Abstract
Growth-regulating factors (GRFs) are plant-specific transcription factors that perform important functions in plant growth and development. Herein, we identified and characterised 24 AhGRF genes in peanut (Arachis hypogaea). AhGRF family genes were divided into six classes with OLQ and WRC domains. Transcriptome expression profile showed that more AhGRF genes, such as AhGRF5a gene, were at higher expression during pod development in Arachis monticola than cultivated species, especially at the pod rapid-expansion stage. AhGRF5a and AhGRF5b genes expressed at higher levels in pods than roots, leaves and stems tissues, existing in the difference between Arachis monticola and H8107. Exogenous GA3 application can activate AhGRF5a and AhGRF5b genes and H8107 line showed more positive response than Arachis monticola species. These results imply that these two AhGRF genes may be active during the peanut pod development.
Keywords: peanut, GRF, gene family, expression analysis
1. Introduction
Transcription factors are highly variable and display functional diversity, and it is a DNA-binding protein that can specifically interact with cis-acting elements in the promoter region of eukaryotic genes. Through their interaction with each other and with other related proteins, it can activate or inhibit the transcription process, and it is the main regulator of gene expression. Plant growth-regulating factors (GRFs) play an important role in the regulation of plant growth and development [1]. GRFs were first discovered in rice (Oryza sativa); expression of OsGRF1 in O. sativa is increased significantly following gibberellin application, revealing its growth regulator function [2]. More recently, with advances in gene sequencing technology, the GRF family has been studied in many plant species including Arabidopsis thaliana, O. sativa, Zea mays, Brassica napus, Cucumis sativus L., Nicotiana tabacum, and other crops. Nine members have been identified in A. thaliana, compared with 12 in O. sativa, 14 in Z. mays, 17 in rape, 35 in cucumber, and 25 in tobacco [3,4,5,6,7]. GRF family proteins contain two conserved domains in the N-terminal region; QLQ (Glu-Leu-Glu, IPR014978) and WRC (Trp-Arg-Cys, IPR014977) [2,4,8]. In addition, most GRF proteins possess short-chain amino acids in the C-terminal region, for example, the TQL (Thr-Glu-Leu) and GGPL (Gly-Gly-Pro-Leu) motifs [9].
GRF proteins play crucial roles in various biological processes, molecular structure and expression levels in different tissues. GRFs are highly expressed in cell proliferation regions such as flowers, leaves and roots [10,11,12,13,14,15,16]. For example, AtGRF9 controls the development of leaves by negatively regulating the proliferation of leaf primordial cells [17]. Overexpression of ZmGRF1 increases the number of cells in leaves of Z. mays, as well as the size of leaves, while overexpression of ZmGRF10 reduces the number of palisade cells and decreases leaf size [18,19,20]. GRFs also perform regulatory roles in biological and abiotic stress [8]. GRF transcription factors are involved in seed development regulation, such as AtGRF1 is closely related to the weight and size of seeds. Overexpression of OsGRF4 can increase grain yield [21,22,23]. Meanwhile GRFs regulate fruit development in tomato [24]. Studies have found that most GRFs are target genes of microRNA396 (miR396), which is involved in the growth and development of various plants along with miR396 [10,12,14,15,23,24,25].
Peanut (Arachis hypogaea) is an allotetraploid (AABB 2n = 4x = 40); the AA subgenome is derived from the diploid wild species Arachis duranensis, and the BB subgenome is derived from the diploid wild species Arachis ipaensis [26]. Cultivated peanut is one of the most economically important oilseed crops and further enhancing the yield and quality is a main goal of peanut breeding programs. Members of the GRF protein family play important roles in plant growth, grain size and stress responses. GRF gene families have been identified in many plant species, but have not yet been reported in peanut. Meanwhile, the genome sequence of two diploid wild ancestors, Arachis monticola and cultivated peanuts, were reported [27,28,29,30], and more and more function genes could be explored and applicated to the peanut breeding [31,32]. Here, 24 AhGRF genes were identified and analysed in terms of phylogenetic relationship, gene structure and expression patterns in various tissues. The results provide a foundation for further function on AhGRF genes in peanut.
2. Results
2.1. Summary of the AhGRF Gene Family in Peanut
A total of 24 AhGRF genes were identified in peanut, named AhGRF1 to AhGRF20 based on their physical locations on chromosomes (Figure S1). The 24 AhGRF genes are distributed on 16 chromosomes. The chromosomes 01, 03, 08, 09, 10, 11, 13, 17, 19 and 20 have only one gene distribution. Two genes on chromosomes 2, 5, 6 and 15, and chromosomes 12 and 16 have three genes, respectively.
The AhGRF genes vary in length from 1365 bp (AhGRF1) to 6278 bp (AhGRF2a), with CDS lengths from 807 bp (AhGRF12b) to 1893 bp (AhGRF11). The number of exons also varies, from two in AhGRF1 to five in others. AhGRF genes encode proteins ranging from 268 (AhGRF12b) to 630 (AhGRF10) amino acids (aa), with an average length of 435 aa, and the molecular weights vary from 29.842 kDa (AhGRF12b) to 65.060 kDa (AhGRF6b). The isoelectric point (pI) ranges from 6.61 (AhGRF17) to 9.94 (AhGRF10), 21 AhGRF members pI > 7, while only AhGRF16c, AhGRF17 and AhGRF20 pI < 7. This may be related to their different roles in the peanut growth and development (Table S1).
