Abstract
Carnivorous plants have the ability to capture and digest small animals as a source of additional nutrients, which allows them to grow in nutrient-poor habitats. Here we report the complete sequences of the plastid genomes of two carnivorous plants of the order Caryophyllales, Drosera rotundifolia and Nepenthes × ventrata. The plastome of D. rotundifolia is repeat-rich and highly rearranged. It lacks NAD(P)H dehydrogenase genes, as well as ycf1 and ycf2 genes, and three essential tRNA genes. Intron losses are observed in some protein-coding and tRNA genes along with a pronounced reduction of RNA editing sites. Only six editing sites were identified by RNA-seq in D. rotundifolia plastid genome and at most conserved editing sites the conserved amino acids are already encoded at the DNA level. In contrast, the N. × ventrata plastome has a typical structure and gene content, except for pseudogenization of the ccsA gene. N. × ventrata and D. rotundifolia could represent different stages of evolution of the plastid genomes of carnivorous plants, resembling events observed in parasitic plants in the course of the switch from autotrophy to a heterotrophic lifestyle.
Keywords: carnivorous plant, plastid genome, gene loss, RNA editing, Caryophyllales
1. Introduction
Carnivorous plants are able to attract, catch, kill, and digest their prey, usually insects, and assimilate amino acids, peptides, and other nutrients resulting from digestion for growth [1,2]. The nutrients obtained are primarily used as a source of nitrogen and phosphorus, enabling these plants to survive in habitats with nutrient-poor soils and in oligotrophic aquatic environments. Nevertheless, all known carnivorous plants retain the ability to photosynthesize and to fix CO2, and they cannot grow heterotrophically with respect to organic carbon. About 700 species of carnivorous plants are currently recognized [3]. Carnivory evolved several times, with at least nine independent origins in five orders of angiosperms (Caryophyllales, Ericales, Lamiales, Oxalidales, and Poales) [4,5,6]. The majority of carnivorous plants belong to the orders Caryophyllales and Lamiales [3].
Another group of plants capable of receiving nutrients from other organisms is direct parasites and mycoheterotrophs, receiving nutrients, respectively, from host plants or through mycorrhizal fungi with which they associate. Contrary to carnivorous plants, parasitic plants depend on organic carbon derived from their hosts. Some parasitic plants retain photosynthetic capabilities, being hemiparasites, while others completely lose the ability to carry out photosynthesis [7]. Plastid genomes (plastomes) of parasitic plants represent model systems for studying the effects of relaxed selective pressure on photosynthetic function. In general, plastid genomes in angiosperms are highly conserved in structure, gene content, and gene order, but structural rearrangements and gene loss appear to be associated with the switch from autotrophy to heterotrophy [8,9,10]. Extensive studies of the plastid genomes of parasitic plants have revealed a reduction in size and gene content compared with their photosynthetic relatives, which correlates with the loss of genes encoding photosynthetic functions ([8,11,12], see [13] for a recent review). Such reduction could be minimal in hemiparasites and “early” parasites such as Corallorhiza striata [14] and Cuscuta sp. [15], extreme in some holoparasitic species [16,17,18,19,20], and even complete, as seen by the loss of the plastome in Rafflesia lagascae [21].
Contrary to parasitic plants, only several complete plastid genomes of carnivorous plants have been sequenced, and the possible effect of carnivorous lifestyle on the plastid genome remains poorly understood. Studies of plastid genomes of Pinguicula ehlersiae, three Utricularia, and seven Genlisea species as representatives of the three genera of the carnivorous family Lentibulariaceae, have revealed the loss and pseudogenization of the NAD(P)H dehydrogenase genes in some species as well as a significant increase of substitution rates and microstructural changes [22,23,24].
To get further insights into the molecular changes associated with the transition to carnivory, we sequenced the plastid genomes of the members of two other families of carnivorous plants, Drosera rotundifolia (Caryophyllales: Droseraceae) and Nepenthes × ventrata (Caryophyllales: Nepenthaceae). Phylogenetic age of both families is estimated in approximately 85 million years ago [3]. The family Droseraceae, comprising the genera Drosera, Dionaea, and Aldrovanda with more than 250 species, is the second most diverse carnivorous family after Lentibulariaceae [25]. D. rotundifolia is one of the most widespread sundew species, common in many areas of the Northern Hemisphere, including Europe, Siberia, and North America. D. rotundifolia typically thrives in wetlands such as bogs, marshes, and fens, but can also grow on open peatlands and wet sands. A typical plant has a size of about 3 to 5 cm. Leaves spread on the soil surface, with long petioles collected in a rosette. The edge and the upper surface of the leaf are covered with reddish glandular hairs in the form of heads on long stalks, which reach a length of 4–5 mm. The hairs secrete a sticky liquid in the form of shiny drops. They are sensitive to irritation, and when an insect hits a leaf, they bend and catch it.
Nepenthaceae comprises the single genus Nepenthes, which includes more than 160 species, most of which inhabit the tropical regions of South-East Asia [26]. Nepenthes use passive pitcher-shaped traps to catch small insects and are probably the most studied carnivorous plants [27]. Nepenthes × ventrata is a natural hybrid between N. alata and N. ventricosa, two species endemic to the Philippines [28]. This is a perennial medium-sized (about 30–50 cm high) plant with red cylindrical pitchers used to catch small insects. Pitchers of Nepenthes have been analyzed by RNA-seq and proteomics for the discovery of genetic traits related to carnivory [29,30].
Here we report the full plastome sequences of D. rotundifolia and N. × ventrata and present a comparative analysis of the plastomes of both species and their carnivorous and non-carnivorous relatives. The plastome of D. rotundifolia is the first one to be sequenced and released to the GenBank (KU168830) in the family Droseraceae. Recently, sequences of plastome of four other species of Droseraceae, Drosera erythrorhiza (GenBank KY651214), Drosera regia (KY679199), Aldrovanda vesiculosa (KY679200), and Dionaea muscipula (KY679201) have been published [31]. For the genus Nepenthes, the complete plastid genome sequences are known for Nepenthes mirabilis (MH286314, MK397879, MK397880, and MK397881), and a near-complete plastome sequence was reported for Nepenthes graciliflora (MH286314) [32,33]. We used these sequences for the comparative analysis. The results of the present study improve our understanding of the evolution of the plastid genomes of carnivorous plants and reveal some similar evolutionary patterns between carnivorous and parasitic plants.
2. Results
2.1. Plastome Size and Gene Content in D. rotundifolia
The plastid genome of D. rotundifolia is 192,912 bp in length and has a typical quadripartite structure with two single-copy regions and an inverted repeat. The increase in genome size compared with typical plastomes of flowering plants is primarily due to the extension of inverted repeats, which are 52,949 bp in length.
The D. rotundifolia plastome was predicted to contain 97 presumably intact unique genes (Table 1), which is somewhat less than its close non-carnivorous relative Fagopyrum esculentum (114 genes; [34]). Consistent with the ability to photosynthesize, the D. rotundifolia plastome contains a typical set of genes encoding photosynthesis-related proteins (photosystems I and II, cytochrome b6f complex, rubisco large subunit, and ATP synthase). The genes for plastid-encoded RNA polymerase and translation initiation factor InfA are also present. The most notable loss is the lack of all 11 genes coding for the NAD(P)H dehydrogenase complex (ndhA–K). Also missing are genes ycf1 and ycf2, thought to be essential in plant plastids.
Table 1.
Summary of genes identified in the D. rotundifolia plastome.
| Function | Genes |
|---|---|
| Photosystem I | psaA, psaB, psaC, psaI, psaJ, ycf3i, ycf4 |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ |
| Cytochrome b6/f complex | petA, petBi, petDi, petG, petL, petN, ccsA |
| ATP synthase | atpA, atpB, atpE, atpFi, atpH, atpI |
| RNA polymerase | rpoA, rpoB, rpoC1i, rpoC2 |
| Ribosomal proteins (large subunit) | rpl2, rpl14, rpl16i, rpl20, rpl22, rpl23, rpl32, rpl33, rpl36 |
| Ribosomal proteins (small subunit) | rps2, rps3, rps4, rps7, rps8, rps11, rps12i, rps14 (x2), rps15, rps16, rps19 |
| Other protein-coding genes | rbcL, infA, matK, cemA, clpP, accD, ycf68 |
| rRNAs | rrn16, rrn23, rrn4.5, rrn5 |
| tRNAs | trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, trnH-GUG, trnI-CAU, trnI-GAUi, trnK-UUU, trnL-CAA, trnL-UAAi, trnL-UAG, trnM-CAU (x2), trnfM-CAU, trnN-GUU, trnP-UGG (x2), trnQ-UUG, trnR-ACG, trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC, trnW-CCA, trnY-GUA |
| Pseudogenes | ψrps18(orf641), ψycf15 |
| Pseudogenes present along with an intact copy | ψinfA, ψpsaB, ψpsaJ, ψpsbI, ψpsbJ, ψrpl2 |
Genes duplicated in inverted repeats were counted once; i denotes intron-containing genes, including trans-spliced rps12.
