Summary
T cell subsets including effector (Teff), regulatory (Treg) and memory (Tmem) cells are characterized by distinct metabolic profiles that influence their differentiation and function. Previous research suggests that engagement of long-chain fatty acid oxidation (LC-FAO) supports Foxp3+ Treg cell and Tmem cell survival. However, evidence for this is mostly based on inhibition of Cpt1a, the rate limiting enzyme for LC-FAO, with the drug etomoxir. Using genetic models to target Cpt1a specifically in T cells, we dissected the role of LC-FAO in primary, memory and regulatory T cell responses. Here we show that the ACC2/Cpt1a axis is largely dispensable for Teff, Tmem or Treg cell formation, and that the effects of etomoxir on T cell differentiation and function are independent of Cpt1a expression. Together our data argue that metabolic pathways other than LC-FAO fuel Tmem or Treg differentiation and suggest alternative mechanisms for the effects of etomoxir that involve mitochondrial respiration.
Keywords: Etomoxir, carnitine palmitoyltransferase, CPT, acetyl CoA carboxylase, AMPK, regulatory T cells, memory T cells, CD8 T cells, fatty acid oxidation
Introduction
In the resting state, naïve T cells have low energy requirements which are preferentially directed to drive basic housekeeping functions and are achieved by oxidation of nutrients in the mitochondria via oxidative phosphorylation (OxPhos). Glucose is an important fuel source for OxPhos and withdrawal of survival signals such as IL-7 causes a decrease in glucose uptake and glycolysis, accompanied by a reduction in mitochondrial membrane potential and loss of cellular ATP that results in atrophy and cell death (Alves et al., 2006; Rathmell et al., 2001; Rathmell et al., 2000; Vander Heiden et al., 2001). A sizable contribution of long-chain fatty acid β-oxidation (LC-FAO) in supporting naïve T cell energy requirements has also been suggested in work by Wong and colleagues, pointing to a flexible use of nutrients in these cells (Wang et al., 2011).
Acquisition of T cell effector functions upon TCR engagement involves a profound reprogramming of cellular metabolism, including an increase in glycolytic activity and enhanced cellular anabolism to support proliferation and effector activity (Almeida et al., 2016; Jones and Thompson, 2007; Lochner et al., 2015; Pearce et al., 2013). In contrast, it has been proposed that Foxp3+ regulatory T (Treg) cells, as well as CD8+ T memory (Tmem) cells, do not rely on glycolysis to the same extent as Teff cells, but instead oxidize long chain fatty acids (LCFAs) such as palmitate to fulfill their energetic needs (Michalek et al., 2011; Pearce et al., 2009; van der Windt et al., 2012).
Fatty acid oxidation (FAO) is a multistep process that involves the degradation of fatty acids by the sequential removal of 2-carbon units from the acyl chain to produce acetyl-CoA, which then enters the mitochondrial tricarboxylic acid (TCA) cycle. In addition, reduced NADH and FADH2 molecules are generated by both FAO and the TCA cycle, and used by the electron transport chain (ETC) to produce ATP. While short- and medium-chain fatty acids (chain length < C12) can freely diffuse into the mitochondrial matrix, the rate of transport, and thus oxidation, of LCFAs (chain length C14 to C18), is regulated by the carnitine palmitoyltransferase system (Fritz and Yue, 1963). CPT1, present in the outer mitochondrial membrane, catalyzes the esterification of long chain acyls with carnitine to form acylcarnitine. This reaction is considered rate-controlling for LC-FAO, and allows the fatty acid moiety to be transported via carnitine-acylcarnitine translocase (CACT) into the mitochondrial matrix where CPT2 regenerates the long-chain acyl-CoA, which can be then oxidized.
Three isoforms of CPT1 have been described with different tissue and subcellular distribution: CPT1A, CPT1B and CPT1C. CPT1A is the main isoform found in lymphocytes and other hematopoietic cells. CPT1 can be inhibited by malonyl-CoA, a product of the enzyme acetyl-CoA carboxylase 2 (ACC2) that is associated with the outer mitochondrial membrane and is mainly expressed in oxidative tissues such as heart and muscle (Abu-Elheiga et al., 2001). Upstream, ACC2 is phosphorylated and inhibited by AMPK, a protein kinase activated under low energy conditions. AMPK also phosphorylates and inhibits ACC1, a different ACC isoform found in the cytosol that produces malonyl-CoA for de novo synthesis of fatty acids (FAS). Under low nutrient conditions, activated AMPK inhibits both ACC1 and ACC2, reducing FAS and releasing the inhibition of CPT1A to allow transport of LCFAs for subsequent FAO and ATP production in the mitochondria.
Previous studies described that CD4+ Treg and CD8+ Tmem cells exhibit high levels of activated AMPK, and that treatment with metformin (an indirect activator of AMPK) can promote CD8+ Tmem cell (Pearce et al., 2009) and Treg cell generation in vivo (Michalek et al., 2011). Given the link between AMPK and ACC inhibition, it has been hypothesized that LC-FAO contributes to Tmem and Treg cell differentiation. In vitro, TGF-β independent differentiation of Treg cells with the AMPK-activator AICAR increases Foxp3 expression (Gualdoni et al., 2016). However, AMPK deficient mice do not present any obvious signs of autoimmunity, nor do they lack Treg cells, suggesting that, in vivo, AMPK is not crucial for Treg cell differentiation (Blagih et al., 2015; Yang et al., 2017). In turn, CD8+ Tmem cells present a more oxidative metabolism than Teff cells, as measured by their increased oxygen consumption rate (OCR), mitochondrial mass, and increased expression of mitochondrial proteins (van der Windt et al., 2012). Furthermore, pharmacological inhibition of LC-FAO using etomoxir, an irreversible inhibitor of CPT1, can prevent Treg cell differentiation (Michalek et al., 2011) and interfere with the proliferation and mitochondrial oxidation of Tmem cells upon secondary activation (van der Windt et al., 2013). At the same time, in certain disease models such as graft-versus-host disease (GvHD), alloreactive T cells display increased FA transport ex vivo and rely primarily on FAO and OxPhos for survival (Byersdorfer et al., 2013). Similarly, treatment of mice with etomoxir reduced EAE severity and inflammation (Shriver and Manchester, 2011), suggesting that LC-FAO, particularly in vivo, might not be an exclusive feature of regulatory or memory T cell subsets (Chiaranunt et al., 2015).
Inhibition of LC-FAO with etomoxir (Ceccarelli et al., 2011), has been widely used to understand the role of FAO in lymphocytes but to date little information is available using genetic tools. Biochemical assays have determined that etomoxir acts as a potent inhibitor of CPT1 with half-maximal inhibitory concentrations in the nanomolar range (IC50= 10 – 700 nM) (Bentebibel et al., 2006; Ceccarelli et al., 2011; Declercq et al., 1987). This raises the possibility of off-target effects of etomoxir on immune cell metabolism when used at high concentrations such as those employed in many studies exploring FAO in immune cells (usually 40–200μM) (Huang et al., 2014; Michalek et al., 2011; van der Windt et al., 2012; van der Windt et al., 2013; Wu et al., 2016). This was recently pointed out in regard to the role of LC-FAO in mediating macrophage polarization (Divakaruni et al., 2018; Nomura et al., 2016). In this work we abrogated Cpt1a expression specifically in T cells using genetic models to elucidate the importance of LC-FAO for T cell homeostasis, development and survival.
Results & Discussion
Cpt1a deletion does not affect T cell homeostasis or activation
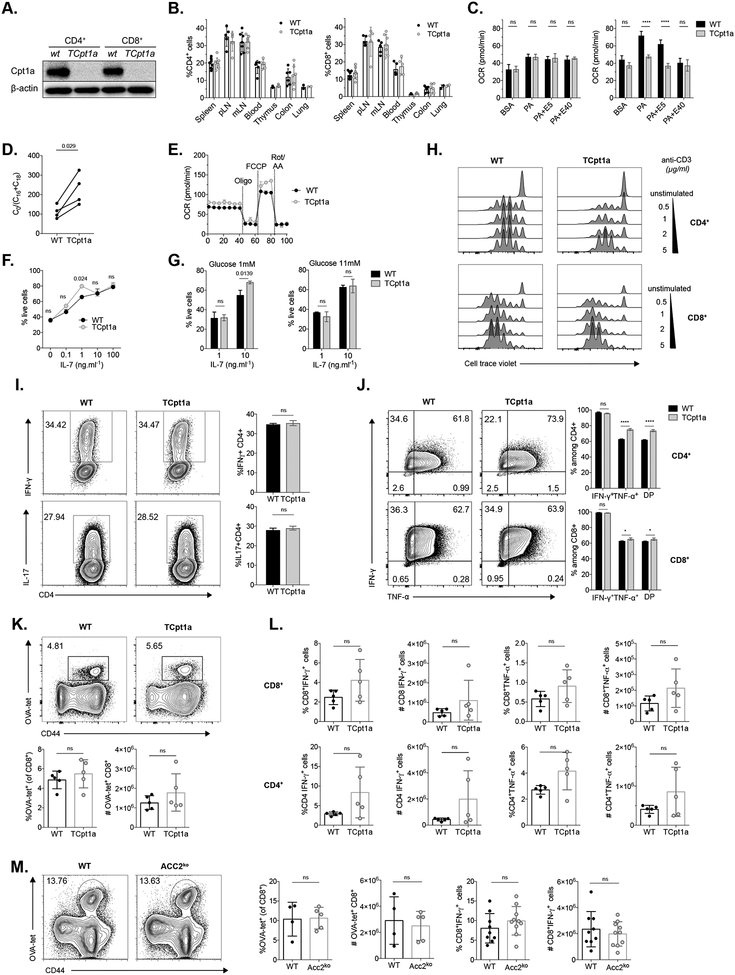
To determine how LC-FAO contributes to T cell homeostasis, we generated mice with a specific deletion of Cpt1a in T cells (Fig 1A), henceforth referred as TCpt1a mice. Analysis of mRNA and protein levels revealed no compensation from other Cpt1 isoforms (Cpt1b or Cpt1c) in naïve CD4+ or CD8+ T cells lacking Cpt1a (Fig S1A–B). TCpt1a mice displayed normal frequencies of CD4+ and CD8+ T cells in both lymphoid and non-lymphoid tissues (Fig 1B and S1C–D), suggesting that CPT1A activity is not crucial for naïve T cell homeostasis. Analysis of cellular respiration using a Seahorse Analyzer revealed that, unlike WT cells, naïve TCpt1a cells were unable to increase their maximal mitochondrial respiration, measured as the oxygen consumption rate (OCR), following treatment with the protonophore FCCP when offered palmitate (PA) as main substrate (Fig 1C). To evaluate CPT1 catalytic activity, we measured the concentration of free carnitine (C0), palmitoylcarnitine (C16) and stearoylcarnitine (C18) in control (WT) and TCpt1a T cells, and calculated the ratio of free carnitine to the sum of conjugated carnitine [C0/(C16 + C18)]. This ratio is increased in patients with CPT1A deficiency and is used for the clinical diagnosis of this condition (Fingerhut et al., 2001). TCpt1a naïve T cells displayed an increased [C0/(C16 + C18)] ratio compared to WT control cells (Fig 1D), as well as reduced 13C-palmitate incorporation into acetylcarnitine (Figure S1E), consistent with reduced Cpt1a activity in these T cells. While deletion of Cpt1a in naïve T cells reduced their ability to oxidize palmitate, overall mitochondrial respiratory capacity was not affected. This was evidenced by basal and maximal OCR similar to control T cells in medium containing glucose and other mitochondrial substrates (Fig 1E), which are oxidized in these cells. In addition, mice with a deletion of ACC2, which have been reported to display increased FAO in several tissues (Abu-Elheiga et al., 2001), display normal T cell homeostasis (Berod et al., 2014).
