This work reports the crystal structure of a previously uncharacterized Fe protein from a methanogenic organism, which provides important insights into the structural properties of the less-characterized, yet highly interesting archaeal nitrogenase enzymes. Moreover, the structure-derived implications for CO2 capture by a surface-exposed [Fe4S4] cluster point to the possibility of developing novel strategies for CO2 sequestration while providing the initial insights into the unique mechanism of FeS-based CO2 activation.
KEYWORDS: CO2 capture, FeS cluster, iron protein, methanogen, nitrogenase
ABSTRACT
Nitrogenase iron (Fe) proteins reduce CO2 to CO and/or hydrocarbons under ambient conditions. Here, we report a 2.4-Å crystal structure of the Fe protein from Methanosarcina acetivorans (MaNifH), which is generated in the presence of a reductant, dithionite, and an alternative CO2 source, bicarbonate. Structural analysis of this methanogen Fe protein species suggests that CO2 is possibly captured in an unactivated, linear conformation near the [Fe4S4] cluster of MaNifH by a conserved arginine (Arg) pair in a concerted and, possibly, asymmetric manner. Density functional theory calculations and mutational analyses provide further support for the capture of CO2 on MaNifH while suggesting a possible role of Arg in the initial coordination of CO2 via hydrogen bonding and electrostatic interactions. These results provide a useful framework for further mechanistic investigations of CO2 activation by a surface-exposed [Fe4S4] cluster, which may facilitate future development of FeS catalysts for ambient conversion of CO2 into valuable chemical commodities.
INTRODUCTION
Iron-sulfur (FeS) proteins utilize a wide array of FeS clusters to play key roles that range from electron transfer and catalysis to structural and regulatory functions in biological systems (1–7). A homodimer carrying a subunit-bridging [Fe4S4] cluster at the protein surface, the iron (Fe) protein of nitrogenase is best known for its function as an obligate electron donor for its catalytic partner during substrate turnover (8, 9). Recently, the Fe protein from a diazotrophic microbe, Azotobacter vinelandii (designated AvNifH) was shown to act as a reductase on its own and catalyze the ambient reduction of CO2 to CO via redox changes of its [Fe4S4] cluster (10). Interestingly, while the cluster of AvNifH is believed to cycle between the [Fe4S4]1+ (reduced) and [Fe4S4]2+ (oxidized) states (11–15) for its function as an electron donor in nitrogenase catalysis, catalytic turnover of CO2 by AvNifH on its own was observed when a strong reductant, europium(II) diethylenetriaminepentaacetic acid (EuII-DTPA; E0′ = 1.14 V at pH 8.0), poised its cluster in the all-ferrous, [Fe4S4]0 state under in vitro conditions (10). Perhaps more interestingly, the Fe protein from a methanogenic microorganism, Methanosarcina acetivorans (designated MaNifH), was capable of reducing CO2 past CO into hydrocarbons under ambient conditions in the presence of EuII-DTPA, further illustrating the unique reactivity of the [Fe4S4] cluster toward CO2 (16, 17). Together, these observations point to the nitrogenase Fe protein as a simple model system for mechanistic investigations of FeS-based CO2 activation and reduction.
Of the two Fe protein species that have been investigated for their reactivity toward CO2, MaNifH is particularly interesting given its ability to convert CO2 to CO and hydrocarbons. Despite its archaeal origin, MaNifH shares a sequence identity of 59% and a sequence homology of 72% with AvNifH. Like AvNifH, MaNifH is a homodimer of ∼60 kDa, and it contains an [Fe4S4] cluster that can adopt three oxidation states upon redox treatments: (i) the oxidized state ([Fe4S4]2+), which is generated upon treatment by indigodisulfonate; (ii) the reduced state ([Fe4S4]1+), which is generated upon treatment by dithionite (DT); and (iii) the “superreduced,” all-ferrous state ([Fe4S4]0), which is generated upon treatment by EuII-DTPA (17). There are differences, however, in the electronic properties of MaNifH and AvNifH, which are reflected by a stronger S = 3/2 contribution to the electron paramagnetic resonance (EPR) spectrum of the reduced MaNifH and a decreased intensity of the parallel mode, g = 16.4 signal in the EPR spectrum of the superreduced MaNifH (17). These differences, along with the lower reduction potential of the [Fe4S4]1+/2+ pair of MaNifH (E0 = −395 mV) than that of AvNifH ([Fe4S4]1+/2+: E0 = −301 mV) (16), may contribute to the difference in the reactivities of MaNifH and AvNifH toward CO2. The redox dependence of this reaction is further illustrated by a substantially decreased CO2-reducing activity of both MaNifH and AvNifH in the presence of dithionite, a weaker reductant than EuII-DTPA, which renders the clusters of these Fe proteins in the catalytically inefficient [Fe4S4]1+ state (10, 16).
The significantly decreased activity of Fe protein in a dithionite-driven reaction could prove advantageous for capturing CO2 in an early stage of CO2 reduction. Here, we report a 2.4-Å crystal structure of MaNifH that was generated in the presence of dithionite and an alternative CO2 source, bicarbonate. Structural analysis of this previously uncharacterized Fe protein from the methanogen nitrogenase family suggests that CO2 is possibly captured in an unactivated, linear conformation on the dithionite-reduced MaNifH; moreover, it reveals the initial coordination of CO2 by a conserved, surface-exposed arginine (Arg) pair in a concerted yet asymmetric manner, which could assist in trapping CO2 near the [Fe4S4] cluster via hydrogen bonding and electrostatic interactions. These results provide a useful framework for further exploration of the mechanism of CO2 activation by Fe proteins, which may enable future development of FeS catalysts for recycling the greenhouse gas CO2 into valuable chemical commodities.
RESULTS
Structural analysis of the dithionite-reduced MaNifH.