The functionality of the accD gene coding for the beta subunit of acetyl-CoA carboxylase is questionable because its predicted protein product contains an ~100 a.a. long N-terminal extension relative to typical AccD proteins (e.g., from Nicotiana tabacum). However, the whole functional domain (COG0777) is present, and the open reading frame remains non-interrupted. AccD protein, involved in fatty-acid synthesis and leaf development [35], is supposed to be essential for maintenance of the plastome in dicots [36].
The clpP gene, encoding the ATP-dependent proteolytic subunit of the Clp peptidase involved in protein metabolism within the plastid [36], is supposed to be essential and is present even in highly reduced plastomes of parasitic plants [16,20]. In the D. rotundifolia plastome, the only sequence corresponding to the N-terminal part of ClpP could be identified between genes psbB and rpl20, where the clpP gene is usually located in plastomes of angiosperms. However, it should be noted that accelerated evolution of ClpP was observed in several parasitic and photosynthetic lineages (see [17,37] for an example) to the extent that its sequence becomes difficult to identify.
The D. rotundifolia plastome encodes 27 tRNA species for all amino acids except alanine and lacks the tRNA genes trnA-UGC, trnG-UCC, and trnV-UAC, usually present in the plastomes of photosynthetic angiosperms. The loss of essential tRNA genes, including the above-mentioned ones, often occurs in the plastid genomes of parasitic plants [11,16,17,20] and it is assumed that the missing tRNA can be imported into the chloroplast from the cytosol.
All genes of ribosomal proteins typical of flowering plants were found in the plastome of D. rotundifolia (Table 1). An exception is the rps18 gene, whose almost complete coding sequence (~100 a.a. residues) is part of the long open reading frame orf641 capable of encoding a 641 a.a. protein. Orf641 is located between genes rpl33 and rpl20, which corresponds to the position of the rps18 gene in typical chloroplast genomes [9]. The predicted protein product of orf641 includes the N-terminal repeat-rich region followed by the almost complete Rps18 sequence, which has an identity of up to 65% with plastid Rps18 proteins. The N-terminal region includes 73 tandemly repeated sequences with GQKQPNI consensus (Figure S1). A GenBank search did not reveal close homologues of the N-terminal part of Orf641. The correctness of the assembly of the orf641 sequence was confirmed via PCR and sequencing of the amplified fragment by the Sanger method.
Along with the loss of some conservative genes, a peculiar feature of the plastid genome of D. rotundifolia is the loss of introns in the remaining genes. Introns are present in atpF, petB, petD, rpl16, rpoC1, ycf3 (two introns), trnI-GAU, and trnL-UAA, but appear to be lost in genes clpP, rpl2, and rps16. The rps12 in the D. rotundifolia plastome is a trans-spliced gene as in most other angiosperms, but it consists of only two rather than three exons, indicating the lack of one (cis-spliced) intron. An unusual feature of the D. rotundifolia plastome is the loss of the intron in the trnK gene, which usually contains the matK gene coding for the maturase for splicing of group IIA introns [38]. The matK gene is preserved in the D. rotundifolia plastome and is located downstream of the intronless trnK gene. The presence of matK correlates with the retention of group IIA introns in atpF and trnI-GAU genes. The loss of the trnK gene and the retention of stand-alone matK has been described in different plant lineages [17,39,40], but the presence of intronless trnK is unusual.
2.2. Structural Rearrangements and Duplications in the D. rotundifolia Plastome
The gene order in chloroplast genomes of flowering plants is highly conserved, and its deviations are usually associated with movement of the boundaries of inverted repeats. However, in some lineages of flowering plants, for example cereals, geranium, and clover, and especially in parasitic plants, the order of genes is significantly different from the standard due to numerous genome rearrangements—translocations, duplications, inversions, and deletions [10,41,42].
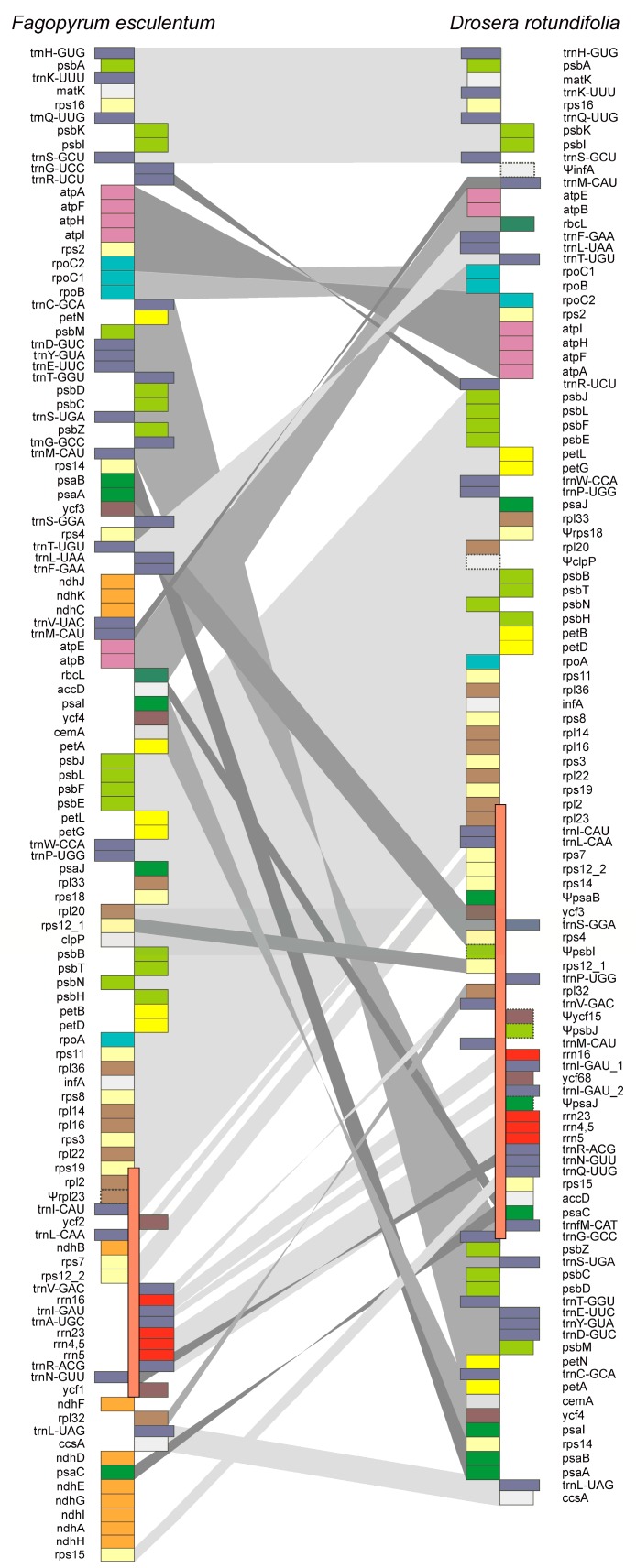
The comparison of the order of genes in the plastid genome of D. rotundifolia relative to the standard showed that the D. rotundifolia plastome, in addition to the above-mentioned deletions of particular genes, is characterized by large-scale rearrangements – inversion, translocation, and duplication of genes (Figure 1). Among these rearrangements are the transfer of psaA-petN genes from the large single-copy region (LSC) to the small one (SSC), insertion of the petA-psaI fragment into this cluster, inversion and translocation of atpA-rpoC2 within the large single-copy region, etc. However, the plastome retains intact structures of the highly conserved S10 operon (rps11, rpl36, infA, rps8, rpl14, rpl16, rps3, rpl22, rps19, rpl2, rpl23) and the rrn gene cluster (rrn16-rrn23-rrn4.5-rrn5). These operons ensuring coordinated expression of the components required for assembly of the ribosome are conserved in the majority of chloroplast genomes.
