Abstract
Enzymes in the cytochrome P450 4 (CYP4) family are involved in the metabolism of fatty acids, xenobiotics, therapeutic drugs, and signaling molecules, including eicosanoids, leukotrienes, and prostanoids. As CYP4 enzymes play a role in the maintenance of fatty acids and fatty-acid-derived bioactive molecules within a normal range, they have been implicated in various biological functions, including inflammation, skin barrier, eye function, cardiovascular health, and cancer. Numerous studies have indicated that genetic variants of CYP4 genes cause inter-individual variations in metabolism and disease susceptibility. Genetic variants of CYP4A11, 4F2 genes are associated with cardiovascular diseases. Mutations of CYP4B1, CYP4Z1, and other CYP4 genes that generate 20-HETE are a potential risk for cancer. CYP4V2 gene variants are associated with ocular disease, while those of CYP4F22 are linked to skin disease and CYP4F3B is associated with the inflammatory response. The present study comprehensively collected research to provide an updated view of the molecular functionality of CYP4 genes and their associations with human diseases. Functional analysis of CYP4 genes with clinical implications is necessary to understand inter-individual variations in disease susceptibility and for the development of alternative treatment strategies.
Keywords: CYP4 genes, genetic polymorphisms, 20-HETE, fatty acid, arachidonic acid, SNPs, molecular functionality, metabolism, lamellar ichthyosis, Bietti’s crystalline dystrophy
1. Introduction
Cytochrome P450s (CYPs) are a superfamily of enzymes located either in the inner membrane of mitochondria or in the endoplasmic reticulum membrane of eukaryotic cells. There are 57 CYP proteins encoded in the human genome, which are responsible for the metabolism of numerous endogenous and exogenous compounds [1,2,3]. CYPs mainly oxidize these compounds to generate more hydrophilic metabolites, enhancing their excretion outside the body and thus playing a major role in the detoxification of toxic chemicals [1]. Generally, CYP families 1, 2, and 3 include major xenobiotic-metabolizing enzymes responsible for their major roles in pharmacogenomics risk, while CYP4 enzymes are involved in the metabolism of fatty acids, with their close links to genetic disease risk. Fatty acid metabolism by CYP4 enzymes is responsible for the elimination of excess free fatty acids from the body, as well as for the synthesis of proper levels of bioactive fatty acid molecules [4]. The present review focused on the CYP4 family of enzymes in terms of their functional roles, genetic variations, and influences on human diseases.
2. Classification and Tissue Distribution of the CYP4 Family
Although there are more than 11 subfamilies of CYP4 in different species, only 6 subfamilies of CYP4 genes have been reported in humans. The human CYP4 subfamilies are CYP4A, B, F, V, X, and Z [2]. Seven CYP4F isoforms, CYP4F2, CYP4F3A, CYP4F3B, CYP4F8, CYP4F11, CYP4F12, and CYP4F22, are clustered on chromosome 19 and are encoded by six genes. CYP4A isoforms include CYP4A11 and CYP4A22 on chromosome 1 [3], and the remaining CYP4 subfamily genes are CYP4B1, CYP4V2, CYP4X1, and CYP4Z1 [4]. The major sites of CYP4A11 expression are the liver and kidney [5]. However, Jarrar et al. found that CYP4A11 protein was also highly expressed in human platelets to a similar level as in the human liver [6]. CYP4A22 expression has been reported in the human liver at very low levels, with poor enzyme activity compared to that of CYP4A11 [7]. Among the seven CYP4F genes, CYP4F2, CYP4F3B, CYP4F11, and CYP4F12 are mainly expressed in the liver and kidney [8,9,10,11,12]. However, their relative contributions to the total amount of CYP4 enzymes in tissues are difficult to determine, as the high structural homology of these four enzymes has hampered the production of specific antibodies for the detection of each enzyme. In addition to the lack of specific antibodies, genetic polymorphisms and differing profiles of up- and downregulation among CYP4 enzymes have further complicated determination of the intrinsic amount of each enzyme in tissues. Currently, mass spectrometry is used to detect target proteins through measurement of specific peptides of the target protein [13,14]. The total amount of CYP4F protein in human liver was estimated as 18–128 pmol/mg liver microsomal protein [15]. One of the most abundant P450s, CYP3A4, was estimated at 64 pmol/mg liver microsomal protein [13], indicating that the contribution of CYP4F to the total P450 level is large. CYP4F3A is expressed in neutrophils and plays a major role in inflammation [16]. CYP4F8 is expressed in the prostate and seminal vesicles [17]. CYP4F22 is expressed in human skin and plays a major role in formation of the skin lipid barrier [18]. CYP4V2 is widely expressed in the liver and ophthalmic tissues and CYP4V2 defect has been linked to ophthalmic diseases, such as Bietti’s crystalline dystrophy [19]. CYP4B1 is expressed mainly in the lung and bladder tissues, and in smaller amounts in the liver [20]. CYP4X1 is expressed in the brain and bronchial airways [21], while CYP4Z1 is expressed in mammary tissue; these proteins are also overexpressed in cancer compared to normal cells [22]. Expression levels of CYP4 proteins are summarized in Table 1.
Table 1.
Substrates and major expression tissues of cytochrome P450 4 (CYP4) enzymes in humans.
| CYP4 Enzyme | Expression Tissues | Substrates | Reference |
|---|---|---|---|
| CYP4A11 | Platelets, liver, kidney | Lauric acid, myristic acid, arachidonic acid | [6,23,24] |
| CYP4A22 | Low level in liver | Arachidonic acid | [25,26] |
| CYP4B1 | Lung, bladder, fat tissues | 2-aminofluorene, 2-naphthylamine, benzidine, arachidonic acid | [27,28] |
| CYP4F2 | Liver, platelet, kidney | Arachidonic acid, lauric acid, vitamin K, leukotriene | [6,29] |
| CYP4F3A | Neutrophils, monocytes, eosinophils |
Leukotriene B4 | [12,30,31] |
| CYP4F3B | Liver, kidney, trachea, gastro intestinal tract |
Eicosapentaenoic acid, arachidonic acid | [12,32] |
| CYP4F8 | Prostate, seminal vesicles, epidermis, hair follicles, sweat glands, corneal epithelium, proximal renal tubules, epithelial linings of gut and urinary tract | Arachidonic acid, prostaglandin H, prostaglandin E2 | [33,34,35] |
| CYP4F11 | Liver, kidney, heart, skeletal muscle, gall bladder, keratinocytes | Vitamin K, erythromycin, arachidonic acid | [11,29,36,37,38] |
| CYP4F12 | Liver, kidney, colon, small intestine, heart, eosinophils, neutrophils | Arachidonic acid, leukotriene B4, ebastine | [8,31,33,39] |
| CYP4F22 | Skin | Ultra-long-chain fatty acid (acylceramide production) | [18] |
| CYP4X1 | Skin, breast, brain, heart, liver, prostate, trachea, aorta | Anandamide, arachidonic acid | [21,40] |
| CYP4V2 | Macrophages, retina cells, cornea cells | Arachidonic acid, lauric acid, eicosapentanoic acid, docosahexanoic acid | [19,41] |
| CYP4Z1 | Mammary tissues, cancer cells | Lauric acid, myristic acid | [21,42] |
3. Role of the CYP4 Family in the Metabolism of Endogenous Compounds
The CYP4 family plays a major role in the metabolism of fatty acids, in most cases through oxidation of fatty acids and subsequent catalysis in the mitochondria to produce cellular energy. CYP4B metabolizes short-chain fatty acids (approximately 7 to 10 carbon fatty acids) [20], while CYP4A and CYP4V metabolize intermediate-chain fatty acids (C10 to 16) [19] and CYP4F catalyzes long-chain fatty acids (C16 to 26), such as prostanoids [43]. Decreasing the expression levels of the CYP4 family was associated with accumulation of fats in tissues such as the liver [44]. Therefore, decreased levels of CYP4 family proteins reduce the capacity for fat removal from tissues. Jarrar et al. [44] found that non-steroidal anti-inflammatory drugs caused fatty livers in treated mice, which were associated with significant downregulation of mouse cyp4a12 gene expression in liver tissues. CYP4F2, 4F3B, 4A11, and 4V2 were found to oxidize arachidonic acid through ω-hydroxylation to 20-hydroxyeicosatetraenoic acid (20-HETE) [6,9,45,46], which is a vasoconstrictor and activator of platelet aggregation [47]. Several studies have reported that CYP4F and CYP4A are overexpressed in cardiovascular diseases, wherein they are correlated with 20-HETE production [48,49,50]. In addition, doxorubicin-induced cardiotoxicity was associated with increased 20-HETE production due to increased mRNA expression of rat CYP4A and CYP4F enzymes [51]. CYP4A11 and 4V2 oxidize saturated fatty acids such as lauric acid [23,41,52]. In addition to the metabolism of arachidonic acid and omega-3 polyunsaturated fatty acids, CYP4F2 has been reported to ω-hydroxylate leukotriene (LTA) 4 [53]. CYP4F3A in white blood cells catalyzes the ω-hydroxylation of leukotriene B4 to 20-hydroxy leukotriene B4, which is an important regulatory step of the inflammatory response [54]. Instead of ω-hydroxylation, CYP4F8 has been reported to hydroxylate prostaglandin (PG) E2 at position 19 [17]. Although epoxyeicosatrienoic acids (EETs) are synthesized by the CYP2C subfamily [55], they can be further ω-hydroxylated by CYP4 enzymes to 20-hydroxyepoxyeicosatrienoic acids (HEETs) [56].
4. Role of the CYP4 Family in the Metabolism of Drugs
The roles of most CYP4 family proteins in the metabolism of drugs and xenobiotic compounds appear to be minor compared to those of CYP1, 2, and 3. However, CYP4F2 metabolizes the ester prodrug of gemcitabine and the antiparasitic pafuramidine [57]. In addition, CYP4A11 exhibited metabolism of the immune suppressant tacrolimus to an inactive form [58]. Although the turnover rates were low compared to those of CYP3A4, CYP4F11 exhibited catalytic activity towards commonly used drugs such as erythromycin, benzphetamine, and chlorpromazine [36,37,59]. CYP4F12 has been reported to slowly metabolize the antihistamine ebastine [60] and the antifungal terfenadine [61]. CYP4 enzymes are indirectly involved in drug metabolism and drug responses. For example, CYP4F2 and CYP4F11 are involved in the metabolism of vitamin K, facilitating vitamin K inactivation and elimination [29,62]. The amount of active vitamin K is important for maintenance of warfarin dosing, as it is metabolized strongly by CYP2C9 [63,64], indicating that CYP4 enzymes are indirectly involved in warfarin dose maintenance. CYP4 enzymes show catalytic activity toward various fatty acids and their metabolites have the potential to act as ligands or activators of nuclear receptors, such as peroxisome proliferator-activated receptors (PPARs) [65,66]. Therefore, drugs targeting the activation or inactivation of PPARs may show altered pharmacokinetics or toxic responses [67,68]. Such indirect involvement may affect the drug response to conditions such as fatty liver diseases, diabetic diseases, and inflammatory diseases.
