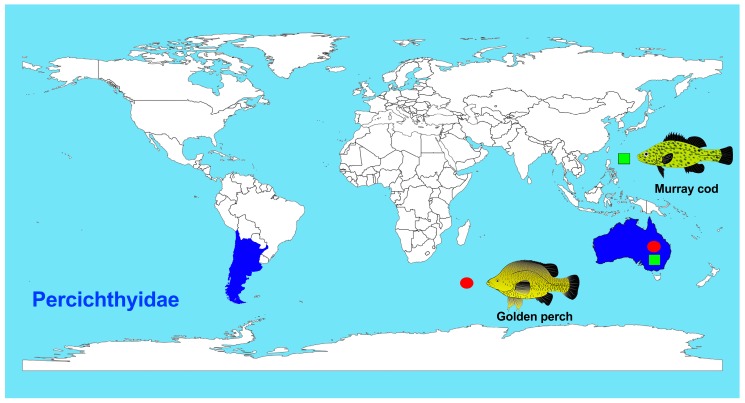
The family Percichthyidae (Order: Centrarchiformes) or temperate perches, consists of 22 freshwater and estuarine species under nine genera, distributed in Australia and South America (Figure 1) [11]. The species investigated here are golden perch (Macquaria ambigua) and Murray cod (Maccullochella peelii), two large-bodied freshwater fish species native to the Murray Darling Basin (MDB) in south-eastern Australia [12,13]. Golden perch also occurs in central Australia in the Lake Eyre Basin and in the central, coastal Queensland Fitzroy Basin, although these populations are considered to be separate cryptic species [14]. Both golden perch and Murray cod have significant indigenous, cultural, recreational and commercial value. Moreover, both species have been bred in hatcheries and extensively stocked in catchments across the MDB for more than 30 years.
Figure 1.
Distribution of nine genera of Percichthyidae covering Australia, Chile and Argentina.
Sex chromosomes in fish have been studied since the early twentieth century, when the presence of heterogametic sex chromosome systems (either XY or WZ) was described [15,16,17]. Studies in fishes have identified diverse sex chromosomal systems (including XX/XY, XX/X0, ZZ/ZW, ZZ/Z0, XX/XY1Y2, X1X1X2X2/X1X2Y, ZZ/ZW1W2, Z1Z1Z2Z2/Z1Z2W), with the XX/XY (male heterogametic) and ZZ/ZW (female heterogametic) systems being predominant [18,19]. The ancestral fish sex chromosome (sex-determining locus) has gone through a complete shift of chromosome pairs because of frequent chromosome rearrangements such as fusion and translocation or transposition [20,21]. Such rearrangement events across the genome, including the sex chromosomes, make teleost fish potentially informative for studies of karyotype and sex chromosome evolution. To date, there have been no published studies on the mode of sex determination in Australian freshwater fish.
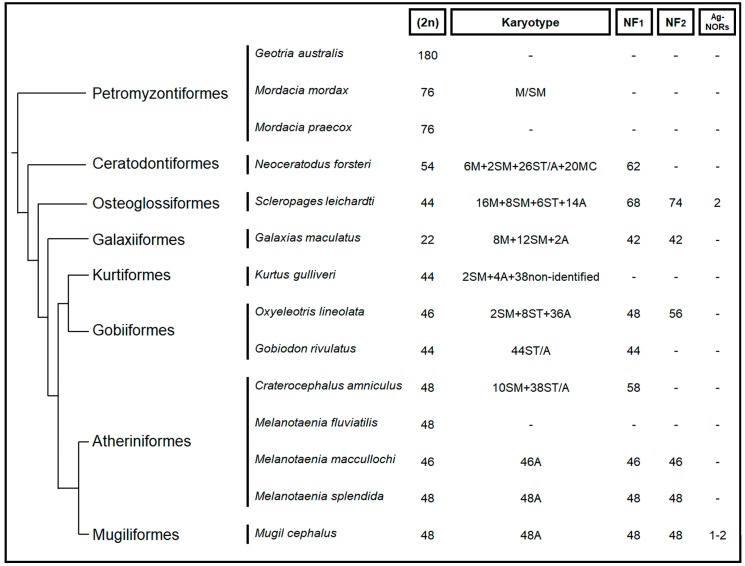
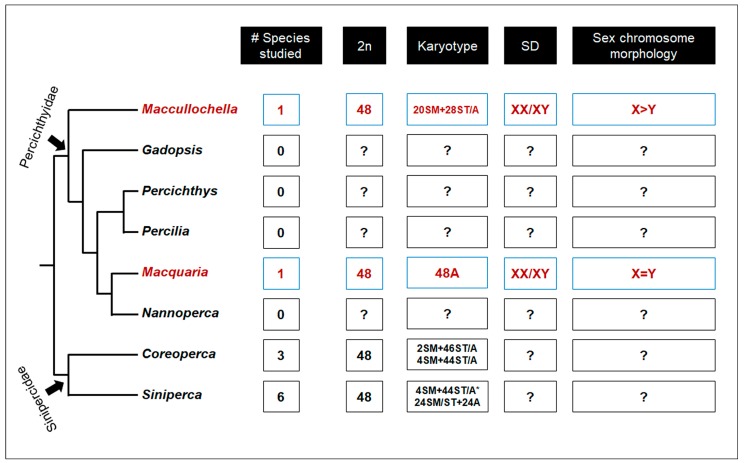
The karyotype data for Australian freshwater fish fauna are scarce (Figure 2) [22,23]. Prior to the current study, there have been no published accounts of karyotype data for any fish from the family Percichthyidae. Molecular characterisation of karyotypes is an important first step towards understanding genome evolution and organisation. In particular, mapping repetitive sequences such as microsatellite associate repeats as well as telomeres can provide an overall landscape of repetitive sequences, as well their role in karyotype evolution, including sex chromosomes. Mapping telomere repeats also provides information about possible chromosome rearrangement events through identification of interstitial telomeric sequences (ITSs).
Figure 2.
Summary of cytogenetic studies of Australian fish showing the nature of chromosomes from Arai [24], Ezaz, et al. [22] and Majtánová, et al. [23]. Phylogeny adopted from Betancur-R, et al. [25]; M = Metacentric; SM = Submetacentric; ST = Sub-telocentric; A = Acrocentric; MC = Micro Chromosome; NF1 = Fundamental arm number, when M and SM are counted as two-armed; NF2 = Fundamental arm number, when M, SM and ST are counted as two-armed; Ag-NORs = Number of transcriptionally active nucleolus organiser regions containing chromosomes.