Figure 1.
Schematic gene order comparison map between the plastid genomes of D. rotundifolia and F. esculentum. The linear representation of the circular mapping genomes using trnH-GUG genes as the starting point does not reflect the actual gene size or spacing between the coding regions. The inverted repeat is indicated by an orange rectangle on the main line; only one copy is shown. Boxes represent protein-encoding and RNA genes; genes transcribed in opposite directions are shown to the left and to the right of the main line. The gray areas between the genes highlight the relative locations of identical genes. Ψ denotes pseudogenes.
It is likely that one of the main drivers of structural rearrangements and an increase in the size of the plastome was the duplication of extended fragments, accompanied by the insertion of their additional copies into other regions of the plastome. An analysis of the presence of repeats revealed that after excluding the large IR, the plastome of D. rotundifolia has many more total repetitive sequences than the plastomes of phylogenetically related species and other carnivorous plants. Repeat sequences account for about 23% of the D. rotundifolia plastome, while this value is less than 5% in the plastomes of other carnivores (Table 2). Increased repeat content is not associated with the accumulation of short tandem repeats often occurring in plastid genomes from slipped-strand mispairing [42,43], since their share in the D. rotundifolia plastome is not higher than in other plants (Table 2).
Table 2.
Repetitive sequence content in plastid genomes.
| Species | Plastome Size (bp) | Repetitive Sequences (bp) | Repetitive Sequences (%) | Tandem Repeats (bp) |
|---|---|---|---|---|
| Caryophyllales; Droseraceae | ||||
| Drosera rotundifolia | 192,912 | 32,380 | 23.13% | 736 |
| Drosera erythrorhiza | 134,391 | 4708 | 4.84% | 164 |
| Drosera regia | 136,810 | 1883 | 1.66% | 175 |
| Dionaea muscipula | 117,589 | 5599 | 4.90% | 460 |
| Aldrovanda vesiculosa | 141,568 | 2514 | 2.20% | 326 |
| Caryophyllales; Nepenthaceae | ||||
| Nepenthes × ventrata | 156,637 | 664 | 0.51% | 82 |
| Nepenthes mirabilis | 156,381 | 5426 | 4.20% | 237 |
| Lamiales; Lentibulariaceae | ||||
| Pinguicula ehlersiae | 147,147 | 893 | 0.74% | 151 |
| Utricularia macrorhiza | 153,228 | 632 | 0.50% | 476 |
| Utricularia reniformis | 139,725 | 682 | 0.59% | 60 |
| Utricularia gibba | 152,113 | 780 | 0.63% | 225 |
| Genlisea margaretae | 141,255 | 500 | 0.43% | 440 |
| Genlisea aurea | 140,010 | 586 | 0.51% | 251 |
| Genlisea filiformis | 140,308 | 622 | 0.54% | 204 |
| Genlisea pygmaea | 140,466 | 467 | 0.40% | 227 |
| Genlisea repens | 140,432 | 467 | 0.40% | 225 |
| Genlisea tuberosa | 140,677 | 563 | 0.49% | 208 |
| Genlisea violacea | 143,416 | 769 | 0.65% | 60 |
| Caryophyllales; Caryophyllaceae | ||||
| Silene noctiflora | 151,639 | 2852 | 2.34% | 225 |
| Silene chalcedonica | 148,081 | 4566 | 3.67% | 135 |
| Silene conica | 147,208 | 1785 | 1.48% | 40 |
| Silene conoidea | 147,896 | 1615 | 1.33% | 151 |
| Silene paradoxa | 151,632 | 2023 | 1.60% | 256 |
| Silene latifolia | 151,736 | 1189 | 0.94% | 20 |
| Silene vulgaris | 151,583 | 1121 | 0.89% | 181 |
| Agrostemma githago | 151,733 | 1297 | 1.03% | 105 |
Reported repeat content excludes one copy of the large IR.
A high repeat content corroborates the presence of additional copies of some protein-coding and tRNA genes. Most of them are retained as truncated pseudogenes (infA, psaB (2×), psbI (2×), psbJ (2×), psaJ (2×)), but three duplicated genes remain intact and could be functional (rps14, trnM-CAU, and trnP-UGG).
2.3. The N. × ventrata Plastome Retains Conserved Structure and Gene Content
The plastid genome of N. × ventrata is 156,637 bp in length and includes a pair of inverted repeats of 25,190 bp separated by 19,208 bp-long and 87,049 bp-long single-copy regions. The predicted gene pool of the N. × ventrata plastid genome is typical for flowering plants and is almost identical to that of F. esculentum [34] except that the rpl23 gene seems to be intact in N. × ventrata, as in most angiosperms [9]. The N. × ventrata plastome was predicted to contain 112 presumably intact unique genes (Table S1), including genes encoding photosynthesis-related functions, plastid-encoded RNA polymerase, and ribosomal proteins, as well as the conserved genes infA, matK, cemA, clpP, accD, ycf1, and ycf2. Unlike that of most carnivorous plants, the N. × ventrata plastome contains a complete set of genes coding for the NAD(P)H dehydrogenase complex (ndhA–K).
A total of 30 tRNA genes, seven of them having additional copies in the inverted repeats, were identified (Table S1). In contrast to D. rotundifolia, the set of tRNA genes in the N. × ventrata plastome is complete and can recognize all the codons present; therefore, no import of nuclear-encoded tRNAs is necessary. An analysis of the N. × ventrata plastome revealed the presence of all introns usually found in plastid genes and, as in most other angiosperms, the rps12 gene is trans-spliced and consists of three exons.
The N. × ventrata plastid genome is colinear to those of tobacco [44] and F. esculentum [34] with respect to the gene order. No evidence of structural rearrangements or increased repeat content was found.
The only notable functional gene loss is the pseudogenization of the ccsA gene due to a 22 bp insertion approximately 507 bp downstream from the start codon, resulting in a frameshift. The mapping of transcriptome sequences obtained in RNA-seq experiments confirmed the transcription of the ccsA pseudogene and the presence of this insertion in the transcripts. It is unlikely that CcsA protein in plastids is replenished due to the ccsA copy which could be transferred to the nuclear genome, since the RNA-seq analysis did not reveal transcripts for CcsA-like proteins carrying a chloroplast transit peptide.
2.4. Identification and Prediction of RNA Editing Sites
RNA editing in the plastids of seed plants is a post-transcriptional modification that changes a cytosine (C) to a uracil (U) nucleotide, producing transcripts that are different from their DNA template [45]. RNA editing can alter the amino acid sequence of proteins and can also introduce new start and stop codons [45]. The average number of editing sites in non-parasitic higher land plants is around 30–40 [46], but some parasitic plants show a pronounced reduction in the number of editing sites [15]. To determine the editotypes of D. rotundifolia and N. × ventrata, we performed an in silico analysis for potential editing sites and identified them experimentally using RNA-seq analysis. The efficiency of editing was determined from the C versus U ratio at the respective position in the transcripts from the RNA-seq data.
Although our RNA sequencing approach based on polyadenylation-dependent sequencing library preparation was not suitable for quantitative analysis of plastid gene expression, a high average genome coverage by RNA-seq reads enabled the identification of the RNA editing sites. The RNA-seq analysis revealed 45 editing sites in 26 genes in the N. × ventrata plastome (Table S2). The most extensively edited appeared to be transcripts of genes rpoC1 (5 sites) and ndhD (5 sites). 28 of these sites were also predicted by PREP-Cp software, indicating that the detected edits restored conserved amino acid residues in the corresponding proteins. Moreover, PREP-Cp predicted 37 additional sites not found in the RNA-seq data, most of which presumably remain unedited. Overall, the number and distribution of RNA editing sites in the N. × ventrata plastome are quite similar to those observed in other angiosperms [46], implying a lack of selection towards the loss of editing sites.
In contrast, only six RNA editing sites were identified by the RNA-seq analysis in the D. rotundifolia plastid genome (Table 3). Five of them were also predicted by PREP-Cp and matched conserved editing sites in genes atpF, rps2 (2 sites), rps14, and rpl23 found in other angiosperms [46]. One more site at codon position 73 in rpl20 could convert serine to leucine, although this position is not conserved in Rpl20. An additional 31 sites were predicted by PREP-Cp but not confirmed by RNA-seq (Table S3).