Figure 1. Cpt1a expression is dispensable for T cell homeostasis and function.
(A) CD4+CD25− and CD8+ cells were sorted from spleen and lymph nodes of WT and TCpt1a mice and Cpt1a deletion was verified by western blot. One of 3 experiments is shown.
(B) Frequencies of CD4+ and CD8+ cells in TCpt1a mice were analyzed by flow cytometry in different organs (pLNs=axillar and inguinal lymph nodes, mLNs=mesenteric lymph nodes). n=2–10 per group, data combined from two experiments are shown.
(C) Palmitate (PA)-induced respiration (oxygen consumption rate, OCR) in CD4+C25− cells from WT and TCpt1a mice. MaxOCR depicts the OCR after injection of 0.8μM FCCP. E5: etomoxir 5μM; E40: etomoxir 40μM. n=4/group, one of two experiments is shown. Graphs show mean ± SEM
(D) Concentrations of free carnitine (C0), palmitoylcarnitine (C16) and stearoylcarnitine (C18) in CD4+CD25− cells from WT and TCpt1a mice were measured, and the ratio [C0/(C16 + C18)] was calculated. Plot shows paired mean values from 4 independent experiments.
(E) OCR of CD4+ naïve WT and TCpt1a cells. Oligomycin (Oligo), FCCP and rotenone and antimycin A (Rote/AA) were injected at the points indicated. n=4/group, one of two experiments is shown. Graphs show mean ± SEM
(F-G) CD4+CD25− T cells were cultured with indicated concentrations of IL-7 and glucose and stained with Annexin V and 7-AAD after 48 hours to evaluate in vitro survival. N=2–3/group, one of two experiments is shown.
(H) T cells from WT and TCpt1a mice were labelled with cell trace violet proliferation dye and stimulated with anti-CD3/CD28 antibodies. Proliferation after 72 hours of one of two experiments is shown.
(I) Cytokine production of naïve CD4+ T cells differentiated under TH1 and TH17-polarizing conditions was measured by flow cytometry. Right, quantitation of IFN-γ- and IL-17-producing CD4+ T cells. n=2–3/group, one of two experiments is shown.
(J) WT and TCpt1a T cells were activated in the presence of anti-CD3/CD28 antibodies and IL-2 for 3 days and TNF-α and IFN-γ production was quantified after stimulation with PMA/ionomycin. n=3/group, one of three experiments is shown.
(K-L) Control (WT) and TCpt1a mice were infected with 1×106 CFU of attenuated L. monocytogenes expressing OVA (attLmOVA) and after 7 days the primary immune response was characterized by quantifying (K) CD8+ antigen-specific T cells and (L) cytokine production in antigen-specific CD8+ (upper panels), and CD4+ T cells (lower panels). Results from one experiment, with n=5 (3F/ 2M) per group.
(M) WT and ACC2ko (n= 10 per group, males) mice were infected with 5×103 CFU of LmOVA and the number of CD8+ OVA-specific cells and IFN-γ production by antigen specific CD8+ cells was quantified 7 dpi.
ns depicts not significant (p>0.05). *P<0.5, **** P<0.0001
To evaluate if LC-FAO contributes to the survival of resting T cells, we cultured CD4+ naïve T cells in the presence of IL-7 and measured viability after 48 hours. As expected, cell viability was proportional to the IL-7 concentration in the culture, but independent of Cpt1a expression, regardless of glucose concentration in the medium (Fig 1F–G). These results suggest that, even under low glucose conditions, LC-FAO is not crucial for the survival of naïve T cells, which may support their bioenergetic needs with other substrates (Jacobs et al., 2010; Wang et al., 2011).
To proliferate and develop effector functions upon activation, T cells engage in anabolic processes that support the synthesis of macromolecules. Cellular ATP production derives mainly from aerobic glycolysis, even when adequate oxygen levels are supplied (Jones and Thompson, 2007), though an increase in OxPhos has been also observed (Tarasenko et al., 2017; van der Windt et al., 2012). Since Teff cells were shown to use LC-FAO in disease models such as GvHD (Byersdorfer et al., 2013), we asked whether Cpt1a expression is required for Teff cell proliferation or differentiation. Naïve T cells were isolated from WT and TCpt1a mice and stimulated with anti-CD28/CD3 antibodies. TCpt1a CD4+ and CD8+ T cells readily proliferated after activation, albeit slightly less than WT cells with weak TCR stimulus (Fig 1H). Additionally, Cpt1a-deficient CD4+ T cells differentiated normally into TH1 and TH17 subsets in vitro when activated under polarizing conditions (Fig 1I). Production of IFN-γ and TNF-α by TCpt1a CD4+ and CD8+ T cells activated in vitro was also similar to their WT counterparts (Fig 1J).
To evaluate the impact of Cpt1a on T cell activation in vivo, we infected control and TCpt1a mice with OVA-expressing Listeria monocytogenes (LmOVA) and analyzed the primary immune response 7 days after infection. Cpt1a deletion did not affect the immune response against LmOVA in terms of activation and expansion of antigen-specific CTLs (Fig 1K), but we did observe a slight increase in the number of cytokine producing CD8+ and CD4+ T cells upon Cpt1a deletion (Fig 1L). However, these findings failed to meet statistical significance. Conversely, ACC2 deletion did not affect IFN-γ production in antigen-specific CD8+ T cells (Fig 1M), consistent with previous research (Lee et al., 2015; Stuve et al., 2018). Taken together, our results suggest that the ACC2/Cpt1a axis is not required for T cell homeostasis or primary immune responses in vivo.
Cpt1 is dispensable for Tmem cell generation
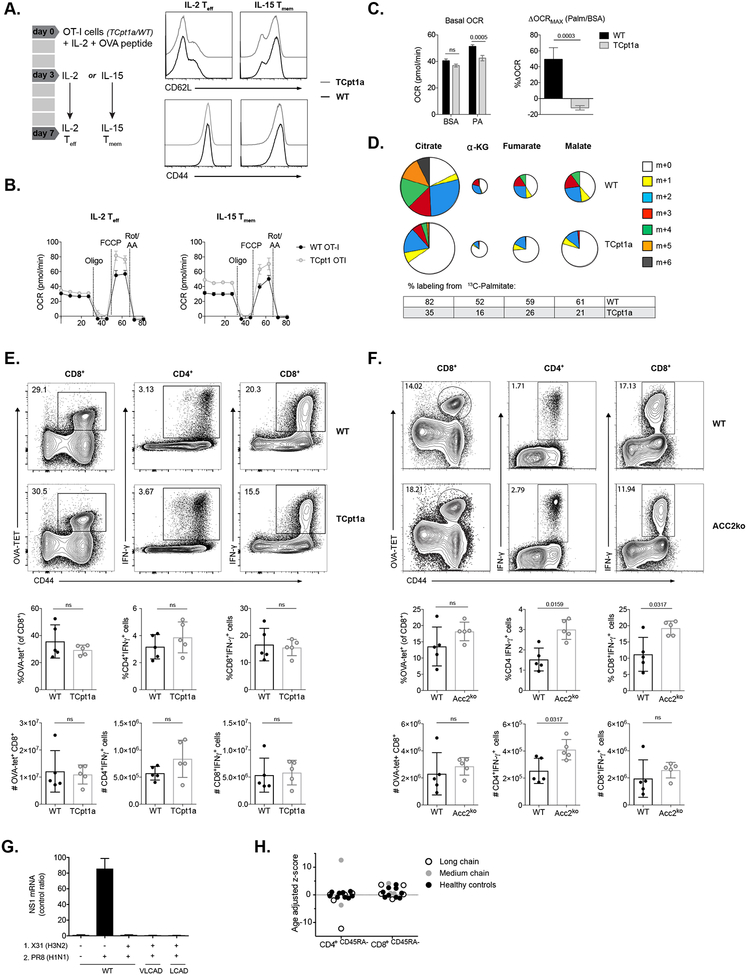
Recent research has suggested a role for LC-FAO and specifically CPT1A in the survival and function of CD8+ Tmem cells after L. monocytogenes infection (van der Windt et al., 2012). Tmem cells differentiated in vivo after infection or in vitro by incubating with IL-15 after antigen stimulation (Carrio et al., 2004) express more Cpt1a and have greater spare respiratory capacity (SRC) than Teff cells (van der Windt et al., 2012). The SRC represents the mitochondrial capacity of the cells to respond to increased energetic demands and a higher SRC can contribute to a “metabolic priming” of Tmem cells, allowing them to respond faster and more vigorously than naïve cells upon antigen stimulation (O’Sullivan et al., 2014; van der Windt et al., 2013). Because inhibition of LC-FAO with etomoxir can severely reduce proliferation and OxPhos of Tmem cells after secondary challenge (van der Windt et al., 2013), it is hypothesized that LC-FAO is critical for CD8+ Tmem cell development and survival (Buck et al., 2015; Chang and Pearce, 2016; Pearce et al., 2013). To explore this, we differentiated WT and TCpt1a CD8+ OT-I cells into memory- or effector-like cells in vitro using established protocols ((Carrio et al., 2004; O’Sullivan et al., 2014), Fig 2A, Fig S2A). As shown for polyclonal naïve T cells, activated Cpt1a-deficient OT-I cells did not exhibit significant upregulation of other Cpt1 isoforms (Fig S2B–C). Deletion of Cpt1a did not affect the expression of memory cell markers (Fig 2A), nor did it reduce the OCR or SRC of either IL-15 Tmem or IL-2 Teff cells (Fig 2B and S2D). Although no differences in basal oxygen consumption were observed between IL-15 Tmem and IL-2 Teff cells, the latter exhibited an increased ECAR, regardless of the presence or absence of Cpt1a (Fig S2E). We could demonstrate a reduction in both basal and maximal palmitate-dependent OCR in Cpt1a-deficient IL-15 Tmem cells, indicating reduced capacity of activated TCpt1a cells to oxidize palmitate (Fig 2C). Interestingly, both control and Cpt1a-deficient T cells were capable of oxidizing the short-chain fatty acid (SCFA) butyrate (Fig S2F–G). Moreover, incubating IL-15 Tmem cells with 13C-palmitate for 24 hours revealed significant labeling of TCA cycle intermediates such as citrate, α-ketoglutarate, fumarate, and malate, indicating that IL-15 Tmem cells can take up and oxidize exogenous FAs (Fig 2D). Conversion of 13C-palmitate to TCA cycle intermediates was significantly reduced in TCpt1a IL-15 Tmem cells (Fig 2D), confirming reduced LC-FAO in these T cells. Importantly, while Cpt1a mediates LC-FAO in T cells (Fig 2D), it was not critical for maintaining the level of TCA cycle intermediates in T cells.
Figure 2. Mitochondrial FAO is not required for memory T cell formation.
(A) CD8+ OT-I T cells were activated with OVA peptide in the presence of IL-2 for 3 days, followed by culture with either IL-2 or IL-15 for 4 days to generate IL-2 Teff and IL-15 Tmem cells. Expression of CD62L and CD44 on T cells of the indicated genotypes is shown. One of two experiments is shown.
(B) OCR of WT and TCpt1a IL-15 Tmem and IL-2 Teff cells. Oligomycin (Oligo), FCCP, and rotenone and antimycin A (Rote/AA) were injected as indicated. Graphs show mean ± SEM.
(C) OCR of in vitro generated IL-15 OT-I Tmem cells assayed with 200 μM BSA-conjugated palmitate under basal conditions (left panel) and in response to FCCP (right panel). Results for maximal OCR are expressed as a percent increase in OCR relative to BSA-treated cells. Graphs show mean ± SEM, one experiment is shown.