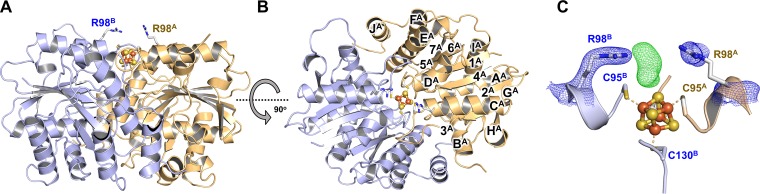
Consistent with the presence of its [Fe4S4] cluster in the +1 oxidation state, MaNifH crystallized in the presence of dithionite had a characteristic brown color. The ∼2.4-Å structure of the dithionite-reduced MaNifH (PDB ID 6NZJ) adopts the same overall conformation as all Fe protein structures reported to date (9, 18–20), with each of its subunits folded as a single α/β-type domain and its [Fe4S4] cluster situated in a surface cavity between the two subunits (Fig. 1A and B). A closer examination of the region surrounding the active site of MaNifH (Fig. 1C) reveals the ligation of the [Fe4S4] cluster by four Cys residues: two from subunit A (Cys95A, Cys130A) and two from subunit B (Cys95B, Cys130B). Interestingly, the electron density omit map (Fo − Fc) of the active site of MaNifH (Fig. 1C, green mesh; also see Fig. S1 in the supplemental material) indicates the presence of additional electron density that lies immediately next to the crystallographic symmetry axis, seemingly held by two pairs of conserved Arg residues (R98A and R98B)—one from each of the two adjacent MaNifH subunit dimers.
FIG 1.
Side (A) and top (B) views of the 2.4-Å crystal structure of MaNifH. The subunits are shown as ribbons (subunit A, light orange; subunit B, light blue). The α-helices (AA-JA) and β-sheets (1A-7A) of subunit A are indicated. The [Fe4S4] cluster is shown in ball-and-stick presentation (Fe, orange; S, yellow). (C) The electron density (2Fo − Fc) of the active site of MaNifH was contoured at 1.5-σ level for the conserved Arg pair (blue meshes), and the omit map (Fo − Fc) of the additional electron density (green mesh) that is unaccounted for in the structure was contoured at 3.0-σ level. The four Cys ligands (C95A, C130A, C95B, C130B) and the conserved Arg pair (R98A, R98B) are shown as sticks.
Structure of MaNifH without additional ligand. The subunits are shown as ribbons and colored light orange (subunit A) and light blue (subunit B) for the asymmetric unit and yellow (subunit A) and dark blue (subunit B) for the symmetry mate. The Fo – Fc map (green mesh) is contoured at 3 σ. The side chains of R98 are shown as sticks. The [Fe4S4] clusters are shown in ball-and-stick presentation and colored as follows: Fe, orange; S, yellow. Download FIG S1, JPG file, 0.8 MB (845.7KB, jpg) .
Copyright © 2019 Rettberg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Modeling the extra electron density in the structure of the dithionite-reduced MaNifH.
Given that the additional electron density may originate from the small molecules in the protein preparations or crystallographic solutions, we then considered possible candidates and modeled water (see Fig. S2A in the supplemental material), carbonate (Fig. S2B), glycerol (Fig. S2C), and CO2 (Fig. S2D), respectively, into this density. Water is an unlikely contributor to this density, as modeling of one water molecule in the asymmetric unit and another in its symmetry mate results in substantial “leftover” electron density in the Fo − Fc omit map (Fig. S2A, green mesh). Carbonate and glycerol, on the other hand, could be modeled as two molecules—each at ∼50% occupancy—at the crystallographic symmetry axis with reasonable R factor values (see Table S1 in the supplemental material). Similarly, CO2 could be modeled with reasonable R factor values at the crystallographic symmetry axis; only in this case, two molecules of CO2—each at 100% occupancy—could be assigned to the asymmetric unit and its symmetry mate, respectively (Table S1). It should be noted that the modeling of two CO2 moieties results in some negative electron density; however, the overall crystallographic statistics are reasonable to support this model (Table S1) despite the difficulty to conclusively assign this ligand near the crystallographic symmetry axis.
Structures of MaNifH modeled with two water molecules (A), two carbonate molecules (B), two glycerol molecules (C), and two CO2 moieties (D), respectively. The subunits are shown as ribbons and colored light orange (subunit A) and light blue (subunit B) for the asymmetric unit and yellow (subunit A) and dark blue (subunit B) for the symmetry mate. The Fo – Fc map (red mesh) is contoured at −3 σ, the Fo – Fc map (green mesh) is contoured at 3 σ, and the 2Fo – Fc map (blue mesh) is contoured at 1.5 σ. One water (A) or CO2 (B) molecule was modeled in the asymmetric unit and the other in the symmetry mate, respectively. The two carbonate (B) or glycerol (C) molecules were placed on the crystallographic axis. The side chains of R98 are shown as sticks. The [Fe4S4] cluster and the water, carbonate, and glycerol moieties are shown in ball-and-stick presentation and colored as follows: Fe, orange; S, yellow; C, gray; O, red. Download FIG S2, JPG file, 2.3 MB (2.4MB, jpg) .
Copyright © 2019 Rettberg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data collection, refinement, and ligand fitting statistics of MaNifH (PDB ID 6NZJ). Shown are the values for the ligand-free model (upper) and the statistics for the models with the plausible ligands CO2, glycerol, and carbonate (lower). Download Table S1, PDF file, 0.1 MB (62KB, pdf) .
Copyright © 2019 Rettberg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DFT calculations of the affinity of CO2 to the dithionite-reduced MaNifH.
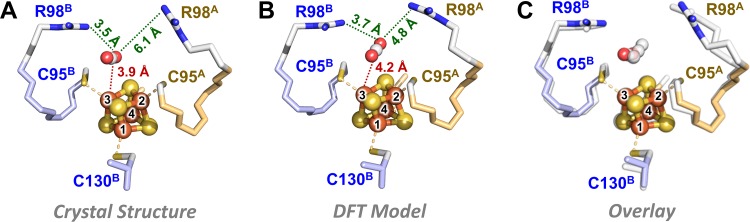
To seek support for the assignment of CO2 as the extra electron density in the crystal structure of MaNifH, we then used density functional theory (DFT) calculations to analyze the CO2 affinity of the [Fe4S4]1+ cluster in MaNifH. Consistent with our previous findings for both AvNifH-bound and synthetic [Fe4S4] clusters (10, 16), CO2 does not interact well with the [Fe4S4]1+ cluster of MaNifH and tends to dissociate from the cluster during the course of structural optimization; however, the two highly conserved Arg residues in MaNifH (R98A, R98B) form a cage-like configuration around the CO2 molecule that assists in trapping it in close proximity to the cluster (see Movie S1 in the supplemental material). Interestingly, the location of the CO2 moiety in the DFT-optimized model is in good agreement with half of the electron density pattern in the structure of MaNifH, except for a slight reorientation of CO2 (Fig. 2). In comparison, DFT optimization reveals protonation of carbonate by R98B, followed by coordination of the resulting bicarbonate in a position parallel to the upper surface of the [Fe4S4] cluster, which is rather distinct from the perpendicular position modeled for carbonate in the crystal structure of MaNifH (see Fig. S3 in the supplemental material). This observation is important, as it provides theoretical support for the assignment of CO2 as a potential ligand in the structure of the dithionite-reduced MaNifH protein. The fact that the MaNifH crystals were generated at a bicarbonate concentration in the same order of magnitude as that used to generate a CO2-bound conformation of CO dehydrogenase (21) provides further support for the assignment of CO2 in the MaNifH structure. In this scenario, the CO2 moiety has its C atom placed at a distance of ∼4 Å from the nearest Fe atom (Fe-3) of the [Fe4S4] cluster, with the NH2+ groups of R98A and R98B assuming the “distal” and “proximal” positions, respectively, to Fe-3 (Fig. 2). This observation suggests a possible role of the conserved Arg pair in capturing CO2 via hydrogen bonding and/or electrostatic interactions, as well as a potentially asymmetric functionality of the two Arg residues in this process.
