Abstract
Adolescent idiopathic scoliosis (AIS) is a complex, three dimensional deformity of the spine that commonly occurs in pubescent girls. Abnormal osteogenic differentiation of mesenchymal stem cells (MSCs) is implicated in the pathogenesis of AIS. However, the biological roles of long noncoding RNAs (lncRNAs) in the regulation of osteogenic differentiation of MSCs are unknown. Through microarray analyses of bone marrow (BM) MSCs from healthy donors and AIS patients, we identified 1483 differentially expressed lncRNAs in AIS BM-MSCs. We defined a novel lncAIS (gene symbol: ENST00000453347) is dramatically downregulated in AIS BM-MSCs. In normal BM-MSCs, lncAIS interacts with NF90 to promote HOXD8 mRNA stability that enhances RUNX2 transcription in BM-MSCs, leading to osteogenic differentiation of normal BM-MSCs. By contrast, lncAIS downregualtion in AIS BM-MSCs cannot recruit NF90 and abrogates HOXD8 mRNA stability, which impedes RUNX2 transcription for osteogenic differentiation. Thereby lncAIS downregualtion in BM-MSCs suppresses osteogenic differentiation that is implicated in the pathogenesis of AIS.
Subject terms: Molecular biology, Cell biology
Introduction
Adolescent idiopathic scoliosis (AIS) is a complex three dimensional deformity of the spine occurring mostly in girls between 10 and 16 years of age during the pubertal growth spurt [1] [2]. The general incidence of AIS is about 2–4% worldwide and ~10% of those diagnosed with AIS need treatment [1]. The main treatment of AIS includes full-time bracing, which may cause back pain along with psychological disorder, and corrective surgery with pedicle screw instrumentation, which inevitably leads to major operative trauma, decreased spinal range of motion, and even permanent catastrophic neurologic or vascular injury in case of screw malposition [3–5]. However, the cause and pathogenesis of AIS remains largely unknown. New therapeutic approaches are likely to come from an improved understanding of the molecular basis of AIS.
AIS patients have abnormal skeletal growth and persistent lower bone mineral density (BMD) compared with sex‑ and age‑matched controls [6, 7]. Reduced bone mass in patients with AIS was first reported by Burner et al. in 1982 [8]. Numerous studies verified that reduced BMD in 27 to 38% of patients with AIS [9–11]. In addition, longitudinal follow-up to skeletal maturity revealed that osteopenia persists in over 80% of girls with AIS [12], suggesting that osteopenia may be a lifelong systematic abnormality of bone metabolism in patients with AIS. Mesenchymal stem cells (MSCs), present in the stroma of all mammalian organs, can differentiate into osteoblasts, adipocytes and chondrocytes [13, 14]. In addition, MSCs are indispensable in both intramembranous and endochondral bone formation [15, 16]. We previously demonstrated that bone marrow (BM) MSCs in AIS patients display decreased osteogenic differentiation ability [17]. Our findings were further confirmed by Park et al.’s study [18]. Given the functional characteristics of MSCs in bone formation and resorption, we thus hypothesized that MSCs is implicated in the pathogenesis of AIS.
Long non-coding RNAs (lncRNAs) are recently recognized as a class of genes, longer than 200 nucleotides (nt) transcripts lacking protein-coding potential [19]. LncRNAs are less conserved in various species but more tissue-specific compared with protein-coding genes. Recent studies revealed that lncRNAs participate in various biological processes, including chromatin modification, transcriptional regulation, imprinting, and nuclear transportation [20]. We recently reported that several lncRNAs are involved in the self-renewal maintenance of liver cancer stem cells [21–23]. Recent studies reported that several lncRNAs modulate their respective nearby protein-coding genes to exert critical roles in the mesendodermal differentiation and heart development [24, 25]. However, the biological roles of lncRNAs in the pathogenesis of AIS are unknown. Here, we show that a novel lncRNA lncAIS (gene symbol: ENST00000453347) is highly expressed in normal BM-MSCs and its downregulation in BM-MSCs is implicated in the pathogenesis of AIS.
Results
LncAIS is downregulated in the BM-MSCs of AIS patients
In order to identify key lncRNAs involved in adolescent idiopathic scoliosis (AIS), we performed microarray analyses in BM-MSCs derived from 5 healthy donors and 12 AIS patients. A total of 1483 lncRNAs (718 upregulated and 765 downregulated) were differentially expressed in normal BM-MSCs versus AIS BM-MSCs (Fig. 1a). Among the top downregulated lncRNAs in AIS BM-MSCs, we focused on an uncharacterized lncRNA that we called lncAIS (gene symbol: ENST00000453347). LncAIS, located on chromosome 1, was composed of four exons with a full-length of 476 nt transcript. Of note, lncAIS was identified as a conserved locus (Fig. 1b). We further verified that lncAIS was significantly downregulated in AIS patient-derived BM-MSCs from 30 samples compared with BM-MSCs from 20 healthy donors (Fig. 1c). Downregulation of lncAIS in AIS BM-MSCs was further validated by Northern blot, with only one transcript in BM-MSCs (Fig. 1d). In addition, lncAIS did not produce any detectable peptides by prediction with coding-potential assessment tool (CPAT) and by in vitro translation assay (Fig. 1e, f). Of note, lncAIS was mainly distributed in the nucleus of human BM-MSCs through cellular fractionation assays (Fig. 1g). Moreover, downregulation of lncAIS in AIS BM-MSCs and nuclear distribution of lncAIS were further validated by RNA fluorescence in situ hybridization (RNA-FISH) (Fig. 1h). Altogether, we reveal that lncAIS is highly expressed in normal human BM-MSCs and dramatically downregulated in AIS patients.
Fig. 1.