2.2. Genes Structure, Conserved Domains and Phylogenetic Analysis of AhGRF
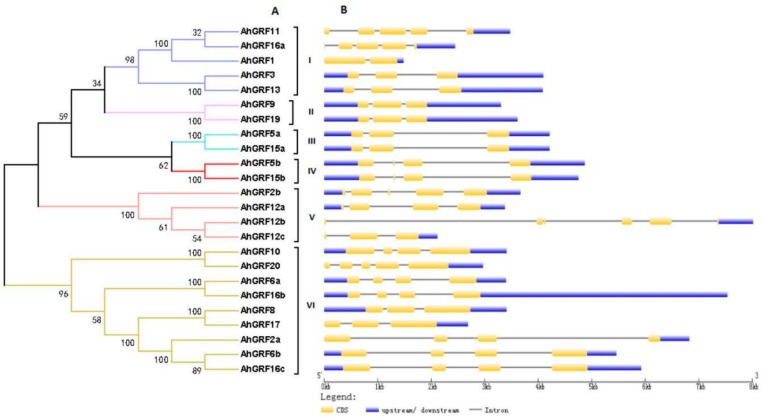
To further investigate the structural characteristics of AhGRF genes, we used NJ method (1000 bootstrap replicates) to construct an evolutionary tree for AhGRF protein sequences, which were divided into six classes (Figure 1).The genes of class II, III and IV have a similar structure and contain three or four exons, whereas class I (AhGRF1) has two exons and V (AhGRF12b) has five exons. Most homologous pairs of genes share high similarity in terms of the length and number of exons/introns, and these features are highly conserved.
Figure 1.
Exon–intron structures of AhGRF genes according to their phylogenetic relationships. (A) An unrooted phylogenetic tree was constructed based on the amino acid sequences. (B) Exon–intron structure analysis of AhGRF genes performed using the online tool GSDS. The lengths of exons and introns of each AhGRF gene are drawn to scale.
Conserved domains in AhGRF family members were predicted (Figure 2). Most AhGRF genes contain four to six conserved domains, such as WRC, QLQ, FFD (Phe-Phe-Asp) and GGPL (Table S2). AhGRF2b, AhGRF12a and AhGRF12c possess two WRC domains, AhGRF5a and AhGRF5b contain similar motifs (Figure S2). The specific distribution of the conserved motifs may lead to functional differences between the AhGRF genes.
Figure 2.
Distribution of conserved domains in AhGRF genes. Ten conserved motifs labeled with different colors were found in the AhGRF sequences using the MEME program. Among them, motif 1, motif 2, motif 4 and motif 6 are the QLQ, WRC, GGPL and FFD conserved domains. The sizes of motifs are proportional to their sequence length.
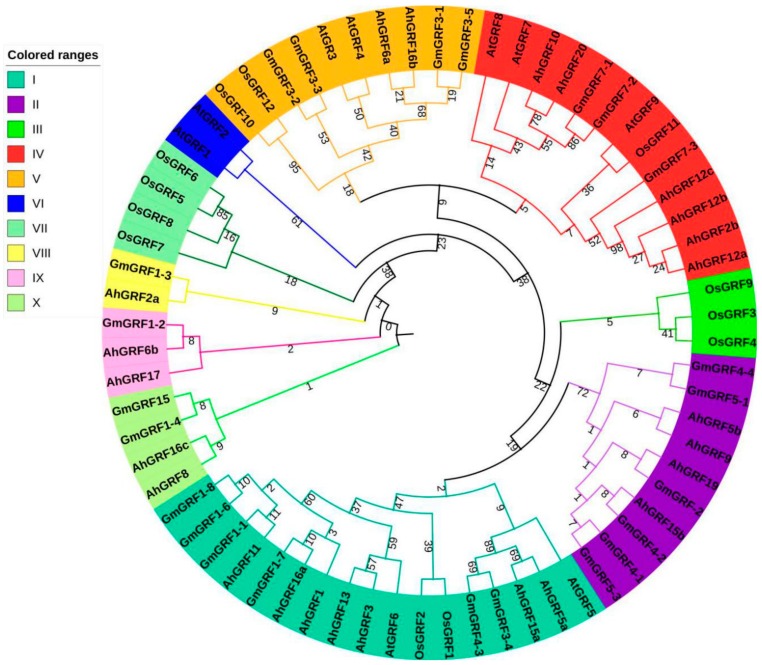
The phylogenetic tree of GRF gene family members from four species was constructed based on full-length AhGRF, AtGRF, OsGRF and GmGRF protein sequences (Figure 3; Table S3). A total of 68 GRFs were clustered into ten subgroups (I–X). AhGRF genes were distributed in seven groups, whereas subgroups III and VII have only OsGRF members. Subgroup VI has only AtGRF members. Among the subgroups, subgroup VIII was relatively small, with only one GRF. By contrast, subgroups I and IV contained the largest number of GRFs (six each), followed by subgroups V, IX and X have two GRFs and subgroup II (four). AhGRF5a and AhGRF15a were clustered together with GmGRF3-4, GmGRF4-3 and AtGRF5 in subgroup I. The phylogenetic tree suggested that the AhGRFs closer with GmGRFs and AtGRFs than OsGRFs, which may be because peanut, soybean and Arabidopsis are dicotyledonous plants.
