Abstract
ArthβDG is a dimeric, cold-adapted β-d-galactosidase that exhibits high hydrolytic and transglycosylation activity. A series of crystal structures of its wild form, as well as its ArthβDG_E441Q mutein complexes with ligands were obtained in order to describe the mode of its action. The ArthβDG_E441Q mutein is an inactive form of the enzyme designed to enable observation of enzyme interaction with its substrate. The resulting three-dimensional structures of complexes: ArthβDG_E441Q/LACs and ArthβDG/IPTG (ligand bound in shallow mode) and structures of complexes ArthβDG_E441Q/LACd, ArthβDG/ONPG (ligands bound in deep mode), and galactose ArthβDG/GAL and their analysis enabled structural characterization of the hydrolysis reaction mechanism. Furthermore, comparative analysis with mesophilic analogs revealed the most striking differences in catalysis mechanisms. The key role in substrate transfer from shallow to deep binding mode involves rotation of the F581 side chain. It is worth noting that the 10-aa loop restricting access to the active site in mesophilic GH2 βDGs, in ArthβDG is moved outward. This facilitates access of substrate to active site. Such a permanent exposure of the entrance to the active site may be a key factor for improved turnover rate of the cold adapted enzyme and thus a structural feature related to its cold adaptation.
Keywords: galactosidase, hydrolysis, reaction mechanism, complex structures, cold-adapted, GH2
1. Introduction
Glycosyl hydrolases (GHs) are sugar processing enzymes, divided into families based on their structures. This is the reason why the lactose processing β-d-galactosidases (βDGs) belong to different GH families: GH1, GH2, GH35, GH42, and GH59. Their common structural feature is presence of a TIM-barrel type catalytic domain followed by a variety of β-architecture domains, which nature and occurrence differ among GH subfamilies [1].
The most studied β-d-galactosidase in the GH2 family is bacterial lacZ βDG from Escherichia coli (EcolβDG) [2,3,4]. It is a large homotetramer, where each monomer consists of 1023 amino acids. Its primary mode of action is to catalyze the hydrolysis of lactose to d-galactose and d-glucose. To achieve its full catalytic efficiency EcolβDG requires the presence of divalent ions such as Mg2+ or Mn2+, which can result in 5−100-fold increase of activation depending on the substrate. Within the EcolβDG catalytic site, two subsites can be distinguished: the first exhibits high specificity for binding the galactose moiety, whereas the second provides a platform for weak binding of different moieties [5]. If an excess of galactose occurs, EcolβDG exhibits transglycosylation activity that results in formation of allolactose, a disaccharide composed of d-galactose and d-glucose moieties linked through a β-(1,6)-glycosidic bond [6,7].
Lactose processing enzymes are commonly used in the dairy industry for production of lactose-free products. Keeping dairy products in refrigerated conditions results in crystallization of lactose that leads to an undesirable grainy texture. That is why lactose removal is used for improving the quality of final products, such as ice-creams and some types of cheeses [8,9].
Another example of an enzyme from GH2 family, commonly used in dairy industry, is yeast βDG from Klyvuromyces lactis (KlyvβDG) [10]. Similar to EcolβDG, it consists of 1032 aa and its functional form is a homotetramer. Not only it catalyzes lactose hydrolysis, but also transglycosylation reaction that results in formation of galactose derivatives, among other alkyl-galactosides [11], gal-mannitol, [12], and bionic acids [13].
However, the usage of a cold-adapted enzyme, exhibiting similar catalytic efficiency to ones already implemented but at lower temperatures (4–18 °C), is highly sought after. Especially for the food industry, removing the need for a heating step not only brings the costs of production down, but it also prevents potential mesophilic contamination and loss of nutritional value of food products due to heating, and production of unwanted products by thermal conversion [14,15].
ArthβDG is an interesting candidate for industrial use. Not only can it hydrolyze lactose at a rate comparable to βDG from Klyvuromyces lactis but it exhibits additional transglycosylation activity [16]. Galactooligosaccharides (GOS) and heterooligosaccharides (HOS) are prebiotics, which are important for human health. That is why, with constantly increasing evidence of their consumption benefits, they found their way into infant nutrition and special nutrition, and more recently have become increasingly present in everyday food products [17,18,19,20,21,22,23,24,25,26].
The modification of transglycosylase activity specificity and efficiency may be achieved by controlling reaction equilibrium or by enzyme engineering. Studies concentrated on introducing mutations into subsites of GHs showed that the modulation of hydrolysis and transglycosylation activities can be achieved by means of knowledge-based enzyme engineering. However, the role of individual amino acids in the active site must be discovered as a basis for successful design of an enzyme with specific, desired activities [27].
ArthβDG is a five-domain protein of molecular weight 110 kDa. The catalytic domain, in form of TIM barrel, is surrounded by three IG-like domains and, as typical for the GH2 family, an N-terminal super β-sandwich domain. Regardless of low sequence identity, this monomer’s architecture is strikingly similar to KlyvβDG and EcolβDG, which enabled determination of catalytic residues E441 and E517. Cold-adapted ArthβDG possesses a functional dimer (not typical for the GH2 family). The dimer is stabilized by head-to-tail interactions between Domains 1 and Domains 5 from neighboring molecules [28]. The same oligomerization state, though shaped differently, was previously described by us for another cold-adapted β-d-galactosidase from Paracoccus sp. 32d for which we had determined crystal structure [29]. Thanks to comparative analysis, the structural features that may play a key part in its cold-adaptation were described. Most interesting was maximization of energy gain from the surface residue–solvent interactions, that was obtained by reduction of oligomerization state and formation of hydrophobic patches on the protein’s surface [30].
The comprehensive structural study of cold-adapted ArthβDG reaction mechanism is a first necessary stage for the knowledge-based enzyme engineering that could lead to creation of an enzyme that would not only hydrolyze lactose, but also effectively convert it to the beneficial for human health GOS and HOS at cold conditions. Usage of native protein would limit us to analysis of substrate binding using substrate analogs which could not by hydrolyzed by the enzyme, such as isopropyl β-d-1-thiogalactopyranoside (IPTG), or less preferable substrates such as ortho-nitrophenyl-β-galactoside (ONPG). However, thanks to designing an inactive mutant in which catalytic E441, acting as acid catalyst, was substituted with the structurally isomorphous glutamate residue, we were able to obtain complexes with the natural substrate lactose bound in both deep and shallow binding modes.
2. Results
2.1. Crystal Structures of ArthβDG and ArthβDG_E441Q Complexes
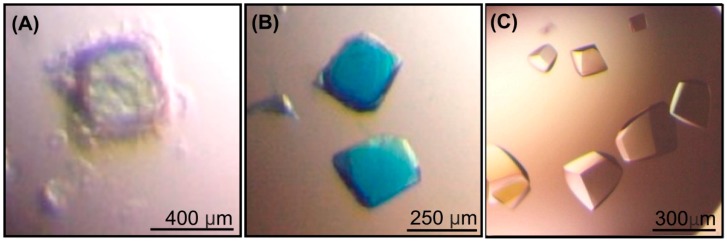
During soaking of ArthβDG crystals with ONPG and X-gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside) a coloration of crystals was observed. It was rapid for crystals soaked with X-gal, as they turned intensely blue within 3 min (Figure 1B).
Figure 1.
The crystals of ArthβDG: after addition of ONPG (A); after 2 h of soaking in X-gal (B). The crystals of ArthβDG_E441Q mutein soaked 24 h with mixture of lactose and galactose (C).