Here, we describe the karyotypes of golden perch and Murray cod and identify their sex chromosome systems. The karyotype data provide an important first step toward better understanding the evolution of Percichthyids fishes specifically, and Australian freshwater fish more broadly. Analyses of sex chromosome systems in these species will provide insight into sex chromosome evolution in fish and provide the basis for developing tools for conservation and management, such as the identification of sex-linked markers and methods for nondestructive sexing. Our study also provides baseline information to understand the evolutionary trends of karyomorphology in this group.
2. Results
2.1. Karyotype and C-Banding of Golden Perch and Murray Cod
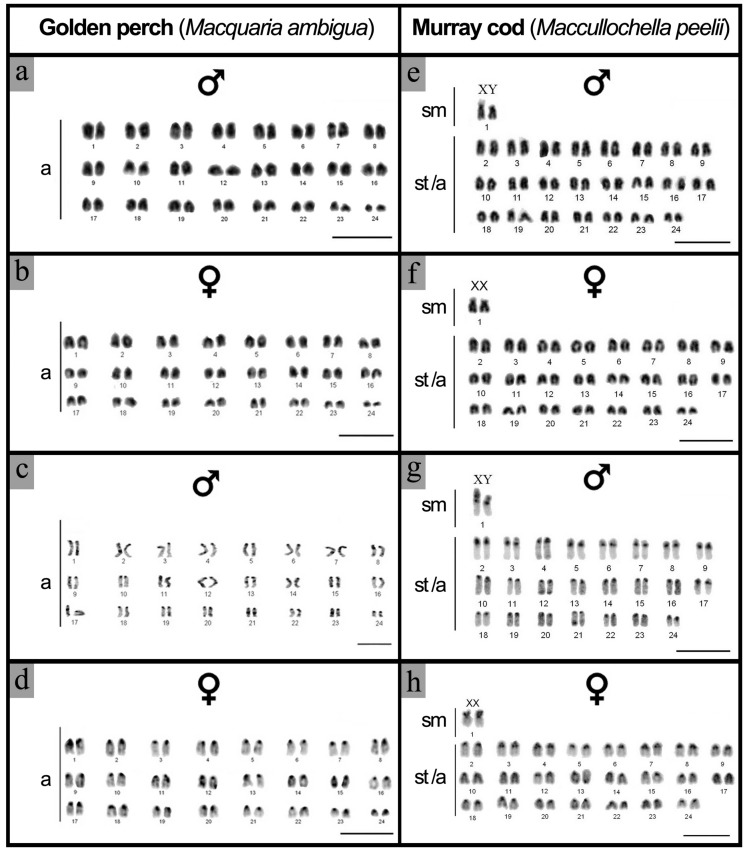
The DAPI (4′,6-diamidino-2-phenylindole)-stained metaphase karyotypes were examined in five individuals from each sex of golden perch and three females and five males from Murray cod. For each individual, a minimum of 60 at metaphase cells were examined (Table 1). For both species, the diploid chromosome complement is 2n = 48. The golden perch karyotype consisted of 24 pairs of acrocentric chromosomes (Figure 3a,b), while the Murray cod karyotype consisted of one pair of submetacentric and 23 pairs of subtelocentric/acrocentric chromosomes (Figure 3e,f). Comparison of the karyotypes from males and females did not reveal any morphologically differentiated sex chromosomes in golden perch (Figure 3a,b). However, heteromorphism was observed between the largest chromosome pair (submetacentric) of Murray cod. In Murray cod, one chromosome from the largest (submetacentric) pair was shorter in the male karyotype, while in females, the largest chromosome pair was equal in size, suggesting a male heterogametic sex determining system (XX/XY) in this species (Figure 3e,f). This heteromorphism was present in all five males examined and was not observed in any of the three females analysed (Table 1).
Table 1.
Number of individuals and cells examined for the two fish species. The number of cells in the column for each group refers to the least number of mitotic chromosomes examined. CGH = Comparative Genomic Hybridisation; FISH = Florescence in situ hybridisation.
| Golden Perch | Murray Cod | |||||||
|---|---|---|---|---|---|---|---|---|
| Female | Male | Female | Male | |||||
| Number of individuals | Number of cells/each | Number of individuals | Number of cells/each | Number of individuals | Number of cells/each | Number of individuals | Number of cells/each | |
| Mitosis | 12 | 60 | 12 | 60 | 3 | 60 | 5 | 60 |
| Karyotyping | 5 | 20 | 5 | 20 | 3 | 20 | 5 | 20 |
| CGH | 3 | 20 | 3 | 20 | 3 | 20 | 3 | 20 |
| C-banding | 3 | 60 | 5 | 60 | 3 | 60 | 5 | 60 |
| FISH- telomeric probe | 3 | 20 | 3 | 20 | 3 | 20 | 3 | 20 |
| FISH-repetitive sequences | 3 | 20 | 3 | 20 | 3 | 20 | 3 | 20 |
| DNA methylation | 3 | 20 | 3 | 20 | 3 | 20 | 3 | 20 |
| Cross-species CGH | - | - | 1 | 20 | - | - | 1 | 20 |
Figure 3.
DAPI (4′,6-diamidino-2-phenylindole)-stained and C-banded metaphase karyotypes of golden perch and Murray cod; (a) golden perch male karyotype; (b) golden perch female karyotype; (c) C-banded karyotype of golden perch male; (d) C-banded karyotype of golden perch female; (e) Murray cod male karyotype; (f) Murray cod female karyotype; (g) C-banded karyotype of Murray cod male; (h) C-banded karyotype of Murray cod female. DAPI-stained metaphases were inverted. Scale bar 5 µm.
C-positive heterochromatin was found in all chromosomes, with C-positive bands near the centromeric regions (Figure 3c,d,g,h) in all animals examined (Table 1). No distinct staining of heterochromatin on any specific chromosome was observed in either males or females of golden perch (Figure 3c,d). On the other hand, a strong C-positive band was observed in the centromeres of all the subtelocentric/acrocentric chromosomes of Murray cod (Figure 3g,h), with a comparatively larger C-positive heterochromatic block on the p arms of the putative sex chromosome pair (heteromorphic pair) (Figure 3g,h).