FIG 2.
Crystal (A) and DFT-optimized (B) structures of MaNifH with the extra electron density modeled as CO2 and (C) an overlay of the two structures. The conserved pair of Arg residues assume “proximal” (R98B) and “distal” (R98A) positions, respectively, to the CO2 moiety and the Fe-3 atom of the cluster (A and B), and CO2 occupies a highly similar position in the crystal structure and the DFT model (C). The [Fe4S4] cluster and CO2 moiety are shown in ball-and-stick presentation and colored as follows: Fe, orange; S, yellow; C, gray; O, red. The Cys ligands and the conserved Arg residues are shown as sticks.
Crystal structure of MaNifH with the extra electron density modeled as carbonate (A) and DFT-optimized structure of MaNifH with carbonate (B) or bicarbonate (C) as the substrate. In the crystal structure, the carbonate moiety assumes a position perpendicular to the upper surface of the [Fe4S4] cluster (A). Based on DFT-based structural optimization, however, carbonate is first protonated by R98B, and the resulting bicarbonate assumes a position parallel to the upper surface of the [Fe4S4] cluster (B). The DFT-optimized structure calculated directly with bicarbonate (C) closely resembles that achieved with carbonate upon protonation (B). Overall, carbonate occupies a different position in the crystal structure than (bi)carbonate in the DFT models, which can be best visualized by the overlay of the carbonate-bound crystal structure with the (bi)carbonate-bound DFT models that originate either from the protonation of carbonate (D) or directly from bicarbonate (E). In both cases, the (bi)carbonate moiety swings ∼90° from a position that is perpendicular to one that is parallel to the upper surface of the [Fe4S4] cluster. Furthermore, the (bi)carbonate moiety rotates ∼90° counterclockwise in a horizontal plane parallel to the upper surface of the cluster. The hydrogen atoms of bicarbonate are omitted from the figure, as they cannot be discerned by X-ray crystallography. The [Fe4S4] cluster and carbonate/bicarbonate moieties are shown in ball-and-stick presentations and colored as follows: Fe, orange; S, yellow; C, gray; O, red. The Cys ligands and the conserved Arg residues are shown as sticks. Download FIG S3, JPG file, 1.4 MB (1.4MB, jpg) .
Copyright © 2019 Rettberg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Course of DFT-based structural optimization (TPSS/def2-SVP/def2-TZVP) of the CO2-captured conformation of MaNifH in the [Fe4S4]1+ state (S = 1/2). The CO2 moiety in the DFT-optimized structure is rendered gray, and the CO2 moiety in the crystal structure is colored as follows: C, gray; O, red. Download Movie S1, MPG file, 7.8 MB (8MB, mpg) .
Copyright © 2019 Rettberg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Examining the role of the conserved Arg pair of MaNifH in CO2 capture.
To test the proposed role of conserved Arg residues in CO2 capture, we performed site-directed mutagenic analysis and mutated R98 of MaNifH to either a His or a Gly. Both R98H and R98G MaNifH variants display the same S = 1/2 EPR signal as the wild-type protein, which is indicative of an unperturbed [Fe4S4] center in the +1 oxidation state (Fig. 3A). However, the R98H variant of MaNifH retains ∼80% CO2-reducing activity, whereas the R98G variant loses ∼85% of this activity (Fig. 3B), consistent with the preservation (i.e., the R→H mutation) or elimination (i.e., the R→G mutation) of the hydrogen bonding ability at the position of R98. The somewhat decreased activity of the R98H variant could be explained by a shorter side chain of His and, consequently, a reduced efficiency of this residue in hydrogen bonding/proton donation than Arg. The slight defect of His in proton donation would also account for a shift of the product profile of the R98H variant (hydrocarbon/CO ratio of 1.9) from hydrocarbon formation to CO formation compared to that of the wild-type MaNifH (hydrocarbon/CO ratio of 2.7), as formation of CO requires fewer protons than that of hydrocarbons.
FIG 3.
Spectroscopic and catalytic features of the wild-type and variant MaNifH. (A) EPR spectra and (B) CO2-reducing activities of wild-type and variant MaNifH. EPR spectra were collected at 10 K. The wild-type and R98H and R98G variant MaNifH are dimers of ∼60 kDa and contain 3.7 ± 0.1, 3.9 ± 0.4, and 3.8 ± 0.2 nmol Fe per nmol protein, respectively. Like the wild-type MaNifH, the R98H and R98G variants display the same [Fe4S4]+ characteristic, S = 1/2 EPR signal in the dithionite-reduced state (A), yet they display disparate activities in CO2 reduction (B). The hydrocarbon/CO ratios (calculated based on total nmol of reduced carbons) of the wild-type MaNifH and R98H variant are 2.7 and 1.9, respectively, suggesting a shift from hydrocarbon formation to CO formation in the latter case.
Proposal of a plausible mechanism for the initial capture of CO2 by MaNifH.
