Abstract
Enteroviruses (family Picornaviridae) comprise a large group of human pathogens against which no licensed antiviral therapy exists. Drug-repurposing screens uncovered the FDA-approved drug fluoxetine as a replication inhibitor of enterovirus B and D species. Fluoxetine likely targets the nonstructural viral protein 2C, but detailed mode-of-action studies are missing because structural information on 2C of fluoxetine-sensitive enteroviruses is lacking. We here show that broad-spectrum anti-enteroviral activity of fluoxetine is stereospecific concomitant with binding to recombinant 2C. (S)-Fluoxetine inhibits with a 5-fold lower 50% effective concentration (EC50) than racemic fluoxetine. Using a homology model of 2C of the fluoxetine-sensitive enterovirus coxsackievirus B3 (CVB3) based upon a recently elucidated structure of a fluoxetine-insensitive enterovirus, we predicted stable binding of (S)-fluoxetine. Structure-guided mutations disrupted binding and rendered coxsackievirus B3 (CVB3) resistant to fluoxetine. The study provides new insights into the anti-enteroviral mode-of-action of fluoxetine. Importantly, using only (S)-fluoxetine would allow for lower dosing in patients, thereby likely reducing side effects.
Keywords: antiviral, enteroviruses, drug repurposing, virus replication, molecular modeling
The genus Enterovirus within the Picornaviridae family includes many medically and socioeconomically important pathogens, which are among the most common infections in mankind. Four enterovirus (EV) species (EV-A, -B, -C, and -D) and three rhinovirus (RV) species (RV-A, -B, and -C) include serotypes that are known to cause human infections, like poliovirus, coxsackie A and B viruses, echoviruses, numbered enteroviruses (e.g., EV-A71 and EV-D68), and rhinovirus. Infections with enteroviruses can cause a broad spectrum of diseases ranging from hand-foot-and-mouth disease to conjunctivitis, aseptic meningitis, severe neonatal sepsis-like disease, and acute flaccid paralysis, whereas infections with rhinoviruses cause the common cold as well as exacerbations of asthma and chronic obstructive pulmonary disease (COPD).1 These viral infections are often self-limiting but can also result in severe complications especially in young children. To date, no antiviral therapy to treat enterovirus infections has been approved and treatment remains limited to supportive care. Worldwide vaccination campaigns have almost eradicated poliomyelitis. However, the vaccines against poliovirus and a recently approved EV-A71 vaccine in China are currently the only ones developed against enteroviruses. Vaccination is likely not a feasible general strategy to prevent enterovirus infections given the enormous amount (>250) of enterovirus (sero)types. Hence, the development of broad-spectrum anti-enteroviral drugs could be a promising alternative.
Enteroviruses are small, nonenveloped, positive-sense, single-stranded RNA viruses with an icosahedral capsid. The genome of ∼7.5 kb encodes a single polyprotein that is autoprocessed into structural proteins (VP1, VP2, VP3, and VP4), nonstructural proteins (2A, 2B, 2C, 3A, 3B, 3C, and 3D), and several functional processing intermediates. The viral nonstructural proteins, particularly the protease 3Cpro and the RNA-dependent RNA polymerase 3Dpol, are attractive targets for antiviral drug development.2
The viral protein 2C is the most conserved nonstructural protein among picornaviruses, which makes it particularly interesting for broad-spectrum anti-enteroviral drug design. The viral 2C protein functions as ATPase,3−5 ATPase-dependent RNA helicase, and an ATPase-independent RNA chaperone;6 all of these enzymatic functions are indispensable for the viral life cycle. The ATPase domain of the protein belongs to the superfamily of SF3 helicases of the AAA+ ATPases and contains Walker A and Walker B motifs and motif C.7 Besides the ATPase domain, 2C harbors an N-terminal membrane-associated helical domain, a cysteine-rich motif, and putative RNA binding motifs. 2C has been implicated in pleiotropic functions such as uncoating,8 cellular membrane rearrangement,9−12 RNA binding,13−15 RNA replication,16−21 immune evasion,22 and encapsidation.23−26 Although 2C has a central role in the viral life cycle, the exact details of its involvement remain poorly understood.
Over the past decades, structurally disparate 2C inhibitors such as guanidine hydrochloride (GuaHCl), 2-(α-hydroxybenzyl)-benzimidazole HBB, MRL-1237, and TBZE-029 have been identified.2,27−30 An emerging concept to discover new antivirals is drug repurposing. This strategy offers an attractive alternative to de novo drug development, as profound pharmacological and toxicological profiles of the compounds are already available. Furthermore, when the repurposed drug can be used at a similar dosage as for the original indication, it may directly enter phase 2 clinical trials,31,32 thereby reducing development cost and time. Several drug-repurposing screens have uncovered FDA-approved drugs as inhibitors of enterovirus replication.2 Some of these compounds are thought to inhibit the nonstructural protein 2C because nonsynonymous resistance mutations occur in 2C. Fluoxetine (Prozac), a selective serotonin reuptake inhibitor (SSRI) that is FDA-approved for the treatment of major depression and anxiety disorders, was identified as a potent inhibitor of EV-B and -D species but EV-A, EV-C, or rhinovirus species remained unaffected.33−35 Besides its anti-enteroviral activity, fluoxetine was also shown to inhibit dengue virus and hepatitis C virus, two members of the Flaviviridae family, where it likely acts as a host-targeting rather than a direct-acting antiviral as it is the case for enteroviruses.36,37 Fluoxetine has already been successfully used to treat an immunocompromised child with life-threatening chronic enterovirus encephalitis,38 underscoring the potential of fluoxetine for the application as an anti-enteroviral compound. Although various 2C inhibitors have been discovered over the years, their mode-of-action is still poorly understood.
Here, we set out to investigate how fluoxetine targets 2C of coxsackievirus B3 (CVB3), a virus model commonly used as a prototype for enterovirus B species. Fluoxetine has one chiral center, resulting in 2 enantiomers, and we experimentally showed that only the S-enantiomer inhibits enterovirus replication by directly binding to the viral protein 2C. On the basis of the recently published crystal structure of the catalytic domain of EV-A71 2C protein,39 a homology model for the corresponding part of CVB3 2C was generated. Two pockets flanking a stretch of amino acids that often mutate to convey resistance against 2C inhibitors (224AGSINA229) were identified. Molecular dynamics simulations predicted a stable interaction for the (S)-fluoxetine in only one of these pockets. Mutations of residues deep in the predicted binding pocket confer resistance to fluoxetine and contribute to the understanding of the antiviral mode-of-action. Thus, we identified for the first time a putative binding pocket for antiviral compounds in the nonstructural enterovirus protein 2C.
1. Results
1.1. (S)-Fluoxetine Inhibits CVB3 Replication by Binding to the Nonstructural Protein 2C
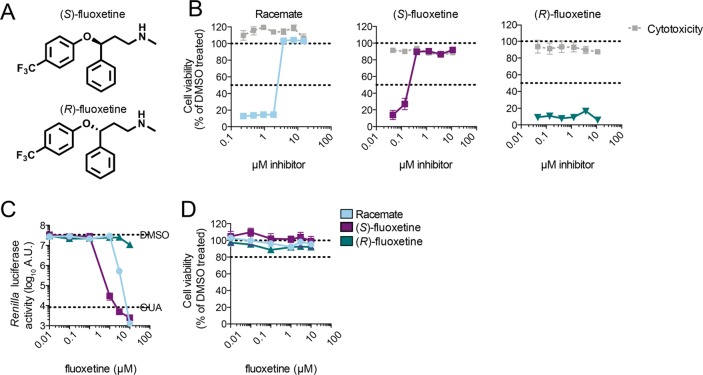
Fluoxetine is clinically used as a racemic mixture (1:1 enantiomeric ratio), and both enantiomers are of equal pharmacological activity toward the serotonin transporter SERT.40 The racemic compound was identified in drug-repurposing screens as an inhibitor of replication of EV-B and EV-D species.33,34 Since fluoxetine has one chiral center, we investigated the antiviral properties of both enantiomers (Figure 1A). Coxsackievirus B3 (CVB3), a member of the EV-B genus, causes a readily observable cytopathic effect (CPE), apparent as rounding, detachment, and eventually dying of the cell. The racemic mixture and both enantiomers, purchased from two different vendors (Sigma-Aldrich and Carbosynth), were tested in a multicycle CPE-reduction assay to elucidate whether the compounds inhibit virus replication and thereby prevent the development of CPE. In parallel, cytotoxicity of the compounds was determined using an calorimetric method for sensitive quantification using the (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) salt (MTS).
Figure 1.
Fluoxetine inhibits CVB3 replication in a stereospecific manner. (A) The two enantiomers of fluoxetine. (B) Multicycle CPE reduction assay to determine the antiviral activity of fluoxetine enantiomers. HeLa R19 cells were treated with serial dilutions of racemate, (S)-, or (R)-fluoxetine and infected with coxsackievirus B3 (CVB3) at an MOI of 0.001. In parallel, cells were treated with the compound only to assess cytotoxicity. After 3 days, cell viability was determined using an MTS assay. Data are from one experiment, representative of at least three independent experiments. (C) In a single cycle assay, HeLa R19 cells were infected with Renilla luciferase (RLuc)-CVB3 reporter virus and treated with serial dilutions of racemate, (S)-, or (R)-fluoxetine, and luciferase activity was determined at 7 h post-infection as a quantitative measure of replication. (D) In parallel, uninfected cells were treated with the compound and cell viability was determined using an MTS assay. Data are from one experiment, representative of two independent experiments.
HeLa R19 cells were infected with CVB3 at a multiplicity of infection (MOI) of 0.001, which yields complete cell death within 3 days of incubation. The racemic mixture inhibited CVB3 with 50% effective concentration (EC50) of 3.2 ± 0.95 μM, while the S-enantiomer inhibited with an EC50 of 0.4 ± 0.15 μM (Figure 1B). In contrast, the R-enantiomer did not show any protection against CVB3 (Figure 1B). To validate these findings in a single cycle assay, HeLa R19 cells were infected with RLuc-CVB3 and the cells were treated with serial dilutions of the corresponding compounds. Cells were lysed at 7 h post-infection, and luciferase activity was measured as a quantitative and sensitive readout for viral replication. At the same time, cytotoxicity of the compounds was determined with an MTS assay, and the CC50 of the compounds in HeLa R19 cells ranges from 23 to 28 μM (Table 1). The racemic mixture and the S-enantiomer exerted antiviral activity with an ∼5-fold higher potency for the S-enantiomer (EC50 of 0.42 ± 0.17 μM) compared to the racemic mixture (EC50 of 2.02 ± 0.94 μM) (Figure 1C). Again, the R-enantiomer did not show an antiviral effect (Figure 1C,D).