2.2. Intraspecies Comparative Genomic Hybridisation (CGH)
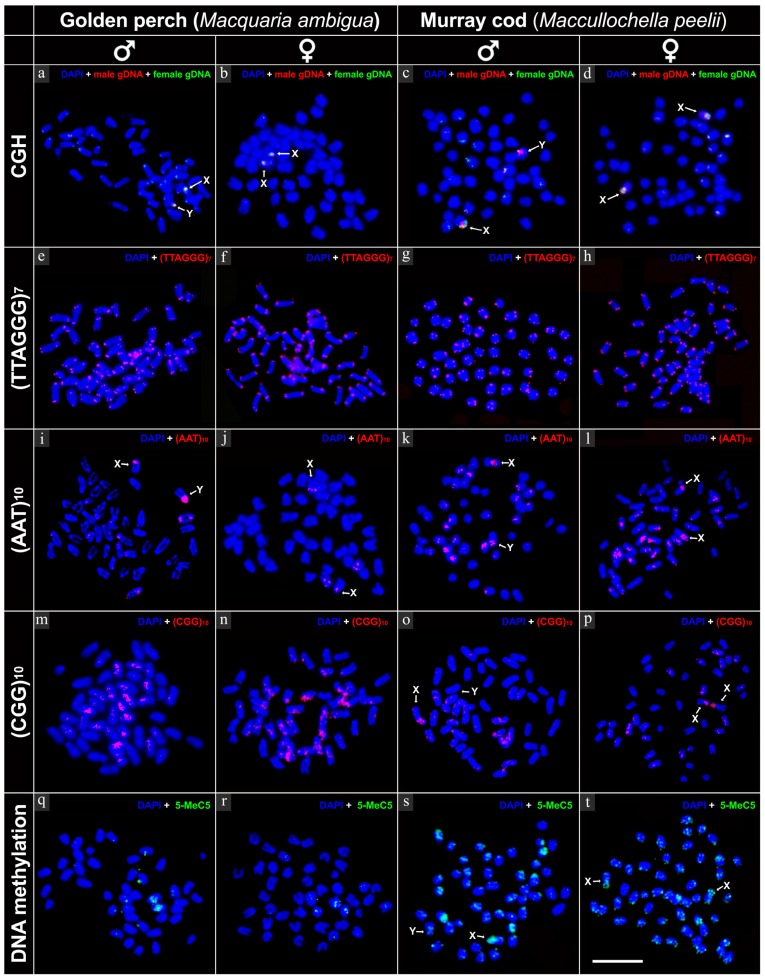
Intraspecies CGH was performed in three females and three males of each species (Table 1). A minimum of 20 cells were examined from each individual (Table 1). A distinct hybridisation signal for fluorescently labelled male genomic DNA was observed in one of the acrocentric pairs from all golden perch males, but not on chromosomes from any females (Figure 4a,b). This male-specific chromosome is therefore a Y chromosome, identifying males as the heterogametic sex in golden perch. For more accuracy, and to control for any sensitivity differences of the different fluorochromes to different filter sets, male and female genomic DNA was reciprocally labelled (reciprocal CGH) and hybridised onto male and female metaphase spreads. A similar pattern of signal was detected, supporting the presence of the Y chromosome. To conclusively identify Y chromosomes in Murray cod, CGH was also performed on samples from both sexes. In addition to dispersed hybridisation signals on all chromosomes, a comparatively strong hybridisation signal for fluorescently labelled male genomic DNA was observed on the short (p) arm of the smaller homologue of the largest chromosome pair in all three males (Figure 4c,d). This hybridisation signal was consistent in the reciprocally labelled experiments, indicating a male heterogametic sex chromosome system (XX/XY) in Murray cod.
Figure 4.
Fluorescence in situ hybridisation signals of genomic DNA (CGH, a–d), telomeric probe (e–h), microsatellite motifs (i–p) and DNA methylation patterns in golden perch and Murray cod (q–t). CGH (a–d): hybridisation signals for fluorescently labelled genomic DNA of golden perch and Murray cod, showing hybridisation signals on X and Y sex chromosome; TTAGGG7 (e–h): showing terminal hybridisation signals in all chromosomes; AAT10 (i–l): hybridisation pattern of repetitive microsatellite motif (AAT)10, showing abundant accumulation on golden perch Y chromosome; CGG10 (m–p): hybridisation signals from repetitive microsatellite motif (CGG)10, showing high accumulation on X chromosome of Murray cod compared with that of the Y chromosome. DNA methylation (q–t): immunofluorescent detection of DNA methylation, showing hypermethylated X chromosome in Murray cod compared with that of the Y chromosome. Scale bar represent 5 µm.
2.3. FISH Mapping of Telomeric Probe and Microsatellite Motifs
In both species, mapping of telomeric sequences (TTAGGG)7 identified terminal hybridisation signals in all chromosomes, with no evidence of interstitial telomeric sequences (ITSs) (Figure 4e–h). In addition, AT-rich (AAT)10 and GC-rich (CGG)10 microsatellite motifs were hybridised across the genome of both species, predominantly near the centromeric region. A strong banding pattern for (AAT)10 was observed on the Y chromosome of golden perch (Figure 4i), with a dominant hybridisation signal evident on both sex chromosomes (X and Y) of Murray cod (Figure 4k,l). For GC-rich (CGG)10 microsatellite motif, a strong banding pattern was observed on the X chromosome of Murray cod (Figure 4o,p) and on both X and Y chromosomes in golden perch (Figure 4m,n).
2.4. Immunofluorescence Detection of DNA Methylation
DNA methylation is one of the active epigenetic marks that actively regulates gene expression, and is responsible for X chromosome inactivation in mammals. In this study, we performed a DNA methylation experiment using anti-5-methylcytosine to characterize global methylation patterns in golden perch and Murray cod. For both species, genome-wide signals were detected, predominantly in the telomeric regions. A comparatively stronger but equal strength hybridisation signal was detected on both X and Y chromosomes in golden perch, however, in Murray cod, the X chromosome was hypermethylated compared with that of the Y chromosome (Figure 4q–t).
2.5. Cross-Species CGH
Cross-species CGH is an effective tool for understanding genomic and sex chromosome homologies in allopolyploids [26,27,28]. The technique has also been used in comparative genome analysis to visualize the chromosomal distribution of conserved DNA sequences between two species. We performed hybridisation of the genomic DNA probe with differential fluorescent labelling on metaphase chromosome of both species. The strongest hybridisation signal was detected near the centromeric region of the chromosomes, with no distinguishable differences in sex chromosomes (Figure S1).
3. Discussion
This study is the first karyotypic report of the two Australian native freshwater fish species from the family Percichthyidae, revealing conserved chromosome numbers, 2n = 48, in both species. However, we detected a substantial variation in chromosome morphologies (e.g., number of submetacentric and subtelocentric/acrocentric chromosomes) between the studied species; suggesting major chromosome rearrangements. We could not, however, conclusively predict the directionality of the evolution of such chromosome rearrangements because of a lack of karyotypic data from outgroup taxa (Figure 5).