5. The CYP4 Family and Inflammation
CYP4 enzymes are involved in inflammation through the metabolism of inflammatory molecules. They metabolize inflammatory mediators such as leukotrienes (LTs) and also produce 20-HETE [53]. While CYP4F11 possesses lower affinity toward leukotriene B4 (LTB4), neutrophilic CYP4F3A has the highest affinity for LTB4 ω-hydroxylation [36]. CYP4F3A metabolizes LTB4 into the inactive form 20-hydroxy leukotriene B4, mediating a critical step in regulation of the inflammatory response. However, CYP4A11 has shown low activity toward LTB4 using in vitro methods [69]. CYP4F3B ω-hydroxylates omega-3 eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) to their 20-hydroxy and 22-hydroxy metabolites, respectively [32], which are lipid mediators that can activate inflammatory PPARs [70]. Studies have shown that hepatic and renal rat CYP4F genes were upregulated under inflammatory conditions following treatment with barium sulfate [71]. On the other hand, rat hepatic CYP4A mRNAs were downregulated in response to lipopolysaccharides used as a model of inflammation [72]. Human CYP4V2 was first identified in inflammatory cell macrophages, and its gene expression was reduced following selective treatment with a PPARγ agonist [41]. Depending on the clinical situation, ω-hydroxylase activity associated with the CYP4 family could be considered as a potential drug target for reducing the inflammatory response, providing a novel mechanism for future anti-inflammatory drugs.
6. The CYP4 Family and Cancers
Induction of CYP4 family members, including CYP4F2, CYP4F3, CYP4A11, and CYP4Z1, has been reported in various types of cancer [73,74]. Upregulation of CYPF2 and CYP4A11 was confirmed through Western blot assays in human thyroid, ovarian, breast, pancreatic, and colon cancer tissues [75]. CYP4Z1 is expressed in mammary tissue and upregulated in breast cancer tissue [74]. These findings suggest that ω-hydroxylase activity may be a biomarker of cancer prognosis. Evaluation of the CYP4 expression profile in hepatocellular carcinoma (HCC) showed that CYP4F2, CYP4F12, and CYP4V2 mRNA levels were negatively correlated with cell-cycle-associated genes, suggesting that these CYP4 genes are favorable prognostic factors in HCC [76]. In addition, expression of CYP4 has been reported to be associated with angiogenesis through production of 20-HETE, which activates vascular endothelial receptors in arteries and thus increases blood supply to cancer cells [77]. Among CYP4 enzymes, CYP4F3B, CYP4A11, and CYP4F2 are major enzymes involved in the generation of 20-HETE, which plays an important role in tumor progression and angiogenesis. Therefore, their tissue expression and omega-hydroxylase activity levels play roles in cancer progression. CYP4B1 metabolizes several protoxic xenobiotics, including 2-aminofluorine, 2-naphthylamine, 4-ipomeanol, and benzidine [78,79,80,81]. Therefore, CYP4B1 involvement in cancers has been suggested based on its expression levels and metabolism of pro-carcinogens in the bladder and lung [78,82]. CYP4B1 may play a role in detoxification or activation in tissues. Sasaki et al. reported that the individuals carrying the CYP4B1*2 allele have an increased risk of bladder cancer [27]. However, it has also been reported that there is no association between the CYP4B1 genotype and the risk of lung cancer in the Japanese population [83]. Downregulation of CYP4B1 proteins represented an unfavorable indicator in patients with urothelial carcinomas of the upper urinary tract and bladder, indicating a protective role of CYP4B1 in patients with urotherial carcinomas [84]. Involvement of CYP4Z1 in breast cancer has been suggested, as it was identified in breast tissue and upregulated in breast carcinoma [74,76]. Therefore, CYP4Z1 was proposed as a biomarker for malignancy and/or progression of ovarian and prostate cancer [85]. It was reported that breast cancer cells exhibited the abnormal translocation of CYP4Z1 protein to the plasma membrane instead of targeting to the intracellular membrane of the endoplasmic reticulum, which caused the CYP4Z1 autoantibody production that might serve as a biomarker for the diagnosis [86]. Expression of CYP4Z1 has been reported to promote angiogenesis and tumor growth by increasing 20-HETE synthesis [74]. However, a recent functional study of CYP4Z1 in a recombinant enzyme system indicated that 20-HETE was not detected in the CYP4Z1 reaction with arachidonic acid, and suggested that CYP4Z1 may modulate breast cancer without direct 20-HETE synthesis [87]. Further studies are needed to clarify the roles of CYP4Z1 in carcinogenesis in various tissues.
7. The CYP4 Family and Cardiovascular Diseases
Several studies have shown that CYP4 family genes are associated with cardiovascular diseases, including hypertension and myocardial infarction, through the production of 20-HETE or perturbation of fatty acid metabolism [88,89]. Multiple aspects of the mechanism underlying the effect of 20-HETE on the cardiovascular complex have been reported. In a metabolomics study in mice, increased 20-HETE levels in the blood (>120-fold) with chronic rofecoxib treatment were associated with reduced bleeding time and increased platelet aggregation [47]. Additionally, 20-HETE has been suggested to mediate androgen-induced hypertension through increasing the level of Cyp4a12 in a mouse study [90], wherein the increased level of Cyp4a12 produced more eicosanoids, which were predicted to mediate androgen-induced hypertension. In the kidney, however, 20-HETE exerts anti-hypertensive effects through inhibition of sodium reabsorption in the proximal tubule and thick ascending limb of Henle [91]. Furthermore, 20-HETE was found to act as a vasoconstrictor of vascular smooth muscle cells by allowing increased calcium entry into cells and enhanced phosphorylation of contractile elements [92,93,94]. Several studies have suggested interplay between 20-HETE and the renin–angiotensin aldosterone system (RAAS) in hypertension. Briefly, angiotensinogen II has been reported to increase renal production of 20-HETE [95], and 20-HETE can activate the RAAS by inducing angiotensin-converting enzyme [96,97]. Further investigations are needed to fully elucidate the mechanistic link between 20-HETE and the RAAS in humans. Rat CYP4A was downregulated in the kidney of hypertensive rats, which was associated with reduced formation of 20-HETE in the kidney and reduction of the diuretic effect [98]. CYP4A was upregulated in studies of doxorubicin-induced cardiotoxicity, where it was associated with myocardial infarction and increased 20-HETE synthesis [51]. Furthermore, Jarrar et al. found that heart cyp4a12 was highly upregulated in mice after cardiac toxicity induced by non-steroidal anti-inflammatory drugs [44]. Thus, targeting of 20-HETE synthesis or modulation of eicosanoid levels through manipulation of CYP4 enzymes can decrease the cardiotoxicity of such drugs. This application should be considered in future development of the drug for cardiovascular health care.
8. Role of the CYP4 Family in Other Diseases
Bietti’s crystalline dystrophy (BCD) is an autosomal recessive disease characterized by the presence of numerous small, yellow or white crystal-like deposits of fatty compounds in the light-sensitive retina tissue [52,99,100]. These deposits damage the retina, resulting in progressive atrophy of the retinal pigment epithelium and progressive vision loss at approximately 40 or 50 years of age [101,102]. The occurrence of BCD is more common in East Asian populations than other ethnic groups [103,104]. BCD is caused by mutations in the CYP4V2 gene, which is comprised of 11 exons encoding a 525 amino acid protein on chromosome 4 [99,105,106]. CYP4V2 is known to metabolize fatty acids, and thus CYP4V2 in the retina is most likely involved in the breakdown and elimination of fatty acids from the retina [52]. Impaired CYP4V2 function due to genetic mutations may affect lipid metabolism and elimination from the retina. The severity and progression of BCD symptoms varies widely among patients. These variations may be influenced by differing levels of defectiveness in CYP4V2 function caused by mutations of different severities. Various mutations in CYP4V2 have been found, including stop codon creation, an amino acid change in an important region, destruction of a splice site, and a frameshift in the CYP4V2 protein-coding cDNA. More than 60 mutations of the CYP4V2 gene have been reported in BCD patients [99,103,105,107,108,109,110,111,112]. A number of mutations of CYP4V2 have significant impacts on CYP4V2 activity. The most common mutation in BCD is an insertion–deletion mutation at the end of intron 6 and the beginning of exon 7 (IVS6-8del17insGC, c.802-8del17/insGC) [103,105,106,108,109,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125]. This mutation causes the deletion of exon 7 in the CYP4V2 protein, resulting in a major structural change and the complete loss of CYP4V2 activity.
Type 3 lamellar ichthyosis, a skin keratinization disease, was found to be caused by genetic mutation of CYP4F22 [126]. Since the discovery that CYP4F22 is one of the causative genes for ichthyosis, the molecular mechanisms underlying the role of CYP4F22 in the etiology of ichthyosis have remained largely unknown until recently. Acylceramide is an important lipid of the skin permeability barrier, and patients with ichthyosis show strongly repressed acylceramide production [127,128,129,130]. Ohno et al. (2015) reported that CYP4F22 is responsible for the generation of acylceramide through ω-hydroxylation of long-chain fatty acids [18]. Recently, a CYP4F22 genetic variant associated with lamellar ichthyosis was reported in a Tunisian family [131]. A missense mutation in exon 8, CYP4F22 Arg243Leu, was suggested to be linked to lamellar ichthyosis and predicted to be a functionally defective variant based on in silico analysis. Genetic screening for CYP4F22 mutations associated with lamellar ichthyosis should be extended in future works.
9. Genetic Variants of the CYP4 Family
9.1. Genetic Variants of CYP4B1
The first screening study for genetic polymorphism of CYP4B1 was performed in French Caucasians and identified the new CYP4B1 alleles CYP4B1*2, *3, *4, and *5 based on the P450 Nomenclature Committee [132]. Among them, CYP4B1*2 caused a frameshift and premature stop codon, resulting in complete loss of CYP4B1 function. Two more alleles with frequencies <1%, CYP4B1*6 and CYP4B1*7, were identified using a denaturing high-performance liquid chromatography method for 192 Japanese individuals [133]. Since CYP4B1 is involved in the metabolism of pro-carcinogens, its association with bladder cancer was investigated in a Japanese population, and subjects carrying the CYP4B1*1/*2 or CYP4B1*2/*2 genotypes exhibited a 1.75-fold increased risk of bladder cancer [27]. This finding might be explained as the loss of function allele CYP4B1*2 providing lower capacity for activation of carcinogenic compounds. However, a lung cancer risk study of CYP4B1*1–*7 showed no association with lung cancer in a Japanese population [83]. Further studies are needed to determine its association with lung cancer using a large cohort. Study of structure–function relationships has been essential to understanding the efficiency of catalytic activity as well as to explaining the varying degrees of molecular defectiveness of the protein mutants. Investigation of local peptide structures on the CYP4B1 protein and their roles in heme stability with catalytic function has been reported [134,135,136], and these data will be important to understand inter-individual variations in the activity of CYP4B1 coding variants.