Different short soaking times (in a range of 10 s to 10 min, 22 crystals tested) and excess of X-gal (from 3 to 20 molar access in respect to protein concentration) gave intensely blue coloration of soaked crystals. This indicates that the enzyme in the crystal was in its active form and performed hydrolysis of X-gal. The structure solved using diffraction data collected from blue crystals was identical with the ArthβDG/GAL obtained by soaking with lactose. It means that X-gal, similarly to lactose, was hydrolyzed by enzyme during soaking of native crystals in ligand solution. Blue color came from 5,5′-dibromo-4,4′-dichloro-indigo—a dimer of the second product (5-bromo-4-chloro-3-hydroxyindol) of X-gal hydrolysis. The blue dye is either deposited in solvent channels of the crystal or randomly bound to the surface of protein. If the dimerized product of X-gal hydrolysis would be bound specifically, it should be detectible in the resulting crystal structure, because it contains two Br atoms, which give strong picks on the electron density maps.
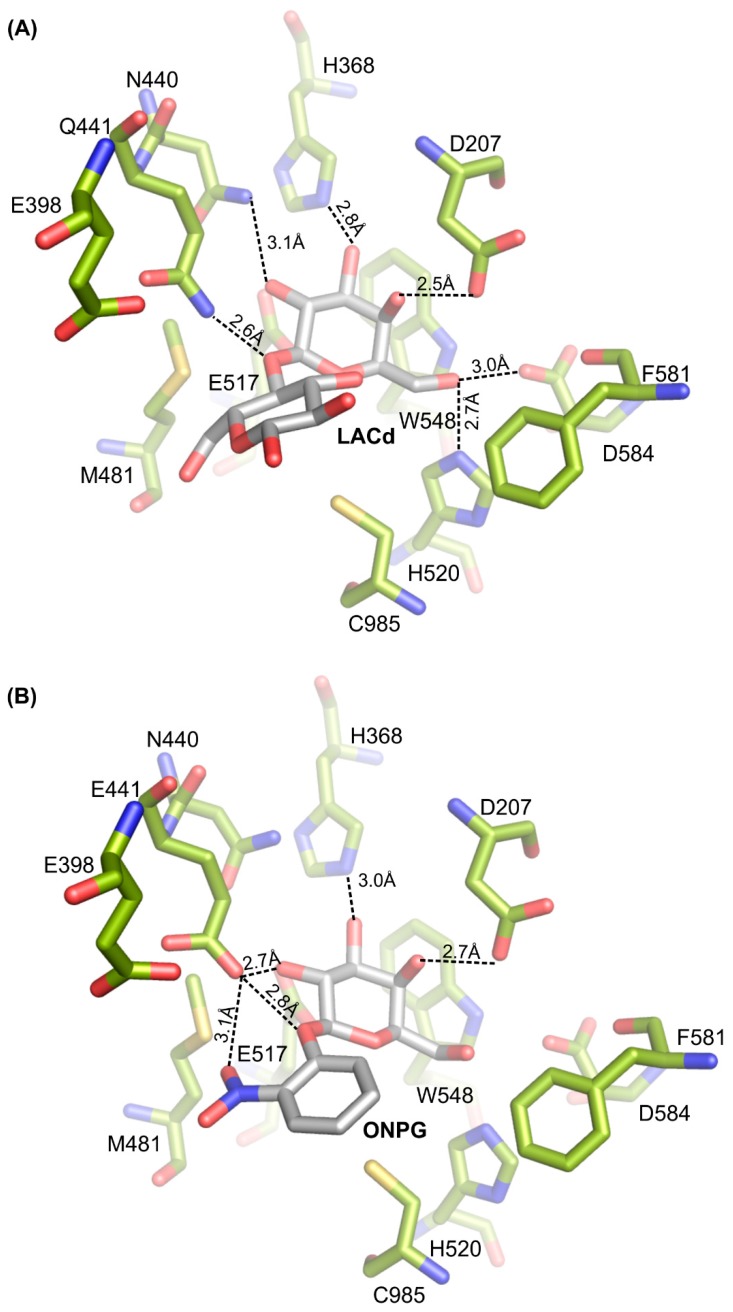
Soaking of ArthβDG crystals with ONPG resulted in a yellow halo around the soaked crystals (Figure 1A); however, the rate of ONPG hydrolysis was probably lower than X-gal as the coloration was visually observed only after more than an hour soaking. For the crystal soaked in ONPG for 15 min, the color was not observed and the complex of ArthβDG with ONPG bound was formed - an intact ONPG was visible in the active site after determining the crystal structure.
Crystals of ArthβDG soaked with lactose underwent fast deterioration, and a very short soaking time was required to obtain crystals still suitable for diffraction experiments. The diffraction data collected after only 1 min of soak with lactose resulted in the crystal structure with galactose. This is evidence that the active enzyme performed hydrolysis of lactose in crystal, in such fast rate, and product of reaction–galactose, bound in active site was visible in electron density maps.
The soaking of ArthβDG_E441Q mutein crystals with lactose yielded no complex in solved crystal structures, as for soaking times up to 6 h no ligand was found in the structure and for longer soaking times crystals were destroyed. We have succeeded in determining the crystal structure of ArthβDG_E441Q complex with lactose after soaking crystals of ArthβDG_E441Q with mixtures of lactose-galactose (Figure 1C) and lactose-fructose for 24 h. After this time, crystals of the mutein complexes were still without visible signs of deterioration and resulted in very good resolution data. The complex structures of lactose bound in shallow and deep mode were obtained while subjecting those crystals to diffraction experiments.
The crystal structures of ArthβDG complexes with galactose (2.1 Å), IPTG (2.2 Å), ONPG (2.8 Å), and its mutein ArthβDG_E441Q (1.8 Å) in complexes with lactose (LAC) bound in shallow mode ArthβDG_E441Q/LACs (1.9 Å) and in deep mode ArthβDG_E441Q/LACd (1.8 Å) were processed in trigonal space group P3121, the same as ArthβDG native structure. Matthew’s volume calculation [31] revealed that no changes in crystal packing were detected, and protein monomer was present in each asymmetric unit. Crystal structures of these complexes were solved in PHENIX by isomorphous replacement using rigid body procedure and the native structure of ArthβDG (PDB ID: 6ETZ) as a model. This allowed us to obtain the molecule in the same position and orientation in all the analyzed structures, which facilitated comparison of not only the structures, but also electron densities. The details for the diffraction data collection and processing are presented in Table 1. Each structure was further refined in PHENIX.REFINE, including TLS parameters [32] defined for each domain. The resulting refinement statistics are given in Table 1.
Table 1.