LncAIS is downregulated in the BM-MSCs of AIS patients. a Differentially expressed lncRNAs were analyzed by microarray using BM-MSCs from healthy donors versus AIS patients. BM-MSCs were derived from 5 healthy donors and 12 AIS patients. b Location of lncAIS in human genome. LncAIS locates on human chromosome 1, comprising 4 exons. c LncAIS transcript was analyzed in normal BM-MSCs and AIS BM-MSCs by real-time qPCR. BM-MSCs were derived from 20 healthy donors and 30 AIS patients. Primers were listed in Table S1. Relative gene expression folds were normalized to endogenous β-actin and counted as means ± S.D. **P < 0.01. d LncAIS expression in normal BM-MSCs and AIS BM-MSCs were examined by Northern blot. A 217 nt probe of lncAIS (11–228nt) was labeled for northern blot analysis. RNAs were extracted from indicated cells. 18S RNA was used as a loading control. BM-MSCs were derived from 3 healthy donors and 3 AIS patients. e LncAIS displayed no coding potential by CPAT analysis. XIST transcript served as a non-coding gene control. GAPDH and RUNX2 served as coding gene controls. f LncAIS transcript was cloned into pcDNA4-mychisplasmid and transfected into 293 T cells for 48 h. Expression of Myc-fused protein was analyzed by immunoblotting with anti-Myc antibody. KLF4 was used as a coding protein control. g Fractionation of BM-MSCs followed by qPCR. BM-MSCs were lyzed followed by nuclear and cytoplasmic fractionation and RNA extraction for qRT-PCR analysis. ACTIN RNA and GAPDH RNA served as positive controls for cytoplasmic gene expression. U1 RNA served as a positive control for nuclear gene expression. N: nuclear fraction. C: cytoplasmic fraction. Primers were listed in Table S1. The data are from three independent experiments using BM-MSCs derived from 3 healthy donors. h LncAIS was visualized in BM-MSCs by RNA-FISH assays followed with immunofluorescence staining. Red: lncAIS probe; Green: Actin; nuclei were counterstained by DAPI. Scale bar, 20 μm. Probes Sequences were listed in Table S1. More than 100 typical cells were observed
LncAIS knockdown suppresses in vitro osteogenic differentiation and in vivo bone formation
To explore the physiological role of lncAIS in AIS pathogenesis, we silenced lncAIS in healthy normal BM-MSCs using lentivirus-mediated short hairpin RNAs (shRNAs). Knockdown efficiency was confirmed by real-time quantitative PCR (Fig. 2a). We found that lncAIS depletion inhibited proliferation of BM-MSCs compared with scrambled control (shCtrl) treated cells (Fig. 2b). However, lncAIS knockdown did not affect migratory capability of BM-MSCs by wound healing assay (Fig. 2c). Moreover, lncAIS depletion did not significantly alter cell apoptosis compared to shCtrl-treated BM-MSCs (Fig. 2d). Of note, lncAIS depletion remarkably suppressed expression levels of self-renewal related genes in BM-MSCs when cells were cultured in MSC maintenance medium (Fig. 2e). In addition, lncAIS depletion substantially downregulated expression levels of osteogenic differentiation genes when cells were cultured in osteogenic differentiation medium (Fig. 2f). Consequently, lncAIS knockdown dramatically suppressed osteogenic differentiation via alkaline phosphatase (ALP) staining (Fig. 2g), as well as decreased mineral deposition by Von Kossa staining (Fig. 2h).
Fig. 2.
LncAIS knockdown suppresses in vitro osteogenic differentiation and in vivo bone formation. a BM-MSCs were infected with lentivirus expressing shlncAIS and cultured MSC media for 3 days, followed by detection of mRNA levels of lncAIS with real-time qPCR. Primers were listed in Supplementary Table S1. Relative gene expression fold changes were normalized to endogenous β-actin and counted as means ± S.D. **P < 0.01. The data are from three independent experiments using BM-MSCs derived from 3 healthy donors. b Cell proliferation of shlncAIS BM-MSCs compared with shCtrl BM-MSCs were tested by Cell Counting Kit-8 (CCK-8) assay. 1 × 103 human BM-MSCs per well were seeded. CCK-8 reagent was loaded 24 h after seeding. OD450 was measured every 2 days. Absorbance changes were counted as means ± S.D. **P < 0.01. Data are from three independent experiments using BM-MSCs derived from 3 healthy donors. c Cell migration of BM-MSCs was measured by wound healing assay. Data are from three independent experiments using BM-MSCs derived from 3 healthy donors. Scale bar, 10 μm. d Apoptosis of BM-MSCs was analyzed by Hoechst 33342/PI double-staining followed by flow cytometry. Data represent three independent experiments using BM-MSCs derived from 3 healthy donors. e BM-MSCs were cultured in MSC maintenance medium. Expression levels of self-renewal related genes were assessed in indicated BM-MSCs by real-time qPCR. Primers were listed in Table S1. Relative gene expression fold changes were normalized to endogenous β-actin and counted as means ± S.D. **P < 0.01. Data represent three independent experiments using BM-MSCs derived from 3 healthy donors. f BM-MSCs were cultured in OriCell MSC osteogenic differentiation medium for 6 days. Expression levels of osteogenic differentiation genes were assessed in indicated BM-MSCs by real-time qPCR. Primers were listed in Supplementary Table S1. Relative gene expression fold changes were normalized to endogenous β-actin and counted as means ± S.D. **P < 0.01. Data represent three independent experiments using BM-MSCs derived from 3 healthy donors. g ALP staining was performed on day 6 of osteogenic differentiation. Scale bar, 10 μm. h Von kossa staining was performed to detect mineral deposition on day 12. Scale bar, 10 μm. Color intensity of mineral deposition was quantified by ImageJ. Intensity changes were counted as means ± S.D. **P < 0.01. i Indicated BM-MSCs were implanted into NOD/SCID mice. Bone matrix formation was measured by Fast Green staining after 8 weeks of implantation. Black arrow denotes bone formation in upper panel. Osteoid areas were quantified by ImageJ. Positive-stained areas were counted as means ± S.D. **P < 0.01. Bone maturity was measured by Alcian Blue staining. N = 6 mice for each group. BM-MSCs were derived from 3 healthy donors. Scale bar, 50 μm
To further determine the role of lncAIS in vivo, we used an ectopic bone formation model in NOD/SCID mice. shCtrl or shlncAIS-infected BM-MSCs were incubated with hydroxyapatite/tricalcium phosphate (HA/TCP) scaffolds and implanted subcutaneously into NOD/SCID mice for 8 weeks. We observed that shCtrl-infected BM-MSCs could form substantial osteoids in the mouse grafts, whereas shlncAIS-infected BM-MSCs formed less osteoids in the mouse grafts (Fig. 2i). Therefore, lncAIS knockdown inhibits osteogenic differentiation in vitro and ectopic bone formation in vivo.