(D) Mass isotopomer distribution (MID) of U-[13C]-palmitate-derived TCA cycle intermediates in WT and TCpt1a IL-15 Tmem cells. Tmem cells were cultured for 24hr in medium containing U-[13C]-palmitate (200 μM), and 13C-isotopomer distribution in citrate, α-ketoglutatrate, fumarate and malate was determined by GC-MS. The percent distribution of each isotopomer for their respective metabolite pool is shown. The overall metabolite pool size is represented by the size of the pie chart. Data are normalized to cell number (n = 3), one of two experiments is shown.
(E) WT and TCpt1a mice were immunized with a sublethal dose of attLmOVA and re-challenged 34 days later. Antigen-specific CD8+ cells (left panels) and cytokine production in antigen specific CD4+ (center panels) and CD8+ (right panels) cells were quantified. Data of one experiment, n=5 per group, female.
(F) WT and ACC2ko mice were infected with 5×103 CFU of LmOVA and re-challenged after 10 weeks. Splenocytes were analyzed as in (E). Data of one experiment, n=5 per group, male
(G) Mice deficient in long-chain acyl-CoA dehydrogenase (LCAD) and very long-chain acyl-CoA dehydrogenase (VLCAD) were infected with influenza virus (X31) and one month later re-infected with adapted human influenza virus (PR8, n=5 per group, male). Expression of viral nucleoprotein (NP) mRNA in the lungs of infected mice was measured.
(H) CD4+ and CD8+ memory T cells in patients with inherited deficiencies in long-chain (N=5) and medium-chain fatty (N=3) acid oxidation and healty controls (N=7) were evaluated by clinical flow cytometry and their age-adjusted Z score calculated.
ns depicts not significant (p>0.05) or numeric values represent p values for that comparison.
Next, we analyzed the in vivo memory response to LmOVA in TCpt1a (Fig 2E) and ACC2ko (Fig 2F) mice, as well as the recall response of CD8+ OT-I memory T cells (Fig S2H). Both the expansion of and cytokine production by Cpt1a- or ACC2ko CD8+ T cells after re-challenge with LmOVA were similar to controls (Fig 2E–F and S2H–I), suggesting that absence of ACC2 or Cpt1a does not affect the development of a normal CD8+ Tmem cell response. Cpt1a-deficient Tmem cells displayed KLRG1 and CD127 expression similar to their WT counterparts (Fig S2J). To further rule out that LC-FAO influences Tmem formation, we evaluated T cell responses in mice deficient in other mitochondrial enzymes required for LC-FAO downstream of CPT1A, such as long-chain acyl-CoA dehydrogenase (LCAD) and very long-chain acyl-CoA dehydrogenase (VLCAD). These mouse models recapitulate the human phenotype of LC-FAO disorders and present increased long-chain acylcarnitine levels in plasma, along with cardiomyopathy and hepatic and myopathic disorders due to reduced FAO that manifest following stress. Both LCAD- and VLCAD-deficient mice generated a protective immune response against H1N1 influenza after infection with mouse-adapted H3N2 influenza (Fig 2G). Consistent with these data, analysis of patients with inherited deficiencies in long-chain and medium-chain FAO showed no significant deficiencies in CD4+ or CD8+ Tmem populations, with the exception of a single patient outlier with long-chain 3-hydroxyacyl-coA dehydrogenase deficiency displaying decreased CD4+ T cell memory (Fig 2H). Overall, our data suggest that LC-FAO is not required for T cell memory formation in mice or in humans.
Treg cell differentiation does not rely on Cpt1a- or AMPK-driven LC-FAO
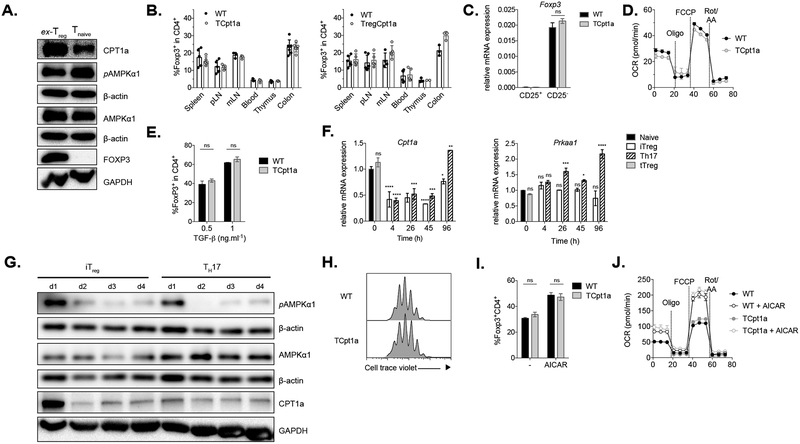
Due to the importance attributed to LC-FAO in Treg biology (Michalek et al., 2011), we next evaluated the role of Cpt1a in Treg cell development and function. Similar to naïve T cells, ex vivo isolated Treg cells express high levels of Cpt1a and activated AMPK (Fig 3A). The frequencies and total numbers of Treg cells in mice lacking Cpt1a in CD4+ cells or in Foxp3+ Treg cells only (using Foxp3-Cre), were comparable across different tissues (Fig 3B and Fig S3A). Ex vivo isolated Treg cells lacking Cpt1a displayed normal Foxp3 expression levels (Fig 3C) and similar mitochondrial oxidative capacity (Fig 3D) relative to their WT counterparts. Moreover, loss of Cpt1a did not affect the suppressive capacity of Treg cells in vitro (Fig S3B).
Figure 3. Treg differentiation and activity is independent of Cpt1a.
(A) Expression of Cpt1a and AMPKα as well as AMPKα phosphorylation at Thr172 was evaluated in T naïve (CD4+CD25−) and ex-vivo Treg (ex-Treg, CD4+CD25+) cells by immunoblot. One of two experiments is shown.
(B) Frequency of Foxp3+ cells (among CD4+ cells) in different tissues from control (WT), TCpt1a (left), and TregCpt1a (right) mice.
(C) Expression of Foxp3 mRNA relative to β-actin was determined in sorted CD4+CD25− and CD4+CD25+ cells from control (WT) and TCpt1a mice. Data combined from two independent experiments.
(D) OCR of ex-vivo isolated Treg cells from control (WT) and TCpt1a mice. Oligomycin (Oligo), FCCP, and rotenone and antimycin A (Rote/AA) were injected as indicated. Graphs show mean ± SEM, n=3. One of two experiments is shown.
(E) Naïve T cells from control (WT) and TCpt1a mice were differentiated under Treg polarizing conditions and Foxp3 expression quantified after 4 days by flow cytometry. n=3, one of two experiments is shown.
(F) WT naïve CD4+ T cells were differentiated under iTreg or TH17-polarizing conditions and the expression of Cpt1a and Prkaa1 mRNA relative to β-actin was quantified by RT-PCR. Statistically significant differences to Tnaive are shown in one of two experiments.
(G) Protein expression of Cpt1a, AMPKα and pAMPKα for cells differentiated as in (F) was evaluated by immunoblot. One of two experiments is shown.
(H-J) WT and TCpt1a naïve T CD4+ cells were activated with plate-bound anti-CD3/CD28 for 19 hours, transferred to an uncoated plate with IL-2 (TGF-β-independent iTreg culture) and their proliferation evaluated after 3 days (H). Alternatively, activated cells were transferred to plates with IL-2 and 300μM AICAR for 2 days (I) Foxp3 expression was quantified by flow cytometry (n=3 per group) and (J) mitochondrial respiration of intact cells was measured as in (D) (graphs show mean ± SEM, n=6 per group). (H-J) One of two experiments is shown.
ns= not significant, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001
Next, we activated naïve control (WT) and TCpt1a CD4+ T cells in the presence of IL-2 and TGF-β to assess the impact of Cpt1a deletion on the differentiation of induced Treg (iTreg) cells in vitro. iTreg cell differentiation was not affected by the lack of Cpt1a (Fig 3E), which was not compensated by increased expression of other Cpt1 isoforms (Fig S3C). In WT cells, expression of Cpt1a during iTreg cell differentiation declined shortly after activation and increased only after 4 days of culture, similar to TH17 cells (Fig 3F, left panel). In contrast, the expression of Prkaa1, the gene that encodes the AMPKα1 subunit was sustained throughout differentiation (Fig 3F, right panel). Protein levels of these genes followed the same pattern, with Cpt1a expression decreasing after the first day of activation (Fig 3G). This result is in accordance with previous reports showing that CD3/CD28 stimulation of primary lymphocytes causes a decrease in Cpt1a protein and mRNA levels, through a mechanism mediated by PI3K/Akt signaling (Deberardinis et al., 2006). Taken together, our results indicate that despite the high expression of Cpt1a in ex-vivo isolated Treg cells, Cpt1a is not functionally required for Treg cell development in vitro or in vivo, nor required for Treg cell suppressive activity.
Foxp3-expressing T cells can also be differentiated by removing TCR and CD28 stimulation after an initial activation (Sauer et al., 2008). This premature termination of receptor signaling results in a reduction of Akt and mTOR activity that resembles the induction of Treg cells by rapamycin. Gualdoni and colleagues found that treatment of TGF-β-independent iTreg cell cultures with AICAR, an agonist of AMPK, augmented the differentiation of Foxp3+ Treg cells, and that the addition of high-dose etomoxir (200μM) could prevent this increase in iTreg cell differentiation, an effect attributed to inhibition of LC-FAO (Gualdoni et al., 2016). In light of these findings, we tested whether abrogation of Cpt1a could influence the differentiation of Treg cells under these experimental conditions. Absence of Cpt1a affected neither the proliferation (Fig 3H) nor Foxp3 expression (Fig 3I) in Treg cells. Addition of AICAR following TCR stimulation increased the percentage of Foxp3+ cells, regardless of Cpt1a expression (Fig 3I). It should be noted that AICAR had a negative effect on T cell viability and proliferation (data not shown). Finally, treatment with AICAR increased basal and maximal OCR of iTreg cultures even in cells lacking Cpt1a expression (Fig 3J). Thus, the effect of AICAR on Treg differentiation appears to be independent of Cpt1a-dependent LC-FAO.
Off-target effects of etomoxir in T cells
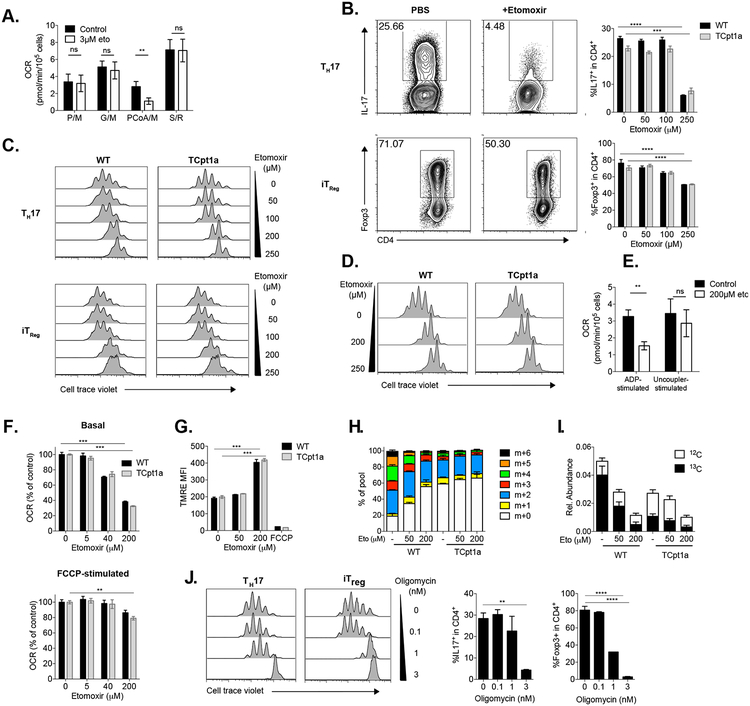
Since our results so far indicate that Cpt1a is not required for Treg or Tmem cell differentiation, we next examined the effect of etomoxir treatment on T cells. Multiple biochemical studies support the notion that etomoxir can efficiently block Cpt1a activity at low micromolar concentrations (Ceccarelli et al., 2011). To evaluate the concentration at which etomoxir has a specific effect on CPT1A in lymphocytes, we treated iTreg cells with 3μM etomoxir. This was followed by cellular membrane permeabilization to perform substrate-specific respirometry by measuring the OCR in the presence of various mitochondrial substrates, providing more direct and in-depth measurement of the mitochondrial respiratory capacity (Divakaruni et al., 2018; Divakaruni et al., 2014). In these experiments, we observed that 3μM etomoxir did not affect the oxidation of pyruvate, glutamate or succinate (substrates that feed the mitochondrial ETC independently of CPT1 activity), but did inhibit respiration when palmitate-CoA and carnitine (the substrates of CPT1, Fig 4A) were offered to permeabilized cells. Similarly, 3μM etomoxir is sufficient to specifically inhibit Cpt1a activity in bone marrow derived macrophages (BMDMs) (Divakaruni et al., 2018). These results indicate that micromolar concentrations of etomoxir are capable of inhibiting CPT1 activity in Treg cells, similar to what has been described for BMDMs and other cell types (Ceccarelli et al., 2011; Divakaruni et al., 2018).