Figure 3.
Phylogenetic tree based on the growth regulating factors (GRFs) protein family in four plants. Ah, peanut; At, Arabidopsis thaliana; OS, rice; Gm, soybean. The number stands for the confidence of the branch.
2.3. Differential Expression Analysis of AhGRF Genes
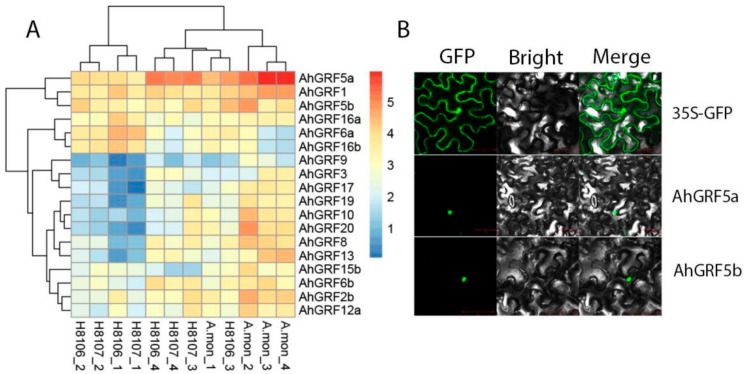
To gain insight into the functions of AhGRF genes, we measured the expression levels of eighteen genes in three varieties during pod development and built a heatmap of the results (Figure 4A; Table S4). More GRFs were expressed at higher levels in the wild species. AhGRF5a was expressed at high levels in Arachis monticola (A.mon) and H8107 species, and had highly expressed in the later stage of pod development. Meanwhile, AhGRF5a and AhGRF5b GFP expression vectors were constructed, and two proteins were localized on the nuclear (Figure 4B).
Figure 4.
Heatmap and subcellular localization of AhGRF genes. (A): Heatmap of AhGRF genes expression in three peanut varieties. A.mon, H8106 and H8107 are peanut recombinant inbred lines (RILs). Pods from four developmental stages were used for expression analysis; 15~DAF (days after flowering) (−1), 25~DAF (−2), 35~DAF (−3) and 45~DAF (−4). Expression values from RNA-seq data were log2-transformed and are displayed as filled blocks coloured blue to red. (B): Subcellular localization of AhGRF proteins in tobacco leaves. AhGRF5a and AhGRF5b fusion proteins, as well as GFP alone. DAF, days after flowering.
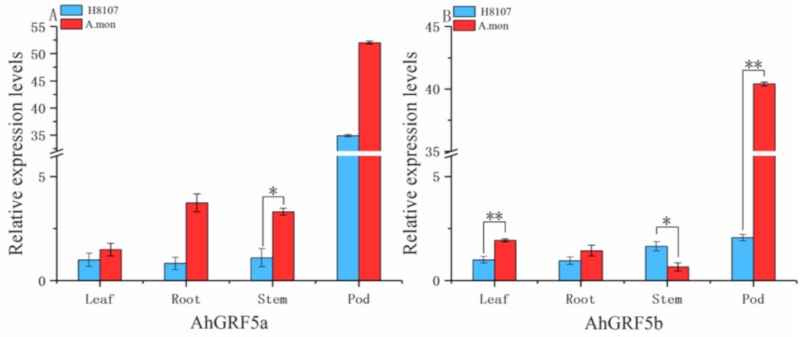
We further investigated the expression levels of AhGRF5a and AhGRF5b in different tissues between A.mon and H8107 (Figure 5), and found that genes expressed at higher levels in pod tissues than roots, leaves and stems tissues, existing in the difference between the two lines. Expression levels of AhGRF5a and AhGRF5b in leaves, roots and pods of A.mon were higher than H8107. In particular, expression of AhGRF5a and AhGRF5b in A.mon pods were 1.72-fold and 17.63-fold greater than in H8107, and AhGRF5b was achieved extremely significant differences. These results suggest that AhGRF5a and AhGRF5b may play the dominant role during the development of pods.
Figure 5.
Expression analysis of AhGRF genes in different tissues. The qRT-PCR analysis of AhGRFs transcript levels was performed on leaves, roots, stems and pods. (A) AhGRF5a and (B) AhGRF5b. Experiments were repeated three times and vertical bars indicate standard errors. * and ** represent significant differences at p < 0.05 and p < 0.01, respectively.
2.4. Responses of AhGRFs to Exogenous GA3 Treatment
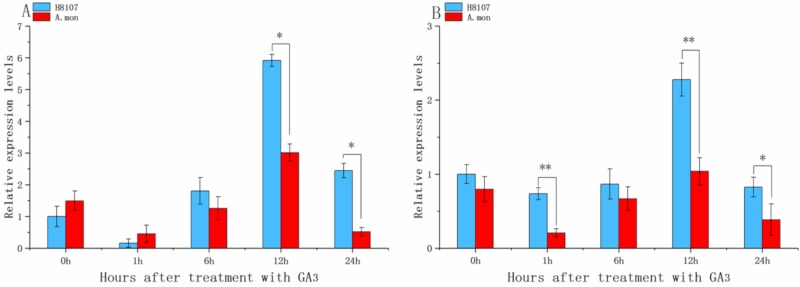
In order to investigate the response of AhGRFs to exogenous gibberellin A3 (GA3) application, we selected two-week-old seedlings spraying GA3 with 100 uM. Exogenous GA3 application can activate AhGRF5a and AhGRF5b genes and H8107 line showed more positive response than A.mon line (Figure 6). The expression levels of AhGRF5a and AhGRF5b were initially increased, then decreased and achieve highest level at 12 h in two lines. Thus, AhGRF5a and AhGRF5b may act as response factors to GA treatment.