To obtain further insights into the mechanism of CO2 capture by nitrogenase Fe proteins, we compared our dithionite-reduced MaNifH structure (PDB ID 6NZJ) that is potentially bound with CO2 with a previously reported, dithionite-reduced AvNifH structure (PDB ID 1G5P) that is free of CO2 (19). Consistent with a high degree of sequence homology between MaNifH and AvNifH, the subunits A and B in MaNifH show Cα deviations of only 0.599 and 0.616 Å, respectively, relative to those in AvNifH, yet the two subunit chains in MaNifH are more similar to each other in terms of secondary structural elements, particularly with respect to the structurally less conserved α-helical regions (see Fig. S4 and S5 in the supplemental material). More strikingly, compared to their counterparts in AvNifH, there is a notable movement of the two subunits of MaNifH with respect to each other, which flattens the surface cavity and consequently “pushes” the [Fe4S4] cluster further toward the surface where a CO2 molecule could be modeled (see Fig. S6 and Movie S2 in the supplemental material). A top-view comparison between the two structures further reveals a “linearization” of helices CA and CB in MaNifH relative to those in AvNifH, which is accompanied by a substantial swing of the Arg pair, R98A and R98B (located at the tips of helices CA and CB), toward the center of the surface cavity (see Fig. S6 and Movie S3 in the supplemental material). Such a movement of the conserved Arg pair could reflect a concerted action of the “distal” R98A and the “proximal” R98B in the initial capture of CO2 in an unactivated, linear conformation near the Fe-3 atom of the [Fe4S4] cluster (Fig. 4). Further activation of CO2 into a bent, carboxylate-like conformation may continue to employ an asymmetric mechanism. Previous DFT calculations of CO2 activation by the catalytically competent, all-ferrous AvNifH (10) led to the proposal of binding of an activated CO2 moiety via coordination of C with Fe-3 of the cluster and coordination of O with the guanidinium group of the “proximal” R100B (corresponding to the “proximal” R98B in MaNifH), with the latter potentially donating protons for the subsequent C-O bond cleavage.
FIG 4.
Comparison of the CO2-free (A) and CO2-captured (B) conformations of Fe protein, showing concerted yet asymmetric movement of a pair of conserved Arg residues that potentially capture CO2 near the [Fe4S4] cluster. The CO2-free and CO2-captured conformations are represented by the homologous AvNifH and MaNifH, respectively. The movement of the “proximal” Arg (R100B in AvNifH and the corresponding R98B in MaNifH) and the “distal” Arg (R100A in AvNifH and the corresponding R98A in MaNifH) is shown from two angles.
Identities between subunits A and B within AvNifH and MaNifH. (A and B) Percentages of identities between individual α-helices (A) and β-sheets (B) of subunits A and B within AvNifH (black) and MaNifH (red). (C and D) Overall percentages of identities between α-helices (C) and β-sheets (D) of subunits A and B within AvNifH (black) and MaNifH (red). Data from Fig. S5 were used to calculate these percentages. Cα deviations of subunit A versus subunit B: AvNifH, 0.356 Å, and MaNifH, 0.334 Å. Download FIG S4, JPG file, 0.8 MB (849.9KB, jpg) .
Copyright © 2019 Rettberg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Subunits A and B of AvNifH and MaNifH. Shown are comparisons between (A and B) primary sequences and (C and D) secondary structures of subunits A (A and C) and B (B and D) of AvNifH and MaNifH. The identical residues in the primary sequences are indicated with *, and the conserved Cys ligands for the [Fe4S4] cluster are shown in red (A and B). The α-helices are colored green, the β-sheets are colored cyan (C and D), and the primary sequences corresponding to these structural elements are highlighted with the corresponding colors (A and B). Download FIG S5, JPG file, 2.8 MB (2.9MB, jpg) .
Copyright © 2019 Rettberg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Structural comparison between AvNifH and MaNifH. Shown are side (left) and top (middle) views of AvNifH (A) and MaNifH (B), with the alignments of helices CB and CA and the positions of the Arg pairs (located at the tips of helices CB and CA) highlighted (right). The movements of Arg residues in MaNifH (B, right) relative to those in AvNifH are indicated by dashed red arrows. Subunits are shown as ribbons (side view) or cylinders (top view) and colored light orange (subunit A) and light blue (subunit B), respectively. The [Fe4S4] cluster and CO2 moiety are shown in ball-and-stick presentation and colored as follows: Fe, orange; S, yellow; C, gray; O, red. Download FIG S6, JPG file, 2.3 MB (2.3MB, jpg) .
Copyright © 2019 Rettberg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Conformational morphing of AvNifH into MaNifH (side view). Both structures are depicted and colored as described in the legend to Fig. S1. The conformational morphing plugin of PyMol 2.1.1 (https://pymol.org) was used to generate the movie, with AvNifH as the start conformation and MaNifH as the end conformation. Refinement cycles, 3; number of output states, 100; interpolation method, RigiMOL. Download Movie S2, MPG file, 7.9 MB (8.1MB, mpg) .
Copyright © 2019 Rettberg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Conformational morphing of AvNifH into MaNifH (top view). The movie was generated as described in the legend to Movie S2. Download Movie S3, MPG file, 6.6 MB (6.7MB, mpg) .
Copyright © 2019 Rettberg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
In light of a plausible asymmetric mechanism of CO2 activation by Fe protein, it is interesting to consider the mechanism proposed for the Ni-dependent CO dehydrogenase in CO2 activation, which involves the action of the Fe/Ni atoms of its heterometallic C-cluster ([NiFe4S4]) as a pair of Lewis acid/base to facilitate scission of a C-O bond (21–24). In the absence of such a heterometal-based asymmetry, it is plausible that activation of CO2 by the homometallic [Fe4S4] cluster would resort to a structure-based asymmetry that enables interactions between O and the guanidinium group of the proximal Arg, as well as binding of C to the nearest Fe-3 atom. It is worth noting that the proposed asymmetric functionality of the conserved Arg pair in CO2 activation is consistent with the previously established regulatory mechanism of nitrogenase activity through ADP-ribosylation of only one of these conserved Arg residues (25), whereas the structure-based suggestion of a single reactive Fe (Fe-3) site for CO2 activation may have certain relevance to the unique Fe site that was identified by earlier Mössbauer studies of the all-ferrous Fe protein (14). While the functions of these asymmetric elements await further elucidation, the current study provides a useful framework for investigating the structural basis of Fe protein-based CO2 capture and activation. Moreover, the strategy utilized by the Fe protein to trap CO2 by a pair of surface-located arginines loosely resembles the approaches that employ nitrogen-based ligands, such as metal-organic frameworks (MOFs) with amine or amide groups (26) or protein amyloid fibers comprising lysines in stacked sheets (27), for CO2 capture and sequestration. The fact that the arginine residues of the Fe protein trap CO2 in the close proximity to a surface-exposed [Fe4S4] cluster for further processing may provide a conceptual basis for the future development of MOF- or protein-based FeS catalysts that couple the capture of CO2 with the recycling of this greenhouse gas into useful chemical commodities.