Table 3.
RNA-editing pattern in D. rotundifolia plastome identified by RNAseq read alignment.
| Gene | Amino Acid Position | Codon * | Amino Acid Change | Editing Frequency | PREP-Cp Editing Score |
|---|---|---|---|---|---|
| atpF | 31 | CcA | P=>L | 98% | 0.86 |
| rps2 | 54 | AcA | T=>I | 72% | 0.71 |
| rps2 | 92 | TcA | S=>L | 93% | 1.00 |
| rps14 | 50 | CcA | P=>L | 67% | 1.00 |
| rpl22 | 73 | TcA | S=>L | 70% | ** |
| rpl23 | 24 | TcT | S=>F | 81% | 0.71 |
* lower-case ‘c’ indicates the edited position; ** PREP-Cp does not analyze this gene.
3. Discussion
3.1. Evolution of the rps18 Gene in D. rotundifolia Plastome
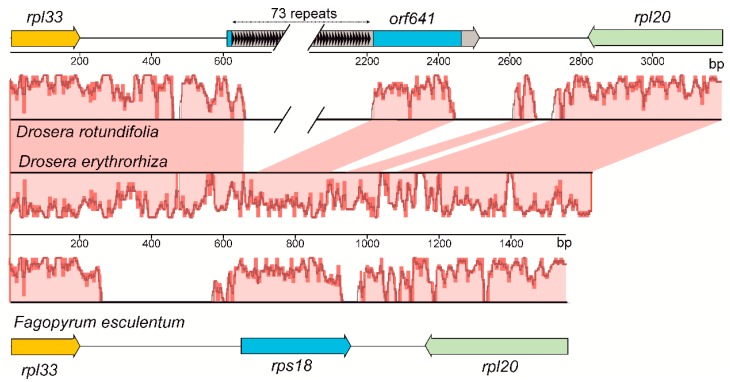
A peculiar feature of the plastid genome of D. rotundifolia is the presence of a long orf641, predicted to encode a protein comprising an N-terminal repeat-rich region linked to a near-complete Rps18-like sequence. Such a gene has not been found in any other plastid genomes, including the recently sequenced plastomes of four other species of Droseraceae, Drosera regia, Drosera erythrorhiza, Aldrovanda vesiculosa, and Dionaea muscipula [31]. The appearance of orf641 is an evolutionarily recent event and is likely the result of the insertion of the repeat-rich region (73 tandem repeats) at the 5′ end of the parental rps18 gene. A comparison of the regions of plastid genomes comprising orf641 and its flanking genes, rpl33 and rpl20, revealed that the repeat-rich part of orf641 is surrounded by sequences conserved in related species, including F. esculentum (Figure 2).
Figure 2.
Comparison of the plastid genome regions comprising genes rpl33, rps18, and rpl20 for D. rotundifolia, D. erythrorhiza, and F. esculentum. Regions between the start of rpl33 and the start of the counter-oriented rpl20 are shown; coordinate numbering for each plastome starts from the beginning of gene rpl33. Genes are indicated by rectangles with arrowheads. Black triangles within the orf641 rectangle indicate 73 copies of the 21 bp-long repeat sequence coding for a peptide with GQKQPNI consensus in D. rotundifolia. The part of orf641 that corresponds to the rps18 sequence is indicated in blue. The central part shows alignment in Mauve; pink areas between D. rotundifolia and D. erythrorhiza sequences highlight homologous regions. Note that the Mauve profile obtained for D. erythrorhiza has been turned upside down for clarity of presentation.
We propose two possible explanations for the origin of the orf641 in D. rotundifolia. One possibility is that the repeat-rich part of orf641 was derived from lateral transfer from another organism. There is abundant evidence of horizontal gene transfer to plant mitochondrial and nuclear genomes [47], which is especially frequent in parasitic lineages [48]. However, to the best of our knowledge, gene transfer from the nuclear genome to the plastid has not been reported for land plants. Alternatively, the orf641 gene in D. rotundifolia could result from recent multiple tandem duplication of repeats in the 5′-terminal region of the parental rps18 gene. An analysis of nucleotide sequences of rps18 genes revealed that accumulation of approximately 21 bp-long repeats in this region occurred in many species of angiosperms (especially in Poaceae and Campanulaceae), although the number of repeats did not exceeded 15 (Table S4). In some species repeats are present also in the 3′-terminal region of rps18. Both explanations leave open the question of the functionality of the Rps18-like part of Orf641. Preservation of the reading frame within the entire gene despite the presence of multiple repeats indicates its functionality, although the presence of a large N-terminal insert in Rps18 may be incompatible with the structure of the ribosome. It is possible that the formation of a protein close to the native Rps18 is possible due to the use of internal start codons within orf641.
3.2. Gene Loss and Genome Rearrangements in the Plastids in Three Families of Carnivorous Plants
Although the set of genes present in the plastid genomes of higher plants is highly conserved, there are numerous cases of the loss of some genes in different lineages. Most frequently lost is the infA gene encoding a translation initiation factor, the loss of which has been described in at least 24 lineages of angiosperms [49]. Examples of the loss of genes of individual ribosomal proteins are also known [50,51,52,53]. In some cases, it has been shown that loss of the chloroplast gene is accompanied by its transfer to the nuclear genome, and the lost protein is replaced by the one synthesized from the nuclear copy due to its acquisition of a signal sequence for transport into chloroplasts [49,52]. Of all the ribosomal proteins genes, only rps18 was likely pseudogenized in the D. rotundifolia plastome, and such a mechanism could deliver an intact Rps18 to chloroplasts. However, the RNA-seq analysis did not reveal transcripts for Rps18-like proteins carrying a chloroplast transit peptide.
However, gene losses in D. rotundifolia are much more extensive and resemble events occurring in plastid genomes of parasitic plants. In the course of transition to a heterotrophic lifestyle NAD(P)H dehydrogenase genes are lost first, followed by genes responsible for photosynthesis (psa, psb, pet, rbcL, etc.), the plastid-encoded RNA polymerase, and ATP synthase genes [13,14]. The last to be lost are genes encoding ribosomal and tRNAs, ribosomal proteins, and some other essential genes (accD, clpP, matK, ycf1, ycf2). Gene losses are accompanied by a decrease in the size of the plastome, loss of introns, genomic rearrangements, and loss of the characteristic quadripartite structure. The analysis of the plastid genomes of D. rotundifolia and N. × ventrata revealed that they have common features with plastid genomes of parasitic plants at different stages of such a process. At the same time, obligate dependence of carnivorous plants on photosynthesis determines preservation of the complete set of photosynthesis genes, as well as the plastid-encoded RNA polymerase, and ATP synthase genes.
Partial or complete loss of 11 genes coding for subunits of the thylakoid NAD(P)H dehydrogenase (NADH) complex from the plastome has been reported for parasitic plants [17,54,55] and some fully autotrophic lineages (e.g., Pinaceae; [56]). The NAD(P)H dehydrogenase complex mediates electron cycling around photosystem I and balances the ratio of ATP and NAD(P)H. The latter is used for carbon fixation in the Calvin cycle [57]. NAD(P)H dehydrogenase is not absolutely required under normal conditions, but is important in stress conditions, such as increased or decreased light intensity, and low CO2 concentrations [58,59]. The loss of NAD(P)H dehydrogenase has been reported in carnivorous Lentibulariaceae [22,23,24]. In Genlisea margaretae, the genes ndhC, D, F, G, H, J, and K have been lost from the plastome, ndhA, B, E, and I remain as pseudogenes, and ndhB is intact [22]. The plastome of Pinguicula ehlersiae contains intact ndhB, ndhA, D, E, G, H, I, J, and K pseudogenes and completely lacks ndhC and F [22]. A different pattern was found in Utricularia reniformis, where the genes ndhC, F, J, and K have been lost from the plastome, and the genes ndhA, B, D, E, G, H, and I reside as truncated pseudogenes [23]. A complete set of ndh genes was found in the plastomes of Utricularia macrorhiza and Utricularia gibba [22,60]. It is likely that the reduced dependence of sundew, some other carnivorous plants on active photosynthesis, and a relatively stable habitat enable the loss of NAD(P)H dehydrogenase.