Figure 4. Etomoxir has off-target effects on T cells independent of Cpt1a.
(A) iTreg cells were treated with 3μM etomoxir or vehicle control, incubated 15 minutes, permeabilized with XF Plasma Membrane Permeabilizer (PMP) and OCR with indicated substrates measured (P/M= pyruvate/malate, G/M= glutamate/malate, PCoA/M=palmitoyl-CoA/malate, S/R= succinate/rotenone). Graphs show mean ± SEM, n=4 per group.
(B-C) Naïve CD4+ control (WT) and TCpt1a T cells were cultured under Treg- and TH17-polarizing conditions in the presence of etomoxir (Eto) at indicated concentrations, and their (B) differentiation and (C) proliferation characterized by flow cytometry. n=3, one of two experiments is shown.
(D) Naïve CD4+ control (WT) and TCpt1a T cells were activated with anti-CD3/CD28 antibodies and further cultured with IL-2 to generate iTreg cells in the presence of etomoxir. Proliferation was determined after 3 days by flow cytometry. One of two experiments is shown.
(E) iTreg cells were incubated with 200 μM etomoxir or vehicle control, permeabilized and incubated with pyruvate/malate, and mitochondrial ADP- and FCCP-stimulated OCR was measured. Graphs show mean ± SEM, n=4 per group.
(F) Naïve control (WT) and TCpt1a T cells were cultured under iTreg polarizing conditions and their OCR analyzed after the addition of etomoxir at 5μM, 40μM, or 200μM before (basal rate, upper panel) and after (lower panel) FCCP treatment. Graphs show mean ± SEM, n=4 per group, one of three experiments is shown.
(G) Naïve control (WT) and TCpt1a T cells were cultured for 3 days under iTreg polarizing conditions in the presence of indicated concentration of etomoxir and incubated with 50nM TMRE to evaluate mitochondrial membrane potential. Cells treated with 40μM FCCP (which depolarizes the mitochondrial membrane) before addition of TMRE were used as controls for the TMRE stain. n=3, one of three experiments is shown.
(H-I) Mass isotopomer distribution (MID) of U-[13C]-palmitate-derived citrate in WT and TCpt1a in vitro generated IL-15 OT-I Tmem cells. Tmem cells were cultured for 24hr in medium containing U-[13C]-palmitate (200μM), with or without 50μM or 200μM etomoxir, and the 13C isotopomer distribution in citrate was determined by GC-MS.The percent distribution of each isotopomer for the citrate pool (H), as well as the relative abundance of unlabeled and U-[13C]-palmitate-labeled citrate (I), is shown. Data are normalized to cell number (mean ± SEM, n = 3). One of two experiments is shown.
(J) WT naïve T cells labelled with CellTrace violet proliferation dye were cultured for 3 days under Treg and TH17-polarizing conditions in the presence of nanomolar concentrations of oligomycin. Proliferation, IL-17A and Foxp3 expression were analyzed by flow cytometry. n=2 per group, one of two experiments is shown.
**** P<0.0001, **P<0.01, *P<0.05.
To test the dose of etomoxir capable of affecting T cell differentiation, we cultured WT and Cpt1a-deficient CD4+ naïve T cells under Treg or TH17-polarizing conditions in the presence of different concentrations of etomoxir. As shown in Fig 4B, only high doses of etomoxir (>100 μM) reduced Foxp3 expression in iTreg cultures, but to similar levels in both WT and TCpt1a cultures. Etomoxir also strongly reduced IL-17 production in both WT and TCpt1a TH17 cells (Fig 4B). Furthermore, we observed a dose-dependent inhibitory effect of etomoxir on iTreg and TH17 cell proliferation, which was independent of Cpt1a expression (Fig 4C). High dose etomoxir similarly inhibited the proliferation of TGF-β-independent iTreg cultures regardless of Cpt1a expression (Fig 4D). Notably, treatment of Treg cells with 200μM etomoxir reduced ADP-stimulated permeabilized cell respiration on pyruvate/malate, substrates that do not require Cpt1a activity to be metabolized (Fig 4E), while not significantly affecting FCCP-stimulated maximum OCR. This inhibitory profile suggests that etomoxir can affect oxidative phosphorylation independently of Cpt1a, similar to what is observed in BMDMs, other cell types, and isolated mitochondria (Divakaruni et al., 2018).
We next assessed the effect of etomoxir on the mitochondrial respiration of intact iTreg cells. Etomoxir doses of 40 and 200 μM (but not 5 μM) reduced basal, ATP-linked OCR in both WT and TCpt1a cells to a similar extent (Fig 4F), in accordance with our data using permeabilized cells (Fig 4A). Additionally, we observed that a high concentration of etomoxir increased the mitochondrial membrane potential (Fig 4G). These observations are compatible with a model in which etomoxir can inhibit the adenine nucleotide translocator (ANT), the enzyme responsible for exchanging the ATP produced in the mitochondrial matrix with ADP from the intermembrane space (Divakaruni et al., 2018). Inhibiting ANT would result in an accumulation of ATP in the matrix, stalling the flow of protons through the mitochondrial complex V and resulting in a net increase of mitochondrial membrane potential and a reduction of cellular OCR. We next evaluated the impact of high-dose etomoxir on the metabolism of 13C-palmitate by IL-15 Tmem cells. Treatment of WT IL-15 Tmem cells with high-dose etomoxir reduced the conversion of 13C-palmitate to citrate, resulting in a similar isotope distribution pattern as observed in TCpt1a cells (Fig 4H). However, unlike genetic Cpt1a deletion, etomoxir treatment dramatically reduced the abundance of citrate (Fig 4I) and other TCA cycle intermediates (Fig S4). This effect of etomoxir on TCA cycle metabolite abundances is similar to the effect of other ETC inhibitors, such as metformin (Griss et al., 2015).
To mimic the proposed off-target effect of etomoxir in activated CD4+ T cells, TH17 and iTreg cultures were differentiated with low nanomolar concentrations of oligomycin, a potent inhibitor of mitochondrial complex V. Treatment with oligomycin reduced cell proliferation and expression of IL-17 and Foxp3 in TH17 and iTreg cultures, respectively (Fig 4J). However, oligomycin affected viability and proliferation of iTreg cultures disproportionally more than TH17 cultures, while etomoxir did not. These results suggest that while inhibition of mitochondrial respiration might not be the sole off-target effect of etomoxir (Divakaruni et al., 2018), this effect alone can account for the inhibition of proliferation and differentiation in T cell cultures.
There are a number of important conclusions from our findings. First, based on genetic models we demonstrate that Teff, Tmem and Treg cell development and function occur normally in the absence of Cpt1a. This indicates that these cell types do not require LC-FAO for their function, or are capable of adapting metabolically to a reduction in LC-FAO. Our data reveal that genetic deletion of Cpt1a in T cells, including in vitro generated Tmem cells, does not significantly alter OCR when multiple respiratory substrates are available, in contrast with previous reports using shRNA against Cpt1a in OT-I TCR transgenic T cells (van der Windt et al., 2012). Whether the latter is the unintended consequence of transducing bulk splenocytes with shRNA constructs targeting Cpt1a or unspecific off-target effects in gene expression, reported for shRNA and related techniques (Jackson and Linsley, 2010), needs further evaluation. Using conditional knockout mice that allow gene targeting with high specificity, we here show for the first time that Cpt1a expression is dispensable for the generation of protective immunological memory in vivo.
Second, unlike previous reports, our experiments indicate that in vitro differentiated IL-15 Tmem cells are capable of oxidizing external palmitate and incorporating palmitate-derived carbon into the TCA cycle (Fig 2D), and that this process is mediated by Cpt1a. These data indicate that LCFAs may serve a previously unappreciated role as a precursor for biomass generation, in addition to ATP production. Third, our data indicate that inhibition of CPT1 activity in T cells occurs at low micromolar concentrations of etomoxir. Higher etomoxir concentrations (>100 μM) reduce the level of TCA cycle intermediates and OxPhos in T cells independent of Cpt1a expression. These data suggest caution in using high doses of etomoxir in cell culture for inhibiting FAO, given the potential for off-target effects of the drug when used at high concentrations, which has also been recently reported by independent research groups (O’Connor et al., 2018; Yao et al., 2018). This observation is particularly important because, in experiments that do not consider substrate specificity, the expected output is the same for the on-target and off-target effects of etomoxir (namely a reduction in OCR). This can lead to incorrect conclusions about the usage and dependency on LC-FAO of T cells and other immune cells (Divakaruni et al., 2018; Raud et al., 2018).
Limitations of study
Intriguingly, our data show that both control and Cpt1a-deficient T cells are capable of oxidizing other fatty acids (such as the SCFA butyrate, Fig S2E–F). It is possible that medium and short-chain fatty acids may serve as additional fuels for oxidative T cell metabolism, although any role for these metabolites in T cell biology remains to be determined. Moreover, since recent work suggests that LC-FAO has an important role in certain T cell populations, such as skin-resident CD8+ Tmem cells (Pan et al., 2017), the effect of Cpt1a deficiency in these subsets represents an exciting developing field of study.
STAR methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagent should be directed to and will be fullfiled by the Lead Contact Luciana Berod (luciana.berod@twincore.de)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
C57BL/6 mice were purchased from Jackson Laboratories or Charles Rivers Laboratories. OT-I mice were purchased from Jackson. The CD4-cre mice (Lee et al., 2001) were crossed to Cpt1alox mice (Schoors et al., 2015) and maintained on a C57BL/6 genetic background. ACC2 knockout mice (Abu-Elheiga et al., 2001) were kept on a C57/129 background. All mice were bred and maintained under specific pathogen-free conditions at the animal facility of the Helmholtz Centre for Infection Research (HZI, Braunschweig, Germany), at TWINCORE (Hannover, Germany) or at McGill University. Animal experiments at Twincore were performed in compliance with the German animal protection law (TierSchG BGBl. I S. 1105; 25.05.1998) and were approved by the Lower Saxony Committee on the Ethics of Animal Experiments as well as the responsible state office (Lower Saxony State Office of Consumer Protection and Food Safety under permit number 33.12-42502-04-10/0140 and 33.19-42502-04-17/2472).
For all experiments, male and female mice, 8–20 weeks were used, and no influence of the mouse gender on the results was presumed. Littermate controls were randomly assigned to experimental procedures. Mice were housed in a pathogen-free facility, caged in groups of 6 mice or less, had access to a pellet based feed, and autoclaved reverse osmosis water. Mice were kept in a temperature (20–24°C) and humidity (45%–70%) controlled environment with a 16 hr light cycle.