MATERIALS AND METHODS
Protein purification and crystallization.
All protein purification steps were carried out anaerobically using Schlenk techniques. His-tagged MaNifH was purified by immobilized metal affinity as described elsewhere (17, 28). Reagents for protein crystallization were purchased from Hampton Research and were thoroughly deaerated by vacuum/Ar-fill cycling before use. All crystals were generated at room temperature in an anaerobic chamber (Coy Laboratory Products), coated with Parabar 10312 oil (Hampton Research) as a cryo-protectant, and flash-frozen in liquid nitrogen for data collection.
MaNifH was crystallized at room temperature by a microbatch method under a layer of Al’s oil (Hampton Research). The purified MaNifH protein was desalted on a G-25 fine column equilibrated with buffer M (10 mM EPPS [pH 8.0], 100 mM NaCl, 10% [vol/vol] glycerol, and 2 mM dithionite [DT]) and then concentrated to 10 mg/ml by Amicon Ultra-4 30-kDa centrifugal filter units. The crystals were grown by evaporating a mixture of 1 μl protein solution and 3 μl precipitant solution (2.3 M ammonium sulfate, 7% [wt/vol] polyethylene glycol 3350 [PEG 3350], 12 mM carbonate, and 2 mM DT) under Al’s oil. The protein solution was brown, indicating that the protein-bound cluster was present in the reduced, +1 state. Brown crystals grew after 2 weeks and were flash-frozen in liquid nitrogen for data collection.
Data collection and structural determination.
The diffraction data of MaNifH crystals were collected at 100 K on beamline 8.2.1 of Advanced Light Source using a wavelength of 0.9774 Å and an ADSC Q315r charge-coupled device (CCD) detector. A total of 501 images were recorded for MaNifH at a distance of 450 mm, with an oscillation angle of 0.25° and an exposure time of 0.25 s. The raw data were indexed and processed using iMosflm and Scala in the CCP4 package (29). Molecular replacement was performed with Phaser in PHENIX (30) using the structure of the Clostridium pasteurianum NifH protein (PDB ID 1CP2) (19) as a search model. The initial model was further improved by cycles of manual building and refinement using Coot and PHENIX (30–32). At the end of the refinement cycle, water, carbonate, glycerol, or CO2 was manually put into the model of MaNifH and further refined for 3 cycles using PHENIX. The stereochemical quality of the final structures was evaluated by MolProbity (33). All molecular graphics were prepared using PyMol (34). Data collection and statistics for refinement and ligand modeling are summarized in Table S1.
Strain construction and activity analyses.
Strains expressing R98H and R98G MaNifH variants were constructed via site-directed mutagenesis of the wild-type Methanosarcina acetivorans nifH sequence carried on a pET14b vector (17), followed by transformation of the resultant plasmids into Escherichia coli strain BL21(DE3). The in vitro CO2-reduction assays were carried out in 9.4-ml assay vials with crimped butyl rubber serum stoppers. Each assay contained, in a total volume of 1.0 ml, 500 mM Tris-HCl (pH 10.0), 0.5 mg Fe protein (wild-type or R98H or R98G variant MaNifH), and 100 mM EuII-DTPA. In addition, the headspace of each assay contained 100% CO2 (for reactions) or 100% Ar (for controls). The assays were assembled without protein and EuII-DTPA and repeatedly flushed and exchanged with CO2, followed by equilibration for 30 min until pH stabilized at ∼8.0. The reaction was initiated upon addition of MaNifH, followed immediately by addition of EuII-DTPA and incubation with continuous shaking at 30°C for 300 min until the reaction was complete. Following the quenching of each assay by 100 μl of 30% trichloroacetic acid, the headspace sample was examined for the production of CO and hydrocarbons as described previously (16).
EPR spectroscopy analyses.
The EPR samples were prepared in a Vacuum Atmospheres glove box and flash-frozen in liquid nitrogen prior to analysis. The DT-reduced samples contained 2 mM DT, 50 mM Tris-HCl (pH 8.0), 500 mM NaCl, and 10% (vol/vol) glycerol. EPR spectra were recorded by an ESP 300 Ez spectrophotometer (Bruker) interfaced with an ESR-9002 liquid-helium continuous-flow cryostat (Oxford Instruments) using a microwave power of 50 mW, a gain of 5 × 104, a modulation frequency of 100 kHz, and a modulation amplitude of 5 G. Five scans were recorded for each EPR sample at a temperature of 10 K and a microwave frequency of 9.62 GHz.
Density functional theory calculations.
The mechanism of CO2, carbonate, and bicarbonate coordination was studied with the DFT programs in the Turbomole package, version 7.0 (35). Atomistic models of the [Fe4S4] cluster and its immediate protein environment were generated from the structure of MaNifH (PDB ID 6NZJ [this work]) in the DT-reduced, [Fe4S4]1+ state.
The models were selected as described previously (10) and contained the [Fe4S4] cluster and C95A, C95B, C130A, C130B, R98A, R98B, F133A, F133B, and the main-chain atoms of the residues A96A, A96B, A97A, G97B, G131A, G131B, G132A, and G132B of MaNifH to account for all interactions of the cluster with the protein backbone. N termini were saturated with acetyl groups according to the crystallographic atom positions. Hydrogen atoms were added to the model with Open Babel (36), assuming protonation of the Arg residues. During structural optimizations, the atoms of the cluster, the side-chain atoms of the cluster-coordinating Cys residues (including Cα), the side-chain atoms of the Arg residues (starting from Cγ), the benzene groups of the Phe residues, and all hydrogen atoms were allowed to spatially relax. All other atoms were kept structurally frozen. The models were treated as open-shell systems in the unrestricted Kohn-Sham framework. Solvent effects were treated implicitly by the conductor-like solvent screening model (COSMO) (37), assuming a dielectric constant of ɛ = 40. The structures were optimized with the TPSS (Tao-Perdew-Staroverov-Scuseria) functional (38). A def2-TZVP basis set (39, 40) was used for the [Fe4S4] cluster, the side-chain atoms of the Cys residues (including Cα atoms), the atoms of the guanidinium groups, and the cluster-bound CO2, carbonate, and bicarbonate moieties. A def2-SVP basis set was assigned to all remaining atoms to accelerate the calculations. Computational time was further reduced by utilizing the resolution-of-the-identity approximation (41, 42). Antiferromagnetic coupling in the FeS cluster was accounted for by the broken symmetry approach (43–45).