9.2. Genetic Variants of CYP4A11, CYP4F2, 4F11, and CYP4F22
Among CYP4 family genes, CYP4A11 and CYP4F2 have been extensively studied in association with warfarin dosage and the cardiovascular complex. Genetic variants of CYP4F2 and CYP4A11 genes are reportedly associated with cardiovascular diseases such as hypertension [137,138,139]. More than 3400 single nucleotide polymorphisms (SNPs) of human CYP4A11 and 5900 SNPs of the CYP4F2 gene have been reported in the NCBI database to date. However, only a small number of the SNPs have been shown to have clinical associations with functional changes. One of the most extensively studied SNPs of CYP4A11 is a variant of rs1126742 that causes an amino acid change of Phe434 to Ser, leading to reduced 20-HETE synthesis from arachidonic acid [140,141]. Since the discovery of the functional role of CYP4A11 in the synthesis of 20-HETE, the association of CYP4A11 polymorphisms with cardiovascular risk has been studied extensively in humans [142,143,144,145,146,147,148]. The US Food and Drug Administration recommends genotyping of CYP4F2 variants for determination of warfarin doses [149,150]. The CYP4F2 genetic variant rs2108622 is a non-synonymous variant that causes a change in the amino acid sequence of valine to methionine and exhibits reduced enzymatic activity toward the metabolism of vitamin K [62]. Since individuals with reduced activity of CYP4F2 for vitamin K inactivation may have higher levels of warfarin than individuals with CYP4F2*1/*1, higher maintenance dosages of warfarin have been recommended for individuals with reduced CYP4F2 alleles [149]. Many studies have attempted to develop an accurate warfarin dosing algorithm using multiple genes, such as CYP2C9, VKORC1, and CYP4F2 [151,152,153,154,155]. Studies regarding CYP4A22 genetic polymorphisms have been limited to certain populations, such as Japanese and French populations [25,156]. The association of CYP4A22 variants with human diseases has still not been investigated, which might be due to low expression levels of the CYP4A22 gene. The CYP4F3 gene undergoes alternative splicing to form the CYP4F3A and CYP4F3B enzymes, depending on the cell type [157]. Genome-wide investigation showed that the functional SNP CYP4F3 rs4646904 was associated with lung cancer, especially in smokers [30]. However, the functionality of this SNP in lung cancer pathology remains unidentified. In addition, a high intake of polyunsaturated fatty acids was associated with reduced risk of ulcerative colitis in patients with CYP4F3 rs4646904 GG/AG, but not those with the AA genotype [158]. Regarding the CYP4F11 gene, Yi et al. found through in vitro methods that CYP4F11 D315N protein showed approximately 50% and 32% decreases in intrinsic clearance of erythromycin and arachidonic acid, respectively, compared to the wild type [37]. The CYP4F11 variant (rs1060463) was associated with small bowel bleeding risk induced by aspirin [159]. Seven variants with amino acid changes in the CYP4F12 gene were identified and functional changes were investigated using ebastine as a substrate [160]. In their report, two coding variants, Val90Ile and Arg188Cys, exhibited significantly decreased activity toward ebastine hydroxylation. The intronic variant CYP4F12 rs11085971, which contains a nucleotide substitution of guanine to thymine, was identified as a candidate oxidative-stress-related genetic marker for the development of type 1 lesions in cerebral cavernous malformation, and could serve as an early objective predictor of disease outcome [161]. Since the discovery of CYP4F22 was linked to its association with lamellar ichthyosis [18], genetic studies of CYP4F22 polymorphisms have been undertaken. A CYP4F22 variant, CYP4F22 Arg243Leu, was associated with lamellar ichthyosis in a Tunisian family [131], and further genetic studies should be conducted in clinical settings.
9.3. Genetic Variants of Other CYP4 Genes
Genetic polymorphism studies of CYP4V2 with respect to BCD are described above. In addition to BCD, genome-wide analysis found that a CYP4V2 genetic variant was strongly associated with deep vein thrombosis [162], which was confirmed later in multiple studies [163,164]. Association of the genetic variant 7234C>A (rs13146272) on exon 6 of the CYP4V2 gene with the risk of deep venous thrombosis and tamoxifen-induced venous thrombosis has been reported [165]. The exact mechanism through which the CYP4V2 gene defect increases the risk of deep vein thrombosis remains poorly understood. This genetic variant substitutes polar glutamine with basic lysine at position 259 of the CYP4V2 amino acid sequence, which may influence its activity. Genetic studies of 4Z1 and 4X1 are scarce, as these genes were recently identified and their physiological roles remain unclear. CYP4X1 was found to convert the endocannabinoid anandamide, an important signaling molecule in the neurovascular cascade, into a single monooxygenated product (14,15-epoxyeicosatrienoic ethanolamide), suggesting a potential role in brain signaling [40]. High levels of mRNA expression of CYP4X1 were found in the skin, brain, heart, liver, prostate, and breast [40] and CYP4Z1 mRNA was preferentially expressed in mammary tissue [21]. Functional analysis of CYP4Z1 indicated that CYP4Z1 has catalytic activity toward lauric and arachidonic acids, but 20-HETE was not detected in arachidonic acid metabolism [87]. Major genetic polymorphisms in CYP4 genes with clinical impact were summarized in Table 2.
Table 2.
Representatives of genetic polymorphisms in CYP4 genes with clinical impact and their frequencies in different ethnic groups.
| Gene | SNP | Location | Mutation | Effect | Frequency a | Functional Effect | ||
|---|---|---|---|---|---|---|---|---|
| European | African | Asian | ||||||
| CYP4A11 | rs1126742 | Exon | A > G | Phe330Ser | 0.15 | 0.36 | 0.25 | It was associated with hypertension in white individuals, most probably through decreased production of 20-HETE in the kidney [137]. |
| CYP4F2 | rs2108622 | Exon | C > T | Val433Met | 0.27 | 0.06 | 0.26 | It reduced the metabolism of vitamin K. Therefore, patients carrying this genetic variant needed a higher dose of warfarin, in order to keep the targeted anticoagulant effect [149,150,151]. |
| CYP4F3 | rs4646904 | Exon | A > G | Val358Val | 0.65 | 0.35 | 0.34 | It was associated with lung cancer, especially in smokers [30] and ulcerative colitis [158]. |
| CYP4F11 | rs200033002 | Exon | C > T | Asp315Asn | 0 | 0 | 0.01 | It decreased the metabolism of erythromycin and arachidonic acid compared to the wild type in vitro [37]. |
| CYP4B1 | rs3215983 | Frameshift variant | AT881–882del | Produces premature stop codon | 0.15 | ND | 0.33 | It was reported to increase the risk of bladder cancer, because it has lower capacity to metabolize the carcinogenic compounds [27]. |
| CYP4V2 | rs13146272 | Exon | C > A | Gln259Lys | 0.36 | 0.4 | 0.6 | It was associated with the risk of deep venous thrombosis and tamoxifen-induced venous thrombosis [162,163,164,165]. |
| CYP4V2 | rs199476197 | Exon | A > C | His331Pro | 0 | 0 | 0.0004 | It decreased CYP4V2 protein expression and activity toward fatty acid metabolism. Therefore, this genetic variant may cause accumulation of fatty acids in the retina [19,166]. |
| CYP4V2 | IVS6-8del17insGC | Intron 6, exon 7 | Insertion/deletion | Exon7 del | ND | ND | ND | It causes deletion of exon 7 in the CYP4V2 protein, resulting in a complete loss of CYP4V2 activity. It is the most common mutations in BCD patients [52,99]. |
a Data regarding the frequency of genetic variants among different ethnic groups were obtained from Ensemble database. ND, not determined. BCD, Bietti’s crystalline dystrophy.
10. Linkage Disequilibrium among CYP4 Genes
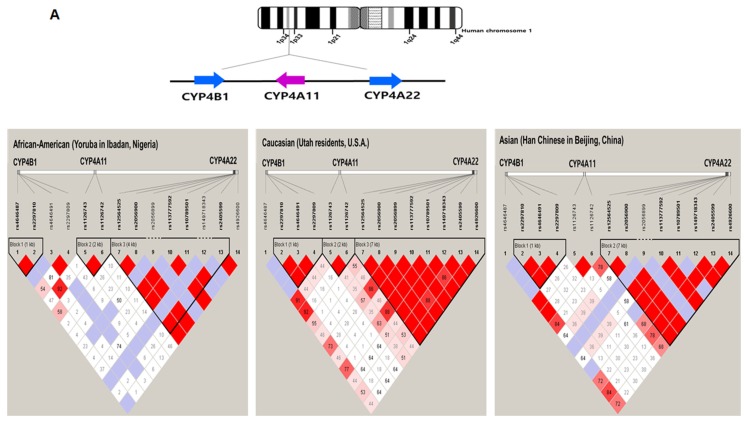
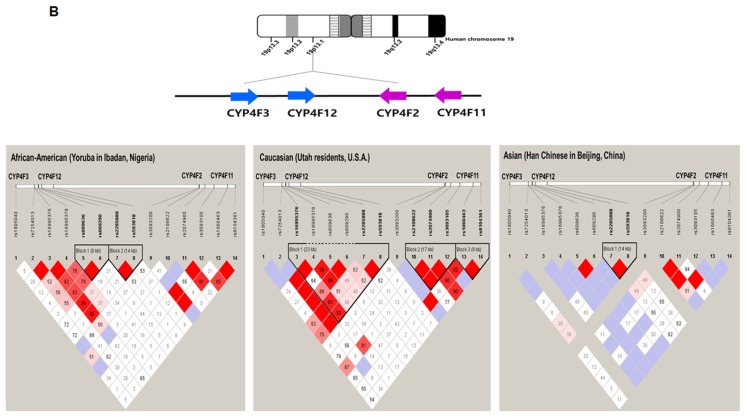
Five CYP4 genes, CYP4A22, CYP4A11, CYP4B1, CYP4X1, and CYP4Z1, are located on chromosome 1 [4]. A number of studies based on next generation sequencing tools and a 1000-genome project have identified SNPs in these genes. However, their functional roles, clinical relationships, and linkage disequilibrium (LD) are poorly characterized. From the 1000-genome database, a total of 14 coding SNPs with > 5% global minimum allele frequency were identified for CYP4A22, CYP4A11, and CYP4B1, and this LD block was analyzed (Figure 1A). Ethnically distinct populations exhibited differing LD blocks and haplotype structures. No strong LD was found among these three CYP4 genes that are clustered on chromosome 1. Six CYP4F genes, including CYP4F2, CYP4F3, CYP4F8, CYP5F11, CYP4F12, and CYP4F22, are located on the same chromatid of chromosome 19 [4]. Using the same method, coding variants with > 5% global frequency were selected from a 1000-genome database and their haplotypes and LD were analyzed (Figure 1B). As illustrated in Figure 1A, ethnically distinct groups showed differing frequencies and LD structures. An LD block covering more than one CYP4 gene was not observed for CYP4F genes in coding variant analysis. Instead, a strong linkage was found between CYP4F2 (rs2074900) and CYP4F11 (rs8104361) in a Western European population. Since CYP4 genes on the same chromosome with highly similar DNA structures can act as a linkage unit or as independent genes, further linkage analysis using more validated SNPs over all regions of CYP4 genes is needed to improve the current knowledge of CYP4 genetics.
Figure 1.
Linkage disequilibrium (LD) plots of CYP4 genetic variants in African, Caucasian, and Asian populations. Populations in Yoruba, Utah, and Beijing represent African, Caucasian, and Asian populations, respectively. The coding single nucleotide polymorphisms (SNPs) with a minor allele frequency (MAF) of 0.05 or greater in the 1000 genome data base were selected to avoid estimation errors in linkage analysis. (A) LD structures of CYP4A11, CYP4A22, and CYP4B1 with common coding SNPs. CYP4A11, CYP4A22, and CYP4B1 are clustered on chromosome 1. The SNPs, shown from left to right within the figure, are as follows: rs4646487, rs2297810, rs4646491, rs2297809, rs1126743, rs1126742, rs12564525, rs2056900, rs2056899, rs113777592, rs10789501, rs149718343, rs2405599, and rs4926600. (B) LD structures of CYP4F2, CYP4F3, CYP4F11, and CYP4F12 using common coding SNPs. CYP4F2, CYP4F3, CYP4F11, and CYP4F12 are clustered on chromosome 19. The SNPs, shown from left to right within the figure, are as follows: rs1805040, rs7254013, rs16995376, rs16995378, rs609636, rs609290, rs2285888, rs593818, rs3093200, rs2108622, rs2074900, rs3093105, rs1060463, and rs8104361. The numbers in squares refer to pairwise LD values, measured as D’ (coefficient of linkage disequilibrium). Red depicts a significant linkage between a pair of SNPs. Numbers inside squares indicate the D’ value multiplied by 100.