Figure 6.
Expression analysis of AhGRF genes in response to treatment with the hormone GA3. qRT-PCR analysis of AhGRF transcript levels was performed on 2-week-old seedlings treated with GA3-0 h, GA3-1 h, GA3-6 h, GA3-12 h, GA3-24 h. (A) AhGRF5a and (B) AhGRF5b. Experiments were repeated three times and vertical bars indicate standard errors. * and ** represent significant differences at p < 0.05 and p < 0.01, respectively.
3. Discussion
Herein, we identified 24 AhGRFs from peanut and explored their characteristics, further clustered into ten classes based on a phylogenetic tree of 68 GRF homologs from four representative plant species. Expression profiles showed that AhGRF5a was expressed at high levels in A.mon and H8107, and their expression levels revealed that AhGRF5a and AhGRF5b expressed higher levels in pod and showed positive response with GA3 treatment.
Early expansion of the GRF gene family can be linked to whole-genome triplication that occurred in the common ancestor of eudicots, and further expansion in this family has occurred through several independent whole-genome duplications in various plant lineages [8]. Interestingly, early study identified 23 AhGRF genes in Arachis duranensis and Arachis ipaensis (Table S5). Further analysis found AhGRF2-3 of wild peanut as AhGRF12a and AhGRF12b in cultivated species. This result may be due to gene expansion during evolution. Previous studies found that GRF genes may exhibit structural and functional differentiation in monocotyledonous and dicotyledonous plants [8], the phylogenetic tree suggested that the AhGRFs were closer with GmGRFs and AtGRFs than OsGRFs, which may be because peanut, soybean and Arabidopsis are dicotyledonous plants. Many GRFs are generally expressed at higher levels in actively growing tissues than in mature tissues, and it also play a role in regulating plant senescence [3,33,34]. GRFs regulate fruit development in tomato and overexpression of BnGRF2a results in increased seed weight and oil content [11,24]. High expression of OsGRF4 modulates tissue and organ size, resulting in larger grains, and enhanced grain yield [4,22,35,36]. Peanut pod development can be divided into two stages: pod expansion and pod filling. In general, the first sign of pod development is seen at 15~DAF (days after flowering), and pods enlarge to reach their maximum size at 35~DAF, at which point typical fruits are produced. Peanut pods mature at 60~DAF [37]. In this study, AhGRF5a and AhGRF15a were clustered together with GmGRF3-4, GmGRF4-3 and AtGRF5. The similarities are 63.27%, 59.02% and 28.45%, respectively. Meanwhile, AhGRF5a expression levels in pods were much higher than in other tissues, and expression analysis showed that AhGRF5a was expressed at high levels during at the pod rapid-expansion stage. This suggests that AhGRF5a may play important roles during seed formation. Knotted1-like homeobox (KNOX) is one of the most important regulators in the development and function of meristematic tissues, where it controls meristem development and restricts cell differentiation. GRFs are upstream repressors of KNOX genes that inhibit GA biosynthesis [38]. BrGRF genes expression is enhanced by GA3 treatment in Chinese cabbage [39]. Here, we measured the expression levels of AhGRF5a and AhGRF5b in two peanut varieties with GA3 treatment, and found that exogenous GA3 application can activate AhGRF5a and AhGRF5b genes. Thus, AhGRF5a and AhGRF5b act as response factors to GA3. And 12 h after GA3 application is an important time node. It can be inferred that GA3 treatment stimulates the expression of KNOX, and high KNOX levels subsequently lead to up-regulation of AhGRF5a and AhGRF5b, which was consistent with previous study in tobacco [40].
Our results provide a foundation for further function of AhGRF genes in peanut. Studies have found that most GRFs are target genes of miR396, which is involved in the growth and development of various plants, but the relationship of miR396 and AhGRFs needs to be explored. We found that AhGRF5a has a high expression in pod. In addition, plant circadian rhythm and other factors may also have an impact on gene response. This present study provides a foundation for a better understanding of the roles of AhGRF5a and AhGRF5b genes. In the future, the results need to be verified widely and potential function be explored in peanut.
4. Materials and Methods
4.1. Analysis of Peanut AhGRF Gene Family Members
The sequences (genomic, CDS and amino acid) and physical locations were downloaded from peanutbase (http://www.peanutbase.org/). SMART (http://smart.embl-heidelberg.de/) was used to confirm the conserved QLQ and WRC domains. Molecular masses of putative AhGRF proteins were calculated using the compute pI/Mw tool in ExPASy (https://web.expasy.org/protparam/).