An interesting finding is the loss of the ycf1 and ycf2 genes in D. rotundifolia, not reported in the plastids of carnivorous plants of the family Lentibulariaceae. In contrast to ndh genes, ycf1 and ycf2 are essential when tested by gene knockout in tobacco [61], but losses of these genes have been reported in plant plastids [9,42] including some parasitic plants [18]. It was proposed that Ycf1 is involved in protein import into chloroplasts [62], but this proposal was questioned later [63].
Plastomes of three Nepenthes species, N. × ventrata, N. graciliflora (GenBank MH286314) and N. mirabilis (GenBank MH346374, MK397881, and MK397880), have retained a standard structure and a set of genes typical for photosynthetic flowering plants. A notable exception is the ccsA, which is a pseudogene in N. × ventrata due to a 22 bp insertion. GenBank searches showed that similar frameshifting insertions are present in the ccsA genes of N. graciliflora and N. mirabilis, indicating that the pseudogenization of ccsA could be a common trait in Nepenthes. The ccsA gene codes for a cytochrome C biogenesis protein and is conserved among photosynthetic plants [9], but it is lost along with other photosynthesis-related genes in achlorophyllous parasitic plants [10,16,19]. The only other known example of the functional loss of ccsA in photosynthetic plants is its pseudogenization in an obligate hemiparasitic species, Viscum album (Santalales: Viscaceae) [64]. Therefore, this loss could be enabled by a reduced dependency of hemiparasitic and carnivorous plants on active photosynthesis.
Changes in the plastid genome of D. rotundifolia resemble those described for hemiparasitic plants—the loss of not only NAD(P)H dehydrogenase but also some other conservative genes, loss of introns, accumulation of repeats, and multiple rearrangements of the genome. However, the photosynthetic apparatus and the functions associated with its operation are fully preserved. A similar pattern of plastome structure and gene content was recently described for four other Droseraceae species, D. erythrorhiza, D. regia, A. vesiculosa, and D. muscipula [31]. All four plastomes lacked all ndh genes. The plastome of D. erythrorhiza also lacks intact ycf1, ycf2, psbK, rpl23, rps16, and possibly rpl32. Losses of protein-coding genes besides ndh in the plastid genomes of three other species were not reported [31]. As in the case of D. rotundifolia, the losses of essential tRNA genes have been reported, namely trnG-UCC and trnV-UAC in D. muscipula, trnA-UGC, trnG-UCC, trnI-GAU, and trnV-GAC in D. erythrorhiza. Plastomes of Droseraceae differ significantly in size, from 117,589 bp in D. muscipula to 192,912 bp in D. rotundifolia (Table 2), mostly due to variation in the size of IR region. The comparison of the order of genes in the plastomes of Droseraceae relative to the standard for angiosperms revealed that all plastomes experienced multiple structural rearrangements, including inversions, translocations, and duplications [31]. The order of genes in all Droseraceae plastomes is different, with the largest number of rearrangements observed in D. rotundifolia. This is consistent with the observation that the D. rotundifolia plastome contains the largest number of repeats compared to other plastomes of Droseraceae (Table 2).
3.3. Reduction of RNA Editing Sites in Plastomes of Some Carnivorous Plants
A notable feature of the D. rotundifolia plastid genome, absent in N. × ventrata, is the reduction of the number of RNA editing sites. Only six sites were identified by RNA-seq, contrary to the 30–40 editing sites typically found in non-parasitic flowering plants. Even considering the loss of ndh genes containing up to half of the editing sites of angiosperm plastid transcripts, the number of sites in the remaining genes is unusually low and they were found only in atpF, rps2, rps14, rpl20, and rpl23, but not in the usually heavily edited genes rpoA, rpoB, rpoC1, rpoC2, accD, and matK. At most conserved editing sites, the conserved amino acids in D. rotundifolia are already encoded at the DNA level, which makes editing superfluous. Interestingly, only 7 editing sites were identified by RNA-seq in the plastome of another carnivorous plant, U. reniformis [23]. Previously, a pronounced reduction of editing sites and a reduction of editing efficiency were found in some parasitic plants [15]. Since RNA editing is a mechanism of post-transcriptional regulation of gene expression in the chloroplast, its reduction in the plastids of parasitic and carnivorous plants could be associated with a decrease in their dependence on active photosynthesis.
3.4. Convergent Plastid Genome Evolution in Carnivorous and Parasitic Plants
Overall, this study revealed the remarkable similarities between the plastid genomes of carnivorous and parasitic plants. Such convergence between carnivorous and photosynthetic parasitic plants was noted by Wicke et al. [22], in which a loss of NAD(P)H dehydrogenase genes and a significant relaxation of purifying selection in ATP synthase complex, photosystem I, and in several other photosynthesis and metabolic genes were observed in three genera of the carnivorous family Lentibulariaceae. However, in terms of structure, besides the losses of the ndh genes, plastomes of these species are collinear in gene order to those in the majority of angiosperms and show no structural rearrangements.
Plastid genomes of the members of the family Droseraceae and especially of D. rotundifolia, revealed more pronounced features typical for parasitic plants such as the loss of some housekeeping genes, the loss of introns and RNA editing sites, accumulation of repeats, and structural rearrangements of plastomes. It can be proposed that the reason for this convergence may be the possibility of obtaining nutrients from other organisms, common to carnivorous and parasitic plants. Even though all carnivorous plants have an obligatory dependence on photosynthesis and primarily use prey as a source of nitrogen, direct uptake of organic carbon from prey has been reported [2,65]. Moreover, feeding on prey increases photosynthetic efficiency in Drosera capensis [66]. The outcome of such lifestyle could be the relaxation of purifying selection in photosynthesis and photosynthesis-related genes further promoting gene loss and structural instability of the plastome, as it occurs in parasitic plant species.
4. Materials and Methods
4.1. Plant Material and DNA Isolation
Plant material of D. rotundifolia was collected from a wetland in the Moscow region, Russia. N. × ventrata plants were grown in the greenhouse of Research Center of Biotechnology RAS, Moscow, Russia. The voucher specimens were deposited in the herbarium of the Institute of Bioengineering, Research Center of Biotechnology RAS (accession numbers DRT-CB1 for D. rotundifolia and NEP-CB1 for N. × ventrata). The same N. × ventrata plant was previously used for the sequencing of the mitochondrial genome [67]. The leaves of several D. rotundifolia plants and leaves of a single N. × ventrata plant were used for the extraction of total genomic DNA using a CTAB-NaCl method [68].
4.2. Sequencing and Assembly of the Plastid Genome of D. rotundifolia
Total genomic DNA of D. rotundifolia was sequenced with a Roche GS FLX Genome Sequencer (Roche, Basel, Switzerland) using the Titanium XL+ protocol for a shotgun genome library. About 81 Mb of cleaned sequences with an average read length of 609 nt was generated. De novo assembly was performed with Newbler Assembler v. 2.9 (454 Life Sciences, Branford, CT, USA) with default settings, which yielded eight long chloroplast DNA contigs with 36-fold average coverage. These contigs were identified based on sequence similarity to chloroplast genomes of angiosperms and high coverage. The complete plastid genome sequence was obtained upon the generation of appropriate PCR fragments covering the gaps between the contigs and their sequencing by the Sanger method on an ABI PRISM 3730 analyzer (Applied Biosystems, Foster City, CA, USA). The list of primers is available in Table S5. To verify the correct assembly of the reconstructed plastid genome, raw reads were mapped against the obtained sequence with GS Reference Mapper (454 Life Sciences).
4.3. Sequencing and Assembly of the Plastid Genome of N. × ventrata
The plastid genome of N. × ventrata was sequenced using the Illumina technique. The sequencing of a TrueSeq DNA library on an Illumina HiSeq 2500 system (Illumina, San Diego, CA, USA) generated 3 million single-end reads with a length of 250 nt. Primer and quality trimming was performed with Cutadapt v. 1.17 [69] and Sickle v. 1.33 (https://github.com/najoshi/sickle), respectively. Cutadapt was used with default settings, and Q33 score was used for Sickle. The reads were de novo assembled using SPAdes v. 3.7.1 [70]. Fifteen contigs representing the plastid genome were identified based on sequence similarity to chloroplast genomes of angiosperms and high average coverage (98×). Contigs were joined to produce a single circular molecule using the Bandage v. 0.8.0 tool [71]. Reads spanning junctions between the single copy regions and inverted repeats were used to infer joins at these sites. The correctness of the assembly of the complete circular plastid genome was verified by mapping Illumina reads back to the assembled sequence using Bowtie 2 [72], and no evidence of misassembly was found.