Figure 5.
Truncated phylogenetic tree of the family Percichthyidae and sister clade from family Sinipercidae. Three monotypic genus, Bostockia, Guyu and Nannatherina were excluded due to unresolved phylogenetic position. Truncated phylogeny based on 10 nuclear genes was adopted from Near, et al. [29]. Red colour represents information derived from the present study. Phylogeny is not to scale. Asterisk (*) in Siniperca genus indicates most frequently observed karyotype [24].
The analysis of patterns of heterochromatin distribution is a useful tool for investigating karyotype diversification in vertebrate species. Within vertebrates, fish tend to lose substantial heterochromatin in their genomes [30]. Our C-banding identified significant C-bands near the centromeric region of the karyotype in both species. There were obvious differences in C-banding patterns in the Murray cod karyotype compared with that of the golden perch. Furthermore, within the Murray cod karyotype, we detected a strong banding pattern in a number of subtelocentric/acrocentric chromosomes. Such diversification in heterochromatin block can be explained by the abundance and variable function of transposable elements.
We used molecular cytogenetic techniques combined with differential banding to search for cryptic sex chromosomes in golden perch and Murray cod. C-banding is effective in revealing sex chromosomes in vertebrates because of the possible large accumulation of heterochromatin in the sex chromosomes [31,32,33]. Along with C-banding we used comparative genome hybridisation (CGH), which has been found to be effective in identifying sex chromosomes in divergent taxa including fish and reptiles [33,34,35]. We identified a male-specific submetacentric chromosome pair as the sex chromosome of Murray cod and a male-specific acrocentric pair of golden perch chromosomes. This suggests that both species have a male heterogametic sex chromosome system (XX/XY). Sex chromosomes of Murray cod can be characterised as having tiny size variation between the X and Y chromosomes (shorter Y), along with an accumulation of heterochromatin on the short arm of both X and Y chromosomes. Sex chromosomes of golden perch are characterised as a homomorphic acrocentric pair, with minor accumulation of heterochromatin at the pericentromeric region.
The mechanism behind sex chromosome evolution remains yet a mystery. Sex chromosomes are thought to have evolved from an autosomal pair through acquisition of a sexually advantageous mutation, which has subsequently been selected in favour of either maleness or femaleness via suppression of recombination, leading to the degeneration of Y or W chromosomes [36,37,38,39]. Accumulation and amplification of repetitive sequences on the sex chromosomes, particularly microsatellites, is thought to be one of the drivers that trigger this process because of their ability to expand rapidly in comparison with other classes of repetitive DNA [40,41,42,43,44]. Compared with birds and mammals, fishes often possess homomorphic sex chromosomes, which have been hypothesized to display high rate of turnover events, where previously established sex chromosomes are being replaced with new sex chromosomes including novel sex-determining genes [45,46]. Our study provides evidence of cryptic sex chromosome turnover between these two species based on the morphological variation, variability in accumulation of heterochromatin and amplification of microsatellite associated repetitive sequences on the X and Y chromosomes. However, many species within the nine genera are yet to be investigated to provide more evidence for the mechanisms and directionality of sex chromosome turnover within the family Percichthyidae.
We observed random accumulation of AT- and GC-rich tri-nucleotide microsatellite motifs ((AAT)10 and (CGG)10) across the genomes of both species, providing insights into the abundance of repetitive sequences including transposable elements. A distinct variation in the amplification of tri-nucleotide motifs (AAT)10 and (CGG)10 could indicate karyotype rearrangement between both species. In general, tandem repeats (microsatellite) of small stretches of DNA motifs are widespread in the genomes of eukaryotes. Furthermore, several di-, tri- and tetra-nucleotide microsatellite repeats have been identified on the sex chromosomes of fishes [43,47,48] and amniotes [3,49,50]. The widespread association of microsatellite proliferation within sex chromosomes suggests that such proliferation is one of the likely mechanisms that drive sex chromosome differentiation via suppression of recombination and possible evolution of differentiated sex chromosomes [41]. Future studies on the accumulation of such repeats in other percichthyids may provide a better understanding of karyotype evolution in this family. In addition, we also detected hybridisation signals from conserved vertebrate telomeric repeat (TTAGGG)7 in terminal locations of all chromosomes in both species, as expected. However, it is intriguing that we did not detect any interstitial telomeric (ITS) signals on any chromosomes in either of the species given the stark morphological differences in chromosome morphologies between these two species. Accumulation of telomeric repeats in nontelomeric regions signifies evidence of ancestral chromosome rearrangement events. Further studies including other species from this family will be required to provide conclusive evidence on the evolution of morphologically divergent yet conserved karyotypes in this group.
Comparative analysis using cross-species CGH revealed clear banding in the centromeric position of chromosomes from both species (Figure S1). This could be because of a shared accumulation of repetitive sequences in both species. Apart from the centromeric sequences, there was no evidence for common shared genomic regions (entire chromosome or any arm of the chromosome), indicating a variant evolutionary trajectory for these species, even though they belong to the same phylogenetic lineage.
4. Materials and Methods
4.1. Specimen Collection and Mitotic Chromosome Preparation
Eight Murray cod (five males and three females) were sourced from a hatchery (Murray Darling Fisheries, Narrandera, Australia), and 24 golden perch (12 male and 12 female) were sourced from the Lachlan River (Murray Darling Basin, Australia). For Murray cod, mitotic chromosomes were obtained from kidney and liver cells after mitotic stimulation with yeast, using the protocol described in Bertollo et al. [51]. Briefly, yeast was conjugated with dextrose, diluted in distilled water (1.25 g yeast/0.75 g dextrose/2.5 mL distilled water), homogenized using a sterile spoon spatula, and incubated at 37 °C for 30 min to activate the yeast cells. The solution was homogenized again, and 200 μL were injected into muscle on the caudal peduncle. The specimen was kept in a clean glass tank under optimal condition for 24 h before sacrifice. The fragments of kidney and liver tissues were first dissociated in RPMI 1640 cell culture medium (Life Technologies, Inc., Scoresby, Victoria, Australia), then 160 μL of colchicine (0.0125%) was added and the culture was incubated for 30 min at room temperature. Finally, the cells were subjected to hypotonic treatment in 0.075 M KCl for 30 min at 37 °C and then fixed in Carnoy’s solution (methanol:acetic acid 3:1). Mitotic chromosomes of golden perch were prepared from regenerated fin clips following the protocol described by Völker and Ráb [52]. The hind most part of the caudal fin was cut using sterile dissecting scissors, after which the fish was maintained under optimal condition for three to four days. The regenerated fin material was cut off and treated with Ringer solution (1.281 M NaCl, 0.025 M KCl, 0.018 M CaCl2, 0.002 M NaHCO3) containing 0.05% colchicine. After two hours of incubation, cells were fixed in Carnoy’s solution.