LncAIS overexpression promotes osteogenic differentiation and bone formation
We next overexpressed lncAIS in normal BM-MSCs by lentivirus. LncAIS overexpression dramatically increased cell proliferation of normal BM-MSCs (Fig. 3a). Consistently, lncAIS overexpression enhanced expression levels of self-renewal related genes and osteogenic differentiation genes (Fig. 3b, c). In addition, lncAIS overexpression also augmented protein levels of the osteogenic marker genes such as osteopontin (OPN), COL1A1 and IBSP compared with vector-transfected BM-MSCs during osteogenic differentiation (Fig. 3d). Consequently, lncAIS overexpression enhanced osteogenic differentiation of BM-MSCs by ALP staining (Fig. 3e) and Von Kossa staining (Fig. 3f). Finally, lncAIS-overexpressed BM-MSCs remarkably increased quantity and size of osteoids in the mouse grafts through in vivo ectopic bone formation model in NOD/SCID mice (Fig. 3g).
Fig. 3.
LncAIS overexpression promotes osteogenic differentiation and bone formation. a BM-MSCs from healthy donors (Normal) were infected with lentivirus expressing lncAIS and cultured in MSC media for 3 days, followed by detection of cell proliferation with CCK-8 kit as in Fig. 2b. Absorbance changes were counted as means ± S.D. **P < 0.01. Data are from three independent experiments using BM-MSCs derived from 3 healthy donors. b Indicated BM-MSCs were cultured as in Fig. 2e. Expression levels of self-renewal related genes were assessed by real-time qPCR. c Indicated BM-MSCs were cultured as in Fig. 2f. Expression levels of osteogenic differentiation genes were assessed by real-time qPCR. Data are from three independent experiments using BM-MSCs derived from 3 healthy donors. d Western blot analysis of osteoblast marker gene expression in indicated cells on day 6 of osteogenic differentiation. Data are from three independent experiments using BM-MSCs derived from 3 healthy donors. e Osteogenic differentiation was induced as in Fig. 2f. ALP staining was performed on day 6 of osteogenic differentiation. Data are from three independent experiments using BM-MSCs derived from 3 healthy donors. Scale bar, 10 μm. f Osteogenic differentiation was induced as in Fig. 2f. Von Kossa staining was performed to detect mineral deposition on day 12. Data are from three independent experiments using BM-MSCs derived from 3 healthy donors. Scale bar, 10 μm. Color intensity of mineral deposition was quantified by ImageJ. Intensity changes were counted as means ± S.D. **P < 0.01. g Ectopic bone formation in vivo was induced as in Fig. 2i. Bone matrix was measured by Fast green staining after 8 weeks of implantation. Black arrow denotes bone formation in upper panel. Osteoid areas were quantified by ImageJ. Positive-stained areas were counted as means ± S.D. **P < 0.01. Bone maturity was measured by Alcian Blue staining. N = 6 mice for each group. BM-MSCs were derived from 3 healthy donors. Scale bar, 50 μm. h BM-MSCs from healthy donors (Normal) or AIS patients (AIS) were infected with lentivirus expressing lncAIS (oelncAIS) or vector control (Vec) and cultured in MSC media for 3 days, followed by detection of expression levels of self-renewal related genes as in Fig. 2e. i BM-MSCs from healthy donors (Normal) or AIS patients (AIS) were infected with lentivirus expressing lncAIS (oelncAIS) or vector control (Vec) and cultured in MSC media for 3 days, followed by detection of expression levels of osteogenic differentiation genes as in Fig. 2f. Data are from three independent experiments using BM-MSCs derived from 3 healthy donors. j BM-MSCs from healthy donors (Normal) or AIS patients (AIS) were infected with lentivirus expressing lncAIS (oelncAIS) or vector control (Vec) and cultured in MSC media for 3 days. Osteogenic differentiation was induced as in Fig. 2f. ALP staining was performed on day 6 of osteogenic differentiation. k BM-MSCs from healthy donors (Normal) or AIS patients (AIS) were infected with lentivirus expressing lncAIS (oelncAIS) or vector control (Vec) and cultured in MSC media for 3 days. Osteogenic differentiation was induced as in Fig. 2f. Von Kossa staining was performed to indicate mineral deposition on day 12. Data are from three independent experiments using BM-MSCs derived from 3 healthy donors. Scale bar, 10 μm
In order to determine the role of lncAIS in the pathologic process of AIS, we enforced lncAIS expression in AIS BM-MSCs by lentivirus infection. Of note, overexpression of lncAIS in AIS BM-MSCs could also rescue the expression levels of self-renewal related genes and osteogenic differentiation genes to normal levels (Fig. 3h, i). Importantly, enforced lncAIS expression in AIS BM-MSCs restored osteogenic differentiation capacity as well (Fig. 3j, k). Collectively, lncAIS promotes BM-MSCs to drive osteogenic differentiation.
LncAIS activates HOXD8 transcription
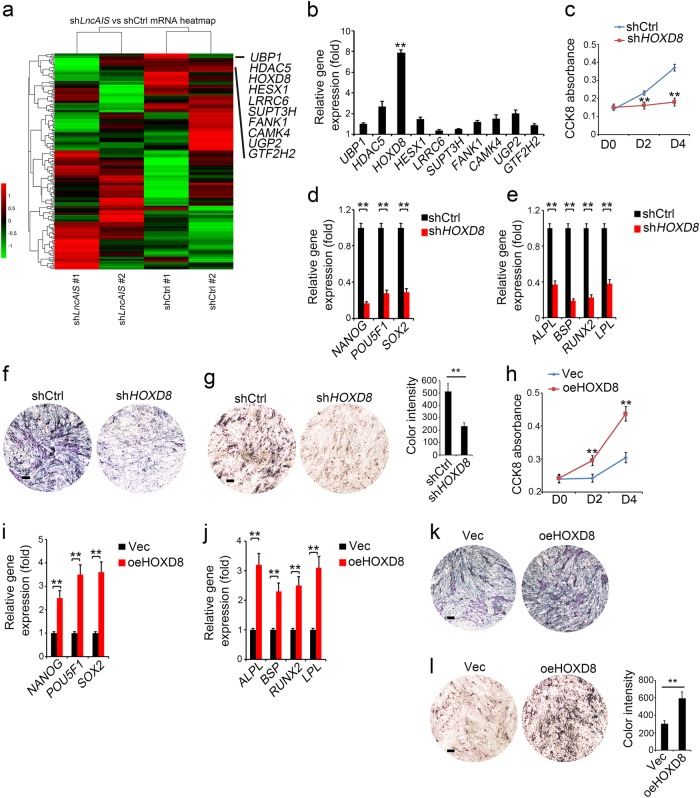
We next performed transcriptome microarray analysis between lncAIS-silenced and shCtrl-treated BM-MSCs. We chose top ten downregulated transcription factors (TFs) in lncAIS-silenced BM-MSCs (Fig. 4a), and detected their expression levels in normal BM-MSCs. Of note, HOXD8, acting as a BM-MSC marker gene, was most highly expressed in BM-MSCs (Fig. 4b). However, the role of HOXD8 in regulating osteogenic differentiation of BM-MSCs has not been characterized. We then depleted HOXD8 in normal BM-MSCs by lentivirus-mediated shRNAs. We observed that HOXD8 depletion inhibited proliferation of BM-MSCs compared with shCtrl treated cells (Fig. 4c). Moreover, HOXD8 depletion also suppressed expression levels of self-renewal related genes and osteogenic differentiation genes in BM-MSCs (Fig. 4d, e). Repressed osteogenic differentiation by HOXD8 depletion was further validated by ALP staining (Fig. 4f) and by Von Kossa staining (Fig. 4g).