Diffraction data collection, processing, and refinement statistics for crystal structures of investigated ArthβDG complexes.
|
ArthβDG_E441Q PDB ID: 6SE8 |
ArthβDG_LACs PDB ID: 6SE9 |
ArthβDG_LACd PDB ID: 6SEA |
ArthβDG_IPTG PDB ID: 6SEB |
ArthβDG_ONPG PDB ID: 6SEC |
ArthβDG_GAL PDB ID: 6SED |
|
|---|---|---|---|---|---|---|
| Diffraction source | P13 PETRA, Hamburg, Germany | BL 14.1 BESSY, Berlin, Germany | BL 14.1 BESSY, Berlin, Germany | BL 14.2 BESSY, Berlin, Germany | BL 14.2 BESSY, Berlin, Germany | BL 14.2 BESSY, Berlin, Germany |
| Wavelength (Å) | 0.976250 | 0.918400 | 0.918400 | 0.918400 | 0.918400 | 0.918400 |
| Temperature (K) | 100 K | 100 K | 100 K | 100 K | 100 K | 100 K |
| Detector | PILATUS 6M | PILATUS 3S 2M | PILATUS 3S 2M | PILATUS 3S 2M | PILATUS 3S 2M | PILATUS 3S 2M |
| Rotation range per image (°) | 0.05 | 0.1 | 0.1 | 0.2 | 0.2 | 0.2 |
| Total rotation range (°) | 160 | 180 | 180 | 180 | 180 | 180 |
| Exposure time per image (s) | 0.1 | 0.2 | 0.2 | 0.3 | 0.3 | 0.3 |
| Space group | P3121 | P3121 | P3121 | P3121 | P3121 | P3121 |
| a, b, c (Å) | 136.8, 136.8, 127.0 | 138.9, 138.9, 127.9 | 138.6, 138.6, 127.4 | 137.1, 137.1, 126.9 | 137.4, 137.4, 126.8 | 136.8, 136.8, 126.8 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 |
| Mosaicity (°) | 0.133 | 0.084 | 0.72 | 0.115 | 0.247 | 0.130 |
| Resolution range (Å) | 50.0–1.8 (1.9–1.8) | 47.0–2.0 (2.1–2.0) | 46.9–1.9 (2.0–1.9) | 46.6–2.2 (2.3–2.2) | 46.6–2.6 ( 2.7–2.6) | 50.0–2.1 (2.2–2.1) |
| No. of unique reflections | 118,383 | 95,079 | 109,583 | 75,085 | 43,328 | 80,260 |
| Completeness (%) | 98.8 (92.5) | 99.4 (97.0) | 99.9 (99.5) | 99.9 (99.7) | 99.4 (98.1) | 99.8 (99.3) |
| Redundancy | 7.75 (6.33) | 10.09 (10.23) | 9.68 (9.85) | 5.60 (5.58) | 6.63 (6.32) | 10.08 (9.66) |
| I/σ (I) | 13.94 (2.12) | 15.23 (1.17) | 13.86 (1.12) | 11.86 (1.01) | 11.75 (1.03) | 16.07 (1.72) |
| Rmeas (%) | 8.6 (64.5) | 10.8 (193.7) | 9.8 (177.8) | 14.2 (170.6) | 16.8 (165.0) | 13.6 (140.6) |
| Overall B factor: Wilson plot/refinement (Å2) |
37.8/30.3 | 47.3/38.8 | 43.7/36.1 | 47.2/41.7 | 60.2/58.3 | 43.1/37.7 |
| No. of reflections: working/test set |
118,348/2101 | 100,463/2091 | 116,171/2088 | 63,550/2101 | 67,872/2112 | 66,731/2101 |
| R/Rfree | 0.135/0.165 | 0.203/0.238 | 0.182/0.205 | 0.159/0.205 | 0.174/0.240 | 0.166/0.204 |
| No. of non-H atoms: Protein/Ligand/Water |
7794/140/826 | 7619/133/400 | 7652/97/671 | 7649/64/617 | 7624/30/135 | 7672/65/605 |
| R.m.s. deviations: Bonds (Å)/Angles (°) |
0.008/0.964 | 0.003/0.602 | 0.010/1.004 | 0.007/0.873 | 0.008/1.008 | 0.002/0.563 |
| Ramachandran plot: Most favored/allowed (%) |
97.4/2.6 | 96.8/3.2 | 97.6/2.4 | 97.1/2.9 | 94.9/5.1 | 97.2/2.8 |
2.2. Active Center of ArthβDG and ArthβDG_E441Q Mutein
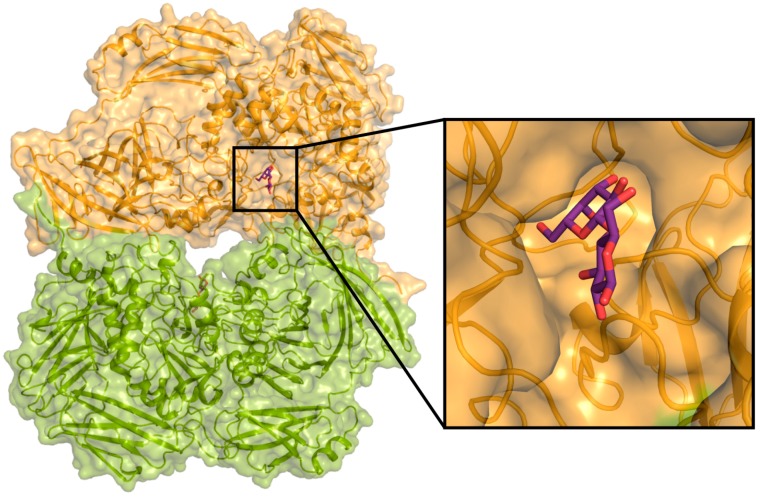
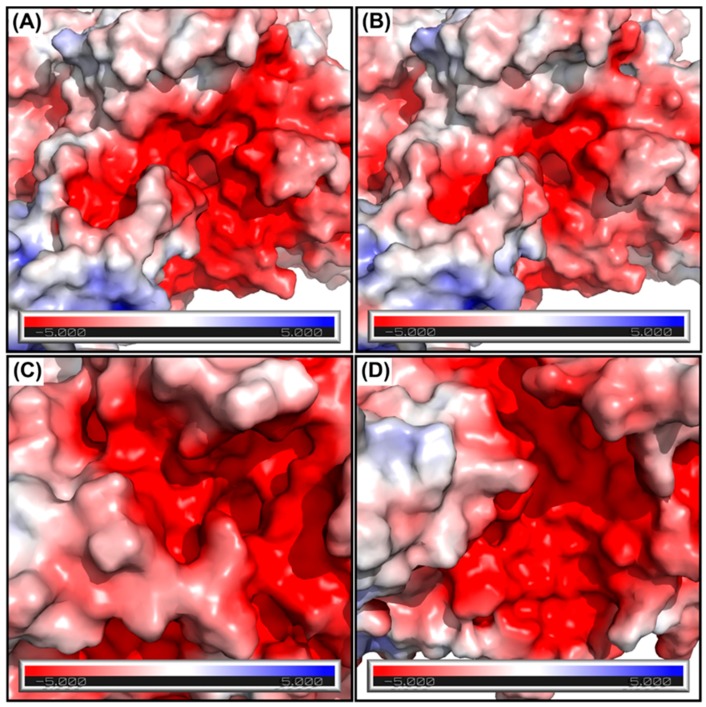
Catalytic site of ArthβDG, located at the bottom of a relatively wide funnel on the top of catalytic Domain 3, is complemented with parts of the chain from Domain 1 and Domain 5 at the entrance of the active cavity (Figure 2). The funnel leading to the active site has a strongly acidic character, which is beneficial for binding of carbohydrate substrates (Figure 3).
Figure 2.
The dimer of ArthβDG_E441Q and the zoom of one of the active site cavities with lactose.
Figure 3.
The surface potential visualization at the active site of ArthβDG (A), ArthβDG_E441Q (B), EcolβDG (C), and KlyvβDG (D).