Cell suspensions were dropped onto glass slides and air-dried. For DAPI (40-6-diamidino-2-phenylindole) staining, slides were mounted with antifade medium Vectashield (Vector Laboratories, Burlingame, CA, USA) containing 1.5 mg/mL DAPI.
4.2. C-Banding
Detection of centromeric heterochromatin (C-banding) was performed following the protocol described by Salvadori, et al. [53]. Briefly, 20–25 µL of cell suspension was dropped on slides and air dried and aged on a 60 °C hot plate for 60 min. Slides were then treated with 0.2 M HCl at room temperature for 20 min and with 5% Ba(OH)2 at 45 °C for 3 min. Then, the slides were incubated in 2× saline sodium citrate (SSC) at 65 °C for 60 min. Finally, the chromosomes were stained with antifade medium Vectashield containing 1.5 mg/mL DAPI.
4.3. Genomic DNA Extraction
Total genomic DNA was extracted from whole blood and fin clips, following the protocol of Ezaz, et al. [54]. Briefly, 25 mg of tissue sample was incubated overnight at 56 °C with lysis buffer (100 mM Tris-HCL, pH 8.0; 10 mM EDTA; 250 mM NaCL), 10% (w/v) SDS (sodium dodecyl sulphate) and proteinase K. Lysed tissue was incubated at 37 °C for 60 min with 10 µL RNAse A (20 mg/mL), followed by a room temperature incubation for 20 min with 500 µL buffered phenol. After the incubation, the mixture was centrifuged at 16,000× g for 12 min, and the supernatant was transferred to a new sterile tube. This step was performed twice, and 500 µL of 24:1 (v/v) chloroform:isoamyl alcohol was added to the solution. The solution was incubated at room temperature for 20 min. After the incubation, the mixture was centrifuged twice again, as previously described, and the supernatant was subsequently transferred to a new sterile tube. Then, 0.6–0.8 volume of chilled isopropanol was added to the solution and shaken vigorously for 60 s. The precipitated DNA was washed with 70% ethanol and dried. Finally, the pallet was resuspended in elution buffer (10 mM Tris-HCl, pH 8.0; 0.1 mM EDTA) for 30 min at 50 °C and kept at 4 °C.
4.4. Comparative Genomic Hybridisation (CGH) and Cross-Species CGH
We followed the procedure of comparative genomic hybridisation and cross-species CGH described by Ezaz, et al. [33] and Symonová, et al. [55]. Briefly, female and male total genomic DNA was labelled by Nick translation (NT), incorporating Spectrum-Green and Spectrum-Red dUTP, respectively, NT kit (Abbott Molecular, Macquarie Park, Australia). Nick translation reaction was incubated at 15 °C for 2 h. After incubation, NT labelled DNA was checked using 1% agarose gel electrophoresis for size fractionation to 200–500bp. Labelled DNA was co-precipitated by overnight incubation at −20 °C with 5–10 µg of salmon sperm DNA, 20 µg glycogen and 3 volumes of chilled 100% ethanol. Precipitation reaction was centrifuged at maximum speed for 30 min, and the supernatant was discarded and air-dried. Depending on the size of pellet, the co-precipitated probe DNA was resuspended in 30–40 µL hybridisation buffer (50% formamide, 10% dextran sulfate, 2× SSC, 40 mmol/L sodium phosphate pH 7.0 and 1× Denhardt’s solution). The hybridisation mixture was denatured at 70 °C for 10 min, then chilled on ice for 2 min. Subsequently, 18 µL of probe mixture was placed as a single drop on a slide and hybridised at 37°C for 3 days in a humid chamber. Slides were washed once at 60 °C in 0.4× SSC, 0.3% Igepal for 2 min, followed by another wash at room temperature in 2× SSC, 0.1% Igepal. Slides were then air-dried and mounted with antifade medium Vectashield containing 1.5 mg/mL DAPI. Images were captured using a Zeiss Axioplan epifluorescence microscope equipped with a CCD (charge-coupled device) camera (RT-Spot), (Zeiss, Oberkochen, Germany) using filters 02, 10 and 15 from the Zeiss fluorescence filter set or the Pinkel filter set (Chroma technologies, filter set 8300, Bellows Falls, VT, USA). ISIS scientific imaging software (Metasystems, Altlussheim, Germany) was used for image capture and analysis, including karyotyping. For FISH and CGH image analysis, multiple functions of ISIS scientific imaging software were used, such as signal normalizing and background correction. Processed images were pseudocoloured and superimposed, using ISIS scientific imaging software.
For cross-species CGH, golden perch gDNA was labelled with Spectrum-Red dUTP while Murray cod gDNA with Spectrum-Green dUTP using NT, as described above. DNA was coprecipitated with 5–10 µg of salmon sperm DNA (as competitor) and 20 µg glycogen (as carrier). Finally, slides with metaphase spread and labelled genomic DNA were hybridised at 37 °C inside a humid chamber for 3 days. Washing of slides and mounting with antifade medium was carried out in the same way as for the CGH experiments.
4.5. Probes for FISH (Fluorescence In Situ Hybridisation) Experiments
The 5’-Cy3-labeled telomeric probe (TTAGGG)7 and oligonucleotides of two microsatellite motifs, (AAT)10 and (CGG)10 were purchased from GeneWorks (Thebarton, South Australia, Australia) to be used as probes. Repetitive DNA mapping FISH was performed under high stringency conditions on metaphase chromosomes, as described by Bonillo, et al. [56]. After denaturation in 70% formamide/2× SSC at 70 °C, chromosomes were dehydrated in a series of ethanol washes. Approximately 500 ng probe for each slide was resuspended in 15 µL hybridisation buffer (50% formamide, 10% dextran sulfate, 2× SSC, 40 mmol/l sodium phosphate pH 7.0 and 1× Denhardt’s solution). The probe mixture was denatured at 70 °C for 10 min, then chilled on ice for 2 min. Denatured probe was subjected to metaphase chromosomes and hybridised at 37 °C for 48 h in a humid chamber. Similar to CGH, slides were washed once at 60 °C in 0.4× SSC, 0.3% Igepal for 2 min, followed by another wash at room temperature in 2×SSC, 0.1% Igepal. Slides were then air-dried and mounted with antifade medium Vectashield containing 1.5 mg/mL DAPI.