VLCADD mice (C57Bl/6-acadvltm1AS) were kindly provided by Dr. Arnold Strauss (Cincinnati Children’s Hospital Medical Center) and LCAD mice (B6.129S6-Acadltm1Uab) were kindly provided by Dr. Philip Wood (Burnham Institute for Medical Research).
Mouse T cell cultures.
For lymphocyte isolation, CD3+ T cells were purified from spleen and peripheral lymph nodes from either female or male mice by negative selection (StemCell Technologies, Vancouver, BC, Canada) as previously described. For isolation of naïve CD4+ lymphocytes, cells were isolated ex vivo from spleens and lymph nodes of mice by enrichment with Dynal Mouse CD4 Negative Isolation Kit (Life Technologies) followed by FACS sorting (FACSAria, BD; XDP or MoFlo, Beckman Coulter) for live CD4+CD25−CD62L+ cells. The purity of the isolated cells was >95%. CD8+ cells were isolated using the Dynabeads Untouched Mouse CD8 Cells Kit (Invitrogen). All cell cultures were carried out at 37°C in a humidified atmosphere containing 5% CO2 in IMDM or RPMI cell culture media supplemented as described below. RPMI 1640 GlutaMAX medium or IMDM GlutaMAX medium (both from Life Technologies) were used for Treg cultures and IMDM for Th17 cell cultures. Medium was supplemented with 10% heat-inactivated FCS (Biochrom), 500 U penicillin-streptomycin (PAA laboratories or Invitrogen) and 50 μM β-mercaptoethanol (Life Technologies or Sigma Aldrich). For TH17 cell induction, 2 × 105 naïve T cells were cultured for 4 days with plate-bound anti-CD3ε (10 μg mL−1, clone 14-2C11; Bio X Cell), anti-CD28 (1 μg.mL−1, clone 37.51; Bio X Cell), anti-IFN-γ (5 μg.mL−1, clone XMG1.2; Bio × Cell), anti-IL-4 (5 μg.mL−1, clone 11B11; Bio × Cell), rhTGF-β1 (2 ng.mL−1; Peprotech), rmIL-6 (5 ng.mL−1; Peprotech) and rmIL-1β (50 ng.mL−1; Peprotech). For TH1 cell induction, 1 × 105 naive T cells were cultured for 4 days with plate-bound anti-CD3ε (10 μg.mL−1), anti-CD28 (1 μg.mL−1), anti-IL-4 (5 μg.mL−1) and rmIL-12 (20 ng.mL−1; Peprotech). For Treg cell induction, 5 × 104 naive T cells were cultured for 4 days in the presence of plate-bound anti-CD3ε (5 μg mL−1), anti-CD28 (1 μg.mL−1), rhIL-2 (200 U.mL−1; Roche Applied Science) and rhTGF-β1 (1 ng.mL−1). On day 2, rhIL-2 (200 U.mL−1) was added again. For TGF-β-independent Treg differentiation naïve T CD4+ cells were activated with plate-bound anti-CD3/CD28 for 19 hours and transferred to an uncoated plate with 200 U.mL−1 IL-2 for 3 days. For proliferation analysis, naive T cells were labelled using 5μM Cell Trace Violet Cell Proliferation Kit (CTV, Life Technologies) and analyzed after 72 hours of activation. For in vitro CD8+ memory cultures, OT-I splenocytes were activated with OVA-peptide (1μM) and IL-2 (50 U/ml) for 3 days, and subsequently cultured in the presence of either IL-2 or IL-15 (10 ng/ml, Peprotech) for an additional 4 days.
For T naïve survival experiments, 1×105 cells were seeded in 96-well plate with supplemented RPMI and the indicated concentration of rmIL-7 (Peprotech). Apoptosis was measured after 48 hours with Annexin V Apoptosis Detection kit eFluor 450 (Invitrogen). Etomoxir sodium salt hydrate was purchased from Sigma-Aldrich and stock solution of 10mM was prepared in PBS.
Human Subjects
Human samples were collected with informed consent via The NIH MINI Study: Metabolism, Infection and Immunity in Inborn Errors of Metabolism (clinicaltrials.gov, ). Approval and oversight for this study was provided by the Institutional Review Board of the National Human Genome Research Institute.). All subjects were initially identified as having a disorder of mitochondrial beta-oxidation via state mandated newborn screening with confirmation by medical geneticists at referral centers. Clinical flow cytometry for memory T cells was performed by the Department of Laboratory Medicine at the NIH Clinical Center. Subjects with fatty acid oxidation disorders (FAOD): N=7 (3 male and 4 female), mean age±SD= 8±3.6 years; healthy controls: N=8, mean age±SD= 9±3.8 years.
METHOD DETAILS
Adoptive transfer of OT-I T cells
CD8+ OT-I T cells were isolated from the spleens of WT and TCpt1a mice using a CD8 negative selection kit (StemCell Technologies). 5,000 OT-I cells were adoptively transferred into mice one day prior to infection with Lm-OVA.
Listeria infections
TCpt1a mice and littermates were immunized i.v. with a sublethal dose of recombinant attenuated Listeria monocytogenes (Lm) expressing OVA (attLmOVA, 2 ×106 CFU) as previously described (Chang et al., 2013; Krawczyk et al., 2007). Splenocytes were isolated from mice 7 dpi, and analyzed for the presence of OVA-specific CD8+ T cells by MHC class I tetramer (Kb/OVA257–264). Cytokine production by CD4+ and CD8+ T cells was analyzed by intracellular staining following peptide re-stimulation (OVA257 or LLO190) as previously described (Jones et al., 2007). For memory re-challenge experiments, mice were immunized with a sublethal dose of recombinant attLmOVA, followed by re-challenge with a lethal dose of recombinant virulent LmOVA (virLmOVA, 1 ×106 CFU) 34 days after primary immunization. Splenocytes were isolated 5 days post re-challenge and analyzed as described above. ACC2ko mice were infected with L. monocytogenes expressing OVA (LmOVA, 5×103), as previously described (Arnold-Schrauf et al., 2014). Splenocytes were isolated after 7 days and cytokine production of CD8+ T cells was analyzed by intracellular staining following restimulation with OVA257 peptide. For memory experiments, mice were re-challenged with 2×105 CFU of LmOVA and splenocytes isolated 3 dpi and cytokine production was evaluated by intracellular stain after 4h stimulation with PMA/iono (for CD4+ T cells) or OVA257 (for CD8+ T cells).
Influenza infection
Mice were infected with influenza virus A/X/31 (X31) and re-infected with adapted human influenza virus A/PR/8/34 (PR8) one month later. Mice (N = 5 / genotype) were exposed to aerosolized (Glas-Col) 500 TCID of X31 or PR8 in 7mL of saline. Expression of viral nucleoprotein (NP) in the lungs of infected mice was determined by real time PCR. For this purpose, RNA was extracted from the tissues using Pure link RNA mini kit (Thermo Fisher Scientific) and was reverse transcribed to cDNA (iScript, Bio-Rad) according to the manufacturer’s instructions. Reactions were cycled and quantitated with an ABI 7500 Fast Real Time PCR System (Applied Biosystems). All assays were performed with N = 3 or more per condition and repeated 3 times. Relative level of viral RNA was normalized to b-actin.
Flow cytometry.
Monoclonal antibodies specific to the following mouse antigens (and labeled with the indicated fluorescent markers) were purchased from Affymetrix/eBioscience: CD25-PE (PC61), CD4-Alexa488 (GK1.5), CD62L-PE-Cy7 (MEL-14), Foxp3-eFluor450, Foxp3-eF660 (FJK-16s), IL-17A-APC and IL-17A-PE-Cy7 (eBio17B7). Kb/OVA tetramer was purchased from MHC Tetramer Production facility, Baylor College of Medicine. For analysis of surface markers, cells were stained in PBS containing 0.25% BSA and 0.02% azide. Dead cells were excluded using the LIVE/DEAD Fixable Dead Cell Stain Kit (Life Technologies) or the Fixable Dye eFluor® 780 and 506 (eBioscience) following the manufacturer’s protocol. For intracellular cytokine staining, cells were stimulated with phorbol 12-myristate 13-acetate (PMA, 0.1 μg mL−1; Sigma-Aldrich) and ionomycin (1μg mL−1; Sigma-Aldrich) for 2 h followed by Brefeldin A (5 μg.mL−1) or monensin for 2 h and stained. Mitochondrial membrane potential was measured using TMRE (20nM, Thermofisher). Cells were acquired on a CyAn ADP (Beckman Coulter), LSR Fortessa or LSR II (Becton Dickinson), and data were analyzed with FlowJo software (Tree Star).
Western blot.
Data in figures 1, 3 and S1: Whole-cell lysates were prepared using lysis buffer (Pierce RIPA buffer, Thermo Scientific) supplemented with complete EASYpack Mini Protease Inhibitor Cocktail and PhosSTOP Phosphatase Inhibitor (both from Roche Applied Science). Cell lysates were separated by SDS-gel electrophoresis and transferred to polyvinylidene fluoride membranes (Merck Millipore). Immunoblotting was performed using anti-CPT1A (clone 8F6AE9, 1:1000), purchased from Abcam, anti-Foxp3 (eBiosciences, eBio7979) anti-AMPK-α (clone 23A3; 1:1000), anti-pAMPK-α (Thr172) (clone 40H9; 1:1000), goat anti-mouse horseradish peroxidase and goat anti-rabbit horseradish peroxidase (cat. no. 7074; 1:2,000) (all from Cell Signaling) and detected using ECL prime (GE Healthcare).
Data in figure S2: Cells were lysed in modified AMPK lysis buffer (50mM Tris•HCl, 5mM NaF, 5mM Na pyrophosphate, 1mM EDTA, 1mM EGTA, 250mM mannitol, 1% (v/v) triton x-100) supplemented with the following protease additives: protease and phosphatase tablets (Roche), DTT (1mM), and benzamidine (1 μg/mL). Cleared lysates were resolved by SDS/PAGE, transferred to nitrocellulose, and incubated with primary antibodies. Primary antibodies used were to CPT1A (Abcam), CPT1B (Abcam), CPT1C (Protein-Tech), as well as Actin (Cell Signaling Technology). HRP-conjugated anti-mouse and anti-rabbit secondary antibodies were obtained from Cell Signaling Technology.
Suppression assay.
CD4+CD25+ tTreg cells were isolated from spleens and lymph nodes and cultured at ratios of 1:1 to 1:16 with 50,000 CTV-labelled CD45.1+CD4+CD25- T cells along with 5,000 GMCSF-DCs cells and 10μg.ml-1 of anti-CD3. Suppression of effector T cell proliferation was determined 90 hours post-stimulation by dilution of the CTV in the CD45.1 population by flow cytometry.
Extracellular flux analysis
Extracellular flux was measured using a Seahorse XFe96 Analyzer (Agilent) using Mito Stress Test kits as per manufacturer instructions with minor modifications.
For measurements of intact cells, 150,000 activated T cells or 300,000 ex-vivo isolated T cells were plated in poly-D-lysine-coated XF96 plates via centrifugation in XF media (non-buffered DMEM 1640 containing 10mM glucose, 2mM L-glutamine and 1mM sodium pyruvate). Respiration was measured in the basal state as well as in response to 1μM oligomycin, 1.5μM FCCP and 0.5μM rotenone with 0.5μM antimycin A.