The active site cavity has an acidic character throughout, which facilitates the binding of the saccharide substrate, which is typically lactose. Such a shape of the active site cavity is observed for other βDGs with transglycosylation activities: EcolβDG and KlyvβDG [4,7,10] as it facilitates the binding of a larger acceptor of galactosyl group, such as galactose, fructose, or salicin. ArthβDG forms a widely open entrance to its catalytic site, which makes it more accessible for the substrate but also promotes product dissociation. Both, easier product dissociation and active sites not being shielded or restricted from solvent may be considered as structural cold-adaptation as it influences the enzymes’ turnover rate.
2.3. Structural Analysis of Reactions’ Mechanism Catalyzed by ArthβDG
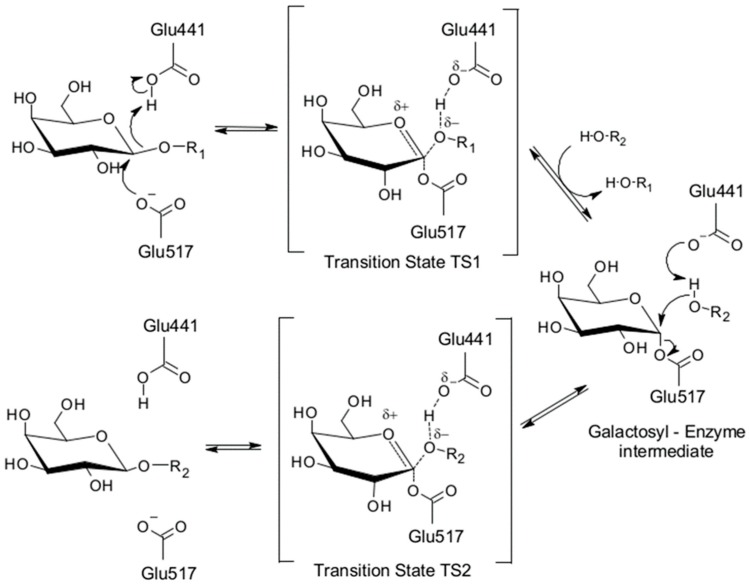
All determined crystal structures showed precisely the changes in the active site of the enzyme in different stages of catalyzed reaction. Visualizing these structural changes helps to understand and explain a classical Koshland double-displacement mechanism occurring during hydrolysis of (1,4)-β-O-glycosidic bond catalyzed by ArthβDG (Figure 4).
Figure 4.
The reaction mechanism of Koshland double displacement with the catalytic residues numbered as for ArthβDG [33].
Determined crystal structures of ArthβDG complexes with specific ligands may be divided into three groups: complexes with substrates and their analogues (early complexes) ArthβDG_E441Q/LACs, ArthβDG/IPTG; the second group are intermediates complexes ArthβDG_E441Q/LACd ArthβDG/ONPG; and the third one is the complex with product ArthβDG/GAL.
The early complexes with substrate show LAC and IPTG binding in shallow mode, intermediate complexes depicts deep binding of substrate that directly precedes formation of galactosyl-enzyme covalent bond, and the product complex allows description of the product release process.
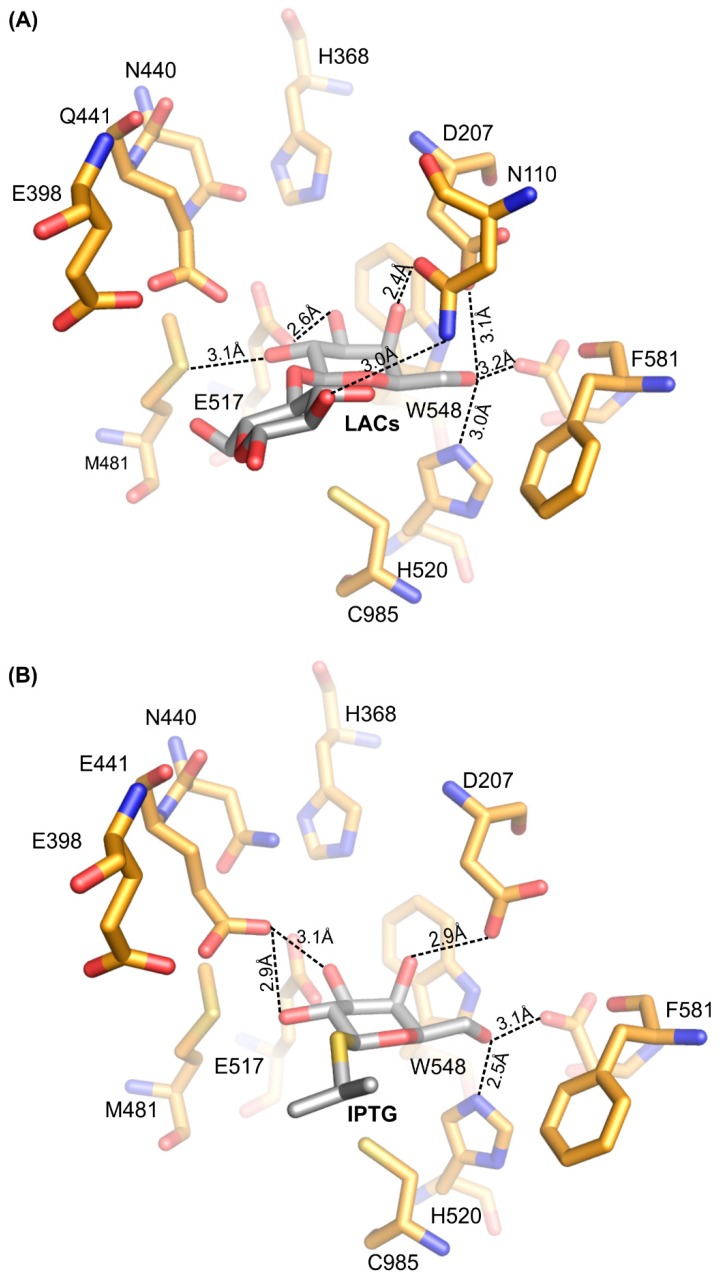
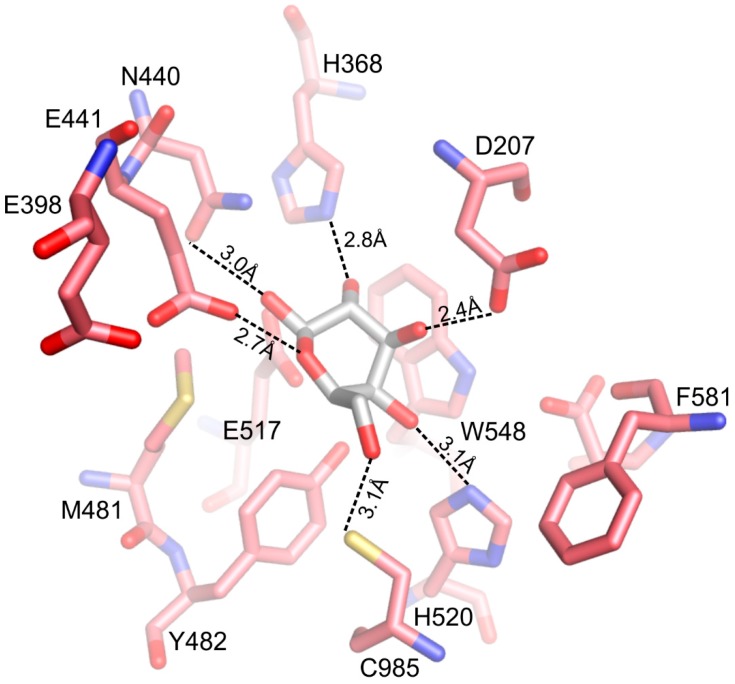
At the early stage of the reaction, the substrate is bound in the shallow binding site where the galactosyl group is stabilized by a number of H-bonds between its hydroxyl groups and residues N110, E441, E517, M481, H520, and H368 via water molecules and by an interaction with a sodium ion. Additionally, the glucosyl moiety of lactose is stabilized by H395 and E398 via a water molecule, even though there is already an interaction between substrate and catalytic residues (E441 and E517), and the position of the substrate does not allow access to O-glycosidic bond (Figure 5).