11. Conclusions and Future Prospects
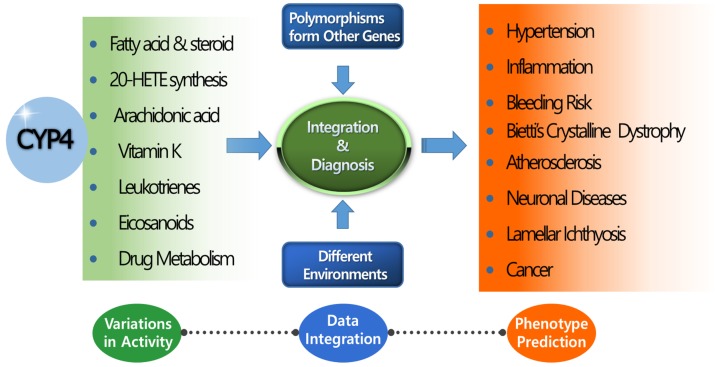
CYP4 enzymes are responsible for the metabolism of fatty acids and play important roles in the homeostasis of fatty acids and fatty-acid-derived biomolecules such as leukotriene, prostanoid, and 20-HETE. Thus, CYP4 enzymes make important contributions to human health, including cardiovascular health, skin barrier maintenance, eye function, and cancer protection. However, the lack of research into certain aspects of the CYP4 family must be overcome. First, a specific antibody for the detection of each CYP4 protein and a specific substrate for each enzyme function must be developed to clearly determine the expression levels of these enzymes in different tissues under various induction, inhibition, and genetic conditions. High similarity of protein structures, overlapping substrates, co-expression in the same tissues, and genetic differences among individuals have interfered with the identification and characterization of CYP4 genes. For targeted therapy and targeted delivery of drugs into cells or specific tissues, accurate measurement of CYP4 activity in tissues is essential. Second, further functional studies of CYP4 genetic variants are needed. A growing number of genetic mutations of CYP4 genes have been identified using high-throughput sequencing techniques. However, most of their functional changes compared to the wild type remain unknown. Only a small number of high-frequency genetic variants with known functional information have been investigated in multiple populations, likely due to their high statistical power, which enables publication. Although in silico tools are useful for the prediction of functional changes, in silico prediction does not yet perfectly reflect in vivo conditions. Therefore, various commercial software programs often provide inconsistent predictions for the same genetic mutations. Development of high-throughput techniques for in vitro functional study and improvement of in silico methods are needed to elucidate the functional changes caused by mutations. Third, globally standardized values for CYP4 activity must be developed for application in artificial intelligence technology and algorithms used for the prediction of CYP4-related human diseases or the progression of disease states. As shown in Figure 2, large variations in CYP4-mediated metabolism, genetic variants of CYP4 and other genes, and differing environmental conditions have been observed among individuals. Data integration to support correct diagnosis in humans is currently not possible, but is the ultimate goal of such research. To achieve this goal, accurate molecular tools for characterization of each CYP4 enzyme, functional information about CYP4 genetic variants, and a standardized system for the application of CYP4 functional values in artificial intelligence or machine-learning tools are needed for personalized health care.
Figure 2.
Correlation of CYP4 genes with phenotypic outcomes. Most CYP4 genes share similar structures and overlapping metabolic substrates. Phenotypic outcome prediction is difficult with a single or few CYP4 genetic studies. Phenotypic outcomes are affected by genetic polymorphisms of various genes and dynamic environmental factors. Fundamental research into CYP4 genes is essential to provide the data integration necessary for more accurate phenotype prediction than can be obtained using conventional methods.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No.2018R1A5A2021242) and by the National Research Foundation of Korea grant funded by the Korea government (NRF-2017R1D1A3B03031007).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Gonzalez F.J., Nebert D.W. Evolution of the P450 gene superfamily: animal-plant ‘warfare’, molecular drive and human genetic differences in drug oxidation. Trends Genet. 1990;6:182–186. doi: 10.1016/0168-9525(90)90174-5. [DOI] [PubMed] [Google Scholar]
- 2.Edson K.Z., Rettie A.E. CYP4 enzymes as potential drug targets: focus on enzyme multiplicity, inducers and inhibitors, and therapeutic modulation of 20-hydroxyeicosatetraenoic acid (20-HETE) synthase and fatty acid omega-hydroxylase activities. Curr. Top. Med. Chem. 2013;13:1429–1440. doi: 10.2174/15680266113139990110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drolet B., Pilote S., Gélinas C. Altered Protein Expression of Cardiac CYP2J and Hepatic CYP2C, CYP4A and CYP4F in a Mouse Model of Type II Diabetes-A Link in the Onset and Development of Cardiovascular Disease? Pharmaceutics. 2017;9:44. doi: 10.3390/pharmaceutics9040044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsu M.H., Savas U., Griffin K.J., Johnson E.F. Human cytochrome p450 family 4 enzymes: function, genetic variation and regulation. Drug Metab. Rev. 2007;39:515–538. doi: 10.1080/03602530701468573. [DOI] [PubMed] [Google Scholar]
- 5.Savas U., Hsu M.H., Johnson E.F. Differential regulation of human CYP4A genes by peroxisome proliferators and dexamethasone. Arch. Biochem. Biophys. 2003;409:212–220. doi: 10.1016/S0003-9861(02)00499-X. [DOI] [PubMed] [Google Scholar]
- 6.Jarrar Y.B., Cho S.A., Oh K.S., Kim D.H., Shin J.G., Lee S.J. Identification of cytochrome P450s involved in the metabolism of arachidonic acid in human platelets. Prostaglandins Leukot Essent. Fatty Acids. 2013;89:227–234. doi: 10.1016/j.plefa.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 7.Hsu M.H., Savas U., Griffin K.J., Johnson E.F. Regulation of human cytochrome P450 4F2 expression by sterol regulatory element-binding protein and lovastatin. J. Biol. Chem. 2007;282:5225–5236. doi: 10.1074/jbc.M608176200. [DOI] [PubMed] [Google Scholar]
- 8.Bylund J., Bylund M., Oliw E.H. cDNA cloning and expression of CYP4F12, a novel human cytochrome. Biochem. Biophys. Res. Commun. 2001;280:892–897. doi: 10.1006/bbrc.2000.4191. [DOI] [PubMed] [Google Scholar]
- 9.Powell P.K., Wolf I., Jin R., Lasker J.M. Metabolism of arachidonic acid to 20-hydroxy-5,8,11, 14-eicosatetraenoic acid by P450 enzymes in human liver: involvement of CYP4F2 and CYP4A11. J. Pharmacol. Exp. Ther. 1998;285:1327–1336. [PubMed] [Google Scholar]
- 10.Lasker J.M., Chen W.B., Wolf I., Bloswick B.P., Wilson P.D., Powell P.K. Formation of 20-hydroxyeicosatetraenoic acid, a vasoactive and natriuretic eicosanoid, in human kidney. Role of Cyp4F2 and Cyp4A11. J. Biol. Chem. 2000;275:4118–4126. doi: 10.1074/jbc.275.6.4118. [DOI] [PubMed] [Google Scholar]
- 11.Cui X., Nelson D.R., Strobel H.W. A novel human cytochrome P450 4F isoform (CYP4F11): cDNA cloning, expression, and genomic structural characterization. Genomics. 2000;68:161–166. doi: 10.1006/geno.2000.6276. [DOI] [PubMed] [Google Scholar]
- 12.Christmas P., Jones J.P., Patten C.J., Rock D.A., Zheng Y., Cheng S.M., Weber B.M., Carlesso N., Scadden D.T. Rettie AE Alternative splicing determines the function of CYP4F3 by switching substrate specificity. J Biol Chem. 2001;276:38166–38172. doi: 10.1074/jbc.M104818200. [DOI] [PubMed] [Google Scholar]
- 13.Kawakami H., Ohtsuki S., Kamiie J., Suzuki T., Abe T., Terasaki T. Simultaneous absolute quantification of 11 cytochrome P450 isoforms in human liver microsomes by liquid chromatography tandem mass spectrometry with in silico target peptide selection. J. Pharm. Sci. 2011;100:341–352. doi: 10.1002/jps.22255. [DOI] [PubMed] [Google Scholar]
- 14.Groer C., Busch D., Patrzyk M., Beyer K., Busemann A., Heidecke C.D., Drozdzik M., Siegmund W., Oswald S. Absolute protein quantification of clinically relevant cytochrome P450 enzymes and UDP-glucuronosyltransferases by mass spectrometry-based targeted proteomics. J. Pharm. Biomed. Anal. 2014;100:393–401. doi: 10.1016/j.jpba.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 15.Jin Y., Zollinger M., Borell H., Zimmerlin A. Patten CJ CYP4F enzymes are responsible for the elimination of fingolimod (FTY720), a novel treatment of relapsing multiple sclerosis. Drug Metab. Dispos. 2011;39:191–198. doi: 10.1124/dmd.110.035378. [DOI] [PubMed] [Google Scholar]
- 16.Christmas P., Carlesso N., Shang H., Cheng S.M., Weber B.M., Preffer F.I., Scadden D.T., Soberman R.J. Myeloid expression of cytochrome P450 4F3 is determined by a lineage-specific alternative promoter. J. Biol. Chem. 2003;278:25133–25142. doi: 10.1074/jbc.M302106200. [DOI] [PubMed] [Google Scholar]
- 17.Bylund J., Hidestrand M., Ingelman-Sundberg M., Oliw E.H. Identification of CYP4F8 in human seminal vesicles as a prominent 19-hydroxylase of prostaglandin endoperoxides. J. Biol. Chem. 2000;275:21844–21849. doi: 10.1074/jbc.M001712200. [DOI] [PubMed] [Google Scholar]
- 18.Ohno Y., Nakamichi S., Ohkuni A., Kamiyama N., Naoe A., Tsujimura H., Yokose U., Sugiura K., Ishikawa J., Akiyama M., et al. Essential role of the cytochrome P450 CYP4F22 in the production of acylceramide, the key lipid for skin permeability barrier formation. Proc. Natl. Acad. Sci. USA. 2015;112:77. doi: 10.1073/pnas.1503491112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakano M., Kelly E.J., Wiek C., Hanenberg H., Rettie A.E. cyp4v2 in Bietti’s crystalline dystrophy: ocular localization, metabolism of omega-3-polyunsaturated fatty acids and functional deficit of the p.H331P variant. Mol. Pharmacol. 2012;82:679–686. doi: 10.1124/mol.112.080085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiek C., Schmidt E.M., Roellecke K., Freund M., Nakano M., Kelly E.J., Kaisers W., Yarov-Yarovoy V., Kramm C.M., Rettie A.E., et al. Identification of amino acid determinants in CYP4B1 for optimal catalytic processing of 4-ipomeanol. Biochem. J. 2015;465:103–114. doi: 10.1042/BJ20140813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Savas U., Hsu M.H., Griffin K.J., Bell D.R., Johnson E.F. Conditional regulation of the human CYP4X1 and CYP4Z1 genes. Arch. Biochem. Biophys. 2005;436:377–385. doi: 10.1016/j.abb.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 22.Rieger M.A., Ebner R., Bell D.R., Kiessling A., Rohayem J., Schmitz M., Temme A., Rieber E.P., Weigle B. Identification of a novel mammary-restricted cytochrome P450, CYP4Z1, with overexpression in breast carcinoma. Cancer Res. 2004;64:2357–2364. doi: 10.1158/0008-5472.CAN-03-0849. [DOI] [PubMed] [Google Scholar]