4.2. AhGRF Gene Structure and Phylogenetic Analysis
Protein sequence motifs were identified using the MEME program (http://meme-suite.org/). AhGRF gene structures were deduced using the Gene Structure Display Server (GSDS2.0, http://gsds.cbi.pku.edu.cn/index.php). Gene chromosomal location mapping was performed using MapGene2chrom webv2 (http://mg2c.iask.in/mg2c_v2.0/). Arabidopsis (http://www.arabidopsis.org/), O. sativa (http://rice.plantbiology.msu.edu/) and soybean (http://plants.ensembl.org/index.html) GRF protein sequences were downloaded from the appropriate databases, and sequence alignment was conducted with DNAMAN (Lynnon Biosoft Inc, San Ramon, CA, USA) software. An unrooted phylogenetic tree was constructed by MEGA5.2 software using the neighbour-joining (NJ) method with 1000 bootstrap replicates.
4.3. Expression Profiling Based on RNA Sequencing (RNA-Seq) Data
Three varieties, A.mon (tetraploid wild species) and recombinant inbred lines (RILs) H8106 and H8107, were selected. The main difference between the two RILs is the pod size: H8106 has medium-sized pods (3.2 cm long1.3 cm wide), and a 100-seed weight of 100 g, while H8107 has super-large pods (5.5 cm 2.07 cm) with a corresponding 100-seed weight of 182 g. The developmental stages of seeds 15~DAF, 25~DAF, 35~DAF, and 45~DAF were showed in Figure S3. Total RNA was extracted and used to purify poly (A) mRNA using Oligotex mRNA midi prep kit (QIAGEN, Germany). Sequencing libraries were generated using NEBNext UltraTM RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA, USA). Raw sequences were transformed into clean reads after data processing. These clean reads were then mapped to the reference genome sequence. Quantification of gene expression levels were estimated by fragments per kilobase of transcript per million fragments (FPKM). The expression of the AhGRFs family genes in these three different varieties at the different developmental stages was obtained. A heat map of AhGRF genes was generated using an R script based on normalised reads FPKM values of all genes transformed to log2 (value + 1).
4.4. Plant Materials and Hormone Treatments
A.mon and H8107 were used for transcriptome expression analysis. To verify the expression patterns of AhGRF genes, root, stem, seedling leaf (five leaf stage) and pod (45 DAF) samples were collected, immediately frozen in liquid nitrogen and stored at −80 °C until RNA extraction. For hormone treatments, seeds were cultivated with 1/2 Hoagland solution and grown with a 16/8 h light/dark photoperiod and 60% relative humidity at 32/25 °C, respectively. Two-week-old seedlings grown on plates were treated by spraying 100 μM GA3. Leaves of seedlings were selected before GA treatment and designated as GA-control check (GA-CK). Leaves of seedlings were collected at 1, 6, 12 and 24 h after GA3 treatment, immediately frozen in liquid nitrogen, and stored at −80 °C until RNA extraction.
4.5. Total RNA Extraction and cDNA Synthesis
RNA was extracted from roots, stems, leaves and pods using a DNAprep Pure Polysaccharide Polyphenol Plant Total RNA Extraction kit (TianGen, Beijing, China). RNA concentration and quality were evaluated with a NanoDrop One spectrophotometer (Thermo Fisher Scientific, Madison, WI, USA) and visualised by standard agarose gel electrophoresis (1%, w/v). Total RNA was then treated with DNAse to remove contaminating genomic DNA. First-strand cDNA was synthesised from 400 ng of DNA-free total RNA using PrimeScript RT Master Mix (Perfect Real Time) with oligo (dT) 20 primer following the manufacturer’s instructions (TaKaRa, Dalian, China).
4.6. Real-Time Quantitative PCR
Using gene-specific primers designed with the NCBI (https://www.ncbi.nlm.nih.gov/), qRT-PCR was carried out using TB Green Premix Ex Taq II (Tli RNaseH Plus) mix (TaKaRa) on a CFX96 Touch Real-Time PCR System (Bio-Rad, Hercules, CA, USA). Specific primers for GRF genes and actin (the housekeeping gene) were designed to amplify about 200~bp [32]. The reaction mixture included 10 μL TB Green mix, 1 μL forward primer, 1 μL reverse primer, 2 μL cDNA template and 6 μL nuclease-free H2O in a final volume of 20 μL. Thermal cycling included an initial denaturation at 95 °C for 30 s, followed by 40 cycles at 95 °C for 5 s and 60 °C for 30 s. The specificity of PCR amplification was monitored by melting curve analysis from 65 °C to 95 °C at 0.5 °C/s. After each PCR run, a dissociation curve was generated to confirm the specificity of the product. Three biological replicates with three technical replicates were performed for each reaction. The 2−ΔΔCT method was employed to calculate the relative expression levels of AhGRF genes. Sequences of primers used for qRT-PCR are listed in Table S6.
4.7. Construction of AhGRF Transient Expression Vectors and Subcellular Localization Studies in Tobacco
To investigate the subcellular localization of the AhGRF proteins, they were transiently expressed as translational GFP (green fluorescent protein) fusion proteins in tobacco (Nicotiana benthamiana) leaf epidermal cells. The full-length coding sequences of AhGRF5a and AhGRF5b were amplified using Q5 high fidelity enzyme (New England Biolabs, Beijing, China) and cloned into ZT4 Blunt Fast Cloning Kit (Zoman Biotechnology, Beijing, China) according to the instruction of manufacturer (Table S7). Then we designed two forward primers AhGRF5a-1 and AhGRF5b-1 containing a homologous recombination sequence (F:5′-ACAAATCTATCTCTCTCGAG-3′ R:5′-GCTCACCATGGATCC-3′) of the vector. The amplification products were using SE Seamless Cloning and Assembly Kit (Zoman Biotechnology, Beijing, China) ligated into the PFGC5941-35S-GFP (35S-GFP) vector. The recombined plasmids were then transformed into Agrobacterium tumefaciens strain EHA105. Agrobacterium transient expression and infiltration was carried out according to previously published protocols [41,42]. Leaves transformed with the 35S-GFP vector alone were used as controls. Two days after infiltration, fluorescence and bright-light images of transiently infected tobacco leaves were obtained using a Laser Scanning Confocal Microscopy (LSM710, Axio Obseror Z1, Zeiss, Germany). The primers used are listed in Table S6.