Figure 5.
Early complexes of ArthβDG with saccharide substrate and substrate analogue: the molecule of lactose (A) and IPTG (B) bound at shallow binding site.
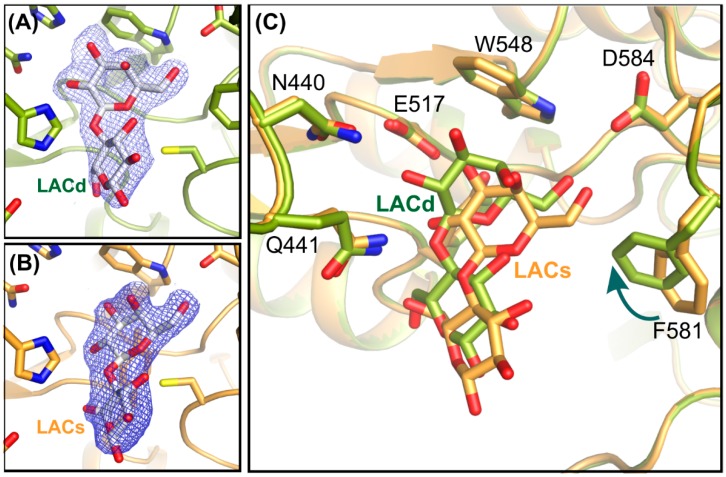
The insertion of substrate into deep binding is associated with movement of F581 phenyl ring, which rotates around Cα-Cβ bond causing shift of aromatic ring by 2.9 Å in the direction of the active center, reducing the volume of the shallow binding site (Figure 6). Surprisingly, no movement of backbone is observed during the transfer of ligand from shallow to deep binding site. During substrate transfer into the deep binding site, the galactosyl moiety, properly positioned in shallow binding stage, is moved deeper into the active site by approximately 2.4 Å being at the same time rotated around an axis perpendicular to the sugar ring by approximately 60°.
Figure 6.
Enzyme active site of shallow and deep binding of lactose. Electron density 2Fo-Fc map of lactose in deep (A) and shallow (B) binding mode (contoured at 1σ). Superposition of enzyme active site in both structures (C).
In the deep binding site of ArthβDG, the galactosyl ring is stabilized by direct interactions of its hydroxyl groups with E441 (Q441 in mutein), E517, H368, D207, sodium ion and N440, D584, H520 (latter observed by deeper bound lactose in complex of ArthβDG_E441Q_LACd). Now in ArthβDG_E441Q_LACd, the NH2 from the amide group of Q441 interacts directly with oxygen from the glycosidic bond of lactose, which is 2.6Å away (Figure 7A). In ArthβDG/ONPG, the carboxyl group of E441 interacts directly with oxygen from the glycosidic bond of ONPG, which is at a distance of 2.8 Å (Figure 7B).
Figure 7.
Late complexes of ArthβDG with substrates: the molecules of lactose (A) and ONPG (B).
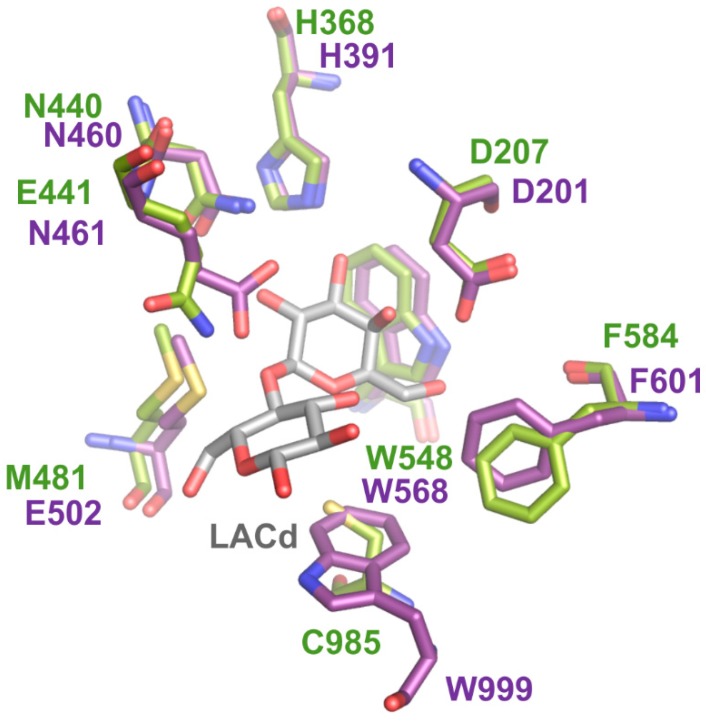
The superposition of active sites of ArthβDG with EcolβDG shows the conservation of amino acids involved in stabilization of the galactosyl moiety. However, these two enzymes differ in the stabilization of a second moiety of β-d-galactoside (Figure 8). If we consider binding of the natural substrate, lactose, the second moiety is glucopyranose. In the active site of EcolβDG, the glucopyranose ring is stabilized by π-stacking interaction with W999. However, in the ArthβDG active site, W999 is substituted by C985. This cysteine residue does not influence stabilization of substrate in shallow binding mode. However, when lactose is bound in deep mode, the center of the glucopyranose ring is at a distance of 4.4 Å from C985 making π-sulphur interaction possible. Thus, substitution of W999 with C985 reduces stabilization of the second moiety of β-d-galactoside during shallow binding mode, but is still creating stabilizing interactions when the substrate is bound in deep mode. Furthermore, this results in creating more space in the close vicinity of the active site by substituting the bulky indol group with a smaller cysteine residue side chain.
Figure 8.
Superposition of catalytic sites of ArthβDG with lactose bound in deep mode (green) and EcolβDG (purple).
After the hydrolysis reaction is completed, the F581 side chain moves back into its previous position, opening the way for galactose molecule evacuation from the active site (Figure 9).
Figure 9.
Complex structure of ArthβDG with galactose in half-chair conformation bound in the active center.
The product, now in half-chair conformation, is still stabilized by a number of interactions: D207, H368, N440, E441, Y482, E517, H520, and C985 (Figure 7). The ‘open’ position of F581 is also observed for unliganded structures of ArthβDG (PDB IDs: 6ETZ) and its mutant ArthβDG_E441Q, suggesting that its movement is dependent upon substrate presence at the shallow binding site.
3. Discussion
GH2 family β-d-galactosidases are sugar configuration-retaining enzymes that follow a classical Koshland double-displacement mechanism. These crystal structures of ArthβDG complexes with ligands enabled characterization of the active site and determined which residues take part in two modes of substrate binding: deep and shallow.