Fig. 4.
LncAIS activates HOXD8 transcription. a Heat map of transcriptome microarray analysis between shlncAIS and shCtrl treated BM-MSCs. Top ten downregulated TFs in shlncAIS BM-MSCs were listed. Data are from two independent experiments using BM-MSCs derived from 2 healthy donors. Each matched shCtrl and shLncAIS was derived from the same healthy BM-MSC donor. b HOXD8 was most highly expressed in BM-MSCs among the top 10 downregulated TFs by real-time qPCR analysis. Data are from three independent experiments using BM-MSCs derived from 3 healthy donors. c BM-MSCs were infected with lentivirus expressing shHOXD8 and cultured in MSC media for 3 days, followed by detection of cell proliferation with CCK-8 kit as in Fig. 2b. Absorbance changes were counted as means ± S.D. **P < 0.01. Data are from three independent experiments using BM-MSCs derived from 3 healthy donors. d Indicated BM-MSCs were cultured as in Fig. 2e. Expression levels of self-renewal related genes were assessed by real-time qPCR. e Indicated BM-MSCs were cultured as in Fig. 2f. Expression levels of osteogenic differentiation genes were assessed by real-time qPCR. Data are from three independent experiments using BM-MSCs derived from 3 healthy donors. f Osteogenic differentiation was induced as in Fig. 2f. ALP staining was performed on day 6 of osteogenic differentiation. g Osteogenic differentiation was induced as in Fig. 2f. Von Kossa staining was performed to indicate mineral deposition on day 12. Data are from three independent experiments using BM-MSCs derived from 3 healthy donors. Scale bar, 10 μm. Color intensity of mineral deposition was quantified by ImageJ. Intensity changes were counted as means ± S.D. **P < 0.01. h BM-MSCs were infected with lentivirus overexpressing HOXD8 (oeHOXD8) and cultured in MSC media for 3 days, followed by detection of cell proliferation with CCK-8 kit as in Fig. 2b. Data are from three independent experiments using BM-MSCs derived from 3 healthy donors. i Indicated BM-MSCs were cultured as in Fig. 2e. Expression levels of self-renewal related genes were assessed by real-time qPCR. j Indicated BM-MSCs were cultured as in Fig. 2f. Expression levels of osteogenic differentiation genes were assessed by real-time qPCR. Data are from three independent experiments using BM-MSCs derived from 3 healthy donors. k Osteogenic differentiation was induced as in Fig. 2f. ALP staining was performed on day 6 of osteogenic differentiation. l Osteogenic differentiation was induced as in Fig. 2f. Von Kossa staining was performed to indicate mineral deposition on day 12. Data are from three independent experiments using BM-MSCs derived from 3 healthy donors. Scale bar, 10 μm. Color intensity of mineral deposition was quantified by ImageJ. Intensity changes were counted as means ± S.D. **P < 0.01
In addition, HOXD8 overexpression dramatically increased cell proliferation rate in normal BM-MSCs (Fig. 4h). Consistently, HOXD8 overexpression promoted expression levels of self-renewal related genes and osteogenic differentiation genes in BM-MSCs (Fig. 4i, j). Consequently, HOXD8 overexpression enhanced osteogenic differentiation of BM-MSCs by ALP staining (Fig. 4k) and Von Kossa staining (Fig. 4l). Altogether, these data suggest that HOXD8 is required for osteogenic differentiation of BM-MSCs.
LncAIS interacts with NF90 to enhance the mRNA stability of HOXD8
LncRNAs are usually positively correlated with regulation of their nearby protein-coding genes [22, 23]. However, we found that depletion of lncAIS in BM-MSCs did not affect the expression levels of its neighboring genes (Fig. 5a), suggesting lncAIS may exert its regulatory roles in trans. We then performed a Biotin-labeled RNA pulldown assay to identify potential lncAIS-associated proteins from BM-MSC lysates. NF90 was identified to bind lncAIS in BM-MSCs (Fig. 5b). NF90, a protein of interleukin enhancer binding factor 3 (ILF3) family, is an RNA-binding protein to regulate gene expression or to stabilize mRNAs [26, 27]. The interaction of lncAIS with NF90 was further confirmed by RNA pulldown (Fig. 5c), and RNA-immunoprecipitation (RIP) assay (Fig. 5d). Moreover, lncAIS was co-localized with NF90 in the nuclei of BM-MSCs (Fig. 5e). These results indicate that lncAIS associates with NF90 protein in BM-MSCs.
Fig. 5.