5. Conclusions
In this study, 24 AhGRF genes were identified from the peanut genome, which were divided into six classes with OLQ and WRC domains. AhGRF genes were at higher expression during pod development in Arachis monticola than H8107. Exogenous GA3 can activate AhGRF5a and AhGRF5b genes, and cultispecies showed more positive response than tetraploid wild species.
Supplementary Materials
The following are available online at https://www.mdpi.com/1422-0067/20/17/4120/s1, Figure S1. Distribution of AhGRF genes in the cultivated peanut genome. The chromosomal position of each AhGRF genes was mapped to the cultivated peanut genome. The chromosome number is indicated above each chromosome. Figure S2. Sequence alignment of WRC and QLQ motifs in AhGRF genes. Figure S3. Morphological changes in three peanut materials during seed development. A.mon (Arachis monticola tetraploid wild species), H8106 and 8107 lines indicate the two peanut recombinant inbred lines (RILs). 15~DAF, 25~DAF, 35~DAF, 45~DAF, and 60~DAF indicate about 15, 25, 35, 45, and 60 days after flowering, respectively. Table S1. Summary of the peanut AhGRF gene family. Table S2. Putative motifs conserved in the amino acid sequences of peanut AhGRF proteins. Table S3. Amino acid sequences of 68 GRF proteins from 4 plant species. Table S4. Expression levels of AhGRF genes in three peanut varieties. Table S5. Summary of the AhGRF genes family in Arachis duranensis and Arachis ipaensis. Table S6. Primers used for qRT-PCR analysis. Table S7. The sequences of AhGRF5a and AhGRF5b. Table S8. The original Real-time quantitative PCR data of AhGRF5a and AhGRF5b.
Author Contributions
D.Y. designed and conceived the research. K.Z. wrote the manuscript and analyzed the data. K.Z., K.L., L.N. and H.J. performed the experiments. X.M., and D.Y. revised the paper. All authors read and approved the manuscript.
Funding
This work was supported by grants from the Key Program of National Natural Science Foundation of China (NSFC)-Henan United Fund (No. U1704232), Key Scientific and Technological Project of Henan Province (No. 161100111000; S2012–05-G03), Innovation Scientists and Technicians Troop Construction Projects of Henan Province (No. 2018JR0001).
Conflicts of Interest
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study, in the collection, analysis, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Jeong H.K., Hirokazu T. Regulation of plant growth and development by the growth-regulating factor and grf-interacting factor duo. J. Exp. Bot. 2015;66:6093–6107. doi: 10.1093/jxb/erv349. [DOI] [PubMed] [Google Scholar]
- 2.Knaap E.V.D., Kim H., Kende H. A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. J. Plant Physiol. 2000;122:695–704. doi: 10.1104/pp.122.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim J.H., Choi D., Kende H. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in arabidopsis. Plant J. 2003;36:94–104. doi: 10.1046/j.1365-313X.2003.01862.x. [DOI] [PubMed] [Google Scholar]
- 4.Choi D., Kim J.H., Kende H. Whole genome analysis of the OsGRF gene family encoding plant-specific putative transcription activators in rice (Oryza sativa L.) J. Plant Cell Physiol. 2004;45:897–904. doi: 10.1093/pcp/pch098. [DOI] [PubMed] [Google Scholar]
- 5.Zhang D.F., Li B., Jia G.Q., Zhang T.F., Dai J.R., Li J.S., Wang S.C. Isolation and characterization of genes encoding GRF transcription factors and GIF transcriptional coactivators in maize (Zea mays L.) J. Plant Sci. 2008;175:809–817. doi: 10.1016/j.plantsci.2008.08.002. [DOI] [Google Scholar]
- 6.Ma J.Q., Jian H.J., Yang B., Lu K., Zhang A.X., Liu P., Li J.N. Genome-wide analysis and expression profiling of the GRF gene family in oilseed rape (Brassica napus, L.) J. Gene. 2017;620:36–45. doi: 10.1016/j.gene.2017.03.030. [DOI] [PubMed] [Google Scholar]
- 7.Zhou Y., Ge L.L., Li G.H., Jiang L.W., Yang Y.G. Characterization and expression analysis of growth regulating factor (GRF) family genes in cucumber. J. Arch. Biol. Sci. 2018;70:629–637. doi: 10.2298/ABS180414024Z. [DOI] [Google Scholar]
- 8.Omidbakhshfard M.A., Proost S., Fujikura U., Roeber B. Growth-regulating factors (GRFs): A small transcription factor family with important functions in plant biology. J. Mol. Plant. 2015;8:998–1010. doi: 10.1016/j.molp.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 9.Ertuğrul F.L.Z., İbrAhim K., Hüseyin T. Genome-wide identification and analysis of growth regulating factor genes in Brachypodium distachyon: In silico approaches. J. Turk. J. Biol. 2017;38:296–306. [Google Scholar]