Data availability.
The structure of DT-reduced MaNifH (PDB ID 6NZJ) has been deposited in the Protein Data Bank (https://www.wwpdb.org) and will be released upon publication.
ACKNOWLEDGMENTS
We thank Andrew Ma and Zihao Chen for technical assistance.
This work was supported by NSF CAREER grant CHE-1651398 (Y.H.).
Footnotes
Citation Rettberg LA, Kang W, Stiebritz MT, Hiller CJ, Lee CC, Liedtke J, Ribbe MW, Hu Y. 2019. Structural analysis of a nitrogenase iron protein from Methanosarcina acetivorans: implications for CO2 capture by a surface-exposed [Fe4S4] cluster. mBio 10:e01497-19. https://doi.org/10.1128/mBio.01497-19.
Contributor Information
Derek R. Lovley, University of Massachusetts Amherst.
Russ Hille, University of California, Riverside, USA.
Thomas Ward, University of Basel, Switzerland.
REFERENCES
- 1.Beinert H, Holm RH, Münck E. 1997. Iron-sulfur clusters: nature's modular, multipurpose structures. Science 277:653–659. doi: 10.1126/science.277.5326.653. [DOI] [PubMed] [Google Scholar]
- 2.Lill R. 2009. Function and biogenesis of iron-sulphur proteins. Nature 460:831–838. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- 3.Burgess BK, Lowe DJ. 1996. Mechanism of molybdenum nitrogenase. Chem Rev 96:2983–3012. doi: 10.1021/cr950055x. [DOI] [PubMed] [Google Scholar]
- 4.Schilter D, Camara JM, Huynh MT, Hammes-Schiffer S, Rauchfuss TB. 2016. Hydrogenase enzymes and their synthetic models: the role of metal hydrides. Chem Rev 116:8693–8749. doi: 10.1021/acs.chemrev.6b00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mühlenhoff U, Hoffmann B, Richter N, Rietzschel N, Spantgar F, Stehling O, Uzarska MA, Lill R. 2015. Compartmentalization of iron between mitochondria and the cytosol and its regulation. Eur J Cell Biol 94:292–308. doi: 10.1016/j.ejcb.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 6.O’Brien E, Holt ME, Thompson MK, Salay LE, Ehlinger AC, Chazin WJ, Barton JK. 2017. The [4Fe4S] cluster of human DNA primase functions as a redox switch using DNA charge transport. Science 355:eaag1789. doi: 10.1126/science.aag1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mettert EL, Kiley PJ. 2015. Fe-S proteins that regulate gene expression. Biochim Biophys Acta 1853:1284–1293. doi: 10.1016/j.bbamcr.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rees DC, Tezcan FA, Haynes CA, Walton MY, Andrade S, Einsle O, Howard JB. 2005. Structural basis of biological nitrogen fixation. Philos Transact A Math Phys Eng Sci 363:971–984. doi: 10.1098/rsta.2004.1539. [DOI] [PubMed] [Google Scholar]
- 9.Georgiadis MM, Komiya H, Chakrabarti P, Woo D, Kornuc JJ, Rees DC. 1992. Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii. Science 257:1653–1659. doi: 10.1126/science.1529353. [DOI] [PubMed] [Google Scholar]
- 10.Rebelein JG, Stiebritz MT, Lee CC, Hu Y. 2017. Activation and reduction of carbon dioxide by nitrogenase iron proteins. Nat Chem Biol 13:147–149. doi: 10.1038/nchembio.2245. [DOI] [PubMed] [Google Scholar]
- 11.Watt GD, Reddy K. 1994. Formation of an all ferrous Fe4S4 cluster in the iron protein component of Azotobacter vinelandii nitrogenase. J Inorg Biochem 53:281–294. doi: 10.1016/0162-0134(94)85115-8. [DOI] [Google Scholar]
- 12.Angove HC, Yoo SJ, Burgess BK, Münck E. 1997. Mössbauer and EPR evidence for an all-ferrous Fe4S4 cluster with S = 4 in the Fe protein of nitrogenase. J Am Chem Soc 119:8730–8731. doi: 10.1021/ja9712837. [DOI] [Google Scholar]
- 13.Musgrave KB, Angove HC, Burgess BK, Hedman B, Hodgson KO. 1998. All-ferrous titanium(III) citrate reduced Fe protein of nitrogenase: an XAS study of electronic and metrical structure. J Am Chem Soc 120:5325–5326. doi: 10.1021/ja980598z. [DOI] [Google Scholar]
- 14.Yoo SJ, Angove HC, Burgess BK, Hendrich MP, Münck E. 1999. Mössbauer and integer-spin EPR studies and spin-coupling analysis of the [4Fe-4S]0 cluster of the Fe protein from Azotobacter vinelandii nitrogenase. J Am Chem Soc 121:2534–2545. doi: 10.1021/ja9837405. [DOI] [Google Scholar]
- 15.Yoo SJ, Angove HC, Burgess BK, Münck E, Peterson J. 1998. Magnetic circular dichroism study of the all-ferrous [4Fe-4S] cluster of the Fe-protein of Azotobacter vinelandii nitrogenase. J Am Chem Soc 120:9704–9705. doi: 10.1021/ja981867o. [DOI] [Google Scholar]
- 16.Stiebritz MT, Hiller CJ, Sickerman NS, Lee CC, Tanifuji K, Ohki Y, Hu Y. 2018. Ambient conversion of CO2 to hydrocarbons by biogenic and synthetic [Fe4S4] clusters. Nat Catal 1:444–451. doi: 10.1038/s41929-018-0079-4. [DOI] [Google Scholar]
- 17.Hiller CJ, Stiebritz MT, Lee CC, Liedtke J, Hu Y. 2017. Tuning electron flux through nitrogenase with methanogen iron protein homologues. Chemistry 23:16152–16156. doi: 10.1002/chem.201704378. [DOI] [PubMed] [Google Scholar]
- 18.Strop P, Takahara PM, Chiu H, Angove HC, Burgess BK, Rees DC. 2001. Crystal structure of the all-ferrous [4Fe-4S]0 form of the nitrogenase iron protein from Azotobacter vinelandii. Biochemistry 40:651–656. doi: 10.1021/bi0016467. [DOI] [PubMed] [Google Scholar]
- 19.Schlessman JL, Woo D, Joshua-Tor L, Howard JB, Rees DC. 1998. Conformational variability in structures of the nitrogenase iron proteins from Azotobacter vinelandii and Clostridium pasteurianum. J Mol Biol 280:669–685. doi: 10.1006/jmbi.1998.1898. [DOI] [PubMed] [Google Scholar]