The sequences of the plastid genomes of D. rotundifolia and N. × ventrata were submitted to GenBank under accession numbers KU168830 and MK758110, respectively.
4.4. Plastid Genome Annotation and Analysis Tools
Plastid genome annotation was performed using DOGMA [73], with further manual correction using similarity searches against previously annotated plastid genomes. Repetitive sequences were identified by comparing each genome to itself with NCBI BLASTN+ v. 2.2.24 (MEGABLAST), using a word size of 7 and an E-value threshold of 1 × 10−6 [74]. In addition, short tandem repeats were identified with Phobos v. 3.3.12 (http://www.ruhr-unibochum.de/ecoevo/cm/cm_phobos.htm). This analysis was restricted to sequences of at least 20 bp in length, containing two or more copies of a perfect repeating unit from 2 to 40 bp in length. One copy of the large IR was removed from each genome prior to repeat analyses. Alignment of plastid genome regions of D. rotundifolia, D. erythrorhiza, and F. esculentum comprising genes rpl33, rps18, and rpl20 was performed using the program Mauve [75], with default settings.
4.5. RNA Editing Analysis
The leaves of several D. rotundifolia plants were collected for transcriptome analysis and pooled. Total RNA was isolated from approximately 300 mg of tissue using an RNeasy Plant Mini kit (Qiagen, Valencia, CA, USA). mRNA library preparation was performed using an NEBNext® mRNA Library Prep Reagent Set for Illumina® according to the manufacturer’s instructions (New England BioLabs, Ipswich, MA, USA). The library was sequenced on Illumina MiSeq according to the manufacturer’s instructions, generating 24,629,238 paired-end reads (2 × 250 bp). A total of 20,964,026 high-quality read pairs were filtered after removal of adapter sequences and quality trimming with Cutadapt v. 1.17 [69] and Sickle v. 1.33 (https://github.com/najoshi/sickle), respectively. RNA-seq read data has been deposited in the NCBI SRA database under accession SRR8948654.
Leaves and pitchers of a single N. × ventrata plant were used for isolation of total RNA using an RNeasy Plant Mini kit (Qiagen, Valencia, CA, USA). mRNA library preparation and sequencing were performed as described above for D. rotundifolia. A total of 23,277,185 read pairs (20,496,137 after filtration) were obtained. RNA-seq read data has been deposited in the NCBI SRA database under accessions SRR8944289-SRR8944300.
Illumina RNA-seq reads were mapped to plastid genomes using HISAT2 v. 2.0.4 [76] with the no-softclip option. To decrease the chance of mapping plastid-like sequences from mitochondrial and nuclear genomes, we filtered out alignments with less than 98% sequence identity using a custom Perl script (available at https://github.com/AVBeletsky/bioinformatics_scripts). A total of 503,433 and 752,633 reads were mapped to plastid genomes of D. rotundifolia and N. × ventrata, respectively. Single nucleotide polymorphisms (SNPs) were detected using FreeBayes v. 1.2.0 [77].
SNPs with more than 10% of reads supporting a non-reference variant and a minimum 10× mapping depth at the SNP site were retained. 78% and 83% of exon sequences of protein-coding genes of D. rotundifolia and N. × ventrata, respectively, had at least 10-fold coverage by RNA-seq reads, so some SNPs may be missed. The SNP effect was annotated using SnpEff v. 4.3i [78]. The SNP table obtained was inspected for C to T nucleotide substitutions in protein-coding genes. SNPs with more than three reads supporting the substitution were considered as RNA editing events.
In addition, the PREP-Cp tool (http://prep.unl.edu/) [79] was used to predict RNA editing sites (with a minimal editing score of 0.7).
Abbreviations
| LSC | Large single-copy |
| SSC | Short single-copy |
| IR | Inverted repeat |
Supplementary Materials
Supplementary Materials can be found at https://www.mdpi.com/1422-0067/20/17/4107/s1. Figure S1: Predicted protein product of orf641. Table S1: Summary of genes identified in the N. × ventrata plastome. Table S2: RNA-editing pattern in N. × ventrata plastome predicted by PREP-Cp and identified by RNAseq read alignment. Table S3: RNA-editing pattern in D. rotundifolia plastome predicted by PREP-Cp. Table S4: Repeat regions in plastidial rps18 genes. Table S5: Primers used for D. rotundifolia plastome assembly.
Author Contributions
Experimental design, E.V.G. and N.V.R.; collection and identification of plant material, E.V.G. and E.Z.K.; genome and transcriptome sequencing, A.V.M., V.V.K, E.V.G.; data analysis, E.V.G., A.V.B., A.V.M., V.V.K., and N.V.R.; manuscript draft preparation, E.V.G. and N.V.R.; manuscript review, N.V.R.; supervision and funding acquisition, N.V.R. and K.G.S. All authors approved the final manuscript.
Funding
This work was supported by the Russian Foundation for Basic Research (grant 14-04-00749, sequencing of plastid genomes) and the Russian Science Foundation (grant 14-24-00175, transcriptome sequencing).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Juniper B.E. The path to plant carnivory. In: Juniper B.E., Southwood T.R.E., editors. Insects and the Plant Surface. Edward Arnold; London, UK: 1986. pp. 195–218. [Google Scholar]
- 2.Adamec L. Mineral nutrition of carnivorous plants: A review. Bot. Rev. 1997;63:273–299. doi: 10.1007/BF02857953. [DOI] [Google Scholar]
- 3.Fleischmann A., Schlauer J., Smith S.A., Givnish T.J. Evolution of carnivory in angiosperms. In: Adamec L., Ellison A., editors. Carnivorous Plants: Physiology, Ecology, and Evolution. Oxford University Press; London, UK: 2017. pp. 22–42. [Google Scholar]
- 4.Albert V.A., Williams S.E., Chase M.W. Carnivorous plants: Phylogeny and structural evolution. Science. 1992;257:1491–1495. doi: 10.1126/science.1523408. [DOI] [PubMed] [Google Scholar]
- 5.Pereira C.G., Almenara D.P., Winter C.E., Fritsch P.W., Lambers H., Oliveira R.S. Underground leaves of Philcoxia trap and digest nematodes. Proc. Natl. Acad. Sci. USA. 2012;109:1154–1158. doi: 10.1073/pnas.1114199109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Givnish T.J. New evidence on the origin of carnivorous plants. Proc. Natl. Acad. Sci. USA. 2015;112:10–11. doi: 10.1073/pnas.1422278112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Westwood J.H., Yoder J.I., Timko M.P., dePamphilis C.W. The evolution of parasitism in plants. Trends Plant. Sci. 2010;15:227–235. doi: 10.1016/j.tplants.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 8.dePamphilis C.W., Palmer J.D. Loss of photosynthetic and chlororespiratory genes from the plastid genome of a parasitic flowering plant. Nature. 1990;348:337–339. doi: 10.1038/348337a0. [DOI] [PubMed] [Google Scholar]
- 9.Wakasugi T., Tsudzuki T., Sugiura M. The genomics of land plant chloroplasts: Gene content and alteration of genomic information by RNA editing. Photosynth. Res. 2001;70:107–118. doi: 10.1023/A:1013892009589. [DOI] [PubMed] [Google Scholar]
- 10.Wicke S., Schneeweiss G.M., dePamphilis C.W., Müller K.F., Quandt D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant. Mol. Biol. 2011;76:273–297. doi: 10.1007/s11103-011-9762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolfe K.H., Morden C.W., Palmer J.D. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc. Natl. Acad. Sci. USA. 1992;89:10648–10652. doi: 10.1073/pnas.89.22.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrett C.F., Freudenstein J.V., Li J., Mayfield-Jones D.R., Perez L., Pires J.C., Santos C. Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Mol. Biol. Evol. 2014;31:3095–3112. doi: 10.1093/molbev/msu252. [DOI] [PubMed] [Google Scholar]
- 13.Wicke S., Naumann J. Molecular evolution of plastid genomes in parasitic flowering plants. Adv. Bot. Res. 2018;85:315–347. [Google Scholar]