4.6. Immunofluorescence Detection of DNA Methylation
Immunostaining for DNA methylation was performed following published protocols [57,58], with a slight modification. Briefly, after dehydrating through an ethanol series (70, 90 and 100% v/v) metaphase chromosomes were denatured in 70% formamide (v/v) in phosphate-buffered saline (PBS: 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM 2.4 KH2PO4,) at 70 °C for 2 min. Slides were then quenched in ice cold 70% (v/v) ethanol for 5 min, then dehydrated in 90% (v/v) and 100% (v/v) ethanol for 3 min each before being air-dried. After incubating with PBS-T (PBS with 0.03 % (v/v) Tween 20) for 3 min, slides were blocked by incubating in PBS-T with 1% (w/v) bovine serum albumin (BSA) for 20 min at room temperature. Metaphase chromosomes were treated with anti-5-methylcytosine primary antibody (1:200 in PBS-T, Clone 10G4, Zymo Research, Irvine, CA, USA) and incubated in a humid chamber at 37 °C for 60 min. Slides were washed twice for 5 min in PBS-T and subjected to 200 μL of the secondary probe (AffiniPure Donkey Anti-Mouse IgG in PBS-T; Jackson ImmunoResearch Laboratories, West Grove, Pa, USA), diluted 1:500 in PBS-T. After 60 min second incubation in a humid chamber at 37 °C, slides were washed twice for 5 min each in PBS-T. Metaphase chromosomes were incubated in 4% (w/v) paraformaldehyde at room temperature for 15 min and washed three times (3 min each) in PBS-T. Air-dried metaphase chromosomes were mounted with DAPI (4’,6-diamidino-2-phenylindole) in Vectashield (Vector Laboratories Inc., Burlingame, CA, USA).
5. Conclusions
The present study demonstrated the successful use of both conventional (mitotic chromosome preparation, C-banding) and advanced (CGH, FISH, cross-species CGH) cytogenetic techniques for two Australian freshwater fishes to characterise karyotypes. This information provides critical baseline data for future cytogenetic analysis of fishes, especially in the family Percichthyidae. The identification of sex chromosomes is a first step in understanding the evolution of sex determination mechanisms in these fishes, which occupy highly hydrologically variable habitats. In addition, information on sex chromosomes provides the basis for developing nondestructive methods for sex identification of fish in hatcheries, allowing management of sex ratios and identifying the probability of sex reversal.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/17/4244/s1.
Author Contributions
T.E. and F.S. conceptualised the study. F.S. and T.E. designed and co-drafted the manuscript. J.D.T. provided golden perch regenerated fins for mitotic chromosome preparation. F.S. and Z.M. prepared chromosomes from golden perch and Murray cod. F.S. performed cytogenetic analyses. F.D., R.T., R.P.D., J.D.T. and Z.M. edited and proofread the manuscript. All authors approved the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Sharma A., Sen S. Chromosome Botany. Science Publishers; Enfield, CT, USA: 2002. [Google Scholar]
- 2.Getlekha N., Cioffi M.d.B., Maneechot N., Bertollo L.A.C., Supiwong W., Tanomtong A., Molina W.F. Contrasting evolutionary paths among Indo-Pacific Pomacentrus species promoted by extensive pericentric inversions and genome organization of repetitive sequences. Zebrafish. 2018;15:45–54. doi: 10.1089/zeb.2017.1484. [DOI] [PubMed] [Google Scholar]
- 3.Matsubara K., Knopp T., Sarre S.D., Georges A., Ezaz T. Karyotypic analysis and FISH mapping of microsatellite motifs reveal highly differentiated XX/XY sex chromosomes in the pink-tailed worm-lizard (Aprasia parapulchella, Pygopodidae, Squamata) Mol. Cytogenet. 2013;6:60. doi: 10.1186/1755-8166-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ráb P., Yano C.F., Lavoué S., Jegede O.I., Bertollo L.A., Ezaz T., Majtánová Z., de Oliveira E.A., Cioffi M.B. Karyotype and Mapping of Repetitive DNAs in the African Butterfly Fish Pantodon buchholzi, the Sole Species of the Family Pantodontidae. Cytogenet. Genome Res. 2016;149:312–320. doi: 10.1159/000450534. [DOI] [PubMed] [Google Scholar]
- 5.Baroiller J.-F., D’Cotta H., Saillant E. Environmental effects on fish sex determination and differentiation. Sex. Dev. 2009;3:118–135. doi: 10.1159/000223077. [DOI] [PubMed] [Google Scholar]
- 6.Hayes T.B. Sex determination and primary sex differentiation in amphibians: Genetic and developmental mechanisms. J. Exp. Zool. 1998;281:373–399. doi: 10.1002/(SICI)1097-010X(19980801)281:5<373::AID-JEZ4>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi Y., Nagahama Y., Nakamura M. Diversity and Plasticity of Sex Determination and Differentiation in Fishes. Sex. Dev. 2013;7:115–125. doi: 10.1159/000342009. [DOI] [PubMed] [Google Scholar]
- 8.Baillie J., Hilton-Taylor C., Stuart S.N. 2004 IUCN Red List of Threatened Species: A Global Species Assessment. IUCN; Gland, Switzerland: 2004. [Google Scholar]
- 9.Nelson J.S., Grande T.C., Wilson M.V. Fishes of the World. John Wiley & Sons; Hoboken, NJ, USA: 2016. [Google Scholar]
- 10.Vörösmarty C.J., Lévêque C., Revenga C., Bos R., Caudill C., Chilton J., Douglas E., Meybeck M., Prager D., Balvanera P. Fresh water. Ecosyst. Hum. Well-Being Curr. State Trends. 2005;1:167–201. [Google Scholar]
- 11.Bray D.J. Temperate Basses, Percichthyidae in Fishes of Australia. [(accessed on 25 June 2018)]; Available online: https://australianmuseum.net.au/learn/animals/fishes/percichthyidae-australian-freshwater-basses-perches-and-cods/
- 12.Allen G.R., Midgley S.H., Allen M. Field Guide to the Freshwater Fishes of Australia. Western Australian Museum; Welshpool, WA, Australia: 2002. [Google Scholar]