- 10.Hewezi T., Maier T.R., Nettleton D., Baum T.J. The Arabidopsis microRNA396-GRF1/GRF3 regulatory module acts as a developmental regulator in the reprogramming of root cells during cyst nematode infection. J. Plant Physiol. 2012;159:321–335. doi: 10.1104/pp.112.193649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J., Hua W., Yang H.L., Zhan G.M., Li R.J., Deng L.B., Wang X.F., Liu G.H., Wang H.Z. The BnGRF2 gene (GRF2-like gene from Brassica napus) enhances seed oil production through regulating cell number and plant photosynthesis. J. Exp. Bot. 2012;63:3727–3740. doi: 10.1093/jxb/ers066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang G., He H., Li Y., Wang F., Yu D.Y. Molecular Mechanism of microRNA396 Mediating Pistil Development in Arabidopsis. J. Plant Physiol. 2013;164:249–258. doi: 10.1104/pp.113.225144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vercruyssen L., Tognetti V.B., Gonzalez N., Van D.J., De M.L., Bielach A., De R.R., Van B.F., Inzé D. Growth Regulating Factor 5 stimulates Arabidopsis chloroplast division, photosynthesis, and leaf longevity. J. Plant Physiol. 2015;167:817–832. doi: 10.1104/pp.114.256180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bao M., Bian H., Zha Y., Li F., Sun Y., Bai B., Chen Z., Wang J., Zhu M., Han N. MiR396a-Mediated basic helix-loop-helix transcription factor bHLH74 Repression Acts as a Regulator for Root Growth in Arabidopsis Seedlings. J. Plant Cell Physiol. 2014;55:1343–1353. doi: 10.1093/pcp/pcu058. [DOI] [PubMed] [Google Scholar]
- 15.Lee S.J., Lee B.H., Jung J.H., Park S.K., Song J.T., Kim J.H. Growth-regulating factor and grf-interacting factor specify meristematic cells of gynoecia and anthers. J. Plant Physiol. 2018;176:717–729. doi: 10.1104/pp.17.00960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horiguchi G., Kim G.T., Tsukaya H. The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 2005;43:68–78. doi: 10.1111/j.1365-313X.2005.02429.x. [DOI] [PubMed] [Google Scholar]
- 17.Omidbakhshfard M.A., Fujikura U., Olas J.J., Xue G.P., Balazadeh S., Mueller-Roeber B. Growth-Regulating Factor 9 negatively regulates arabidopsis leaf growth by controlling ORG3 and restricting cell proliferation in leaf primordia. J. PLoS Genet. 2018;14:1007484. doi: 10.1371/journal.pgen.1007484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelissen H., Eechout D., Demuynck K., Persiau G., Walton A., van Bel M., Vervoort M., Candaele J., De Block J., De Jaeger G. Dynamic changes in ANGUSTIFOLIA3 complex composition reveal a growth regulatory mechanism in the maize leaf. Plant Cell. 2015;27:1605–1619. doi: 10.1105/tpc.15.00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu L., Zhang D., Xue M., Qian J., He Y., Wang S. Overexpression of the maize GRF10, an endogenous truncated growth-regulating factor protein, leads to reduction in leaf size and plant height. J. Integr. Plant Biol. 2014;56:1053–1063. doi: 10.1111/jipb.12220. [DOI] [PubMed] [Google Scholar]
- 20.Kim J.H., Lee B.H. Growth-regulating factor4ofarabidopsis thalianais required for development of leaves, cotyledons, and shoot apical meristem. J. Plant Biol. 2006;49:463–468. doi: 10.1007/BF03031127. [DOI] [Google Scholar]
- 21.Daele I.V., Gonzalez N., Vercauteren I., Smet L.D., Inzé D., Roldan-Ruiz I., Vuylsteke M. A comparative study of seed yield parameters in Arabidopsis thaliana mutants and transgenics. Plant Biotechnol. J. 2012;10:488–500. doi: 10.1111/j.1467-7652.2012.00687.x. [DOI] [PubMed] [Google Scholar]
- 22.Sun P., Zhang W., Wang Y., He Q., Shu F., Liu H., Wang J., Wang J., Yuan L., Deng H. OsGRF4 controls grain shape, panicle length and seed shattering in rice. J. Integr. Plant Biol. 2016;58:836–847. doi: 10.1111/jipb.12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duan P., Ni S., Wang J., Zhang B., Xu R., Wang Y., Chen H., Zhu X., Li Y. Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. J. Nat. Plants. 2015:203. doi: 10.1038/nplants.2015.203. [DOI] [PubMed] [Google Scholar]
- 24.Cao D., Wang J., Ju Z., Liu Q., Li S., Tian H., Fu D., Zhu H., Luo Y., Zhu B. Regulations on growth and development in tomato cotyledon, flower and fruit via destruction of miR396 with short tandem target mimic. J. Plant Sci. 2016;247:1–12. doi: 10.1016/j.plantsci.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 25.Jones-Rhoades M.W., Bartel D.P. Computational Identification of Plant MicroRNAs and Their Targets, Including a Stress-Induced miRNA. Mol. Cell. 2004;214:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 26.Dhillon S.S., Rake A.V., Miksche J. Reassociation kinetics and cytophotometric characterization of peanut (Arachis hypogaea L.) DNA. J. Plant Physiol. 1980;65:1121–1127. doi: 10.1104/pp.65.6.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bertioli D.J., Cannon S.B., Froenicke L., Huang G., Farmer A.D., Cannon E.K., Liu X., Gao D., Clevenger J., Ozias-Akins P. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 2016;48:438–446. doi: 10.1038/ng.3517. [DOI] [PubMed] [Google Scholar]