LncAIS interacts with NF90 to enhance the mRNA stability of HOXD8. a LncAIS depletion did not alter the expression of neighboring genes assessed by RT-qPCR. Relative gene expression fold changes were calculated as means ± S.D. Primers were listed in Table S1. Data are from three independent experiments using BM-MSCs derived from 3 healthy donors. b Biotin-RNA pulldowns were performed with lysates of BM-MSCs using full-length lncAIS transcript (Sense) and antisense sequence control followed by mass spectrometry. c The interaction of lncAIS with NF90 was verified by RNA pulldown assay, followed with immunoblot analysis by anti-NF90 antibody. Data are from three independent experiments using BM-MSCs derived from 3 healthy donors. d The interaction of lncAIS with NF90 was verified by immunoprecipitation (RIP) assay. BM-MSC lysates were incubated with anti-NF90 antibody, followed by RIP assay. RNA was extracted and reversely transcribed. LncAIS transcript was analyzed by real-time qPCR. Primers were listed in Table S1. Data are from three independent experiments using BM-MSCs derived from 3 healthy donors. e BM-MSCs were probed with lncAIS by RNA-FISH, followed by immunofluorescence staining for NF90. Red: lncAIS probe; Green: NF90; Nuclei were counterstained by DAPI. Scale bar, 50 μm. More than 100 typical cells were observed. Data are from three independent experiments using BM-MSCs derived from 3 healthy donors. f BM-MSCs were infected with lentivirus expressing shNF90 and cultured in MSC media for 3 days. Indicated BM-MSCs were cultured as in Fig. 2e. Expression levels of self-renewal related genes were assessed by real-time qPCR. g Indicated BM-MSCs were cultured as in Fig. 2f. Expression levels of osteogenic differentiation genes were assessed by real-time qPCR. Data are from three independent experiments using BM-MSCs derived from 3 healthy donors. h Osteogenic differentiation was induced as in Fig. 2f. ALP staining was performed on day 6 of osteogenic differentiation. i Osteogenic differentiation was induced as in Fig. 2f. Von Kossa staining was performed to indicate mineral deposition on day 12. Data are from three independent experiments using BM-MSCs derived from 3 healthy donors. Scale bar, 10 μm. Color intensity of mineral deposition was quantified by ImageJ. Intensity changes were counted as means ± S.D. **P < 0.01. j Interactions of indicated mRNA with NF90 in BM-MSCs were analyzed by RIP assay. BM-MSC lysates were incubated with anti-NF90 antibody, followed by RNA immunoprecipitation and real-time qPCR. Primers were listed in Table S1. Data are from three independent experiments using BM-MSCs derived from 3 healthy donors. k The interaction of HOXD8 mRNA with NF90 in BM-MSCs was verified by RIP assay. BM-MSC lysates were incubated with anti-NF90 antibody, assayed as in (k). Data are from three independent experiments using BM-MSCs derived from 3 healthy donors. l HOXD8 mRNA stability from indicated BM-MSCs were measured by real-time qPCR at indicated time points after Act D treatment. Data were shown as means ± S.D. **P < 0.01. Data are from three independent experiments using BM-MSCs derived from 3 healthy donors. NS no significance
How NF90 regulates osteogenic differentiation and AIS pathogenesis has not been defined yet. We then depleted NF90 in normal BM-MSCs by lentivirus-mediated shRNAs. We observed that NF90 depletion inhibited expression levels of self-renewal related genes and osteogenic differentiation genes in BM-MSCs (Fig. 5f, g). Moreover, repressed osteogenic differentiation was further validated by ALP staining (Fig. 5h) and Von Kossa staining (Fig. 5i). These data suggest that NF90 is required for osteogenic differentiation of BM-MSCs.
It has been reported that NF90 interacts with the 3’-UTR region of PARP1 mRNA to maintain its mRNA stability [27]. Notably, among the top ten downregulated transcription factors in lncAIS silenced BM-MSCs (Fig. 4a), NF90 specifically bound to HOXD8 mRNA (Fig. 5J). In addition, lncAIS depletion in BM-MSCs abrogated the interaction of NF90 with the 3’-UTR region of HOXD8 mRNA (Fig. 5k) and consequently caused HOXD8 mRNA decay (Fig. 5l). Consistently, the interaction of NF90 with the 3’-UTR region of HOXD8 mRNA was undetectable in AIS BM-MSCs (Fig. 5k), and abrogated the stability of HOXD8 mRNA as well (Fig. 5l). We conclude that lncAIS interacts with NF90 that maintains HOXD8 mRNA stability in normal BM-MSCs.
HOXD8 initiates RUNX2 expression to drive osteogenic differentiation
We wanted to explore the mechanism by which lncAIS regulated osteogenic differentiation of BM-MSCs through HOXD8. RUNX2 is a key transcription factor for osteogenic differentiation [28]. From our above data, we noticed that lncAIS knockdown significantly decreased RUNX2 expression (Fig. 2f). Intriguingly, HOXD8, RNA polymerase II and transcription active marker H3K4me3 were all enriched onto the same region of RUNX2 gene (Fig. 6a). In addition, depletion of HOXD8 suppressed RUNX2 expression in BM-MSCs (Fig. 6b). By contrast, overexpression of HOXD8 in HOXD8 silenced BM-MSCs could restore RUNX2 expression (Fig. 6b). These results suggest that the RUNX2 gene is a downstream target for HOXD8.
Fig. 6.
HOXD8 initiates RUNX2 expression to drive osteogenic differentiation. a HOXD8 together with RNA polymerase II (Pol II) and H3K4me3 were highly enriched on RUNX2 promoter by ChIP assays. Data are from three independent experiments using BM-MSCs derived from 3 healthy donors. b BM-MSCs were infected with indicated lentivirus and RUNX2 expression was assessed by qPCR. Data are from three independent experiments using BM-MSCs derived from 3 healthy donors. c Normal BM-MSCs were infected with indicated lentivirus and cultured in MSC media for 3 days. Indicated BM-MSCs were cultured as in Fig. 2e. Expression levels of self-renewal related genes were assessed by real-time qPCR. Data are from three independent experiments using BM-MSCs derived from 3 healthy donors. d Normal BM-MSCs were infected with indicated lentivirus and cultured in MSC media for 3 days. Indicated BM-MSCs were cultured as in Fig. 2f. Expression levels of osteogenic differentiation genes were assessed by real-time qPCR. Data are from three independent experiments using BM-MSCs derived from 3 healthy donors. e Normal BM-MSCs were infected with indicated lentivirus and cultured in MSC media for 3 days. Osteogenic differentiation was induced as in Fig. 2f. ALP staining was performed on day 6 of osteogenic differentiation. f Normal BM-MSCs were infected with indicated lentivirus and cultured in MSC media for 3 days. Osteogenic differentiation was induced as in Fig. 2f. Von Kossa staining was performed to indicate mineral deposition on day 12. Data are from three independent experiments using BM-MSCs derived from 3 healthy donors. Scale bar, 10 μm. Color intensity of mineral deposition was quantified by Image J. Intensity changes were counted as means ± S.D. **P < 0.01. g Indicated BM-MSCs were infected with lentivirus and cultured as in Fig. 2f. ALP staining was performed on day 6 of osteogenic differentiation. h Indicated BM-MSCs were infected with lentivirus and cultured as in Fig. 2f. Von Kossa staining was performed to indicate mineral deposition on day 12. Data are from three independent experiments using BM-MSCs derived from 3 healthy donors. Scale bar, 10 μm. Color intensity of mineral deposition was quantified by Image J. Intensity changes were counted as means ± S.D. **P < 0.01
Of note, restoration of either HOXD8 or RUNX2 in lncAIS-silenced BM-MSCs could well rescue expression levels of self-renewal related genes and osteogenic differentiation genes (Fig. 6c, d). In addition, restoration of either HOXD8 or RUNX2 in lncAIS-silenced BM-MSCs could well recover osteogenic differentiation capacity to normal levels (Fig. 6e, f). Importantly, enforced expression of either HOXD8 or RUNX2 in AIS BM-MSCs could also restore osteogenic differentiation capacity to the normal level (Fig. 6g, h). Taken together, lncAIS interacts with NF90 to promote HOXD8 mRNA stability that enhances RUNX2 transcription in BM-MSCs, leading to osteogenic differentiation of normal BM-MSCs. By contrast, lncAIS downregualtion in AIS BM-MSCs cannot recruit NF90 to promote HOXD8 mRNA stability, which impedes RUNX2 transcription for osteogenic differentiation.