Table 1. Antiviral Activity of Stereoisomers of Fluoxetinea.
| virus | species | strain | racemate | (S)-fluoxetine | (R)-fluoxetine | SIRacemic | SI(S)-fluoxetine |
|---|---|---|---|---|---|---|---|
| EV-A71 | EV-A | BrCr | NA | NA | NA | NA | NA |
| CVB3 | EV-B | Nancy | 2.02 ± 0.52 | 0.42 ± 0.17 | NA | 14.51 | 71.56 |
| PV-1 | EV-C | Sabin1 | NA | NA | NA | NA | NA |
| EV-D68 | EV-D | Fermon | 1.85 ± 0.10 | 0.67 ± 0.22 | NA | 21.72 | 42.73 |
| HRV-A2 | RV-A | NA | 7.95 ± 0.39 | NA | NA | 3.60 | |
| HRV-B14 | RV-B | NA | 6.34 ± 1.02 | NA | NA | 4.52 | |
| CC50 | 29.32 ± 0.35 | 28.63 ± 1.02 | 23.63 ± 1.40 |
Shown are EC50 and CC50 values in μM. Data represents mean values ± SD calculated from at least three different experiments. NA = not active. SI = selectivity index (CC50/EC50).
1.2. Antiviral Effect of (S)-Fluoxetine against Other Enteroviruses
Previously, it was shown that the racemic mixture of fluoxetine inhibits enterovirus B and D replication prototyped by CVB3 (strain Nancy) and EV-D68 (strain Fermon).33,34 As the S-enantiomer is more active toward CVB3 than the racemic mixture, we reassessed the antiviral activity of (S)-fluoxetine against a panel of enteroviruses. As expected, the racemic mixture is only active against enterovirus B and D, e.g., CVB3 and EV-D68 (strain Fermon) species (Table 1).34,41 (S)-Fluoxetine is more potent than the racemic mixture not only toward CVB3 but also toward EV-D68. Strikingly, while the racemic mixture does not confer any antiviral effect against rhinoviruses, the S-enantiomer inhibits rhinovirus 2 (HRV-2) and HRV-14 replication, with an EC50 of 7.95 ± 0.39 and 6.34 ± 1.02 μM, respectively (Table 1). The S-enantiomer was not active against EV-A71 (strain BrCr) and poliovirus (strain Sabin) at a concentration up to 30 μM. Higher concentrations could not be reached due to cytotoxicity of (S)-fluoxetine. We cannot exclude that (S)-fluoxetine would also inhibit other enteroviruses at higher concentrations in other systems in which the CC50 is much higher.
1.3. Fluoxetine Directly Binds to Recombinant 2C Protein in Vitro
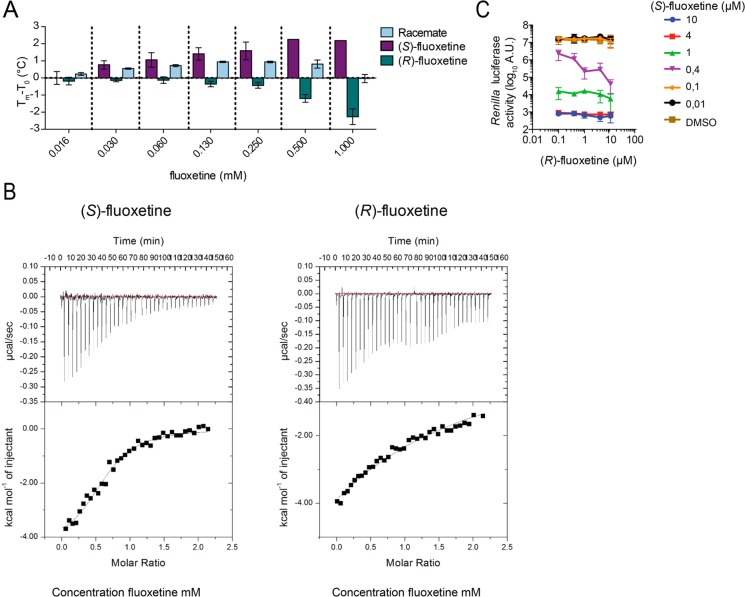
To gain further insights into the fluoxetine mode-of-action, we next investigated whether it directly binds to 2C protein. Production and purification of full-length 2C protein usually leads to a polydisperse preparation, which is problematic for binding assays. By removing the first 36 amino acids of the N-terminus, a homogeneous preparation of monomeric protein can be obtained and used for binding assays, namely, thermal shift assay (TSA) and isothermal titration calorimetry (ITC).35 First, TSA was used to detect a direct binding of the racemic mixture and the enantiomers of fluoxetine to CVB3 2C. In this assay, the increase of the melting temperature (Tm) of the protein reflects the binding of a ligand. The racemic mixture was able to induce a dose-dependent increase of the Tm of 2C protein in a range of 10 to 250 μM (Figure 2A) while at higher concentrations the Tm decreased. In contrast, the S-enantiomer of fluoxetine was able to thermally stabilize the 2C protein in a concentration-dependent manner (Figure 2A) with no destabilization at high concentrations of compound, suggestive of a direct binding of (S)-fluoxetine to the protein. As a positive control, we used dibucaine, an established 2C inhibitor.35
Figure 2.
(S)-Fluoxetine binds to the nonstructural protein 2C in vitro. (A) The binding of racemate, (S)-, and (R)-fluoxetine to recombinant CVB3 2C was determined by the thermal shift assay. The thermal stabilization of 2C by racemic and (S)-fluoxetine, represented by an increase in melting temperature, indicates binding of the compounds to 2C. (B) The binding of (S)- and (R)-fluoxetine to 2C were determined by isothermal calorimetry. As a positive control, the known 2C inhibitor dibucaine was used.35 Raw data are depicted at the top, and the integrated data are depicted at the bottom. Data are shown fitted to a one-site binding model. (C) HeLa R19 cells were infected with RLuc-CVB3, and the cells were treated with a fixed concentration of (S)-fluoxetine combined with serial dilutions of (R)-fluoxetine. Error bars depict standard error of the mean calculated from biological triplicates. Data are representative of two independent experiments.
Interestingly, the R-enantiomer appeared to have a destabilizing effect on the protein at high concentrations (Figure 2A). The unexpected pattern of the racemic mixture may be explained by the collective effects of (S)-fluoxetine (stabilization) and (R)-fluoxetine (destabilization). To confirm these results, we next quantified the binding of (S)- and (R)-fluoxetine to 2C protein by ITC. The S-enantiomer bound to 2C with a dissociation equilibrium constant (Kd) of ∼9.5 μM (Figure 2B). 2C partly aggregated during the titration of (R)-fluoxetine, in agreement with the results obtained by TSA. It was therefore not possible to retrieve a precise Kd, but when fitting the experimental data to a one-site binding model, the observed Kd was higher than 200 μM (Figure 2B).
Thus, together with the data obtained in the antiviral assay, binding of (S)-fluoxetine is likely responsible for the 2C-mediated antiviral effect. However, given the difference of binding of (S)-fluoxetine to the recombinant protein (10 μM) and the EC50 in cell-based assays (<1 μM), off-target effects cannot be formally ruled out. Still, (R)-fluoxetine did affect the 2C protein in the TSA and ITC at high concentrations. Therefore, we investigated whether (R)-fluoxetine can exert any additional effect on virus replication in combination with (S)-fluoxetine. To this end, the effect of different concentrations of (S)-fluoxetine combined with increasing concentrations of the R-enantiomer on CVB3 replication was determined. At high concentrations of (S)-fluoxetine (4 and 10 μM), replication of CVB3 was completely inhibited, and as expected, no additional effect of (R)-fluoxetine could be observed (Figure 2C). At low concentrations of (S)-fluoxetine, CVB3 replication was not impaired, nor could an additional effect of the R-enantiomer be observed (Figure 2C), in line with the absence of antiviral activity of (R)-fluoxetine alone (Figure 1B,C). Strikingly, when cells were treated with the approximate EC50 concentration of (S)-fluoxetine (0.4 μM), (R)-fluoxetine exerted a clear dose-dependent inhibition of virus replication (Figure 2C). Hence, (R)-fluoxetine can apparently exert weak antiviral activity that could come from the destabilization of the 2C protein or from the broad-spectrum antiviral effect induced by the targeting of a cellular partner.36,37
1.4. Fragment Screening Identifies Key Parts of Fluoxetine Involved in Targeting 2C
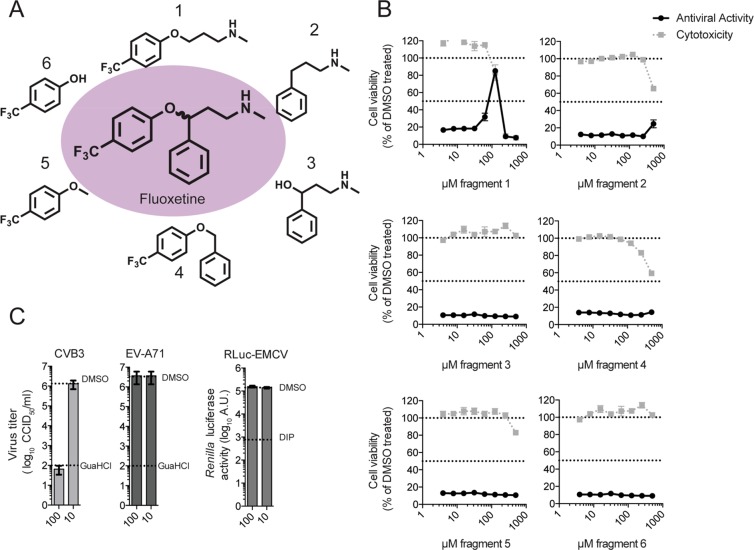
To gain more information about which chemical moieties of fluoxetine are involved in exerting the antiviral activity, we tested six different fragments of fluoxetine (Figure 3A) in a CPE reduction assay using CVB3. Fragments 1, 2, and 4 were synthesized, whereas fragments 3, 5, and 6 were purchased and tested as received without further purification. Only fragment 1 showed a weak antiviral activity in the multicycle assay at concentrations close to cytotoxic concentrations (Figure 3B). To exclude that inhibition of the virus is caused by an unspecific cytotoxic effect, the fragments were also tested in a single cycle assay using a RLuc-CVB3. HeLa R19 cells were infected with RLuc-CVB3, treated with 100 or 10 μM of each fragment, respectively, and luciferase activity at 7 h post-infection was determined as a quantitative and sensitive readout for virus replication. In this single cycle assay, fragment 1 showed an antiviral effect at 100 μM (Supplementary Figure 1A). In parallel, acute cytotoxicity of the fragments was excluded using an MTS assay (Supplementary Figure 1B). To further affirm that the observed weak antiviral activity of fragment 1 is specific and not due to cytotoxicity, we tested whether two other fluoxetine-insensitive viruses were affected by fragment 1. Cells were infected with EV-A71 or Renilla luciferase-expressing encephalomyocarditis virus (RLuc-EMCV),42,43 and as positive controls, guanidium chloride (GuaHCl), a replication inhibitor for EV-A71 and CVB3, and dipyridamole (DIP), a replication inhibitor for EMCV, were used. The virus titers and replication were determined by end point titration and a luciferase assay, respectively. Both viruses were not inhibited by fragment 1 at 100 μM (Figure 3C), indicating that the inhibitory effect of fragment 1 on CVB3 replication is specific.
Figure 3.