- 14.Barrett C.F., Davis J.I. The plastid genome of the mycoheterotrophic Corallorhiza striata (Orchidaceae) is in the relatively early stages of degradation. Am. J. Bot. 2012;99:1513–1523. doi: 10.3732/ajb.1200256. [DOI] [PubMed] [Google Scholar]
- 15.Funk H.T., Berg S., Krupinska K., Maier U.G., Krause K. Complete DNA sequences of the plastid genomes of two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii. BMC Plant. Biol. 2007;7:45. doi: 10.1186/1471-2229-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delannoy E., Fujii S., des Francs-Small C.C., Brundrett M., Small I. Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol. Biol. Evol. 2011;28:2077–2086. doi: 10.1093/molbev/msr028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wicke S., Müller K.F., de Pamphilis C.W., Quandt D., Wickett N.J., Zhang Y., Renner S.S., Schneeweiss G.M. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell. 2013;25:3711–3725. doi: 10.1105/tpc.113.113373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bellot S., Renner S.S. The plastomes of two species in the endoparasite genus Pilostyles (Apodanthaceae) each retain just five or six possibly functional genes. Genome Biol. Evol. 2015;8:189–201. doi: 10.1093/gbe/evv251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lam V.K., Soto Gomez M., Graham S.W. The highly reduced plastome of mycoheterotrophic Sciaphila (Triuridaceae) is colinear with its green relatives and is under strong purifying selection. Genome Biol. Evol. 2015;7:2220–2236. doi: 10.1093/gbe/evv134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ravin N.V., Gruzdev E.V., Beletsky A.V., Mazur A.M., Prokhortchouk E.B., Filyushin M.A., Kochieva E.Z., Kadnikov V.V., Mardanov A.V., Skryabin K.G. The loss of photosynthetic pathways in the plastid and nuclear genomes of the non-photosynthetic mycoheterotrophic eudicot Monotropa hypopitys. BMC Plant Biol. 2016;16(3):238. doi: 10.1186/s12870-016-0929-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Molina J., Hazzouri K.M., Nickrent D., Geisler M., Meyer R.S., Pentony M.M., Flowers J.M., Pelser P., Barcelona J., Inovejas S.A., et al. Possible loss of the chloroplast genome in the parasitic flowering plant Rafflesia lagascae (Rafflesiaceae) Mol. Biol. Evol. 2014;31:793–803. doi: 10.1093/molbev/msu051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wicke S., Schäferhoff B., dePamphilis C.W., Müller K.F. Disproportional plastome-wide increase of substitution rates and relaxed purifying selection in genes of carnivorous Lentibulariaceae. Mol. Biol. Evol. 2014;31:529–545. doi: 10.1093/molbev/mst261. [DOI] [PubMed] [Google Scholar]
- 23.Silva S.R., Diaz Y.C., Penha H.A., Pinheiro D.G., Fernandes C.C., Miranda V.F., Michael T.P., Varani A.M. The chloroplast genome of Utricularia reniformis sheds light on the evolution of the ndh gene complex of terrestrial carnivorous plants from the Lentibulariaceae family. PLoS ONE. 2016;11:e0165176. doi: 10.1371/journal.pone.0165176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silva S.R., Michael T.P., Meer E.J., Pinheiro D.G., Varani A.M., Miranda V.F.O. Comparative genomic analysis of Genlisea (corkscrew plants-Lentibulariaceae) chloroplast genomes reveals an increasing loss of the ndh genes. PLoS ONE. 2018;13:e0190321. doi: 10.1371/journal.pone.0190321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fleischmann A., Cross A.T., Gibson R., Gonella P.M., Dixon K.W. Systematics and evolution of Droseraceae. In: Adamec L., Ellison A., editors. Carnivorous Plants: Physiology, Ecology, and Evolution. Oxford University Press; London, UK: 2017. pp. 45–57. [Google Scholar]
- 26.Meimberg H., Heubl G. Introduction of a nuclear marker for phylogenetic analysis of Nepenthaceae. Plant Biol. 2006;8:831–840. doi: 10.1055/s-2006-924676. [DOI] [PubMed] [Google Scholar]
- 27.Mithöfer A. Carnivorous pitcher plants: Insights in an old topic. Phytochemistry. 2011;72:1678–1682. doi: 10.1016/j.phytochem.2010.11.024. [DOI] [PubMed] [Google Scholar]
- 28.Fleming R. Hybrid Nepenthes. Carniv. Plant Newslett. 1979;8:10–12. [Google Scholar]
- 29.Hatano N., Hamada T. Proteome analysis of pitcher fluid of the carnivorous plant Nepenthes alata. J. Proteome Res. 2008;7:809–816. doi: 10.1021/pr700566d. [DOI] [PubMed] [Google Scholar]
- 30.Wan Zakaria W.N., Loke K.K., Goh H.H., Mohd Noor N. RNA-seq analysis for plant carnivory gene discovery in Nepenthes × ventrata. Genom. Data. 2016;7:18–19. doi: 10.1016/j.gdata.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nevill P.G., Howell K.A., Cross A.T., Williams A.V., Zhong X., Tonti-Filippini J., Boykin L.M., Dixon K.W., Small I. Plastome-wide rearrangements and gene losses in carnivorous Droseraceae. Genome Biol. Evol. 2019;11:472–485. doi: 10.1093/gbe/evz005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu Z.-X., Wang J.-H., Chen C.-R., Zhao K.-K., Wang H.-F. Complete plastome sequence of Nepenthes mirabilis (Nepenthaceae): A ‘vulnerable’ herb in China. Mitochondrial DNA B. 2018;3:732–733. doi: 10.1080/23802359.2018.1483765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yao G., Jin J.J., Li H.T., Yang J.B., Mandala V.S., Croley M., Mostow R., Douglas N.A., Chase M.W., Christenhusz M.J.M., et al. Plastid phylogenomic insights into the evolution of Caryophyllales. Mol. Phylogenet. Evol. 2019;134:74–86. doi: 10.1016/j.ympev.2018.12.023. [DOI] [PubMed] [Google Scholar]
- 34.Logacheva M.D., Samigullin T.H., Dhingra A., Penin A.A. Comparative chloroplast genomics and phylogenetics of Fagopyrum esculentum ssp. ancestrale -a wild ancestor of cultivated buckwheat. BMC Plant. Biol. 2008;8:59. doi: 10.1186/1471-2229-8-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kode V., Mudd E.A., Iamtham S., Day A. The tobacco plastid accD gene is essential and is required for leaf development. Plant J. 2005;44:237–244. doi: 10.1111/j.1365-313X.2005.02533.x. [DOI] [PubMed] [Google Scholar]
- 36.Krause K. Plastid genomes of parasitic plants: A trail of reductions and losses. In: Bullerwell C.E., editor. Organelle genetics. Springer; Berlin, Germany: 2012. pp. 79–103. [Google Scholar]
- 37.Sloan D.B., Triant D.A., Forrester N.J., Bergner L.M., Wu M., Taylor D.R. A recurring syndrome of accelerated plastid genome evolution in the angiosperm tribe Sileneae (Caryophyllaceae) Mol. Phylogenet. Evol. 2014;72:82–89. doi: 10.1016/j.ympev.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 38.Zoschke R., Nakamura M., Liere K., Sugiura M., Börner T., Schmitz-Linneweber C. An organellar maturase associates with multiple group II introns. Proc. Natl. Acad. Sci. USA. 2010;107:3245–3250. doi: 10.1073/pnas.0909400107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duffy A.M., Kelchner S.A., Wolf P.G. Conservation of selection on matK following an ancient loss of its flanking intron. Gene. 2009;438:17–25. doi: 10.1016/j.gene.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 40.McNeal J.R., Kuehl J.V., Boore J.L., Leebens-Mack J., dePamphilis C.W. Parallel loss of plastid introns and their maturase in the genus Cuscuta. PLoS ONE. 2009;4:e5982. doi: 10.1371/journal.pone.0005982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chumley T.W., Palmer J.D., Mower J.P., Fourcade H.M., Calie P.J., Boore J.L., Jansen R.K. The complete chloroplast genome sequence of Pelargonium x hortorum: Organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol. Biol. Evol. 2006;23:2175–2190. doi: 10.1093/molbev/msl089. [DOI] [PubMed] [Google Scholar]
- 42.Cai Z., Guisinger M., Kim H.G., Ruck E., Blazier J.C., McMurtry V., Kuehl J.V., Boore J., Jansen R.K. Extensive reorganization of the plastid genome of Trifolium subterraneum (Fabaceae) is associated with numerous repeated sequences and novel DNA insertions. J. Mol. Evol. 2008;67:696–704. doi: 10.1007/s00239-008-9180-7. [DOI] [PubMed] [Google Scholar]