For measuring permeabilized cells, the plasma membrane of cells was selectively permeabilized with recombinant, mutant perfringolysin O [rPFO; commercially XF Plasma Membrane Permeabilizer (XF PMP, Agilent Technologies]. Experiments were conducted essentially as previously described except medium lacked BSA to prevent buffering of etomoxir (Divakaruni et al, 2014a). Cells were initially offered 3nM rPFO, respiratory substrates, and 4mM ADP. The ADP-stimulated respiration rate was measured, and rates were also measured in response to 2μM oligomycin, sequential additions of 400nM FCCP, and 0.2μM rotenone with 1μM antimycin A. Substrate concentrations were as follows: 5mM glutamate with 5mM malate (G/M), 10mM pyruvate with 1mM malate (P/M), 40μM palmitoyl CoA with 0.5 mM carnitine and 1 mM malate (PCoA/M), or 10mM succinate with 2μM rotenone (S/R). Etomoxir was added to intact cells 15 min prior to cell permeabilization to form the covalent inhibitor of CPT-1 etomoxiryl CoA (Divakaruni et al., 2018)
Oxidation of exogenous LCFAs was measured using PA-BSA as a substrate. For data in figure 1, CD4+CD25- T cells (4×105) isolated from spleen and lymph nodes were incubated for 15 min in Seahorse Minimal DMEM medium with 0.5mM L-carnitine, 2.5mM glucose and 2mM HEPES. 5 or 40μM etomoxir was added in corresponding wells. Palmitate-BSA reagent (Affymetrix) was added to a final concentration of 167μM total palmitate (bound in a 6:1 molar ratio to BSA) and OCR was measured under basal conditions and after injection of 0.8μM of FCCP. Results were shown after substracting non-mitochondrial OCR calculated after injection of 0.5μM Rotenone + 0.5μM Antimycin A. For data in figure 2, oxidation of exogenous LCFAs was measured using palmitate-BSA as a substrate. In vitro generated IL-15 CD8 Tmem cells (2×105) were incubated for 15 min with fatty-acid oxidation assay medium (111mM NaCl, 4.7mM KCl, 1.25mM CaCl2, 2.0mM MgSO4, 1.2mM Na2HPO4, 2.5mM glucose, 0.5mM carnitine and 5mM HEPES). Afterwards, BSA (34μM) or palmitate-BSA (200μM palmitate conjugated with 34μM BSA) was added to the medium, and the OCR was measured under basal conditions and in response to 1μM oligomycin, 1.5μM fluorocarbonyl cyanide phenylhydrazone (FCCP), and 100nM rotenone + 1μM antimycin A. Results were normalized to data from control cells in the presence of BSA.
RT-PCR analysis
Total RNA was isolated from T cells using Qiagen RNeasy Kit (Qiagen) or lysis in TRIzol reagent (Invitrogen) followed by RNA isolation with Direct-zol™ RNA Kit (Zymo Research). cDNA was synthesized with SuperScript™ III Reverse Transcriptase (Invitrogen) according to manufacturer’s instructions using Oligo(dT) 12–18 Primers (Invitrogen) or High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems/Life Technologies). Quantitative PCR (qPCR) was either performed using SYBR Green qPCR Bioline SensiFAST SYBR® Low-ROX Kit (Bioline) and Agilent Technologies AriaMX Realtime PCR or IQ SYBR Green Supermix (Bio-rad) and measured in a LightCycler® 480 System - High-Throughput Real-Time PCR (Roche). Ex-vivo isolated Tregs for this assay were obtained from DEREG mice that express GFP under the control of the Foxp3 gene locus (Lahl et al., 2007).
Carnitine and acylcarnitine quantification
Methanol (HPLC grade) and water (HPLC grade) were purchased from J. T. Baker Chemicals (Center Valley, PA). Biocrates solvent I for FIA solvent preparation was obtained from Biocrates Life Sciences (Innsbruck, Austria). L-carnitine-(N-methyl-d3) and palmitoyl-L-carnitine-(N-methyl-d3) were purchased from Cambridge Isotope Laboratories (Tewksbury, MA). Stearoyl-L-carnitine-(N-methyl-d3) was obtained from Sigma-Aldrich (St. Louis, MO). For sample preparation, cell pellets (2.5 million cells) were extracted three times by adding 500 μl methanol at room temperature, vortexing for 2 min, and ultrasonication for 15 min. After centrifugation (13000 rpm, 5 min, 4 °C), the collected supernatants were dried in a speedvac. For FIA-MS/MS analysis the residue was reconstituted using 40 μl of 90% methanol/10% water. The FIA solvent also contained the internal standards, resulting in final concentrations of 50 ng/ml for L-carnitine-(N-methyl-d3), 0.9 ng/ml for palmitoyl-L-carnitine-(N-methyl-d3) and 0.3 ng/ml for stearoyl-L-carnitine-(N-methyl-d3). 20 μl were injected for FIA-MS/MS analysis.
Flow injection analysis/tandem mass spectrometry (FIA-MS/MS) was adapted from the Biocrates p180 kit (Biocrates Life Sciences, Innsbruck, Austria) and carried out on a triple quadrupole linear ion trap mass spectrometer (QTRAP6500, AB Sciex, Framingham, MA), equipped with electrospray ionization (ESI) in positive ionization mode. A 1290 series UPLC system (Agilent, Santa Clara, CA) was used for flow injection by directly connecting the autosampler to the Turbo V ion spray source via PEEK tubing and using FIA solvent as a mobile phase (45 μl/min). The source settings were: ion spray voltage, 5500 V; heater temperature, 175 °C; source gas 1, 40 psi; source gas 2, 50 psi and curtain gas, 40 psi. Entrance potential 10.00 V, collision exit potential 15.00 V, dwell time 25 ms.
| Analyte | Q1 mass [Da] | Q3 mass [Da] |
|---|---|---|
| C0 | 162.1 | 85.1 |
| d3-C0 | 165.1 | 85.1 |
| C2 | 204,1 | 85,1 |
| 13C2 | 206.1 | 85.1 |
| C16 | 400.3 | 85.1 |
| d3-C16 | 403.3 | 85.1 |
| C18 | 428.4 | 85.1 |
| d3-C18 | 431.4 | 85.1 |
Concentrations for free carnitine (C0), acetylcarnitine (C2), 13C-acetylcarnitine (13C2), C16 and C18 acylcarnitines were calculated by single point calibration using carnitine-d3 as internal standard for carnitine, acetylcarnitine and 13C-acetylcarnitine and C18 acylcarnitine-d3 as internal standard for C18 acylcarnitines. C16 acylcarnitine was quantified using either C16 acylcarnitine-d3 or C18 acylcarnitine-d3. The ratio of free carnitine to the sum of palmitoylcarnitine and stearoylcarnitine [C0/(C16 + C18)] was calculated, because it is a clinical marker for CPT-I deficiency (Fingerhut et al., Clinical Chemistry 2001). Data processing was done using Analyst 1.6.2 software (AB Sciex).
GC-MS analysis of 13C metabolites:
Gas chromatography coupled to mass spectrometry (GC-MS) was performed on T cells using previously described methods (Blagih et al., 2015; Griss et al., 2015; Vincent et al., 2015). Briefly, in vitro generated IL-15 Tmem cells were cultured in glucose and glutamine-free TCM supplemented with 12mM glucose, 6mM glutamine, 10% FBS, 0.5mM carnitine, 10ng/mL IL-15 and 200uM uniformly labeled [13C]-palmitate (Sigma Aldrich). T cells (6–8 ×106 per well in 6-well plates) were cultured in 13C-containing medium for 24 hours. Metabolites were extracted using ice-cold 80% methanol, sonicated, and D-myristic acid added (750ng/sample) as an internal standard. Dried samples were dissolved in 30 μL methozyamine hydrochloride (10mg/ml) in pyridine and derivatized as tert-butyldimethylsily (TBDMS) esters using 70 μL N-(tert-butyldimethylsilyl)-N-methyltrifluoroacetamide (MTBSTFA) (Faubert et al., 2014). For metabolite analysis, an Agilent 5975C GC/MS equipped with a DB-5MS+DG (30 m × 250 μm × 0.25 μm) capillary column (Agilent J&W, Santa Clara, CA, USA) was used. All data were collected by electron impact set at 70 eV. A total of 1 μL of the derivatized sample was injected in the GC in splitless mode with inlet temperature set to 280°C, using helium as a carrier gas with a flow rate of 1.5512 mL/min (rate at which myristic acid elutes at 17.94 min). The quadrupole was set at 150°C and the GC/MS interface at 285°C. The oven program for all metabolite analyses started at 60°C held for 1 min, then increasing at a rate of 10°C/min until 320°C. Bake-out was at 320°C for 10 min. Sample data were acquired both in scan (1–600 m/z) and selected ion monitoring (SIM) modes. Mass isotopomer distribution for cellular metabolites was determined using a custom algorithm developed at McGill University (McGuirk et al., 2013). Briefly, the atomic composition of the TBDMS-derivatized metabolite fragments (M-57) was determined, and matrices correcting for natural contribution of isotopomer enrichment were generated for each metabolite. After correction for natural abundance, a comparison was made between non-labeled metabolite abundances (12C) and metabolite abundances which were synthesized from the 13C tracer. Metabolite abundance was expressed relative to the internal standard (D-myristic acid) and normalized to cell number.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical parameters can be found in the figure legends. Data were analyzed using GraphPad Prism 7.0 or 6.0. Statistical analyses were performed as follows: two-way ANOVA followed by Sidak multiple comparison was used to analyze experiments with 2 variables and 3 or more groups, and one-way ANOVA followed to Duvett’s comparison to a control was used for experiments with one variable and 3 or more groups; in vitro experiments with two groups were analyzed with Student’s T test; in vivo experiments with two groups were analyzed with Mann-Whitney test, due to small sample size. In all cases, statistical significance was considered when p<0.05. Graphs show mean values and error bars represent standard deviation unless specified otherwise in the figure legends. P-values were represented as follows: *p < 0.05; **, p <0.01; ***, p<0.001, **** p<0.0001