Fragment screening identifies key parts of fluoxetine involved in targeting 2C. (A) Molecular structures of fluoxetine and the different fragments that were used. (B) The CPE-reduction assay as performed in Figure 1B. (C) HeLa R19 cells were infected with CVB3, EV-A71, or RLuc-EMCV in order to exclude an unspecific antiviral effect of fragment 1. The replication inhibitor guanidium chloride (GuaHCl, 2 mM) was used in the case of CVB3 and EV-A71 as positive control. Dipyridamole (DIP, 100 μM) was used as the positive control replication inhibitor during RLuc-EMCV infection.
Finally, we investigated binding of the fluoxetine fragments to recombinant 2C using a thermal shift assay (TSA). The binding of low molecular weight molecules usually has a moderate effect on protein stability (<1 °C in protein stabilization), and concentrations should be >100 μM to observe protein stabilization.44 Therefore, we tested the fragments in the TSA at a concentration range from 100 to 400 μM. Neither fragment 1 nor any other fragment was able to stabilize the 2C protein at the indicated concentrations (Supplementary Figure 1C). At present, we can only speculate why fragment 1 exerts weak antiviral activity but does not stabilize 2C in the TSA. Possibly, the amount of recombinant 2C protein in the in vitro assay is higher than in an infection setting, which would require a higher concentration of the compound to induce a shift in the melting temperature of 2C. It can however be noticed that the effect of fragments on Tm is usually very modest because the binding energy is weak.44 Taken together, fragment 1 was able to inhibit CVB3 replication, but a direct binding could not be observed in the TSA.
1.5. Identification of a Potential Binding Pocket for Fluoxetine on 2C
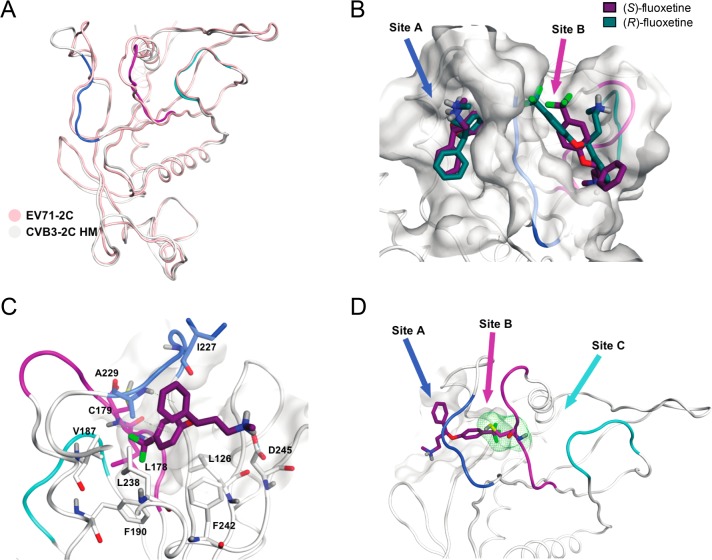
After having investigated chemical properties of fluoxetine that are important for its antiviral activity, we next wanted to further characterize the possible intermolecular interactions between (S)-fluoxetine and 2C. Because structures of 2C of fluoxetine-sensitive viruses were not available, we resorted to an in silico modeling approach. We first generated a homology model of CVB3 2C (Figure 4A) on the basis of the crystal structure of a part of 2C of the fluoxetine-insensitive virus EV-A71.39 The crystallized part of EV-A71 2C covers amino acids 116–329. In this region, the sequence identity and similarity between EV-A71 and CVB3 2C is 62% and 80%, respectively.
Figure 4.
Predicted fluoxetine binding sites on CVB3 2C protein. (A) Homology model of the 2C protein of CVB3 built on the crystal structure of EV-A71. Ribbon and carbon atoms of the 224AGSINA229 loop are in blue, the 175–183 loop, in violet, and the 158–164 loop, in cyan. (B) S- and R-enantiomers of fluoxetine docked into sites A and B of the homology model. (C) View of (S)-fluoxetine in site A as identified in the molecular dynamics simulations, comprising the residues L126, L178, C179, V187, F190, I227, A229, L238, F242, and D245. The trifluoromethyl moiety of fluoxetine is buried deep inside the hydrophobic pocket. (D) Three possible entrances of (S)-fluoxetine to reach C179 (green line surface).
Enterovirus 2C belongs to a family of AAA+ ATPases, which oligomerize in hexameric ring structures in which the ATP is coordinated by two monomers.39 Such ring structures were observed in low resolution electron microscopy structures of 2C proteins from poliovirus and foot-and-mouth disease virus, a picornavirus from the aphthovirus genus.45,46 The published EV-A71 2C structure (PDB: 5GRB) shows a C-terminal interaction between two adjacent 2C molecules to form a bipartite binding site for the ATP. In total, there are six protein chains in the crystal structure of which only chains A and B are fully resolved, whereas all the others have at least one gap.39 The cocrystallized ATP molecules adopt different conformations for each chain, and only chains A and F crystallize in a conformation resembling a bipartite binding site with both monomers forming hydrogen bonds with the ATP. Therefore, chain A was used as a template to generate the homology model of CVB3 2C (Figure 4A). Superimposition onto the EV-A71 2C structure resulted in a CVB3 2C model with a reasonably good fit and an overall root mean square deviation (RMSD) of 0.30 Å from the template with the highest deviation for residues N257 (1.65 Å) and D274 (2.08 Å).
The CVB3 2C homology model was used to search for pockets in which fluoxetine may bind. Because mutations in the flexible 224AGSINA229 loop confer resistance to fluoxetine,34 we focused on pockets near this loop. Two potential binding pockets flanking the 224AGSINA229 loop were identified, termed site A and site B (Figure 4B). Site A faces away from the ATP binding site and is confined by the 224AGSINA229 loop on one side and hydrophilic residues (D245, R295, and R296) on the other side. Site A is a deep, lipophilic pocket, whereas site B is a rather shallow pocket and lies between the 224AGSINA229 loop and the 175–183 loop, which is downstream of the Walker B motif. In the crystal structure of EV-A71 and in the CVB3 homology model, both sites might considerably change their shapes due to loop movements. However, site A is not affected to the point of blocking the binding of fluoxetine, while site B is. Both enantiomers of fluoxetine were docked into each of the two pockets for further computational investigations (Figure 4B). In site A, both enantiomers docked in proximity of the residues A229 and I227 of the 224AGSINA229 loop. The 4-(trifluoromethyl)benzene moiety occupied the hydrophobic bottom of the pocket consisting of the residues L178, C179, V187, and F190. Instead, in site B, both enantiomers were mostly exposed to the solvent.
1.5.1. Molecular Dynamics Simulations Reveal Stable Positioning of (S)-Fluoxetine in Pocket A
After the selection of the possible pockets near the AGSINA motif, the binding sites containing both enantiomers were subjected to molecular dynamics simulations. The 224AGSINA229 loop is thought to be flexible, thereby alternating the shape of the two identified pockets, resulting in different predicted binding modes compared to the docking. To evaluate the stability of the fluoxetine enantiomers docked into the pockets and to address why mutations in this loop can cause resistance toward fluoxetine, the docking models of (S)-fluoxetine and (R)-fluoxetine in site A and site B were subjected to molecular dynamics simulations. For each enantiomer docked into either site, three independent molecular dynamic simulations of 100 ns were performed. The calculated binding energies of (S)- or (R)-fluoxetine into site A and site B from the molecular dynamics simulations are listed in Table 2. The simulations showed that (S)-fluoxetine bound stronger to 2C than (R)-fluoxetine in both site A and site B. We observed that (R)-fluoxetine dissociated from pocket A as well as pocket B in one of the three independent simulations, suggesting that (R)-fluoxetine cannot engage in stable interaction with 2C. In site B, movement of the 224AGSINA229 loop was observed, making the hydrophobic pocket accessible for the 4-(trifluoromethyl)benzene moiety of fluoxetine. However, visual inspection and the binding energies of the molecular dynamics suggested that site A provides a more stable binding pocket for (S)-fluoxetine, defined by the residues L126, L178, V187, F190, L238, I227, A229, and F242 (Figure 4C).
Table 2. Binding Energies of the Protein Ligand Complexes during MD Simulationsa.
| compound | MD1 | MD2 | MD3 | |
|---|---|---|---|---|
| site A | (R)-fluoxetine | –29.71* | –42.70 | –19.85* |
| (S)-fluoxetine | –41.63 | –42.28 | –41.54 | |
| site B | (R)-fluoxetine | –29.59* | –42.95 | –27.38 |
| (S)-fluoxetine | –34.79 | –29.88 | –30.83* |
Values indicated are calculated ΔGbinding average values over 100 ns of each of the three independent molecular dynamics simulations (MD) for the indicated ligands in site A or B (kJ/mol). The most favorable complex according to the simulations is (S)-fluoxetine in site A. For all the other complexes in at least one simulation, the ligand dissociated from the protein (indicated by *).
In each independent repetition of the molecular dynamics simulation on (S)-fluoxetine in site A, the compound was oriented such that the 4-(trifluoromethyl)benzene moiety occupied the hydrophobic pocket (Figure 4C). During the molecular dynamics simulations, a hydrogen bond repeatedly formed between the amino acid D245 and the positively charged amino group of (S)-fluoxetine (Figure 4C). We conclude, in line with the antiviral data and binding assays from the in silico approach, that (S)-fluoxetine binds stronger to 2C than the R-enantiomer. Furthermore, on the basis of our in silico analyses, we conclude that (S)-fluoxetine most likely binds 2C to site A.
1.5.2. Mutations in the Identified Binding Pocket Confer Resistance to (S)-Fluoxetine
The triple amino acid substitution A224V–I227V–A229V (AVIVAV) in CVB3 2C, clustered in the 224AGSINA229 region, was previously shown to confer resistance to the racemic fluoxetine mixture34 and to several other 2C inhibitors.29 Likewise, the triple mutant provided resistance to (S)-fluoxetine (Figure 5A). This resistance is specific, as AVIVAV mutations did not confer resistance to BF738735, a compound that inhibits enterovirus replication via a different mechanism, namely, by targeting the cellular protein PI4KIIIβ, which is essential for enterovirus replication.47
Figure 5.
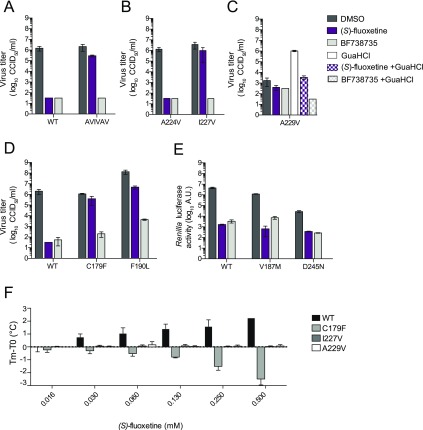
Mutations in the identified binding pocket confer resistance toward (S)-fluoxetine. (A) HeLa R19 cells were infected with WT CVB3 or the AVIVAV mutant (A224V–I227V–A229V triple mutant) at an MOI of 0.1 and treated with 1 μM (S)-fluoxetine or 1 μM BF738735 as a control replication inhibitor that acts via the host protein PI4KBIIIβ.48 Eight hours post-infection, cells were freeze–thawed and virus titers were determined by end point titration. Means and standard deviations were calculated from biological triplicates. (B) The individual A224V and I227V mutations were tested for resistance toward (S)-fluoxetine as in (A). (C) The guanidine chloride-dependent virus A229V was tested for dependency on (S)-fluoxetine. Experiments were performed similar to those in (A). Because the A229V virus is dependent on GuaHCl, sensitivity to the inhibitors was also tested in the presence of 1 mM GuaHCl (blocked bars). (D) Residues in the hydrophobic binding pocket were mutated (C179F and F190L) and tested for resistance toward (S)-fluoxetine as in (A). (E) Residues at the surface of the hydrophobic binding pocket (V187 M and D245N) were substituted in the Renilla luciferase virus, and sensitivity to (S)-fluoxetine was determined as in Figure 1C. (F) The binding of (S)-fluoxetine to recombinant WT CVB3 2C or 2C harboring the resistance mutations C179F, I227V, and A229V was tested using the thermal shift assay as in Figure 2A. In all panels, data are shown from one experiment representative of at least two independent experiments. Error bars depict the standard error of the mean calculated from biological triplicates.