- 20.Schindelin H, Kisker C, Schlessman JL, Howard JB, Rees DC. 1997. Structure of ADP × AIF4−-stabilized nitrogenase complex and its implications for signal transduction. Nature 387:370–376. doi: 10.1038/387370a0. [DOI] [PubMed] [Google Scholar]
- 21.Jeoung JH, Fesseler J, Goetzl S, Dobbek H. 2014. Carbon monoxide. Toxic gas and fuel for anaerobes and aerobes: carbon monoxide dehydrogenases. Met Ions Life Sci 14:37–69. doi: 10.1007/978-94-017-9269-1_3. [DOI] [PubMed] [Google Scholar]
- 22.Fesseler J, Jeoung JH, Dobbek H. 2015. How the [NiFe4S4] cluster of CO dehydrogenase activates CO2 and NCO. Angew Chem Int Ed Engl 54:8560–8564. doi: 10.1002/anie.201501778. [DOI] [PubMed] [Google Scholar]
- 23.Kung Y, Drennan CL. 2011. A role for nickel-iron cofactors in biological carbon monoxide and carbon dioxide utilization. Curr Opin Chem Biol 15:276–283. doi: 10.1016/j.cbpa.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Can M, Armstrong FA, Ragsdale SW. 2014. Structure, function, and mechanism of the nickel metalloenzymes, CO dehydrogenase, and acetyl-CoA synthase. Chem Rev 114:4149–4174. doi: 10.1021/cr400461p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nordlund S, Högbom M. 2013. ADP-ribosylation, a mechanism regulating nitrogenase activity. FEBS J 280:3484–3490. doi: 10.1111/febs.12279. [DOI] [PubMed] [Google Scholar]
- 26.Kazemi S, Safarifard V. 2018. Carbon dioxide capture in MOFs: the effect of ligand functionalization. Polyhedron 154:236–251. doi: 10.1016/j.poly.2018.07.042. [DOI] [Google Scholar]
- 27.Li D, Furukawa H, Deng H, Liu C, Yaghi OM, Eisenberg DS. 2014. Designed amyloid fibers as materials for selective carbon dioxide capture. Proc Natl Acad Sci U S A 111:191–196. doi: 10.1073/pnas.1321797111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sickerman NS, Hu Y, Ribbe MW. 2017. Nitrogenase assembly: strategies and procedures. Methods Enzymol 595:261–302. doi: 10.1016/bs.mie.2017.07.006. [DOI] [PubMed] [Google Scholar]
- 29.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. 2012. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, Snoeyink J, Richardson JS, Richardson DC. 2007. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schrodinger, LLC. 2017. The PyMol molecular graphics system, version 2.0. Schrödinger, LLC, New York, NY: https://pymol.org. [Google Scholar]
- 35.Ahlrichs R, Bär M, Häser M, Horn H, Kölmel C. 1989. Electronic structure calculations on workstation computers: the program system turbomole. Chem Phys Lett 162:165–169. doi: 10.1016/0009-2614(89)85118-8. [DOI] [Google Scholar]
- 36.O'Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. 2011. Open Babel: an open chemical toolbox. J Cheminform 3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klamt A, Schüürmann G. 1993. COSMO: a new approach to dielectric screening in solvents with explicit expressions for the screening energy and its gradient. J Chem Soc Perkin Trans 2 1993:799–805. doi: 10.1039/P29930000799. [DOI] [Google Scholar]
- 38.Tao J, Perdew JP, Staroverov VN, Scuseria GE. 2003. Climbing the density functional ladder: nonempirical meta-generalized gradient approximation designed for molecules and solids. Phys Rev Lett 91:146401. doi: 10.1103/PhysRevLett.91.146401. [DOI] [PubMed] [Google Scholar]
- 39.Schäfer A, Huber C, Ahlrichs R. 1994. Fully optimized contracted Gaussian basis sets of triple zeta valence quality for atoms Li to Kr. J Chem Phys 100:5829–5835. doi: 10.1063/1.467146. [DOI] [Google Scholar]
- 40.Weigend F, Ahlrichs R. 2005. Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: design and assessment of accuracy. Phys Chem Chem Phys 7:3297–3305. doi: 10.1039/b508541a. [DOI] [PubMed] [Google Scholar]
- 41.Eichkorn K, Weigend F, Treutler O, Ahlrichs R. 1997. Auxiliary basis sets for main row atoms and transition metals and their use to approximate Coulomb potentials. Theor Chem Acc 97:119–124. doi: 10.1007/s002140050244. [DOI] [Google Scholar]
- 42.Weigend F. 2006. Accurate Coulomb-fitting basis sets for H to Rn. Phys Chem Chem Phys 8:1057–1065. doi: 10.1039/b515623h. [DOI] [PubMed] [Google Scholar]
- 43.Noodleman L. 1981. Valence bond description of antiferromagnetic coupling in transition metal dimers. J Chem Phys 74:5737–5743. doi: 10.1063/1.440939. [DOI] [Google Scholar]
- 44.Noodleman L, Post D, Baerends E. 1982. Symmetry breaking and ionization from symmetry equivalent inner shells and lone pairs in Xα theory. Chem Phys 64:159–166. doi: 10.1016/0301-0104(82)85012-X. [DOI] [Google Scholar]
- 45.Noodleman L, Peng CY, Case DA, Mouesca JM. 1995. Orbital interactions, electron delocalization and spin coupling in iron-sulfur clusters. Coord Chem 144:199–244. doi: 10.1016/0010-8545(95)07011-L. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Structure of MaNifH without additional ligand. The subunits are shown as ribbons and colored light orange (subunit A) and light blue (subunit B) for the asymmetric unit and yellow (subunit A) and dark blue (subunit B) for the symmetry mate. The Fo – Fc map (green mesh) is contoured at 3 σ. The side chains of R98 are shown as sticks. The [Fe4S4] clusters are shown in ball-and-stick presentation and colored as follows: Fe, orange; S, yellow. Download FIG S1, JPG file, 0.8 MB (845.7KB, jpg) .