Discussion
AIS is a complex, three dimensional deformity of the spine that commonly occurs in pubescent girls. We and others showed that MSCs in AIS patients display decreased osteogenic differentiation ability [17, 18, 29, 30]. Thus abnormal osteogenic differentiation of MSCs is implicated in the pathogenesis of AIS. In this study, we performed microarray analyses in BM-MSCs from 5 healthy donors and 12 AIS patients and identified 1483 differentially expressed lncRNAs in AIS BM-MSCs. We defined a novel lncAIS (gene symbol: ENST00000453347) is dramatically downregulated in AIS BM-MSCs. In normal BM-MSCs, lncAIS interacts with NF90 to promote HOXD8 mRNA stability that enhances RUNX2 transcription in BM-MSCs, leading to osteogenic differentiation of normal BM-MSCs. By contrast, lncAIS downregualtion in AIS BM-MSCs cannot recruit NF90 and abrogates HOXD8 mRNA stability, which impedes RUNX2 transcription for osteogenic differentiation. Thus lncAIS downregualtion in BM-MSCs suppresses osteogenic differentiation that is implicated in the pathogenesis of AIS.
Around one-third AIS patients are associated with poor bone mineral density (BMD) [31, 32]. Low BMD has been reported to be a key prognostic factor of curve progression in girls with AIS [11, 33, 34]. It has been considered that osteopenia may be a primary contributing factor to the spinal deformity of AIS. We and others showed that MSCs from AIS patients undergo abnormal differentiation during development of osteoblasts, chondrocytes and adipocytes [17, 18, 29, 30], suggesting that abnormal osteogenic differentiation of MSCs is implicated in the pathogenesis of AIS. However, how MSCs is abnormally regulated in the patients with AIS remains elusive.
LncRNAs have been reported to play widespread roles in gene regulation and other cellular processes [19, 35]. LncRNAs exert their functions via diverse mechanisms, including cotranscriptional regulation, modulation of gene expression, scaffolding of nuclear or cytoplasmic complexes, and pairing with other RNAs [36]. We previously demonstrated that peripheral blood of AIS patients manifests differentially expressed 139 lncRNAs and 546 mRNAs compared to healthy controls [37]. In this study we showed that AIS BM-MSCs harbor 1483 differentially expressed lncRNAs, with 718 upregulated and 765 downregulated. We found that lncAIS downregualtion cannot recruit NF90 and consequently inhibits RUNX2 expression in AIS BM-MSCs, which suppresses osteogenic differentiation leading to osteopenia. We firstly revealed that lncAIS is implicated in the pathogenesis of AIS.
Of note, lncRNAs can function in cis in regulating expression of neighboring genes or in trans through various modes [38]. Here we showed that lncAIS depletion in normal BM-MSCs does not change the expression levels of its neighboring genes, indicating lncAIS exerts its regulatory role in trans, but not in cis. Intriguingly, lncAIS associates with a RNA binding protein NF90. ILF3 family members, including NF90 and NF110, were originally identified from a protein complex that regulates interleukin-2 (IL-2) transcription [39, 40], which are the products of differential splicing of ILF3 gene. NF90 and NF110 proteins are identical at their N-terminal and central regions but different at their C-terminal regions [41]. Through the ability to bind both DNA and RNA, ILF3 family members can regulate transcription [40, 42, 43], translation [44], mRNA stability [45, 46] and primary microRNA processing [47]. Herein we demonstrated that NF90 specifically binds to HOXD8 mRNA and sustains its stability in normal BM-MSCs. Consequently, HOXD8 protein can initiate RUNX2 transcription that drives MSCs into osteogenic differentiation.
Homeobox genes encode homeodomain-containing transcription factors that determine the positional identity along the anterior-posterior body axis of animal embryos [48]. They are widely expressed in the adult tissues [49, 50]. In humans, 39 known HOX transcription factors reside in four separate clusters HOXA to D, which are located on four different chromosomes. HOX genes modulate cell differentiation during embryonic development in many different lineages and developmental pathways [51]. Ackema et al. reported that mouse MSCs from different organs are characterized by distinct topographic HOX codes [52]. Chang et al. revealed that fibroblasts from different anatomic sites across the human body express distinct HOX patterns [53]. These studies suggest that the typical HOX code of a cell reflects the specific expression of functionally active HOX genes in distinct tissues. Herein we showed that HOXD8 gene is highly expressed in human BM-MSCs, whose mRNA stability is modulated by NF90 recruited by lncAIS. HOXD8 protein can trigger RUNX2 expression for osteogenic differentiation of normal MSCs. RUNX2 is a master transcription modulator of osteoblasts differentiation, which plays a fundamental role in modulating osteoblasts maturation and homeostasis [54, 55]. Runx2-deficient mice display weakly calcified bone without detectable osteoblasts [56]. It has been reported that RUNX2 promotes bone formation through upregulation of expression of bone matrix formation, including type I collagen, osteopontin, osteocalcin, bone sialoprotein and fibronectin [57, 58]. A recent study showed that there are positive correlations between low RUNX2 expression and BMD of lumbar spine and femoral neck of AIS patients. [59] We found that RUNX2 is a downstream target of HOXD8 in normal BM-MSCs that drives osteogenic differentiation. By contrast, in AIS BM-MSCs, lncAIS downregualtion fails to recruit NF90 and abrogates HOXD8 mRNA stability, which impedes RUNX2 transcription for osteogenic differentiation.