The large rotation of the galactosyl residue during deep binding would most probably result in forming π-stacking interaction with W548. Such a form of intermediate stabilization was described for EcolβDG [7]. It should be noted that a tryptophan residue is the preferred aromatic amino acid for binding of carbohydrates [34,35] and is frequently present in carbohydrate binding domains of proteins. In the case of ArthβDG, only one tryptophan, W548, is located in the bottom of the active site. Additionally, three tryptophan residues are present at the entrance to the catalytic pocket (W402, W470, and W773), where they may form platforms for initial sugar binding. Another amino acid which is considered to play an important role in carbohydrate binding is histidine. The active site of ArthβDG contains several histidine residues: H334, H368, H395, H520, and H553. Among them, H368 and H520 are directly involved in stabilizing the galactosyl moiety. H520 is primarily involved in stabilization of hydroxyl group O6 of the galactosyl moiety during shallow binding of substrate. When the substrate is moved deeper into catalytic site, H368 stabilizes the position of hydroxyl group O3. It must be noted that the catalytic site architecture of ArthβDG is composed such a way that only a sugar moiety with a proper conformation of hydroxyls O2, O3, and O4 can be effectively bound in the active site. Hence, residues forming H-bonds with hydroxyl groups in these positions H368, N440, and D207, play a crucial role in enzyme’s specificity.
It is worth noting that a typical chair conformation of the galactosyl ring in substrate (1C4) is changing to a half-chair (3H4)hkkkkHhhh conformation in the still bound product of the half-reaction. There are many conformations of pyranose ring possible in solution; however, some of them are more stable than others. In the case of lactose, it usually has a relaxed chair conformation in solution. The double displacement mechanism, in which lactose is hydrolyzed by retaining galactosidases, such as ArthβDG, undergoes formation of two oxocarbenium ion-like transition states (Figure 4). Such transition states must be formed with sp2 hybridization and formation of a positive charge on anomeric carbon atom of the substrate. Only a few conformations of galactosyl moiety allow sp2 hybridization on anomeric carbon, one of which is half-chair conformation 3H4, observed for the galactose bound in active site of ArthβDG [36].
The rotation of F581 (F601 in EcolβDG) was described as one of the factors associated with the deep binding mode, together with 10 Å movement of a 10-aa loop from Domain 5. However, in the case of ArthβDG, the movement of F581 and D207 are the only conformational changes accompanying the reaction mechanism. In fact, the 10-aa loop in ArthβDG is stabilized by a number of strong interactions with other parts of Domain 5, in a position allowing better access of the substrate to the active site. It should be noted that it is one of the regions of ArthβDG in which the backbone differed significantly from homologous structures. These facts lead us to consider this permanent exposure of the entrance to the active site as a structural adaptation towards activity in cold conditions. Fewer structural hindrances for substrate entering and product leaving the active site can result in a higher turnover rate. Analysis of these obtained crystal structures shows that ArthβGD forms a widely open entrance to its catalytic site, which makes it more accessible for the saccharide substrate and promotes product dissociation.
Both galactosyl binding sites, shallow and deep ones, form a net of H-bonds that stabilize this part of the substrate. On the other hand, the glucosyl moiety of lactose, or the non-galactose moieties of IPTG and ONPG, are hardly stabilized by any interactions during shallow binding. The EcolβDG W999 is substituted at ArthβDG with a cysteine residue which may stabilize the second sugar ring of the substrate by π-sulphur interactions during deep binding, however such a substitution would render the enzyme less specific toward binding a sugar moiety at this position. Thus, not only disaccharides, but also other galactosides are processed by ArthβDG. The enzyme’s lack of preference for the second moiety in galactoside may be the main reason for its ability to hydrolyze a wide variety of substrates, as well as for its ability to transfer galactosyl group to a variety of acceptors [16] resulting in an interesting range of potentially useful heterooligosaccharides.
4. Materials and Method
4.1. Site-Directed Mutagenesis of Gene Encoding ArthβDG
The gene encoding the ArthβDG enzyme, which was previously cloned into the pBAD/Myc-His A expression vector [16], has been mutated in a site-specific manner using the Q5 Site-Directed Mutagenesis Kit (NEB, Ipswich, MA, USA) following the manufacturer’s protocol. For this purpose, a pair of mutagenic primers was designed and synthesized (Genomed, Warszawa, Poland). Primer ForBglAr32cBm441: 5′GTCCCTGGGCAACCAGGCGGCACCGG3′ and primer RevBglAr32m441: 5′CACATGACCACCGAGGCGTGGTTCTTGTCGCGC3′ allowed us to introduce a point mutation at 1321 nucleotide position in the gene substituting G with C resulting in the substitution of glutamic acid (E) residue with glutamate (Q) residue in the 441 position of the amino acid chain of ArthβDG. Hence, the product of mutated gene expression has been called ArthβDG_E441Q. In theory, this amino acid change should abolish β-d-galactosidase activity of mutein ArthβDG_E441Q.
PCR cycling conditions were as follows: (1) Initial DNA denaturation at 98 °C for 30 s; then (2) 25 cycles of PCR product amplification consisting of 10 s of DNA denaturation at 98 °C, 20 s of mutagenic primers annealing at 70 °C, and 3 min 20 s of PCR product extension at 72 °C; and (3) the final PCR product extension at 72 °C for 7 min. After PCR, the amplified DNA product was directly added to unique Kinase-Ligase-DpnI (KLD) enzymes mix. Then the product of KLD reaction (5 min at room temperature) was directly used to transform NEB 5-alpha chemically competent E.coli cells (the lacZ deletion mutant, ∆ (lacZ) M15). After that, transformants were spread on Luria–Bertani agar plates (10 g L−1 of peptone K, 5 g L−1 of yeast extract, 10 g L−1 of NaCl, and 15 g L−1 of agar) supplemented with ampicillin (100 μg mL−1), X-Gal (40 μg mL−1) and l-arabinose (200 μg mL−1). After plate incubation—firstly at 37 °C for 12 h, and then at 22 °C for next 12 h—a few recombinant colonies without β-d-galactosidase activity were chosen for further studies. Plasmids isolated using the ExtractMe Plasmid DNA Kit (Blirt, Gdansk, Poland) from selected recombinants were sequenced (Genomed, Warszawa, Poland) and analyzed (blast2go on-line tool). Recombinant plasmid pBAD-Bgal32cB_E441Q(A) harboring the properly mutated Arthrobacter sp. 32cB β-d-galactosidase gene under the control of the PBAD promoter was used for effective production of ArthβDG_E441Q mutein in E. coli host [16].
4.2. Expression and Purification of ArthβDG and ArthβDG_E441Q
Heterologous expressions of recombinant ArthβDG and ArthβDG_E441Q proteins were performed in the E. coli LMG 194 cells transformed with pBAD-Bgal32cB and pBAD-Bgal32cB_E441Q plasmids, respectively, as previously described. [25] Both proteins were purified by two ion-exchange chromatography steps (weak anion exchanger and strong anion exchanger), followed by a size-exclusion chromatography step.
The fractions containing ArthβDG were identified by SDS-PAGE electrophoresis run on 10% SDS-polyacrylamide gel and by enzymatic activity assay with ONPG as a substrate [25], whereas the fractions containing ArthβDG_E441Q were identified by SDS-PAGE only, due to lack of enzymatic activity. The sample buffer was changed into 0.05 M HEPES pH 7.0 and the samples were concentrated using 50 kDa cut-off membrane Vivaspin filters (Sartorius, Goettingen, Germany) up to the protein concentration of 15 mg/mL.