For the data obtained from human subjects, since the subjects with fatty acid oxidation disorders differ in age, it was necessary to normalize to healthy controls. Mean and SD were calculated for age matched healthy controls and were used to calculate Z scores for that age group. No statistical analysis was performed due to the limited number of subjects.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Hamster monoclonal anti-mouse CD3 | BioXCell, eBioscience | BE0001-1 |
| Hamster monoclonal anti-mouse CD28 | BioXCell, eBioscience | BE00015-1 |
| Hamster monoclonal anti-mouse IFN-gamma | BioXCell | BE0055 |
| Rat monoclonal anti-mouse IL-4 | BioXCell | BE0045 |
| Rat monoclonal anti-mouse CD25 | eBioscience | Clone PC61.5 |
| Rat monoclonal anti-mouse CD4 | eBioscience | Clone GK1.5 |
| Rat monoclonal anti-mouse CD62L | eBioscience | Clone MEL-14 |
| Rat monoclonal anti-mouse Foxp3 | eBioscience | Clone FJK-16s |
| Rat monoclonal anti-mouse IL-17A | eBioscience | Clone eBio17B7 |
| Rat monoclonal anti-mouse TNF-alpha | eBioscience | Clone MP6-XT22 |
| Rat monoclonal anti-mouse IFN-gamma | eBioscience | Clone XMG1.2 |
| Rat monoclonal anti-mouse CD8 | eBioscience | Clone 53-6.7 |
| Mouse monoclonal anti-mouse CD45.2 | eBioscience | Clone 104 |
| Rat monoclonal anti-mouse CD127 | BioLegend | Clone A7R34 |
| Hamster monoclonal anti-mouse KLRG1 | eBioscience | Clone 2F1 |
| Rat monoclonal anti-mouse CD44 | eBioscience | Clone IM7 |
| Mouse monoclonal anti-human/mouse CPT1A | Abcam | Clone 8F6AE9 |
| Mouse monoclonal anti-human/mouse Foxp3 | eBioscience | eBio7979 |
| AMPKα (23A3) Rabbit mAb | Cell Signaling | #2603 |
| Phospho-AMPKα (Thr172) Rabbit mAb | Cell Signaling | #2535 |
| Anti-rabbit IgG, HRP-linked Antibody | Cell Signaling | #7074 |
| Anti-mouse IgG, HRP-linked Antibody | Cell Signaling | #7076 |
| Rabbit polyclonal anti-human/mouse CPT1B | Abcam | # ab104662 |
| Rabbit polyclonal anti-human/mouse CPT1C | Proteintech | # 12969-1-AP |
| Rabbit polyclonal anti-human/mouse beta-actin | Cell Signaling | # 4967 |
| H-2Kb/OVA257 Tetramer | Baylor College of Medicine | |
| Bacterial and Virus Strains | ||
| Recombinant Listeria monocytogenes (rLM-OVA) | Derived from Lm strain 10403S | |
| recombinant attenuated Listeria monocytogenes (Lm) deleted for actA and expressing OVA (attLmOVA) | ||
| recombinant virulent LmOVA (virLmOVA) | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| RPMI 1640 GlutaMAX | Thermo Fisher | 61870-010 |
| RPMI Medium 1640 (no glucose) | Thermo Fisher | 11879-020 |
| IMDM GlutaMAX | Thermo Fisher | 3198-022 |
| Heat-inactivated FCS | Biochrom | S 0115 - Lot# 1289W |
| Penicillin-streptomycin | Millipore | A2213 |
| 2-mercaptoethanol | Gibco | #21985-023 |
| recombinant human TGF-beta | Peprotech | 100-21 |
| recombinant murine IL-6 | Peprotech | 216-16 |
| recombinant murine IL-1beta | Peprotech | 211-11B |
| recombinant human IL-2 | Roche | C-149447 |
| recombinant murine IL-12 | Peprotech | 210-12 |
| recombinant murine IL-7 | Peprotech | 217-17 |
| Etomoxir sodium salt hydrate | Sigma Aldrich | E1905 |
| Phorbol 12-myristate 13-acetate (PMA) | Sigma-Aldrich | P-8139 |
| Ionomycin | Sigma-Aldrich | 10634 |
| Tetramethylrhodamine, Ethyl Ester, Perchlorate (TMRE) | Invitrogen | T669 |
| Pierce RIPA Buffer | Thermo Scientific | |
| Oligomycin | Sigma Aldrich | 75351 |
| FCCP | Cayman | 15218 |
| Rotenone | Cayman | 13955 |
| Antimycin A | Sigma Aldrich | A8674 |
| XF Base Medium Minimal DMEM without Phenol Red | Agilent | 103335-100 |
| XF Plasma Membrane Permeabilizer | Agilent | |
| L-carnitine | Sigma Aldrich | C0158 |
| Glucose ≥99,5 % D(+) | Roth | HN06.1 |
| L-Glutamine | Biochrom AG | M11-004 |
| Sodium Pyruvate | Gibco | #11360-070 |
| Oligo(dT)18-22 Primers | Invitrogen | |
| IQ SYBR Green Supermix | Bio-Rad | #170-8882 |
| Palmitic acid | Sigma Aldrich | P0500 |
| Palmitic acid (U-13C16) | Cambridge Isotope Laboratories | CLM-409-0.5 |
| Poly-D-lysine hydrobromide | Sigma Aldrich | P6407 |
| RNAse OUT Ribonuclease inhibitor (recombinant | Invitrogen | #10777-019 |
| Albumin fraction V fatty acid free | Roth | 0052.1 |
| Recombinant murine IL-15 | Peprotech | #3210-15 |
| Fixable Viability Dye eFluor 506 | eBioscience | #65-0866-14 |
| Fixable Viability Dye eFluor 780 | eBioscience | #65-0865-14 |
| Cell Proliferation Dye eFluor 450 | eBioscience | #65-0842-85 |
| Sodium Palmitate | Sigma Aldrich | P9767 |
| (U-13C16) Sodium Palmitate | Sigma Aldrich | 700258 |
| Sodium Butyrate | Sigma Aldrich | B5887 |
| BD GolgiStop | BD Biosciences | #51-2092KZ |
| Foxp3/transcription factor staining buffer set | eBioscience | #00-5523-00 |
| OVA(257-264) SIINFEKL peptide | Bio-Synthesis Inc. | custom |
| LLO(91–99) GYKDGNEYI peptide | Bio-Synthesis Inc. | custom |
| Critical Commercial Assays | ||
| EasySep Mouse T Cell Isolation Kit | StemCell technologies | #19851 |
| EasySep Mouse CD4 T cell isolation kit | StemCell technologies | #19852 |
| EasySep Mouse CD8 T cell isolation kit | StemCell technologies | #19853 |
| Dynabeads Untouched Mouse CD4 Cells Kit | Invitrogen | 11415D |
| Dynabeads Untouched Mouse CD8 Cells Kit | Invitrogen | 11417D |
| Cell Trace Violet Cell Proliferation Kit | Invitrogen | C34571 |
| eBioscience™ Annexin V Apoptosis Detection Kit eFluor™ 450 | eBioscience | 888006-72 |
| LIVE/DEAD™ Fixable Aqua Dead Cell Stain Kit, for 405 nm excitation | Invitrogen | L34957 |
| Seahorse XF Palmitate-BSA FAO Substrate | Agilent Technologies | #102720-100 |
| Seahorse XFe96 FluxPak | Agilent technologies | #102416-100 |
| Qiagen RNAeasy Kit | Qiagen | #74106 |
| SuperScript III Reverse Transcriptase | Invitrogen | #18080-044 |
| High Capacity cDNA Reverse Transcription Kit | Applied Biosystems | #4368814 |
| SensiFAST SYBR Lo-Rox Mix | Bioloine | #BIO-94050 |
| Experimental Models: Organisms/Strains | ||
| Cpt1afl/fl mice: C57BL/6NTac.(129S6/SvEvTac;C57BL/6NCr)F1-Cpt1atm1 Pec | Schoors et al., Nature 520(7546):192-7 (2015) | |
| ACC2ko mice: B6.Cg-Acacb(tm1Sjw) | Baylor College of Medicine, Houston, texas, USA (Abu-Elheiga et al, 2001, Science, 291, 2613–2616) | |
| LCAD mice: B6.129S6-Acadltm1Uab | Dr. Philip Wood, Burnham Institute for Medical Research(Kurtz et al, PNAS, 1998, 22;95(26):15592-7) | |
| VLCADD mice: C57Bl/6-acadvltm1AS | Dr. Arnold Strauss Cincinnati Children’s Hospital Medical Center (Exil, et al 2003, Circulation Research. 2003;93:448–455) | |
| Oligonucleotides | ||
|
Cpt1a primer Fwd: 5’ CTCCGCCTGAGCCATGAAG 3’ Rev: 5’ CACCAGTGATGATGCCATTCT 3’ |
PrimerBank | ID 27804309a1 |
|
Cpt1b primer Fwd: 5’ GCACACCAGGCAGTAGCTTT 3’ Rev:5’ CAGGAGTTGATTCCAGACAGGTA 3’ |
PrimerBank | ID 6753512a1 |
|
Cpt1c primer Fwd: 5’ GATTTTCTGTACGTCACCCCC 3’ Rev: 5’ AGGTGGTCTTTTTCCACCCC 3’ |
This paper | |
|
Actb primer Fwd: 5’ TGT TAC CAA CTG GGA CGA CA 3’ Rev: 5’ GGG GTG TTG AAG GTC TCA AA 3’ |
This paper | |
|
Foxp3 primer Fwd: 5’ AGAAGCTGGGAGCTATGCAG 3’ Rev: 5’ GCTACGATGCAGCAAGAGC 3’ |
This paper | |
|
Cpt1a primer - IL-2/IL-15 cultures Fwd: 5’ CCAACGGGCTCATCTTCTAATC 3’ Rev: 5’ TGGTACAAAGCACCCATTAC 3’ |
Invitrogen | custom |
|
Cpt1b primer - IL-2/IL-15 cultures Fwd: 5’ ATTCTGTGCGGCCCTTATT 3’ Rev: 5’ TGACTTGAGCACCAGGTATTT 3’ |
Invitrogen | custom |
|
Cpt1c primer - IL-2/IL-15 cultures Fwd: 5’ GTTTCCTCTGGAGGTGGATTT 3’ Rev: 5’ GCTGGAGATATGGAAGGTGATT 3’ |
Invitrogen | custom |
|
OGDH primer - IL-2/IL-15 cultures Fwd: 5’ AATGAGGAGAAGCGGACCTT 3’ Rev: 5’ TCCACTCCATTTGCACTTGA 3’ |
Invitrogen | custom |
|
Prkaa1 primer - Fwd: 5’ CAGAGATCGGGATCCATCAG 3’ Rev: 5’ CCTGGTCTTGGAGCTACGTC 3’ |
This paper | |
| PR8 NP primer 5’ CAGCCTAATCAGACCAAATG 3’ 5’ TACCTGGTTCTCAGTTCAAG 3’ |
Tarasenko et al 2017 Cell metabolism | |
| Software and Algorithms | ||
| FlowJo 9.9.5 | FlowJo LLC | www.flowjo.com |
| GraphPad Prism V6 or V7 | GraphPad Software | www.graphpad.com |
| Other | ||
Acknowledgments
B.R. was supported by the Deutscher Akademischer Austauschdienst (DAAD) and the Centre for Infection Biology of the Hannover Biomedical Research School. L.B. was funded by the Ellen-Schmidt Program from the Medical School Hannover. P.S. was supported by the International Research Training Group 1273 from the Deutsche Forschungsgemeinschaft (DFG). M.L. is supported by a grant from the DFG (LO1415 7–1). This work was supported by fellowships from HiLF (MHH) to L.B. and DFG (to L.B. and T.S.), and the CIHR (to A.H.W.) and FRQS (to D.G.R. and E.H.M.), and an operating grant from the CIHR (MOP-142259 to R.G.J.). We would like to acknowledge the assistance of the Cell Sorting Core Facility of the Hannover Medical School, supported in part by Braukmann-Wittenberg-Herz-Stiftung and Deutsche Forschungsgemeinschaft. R.J. thanks Mya Steadman and the Transgenic and Flow Cytometry Facilities at the McGill Goodman Cancer Research Centre for technical support. RF and MB thank Ulrike Beutling and Heike Overwin for technical support.