- 23.Powell P.K., Wolf I., Lasker J.M. Identification of CYP4A11 as the major lauric acid omega-hydroxylase in human liver microsomes. Arch. Biochem. Biophys. 1996;335:219–226. doi: 10.1006/abbi.1996.0501. [DOI] [PubMed] [Google Scholar]
- 24.Crespi C.L., Chang T.K., Waxman D.J. Determination of CYP4A11-catalyzed lauric acid 12-hydroxylation by high-performance liquid chromatography with radiometric detection. Methods Mol. Biol. 2006;320:137–143. doi: 10.1385/1-59259-998-2:143. [DOI] [PubMed] [Google Scholar]
- 25.Lino Cardenas C.L., Renault N., Farce A., Cauffiez C., Allorge D., Lo-Guidice J.M., Lhermitte M., Chavatte P., Broly F., Chevalier D. Genetic polymorphism of CYP4A11 and CYP4A22 genes and in silico insights from comparative 3D modelling in a French population. Gene. 2011;487:10–20. doi: 10.1016/j.gene.2011.07.015. [DOI] [PubMed] [Google Scholar]
- 26.Gajendrarao P., Krishnamoorthy N., Sakkiah S., Lazar P., Lee K.W. Molecular modeling study on orphan human protein CYP4A22 for identification of potential ligand binding site. J. Mol. Graph. Model. 2010;28:524–532. doi: 10.1016/j.jmgm.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 27.Sasaki T., Horikawa M., Orikasa K., Sato M., Arai Y., Mitachi Y., Mizugaki M., Ishikawa M., Hiratsuka M. Possible relationship between the risk of Japanese bladder cancer cases and the CYP4B1 genotype. Jpn. J. Clin. Oncol. 2008;3:634–640. doi: 10.1093/jjco/hyn081. [DOI] [PubMed] [Google Scholar]
- 28.Ashkar S., Mesentsev A., Zhang W.X., Mastyugin V., Dunn M.W. Laniado-Schwartzman M. Retinoic acid induces corneal epithelial CYP4B1 gene expression and stimulates the synthesis of inflammatory 12-hydroxyeicosanoids. J. Ocul. Pharmacol. Ther. 2004;20:65–74. doi: 10.1089/108076804772745473. [DOI] [PubMed] [Google Scholar]
- 29.Edson K.Z., Prasad B., Unadkat J.D., Suhara Y., Okano T., Guengerich F.P., Rettie A.E. Cytochrome P450-dependent catabolism of vitamin K: omega-hydroxylation catalyzed by human CYP4F2 and CYP4F11. Biochemistry. 2013;52:8276–8285. doi: 10.1021/bi401208m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin J., Liu H., Liu Z., Owzar K., Han Y., Su L., Wei Y., Hung R.J., Brhane Y., McLaughlin J. Pathway-analysis of published genome-wide association studies of lung cancer: A potential role for the CYP4F3 locus. Mol. Carcinog. 2017;56:1663–1672. doi: 10.1002/mc.22622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kikuta Y., Mizomoto J., Strobel H.W., Ohkawa H. Expression and physiological function of CYP4F subfamily in human eosinophils. Biochim. Biophys Acta. 2007;1771:1439–1445. doi: 10.1016/j.bbalip.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 32.Harmon S.D., Fang X., Kaduce T.L., Hu S., Raj Gopal V., Falck J.R., Spector A.A. Oxygenation of omega-3 fatty acids by human cytochrome P450 4F3B: effect on 20-hydroxyeicosatetraenoic acid production. Prostaglandins Leukot. Essent. Fatty Acids. 2006;75:169–177. doi: 10.1016/j.plefa.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 33.Stark K., Wongsud B., Burman R., Oliw E.H. Oxygenation of polyunsaturated long chain fatty acids by recombinant CYP4F8 and CYP4F12 and catalytic importance of Tyr-125 and Gly-328 of CYP4F8. Arch. Biochem. Biophys. 2005;441:174–181. doi: 10.1016/j.abb.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 34.Bylund J., Finnstrom N., Oliw E.H. Gene expression of a novel cytochrome P450 of the CYP4F subfamily in human seminal vesicles. Biochem. Biophys. Re.s Commun. 1999;261:169–174. doi: 10.1006/bbrc.1999.1011. [DOI] [PubMed] [Google Scholar]
- 35.Stark K., Törmä H., Cristea M., Oliw E.H. Expression of CYP4F8 (prostaglandin H 19-hydroxylase) in human epithelia and prominent induction in epidermis of psoriatic lesions. Arch. Biochem. Biophys. 2003;409:188–196. doi: 10.1016/S0003-9861(02)00511-8. [DOI] [PubMed] [Google Scholar]
- 36.Kalsotra A., Turman C.M., Kikuta Y., Strobel H.W. Expression and characterization of human cytochrome P450 4F11: Putative role in the metabolism of therapeutic drugs and eicosanoids. Toxicol. Appl. Pharmacol. 2004;199:295–304. doi: 10.1016/j.taap.2003.12.033. [DOI] [PubMed] [Google Scholar]
- 37.Yi M., Cho S.A., Min J., Kim D.H., Shin J.G., Lee S.J. Functional characterization of a common CYP4F11 genetic variant and identification of functionally defective CYP4F11 variants in erythromycin metabolism and 20-HETE synthesis. Arch. Biochem. Biophys. 2017;620:43–51. doi: 10.1016/j.abb.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y. Gene regulation of CYP4F11 in human keratinocyte HaCaT cells. Drug Metab. Dispos. 2010;38:100–107. doi: 10.1124/dmd.109.029025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hashizume T., Imaoka S., Hiroi T., Terauchi Y., Fujii T., Miyazaki H., Kamataki T., Funae Y. cDNA cloning and expression of a novel cytochrome p450 (cyp4f12) from human small intestine. Biochem. Biophys. Res. Commun. 2001;280:1135–1141. doi: 10.1006/bbrc.2000.4238. [DOI] [PubMed] [Google Scholar]
- 40.Stark K., Dostalek M., Guengerich F.P. Expression and purification of orphan cytochrome P450 4X1 and oxidation of anandamide. Febs. J. 2008;275:3706–3717. doi: 10.1111/j.1742-4658.2008.06518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yi M., Shin J.G., Lee S.J. Expression of CYP4V2 in human THP1 macrophages and its transcriptional regulation by peroxisome proliferator-activated receptor gamma. Toxicol. Appl. Pharmacol. 2017;330:100–106. doi: 10.1016/j.taap.2017.07.009. [DOI] [PubMed] [Google Scholar]
- 42.Zollner A., Dragan C.A., Pistorius D., Müller R., Bode H.B., Peters F.T., Maurer H.H., Bureik M. Human CYP4Z1 catalyzes the in-chain hydroxylation of lauric acid and myristic acid. Biol. Chem. 2009;390:313–317. doi: 10.1515/BC.2009.030. [DOI] [PubMed] [Google Scholar]
- 43.Kim W.Y., Lee S.J., Min J., Oh K.S., Kim D.H., Kim H.S., Shin J.G. Identification of novel CYP4F2 genetic variants exhibiting decreased catalytic activity in the conversion of arachidonic acid to 20-hydroxyeicosatetraenoic acid (20-HETE) Prostaglandins Leukot Essent. Fatty Acids. 2018;131:6–13. doi: 10.1016/j.plefa.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 44.Jarrar Y.B., Jarrar Q., Abed A., Abu-Shalhoob M. Effects of nonsteroidal anti-inflammatory drugs on the expression of arachidonic acid-metabolizing Cyp450 genes in mouse hearts, kidneys and livers. Prostaglandins Other Lipid Mediat. 2019;141:14–21. doi: 10.1016/j.prostaglandins.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 45.Nakano M., Kelly E.J., Rettie A.E. Expression and characterization of CYP4V2 as a fatty acid omega-hydroxylase. Drug Metab. Dispos. 2009;37:2119–2122. doi: 10.1124/dmd.109.028530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Antoun J., Goulitquer S., Amet Y., Dreano Y., Salaun J.P., Corcos L., Plée-Gautier E. CYP4F3B is induced by PGA1 in human liver cells: a regulation of the 20-HETE synthesis. J. Lipid Res. 2008;49:2135–21341. doi: 10.1194/jlr.M800043-JLR200. [DOI] [PubMed] [Google Scholar]
- 47.Liu J.Y., Li N., Yang J., Li N., Qiu H., Ai D., Chiamvimonvat N., Zhu Y., Hammock B.D. Metabolic profiling of murine plasma reveals an unexpected biomarker in rofecoxib-mediated cardiovascular events. Proc. Natl. Acad. Sci. USA. 2010;107:17017–17022. doi: 10.1073/pnas.1011278107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Costa T.J., Ceravolo G.S., Echem C., Hashimoto C.M., Costa B.P., Santos-Eichler R.A., Oliveira M.A., Jiménez-Altayó F., Akamine E.H., Dantas A.P., et al. Detrimental Effects of Testosterone Addition to Estrogen Therapy Involve Cytochrome P-450-Induced 20-HETE Synthesis in Aorta of Ovariectomized Spontaneously Hypertensive Rat (SHR), a Model of Postmenopausal Hypertension. Front. Physiol. 2018;9:490. doi: 10.3389/fphys.2018.00490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pavek P., Dvorak Z. Xenobiotic-induced transcriptional regulation of xenobiotic metabolizing enzymes of the cytochrome P450 superfamily in human extrahepatic tissues. Curr. Drug Metab. 2008;9:129–143. doi: 10.2174/138920008783571774. [DOI] [PubMed] [Google Scholar]
- 50.Joseph G. Elevated 20-HETE impairs coronary collateral growth in metabolic syndrome via endothelial dysfunction. Am. J. Physiol. Heart Circ. Physiol. 2017;312:H528–H540. doi: 10.1152/ajpheart.00561.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zordoky B.N., Anwar-Mohamed A., Aboutabl M.E., El-Kadi A.O. Acute doxorubicin cardiotoxicity alters cardiac cytochrome P450 expression and arachidonic acid metabolism in rats. Toxicol. Appl. Pharmacol. 2010;242:38–46. doi: 10.1016/j.taap.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 52.Kelly E.J., Nakano M., Rohatgi P., Yarov-Yarovoy V., Rettie A.E. Finding homes for orphan cytochrome P450s: CYP4V2 and CYP4F22 in disease states. Mol. Interv. 2011;11:124–132. doi: 10.1124/mi.11.2.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kikuta Y., Kusunose E., Kusunose M. Characterization of human liver leukotriene B(4) omega-hydroxylase P450 (CYP4F2) J. Biochem. 2000;127:1047–1052. doi: 10.1093/oxfordjournals.jbchem.a022696. [DOI] [PubMed] [Google Scholar]
- 54.Kikuta Y., Kusunose E., Endo K., Yamamoto S., Sogawa K., Fujii-Kuriyama Y., Kusunose M. A novel form of cytochrome P-450 family 4 in human polymorphonuclear leukocytes. cDNA cloning and expression of leukotriene B4 omega-hydroxylase. J. Biol. Chem. 1993;268:9376–9380. [PubMed] [Google Scholar]
- 55.Spector A.A. Arachidonic acid cytochrome P450 epoxygenase pathway. J. Lipid Res. 2009;50:S52–S56. doi: 10.1194/jlr.R800038-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Le Quere V., Plée-Gautier E., Potin P., Madec S., Salaün J.P. Human CYP4F3s are the main catalysts in the oxidation of fatty acid epoxides. J. Lipid Res. 2004;45:1446–1458. doi: 10.1194/jlr.M300463-JLR200. [DOI] [PubMed] [Google Scholar]