4.3. ArthβDG Crystallization and Diffraction Data Collection
Crystals of ArthβDG and ArthβDG_E441Q mutein were grown using the same optimization matrix of 25–45% TacsimateTM and pH ranges between 6.0–8.0. All the drops were set up using a seed stock prepared from crystals of ArthβDG grown at 35% TacsimateTM pH 7.0 and diluted 10,000 times. Numerous attempts of co-crystallization with ligands were undertaken but no crystal structures of desired complexes were obtained. Furthermore, addition of natural substrate, lactose, prevented formation of ArthβDG_E441Q crystals even at very low concentration of added ligand. Crystal structures of investigated ArthβDG and ArthβDG_E441Q complexes were obtained by soaking of native and mutant crystals with desired ligand or ligands mixture. Soaking was performed by adding powder of ligand directly to the crystallization drop. The soaking experiments were performed for 15 min, 30 min, 1 h, 2 h, 6 h, 14 h, and 24 h prior to flash-freezing. The crystals, prior to mounting and flash-freezing, were protected with 60% TacsimateTM of pH corresponding to crystallization conditions [37].
High-resolution diffraction data were collected using synchrotron sources on beamlines 14.1 and 14.2 at BESSY, Berlin, Germany and P13 beamline at PETRA, DESY Hamburg, Germany. The diffraction images were collected with fine slicing 0.1° and diffraction data were processed using XDSapp [38]. Crystal structures were solved and refined using the PHENIX program suite [39]. As a model, the structure of ArthβDG (PDB ID: 6ETZ) was used.
Acknowledgments
We thank HZB for the allocation of synchrotron radiation beamtime at BL 14.1, BL 14.2, and PETRA synchrotron at P13.
Abbreviations
| ArthβDG | β-d-galactosidase from Arthrobacter sp. 32cB |
| EcolβDG | β-d-galactosidase from Escherichia coli |
| KlyvβDG | β-d-galactosidase from Klyvuromyces lactis |
| GAL | galactose |
| GOS | galactooligosaccharides |
| HOS | heterooligosaccharides |
| IPTG | isopropyl β-d-1-thiogalactopyranoside |
| Lac | Lactose |
| ONPG | ortho-nitrophenyl-β-galactoside |
| X-gal | 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside |
Author Contributions
M.R. performed crystallization; M.R. and A.B. performed synchrotron diffraction data collection, processing, structure solving, and carried out structural analysis; M.R. purified enzyme, refined the structures; M.R. and A.B. prepared the manuscript; M.W. performed native ArthβDG enzyme expression in E. coli and determined the purification protocol; H.C. designed and performed site-direct mutagenesis experiment resulted in a gene encoding ArthβDG_E441Q mutein; A.W.-W. performed ArthβDG-E441Q mutein expression in E.coli; A.B. coordinated the project.
Funding
This research was funded by National Science Centre of Poland grant number 2016/21/B/ST5/00555 (A.B.) and 2018/28/T/ST5/00233 scholarship (M.R.).
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- 1.Talens-Perales D., Górska A., Huson D.H., Polaina J., Marín-Navarro J. Analysis of Domain Architecture and Phylogenetics of Family 2 Glycoside Hydrolases (GH2) PLoS ONE. 2016;11:e0168035. doi: 10.1371/journal.pone.0168035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohn M., Monod J. Purification and properties of the beta-galactosidase (lactase) of Escherichia coli. Biochim. Biophys. Acta. 1951;7:153–174. doi: 10.1016/0006-3002(51)90013-3. [DOI] [PubMed] [Google Scholar]
- 3.Jacobson R.H., Zhang X.J., DuBose R.F., Matthews B.W. Three-dimensional structure of beta-galactosidase from E. coli. Nature. 1994;369:761–766. doi: 10.1038/369761a0. [DOI] [PubMed] [Google Scholar]
- 4.Juers D.H., Matthews B.W., Huber R.E. LacZ beta-galactosidase: Structure and function of an enzyme of historical and molecular biological importance. Protein Sci. 2012;21:1792–1807. doi: 10.1002/pro.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brás N.F., Fernandes P.A., Ramos M.J. QM/MM Studies on the β-Galactosidase Catalytic Mechanism: Hydrolysis and Transglycosylation Reactions. J. Chem. Theory Comput. 2010;6:421–433. doi: 10.1021/ct900530f. [DOI] [PubMed] [Google Scholar]
- 6.Juers D.H., Rob B., Dugdale M.L., Rahimzadeh N., Giang C., Lee M., Matthews B.W., Huber R.E. Direct and indirect roles of His-418 in metal binding and in the activity of beta-galactosidase (E. coli) Protein Sci. 2009;18:1281–1292. doi: 10.1002/pro.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Juers D.H., Heightman T.D., Vasella A., McCarter J.D., Mackenzie L., Withers S.G., Matthews B.W. A structural view of the action of Escherichia coli (lacZ) beta-galactosidase. Biochemistry. 2001;40:14781–14794. doi: 10.1021/bi011727i. [DOI] [PubMed] [Google Scholar]
- 8.Harju M. Milk sugars and minerals as ingredients. Int. J. Dairy Technol. 2001;54:61–63. doi: 10.1046/j.1471-0307.2001.00016.x. [DOI] [Google Scholar]
- 9.Harju M., Kallioinen H., Tossavainen O. Lactose hydrolysis and other conversions in dairy products: Technological aspects. Int. Dairy J. 2012;22:104–109. doi: 10.1016/j.idairyj.2011.09.011. [DOI] [Google Scholar]
- 10.Pereira-Rodriguez A., Fernandez-Leiro R., Gonzalez-Siso M.I., Cerdan M.E., Becerra M., Sanz-Aparicio J. Structural basis of specificity in tetrameric Kluyveromyces lactis beta-galactosidase. J. Struct. Biol. 2012;177:392–401. doi: 10.1016/j.jsb.2011.11.031. [DOI] [PubMed] [Google Scholar]
- 11.Stevenson D.E., Stanley R.A., Furneaux R.H. Optimization of alkyl beta-D-galactopyranoside synthesis from lactose using commercially available beta-galactosidases. Biotechnol. Bioeng. 1993;42:657–666. doi: 10.1002/bit.260420514. [DOI] [PubMed] [Google Scholar]
- 12.Klewicki R., Belina I., Wojciechowska A., Klewicka E., Sójka M. Synthesis of Galactosyl Mannitol Derivative Using β-Galactosidase from Kluyveromyces lactis. Pol. J. Food Nutr. Sci. 2017;67:33–39. doi: 10.1515/pjfns-2016-0002. [DOI] [Google Scholar]
- 13.Wojciechowska A., Klewicki R., Sójka M., Klewicka E. Synthesis of the Galactosyl Derivative of Gluconic Acid with the Transglycosylation Activity of β-Galactosidase. Food Technol. Biotechnol. 2017;55:258–265. doi: 10.17113/ftb.55.02.17.4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cavicchioli R., Charlton T., Ertan H., Omar S.M., Siddiqui K.S., Williams T.J. Biotechnological uses of enzymes from psychrophiles. Microb. Biotechnol. 2011;4:449–460. doi: 10.1111/j.1751-7915.2011.00258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cavicchioli R., Siddiqui K.S., Andrews D., Sowers K.R. Low-temperature extremophiles and their applications. Curr. Opin. Biotechnol. 2002;13:253–261. doi: 10.1016/S0958-1669(02)00317-8. [DOI] [PubMed] [Google Scholar]