- 13.Lintermans M. Fishes of the Murray-Darling Basin: An Introductory Guide. Murray-Darling Basin Commisssion; Canbera, ACT, Australia: 2007. [Google Scholar]
- 14.Beheregaray L.B., Pfeiffer L.V., Attard C.R.M., Sandoval-Castillo J., Domingos F.M.C.B., Faulks L.K., Gilligan D.M., Unmack P.J. Genome-wide data delimits multiple climate-determined species ranges in a widespread Australian fish, the golden perch (Macquaria ambigua) Mol. Phylogenet. Evolut. 2017;111:65–75. doi: 10.1016/j.ympev.2017.03.021. [DOI] [PubMed] [Google Scholar]
- 15.Goodrich H. Mendelian inheritance in fish. Q. Rev. Biol. 1929;4:83–99. doi: 10.1086/394324. [DOI] [Google Scholar]
- 16.Gordon M. Genetics of Platypoecilus III. Inheritance of Sex and Crossing over of the Sex Chromosomes in the Platyfish. Genetics. 1937;22:376–392. doi: 10.1093/genetics/22.3.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huxley J.S. Note on an alternating preponderance of males and females in fish, and its possible significance. J. Genet. 1920;10:265–276. doi: 10.1007/BF02984298. [DOI] [Google Scholar]
- 18.Devlin R.H., Nagahama Y. Sex determination and sex differentiation in fish: An overview of genetic, physiological, and environmental influences. Aquaculture. 2002;208:191–364. doi: 10.1016/S0044-8486(02)00057-1. [DOI] [Google Scholar]
- 19.Moreira-Filho O., Bertollo L.A., Galetti Jr P.M. Distribution of sex chromosome mechanisms in neotropical fish and description of a ZZ/ZW system in Parodon hilarii (Parodontidae) Caryologia. 1993;46:115–125. doi: 10.1080/00087114.1993.10797253. [DOI] [Google Scholar]
- 20.Steinemann M., Steinemann S., Lottspeich F. How Y chromosomes become genetically inert. Proc. Natl. Acad. Sci. USA. 1993;90:5737–5741. doi: 10.1073/pnas.90.12.5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White M.J.D. Animal Cytology and Evolution. CUP Archive; Cambridge, UK: 1977. [Google Scholar]
- 22.Ezaz T., Berra T.M., Graves J.A.M. Karyotype of the Australian nurseryfish, Kurtus gulliveri (Kurtidae: Perciformes) Chromosome Sci. 2006;9:85–88. [Google Scholar]
- 23.Majtánová Z., Moy K.G., Unmack P.J., Ráb P., Ezaz T. Characterization of the karyotype and accumulation of repetitive sequences in Australian Darling hardyhead Craterocephalus amniculus (Atheriniformes, Teleostei) Peer J. Prepr. 2019;7:e27688v27681. doi: 10.7717/peerj.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arai R. Fish. Karyotypes: A Check List. Springer Science & Business Media; Berlin, Germany: 2011. [Google Scholar]
- 25.Betancur-R R., Wiley E.O., Arratia G., Acero A., Bailly N., Miya M., Lecointre G., Orti G. Phylogenetic classification of bony fishes. BMC Evolut. Biol. 2017;17:162. doi: 10.1186/s12862-017-0958-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barby F.F., Bertollo L.A.C., de Oliveira E.A., Yano C.F., Hatanaka T., Ráb P., Sember A., Ezaz T., Artoni R.F., Liehr T. Emerging patterns of genome organization in Notopteridae species (Teleostei, Osteoglossiformes) as revealed by Zoo-FISH and Comparative Genomic Hybridization (CGH) Sci. Rep. 2019;9:1112. doi: 10.1038/s41598-019-38617-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ráb P., Rábová M., Pereira C.S., Collares-Pereira M.J., Pelikánová Š. Chromosome studies of European cyprinid fishes: Interspecific homology of leuciscine cytotaxonomic marker—The largest subtelocentric chromosome pair as revealed by cross-species painting. Chromosome Res. 2008;16:863. doi: 10.1007/s10577-008-1245-3. [DOI] [PubMed] [Google Scholar]
- 28.Viana P.F., Ezaz T., de Bello Cioffi M., Jackson Almeida B., Feldberg E. Evolutionary Insights of the ZW Sex Chromosomes in Snakes: A New Chapter Added by the Amazonian Puffing Snakes of the Genus Spilotes. Genes. 2019;10:288. doi: 10.3390/genes10040288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Near T.J., Sandel M., Kuhn K.L., Unmack P.J., Wainwright P.C., Smith W.L. Nuclear gene-inferred phylogenies resolve the relationships of the enigmatic Pygmy Sunfishes, Elassoma (Teleostei: Percomorpha) Mol. Phylogenet. Evolut. 2012;63:388–395. doi: 10.1016/j.ympev.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 30.Molina W.F. Chromosomal changes and stasis in marine fish groups. Fish. Cytogenet. 2007;31:69–110. [Google Scholar]
- 31.Abramyan J., Ezaz T., Graves J.A.M., Koopman P. Z and W sex chromosomes in the cane toad (Bufo marinus) Chromosome Res. 2009;17:1015. doi: 10.1007/s10577-009-9095-1. [DOI] [PubMed] [Google Scholar]
- 32.Almeida-Toledo L., Foresti F., Daniel M., Toledo-Filho S. Sex chromosome evolution in fish: The formation of the neo-Y chromosome in Eigenmannia (Gymnotiformes) Chromosoma. 2000;109:197–200. doi: 10.1007/s004120050428. [DOI] [PubMed] [Google Scholar]
- 33.Ezaz T., Quinn A.E., Miura I., Sarre S.D., Georges A., Graves J.A.M. The dragon lizard Pogona vitticeps has ZZ/ZW micro-sex chromosomes. Chromosome Res. 2005;13 doi: 10.1007/s10577-005-1010-9. [DOI] [PubMed] [Google Scholar]
- 34.Matsubara K., Sarre S.D., Georges A., Matsuda Y., Graves J.A.M., Ezaz T. Highly differentiated ZW sex microchromosomes in the Australian Varanus species evolved through rapid amplification of repetitive sequences. PLoS ONE. 2014;9:e95226. doi: 10.1371/journal.pone.0095226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Traut W., Winking H. Meiotic chromosomes and stages of sex chromosome evolution in fish: Zebrafish, platyfish and guppy. Chromosome Res. 2001;9:659–672. doi: 10.1023/A:1012956324417. [DOI] [PubMed] [Google Scholar]
- 36.Charlesworth B. The evolution of sex chromosomes. Science. 1991;251:1030–1033. doi: 10.1126/science.1998119. [DOI] [PubMed] [Google Scholar]