- 57.Wang Y., Li Y., Lu J., Qi H., Cheng I., Zhang H. Involvement of CYP4F2 in the Metabolism of a Novel Monophosphate Ester Prodrug of Gemcitabine and Its Interaction Potential In Vitro. Molecules. 2018;23:1195. doi: 10.3390/molecules23051195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang J., Li K., Zhang X., Teng D., Ju M., Jing Y., Zhao Y. The correlation between the expression of genes involved in drug metabolism and the blood level of tacrolimus in liver transplant receipts. Sci. Rep. 2017;7:3429. doi: 10.1038/s41598-017-02698-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang Z., Salamanca-Pinzón S.G., Wu Z.L., Xiao Y., Guengerich F.P. Human cytochrome P450 4F11: heterologous expression in bacteria, purification and characterization of catalytic function. Arch. Biochem. Biophys. 2010;494:86–93. doi: 10.1016/j.abb.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hashizume T., Imaoka S., Mise M., Terauchi Y., Fujii T., Miyazaki H., Kamataki T., Funae Y. Involvement of CYP2J2 and CYP4F12 in the metabolism of ebastine in human intestinal microsomes. J. Pharmacol. Exp. Ther. 2002;300:298–304. doi: 10.1124/jpet.300.1.298. [DOI] [PubMed] [Google Scholar]
- 61.Evangelista E.A., Kaspera R., Mokadam N.A., Jones J.P., Totah R.A. Activity, inhibition, and induction of cytochrome P450 2J2 in adult human primary cardiomyocytes. Drug Metab. Dispos. 2013;41:2087–2094. doi: 10.1124/dmd.113.053389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McDonald M.G., Rieder M.J., Nakano M., Hsia C.K., Rettie A.E. CYP4F2 is a vitamin K1 oxidase: An explanation for altered warfarin dose in carriers of the V433M variant. Mol. Pharmacol. 2009;75:1337–1346. doi: 10.1124/mol.109.054833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kaminsky L.S., Zhang Z.Y. Human P450 metabolism of warfarin. Pharmacol. Ther. 1997;73:67–74. doi: 10.1016/S0163-7258(96)00140-4. [DOI] [PubMed] [Google Scholar]
- 64.Hermans J.J., Thijssen H.H. Human liver microsomal metabolism of the enantiomers of warfarin and acenocoumarol: P450 isozyme diversity determines the differences in their pharmacokinetics. Br. J. Pharmacol. 1993;110:482–490. doi: 10.1111/j.1476-5381.1993.tb13836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Souza-Mello V. Peroxisome proliferator-activated receptors as targets to treat non-alcoholic fatty liver disease. World J. Hepatol. 2015;7:1012–1019. doi: 10.4254/wjh.v7.i8.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hong F., Xu P., Zhai Y. The Opportunities and Challenges of Peroxisome Proliferator-Activated Receptors Ligands in Clinical Drug Discovery and Development. Int. J. Mol. Sci. 2018;19:2189. doi: 10.3390/ijms19082189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hardwick J.P., Osei-Hyiaman D., Wiland H., Abdelmegeed M.A., Song B.J. PPAR/RXR Regulation of Fatty Acid Metabolism and Fatty Acid omega-Hydroxylase (CYP4) Isozymes: Implications for Prevention of Lipotoxicity in Fatty Liver Disease. PPAR Res. 2009;2009:952734. doi: 10.1155/2009/952734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peraza M.A., Burdick A.D., Marin H.E., Gonzalez F.J., Peters J.M. The toxicology of ligands for peroxisome proliferator-activated receptors (PPAR) Toxicol. Sci. 2006;90:269–295. doi: 10.1093/toxsci/kfj062. [DOI] [PubMed] [Google Scholar]
- 69.Kikuta Y., Kusunose E., Kondo T., Yamamoto S., Kinoshita H., Kusunose M. Cloning and expression of a novel form of leukotriene B4 omega-hydroxylase from human liver. FEBS Lett. 1994;348:70–74. doi: 10.1016/0014-5793(94)00587-7. [DOI] [PubMed] [Google Scholar]
- 70.Edwards I.J., O’Flaherty J.T. Omega-3 Fatty Acids and PPARgamma in Cancer. PPAR Res. 2008;2008:358052. doi: 10.1155/2008/358052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kalsotra A., Cui X., Antonovic L., Robida A.M., Morgan E.T., Strobel H.W. Inflammatory prompts produce isoform-specific changes in the expression of leukotriene B(4) omega-hydroxylases in rat liver and kidney. FEBS Lett. 2003;555:236–242. doi: 10.1016/S0014-5793(03)01240-7. [DOI] [PubMed] [Google Scholar]
- 72.Muntane J. Effect of carrageenan-induced granuloma on hepatic cytochrome P-450 isozymes in rats. Inflammation. 1995;19:143–156. doi: 10.1007/BF01534458. [DOI] [PubMed] [Google Scholar]
- 73.Alexanian A., Sorokin A. Targeting 20-HETE producing enzymes in cancer - rationale, pharmacology, and clinical potential. Onco. Targets Ther. 2013;6:243–255. doi: 10.2147/OTT.S31586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu W., Chai H., Li Y., Zhao H., Xie X., Zheng H., Wang C., Wang X., Yang G., Cai X., et al. Increased expression of CYP4Z1 promotes tumor angiogenesis and growth in human breast cancer. Toxicol. Appl. Pharmacol. 2012;264:73–83. doi: 10.1016/j.taap.2012.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alexanian A., Miller B., Roman R.J., Sorokin A. 20-HETE-producing enzymes are up-regulated in human cancers. Cancer Genom. Proteom. 2012;9:163–169. [PMC free article] [PubMed] [Google Scholar]
- 76.Eun H.S., Cho S.Y., Lee B.S., Seong I.O., Kim K.H. Profiling cytochrome P450 family 4 gene expression in human hepatocellular carcinoma. Mol. Med. Rep. 2018;18:4865–4876. doi: 10.3892/mmr.2018.9526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shibuya M. Vascular endothelial growth factor and its receptor system: physiological functions in angiogenesis and pathological roles in various diseases. J. Biochem. 2013;153:13–19. doi: 10.1093/jb/mvs136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Imaoka S., Yoneda Y., Sugimoto T., Hiroi T., Yamamoto K., Nakatani T., Funae Y. CYP4B1 is a possible risk factor for bladder cancer in humans. Biochem. Biophys. Res. Commun. 2000;277:776–780. doi: 10.1006/bbrc.2000.3740. [DOI] [PubMed] [Google Scholar]
- 79.Vanderslice R.R., Boyd J.A., Eling T.E., Philpot R.M. The cytochrome P-450 monooxygenase system of rabbit bladder mucosa: enzyme components and isozyme 5-dependent metabolism of 2-aminofluorene. Cancer Res. 1985;45:5851–5858. [PubMed] [Google Scholar]
- 80.Imaoka S., Yoneda Y., Matsuda T., Degawa M., Fukushima S., Funae Y. Mutagenic activation of urinary bladder carcinogens by CYP4B1 and the presence of CYP4B1 in bladder mucosa. Biochem. Pharmacol. 1997;54:677–683. doi: 10.1016/S0006-2952(97)00216-5. [DOI] [PubMed] [Google Scholar]
- 81.Hsu H., Rainov N.G., Quinones A., Eling D.J., Sakamoto K.M., Spear M.A. Combined radiation and cytochrome CYP4B1/4-ipomeanol gene therapy using the EGR1 promoter. Anticancer Res. 2003;23:2723–2728. [PubMed] [Google Scholar]
- 82.Choudhary D., Jansson I., Stoilov I., Sarfarazi M., Schenkman J.B. Expression patterns of mouse and human CYP orthologs (families 1-4) during development and in different adult tissues. Arch. Biochem. Biophys. 2005;436:50–61. doi: 10.1016/j.abb.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 83.Tamaki Y., Arai T., Sugimura H., Sasaki T., Honda M., Muroi Y., Matsubara Y., Kanno S., Ishikawa M., Hirasawa N., et al. Association between cancer risk and drug-metabolizing enzyme gene (CYP2A6, CYP2A13, CYP4B1, SULT1A1, GSTM1 and GSTT1) polymorphisms in cases of lung cancer in Japan. Drug Metab. Pharmacokinet. 2011;26:516–522. doi: 10.2133/dmpk.DMPK-11-RG-046. [DOI] [PubMed] [Google Scholar]
- 84.Lin J.T. Downregulation of the cytochrome P450 4B1 protein confers a poor prognostic factor in patients with urothelial carcinomas of upper urinary tracts and urinary bladder. Apmis. 2019;127:170–180. doi: 10.1111/apm.12939. [DOI] [PubMed] [Google Scholar]
- 85.Downie D., McFadyen M.C., Rooney P.H., Cruickshank M.E., Parkin D.E., Miller I.D., Telfer C., Melvin W.T., Murray G.I. Profiling cytochrome P450 expression in ovarian cancer: identification of prognostic markers. Clin. Cancer Res. 2005;11:7369–7375. doi: 10.1158/1078-0432.CCR-05-0466. [DOI] [PubMed] [Google Scholar]
- 86.Khayeka-Wandabwa C., Ma X., Cao X., Nunna V., Pathak J.L., Bernhardt R., Cai P., Bureik M. Plasma membrane localization of CYP4Z1 and CYP19A1 and the detection of anti-CYP19A1 autoantibodies in humans. Int. Immunopharmacol. 2019;73:64–71. doi: 10.1016/j.intimp.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 87.McDonald M.G., Ray S., Amorosi C.J., Sitko K.A., Kowalski J.P., Paco L., Nath A., Gallis B., Totah R.A., Dunham M.J., et al. Expression and Functional Characterization of Breast Cancer-Associated Cytochrome P450 4Z1 in Saccharomyces cerevisiae. Drug Metab. Dispos. 2017;45:1364–1371. doi: 10.1124/dmd.117.078188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu C.C., Schwartzman M.L. The role of 20-HETE in androgen-mediated hypertension. Prostaglandins Other Lipid Mediat. 2011;96:45–53. doi: 10.1016/j.prostaglandins.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rocic P., Schwartzman M.L. 20-HETE in the regulation of vascular and cardiac function. Pharmacol. Ther. 2018;192:74–87. doi: 10.1016/j.pharmthera.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu C.C., Ei S., Cheng J., Ding Y., Weidenhammer A., Garcia V., Zhang F., Gotlinger K., Manthati V.L., Falck J.R., et al. Androgen-sensitive hypertension associates with upregulated vascular CYP4A12-20-HETE synthase. J. Am. Soc. Nephrol. 2013;24:1288–1296. doi: 10.1681/ASN.2012070714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang C., Booz G.W., Yu Q., He X., Wang S., Fan F. Conflicting roles of 20-HETE in hypertension and renal end organ damage. Eur. J. Pharmacol. 2018;833:190–200. doi: 10.1016/j.ejphar.2018.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fan F., Sun C.W., Maier K.G., Williams J.M., Pabbidi M.R., Didion S.P., Falck J.R., Zhuo J., Roman R.J. 20-Hydroxyeicosatetraenoic acid contributes to the inhibition of K+ channel activity and vasoconstrictor response to angiotensin II in rat renal microvessels. PLoS ONE. 2013;8:e82482. doi: 10.1371/journal.pone.0082482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gebremedhin D., Lange A.R., Narayanan J., Aebly M.R., Jacobs E.R., Harder D.R. Cat cerebral arterial smooth muscle cells express cytochrome P450 4A2 enzyme and produce the vasoconstrictor 20-HETE which enhances L-type Ca2+ current. J. Physiol. 1998;507:771–781. doi: 10.1111/j.1469-7793.1998.771bs.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Roman R.J. P-450 metabolites of arachidonic acid in the control of cardiovascular function. Physiol. Rev. 2002;82:131–185. doi: 10.1152/physrev.00021.2001. [DOI] [PubMed] [Google Scholar]