Copyright © 2019 Rettberg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Structures of MaNifH modeled with two water molecules (A), two carbonate molecules (B), two glycerol molecules (C), and two CO2 moieties (D), respectively. The subunits are shown as ribbons and colored light orange (subunit A) and light blue (subunit B) for the asymmetric unit and yellow (subunit A) and dark blue (subunit B) for the symmetry mate. The Fo – Fc map (red mesh) is contoured at −3 σ, the Fo – Fc map (green mesh) is contoured at 3 σ, and the 2Fo – Fc map (blue mesh) is contoured at 1.5 σ. One water (A) or CO2 (B) molecule was modeled in the asymmetric unit and the other in the symmetry mate, respectively. The two carbonate (B) or glycerol (C) molecules were placed on the crystallographic axis. The side chains of R98 are shown as sticks. The [Fe4S4] cluster and the water, carbonate, and glycerol moieties are shown in ball-and-stick presentation and colored as follows: Fe, orange; S, yellow; C, gray; O, red. Download FIG S2, JPG file, 2.3 MB (2.4MB, jpg) .
Copyright © 2019 Rettberg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data collection, refinement, and ligand fitting statistics of MaNifH (PDB ID 6NZJ). Shown are the values for the ligand-free model (upper) and the statistics for the models with the plausible ligands CO2, glycerol, and carbonate (lower). Download Table S1, PDF file, 0.1 MB (62KB, pdf) .
Copyright © 2019 Rettberg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Crystal structure of MaNifH with the extra electron density modeled as carbonate (A) and DFT-optimized structure of MaNifH with carbonate (B) or bicarbonate (C) as the substrate. In the crystal structure, the carbonate moiety assumes a position perpendicular to the upper surface of the [Fe4S4] cluster (A). Based on DFT-based structural optimization, however, carbonate is first protonated by R98B, and the resulting bicarbonate assumes a position parallel to the upper surface of the [Fe4S4] cluster (B). The DFT-optimized structure calculated directly with bicarbonate (C) closely resembles that achieved with carbonate upon protonation (B). Overall, carbonate occupies a different position in the crystal structure than (bi)carbonate in the DFT models, which can be best visualized by the overlay of the carbonate-bound crystal structure with the (bi)carbonate-bound DFT models that originate either from the protonation of carbonate (D) or directly from bicarbonate (E). In both cases, the (bi)carbonate moiety swings ∼90° from a position that is perpendicular to one that is parallel to the upper surface of the [Fe4S4] cluster. Furthermore, the (bi)carbonate moiety rotates ∼90° counterclockwise in a horizontal plane parallel to the upper surface of the cluster. The hydrogen atoms of bicarbonate are omitted from the figure, as they cannot be discerned by X-ray crystallography. The [Fe4S4] cluster and carbonate/bicarbonate moieties are shown in ball-and-stick presentations and colored as follows: Fe, orange; S, yellow; C, gray; O, red. The Cys ligands and the conserved Arg residues are shown as sticks. Download FIG S3, JPG file, 1.4 MB (1.4MB, jpg) .
Copyright © 2019 Rettberg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Course of DFT-based structural optimization (TPSS/def2-SVP/def2-TZVP) of the CO2-captured conformation of MaNifH in the [Fe4S4]1+ state (S = 1/2). The CO2 moiety in the DFT-optimized structure is rendered gray, and the CO2 moiety in the crystal structure is colored as follows: C, gray; O, red. Download Movie S1, MPG file, 7.8 MB (8MB, mpg) .
Copyright © 2019 Rettberg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Identities between subunits A and B within AvNifH and MaNifH. (A and B) Percentages of identities between individual α-helices (A) and β-sheets (B) of subunits A and B within AvNifH (black) and MaNifH (red). (C and D) Overall percentages of identities between α-helices (C) and β-sheets (D) of subunits A and B within AvNifH (black) and MaNifH (red). Data from Fig. S5 were used to calculate these percentages. Cα deviations of subunit A versus subunit B: AvNifH, 0.356 Å, and MaNifH, 0.334 Å. Download FIG S4, JPG file, 0.8 MB (849.9KB, jpg) .
Copyright © 2019 Rettberg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Subunits A and B of AvNifH and MaNifH. Shown are comparisons between (A and B) primary sequences and (C and D) secondary structures of subunits A (A and C) and B (B and D) of AvNifH and MaNifH. The identical residues in the primary sequences are indicated with *, and the conserved Cys ligands for the [Fe4S4] cluster are shown in red (A and B). The α-helices are colored green, the β-sheets are colored cyan (C and D), and the primary sequences corresponding to these structural elements are highlighted with the corresponding colors (A and B). Download FIG S5, JPG file, 2.8 MB (2.9MB, jpg) .
Copyright © 2019 Rettberg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Structural comparison between AvNifH and MaNifH. Shown are side (left) and top (middle) views of AvNifH (A) and MaNifH (B), with the alignments of helices CB and CA and the positions of the Arg pairs (located at the tips of helices CB and CA) highlighted (right). The movements of Arg residues in MaNifH (B, right) relative to those in AvNifH are indicated by dashed red arrows. Subunits are shown as ribbons (side view) or cylinders (top view) and colored light orange (subunit A) and light blue (subunit B), respectively. The [Fe4S4] cluster and CO2 moiety are shown in ball-and-stick presentation and colored as follows: Fe, orange; S, yellow; C, gray; O, red. Download FIG S6, JPG file, 2.3 MB (2.3MB, jpg) .
Copyright © 2019 Rettberg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Conformational morphing of AvNifH into MaNifH (side view). Both structures are depicted and colored as described in the legend to Fig. S1. The conformational morphing plugin of PyMol 2.1.1 (https://pymol.org) was used to generate the movie, with AvNifH as the start conformation and MaNifH as the end conformation. Refinement cycles, 3; number of output states, 100; interpolation method, RigiMOL. Download Movie S2, MPG file, 7.9 MB (8.1MB, mpg) .
Copyright © 2019 Rettberg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Conformational morphing of AvNifH into MaNifH (top view). The movie was generated as described in the legend to Movie S2. Download Movie S3, MPG file, 6.6 MB (6.7MB, mpg) .
Copyright © 2019 Rettberg et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The structure of DT-reduced MaNifH (PDB ID 6NZJ) has been deposited in the Protein Data Bank (https://www.wwpdb.org) and will be released upon publication.