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- Abu-Elheiga L, Matzuk MM, Abo-Hashema KA, and Wakil SJ (2001). Continuous fatty acid oxidation and reduced fat storage in mice lacking acetyl-CoA carboxylase 2. Science 291, 2613–2616. [DOI] [PubMed] [Google Scholar]
- Almeida L, Lochner M, Berod L, and Sparwasser T (2016). Metabolic pathways in T cell activation and lineage differentiation. Semin Immunol 28, 514–524. [DOI] [PubMed] [Google Scholar]
- Alves NL, Derks IA, Berk E, Spijker R, van Lier RA, and Eldering E (2006). The Noxa/Mcl-1 axis regulates susceptibility to apoptosis under glucose limitation in dividing T cells. Immunity 24, 703–716. [DOI] [PubMed] [Google Scholar]
- Arnold-Schrauf C, Dudek M, Dielmann A, Pace L, Swallow M, Kruse F, Kuhl AA, Holzmann B, Berod L, and Sparwasser T (2014). Dendritic cells coordinate innate immunity via MyD88 signaling to control Listeria monocytogenes infection. Cell Rep 6, 698–708. [DOI] [PubMed] [Google Scholar]
- Bentebibel A, Sebastian D, Herrero L, Lopez-Vinas E, Serra D, Asins G, Gomez-Puertas P, and Hegardt FG (2006). Novel effect of C75 on carnitine palmitoyltransferase I activity and palmitate oxidation. Biochemistry 45, 4339–4350. [DOI] [PubMed] [Google Scholar]
- Berod L, Friedrich C, Nandan A, Freitag J, Hagemann S, Harmrolfs K, Sandouk A, Hesse C, Castro CN, Bahre H, et al. (2014). De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nature medicine 20, 1327–1333. [DOI] [PubMed] [Google Scholar]
- Blagih J, Coulombe F, Vincent EE, Dupuy F, Galicia-Vazquez G, Yurchenko E, Raissi TC, van der Windt GJ, Viollet B, Pearce EL, et al. (2015). The energy sensor AMPK regulates T cell metabolic adaptation and effector responses in vivo. Immunity 42, 41–54. [DOI] [PubMed] [Google Scholar]
- Buck MD, O’Sullivan D, and Pearce EL (2015). T cell metabolism drives immunity. J Exp Med 212, 1345–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byersdorfer CA, Tkachev V, Opipari AW, Goodell S, Swanson J, Sandquist S, Glick GD, and Ferrara JL (2013). Effector T cells require fatty acid metabolism during murine graft-versus-host disease. Blood 122, 3230–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrio R, Bathe OF, and Malek TR (2004). Initial antigen encounter programs CD8+ T cells competent to develop into memory cells that are activated in an antigen-free, IL-7- and IL-15-rich environment. J Immunol 172, 7315–7323. [DOI] [PubMed] [Google Scholar]
- Ceccarelli SM, Chomienne O, Gubler M, and Arduini A (2011). Carnitine palmitoyltransferase (CPT) modulators: a medicinal chemistry perspective on 35 years of research. Journal of medicinal chemistry 54, 3109–3152. [DOI] [PubMed] [Google Scholar]
- Chang CH, Curtis JD, Maggi LB Jr., Faubert B, Villarino AV, O’Sullivan D, Huang SC, van der Windt GJ, Blagih J, Qiu J, et al. (2013). Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 153, 1239–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CH, and Pearce EL (2016). Emerging concepts of T cell metabolism as a target of immunotherapy. Nat Immunol 17, 364–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaranunt P, Ferrara JL, and Byersdorfer CA (2015). Rethinking the paradigm: How comparative studies on fatty acid oxidation inform our understanding of T cell metabolism. Mol Immunol 68, 564–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deberardinis RJ, Lum JJ, and Thompson CB (2006). Phosphatidylinositol 3-kinase-dependent modulation of carnitine palmitoyltransferase 1A expression regulates lipid metabolism during hematopoietic cell growth. J Biol Chem 281, 37372–37380. [DOI] [PubMed] [Google Scholar]
- Declercq PE, Falck JR, Kuwajima M, Tyminski H, Foster DW, and McGarry JD (1987). Characterization of the mitochondrial carnitine palmitoyltransferase enzyme system. I. Use of inhibitors. J Biol Chem 262, 9812–9821. [PubMed] [Google Scholar]
- Divakaruni AS, Hsieh WY, Minarrieta L, Duong TN, Kim KK, Desousa BR, Andreyev AY, Bowman CE, Caradonna K, Dranka BP, et al. (2018). Etomoxir inhibits macrophage polarization by disrupting CoA homeostasis. [DOI] [PMC free article] [PubMed]
- Divakaruni AS, Rogers GW, and Murphy AN (2014). Measuring Mitochondrial Function in Permeabilized Cells Using the Seahorse XF Analyzer or a Clark-Type Oxygen Electrode. Curr Protoc Toxicol 60, 25 22 21–16. [DOI] [PubMed] [Google Scholar]
- Fingerhut R, Roschinger W, Muntau AC, Dame T, Kreischer J, Arnecke R, Superti-Furga A, Troxler H, Liebl B, Olgemoller B, et al. (2001). Hepatic carnitine palmitoyltransferase I deficiency: acylcarnitine profiles in blood spots are highly specific. Clin Chem 47, 1763–1768. [PubMed] [Google Scholar]
- Fritz IB, and Yue KT (1963). Long-Chain Carnitine Acyltransferase and the Role of Acylcarnitine Derivatives in the Catalytic Increase of Fatty Acid Oxidation Induced by Carnitine. J Lipid Res 4, 279–288. [PubMed] [Google Scholar]
- Griss T, Vincent EE, Egnatchik R, Chen J, Ma EH, Faubert B, Viollet B, DeBerardinis RJ, and Jones RG (2015). Metformin Antagonizes Cancer Cell Proliferation by Suppressing Mitochondrial-Dependent Biosynthesis. PLoS biology 13, e1002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualdoni GA, Mayer KA, Goschl L, Boucheron N, Ellmeier W, and Zlabinger GJ (2016). The AMP analog AICAR modulates the Treg/Th17 axis through enhancement of fatty acid oxidation. FASEB J 30, 3800–3809. [DOI] [PubMed] [Google Scholar]
- Huang SC, Everts B, Ivanova Y, O’Sullivan D, Nascimento M, Smith AM, Beatty W, Love-Gregory L, Lam WY, O’Neill CM, et al. (2014). Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat Immunol 15, 846–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, and Linsley PS (2010). Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nature reviews. Drug discovery 9, 57–67. [DOI] [PubMed] [Google Scholar]
- Jacobs SR, Michalek RD, and Rathmell JC (2010). IL-7 is essential for homeostatic control of T cell metabolism in vivo. J Immunol 184, 3461–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RG, Bui T, White C, Madesh M, Krawczyk CM, Lindsten T, Hawkins BJ, Kubek S, Frauwirth KA, Wang YL, et al. (2007). The proapoptotic factors Bax and Bak regulate T Cell proliferation through control of endoplasmic reticulum Ca(2+) homeostasis. Immunity 27, 268–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RG, and Thompson CB (2007). Revving the engine: signal transduction fuels T cell activation. Immunity 27, 173–178. [DOI] [PubMed] [Google Scholar]
- Krawczyk CM, Shen H, and Pearce EJ (2007). Memory CD4 T cells enhance primary CD8 T-cell responses. Infect Immun 75, 3556–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahl K, Loddenkemper C, Drouin C, Freyer J, Arnason J, Eberl G, Hamann A, Wagner H, Huehn J, and Sparwasser T (2007). Selective depletion of Foxp3+ regulatory T cells induces a scurfy-like disease. J Exp Med 204, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Walsh MC, Hoehn KL, James DE, Wherry EJ, and Choi Y (2015). Acetyl CoA Carboxylase 2 Is Dispensable for CD8+ T Cell Responses. PLoS One 10, e0137776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, et al. (2001). A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 15, 763–774. [DOI] [PubMed] [Google Scholar]
- Lochner M, Berod L, and Sparwasser T (2015). Fatty acid metabolism in the regulation of T cell function. Trends Immunol 36, 81–91. [DOI] [PubMed] [Google Scholar]
- Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, Sullivan SA, Nichols AG, and Rathmell JC (2011). Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol 186, 3299–3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M, Liu J, Rovira II, Gonzalez-Hurtado E, Lee J, Wolfgang MJ, and Finkel T (2016). Fatty acid oxidation in macrophage polarization. Nat Immunol 17, 216–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor RS, Guo L, Ghassemi S, Snyder NW, Worth AJ, Weng L, Kam Y, Philipson B, Trefely S, Nunez-Cruz S, et al. (2018). The CPT1a inhibitor, etomoxir induces severe oxidative stress at commonly used concentrations. Scientific reports 8, 6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan D, van der Windt GJ, Huang SC, Curtis JD, Chang CH, Buck MD, Qiu J, Smith AM, Lam WY, DiPlato LM, et al. (2014). Memory CD8(+) T cells use cell-intrinsic lipolysis to support the metabolic programming necessary for development. Immunity 41, 75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Tian T, Park CO, Lofftus SY, Mei S, Liu X, Luo C, O’Malley JT, Gehad A, Teague JE, et al. (2017). Survival of tissue-resident memory T cells requires exogenous lipid uptake and metabolism. Nature 543, 252–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EL, Poffenberger MC, Chang CH, and Jones RG (2013). Fueling immunity: insights into metabolism and lymphocyte function. Science 342, 1242454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, Jones RG, and Choi Y (2009). Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature 460, 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathmell JC, Farkash EA, Gao W, and Thompson CB (2001). IL-7 enhances the survival and maintains the size of naive T cells. J Immunol 167, 6869–6876. [DOI] [PubMed] [Google Scholar]
- Rathmell JC, Vander Heiden MG, Harris MH, Frauwirth KA, and Thompson CB (2000). In the absence of extrinsic signals, nutrient utilization by lymphocytes is insufficient to maintain either cell size or viability. Molecular cell 6, 683–692. [DOI] [PubMed] [Google Scholar]
- Raud B, McGuire PJ, Jones RG, Sparwasser T, and Berod L (2018). Fatty acid metabolism in CD8(+) T cell memory: Challenging current concepts. Immunol Rev 283, 213–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer S, Bruno L, Hertweck A, Finlay D, Leleu M, Spivakov M, Knight ZA, Cobb BS, Cantrell D, O’Connor E, et al. (2008). T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. Proc Natl Acad Sci U S A 105, 7797–7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoors S, Bruning U, Missiaen R, Queiroz KC, Borgers G, Elia I, Zecchin A, Cantelmo AR, Christen S, Goveia J, et al. (2015). Fatty acid carbon is essential for dNTP synthesis in endothelial cells. Nature 520, 192–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriver LP, and Manchester M (2011). Inhibition of fatty acid metabolism ameliorates disease activity in an animal model of multiple sclerosis. Scientific reports 1, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuve P, Minarrieta L, Erdmann H, Arnold-Schrauf C, Swallow M, Guderian M, Krull F, Holscher A, Ghorbani P, Behrends J, et al. (2018). De Novo Fatty Acid Synthesis During Mycobacterial Infection Is a Prerequisite for the Function of Highly Proliferative T Cells, But Not for Dendritic Cells or Macrophages. Front Immunol 9, 495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasenko TN, Pacheco SE, Koenig MK, Gomez-Rodriguez J, Kapnick SM, Diaz F, Zerfas PM, Barca E, Sudderth J, DeBerardinis RJ, et al. (2017). Cytochrome c Oxidase Activity Is a Metabolic Checkpoint that Regulates Cell Fate Decisions During T Cell Activation and Differentiation. Cell Metab 25, 1254–1268 e1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Windt GJ, Everts B, Chang CH, Curtis JD, Freitas TC, Amiel E, Pearce EJ, and Pearce EL (2012). Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity 36, 68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Windt GJ, O’Sullivan D, Everts B, Huang SC, Buck MD, Curtis JD, Chang CH, Smith AM, Ai T, Faubert B, et al. (2013). CD8 memory T cells have a bioenergetic advantage that underlies their rapid recall ability. Proc Natl Acad Sci U S A 110, 14336–14341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Plas DR, Rathmell JC, Fox CJ, Harris MH, and Thompson CB (2001). Growth factors can influence cell growth and survival through effects on glucose metabolism. Mol Cell Biol 21, 5899–5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, McCormick LL, Fitzgerald P, Chi H, Munger J, et al. (2011). The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35, 871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Sanin DE, Everts B, Chen Q, Qiu J, Buck MD, Patterson A, Smith AM, Chang C-H, and Liu Z (2016). Type 1 interferons induce changes in core metabolism that are critical for immune function. Immunity 44, 1325–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Blanco DB, Neale G, Vogel P, Avila J, Clish CB, Wu C, Shrestha S, Rankin S, Long L, et al. (2017). Homeostatic control of metabolic and functional fitness of Treg cells by LKB1 signalling. Nature 548, 602–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao CH, Liu GY, Wang R, Moon SH, Gross RW, and Patti GJ (2018). Identifying off-target effects of etomoxir reveals that carnitine palmitoyltransferase I is essential for cancer cell proliferation independent of beta-oxidation. PLoS biology 16, e2003782. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.