- 95.Alonso-Galicia M., Maier K.G., Greene A.S., Cowley A.W., Jr., Roman R.J. Role of 20-hydroxyeicosatetraenoic acid in the renal and vasoconstrictor actions of angiotensin II. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2002;283:R60–R68. doi: 10.1152/ajpregu.00664.2001. [DOI] [PubMed] [Google Scholar]
- 96.Garcia V., Shkolnik B., Milhau L., Falck J.R., Schwartzman M.L. 20-HETE Activates the Transcription of Angiotensin-Converting Enzyme via Nuclear Factor-kappaB Translocation and Promoter Binding. J. Pharmacol. Exp. Ther. 2013;56:525–533. doi: 10.1124/jpet.115.229377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sodhi K., Wu C.C., Cheng J., Gotlinger K., Inoue K., Goli M., Falck J.R., Abraham N.G. Schwartzman ML CYP4A2-induced hypertension is 20-hydroxyeicosatetraenoic acid- and angiotensin II-dependent. Hypertension. 2010;56:871–878. doi: 10.1161/HYPERTENSIONAHA.110.154559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gerhold D., Bagchi A., Lu M., Figueroa D., Keenan K., Holder D., Wang Y., Jin H., Connolly B., Austin C., et al. Androgens drive divergent responses to salt stress in male versus female rat kidneys. Genomics. 2007;89:731–744. doi: 10.1016/j.ygeno.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 99.Ng D.S., Lai T.Y., Ng T.K., Pang C.P. Genetics of Bietti Crystalline Dystrophy. Asia Pac. J. Ophthalmol. (Phila.) 2016;5:245–252. doi: 10.1097/APO.0000000000000209. [DOI] [PubMed] [Google Scholar]
- 100.Fong A.M., Koh A., Lee K., Ang C.L. Bietti’s crystalline dystrophy in Asians: clinical, angiographic and electrophysiological characteristics. Int. Ophthalmol. 2009;29:459–470. doi: 10.1007/s10792-008-9266-7. [DOI] [PubMed] [Google Scholar]
- 101.Mansour A.M., Uwaydat S.H., Chan C.C. Long-term follow-up in Bietti crystalline dystrophy. Eur. J. Ophthalmol. 2007;17:680–682. doi: 10.1177/112067210701700434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vargas M., Mitchell A., Yang P., Weleber R. In: Bietti Crystalline Dystrophy. Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J.H., Stephens K., Amemiya A., editors. University of Washington; Seattle, WA, USA: 2019. GeneReviews® [Internet] [Google Scholar]
- 103.Lin J., Nishiguchi K.M., Nakamura M., Dryja T.P., Berson E.L., Miyake Y. Recessive mutations in the CYP4V2 gene in East Asian and Middle Eastern patients with Bietti crystalline corneoretinal dystrophy. J. Med. Genet. 2005;42:e38. doi: 10.1136/jmg.2004.029066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mataftsi A., Zografos L., Millá E., Secrétan M., Munier F.L. Bietti’s crystalline corneoretinal dystrophy: a cross-sectional study. Retina. 2004;24:416–426. doi: 10.1097/00006982-200406000-00013. [DOI] [PubMed] [Google Scholar]
- 105.Shan M., Dong B., Zhao X., Wang J., Li G., Yang Y., Li Y. Novel mutations in the CYP4V2 gene associated with Bietti crystalline corneoretinal dystrophy. Mol. Vis. 2005;11:738–743. [PubMed] [Google Scholar]
- 106.Li A., Jiao X., Munier F.L., Schorderet D.F., Yao W., Iwata F., Hayakawa M., Kanai A., Shy Chen M., Alan Lewis R., et al. Bietti crystalline corneoretinal dystrophy is caused by mutations in the novel gene CYP4V2. Am. J. Hum. Genet. 2004;74:817–826. doi: 10.1086/383228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zenteno J.C., Ayala-Ramirez R., Graue-Wiechers F. Novel CYP4V2 gene mutation in a Mexican patient with Bietti’s crystalline corneoretinal dystrophy. Curr. Eye Res. 2008;33:313–318. doi: 10.1080/02713680801983217. [DOI] [PubMed] [Google Scholar]
- 108.Lee K.Y., Koh A.H., Aung T., Yong V.H., Yeung K., Ang C.L., Vithana E.N. Characterization of Bietti crystalline dystrophy patients with CYP4V2 mutations. Invest. Ophthalmol. Vis. Sci. 2005;46:3812–3816. doi: 10.1167/iovs.05-0378. [DOI] [PubMed] [Google Scholar]
- 109.Gekka T., Hayashi T., Takeuchi T., Goto-Omoto S., Kitahara K. CYP4V2 mutations in two Japanese patients with Bietti’s crystalline dystrophy. Ophthalmic. Res. 2005;37:262–269. doi: 10.1159/000087214. [DOI] [PubMed] [Google Scholar]
- 110.Wada Y., Itabashi T., Sato H., Kawamura M., Tada A., Tamai M. Screening for mutations in CYP4V2 gene in Japanese patients with Bietti’s crystalline corneoretinal dystrophy. Am. J. Ophthalmol. 2005;139:894–899. doi: 10.1016/j.ajo.2004.11.065. [DOI] [PubMed] [Google Scholar]
- 111.Jin Z.B., Ito S., Saito Y., Inoue Y., Yanagi Y., Nao I.N. Clinical and molecular findings in three Japanese patients with crystalline retinopathy. Jpn. J. Ophthalmol. 2006;50:426–431. doi: 10.1007/s10384-006-0350-0. [DOI] [PubMed] [Google Scholar]
- 112.Lai T.Y., Ng T.K., Tam P.O., Yam G.H., Ngai J.W., Chan W.M., Liu D.T., Lam D.S., Pang C.P. Genotype phenotype analysis of Bietti’s crystalline dystrophy in patients with CYP4V2 mutations. Invest. Ophthalmol. Vis. Sci. 2007;48:5212–5220. doi: 10.1167/iovs.07-0660. [DOI] [PubMed] [Google Scholar]
- 113.Li Q., Li Y., Zhang X., Xu Z., Zhu X., Ma K., She H., Peng X. Utilization of fundus autofluorescence, apectral domain optical coherence tomography and enhanced depth imaging in the characterization of Bietti crystalline dystrophy in different stages. Retina. 2015;35:2074–2084. doi: 10.1097/IAE.0000000000000592. [DOI] [PubMed] [Google Scholar]
- 114.Gocho K. High-Resolution Imaging of Patients with Bietti Crystalline Dystrophy with CYP4V2 Mutation. J. Ophthalmol. 2014;2014:283603. doi: 10.1155/2014/283603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Halford S., Liew G., Mackay D.S., Sergouniotis P.I., Holt R., Broadgate S., Volpi E.V., Ocaka L., Robson A.G., Holder G.E., et al. Detailed phenotypic and genotypic characterization of bietti crystalline dystrophy. Ophthalmology. 2014;121:1174–1184. doi: 10.1016/j.ophtha.2013.11.042. [DOI] [PubMed] [Google Scholar]
- 116.Yin H., Jin C., Fang X., Miao Q., Zhao Y., Chen Z., Su Z., Ye P., Wang Y., Yin J. Molecular analysis and phenotypic study in 14 Chinese families with Bietti crystalline dystrophy. PLoS ONE. 2014;9:94960. doi: 10.1371/journal.pone.0094960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Xiao X., Mai G., Li S., Guo X., Zhang Q. Identification of CYP4V2 mutation in 21 families and overview of mutation spectrum in Bietti crystalline corneoretinal dystrophy. Biochem. Biophys. Res. Commun. 2011;409:18–186. doi: 10.1016/j.bbrc.2011.04.112. [DOI] [PubMed] [Google Scholar]
- 118.Meng X.H. Identification of novel CYP4V2 gene mutations in 92 Chinese families with Bietti’s crystalline corneoretinal dystrophy. Mol. Vis. 2014;20:1806–1814. [PMC free article] [PubMed] [Google Scholar]
- 119.Astuti G.D. Novel insights into the molecular pathogenesis of CYP4V2-associated Bietti’s retinal dystrophy. Mol. Genet. Genomic. Med. 2015;3:14–29. doi: 10.1002/mgg3.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nakamura M., Lin J., Nishiguchi K., Kondo M., Sugita J., Miyake Y. Bietti crystalline corneoretinal dystrophy associated with CYP4V2 gene mutations. Adv. Exp. Med. Biol. 2006;572:49–53. doi: 10.1007/0-387-32442-9_8. [DOI] [PubMed] [Google Scholar]
- 121.Chung J.K., Shin J.H., Jeon B.R., Ki C.S., Park T.K. Optical coherence tomographic findings of crystal deposits in the lens and cornea in Bietti crystalline corneoretinopathy associated with mutation in the CYP4V2 gene. Jpn. J. Ophthalmol. 2013;57:447–450. doi: 10.1007/s10384-013-0256-6. [DOI] [PubMed] [Google Scholar]
- 122.Liu D.N., Liu Y., Meng X.H., Yin Z.Q. The characterization of functional disturbances in Chinese patients with Bietti’s crystalline dystrophy at different fundus stages. Graefes Arch. Clin. Exp. Ophthalmol. 2012;250:191–200. doi: 10.1007/s00417-011-1809-3. [DOI] [PubMed] [Google Scholar]
- 123.Tian R., Wang S.R., Wang J., Chen Y.X. Novel CYP4V2 mutations associated with Bietti crystalline corneoretinal dystrophy in Chinese patients. Int. J. Ophthalmol. 2015;8:465–469. doi: 10.3980/j.issn.2222-3959.2015.03.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wada Y. Specular microscopic findings of corneal deposits in patients with Bietti’s crystalline corneal retinal dystrophy. Br. J. Ophthalmol. 1999;83:1095. doi: 10.1136/bjo.83.9.1088k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yokoi Y., Nakazawa M., Mizukoshi S., Sato K., Usui T., Takeuchi K. Crystal deposits on the lens capsules in Bietti crystalline corneoretinal dystrophy associated with a mutation in the CYP4V2 gene. Acta Ophthalmol. 2010;88:607–609. doi: 10.1111/j.1755-3768.2009.01529.x. [DOI] [PubMed] [Google Scholar]
- 126.Lefevre C., Bouadjar B., Ferrand V., Tadini G., Mégarbané A., Lathrop M., Prud’homme J.F., Fischer J. Mutations in a new cytochrome P450 gene in lamellar ichthyosis type 3. Hum. Mol. Genet. 2006;15:767–776. doi: 10.1093/hmg/ddi491. [DOI] [PubMed] [Google Scholar]
- 127.Wertz P.W., Cho E.S., Downing D.T. Effect of essential fatty acid deficiency on the epidermal sphingolipids of the rat. Biochim. Biophys. Acta. 1983;753:350–355. doi: 10.1016/0005-2760(83)90058-9. [DOI] [PubMed] [Google Scholar]
- 128.Imokawa G. Decreased level of ceramides in stratum corneum of atopic dermatitis: an etiologic factor in atopic dry skin? J. Invest. Dermatol. 1991;96:523–526. doi: 10.1111/1523-1747.ep12470233. [DOI] [PubMed] [Google Scholar]
- 129.Ishikawa J., Narita H., Kondo N., Hotta M., Takagi Y., Masukawa Y., Kitahara T., Takema Y., Koyano S., Yamazaki S. Changes in the ceramide profile of atopic dermatitis patients. J. Invest. Dermatol. 2010;130:2511–2514. doi: 10.1038/jid.2010.161. [DOI] [PubMed] [Google Scholar]
- 130.Janssens M., van Smeden J., Gooris G.S., Bras W., Portale G., Caspers P.J., Vreeken R.J., Hankemeier T., Kezic S., Wolterbeek R., et al. Increase in short-chain ceramides correlates with an altered lipid organization and decreased barrier function in atopic eczema patients. J. Lipid Res. 2012;53:2755–2766. doi: 10.1194/jlr.P030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sayeb M., Riahi Z., Laroussi N., Bonnet C., Romdhane L., Mkaouar R., Zaouak A., Marrakchi J., Abdessalem G., Messaoud O. A Tunisian family with a novel mutation in the gene CYP4F22 for lamellar ichthyosis and co-occurrence of hearing loss in a child due to mutation in the SLC26A4 gene. Int. J. Dermatol. 2019 doi: 10.1111/ijd.14452. [DOI] [PubMed] [Google Scholar]