To dissect the contribution of the individual mutations to the resistance, several CVB3 mutant viruses containing the single mutations A224V, I227V, or A229V were made. The A224V mutation alone did not confer resistance to the compounds (Figure 5B). The single mutation I227V provided a high level of resistance toward (S)-fluoxetine but not the control compound BF738735 (Figure 5B). This is in concordance with a recently published report, which raised resistant mutants toward racemic fluoxetine and identified the mutations I227V and N228S.41 The A229V single mutant virus was previously described to be dependent on all structurally divergent 2C inhibitors tested (GuaHCl, HBB, TBZE-029, and MRL-1237),29 meaning that these mutant viruses are not inhibited by the compounds but instead strictly require them for efficient replication. Strikingly, the A229V virus was not dependent on (S)-fluoxetine at the concentration tested (Figure 5C). To test whether the A229V virus is resistant to (S)-fluoxetine, we assessed replication in the presence of both GuaHCl (to allow replication) and (S)-fluoxetine. However, the A229V mutation does not confer resistance to (S)-fluoxetine.
To find experimental support for the predicted binding pocket, we next investigated the importance of key residues in the site A pocket by mutational analysis. We first mutated two amino acids that are located deep within site A and tested whether they could confer resistance to (S)-fluoxetine. These mutations are C179F, which, like C179Y, provides resistance toward several structurally different 2C inhibitors,41 and F190L, which raises resistance to a novel 2C inhibitor.49 In line with our model, viruses containing the C179F or the F190L mutation in 2C were highly resistant to (S)-fluoxetine (Figure 5D). We next investigated two amino acids that are located near the edge of site A. V187 was substituted by an M, because the corresponding amino acid 187 in the fluoxetine-insensitive virus EV-A71 is a methionine and we hypothesized that this methionine may contribute to the insensitivity of EV-A71 to fluoxetine. The amino acid D245 displayed repeated interactions with the positively charged amino group of (S)-fluoxetine in the molecular dynamics simulations (Figure 4C). We therefore decided to remove the charge of this residue by replacing it with asparagine. However, neither the V187 M nor the D245N mutations provided resistance to (S)-fluoxetine (Figure 5E), which seems to contradict our computation-based hypothesis that (S)-fluoxetine binds to site A interacting with C187 and D245. Fluoxetine may bind in a slightly different conformation within the pocket that does not involve interactions with these two residues; thus, mutations would not confer resistance. That would still explain why the C179F and F190L mutations confer resistance. Alternatively, fluoxetine may access residues C179 and F190 from a third entrance site (Site C), schematically depicted in Figure 4D (cyan arrow), and therefore is unaffected by the mutations in site A and the 224AGSINA229 loop. However, neither in the homology model nor in the EV-A71 crystal structure does the supposed site C present an obvious cavity for the potential ligand entrance toward the residues C179 and F190. Because of flexibilities in the loops ranging from amino acid residue 158 to 163, 175 to 183, and 224 to 229, other conformations likely exist so that site C may become accessible. In that case, the occurrence of mutations in the 224AGSINA229 loop and how they can confer resistance are more difficult to explain. We propose the hypothesis that conformational changes over a longer range could affect the shape of the pocket, and therefore, slight changes in the distant 224AGSINA229 loop might be sufficient to convey resistance. Clearly, actual crystallographic data of (S)-fluoxetine bound to 2C is needed to definitively resolve this point.
Finally, we wanted to address whether resistance mutations in 2C affect binding of (S)-fluoxetine to 2C. Recombinant 2C proteins harboring the resistance mutations I227V or C179F or the A229V mutation were produced, and binding was tested by TSA (Figure 5F). (S)-Fluoxetine was not able to stabilize any of the mutated 2C proteins from thermal denaturation, implying that the substitutions abrogated fluoxetine binding to 2C. Unexpectedly, the C179F substitution made 2C more sensitive to thermal denaturation in the presence of (S)-fluoxetine when compared to the C179F mutant without compound (ΔTm of −2.5 °C). The A229V substitution nullified thermal stabilization of 2C by (S)-fluoxetine, suggesting that this mutation abrogated binding of (S)-fluoxetine. However, the GuaHCl dependency of the A229V virus required the combination of GuaHCl and (S)-fluoxetine in the TSA experiment, which may add confounding effects (Figure 5C).
In summary, mutations at position I227 in the 224AGSINA229 loop as well as the C179F and F190L mutations at the bottom of the binding site A confer resistance to (S)-fluoxetine, while the V187 M and D245N mutations, which are more at the edge of the pocket, do not.
2. Discussion
Enteroviruses are a major global health burden, but currently, no antiviral therapy is available. The high degree of conservation makes the enterovirus 2C protein an attractive target for the development of broad-spectrum enterovirus inhibitors (Supplementary Figure 3). Several compounds, including a number of repurposed drugs, have been identified as inhibitors of enterovirus replication by targeting 2C (reviewed in ref (2)), but to date, the molecular mechanisms underlying the antiviral effects are lacking. Fluoxetine, one of the identified compounds, is an FDA-approved drug that is used as a highly selective inhibitor of SERT for the treatment of major depression and anxiety disorders. In this study, we provide new insights into how the repurposed drug fluoxetine acts as an antiviral compound against CVB3.
Fluoxetine has one chiral center, and only the S-enantiomer of fluoxetine has anti-enteroviral activity and binds to 2C in vitro. In previous studies, the racemic mixture of fluoxetine inhibited EV-B and EV-D species but not EV-A, EV-C, and rhinoviruses.33,34 Here, we show that the S-enantiomer, but not the R-enantiomer, has a clearly increased antiviral potency compared to the racemic mixture against CVB3 and also EV-D68, while the cytotoxicity in cell culture is comparable (Table 1). Unexpectedly, we observed that the S-enantiomer also exerts antiviral activity against rhinoviruses. Presumably, the antiviral activity of the racemic mixture against rhinoviruses is so weak that it cannot be separated from the cytotoxicity effects. We cannot exclude that (S)-fluoxetine would also inhibit EV-A or EV-C species at even higher concentrations, but this cannot be tested in the current model systems due to cytotoxicity. Another explanation for why EV-A and EV-C species are not sensitive to fluoxetine is that there could be strain-specific differences in the sensitivity toward inhibitors. For example, we also show that (S)-fluoxetine does not inhibit EV-A71 (strain BrCr). It is unknown why EV-A71 is resistant to (S)-fluoxetine; further in-depth investigations are needed to gain a better understanding of the underlying mechanism of resistance of EV-A71. However, a different EV-A71 strain was shown to be sensitive to racemic fluoxetine.41 Thus, it is possible that different EV-A and EV-C strains are sensitive toward (S)-fluoxetine.
The in vivo efficacy of fluoxetine toward enterovirus infections has until now been relatively poorly studied. Recent outbreaks of acute flaccid myelitis (AFM) in the US are, at least in part, associated with EV-D68.50 In a mouse model for EV-D68 associated paralysis, fluoxetine did not have an effect on motor impairment of mice or viral load in muscle and spinal cord but instead seemed to slightly aggravate the disease.51 Because in human AFM cases treatment options other than supportive care are lacking, several clinicians have tried the off-label use of fluoxetine to treat pediatric patients. A retrospective study of safety and efficacy of fluoxetine to treat AFM revealed no beneficial effect of fluoxetine.52 Instead, fluoxetine-treated patients had somewhat more severe symptoms, suggesting a negative effect of fluoxetine on AFM, in line with the mouse model.51 However, this retrospective study had some limitations that make it difficult to draw definitive conclusions. For example, patients had been treated with different dosing regimens. Furthermore, fluoxetine treatment was started only after onset of AFM symptoms when irreversible neuronal damage may already have been inflicted. Moreover, viral loads were not tested, leaving the question unanswered whether the virus was still actively replicating at the time of treatment and, if so, whether fluoxetine had any effect on viral loads. In contrast, in a pancreatitis mouse model, the racemic fluoxetine mixture reduced the levels of CVB4 infectious particles in heart and pancreas and reduced pancreatitis severity when it was given 1 day prior to CVB4 infection and subsequently every day post-infection.53 Finally, in a case study of an immunocompromised child with chronic EV-B induced encephalitis, the off-label use of fluoxetine eliminated the virus and led to recovery of the patient.38 These seemingly opposing results stress the importance of further in vivo studies of fluoxetine to combat different enteroviruses and treat different types of enterovirus-associated diseases. The plasma concentration of fluoxetine ranges from 91 to 300 ng/mL after 30 days of dosing 40 mg/day.54 The metabolite norfluoxetine, which also shows antiviral activity,33 reaches a plasma concentration from 72 to 258 ng/mL. The slow elimination of fluoxetine together with the metabolite norfluoxetine should reach a sufficient plasma concentration that corresponds with the EC50 in cell culture (∼1 μM, which corresponds to ∼150 ng/mL).
In contrast to the SSRI activity, leading to the antidepressant effect of fluoxetine, the 2C-mediated antiviral activity of fluoxetine is stereoselective, lending further support to the idea that the antiviral activity is unrelated to the known SSRI activity of fluoxetine. This implies that, when fluoxetine is used as an antiviral treatment, any potential SSRI-related side effects can be reduced by providing only (S)-fluoxetine, which is then at lower overall concentration. However, when only (S)-fluoxetine is used, the advantages of drug repurposing of fluoxetine, which is licensed as a racemic mixture, are nullified and new safety studies are needed. Fragment 1, which contains the 4-(trifluoromethyl)benzene moiety and the amino group, gave the first indication for the importance of these chemical features for the antiviral effect. From there on, structure activity relationship studies could help to design new molecules with even a stronger antiviral effect but reduced or suppressed SERT inhibition.
To investigate the mode-of-action of how fluoxetine binds to enterovirus 2C, we built a homology model of CVB3 on the basis of the crystal structure of the fluoxetine-insensitive EV-A71 2C. In the predicted binding model, the 4-(trifluoromethyl)benzene group of fluoxetine occupies the deep hydrophobic pocket close to residues L126, L178, V187, F190, L238, I227, A229, and F242. Mutational studies to test the binding hypothesis were designed, and mutations at the bottom of the predicted pocket reduced (S)-fluoxetine binding by providing resistance. The mutations on the borders of the pocket did not confer resistance to (S)-fluoxetine; therefore, we cannot exclude alternative entrance sites to the hydrophobic cavity. Crystallographic data is needed to yield ultimate clarity about the binding mode.