- 28.Bertioli D.J., Jenkins J., Clevenger J., Dudchenko O., Gao D., Seijo G., Bertioli D.J., Jenkins J., Clevenger J., Schmutz J. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 2019;51:877–884. doi: 10.1038/s41588-019-0405-z. [DOI] [PubMed] [Google Scholar]
- 29.Yin D.M., Ji C.M., Ma X.L., Li H., Zhang W.K., Li S., Liu F.Y., Zhao K.K., Li F.P., Li K., et al. Genome of an allotetraploid wild peanut Arachis monticola: A de novo assembly. GigaScience. 2018;7 doi: 10.1093/gigascience/giy066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhuang W.J., Chen H., Yang M., Wang J., Pandey M.K., Zhang C., Chang W.C., Zhang L., Zhang X., Varshney R.K. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 2019;51:865–876. doi: 10.1038/s41588-019-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin D.M., Hu X.F., Zhang X.G., Sun J.J., Wang Y., Cui D.Q. Temporal and spatial expression of oleate desaturase gene with different O/L values in peanut varieties. Chin. J. Oil Crop Sci. 2013;35:137–141. [Google Scholar]
- 32.Wang Y., Zhang X.G., Zhao M.Y., Prakash C.S., He G., Yin D. Insights into the novel members of the FAD2 gene family involved in high-oleate fluxes in peanut. J. Genome. 2015;58:375–383. doi: 10.1139/gen-2015-0008. [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez R.E., Mecchia M.A., Debernardi J.M., Schommer C., Weigel D., Palatnik J.F. Control of cell proliferation in Arabidopsis thaliana by microrna mir396. J. Dev. 2010;137:103–112. doi: 10.1242/dev.043067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Debernardi J.M., Mecchia M.A., Vercruyssen L., Smaczniak C., Kaufmann K., Inze D., Rodriguez R.E., Palatnik J.F. Post-transcriptional control of\r, GRF\r, transcription factors by microRNA miR396 and GIF co-activator affects leaf size and longevity. Plant J. 2014;79:413–426. doi: 10.1111/tpj.12567. [DOI] [PubMed] [Google Scholar]
- 35.Che R., Tong H., Shi B., Liu Y., Fang S., Liu D., Xiao Y., Hu B., Liu L., Chu C. Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat Plants. 2015;2:15195. doi: 10.1038/nplants.2015.195. [DOI] [PubMed] [Google Scholar]
- 36.Li S., Gao F., Xie K., Zeng X., Cao Y., Zeng J., He Z., Ren Y., Li W., Deng Q., et al. The OsmiR396c-OsGRF4-OsGIF1 regulatory module determines grain size and yield in Rice. Plant Biotechnol. J. 2016;14:2134–2146. doi: 10.1111/pbi.12569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma X.L., Zhang X.G., Zhao K.K., Li F.P., Li K., Ning L.L., He J.L., Xin Z.Y., Yin D.M. Small RNA and Degradome Deep Sequencing Reveals the Roles of microRNAs in Seed Expansion in Peanut (Arachis hypogaea L.) Front. Plant Sci. 2018;9:349. doi: 10.3389/fpls.2018.00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuijt S.J.H., Greco R., Agalou A., Shao J., Hoen C.C., Overnäs E., Osnato M., Curiale S., Meynard D., Ouwerkerk P.B. Interaction between the GROWTH-REGULATING FACTOR and KNOTTED1-LIKE HOMEOBOX Families of Transcription Factors. Plant Physiol. 2014;164:1952–1966. doi: 10.1104/pp.113.222836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang F., Qiu N., Ding Q., Li J., Zhang Y., Li H., Cao J. Genome-wide identification and analysis of the growth-regulating factor family in Chinese cabbage (Brassica rapa L. ssp. pekinensis) BMC Genom. 2014;15:807. doi: 10.1186/1471-2164-15-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang J., Li Z., Jin J., Xie X., Zhang H., Chen Q., Luo Z., Yang J. Genome-wide identification and analysis of the growth-regulating factor family in tobacco (Nicotiana tabacum) Gene. 2017;639:117–127. doi: 10.1016/j.gene.2017.09.070. [DOI] [PubMed] [Google Scholar]
- 41.Sparkes I., Runions J., Kearns A., Hawes C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006;1:2019–2025. doi: 10.1038/nprot.2006.286. [DOI] [PubMed] [Google Scholar]
- 42.Zhang W., Wang S., Yu F., Tang J., Shan X., Bao K., Yu L., Wang H., Fei Z.J., Li J.B. Genome-wide characterization and expression profiling of SWEET genes in cabbage (Brassica oleracea var. capitata L.) reveal their roles in chilling and clubroot disease responses. J. BMC Genom. 2019;20:1471–2164. doi: 10.1186/s12864-019-5454-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.