In summary, lncAIS plays a critical role in the regulation of osteogenic differentiation of BM-MSCs and downregulation of lncAIS is involved in the pathogenesis of AIS. Our findings reveal that lncRNAs may represent an additional layer of regulation of AIS pathogenesis that can be act as important biomarkers for AIS. Therefore transplantation of MSCs with lncAIS overexpression may provide a potential therapy for AIS treatment.
Materials and Methods
Antibodies and reagents
Antibody against H3K4me3 (9751) was purchased from Cell Signaling Technology (Danvers, USA). Antibodies against human HOXD8 (ab228450) and NF90 (ab89100) were purchased from Abcam (Cambridge, USA). Antibody against human RNA-polymerase II (clone 4H8) was purchased from Active Motif (Carlsbad, USA). Antibody against β-actin (clone AC-74) was from Sigma-Aldrich (St. Louis, USA). Antibody against Myc (clone 9E10) was from Santa Cruz Biotechnology (Santa Cruz, USA). Antibodies against COL1A1 (BA0325), IBSP (BA2329) and OPN (PA1432) were from Boster (Wuhan, China). Secondary antibodies conjugated with Alexa-594 were purchased from Molecular probes Inc (Eugene, USA). Streptavidin beads were from Sigma-Aldrich (St. Louis, USA). Protein A/G beads were from Santa Cruz Biotechnology (Santa Cruz, USA). Alkaline phosphatase detection kit was purchased from Millipore (Millerica, USA). SuperReal premix plus qPCR buffer was from TIANGEN Biotech (Beijing, China). OriCell BM-MSC osteogenic differentiation Kit was from Cyagen (HUXMA-90021, China). Von Kossa staining kit was from Genmed Scientifics (Shanghai, China). Cell counting Kit-8 (CCK-8) was from Dojindo (Kumamoto, Japan). Fast Green staining kit and Alcian Blue staining kit were from Xinhualvyuan Biotechnology (Beijing, China).
Patients and specimens
Bone marrow (BM) aspirates were obtained from 42 AIS patients (mean age 14.5 years, range 12–17), and 25 healthy donors (mean age 14.9 years, range 12–17). In the AIS group, all of the patients underwent full clinical and radiological examinations to rule out other causes of scoliosis and to ascertain the diagnosis of AIS. In the control group, each of the 25 age- and gender-matched subjects had a straight spine and a normal forward bending test on the physical examination. They were confirmed to be free of any associated medical diseases or spinal deformities when entered to the study. The study was approved by the Ethics Committee of Chinese Academy of Medical Sciences and Peking Union Medical College Hospital. Written informed consents were obtained from all subjects and their parents before entering the study.
Cell isolation, culture and osteogenic differentiation assay
Human bone marrow tissues were collected from AIS patients and healthy donors. All experiments followed the procedures approved by the Ethics Committee of Chinese Academy of Medical Sciences and Peking Union Medical College Hospital. Human BM-MSCs were isolated and cultured as previously described. [29] Human 293 T cells were cultured with DMEM supplemented with 10% FBS and 100 U/ml penicillin and 100 mg/ml streptomycin. Lentivirus was produced in 293 T cells using the standard protocols. Transfection was performed using lipofectin (Invitrogen). For shRNA knockdown and overexpression experiments, target sequences were constructed into pSicoR plasmid. Lentivirus was produced by 293 T cells. Most efficient shRNA among 3 shRNA constructs was screened for following experiments. Three independent knockdown cell lines were used for biological replicates in each assay. At least four independent experiments were performed as biological replicates. shRNA sequences were listed in Table S2. To induce osteogenic differentiation, third-passage BM-MSCs were seeded in six-well plates and treated with osteogenesis induction medium according to the manufacturer’s protocol. The medium was changed every 3 days.
Wound healing assay
Dishes were coated with 0.1% gelatin (v/v) for 1 h at 37 °C. 1 × 106 BM-MSC cells were plated to create a confluent monolayer. Cells were cultured to adhere and spread completely. Wound was created by manually scraping the cell monolayer with a p200 pipet tip. First images were acquired using reference point marks. Cells were cultured in a tissue culture incubator for 24 h. Second images were acquired by matching the photographed region of first image.
Coding potential analysis
Coding potential of lncAIS was analyzed by Coding-Potential Assessment Tool (CPAT) at the website of http://lilab.research.bcm.edu/cpat/ according to the manufacturer’s instructions. XIST transcript served as a non-coding gene control. GAPDH and RUNX2 served as coding gene controls.
Cellular fractionation assay
BM-MSCs were lyzed followed by nuclear and cytoplasmic fractionation using NE-PER Nuclear and cytoplasmic Extraction Kit (Pierce) according to the manufacturer’s instructions. RNA was extracted using TRIzol Reagent (Invitrogen), followed by purification with an RNeasy kit (Qiagen, Valencia, CA, USA). Reverse transcription was performed by M-MLV reverse transcriptase (Promega) and qRT-PCR analysis. ACTIN RNA and GAPDH RNA served as positive controls for cytoplasmic gene expression. U1 RNA served as a positive control for nuclear gene expression.
ALP and Von Kossa staining
ALP staining was monitored using an ALP staining kit according to the manufacturer’s protocol. Mineral deposition was monitored using a Von Kossa staining kit according to the manufacturer’s protocol. Images were obtained with Nikon EclipseTi microscope (Nikon, Japan). Color intensity of mineral deposition was quantified by ImageJ.
Northern blot
Total RNA was extracted from BM-MSCs with TRIzol. 10 μg RNA from each sample was subjected to formaldehyde denaturing agarose electrophoresis followed by transferring to positively charged NC film with 20 × SSC buffer (3.0 M NaCl and 0.3 M sodium citrate, pH 7.0). Membrane was UV cross-linked and incubated with biotin-labeled RNA probes (lncAIS 11–228 nt) generated by in vitro transcription. Biotin signals were detected with HRP-conjugated streptavidin for the Cheniluminescent Nucleic Acid Detection Module according to the manufacturer’s instruction [60].