- 37.Graves J.A.M. Weird animal genomes and the evolution of vertebrate sex and sex chromosomes. Annu. Rev. Genet. 2008;42:565–586. doi: 10.1146/annurev.genet.42.110807.091714. [DOI] [PubMed] [Google Scholar]
- 38.Muller H.J. A gene for the fourth chromosome of Drosophila. J. Exp. Zool. 1914;17:325–336. doi: 10.1002/jez.1400170303. [DOI] [Google Scholar]
- 39.Ohno S. Sex chromosome and sex-linked genes. Chromosoma. 1967;23:1–9. doi: 10.1007/BF00293307. [DOI] [Google Scholar]
- 40.De Freitas N.L., Al-Rikabi A.B., Bertollo L.A.C., Ezaz T., Yano C.F., de Oliveira E.A., Hatanaka T., de Bello Cioffi M. Early stages of XY sex chromosomes differentiation in the fish Hoplias malabaricus (Characiformes, Erythrinidae) revealed by DNA repeats accumulation. Cur. Genomics. 2018;19:216–226. doi: 10.2174/1389202918666170711160528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ezaz T., Deakin J.E. Repetitive sequence and sex chromosome evolution in vertebrates. Adv. Evolut. Biol. 2014;1:1–9. doi: 10.1155/2014/104683. [DOI] [Google Scholar]
- 42.Kejnovský E., Michalovova M., Steflova P., Kejnovska I., Manzano S., Hobza R., Kubat Z., Kovarik J., Jamilena M., Vyskot B. Expansion of microsatellites on evolutionary young Y chromosome. PLoS ONE. 2013;8:e45519. doi: 10.1371/journal.pone.0045519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Viana P.F., Ezaz T., Marajó L., Ferreira M., Zuanon J., Cioffi M.B., Bertollo L.A., Gross M.C., Feldberg E. Genomic Organization of Repetitive DNAs and Differentiation of an XX/XY Sex Chromosome System in the Amazonian Puffer Fish, Colomesus asellus (Tetraodontiformes) Cytogenet. Genome Res. 2017;153:41–49. doi: 10.1159/000484423. [DOI] [PubMed] [Google Scholar]
- 44.Yano C.F., Bertollo L.A.C., Liehr T., Troy W.P., Cioffi M.d.B. W chromosome dynamics in Triportheus species (Characiformes, Triportheidae): An ongoing process narrated by repetitive sequences. J. Hered. 2016;107:342–348. doi: 10.1093/jhered/esw021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schartl M. Sex chromosome evolution in non-mammalian vertebrates. Cur. Opin. Genet. Dev. 2004;14:634–641. doi: 10.1016/j.gde.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 46.Volff J.-N., Nanda I., Schmid M., Schartl M. Governing sex determination in fish: Regulatory putsches and ephemeral dictators. Sex. Dev. 2007;1:85–99. doi: 10.1159/000100030. [DOI] [PubMed] [Google Scholar]
- 47.Cioffi M., Kejnovsky E., Bertollo L. The chromosomal distribution of microsatellite repeats in the genome of the wolf fish Hoplias malabaricus, focusing on the sex chromosomes. Cytogenet. Genome Res. 2011;132:289–296. doi: 10.1159/000322058. [DOI] [PubMed] [Google Scholar]
- 48.Nanda I., Feichtinger W., Schmid M., Schröder J.H., Zischler H., Epplen J.T. Simple repetitive sequences are associated with differentiation of the sex chromosomes in the guppy fish. J. Mol. Evolut. 1990;30:456–462. doi: 10.1007/BF02101117. [DOI] [Google Scholar]
- 49.Gamble T., Geneva A.J., Glor R.E., Zarkower D. Anolis sex chromosomes are derived from a single ancestral pair. Evolution. 2014;68:1027–1041. doi: 10.1111/evo.12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pokorná M., Kratochvíl L., Kejnovský E. Microsatellite distribution on sex chromosomes at different stages of heteromorphism and heterochromatinization in two lizard species (Squamata: Eublepharidae: Coleonyx elegans and Lacertidae: Eremias velox) BMC Genet. 2011;12 doi: 10.1186/1471-2156-12-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bertollo L., Cioffi M., Moreira-Filho O. Direct chromosome preparation from freshwater teleost fishes. In: Ozouf-Costaz C., Pisano E., Foresti F., Almeida Toledo L.F., editors. Fish Cytogenetic Techniques (Chondrichthyans and Teleosts) CRC Press; Boca Raton, FL, USA: 2015. pp. 21–26. [Google Scholar]
- 52.Völker M., Ráb P. Direct chromosome preparation from regenerating fish fin tissue. In: Ozouf-Costaz C., Pisano E., Foresti F., Almeida Toledo L.F., editors. Fish Cytogenetic Techniques (Ray-Fin Fishes and Chondrichthyans) CRC Press; Boca Raton, FL, USA: 2015. pp. 37–41. [Google Scholar]
- 53.Salvadori S., Coluccia E., Deiana A.M. Fish Cytogenetic Techniques: Ray-Fin Fishes and Chondrichthyans. CRC Press; Boca Raton, FL, USA: 2015. C-Banding; pp. 88–91. [Google Scholar]
- 54.Ezaz M.T., McAndrew B., Penman D. Spontaneous diploidization of the maternal chromosome set in Nile tilapia (Oreochromis niloticus L.) eggs. Aquac. Res. 2004;35:271–277. doi: 10.1111/j.1365-2109.2004.01010.x. [DOI] [Google Scholar]
- 55.Symonová R., Sember A., Majtánová Z., Ráb P. Characterization of fish genomes by GISH and CGH. In: Ozouf-Costaz C., Pisano E., Foresti F., Almeida Toledo L.F., editors. Fish Cytogenetic Techniques: Ray-Fin Fishes and Chondrichthyans. CRC Press; Boca Raton, FL, USA: 2015. pp. 118–131. [Google Scholar]
- 56.Bonillo C., Coutanceau J., D’Cotta H., Ghigliotti L., Ozouf-Costaz C., Pisano E. Standard fluorescence in situ hybridization procedures. In: Ozouf-Costaz C., Pisano E., Foresti F., Almeida Toledo L.F., editors. Fish Cytogenetic Techniques: Ray-Fin Fishes and Chondrichthyans. CRC Press; Boca Raton, FL, USA: 2015. pp. 103–118. [Google Scholar]
- 57.Domaschenz R., Livernois A.M., Rao S., Ezaz T., Deakin J.E. Immunofluorescent staining reveals hypermethylation of microchromosomes in the central bearded dragon, Pogona vitticeps. Mol. Cytogenet. 2015;8:104. doi: 10.1186/s13039-015-0208-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ingles E.D., Deakin J.E. Global DNA Methylation patterns on marsupial and devil facial tumour chromosomes. Mol. Cytogenet. 2015;8:74. doi: 10.1186/s13039-015-0176-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.