2C is a member of the SF3 helicases of the AAA+ ATPase superfamily and contains Walker A and B motifs, which are important for recognizing the triphosphate of ATP, and a short motif C that is located on top of Walker B.7 The residues L178 and C179 in the predicted binding pocket are located within the Walker B motif or immediately thereafter. Furthermore, the 224AGSINA229 loop, which harbors resistance mutations and lines the edge of the predicted pocket, is directly C-terminal to the motif C. Residues L238, F242, and D245 are in proximity to the arginine fingers (R240 and R241), which play an important role in the ATPase domain and are required for ATP hydrolysis. Hence, it is conceivable that fluoxetine inhibits ATPase activity allosterically through relatively short-range effects although alternative modes, e.g., through long-range effects, cannot be excluded. Still, the exact molecular details of how fluoxetine inhibits the ATPase activity of 2C remain to be determined.
The 224AGSINA229 loop forms a hot spot for resistance mutations against fluoxetine and many other 2C inhibitors, while fewer mutations have been found in the predicted fluoxetine binding pocket. The residues in the pocket are evolutionarily highly conserved, implying that there is little room for variation that could induce resistance in the pocket residues without affecting virus fitness. In contrast, the 224–229 loop diverges more between enterovirus species, suggesting that the loop allows for more sequence diversity that could yield resistance (Supplementary Figure 3). The 224AGSINA229 loop is conserved between the fluoxetine-sensitive viruses CVB3 and EV-D68, but the motif differs in the fluoxetine resistant viruses EV-A71 and poliovirus (Supplementary Figure 3). The corresponding 224AGSINA229 loops might be more rigid in these latter viruses, thereby interfering with fluoxetine binding. Mutations in the 224AGSINA229 loop confer resistance not only against fluoxetine but also against several other compounds, including TBZE-029, HBB, MRL-1237, and GuaHCl.29 Resistance mutations may favor a conformation of the 224AGSINA229 loop in which the binding pocket is not accessible for fluoxetine anymore. Furthermore, the mutations may change the flexibility of the loop in such a way that the inhibitory effect of fluoxetine is circumvented. Further biophysical studies are needed to decipher how the resistance mutations affect the overall stability of the protein or the accessibility of the binding pocket. However, addressing the possible role of S-fluoxetine in the hexamerization of 2C is presently not possible because the production of homogeneous 2C protein in its biologically relevant oligomerization state has not yet been achieved.
In conclusion, this study sheds new light onto how the 2C inhibitor fluoxetine may target the enterovirus 2C protein. In particular, the discovery of the stereoselective activity will fuel further mode-of-action studies and support the rational design of novel, fluoxetine-derived broad-spectrum enterovirus inhibitors.
3. Methods
3.1. Cells and Reagents
Buffalo Green Monkey cells (BGM) and HeLa R19 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Lonza) supplemented with 10% fetal bovine serum (FBS, Lonza). Huh7-Lunet 7/T7, a stable cell pool expressing T7 RNA polymerase and blasticidin S-deaminase,55 was cultured in DMEM supplemented with 10% FBS and 10 μg/mL blasticidin (Sigma-Aldrich). All cell lines were grown at 37 °C in 5% CO2. Guanidine hydrochloride (GuaHCl) was purchased from Sigma-Aldrich. A racemic mixture of fluoxetine was purchased from Sigma-Aldrich. The S- and R-enantiomers were purchased either from Sigma-Aldrich or Carbosynth. BF738735 was provided by Galapagos NV.48 Dibucaine was purchased from Sigma-Aldrich. GuaHCl was dissolved in water at 2 M stock concentration, and all other compounds were dissolved in DMSO at 10 mM stock concentration.
3.2. Viruses
EV-A71 (strain BrCr), PV1 (strain Sabin, ATCC), and EV-D68 (strain Fermon) were obtained from the National Institute for Public Health and Environment (RIVM) in The Netherlands. HRV-2 and HRV-14 were obtained from Joachim Seipelt from the Medical University of Vienna in Austria. RLuc-CVB3, which contains a Renilla luciferase gene upstream of the capsid coding region, was obtained by transfecting Huh7-Lunet 7/T7 cells with MluI-linearized pRLuc-53CB3/T7 plasmid as described.56 RLuc-encephalomyocarditis virus (EMCV, strain mengovirus), encoding a Renilla luciferase gene upstream of the capsid-coding region, was described before.42,43 CVB3 (strain Nancy) and CVB3 2C mutant viruses were obtained by transfecting BGM cells with RNA transcripts derived from the full-length infectious clones p53CB3/T7 as described in ref (57). The mutations 2C[A224V], 2C[I227V], 2C[A229V], 2C[A224V/I227V], 2C[A224V/A229V], 2C[I227V/A229V], 2C[A224V/I227V/A229V], 2C[C179F], and 2C[F190L] were introduced into the p53CB3/T7 infectious clone and 2C[V187M] and 2C[D245N] were introduced into the pRLuc-53CB3/T7 using side directed mutagenesis. In vitro transcribed RNA transcripts were transfected into HeLa R19 cells to obtain virus. To ensure that the introduced mutations are retained in the generated virus, viral RNA was isolated with the NucleoSpin RNA Virus kit (Macherey-Nagel) according to the manufacturer’s protocol, and the presence of the desired mutations was confirmed by Sanger sequencing. Virus titers were determined by end point dilution titration, calculated according to the method of Reed and Muench,58 and expressed as 50% cell culture infective dose (CCID50).
3.3. Single-Cycle Virus Infection
Virus infections were performed by incubating subconfluent HeLa R19 cells with virus at a multiplicity of infection (MOI) of 0.1 at 37 °C for 30 min. Next, the medium was removed, and fresh (compound-containing) medium was added to the cells. At the indicated time points, the medium was discarded and cells were lysed. For measurements of infectious particles, virus was released from the cells by three freeze–thawing cycles. Virus titers were determined by the end-point dilution assay and calculated by the method of Reed and Muench.58 In the case of infection with RLuc-CVB3 and RLuc-EMCV, cells were lysed 6–7 h post-infection and the Renilla luciferase Assay System (Promega) was used to determine the luciferase activity. Where indicated, cell viability was determined in parallel using the AQueous One Solution Cell Proliferation Assay (Promega) according to the manufacturer’s protocol. Optical density at 490 nm was determined using a microplate reader.
3.4. Multicycle Virus Infection
Subconfluent layers of HeLa R19 cells were seeded in 96-wells and treated with serial dilutions of the corresponding compounds. Cells were infected with CVB3 at the lowest possible MOI (MOI of 0.001) resulting in full CPE within 3 days. Subsequently, the cells were incubated at 37 °C for 3 days until full CPE was observed in the virus infected untreated cell controls. Cell viability was determined in parallel using the AQueous One Solution Cell Proliferation Assay (Promega) according to the manufacturer’s protocol. The optical density at 490 nm was determined using a microplate reader. Raw OD values were converted to the percentage of untreated and uninfected cell control after subtraction of the background.
3.5. Binding of Fluoxetine to Recombinant WT and Mutant 2C Proteins
The DNA fragment coding for CVB3 2C (amino acids 37 to 329) was cloned downstream of a cleavable thioredoxin-hexahistidine tag. Mutations were introduced into the 2C coding sequence by PCR-based site-directed mutagenesis. The recombinant WT and mutant proteins were produced in Escherichia coli T7 Express (New England BioLabs) at 17 °C. Protein purification and tag removal were performed under nondenaturing conditions as previously described.35 The final size-exclusion chromatography step was performed with a buffer containing 10 mM HEPES and 300 mM NaCl (pH 7.5). The binding of fluoxetine or corresponding fragments on WT and mutant 2C proteins was monitored by the fluorescence-based thermal shift assay (TSA) using a Bio-Rad CFX Connect. TSA plates were prepared by dispensing into each well the 2C protein (final concentration of 15 μM in 50 mM Tris, 300 mM NaCl, pH 8), which was mixed with 1 μL of fragment or fluoxetine (from 20 mM stock in 100% DMSO, 1 mM final concentration in 4% DMSO) and a SYPRO orange solution in concentrations recommended by the manufacturer in a final volume of 25 μL. The experiments were performed under a temperature gradient ranging from 20 to 95 °C (incremental steps of 0.2 °C/12 s). The denaturation of the proteins was monitored by following the increase of the fluorescence emitted by SYPRO orange that binds exposed hydrophobic regions of the denatured protein. The melting temperature (Tm) was calculated as the mid-log of the transition phase from the native to the denatured protein using a Boltzmann model (Origin software). The reference unfolding temperature of proteins in 4% DMSO (T0) was subtracted from the values in the presence of fragment (Tm) to obtain thermal shifts, ΔTm = Tm – T0.
The binding of (S)- and (R)-fluoxetine to WT CVB3 2C was further characterized by isothermal titration calorimetry (ITC) using a MicroCal iTC200 instrument (Malvern). Experiments were carried out at 20 °C in a solution containing 10 mM HEPES, 300 mM NaCl, and 0.8% DMSO (pH 7.5). The 2C protein concentration in the cell was 80 μM whereas the fluoxetine concentration in the syringe was 400 μM. For (R)-fluoxetine, two injections were necessary. Heats of dilution were measured by injecting the ligand into the protein solution. Titration curves were fitted by using MicroCal Origin software, assuming one set of sites, and enthalpy changes (ΔH), dissociation equilibrium constants (Kd), and stoichiometry were extracted.
3.6. Molecular Modeling
The computational studies were carried out on a 1.80 GHz Intel Xeon (8 cores) processor-based system, running Ubuntu 14.04 LTS, using a Molecular Operating Environment (MOE) 2015.10 (Chemical Computing Group Inc. 2016) and Maestro v11.4 (Schrödinger LLC, New York, NY, 2017). The homology model was generated with an MOE using an integrated sequence alignment and structure preparation tools for the template. Preparation of the structure for docking and molecular dynamic simulations and subsequent data analysis were carried out with Maestro. Docking experiments were performed using the GlideSP module in Maestro, running the default settings. The molecular dynamics simulations were performed using the Desmond package (Desmond Molecular Dynamics System, D. E. Shaw Research, New York, NY, 2018; Maestro-Desmond Interoperability Tools, Schrödinger, New York, NY, 2018). Pictures of molecular modeling were prepared using MOE.
3.7. Homology Modeling
The protein sequence of CVB3 (strain Nancy) 2C protein was downloaded from Uniprot (ID: P03313 amino acids 1101–1429). The structures reported in ref (39), especially 5GRB, were used as a starting point in this study and were retrieved from the Protein Data Bank. 5GRB contains ATPγS and was the structure used for the computational studies. The sequence of CVB3 was aligned to the sequence of the crystallized EV-A71 using MOE. 5GRB chain A was used as a structural template for the homology model. The homology model was built with the Amber12: EHT force field.59,60 Automatic detection of disulfide bridges was disabled. Ten intermediate models were generated and refined using a medium refinement by molecular mechanics (highly tethered minimization to relieve steric strains). The final model was calculated using Coulomb and Generalized Born/Volume Integral (GB/VI) interaction energies61 and was not further refined.