- 43.Levinson G., Gutman G.A. Slipped-strand mispairing: A major mechanism for DNA sequence evolution. Mol. Biol. Evol. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- 44.Shinozaki K., Ohme M., Tanaka M., Wakasugi T., Hayashida N., Matsubayashi T., Zaita N., Chunwongse J., Obokata J., Yamaguchi-Shinozaki K., et al. The complete nucleotide sequence of the tobacco chloroplast genome: Its gene organization and expression. EMBO J. 1986;5:2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bock R. Sense from nonsense: How the genetic information of chloroplasts is altered by RNA editing. Biochimie. 2000;82:549–557. doi: 10.1016/S0300-9084(00)00610-6. [DOI] [PubMed] [Google Scholar]
- 46.Zeng W.H., Liao S.C., Chang C.C. Identification of RNA editing sites in chloroplast transcripts of Phalaenopsis aphrodite and comparative analysis with those of other seed plants. Plant Cell Physiol. 2007;48:362–368. doi: 10.1093/pcp/pcl058. [DOI] [PubMed] [Google Scholar]
- 47.Sanchez-Puerta M.V. Involvement of plastid, mitochondrial and nuclear genomes in plant-to-plant horizontal gene transfer. Acta Soc. Bot. Pol. 2014;83:317–323. doi: 10.5586/asbp.2014.041. [DOI] [Google Scholar]
- 48.Davis C.C., Xi Z. Horizontal gene transfer in parasitic plants. Curr. Opin. Plant. Biol. 2015;26:14–19. doi: 10.1016/j.pbi.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 49.Millen R.S., Olmstead R.G., Adams K.L., Palmer J.D., Lao N.T., Heggie L., Kavanagh T.A., Hibberd J.M., Gray J.C., Morden C.W., et al. Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell. 2001;13:645–658. doi: 10.1105/tpc.13.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gantt J.S., Baldauf S.L., Calie P.J., Weeden N.F., Palmer J.D. Transfer of rpl22 to the nucleus greatly preceded its loss from the chloroplast and involved the gain of an intron. EMBO J. 1991;10:3073–3078. doi: 10.1002/j.1460-2075.1991.tb07859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jansen R.K., Cai Z., Raubeson L.A., Daniell H., Depamphilis C.W., Leebens-Mack J., Müller K.F., Guisinger-Bellian M., Haberle R.C., Hansen A.K., et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. USA. 2007;104:19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ueda M., Fujimoto M., Arimura S., Murata J., Tsutsumi N., Kadowaki K. Loss of the rpl32 gene from the chloroplast genome and subsequent acquisition of a preexisting transit peptide within the nuclear gene in Populus. Gene. 2007;402:51–56. doi: 10.1016/j.gene.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 53.Ueda M., Nishikawa T., Fujimoto M., Takanashi H., Arimura S., Tsutsumi N., Kadowaki K. Substitution of the gene for chloroplast RPS16 was assisted by generation of a dual targeting signal. Mol. Biol. Evol. 2008;25:1566–1575. doi: 10.1093/molbev/msn102. [DOI] [PubMed] [Google Scholar]
- 54.McNeal J.R., Kuehl J.V., Boore J.L., de Pamphilis C.W. Complete plastid genome sequences suggest strong selection for retention of photosynthetic genes in the parasitic plant genus Cuscuta. BMC Plant Biol. 2007;7:57. doi: 10.1186/1471-2229-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim H.T., Kim J.S., Moore M.J., Neubig K.M., Williams N.H., Whitten W.M., Kim J.H. Seven new complete plastome sequences reveal rampant independent loss of the ndh gene family across orchids and associated instability of the inverted repeat/small single-copy region boundaries. PLoS ONE. 2015;10:e0142215. doi: 10.1371/journal.pone.0142215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wakasugi T., Tsudzuki J., Ito S., Nakashima K., Tsudzuki T., Sugiura M. Loss of all ndh genes as determined by sequencing the entire chloroplast genome of the black pine Pinus thunbergii. Proc. Natl. Acad. Sci. USA. 1994;91:9794–9798. doi: 10.1073/pnas.91.21.9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rumeau D., Peltier G., Cournac L. Chlororespiration and cyclic electron flow around PSI during photosynthesis and plant stress response. Plant Cell Environ. 2007;30:1041–1051. doi: 10.1111/j.1365-3040.2007.01675.x. [DOI] [PubMed] [Google Scholar]
- 58.Horváth E.M., Peter S.O., Joët T., Rumeau D., Cournac L., Horváth G.V., Kavanagh T.A., Schäfer C., Peltier G., Medgyesy P. Targeted inactivation of the plastid ndhB gene in tobacco results in an enhanced sensitivity of photosynthesis to moderate stomatal closure. Plant Physiol. 2000;123:1337–1350. doi: 10.1104/pp.123.4.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ruhlman T.A., Chang W.J., Chen J.J., Huang Y.T., Chan M.T., Zhang J., Liao D.C., Blazier J.C., Jin X., Shih M.C., et al. NDH expression marks major transitions in plant evolution and reveals coordinate intracellular gene loss. BMC Plant Biol. 2015;15:100. doi: 10.1186/s12870-015-0484-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ibarra-Laclette E., Lyons E., Hernández-Guzmán G., Pérez-Torres C.A., Carretero-Paulet L., Chang T.H., Lan T., Welch A.J., Juárez M.J., Simpson J., et al. Architecture and evolution of a minute plant genome. Nature. 2013;498:94–98. doi: 10.1038/nature12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Drescher A., Ruf S., Calsa T., Jr., Carrer H., Bock R. The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J. 2000;22:97–104. doi: 10.1046/j.1365-313x.2000.00722.x. [DOI] [PubMed] [Google Scholar]
- 62.Kikuchi S., Bédard J., Hirano M., Hirabayashi Y., Oishi M., Imai M., Takase M., Ide T., Nakai M. Uncovering the protein translocon at the chloroplast inner envelope membrane. Science. 2013;339:571–574. doi: 10.1126/science.1229262. [DOI] [PubMed] [Google Scholar]
- 63.Bölter B., Soll J. Ycf1/Tic214 is not essential for the accumulation of plastid proteins. Mol. Plant. 2017;10:219–221. doi: 10.1016/j.molp.2016.10.012. [DOI] [PubMed] [Google Scholar]
- 64.Petersen G., Cuenca A., Seberg O. Plastome evolution in hemiparasitic mistletoes. Genome Biol. Evol. 2015;7:2520–2532. doi: 10.1093/gbe/evv165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rischer H., Hamm A., Bringmann G. Nepenthes insignis uses a C2-portion of the carbon skeleton of L-alanine acquired via its carnivorous organs, to build up the allelochemical plumbagin. Phytochem. 2002;59:603–609. doi: 10.1016/S0031-9422(02)00003-1. [DOI] [PubMed] [Google Scholar]
- 66.Pavlovič A., Krausko M., Libiaková M., Adamec L. Feeding on prey increases photosynthetic efficiency in the carnivorous sundew Drosera capensis. Ann. Bot. 2014;113:69–78. doi: 10.1093/aob/mct254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gruzdev E.V., Mardanov A.V., Beletsky A.V., Ravin N.V., Skryabin K.G. The complete mitochondrial genome of the carnivorous flowering plant Nepenthes x ventrata. Mitochondrial DNA B. 2018;3:1259–1260. doi: 10.1080/23802359.2018.1532353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Murray M.G., Thompson W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 70.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wick R.R., Schultz M.B., Zobel J., Holt K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics. 2015;31:3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Langmead B., Salzberg S. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wyman S.K., Jansen R.K., Boore J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- 74.Zhang Z., Schwartz S., Wagner L., Miller W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 75.Darling A.C.E., Mau B., Blattner F.R., Perna N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim D., Langmead B., Salzberg S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Garrison E., Marth G. Haplotype-based variant detection from short-read sequencing. [(accessed on 21 August 2019)];arXiv. 2012 Available online: https://arxiv.org/abs/1207.3907.1207.3907 [Google Scholar]
- 78.Cingolani P., Platts A., Wang le L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mower J.P. PREP-Mt: Predictive RNA editor for plant mitochondrial genes. BMC Bioinform. 2005;6:96. doi: 10.1186/1471-2105-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.