- 132.Lo-Guidice J.M., Allorge D., Cauffiez C., Chevalier D., Lafitte J.J., Lhermitte M., Broly F. Genetic polymorphism of the human cytochrome P450 CYP4B1: evidence for a non-functional allelic variant. Pharmacogenetics. 2002;12:367–374. doi: 10.1097/00008571-200207000-00004. [DOI] [PubMed] [Google Scholar]
- 133.Hiratsuka M., Nozawa H., Konno Y., Saito T., Konno S., Mizugaki M. Human CYP4B1 gene in the japanese population analyzed by denaturing HPLC. Drug Metab. Pharmacokinet. 2004;19:114–119. doi: 10.2133/dmpk.19.114. [DOI] [PubMed] [Google Scholar]
- 134.Jennings G.K., Hsu M.H., Shock L.S., Johnson E.F., Hackett J.C. Noncovalent interactions dominate dynamic heme distortion in cytochrome P450 4B1. J. Biol. Chem. 2018;293:11433–11446. doi: 10.1074/jbc.RA118.004044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Scott E.E. Omega-versus (omega-1)-hydroxylation: Cytochrome P450 4B1 sterics make the call. J. Biol. Chem. 2017;292:5622–5623. doi: 10.1074/jbc.H117.775494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hsu M.H. The Crystal Structure of Cytochrome P450 4B1 (CYP4B1) Monooxygenase Complexed with Octane Discloses Several Structural Adaptations for omega-Hydroxylation. J. Biol. Chem. 2017;292:5610–5621. doi: 10.1074/jbc.M117.775494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sirotina S., Ponomarenko I., Kharchenko A., Bykanova M., Bocharova A., Vagaytseva K., Stepanov V., Churnosov M., Solodilova M., Polonikov A. A Novel Polymorphism in the Promoter of the CYP4A11 Gene Is Associated with Susceptibility to Coronary Artery Disease. Dis. Markers. 2018;2018:5812802. doi: 10.1155/2018/5812802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yan H.Q., Yuan Y., Zhang P., Huang Z., Chang L., Gui Y.K. CYP4F2 gene single nucleotide polymorphism is associated with ischemic stroke. Genet. Mol. Res. 2015;14:659–664. doi: 10.4238/2015.January.30.8. [DOI] [PubMed] [Google Scholar]
- 139.Fava C. The V433M variant of the CYP4F2 is associated with ischemic stroke in male Swedes beyond its effect on blood pressure. Hypertension. 2008;52:373–380. doi: 10.1161/HYPERTENSIONAHA.108.114199. [DOI] [PubMed] [Google Scholar]
- 140.Gainer J.V., Bellamine A., Dawson E.P., Womble K.E., Grant S.W., Wang Y., Cupples L.A., Guo C.Y., Demissie S., O’Donnell C.J. Functional variant of CYP4A11 20-hydroxyeicosatetraenoic acid synthase is associated with essential hypertension. Circulation. 2005;111:63–69. doi: 10.1161/01.CIR.0000151309.82473.59. [DOI] [PubMed] [Google Scholar]
- 141.Mayer B., Lieb W., Götz A., König I.R., Aherrahrou Z., Thiemig A., Holmer S., Hengstenberg C., Doering A., Loewel H., et al. Association of the T8590C polymorphism of CYP4A11 with hypertension in the MONICA Augsburg echocardiographic substudy. Hypertension. 2005;46:766–771. doi: 10.1161/01.HYP.0000182658.04299.15. [DOI] [PubMed] [Google Scholar]
- 142.Mayer B., Lieb W., Götz A., König I.R., Kauschen L.F., Linsel-Nitschke P., Pomarino A., Holmer S., Hengstenberg C., Doering A. Association of a functional polymorphism in the CYP4A11 gene with systolic blood pressure in survivors of myocardial infarction. J. Hypertens. 2006;24:1965–1970. doi: 10.1097/01.hjh.0000244944.34546.8e. [DOI] [PubMed] [Google Scholar]
- 143.Yu K., Zhang T., Li X. Genetic role of CYP4A11 polymorphisms in the risk of developing cardiovascular and cerebrovascular diseases. Ann. Hum. Genet. 2018;82:371–380. doi: 10.1111/ahg.12280. [DOI] [PubMed] [Google Scholar]
- 144.Liang J.Q., Yan M.R., Yang L., Suyila Q., Cui H.W., Su X.L. Association of a CYP4A11 polymorphism and hypertension in the Mongolian and Han populations of China. Genet. Mol. Res. 2014;13:508–517. doi: 10.4238/2014.January.21.20. [DOI] [PubMed] [Google Scholar]
- 145.Yan H.C., Liu J.H., Li J., He B.X., Yang L., Qiu J., Li L., Ding D.P., Shi L., Zhao S.J. Association between the CYP4A11 T8590C variant and essential hypertension: new data from Han Chinese and a meta-analysis. PLoS ONE. 2013;8:e80072. doi: 10.1371/journal.pone.0080072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Williams J.S., Hopkins P.N., Jeunemaitre X., Brown N.J. CYP4A11 T8590C polymorphism, salt-sensitive hypertension, and renal blood flow. J. Hypertens. 2011;29:1913–1918. doi: 10.1097/HJH.0b013e32834aa786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ding H., Cui G., Zhang L., Xu Y., Bao X., Tu Y., Wu B., Wang Q., Hui R., Wang W. Association of common variants of CYP4A11 and CYP4F2 with stroke in the Han Chinese population. Pharmacogenet Genomics. 2010;20:187–194. doi: 10.1097/FPC.0b013e328336eefe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Laffer C.L. The T8590C polymorphism of CYP4A11 and 20-hydroxyeicosatetraenoic acid in essential hypertension. Hypertension. 2008;51:767–772. doi: 10.1161/HYPERTENSIONAHA.107.102921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Caldwell M.D., Awad T., Johnson J.A., Gage B.F., Falkowski M., Gardina P., Hubbard J., Turpaz Y., Langaee T.Y., Eby C. et al. CYP4F2 genetic variant alters required warfarin dose. Blood. 2008;111:4106–4112. doi: 10.1182/blood-2007-11-122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Dean L. Warfarin Therapy and VKORC1 and CYP Genotype. In: Pratt V., editor. Medical Genetics Summaries. Bethesda; Rockville, MD, USA: 2012. [PubMed] [Google Scholar]
- 151.Borgiani P., Ciccacci C., Forte V., Sirianni E., Novelli L., Bramanti P., Novelli G. CYP4F2 genetic variant (rs2108622) significantly contributes to warfarin dosing variability in the Italian population. Pharmacogenomics. 2009;10:261–266. doi: 10.2217/14622416.10.2.261. [DOI] [PubMed] [Google Scholar]
- 152.Takeuchi F., McGinnis R., Bourgeois S., Barnes C., Eriksson N., Soranzo N., Whittaker P., Ranganath V., Kumanduri V., McLaren W. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009;5:e1000433. doi: 10.1371/journal.pgen.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cohen J.L., Thompson E., Sinvani L., Kozikowski A., Qiu G., Pekmezaris R., Spyropoulos A.C., Wang J.J. Assessment of warfarin algorithms for hospitalized adults: searching for a safe dosing strategy. J. Thromb. Thrombolysis. 2019 doi: 10.1007/s11239-019-01902-0. [DOI] [PubMed] [Google Scholar]
- 154.Sasano M., Ohno M., Fukuda Y., Nonen S., Hirobe S., Maeda S., Miwa Y., Yokoyama J., Nakayama H., Miyagawa S., et al. Verification of pharmacogenomics-based algorithms to predict warfarin maintenance dose using registered data of Japanese patients. Eur. J. Clin. Pharmacol. 2019;75:901–911. doi: 10.1007/s00228-019-02656-7. [DOI] [PubMed] [Google Scholar]
- 155.Horne B.D., Lenzini P.A., Wadelius M., Jorgensen A.L., Kimmel S.E., Ridker P.M., Eriksson N., Anderson J.L., Pirmohamed M., Limdi N.A., et al. Pharmacogenetic warfarin dose refinements remain significantly influenced by genetic factors after one week of therapy. Thromb. Haemost. 2012;107:232–240. doi: 10.1160/TH11-06-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hiratsuka M., Nozawa H., Katsumoto Y., Moteki T., Sasaki T., Konno Y., Mizugaki M. Genetic polymorphisms and haplotype structures of the CYP4A22 gene in a Japanese population. Mutat. Res. 2006;599:98–104. doi: 10.1016/j.mrfmmm.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 157.Nelson D.R., Zeldin D.C., Hoffman S.M., Maltais L.J., Wain H.M., Nebert D.W. Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Pharmacogenetics. 2004;14:1–18. doi: 10.1097/00008571-200401000-00001. [DOI] [PubMed] [Google Scholar]
- 158.Ananthakrishnan A.N., Khalili H., Song M., Higuchi L.M., Lochhead P., Richter J.M., Chan A.T. Genetic Polymorphisms in Fatty Acid Metabolism Modify the Association Between Dietary n3: n6 Intake and Risk of Ulcerative Colitis: A Prospective Cohort Study. Inflamm. Bowel. Dis. 2017;23:1898–1904. doi: 10.1097/MIB.0000000000001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Shiotani A., Murao T., Fujita Y., Fujimura Y., Sakakibara T., Nishio K., Haruma K. Novel single nucleotide polymorphism markers for low dose aspirin-associated small bowel bleeding. PLoS ONE. 2013;8:e84244. doi: 10.1371/journal.pone.0084244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Cauffiez C., Klinzig F., Rat E., Tournel G., Allorge D., Chevalier D., Pottier N., Lovecchio T., Colombel J.F., Lhermitte M., et al. P4F12 genetic polymorphism: identification and functional characterization of seven variant allozymes. Biochem. Pharmacol. 2004;68:2417–2425. doi: 10.1016/j.bcp.2004.08.025. [DOI] [PubMed] [Google Scholar]
- 161.Choquet H., Trapani E., Goitre L., Trabalzini L., Akers A., Fontanella M., Hart B.L., Morrison L.A., Pawlikowska L., Kim H. Cytochrome P450 and matrix metalloproteinase genetic modifiers of disease severity in Cerebral Cavernous Malformation type 1. Free Radic. Biol. Med. 2016;92:100–109. doi: 10.1016/j.freeradbiomed.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Austin H., De Staercke C., Lally C., Bezemer I.D., Rosendaal F.R., Hooper W.C. New gene variants associated with venous thrombosis: A replication study in White and Black Americans. J. Thromb. Haemost. 2011;9:489–495. doi: 10.1111/j.1538-7836.2011.04185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Fiatal S. Genetic profiling revealed an increased risk of venous thrombosis in the Hungarian Roma population. Thromb. Res. 2019;179:37–44. doi: 10.1016/j.thromres.2019.04.031. [DOI] [PubMed] [Google Scholar]
- 164.Yue Y., Sun Q., Man C., Fu Y. Association of the CYP4V2 polymorphism rs13146272 with venous thromboembolism in a Chinese population. Clin. Exp. Med. 2019;19:159–166. doi: 10.1007/s10238-018-0529-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Glurich I., Chyou P.H., Engel J.M., Cross D.S., Onitilo A.A. Tamoxifen-induced venothromboembolic events: exploring validation of putative genetic association. Clin. Med. Res. 2013;11:16–25. doi: 10.3121/cmr.2012.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Song Y., Mo G., Yin G. A novel mutation in the CYP4V2 gene in a Chinese patient with Bietti’s crystalline dystrophy. Int. Ophthalmol. 2013;33:269–276. doi: 10.1007/s10792-012-9686-2. [DOI] [PubMed] [Google Scholar]