After the generation of the homology model, the structure was revised using the Structure Preparation function in MOE. In order to further evaluate the quality of the homology model for future studies, the phi/psi angles were analyzed in the Ramachandran plot using the Protein Geometry tool of MOE. Identified outliers were investigated and, if relevant, corrected manually. Then, the validation of the model was carried out using RAMPAGE Ramachandran plot analysis.620 Amino acid environment analysis was carried out using the SAVES server v3.0 (http://servicesn.mbi.ucla.edu/SAVES/) comprising Verify 3D.62−65
3.8. Site Finder
The site identification tool Site Finder, which comprised the software suite MOE, was run on the prepared CVB3 homology model to identify possible active sites for the known 2C targeting compounds. Two binding pockets (termed site A and site B) were selected on the basis of their vicinity to known mutations in the 224AGSINA229 loop that convey resistance against several known 2C inhibitors including fluoxetine.29,34,35 For both pockets, a set of dummy atoms was created in the positions of the alpha spheres that are used to determine pockets in Site Finder.
3.9. Docking
After the identification of site A and site B, both the compounds and the protein were prepared for the docking with Glide. The homology model of CVB3 was prepared with the Protein Preparation Wizard embedded in Maestro. For each binding site, a grid box for the positioning of the molecules during the docking was generated setting the centers of the boxes to the coordinates of representative dummy atoms generated by Site Finder. The stereochemistry on the chiral center of fluoxetine was defined using the molecule builder in MOE, and the two enantiomers were saved in separate .sdf files. Both of them were subjected to the ligand preparation protocol (ligprep) in Maestro creating up to 32 conformations each. Then, all conformations obtained for (R)- and (S)-fluoxetine were docked with Maestro Glide in standard precision (SP) mode into each binding site. The poses were inspected for their fit within the pockets and their interactions with the protein. The best protein–ligand complexes for each site and each enantiomer were saved and prepared for molecular dynamics (MD) simulations.
3.10. Molecular Dynamics Simulations
All MD simulations were performed using Desmond, part of the Maestro v11.4 simulation package (Schrödinger LLC, New York, NY, 2017). OPLS3 was used as the force field. The complexes of (R)-fluoxetine and (S)-fluoxetine docked to 2C of CVB3 were placed in a cubic box (buffer 10 Å) using the TIP3P water model. The negative charges on the protein were neutralized adding Na+ atoms to the system. Magnesium chloride (10 mM) was added to the box to simulate physiological conditions. Before the MD simulation, the system was first equilibrated for 112 ps at 10 K in an NVT ensemble and then simulated for 48 ps at a constant pressure of 1 atm using the NPT ensemble. All MD simulations were performed for 100 ns at constant temperature (300 K) and pressure recording snapshots every 160 ps.
The estimated ΔGbinding was calculated using the Desmond command-line script thermal_mmgbsa.py. After splitting the trajectory file of the MD simulation into snapshots, the script is calculating the average computed binding energy of the ligand (Table 2 and Supplementary Figure 2).
3.11. Chemistry
All solvents and reagents used were obtained from commercial sources unless otherwise indicated. All reactions were performed under a nitrogen atmosphere. 1H and 13C NMR spectra were recorded with a Bruker Avance DPX500 spectrometer operating at 500 MHz for 1H and 125 MHz for 13C with Me4Si as internal standard. Deuterated dimethyl sulfoxide (DMSO) was used as the solvent for NMR experiments. 1H chemical shift values (δ) are referenced to the residual nondeuterated components of the NMR solvents (δ = 2.50 ppm for DMSO). The 13C chemical shifts (δ) are referenced to DMSO (central peak, δ = 39.5 ppm). Thin layer chromatography (TLC) was performed on Silica gel plates (Merck Kieselgel 60 F254), which were developed by the ascending method. Column chromatography was performed on an Isolera Biotage system. Purity of synthesized compounds was determined by UPLC-UV-MS analysis (Waters UPLC system with both Diode Array detection and Electrospray (+’ve and −’ve ion) MS detection). The purity of all compounds was determined to be >95% by UPLC using the eluents H2O containing 0.1% trifluoroacetic acid (eluent A) and acetonitrile containing 0.1% trifluoroacetic acid (eluent B) at the following conditions: Waters Acquity UPLC BEH C18, 1.7 μm, 2.1 × 50 mm column; 0.5 mL/min; column temperature, 40 °C; sample diluent, acetonitrile; sample concentration, 10 μg/mL; injection volume, 2 μL; gradient, 90% eluent A (0.1 min), 90–0% eluent A (1.5 min), 0% eluent A (1.4 min), and 90% eluent A (0.1 min) (method 1).
3.12. Synthesis of 1-(3-Bromopropoxy)-4-(trifluoromethyl)benzene (Intermediate)
To a solution of 1,3-dibromopropane (6.17 mmol) and potassium carbonate (4.63 mmol) in DMF (3 mL), 4-(trifluoromethyl)phenol (3.08 mmol) in DMF (1.2 mL) was added dropwise; the reaction mixture was stirred at room temperature for 2 h and then heated to 70 °C for 2 h. The mixture was filtrated, diluted with ethyl acetate (10 mL), and washed with water (3 × 10 mL). The organic layer was dried over sodium sulfate and evaporated under reduced pressure. The residue was purified by flash column chromatography and eluted with n-hexane/EtOAc (100:0 v/v) increasing to n-hexane/EtOAc (70:30 v/v). 207 mg of 1-(3-bromopropoxy)-4-(trifluoromethyl)benzene was obtained in 47% yield as a yellow oil. 1H NMR (DMSO) δ 7.65 (d, J = 8.7 Hz, 2H), 7.14 (d, J = 8.7 Hz, 2H), 4.17 (t, J = 6.0 Hz, 2H), 3.68 (t, J = 6.6 Hz, 2H), 2.28 (p, J = 6.3 Hz, 2H). 19F NMR (DMSO) δ −59.83 (s, 3F). 13C NMR (DMSO) δ 161.67, 127.44 (m), 125.02 (q, J = 271.0 Hz), 121.73 (q, J = 32.1 Hz), 115.44, 32.08, 31.50.
3.13. Synthesis of Fragment 1: N-Methyl-3-(4-(trifluoromethyl)phenoxy)propan-1-amine
To a round-bottom flask containing methylamine in absolute ethanol (2 mL) cooled to 0 °C, a solution of 1-(3-bromopropoxy)-4-(trifluoromethyl)benzene (0.80 mmol) in absolute EtOH (0.8 mL) was added dropwise. The reaction mixture was stirred at room temperature overnight. The mixture was filtrated. The obtained residue was dissolved in DCM (10 mL). The organic layer was washed with saturated aqueous NaHCO3 and brine, dried over Na2SO4, and concentrated in vacuo. The residue was treated with HCl in diethyl ether. The resulting solid was then filtered and washed with diethyl ether to give 131 mg of N-methyl-3-(4-(trifluoromethyl)phenoxy)propan-1-amine hydrochloride salt as a white powder in a yield of 69%. 1H NMR (DMSO) δ 8.65 (s, 2H), 7.68 (d, J = 8.6 Hz, 2H), 7.13 (d, J = 8.6 Hz, 2H), 4.16 (t, J = 6.1 Hz, 2H), 3.06 (d, J = 7.4 Hz, 2H), 2.58 (s, 3H), 2.13–2.04 (m, 2H). 19F NMR (DMSO) δ −59.78. 13C NMR (DMSO) δ 161.58, 127.43 (q, J = 3.7 Hz), 125.02 (q, J = 271.1 Hz), 121.76 (q, J = 32.2 Hz), 115.48, 65.58, 46.14, 33.07, 25.71. UPLC: retention time = 1.521 min, MS [ESI, m/z]: 234.1 [M + Na]+.
3.14. Synthesis of Fragment 2: N-Methyl-3-phenylpropan-1-amine
To a round-bottom flask containing methylamine in absolute ethanol (2.18 mL), a solution of (3-bromopropyl)benzene (1.25 mmol) in absolute EtOH (0.87 mL) was added dropwise at 0 °C. The reaction mixture was stirred at room temperature overnight. The solid residue was filtered and washed with absolute EtOH to give 195 mg of N-methyl-3-phenylpropan-1-amine in a yield of 75% as a white powder. 1H NMR (DMSO) δ 8.48 (s, 2H), 7.35–7.27 (m, 2H), 7.26–7.17 (m, 3H), 2.91–2.85 (m, 1H), 2.65 (t, J = 7.7 Hz, 2H), 2.55 (s, 3H), 1.94–1.85 (m, 2H). 13C NMR (DMSO) δ 141.13, 128.92, 128.73, 126.57, 48.30, 32.88, 32.32, 27.57.
3.15. Synthesis of Fragment 4: 1-(Benzyloxy)-4-(trifluoromethyl)benzene
To a solution of 4-(trifluoromethyl)phenol (1.233 mmol), in DMF (3 mL), 1.553 mmol of (bromomethyl)benzene (1.553 mmol) and potassium carbonate (4.932 mmol) were added. The obtained mixture was stirred at 105 °C for 4 h. After the reaction completion, the mixture was filtrated, diluted with ethyl acetate (10 mL), and washed with water (3 × 10 mL). The organic layer was dried over sodium sulfate and evaporated under reduced pressure. The residue was purified by flash column chromatography and eluted with n-hexane/DCM (100:0 v/v increasing to 0:100 v/v), obtaining 212 mg of 1-(benzyloxy)-4-(trifluoromethyl)benzene in a yield of 75% as a white powder. 1H NMR (DMSO) δ 7.66 (d, J = 8.5 Hz, 2H), 7.49–7.44 (m, 2H), 7.44–7.38 (m, 2H), 7.38–7.32 (m, 1H), 7.20 (d, J = 8.5 Hz, 2H), 5.20 (s, 2H). 19F NMR (DMSO) δ −59.81. 13C NMR (DMSO) δ 161.64, 136.88, 128.98, 128.50, 128.25, 127.42 (q, J = 3.7 Hz), 125.02 (q, J = 271.1 Hz), 121.74 (q, J = 32.1 Hz), 115.77, 70.00.
3.16. Purchased Fragments
Fragment 3 (3-(methylamino)-1-phenylpropan-1-ol) and fragment 6 (4-(trifluoromethyl)phenol) were purchased from Sigma-Aldrich; they possess a purity grade of >97% and were used as received. Fragment 3 was obtained as a racemic mixture. Fragment 5 (4-(trifluoromehtyl)anisol) was ordered from Alfa Aesar (>98% pure). The fragments were dissolved in DMSO at a stock concentration of 100 mM.
3.17. Calculations
The concentration of the compound that inhibits virus-induced cell death by 50% (50% effective concentration [EC50]) was calculated by nonlinear regression analysis. Cytotoxicity of the compounds was assessed in a similar setup, and 50% cytotoxic concentration (CC50) values were derived from cell viability values determined with an MTS assay. Each experiment was performed at least in triplicate. The nonlinear regression and the graphs were made with GraphPad Prism Version 6.