- 16.Pawlak-Szukalska A., Wanarska M., Popinigis A.T., Kur J. A novel cold-active β-d-galactosidase with transglycosylation activity from the Antarctic Arthrobacter sp. 32cB–Gene cloning, purification and characterization. Process Biochem. 2014;49:2122–2133. doi: 10.1016/j.procbio.2014.09.018. [DOI] [Google Scholar]
- 17.Boehm G., Fanaro S., Jelinek J., Stahl B., Marini A. Prebiotic concept for infant nutrition. Acta Paediatr. 2003;92:64–67. doi: 10.1111/j.1651-2227.2003.tb00648.x. [DOI] [PubMed] [Google Scholar]
- 18.Lee L.Y., Bharani R., Biswas A., Lee J., Tran L.A., Pecquet S., Steenhout P. Normal growth of infants receiving an infant formula containing Lactobacillus reuteri, galacto-oligosaccharides, and fructo-oligosaccharide: A randomized controlled trial. Matern. Health Neonatol. Perinatol. 2015;1:9. doi: 10.1186/s40748-015-0008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li M., Monaco M.H., Wang M., Comstock S.S., Kuhlenschmidt T.B., Fahey G.C., Jr., Miller M.J., Kuhlenschmidt M.S., Donovan S.M. Human milk oligosaccharides shorten rotavirus-induced diarrhea and modulate piglet mucosal immunity and colonic microbiota. ISME J. 2014;8:1609–1620. doi: 10.1038/ismej.2014.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes C., Davoodi-Semiromi Y., Colee J.C., Culpepper T., Dahl W.J., Mai V., Christman M.C., Langkamp-Henken B. Galactooligosaccharide supplementation reduces stress-induced gastrointestinal dysfunction and days of cold or flu: A randomized, double-blind, controlled trial in healthy university students. Am. J. Clin. Nutr. 2011;93:1305–1311. doi: 10.3945/ajcn.111.014126. [DOI] [PubMed] [Google Scholar]
- 21.Kunz C., Rudloff S. Biological functions of oligosaccharides in human milk. Acta Paediatr. 1993;82:903–912. doi: 10.1111/j.1651-2227.1993.tb12597.x. [DOI] [PubMed] [Google Scholar]
- 22.Torres D.P., Gonçalves M., Teixeira J.A., Rodrigues L.R. Galacto-Oligosaccharides: Production, Properties, Applications, and Significance as Prebiotics. Compr. Rev. Food Sci. Food Saf. 2010;9:438–454. doi: 10.1111/j.1541-4337.2010.00119.x. [DOI] [PubMed] [Google Scholar]
- 23.McVeagh P., Miller J.B. Human milk oligosaccharides: Only the breast. Acta Paediatr. 1997;33:281–286. doi: 10.1111/j.1440-1754.1997.tb01601.x. [DOI] [PubMed] [Google Scholar]
- 24.Musilova S., Rada V., Vlkova E., Bunesova V. Beneficial effects of human milk oligosaccharides on gut microbiota. Benef. Microbes. 2014;5:273–283. doi: 10.3920/BM2013.0080. [DOI] [PubMed] [Google Scholar]
- 25.Mussatto S.I., Mancilha I.M. Non-digestible oligosaccharides: A review. Carbohydr. Polym. 2007;68:587–597. doi: 10.1016/j.carbpol.2006.12.011. [DOI] [Google Scholar]
- 26.Oliveira D.L., Wilbey R.A., Grandison A.S., Roseiro L.B. Milk oligosaccharides: A review. Diary Technol. 2015;68:305–321. doi: 10.1111/1471-0307.12209. [DOI] [Google Scholar]
- 27.Manas N.H.A., Illias R.M., Mahadi N.M. Strategy in manipulating transglycosylation activity of glycosyl hydrolase for oligosaccharide production. Crit. Rev. Biotechnol. 2018;38:272–293. doi: 10.1080/07388551.2017.1339664. [DOI] [PubMed] [Google Scholar]
- 28.Rutkiewicz-Krotewicz M., Pietrzyk-Brzezinska A.J., Wanarska M., Cieslinski H., Bujacz A. In Situ Random Microseeding and Streak Seeding Used for Growth of Crystals of Cold-Adapted beta-D-Galactosidases: Crystal Structure of beta DG from Arthrobacter sp. 32cB. Crystals. 2018;8:13. doi: 10.3390/cryst8010013. [DOI] [Google Scholar]
- 29.Rutkiewicz-Krotewicz M., Pietrzyk-Brzezinska A.J., Sekula B., Cieśliński H., Wierzbicka-Woś A., Kur J., Bujacz A. Structural studies of a cold-adapted dimeric β-D-galactosidase from Paracoccus sp. 32d. Acta Cryst. D Struct. Biol. 2016;72:1049–1061. doi: 10.1107/S2059798316012535. [DOI] [PubMed] [Google Scholar]
- 30.Rutkiewicz M., Bujacz A., Bujacz G. Structural features of cold-adapted dimeric GH2 β-D-galactosidase from Arthrobacter sp. 32cB. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2019;1867:776–786. doi: 10.1016/j.bbapap.2019.06.001. [DOI] [PubMed] [Google Scholar]
- 31.Matthews B.W. Solvent content of protein crystals. J. Mol. Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 32.Winn M.D., Isupov M.N., Murshudov G.N. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Cryst. D. 2001;57:122–133. doi: 10.1107/S0907444900014736. [DOI] [PubMed] [Google Scholar]
- 33.Sinnott M.L., Souchard I.J.L. The mechanism of action of β-galactosidase. Effect of aglycone nature and α-deuterium substitution on the hydrolysis of aryl galactosides. Biochem. J. 1973;133:89–98. doi: 10.1042/bj1330089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hudson K.L., Bartlett G.J., Diehl R.C., Agirre J., Gallagher T., Kiessling L.L., Woolfson D.N. Carbohydrate-Aromatic Interactions in Proteins. J. Am. Chem. Soc. 2015;137:15152–15160. doi: 10.1021/jacs.5b08424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malecki P.H., Vorgias C.E., Petoukhov M.V., Svergun D.I., Rypniewski W. Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution. Acta Cryst. D Biol. Cryst. 2014;70:676–684. doi: 10.1107/S1399004713032264. [DOI] [PubMed] [Google Scholar]
- 36.Ardevol A., Rovira C. Reaction Mechanisms in Carbohydrate-Active Enzymes: Glycoside Hydrolases and Glycosyltransferases. Insights from ab Initio Quantum Mechanics/Molecular Mechanics Dynamic Simulations. J. Am. Chem. Soc. 2015;137:7528–7547. doi: 10.1021/jacs.5b01156. [DOI] [PubMed] [Google Scholar]
- 37.Bujacz G., Wrzesniewska B., Bujacz A. Cryoprotection properties of salts of organic acids: A case study for a tetragonal crystal of HEW lysozyme. Acta Cryst. D. 2010;66:789–796. doi: 10.1107/S0907444910015416. [DOI] [PubMed] [Google Scholar]
- 38.Sparta K.M., Krug M., Heinemann U., Mueller U., Weiss M.S. XDSAPP2.0. J. Appl. Crystallogr. 2016;49:1085–1092. doi: 10.1107/S1600576716004416. [DOI] [Google Scholar]
- 39.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.-W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]