Acknowledgments
This work was supported by research grants from The Netherlands Organisation for Scientific Research (NWO-ECHO-711.017.002 to F.J.M.v.K. and J.R.P.M.S.; NWO-VICI-91812628 to F.J.M.v.K.) and the European Union (Horizon 2020 Marie Skłodowska-Curie ETN “ANTIVIRALS”, grant agreement number 642434 to T.L., B.C., A.B., and F.J.M.v.K.).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsinfecdis.9b00179.
Supplementary Figure 1, antiviral activity and binding of fragments to 2C; Supplementary Figure 2, root mean square deviation (RMSD) of the protein ligand complexes during MD simulations; Supplementary Figure 3, multiple sequence alignment of 2C proteins from different enteroviruses (PDF)
Author Present Address
† J.R.P.M.S.: Viroclinics, Rotterdam, The Netherlands.
Author Present Address
‡ R.U.: The Francis Crick Institute, London, United Kingdom.
Author Contributions
∥ L.B., R.M., and B.Z. contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Tapparel C.; Siegrist F.; Petty T. J.; Kaiser L. (2013) Picornavirus and enterovirus diversity with associated human diseases. Infect., Genet. Evol. 14, 282–93. 10.1016/j.meegid.2012.10.016. [DOI] [PubMed] [Google Scholar]
- Bauer L.; Lyoo H.; van der Schaar H. M.; Strating J. R.; van Kuppeveld F. J. (2017) Direct-acting antivirals and host-targeting strategies to combat enterovirus infections. Curr. Opin. Virol. 24, 1–8. 10.1016/j.coviro.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez P. L.; Carrasco L. (1993) Poliovirus protein 2C has ATPase and GTPase activities. J. Biol. Chem. 268 (11), 8105–8110. [PubMed] [Google Scholar]
- Pfister T.; Wimmer E. (1999) Characterization of the nucleoside triphosphatase activity of poliovirus protein 2C reveals a mechanism by which guanidine inhibits poliovirus replication. J. Biol. Chem. 274 (11), 6992–7001. 10.1074/jbc.274.11.6992. [DOI] [PubMed] [Google Scholar]
- Klein M.; Eggers H. J.; Nelsen-Salz B. (1999) Echovirus 9 strain barty non-structural protein 2C has NTPase activity. Virus Res. 65 (2), 155–60. 10.1016/S0168-1702(99)00110-0. [DOI] [PubMed] [Google Scholar]
- Xia H.; Wang P.; Wang G. C.; Yang J.; Sun X.; Wu W.; Qiu Y.; Shu T.; Zhao X.; Yin L.; Qin C. F.; Hu Y.; Zhou X. (2015) Human Enterovirus Nonstructural Protein 2CATPase Functions as Both an RNA Helicase and ATP-Independent RNA Chaperone. PLoS Pathog. 11 (7), e1005067. 10.1371/journal.ppat.1005067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton M. R.; Dillingham M. S.; Wigley D. B. (2007) Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 76, 23–50. 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- Li J. P.; Baltimore D. (1990) An intragenic revertant of a poliovirus 2C mutant has an uncoating defect. J. Virol. 64 (3), 1102–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho M. W.; Teterina N.; Egger D.; Bienz K.; Ehrenfeld E. (1994) Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology 202 (1), 129–45. 10.1006/viro.1994.1329. [DOI] [PubMed] [Google Scholar]
- Aldabe R.; Carrasco L. (1995) Induction of membrane proliferation by poliovirus proteins 2C and 2BC. Biochem. Biophys. Res. Commun. 206 (1), 64–76. 10.1006/bbrc.1995.1010. [DOI] [PubMed] [Google Scholar]
- Teterina N. L.; Gorbalenya A. E.; Egger D.; Bienz K.; Ehrenfeld E. (1997) Poliovirus 2C protein determinants of membrane binding and rearrangements in mammalian cells. J. Virol. 71 (12), 8962–8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhy D. A.; Giddings T. H. Jr.; Kirkegaard K. (2000) Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74 (19), 8953–65. 10.1128/JVI.74.19.8953-8965.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee R.; Echeverri A.; Dasgupta A. (1997) Poliovirus-encoded 2C polypeptide specifically binds to the 3′-terminal sequences of viral negative-strand RNA. J. Virol. 71 (12), 9570–9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee R.; Dasgupta A. (2001) Interaction of picornavirus 2C polypeptide with the viral negative-strand RNA. J. Gen. Virol. 82 (11), 2621–2627. 10.1099/0022-1317-82-11-2621. [DOI] [PubMed] [Google Scholar]
- Banerjee R.; Tsai W.; Kim W.; Dasgupta A. (2001) Interaction of poliovirus-encoded 2C/2BC polypeptides with the 3′ terminus negative-strand cloverleaf requires an intact stem-loop b. Virology 280 (1), 41–51. 10.1006/viro.2000.0770. [DOI] [PubMed] [Google Scholar]
- Li J. P.; Baltimore D. (1988) Isolation of poliovirus 2C mutants defective in viral RNA synthesis. J. Virol. 62 (11), 4016–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teterina N. L.; Kean K. M.; Gorbalenya A. E.; Agol V. I.; Girard M. (1992) Analysis of the functional significance of amino acid residues in the putative NTP-binding pattern of the poliovirus 2C protein. J. Gen. Virol. 73 (8), 1977–1986. 10.1099/0022-1317-73-8-1977. [DOI] [PubMed] [Google Scholar]
- Tolskaya E. A.; Romanova L. I.; Kolesnikova M. S.; Gmyl A. P.; Gorbalenya A. E.; Agol V. I. (1994) Genetic studies on the poliovirus 2C protein, an NTPase. A plausible mechanism of guanidine effect on the 2C function and evidence for the importance of 2C oligomerization. J. Mol. Biol. 236 (5), 1310–23. 10.1016/0022-2836(94)90060-4. [DOI] [PubMed] [Google Scholar]
- Barton D. J.; Flanegan J. B. (1997) Synchronous replication of poliovirus RNA: initiation of negative-strand RNA synthesis requires the guanidine-inhibited activity of protein 2C. J. Virol. 71 (11), 8482–8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teterina N. L.; Levenson E.; Rinaudo M. S.; Egger D.; Bienz K.; Gorbalenya A. E.; Ehrenfeld E. (2006) Evidence for functional protein interactions required for poliovirus RNA replication. J. Virol. 80 (11), 5327–37. 10.1128/JVI.02684-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W. F.; Yang S. Y.; Wu B. W.; Jheng J. R.; Chen Y. L.; Shih C. H.; Lin K. H.; Lai H. C.; Tang P.; Horng J. T. (2007) Reticulon 3 binds the 2C protein of enterovirus 71 and is required for viral replication. J. Biol. Chem. 282 (8), 5888–98. 10.1074/jbc.M611145200. [DOI] [PubMed] [Google Scholar]
- Zheng Z.; Li H.; Zhang Z.; Meng J.; Mao D.; Bai B.; Lu B.; Mao P.; Hu Q.; Wang H. (2011) Enterovirus 71 2C protein inhibits TNF-alpha-mediated activation of NF-kappaB by suppressing IkappaB kinase beta phosphorylation. J. Immunol. 187 (5), 2202–12. 10.4049/jimmunol.1100285. [DOI] [PubMed] [Google Scholar]
- Vance L. M.; Moscufo N.; Chow M.; Heinz B. A. (1997) Poliovirus 2C region functions during encapsidation of viral RNA. J. Virol. 71 (11), 8759–8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verlinden Y.; Cuconati A.; Wimmer E.; Rombaut B. (2000) The antiviral compound 5-(3,4-dichlorophenyl) methylhydantoin inhibits the post-synthetic cleavages and the assembly of poliovirus in a cell-free system. Antiviral Res. 48 (1), 61–9. 10.1016/S0166-3542(00)00119-4. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Wang C.; Mueller S.; Paul A. V.; Wimmer E.; Jiang P. (2010) Direct interaction between two viral proteins, the nonstructural protein 2C and the capsid protein VP3, is required for enterovirus morphogenesis. PLoS Pathog. 6 (8), e1001066. 10.1371/journal.ppat.1001066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.; Ma H. C.; Wimmer E.; Jiang P.; Paul A. V. (2014) A C-terminal, cysteine-rich site in poliovirus 2C(ATPase) is required for morphogenesis. J. Gen. Virol. 95 (6), 1255–1265. 10.1099/vir.0.062497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Linden L.; Wolthers K. C.; van Kuppeveld F. J. (2015) Replication and Inhibitors of Enteroviruses and Parechoviruses. Viruses 7 (8), 4529–4562. 10.3390/v7082832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadaschik D.; Klein M.; Zimmermann H.; Eggers H. J.; Nelsen-Salz B. (1999) Dependence of echovirus 9 on the enterovirus RNA replication inhibitor 2-(alpha-Hydroxybenzyl)-benzimidazole maps to nonstructural protein 2C. J. Virol. 73 (12), 10536–10539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Palma A. M.; Heggermont W.; Lanke K.; Coutard B.; Bergmann M.; Monforte A. M.; Canard B.; De Clercq E.; Chimirri A.; Purstinger G.; Rohayem J.; van Kuppeveld F.; Neyts J. (2008) The thiazolobenzimidazole TBZE-029 inhibits enterovirus replication by targeting a short region immediately downstream from motif C in the nonstructural protein 2C. J. Virol. 82 (10), 4720–30. 10.1128/JVI.01338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu H.; Agoh M.; Agoh Y.; Yoshida H.; Yoshii K.; Yoneyama T.; Hagiwara A.; Miyamura T. (2000) Mutations in the 2C region of poliovirus responsible for altered sensitivity to benzimidazole derivatives. J. Virol. 74 (9), 4146–54. 10.1128/JVI.74.9.4146-4154.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburn T. T.; Thor K. B. (2004) Drug repositioning: identifying and developing new uses for existing drugs. Nat. Rev. Drug Discovery 3 (8), 673–83. 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- Mercorelli B.; Palu G.; Loregian A. (2018) Drug Repurposing for Viral Infectious Diseases: How Far Are We?. Trends Microbiol. 26 (10), 865–876. 10.1016/j.tim.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J.; Quinn K. K.; Kye S.; Cooper P.; Damoiseaux R.; Krogstad P. (2012) Fluoxetine Is a Potent Inhibitor of Coxsackievirus Replication. Antimicrob. Agents Chemother. 56 (9), 4838–4844. 10.1128/AAC.00983-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulferts R.; van der Linden L.; Thibaut H. J.; Lanke K. H.; Leyssen P.; Coutard B.; De Palma A. M.; Canard B.; Neyts J.; van Kuppeveld F. J. (2013) Selective serotonin reuptake inhibitor fluoxetine inhibits replication of human enteroviruses B and D by targeting viral protein 2C. Antimicrob. Agents Chemother. 57 (4), 1952–6. 10.1128/AAC.02084-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulferts R.; de Boer M.; van der Linden L.; Bauer L.; Lyoo H. R.; Mate M. J.; Lichiere J.; Canard B.; Lelieveld D.; Omta W.; Egan D.; Coutard B.; van Kuppeveld F. J. (2016) Screening of a library of FDA-approved drugs identifies several enterovirus replicaton inhibitors that target viral protein 2C. Antimicrob. Agents Chemother. 60, 2627. 10.1128/AAC.02182-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young K. C.; Bai C. H.; Su H. C.; Tsai P. J.; Pu C. Y.; Liao C. S.; Lin Y. M.; Lai H. W.; Chong L. W.; Tsai Y. S.; Tsao C. W. (2014) Fluoxetine a novel anti-hepatitis C virus agent via ROS-, JNK-, and PPARbeta/gamma-dependent pathways. Antiviral Res. 110, 158–67. 10.1016/j.antiviral.2014.08.002. [DOI] [PubMed] [Google Scholar]
- Medigeshi G. R.; Kumar R.; Dhamija E.; Agrawal T.; Kar M. (2016) N-Desmethylclozapine, Fluoxetine, and Salmeterol Inhibit Postentry Stages of the Dengue Virus Life Cycle. Antimicrob. Agents Chemother. 60 (11), 6709–6718. 10.1128/AAC.01367-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gofshteyn J.; Cardenas A. M.; Bearden D. (2016) Treatment of Chronic Enterovirus Encephalitis With Fluoxetine in a Patient With X-Linked Agammaglobulinemia. Pediatr Neurol 64, 94. 10.1016/j.pediatrneurol.2016.06.014. [DOI] [PubMed] [Google Scholar]
- Guan H.; Tian J.; Qin B.; Wojdyla J. A.; Wang B.; Zhao Z.; Wang M.; Cui S. (2017) Crystal structure of 2C helicase from enterovirus 71. Sci. Adv. 3 (4), e1602573. 10.1126/sciadv.1602573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson D. W.; Jones N. D.; Swartzendruber J. K.; Yang K. S.; Wong D. T. (1988) Molecular structure of fluoxetine hydrochloride, a highly selective serotonin-uptake inhibitor. J. Med. Chem. 31 (1), 185–9. 10.1021/jm00396a030. [DOI] [PubMed] [Google Scholar]
- Zuo J.; Kye S.; Quinn K. K.; Cooper P.; Damoiseaux R.; Krogstad P. (2016) Discovery of Structurally Diverse Small-Molecule Compounds with Broad Antiviral Activity against Enteroviruses. Antimicrob. Agents Chemother. 60 (3), 1615–26. 10.1128/AAC.02646-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albulescu L.; Wubbolts R.; van Kuppeveld F. J.; Strating J. R. (2015) Cholesterol shuttling is important for RNA replication of coxsackievirus B3 and encephalomyocarditis virus. Cell. Microbiol. 17, 1144. 10.1111/cmi.12425. [DOI] [PubMed] [Google Scholar]
- Strating J. R.; van der Linden L.; Albulescu L.; Bigay J.; Arita M.; Delang L.; Leyssen P.; van der Schaar H. M.; Lanke K. H.; Thibaut H. J.; Ulferts R.; Drin G.; Schlinck N.; Wubbolts R. W.; Sever N.; Head S. A.; Liu J. O.; Beachy P. A.; De Matteis M. A.; Shair M. D.; Olkkonen V. M.; Neyts J.; van Kuppeveld F. J. (2015) Itraconazole inhibits enterovirus replication by targeting the oxysterol-binding protein. Cell Rep. 10 (4), 600–15. 10.1016/j.celrep.2014.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B.; Decroly E.; Li C.; Sharff A.; Lescar J.; Bricogne G.; Barral K. (2014) Assessment of Dengue virus helicase and methyltransferase as targets for fragment-based drug discovery. Antiviral Res. 106, 61–70. 10.1016/j.antiviral.2014.03.013. [DOI] [PubMed] [Google Scholar]
- Adams P.; Kandiah E.; Effantin G.; Steven A. C.; Ehrenfeld E. (2009) Poliovirus 2C protein forms homo-oligomeric structures required for ATPase activity. J. Biol. Chem. 284 (33), 22012–21. 10.1074/jbc.M109.031807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney T. R.; Cisnetto V.; Bose D.; Bailey M.; Wilson J. R.; Zhang X.; Belsham G. J.; Curry S. (2010) Foot-and-mouth disease virus 2C is a hexameric AAA+ protein with a coordinated ATP hydrolysis mechanism. J. Biol. Chem. 285 (32), 24347–59. 10.1074/jbc.M110.129940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Schaar H. M.; Leyssen P.; Thibaut H. J.; de Palma A.; van der Linden L.; Lanke K. H.; Lacroix C.; Verbeken E.; Conrath K.; Macleod A. M.; Mitchell D. R.; Palmer N. J.; van de Poel H.; Andrews M.; Neyts J.; van Kuppeveld F. J. (2013) A novel, broad-spectrum inhibitor of enterovirus replication that targets host cell factor phosphatidylinositol 4-kinase IIIbeta. Antimicrob. Agents Chemother. 57 (10), 4971–81. 10.1128/AAC.01175-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod A. M.; Mitchell D. R.; Palmer N. J.; Van de Poel H.; Conrath K.; Andrews M.; Leyssen P.; Neyts J. (2013) Identification of a series of compounds with potent antiviral activity for the treatment of enterovirus infections. ACS Med. Chem. Lett. 4 (7), 585–9. 10.1021/ml400095m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaut H. J., Lee C. K., Coutard B., Van der Linden L., Canard B., De Palma A. M., Van Kuppeveld F., Jung Y. S., and Neyts J.. A novel class of highly potent small molecule inhibitors of entero/rhinovirus replication that target the non structural protein 2C, https://rega.kuleuven.be/cmt/jn/poster/2013/2013_ht/Poster%20ICAR%202013%20MOA%20KRICT.pdf, 2013 (accessed February 22, 2019).
- Holm-Hansen C. C.; Midgley S. E.; Fischer T. K. (2016) Global emergence of enterovirus D68: a systematic review. Lancet Infect. Dis. 16 (5), e64–e75. 10.1016/S1473-3099(15)00543-5. [DOI] [PubMed] [Google Scholar]
- Hixon A. M.; Clarke P.; Tyler K. L. (2017) Evaluating Treatment Efficacy in a Mouse Model of Enterovirus D68-Associated Paralytic Myelitis. J. Infect. Dis. 216 (10), 1245–1253. 10.1093/infdis/jix468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messacar K.; Sillau S.; Hopkins S. E.; Otten C.; Wilson-Murphy M.; Wong B.; Santoro J. D.; Treister A.; Bains H. K.; Torres A.; Zabrocki L.; Glanternik J. R.; Hurst A. L.; Martin J. A.; Schreiner T.; Makhani N.; DeBiasi R. L.; Kruer M. C.; Tremoulet A. H.; Van Haren K.; Desai J.; Benson L. A.; Gorman M. P.; Abzug M. J.; Tyler K. L.; Dominguez S. R. (2019) Safety, tolerability, and efficacy of fluoxetine as an antiviral for acute flaccid myelitis. Neurology 92 (18), e2118–e2126. 10.1212/WNL.0000000000006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkahla M. A.; Alidjinou E. K.; Sane F.; Desailloud R.; Hober D. (2018) Fluoxetine can inhibit coxsackievirus-B4 E2 in vitro and in vivo. Antiviral Res. 159, 130–133. 10.1016/j.antiviral.2018.10.002. [DOI] [PubMed] [Google Scholar]
- Amsterdam J. D.; Fawcett J.; Quitkin F. M.; Reimherr F. W.; Rosenbaum J. F.; Michelson D.; Hornig-Rohan M.; Beasley C. M. (1997) Fluoxetine and norfluoxetine plasma concentrations in major depression: a multicenter study. Am. J. Psychiatry 154 (7), 963–969. [DOI] [PubMed] [Google Scholar]
- Backes P.; Quinkert D.; Reiss S.; Binder M.; Zayas M.; Rescher U.; Gerke V.; Bartenschlager R.; Lohmann V. (2010) Role of annexin A2 in the production of infectious hepatitis C virus particles. J. Virol. 84 (11), 5775–89. 10.1128/JVI.02343-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanke K. H.; van der Schaar H. M.; Belov G. A.; Feng Q.; Duijsings D.; Jackson C. L.; Ehrenfeld E.; van Kuppeveld F. J. (2009) GBF1, a guanine nucleotide exchange factor for Arf, is crucial for coxsackievirus B3 RNA replication. J. Virol. 83 (22), 11940–9. 10.1128/JVI.01244-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels E.; Duijsings D.; Lanke K. H.; van Dooren S. H.; Jackson C. L.; Melchers W. J.; van Kuppeveld F. J. (2006) Effects of picornavirus 3A Proteins on Protein Transport and GBF1-dependent COP-I recruitment. J. Virol. 80 (23), 11852–60. 10.1128/JVI.01225-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L. J.; Muench H. (1938) A simple method of estimating fifty percent endpoints. Am. J. Epidemiol. 27 (3), 493–497. 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- Case D. A., Darden T. A., Cheatham T. E. III, Simmerling C. L., Wang J., Duke R. E., Luo R., Walker R. C., Zhang W., Merz K. M., Roberts B., Hayik S., Roitberg A., Seabra G., Swails J., Götz A. W., Kolossváry I., Wong K. F., Paesani F., Vanicek J., Wolf R. M., Liu J., Wu X., Brozell S. R., Steinbrecher T., Gohlke H., Cai Q., Ye X., Wang J., Hsieh M.-J., Cui G., Roe D. R., Mathews D. H., Seetin M. G., Salomon-Ferrer R., Sagui C., Babin V., Luchko T., Gusarov S., Kovalenko A., and Kollman P. A. (2012) AMBER 12, http://ambermd.org/doc12/Amber12.pdf (accessed 11.03.2019). [Google Scholar]
- Gerber P. R.; Muller K. (1995) MAB, a generally applicable molecular force field for structure modelling in medicinal chemistry. J. Comput.-Aided Mol. Des. 9 (3), 251–68. 10.1007/BF00124456. [DOI] [PubMed] [Google Scholar]
- Labute P. (2008) The generalized Born/volume integral implicit solvent model: estimation of the free energy of hydration using London dispersion instead of atomic surface area. J. Comput. Chem. 29 (10), 1693–8. 10.1002/jcc.20933. [DOI] [PubMed] [Google Scholar]
- Lovell S. C.; Davis I. W.; Arendall W. B.; de Bakker P. I. W.; Word J. M.; Prisant M. G.; Richardson J. S.; Richardson D. C. (2003) Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins: Struct., Funct., Genet. 50, 437–450. 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- Bowie J. U.; Luthy R.; Eisenberg D. (1991) A method to identify protein sequences that fold into a known three-dimensional structure. Science 253 (5016), 164–70. 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- Luthy R.; Bowie J. U.; Eisenberg D. (1992) Assessment of protein models with three-dimensional profiles. Nature 356 (6364), 83–5. 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- Colovos C.; Yeates T. O. (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 2 (9), 1511–9. 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontius J.; Richelle J.; Wodak S. J. (1996) Deviations from standard atomic volumes as a quality measure for protein crystal structures. J. Mol. Biol. 264 (1), 121–36. 10.1006/jmbi.1996.0628. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.