RNA FISH
Fluorescence-conjugated lncAIS probes were generated according to protocols from Biosearch Technologies. LncAIS probe sets for RNA FISH were listed in Table S1. BM-MSCs were hybridized with DNA probe sets then stained with indicated antibodies. Images were obtained with Olympus FV1200 laser scanning confocal microscopy (Olympus, Japan).
Microarray analysis
RNA was extracted from BM MSCs using TRIzol Reagent (Invitrogen), according to the manufacturer’s instructions, followed by purification with an RNeasy kit (Qiagen, Valencia, CA, USA). cDNA was generated using One-Cycle Target Labeling and Control Reagents (Affymetrix, SantaClara, CA, USA), and cRNA was created with a GeneChip WT Labeling Kit (Affymetrix, Santa Clara, CA, USA). Biotin-labeled, fragmented (≤200nt) cRNA was hybridized for 16 h at 45 °C to Affymetrix GeneChip Human transcript array 2.0 (Affymetrix). GeneChips were washed and stained in the Affymetrix Fluidics Station 450. GeneChips were scanned by using Affymetrix® GeneChip Command Console (AGCC) which installed in GeneChip® Scanner 3000 7 G. The data were analyzed with Robust Multichip Analysis (RMA) algorithm using Affymetrix default analysis settings and global scaling as normalization method. Values presented are log2 RMA signal intensity. Microarray data were deposited in GEO with an accession number (GSE110359).
RNA pulldown assay
Biotin-labeled lncAIS full-length (sense) and antisense RNA were obtained with Biotin RNA labeling Mix (Roche) in vitro followed by incubation with nuclear extracts separated from BM-MSCs. RNA-binding proteins were pulled down by streptavidin beads. Pulldown components were separated with SDS-PAGE followed by silver staining. Differential bands enriched by lncAIS were analyzed by LTQ Orbitrap XL mass spectrometry or immunobloted with indicated antibodies.
RNA immunoprecipitation (RIP) assay
BM-MSCs were treated with 1% formaldehyde then dissolved with RNase-free RIPA buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.5% sodium desoxycholate, 0.1% SDS, 5 mM EDTA, 2 mM PMSF, 20 mg/ml aprotinin, 20 mg/ml leupeptin, 10 mg/ml pepstatin A, 150 mM benzamidine, 1% Nonidet P-40 and RNase inhibitors). Samples were sonicated on ice and centrifuged. Supernatants were pre-cleared and incubated with indicated antibodies, followed by protein A/G beads immunoprecipitation. Total RNA was extracted from eluents. LncAIS enrichment was analyzed by qPCR. Primers were listed in Table S1.
Chromatin immunoprecipitation (ChIP) assay
Quantitative ChIP was performed according to a standard protocol (Upstate). Sheared chromatin (sonicated to 200–500 bp) from BM-MSCs (2 × 106) fixed in 1% formaldehyde was incubated with 4 μg antibody overnight at 4 °C followed by immunoprecipitation with salmon sperm DNA/protein agarose beads. After washing, elution and cross-link reversal, DNA from each ChIP sample and corresponding input sample were purified and analyzed using qPCR. Primers were listed in Table S1.
Ectopic bone formation in vivo
A total of 2 × 106 human BM-MSCs were incubated with about 100 mg wetted HA/TCP ceramic powder (National Engineering Research Center for Biomaterials, Chengdu, China) at 37 °C overnight. Cells were implanted subcutaneously into the dorsal surface of 8-week-old NOD/SCID mice. Implants were harvested after 8 weeks, fixed in 4% paraformaldehyde, decalcified in 10% EDTA, embedded in paraffin followed by section and staining. Osteoid tissues were dyed green by fast green staining. Cartilage tissues were dyed blue by Alcian blue staining to indicate bone maturity.
Statistical analysis
An unpaired Student’s t test was used as statistical analysis in this study. Statistical calculations were performed by using Microsoft Excel or SPSS 13. P values were significant when P < 0.05.
Electronic supplementary material
Acknowledgements
We thank Jing Li (Cnkingbio Company Ltd, Beijing, China) for technical support. We thank Peng Xue, Xiang Ding, Yan Teng, Yihui Xu, Junying Jia, Xudong Zhao and Xiaofei Guo for technical support. This work was supported by the National Natural Science Foundation of China (81272054, 81171673, 91640203), Beijing Talent Fund (2015000021223ZK27), Beijing Nova program Grant (2014A019), Beijing High-level Innovative Entrepreneurial Talent Fund, Beijing Natural Science Foundation (7181006). Peking Union Medical College Youth Fund, PUMC Nova program Grant of Chinese academy of medical sciences.
Author contributions
QZ, BY and SH designed and performed experiments; QZ and BY analyzed the data and wrote the paper; YD, RZ, JL, ZW, NL, YZ, HL, SW, and Y.Y. analyzed the data; JL, SL, and HZ performed some experiments; ZF organized, designed, and wrote the paper, and GQ and JZ initiated the study and analyzed the data.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by R. De Maria
These authors contributed equally: Qianyu Zhuang, Buqing Ye, Shangyi Hui.
These authors contributed equally: Zusen Fan, Guixing Qiu, Jianguo Zhang.
Contributor Information
Zusen Fan, Email: fanz@moon.ibp.ac.cn.
Jianguo Zhang, Email: jgzhang_pumch@yahoo.com.
Electronic supplementary material
The online version of this article (10.1038/s41418-018-0240-2) contains supplementary material, which is available to authorized users.
References
- 1.Weinstein SL, Dolan LA, Cheng JC, Danielsson A, Morcuende JA. Adolescent idiopathic scoliosis. Lancet. 2008;371:1527–37. doi: 10.1016/S0140-6736(08)60658-3. [DOI] [PubMed] [Google Scholar]
- 2.Cheng JC, Castelein RM, Chu WC, Danielsson AJ, Dobbs MB, Grivas TB, et al. Adolescent idiopathic scoliosis. Nat Rev Dis Prim. 2015;1:30. doi: 10.1038/nrdp.2015.30. [DOI] [PubMed] [Google Scholar]
- 3.Lou EH, Hill DL, Raso JV, Moreau M, Hedden D. How quantity and quality of brace wear affect the brace treatment outcomes for AIS. Eur Spine J. 2016;25:495–9. doi: 10.1007/s00586-015-4233-2. [DOI] [PubMed] [Google Scholar]
- 4.Sun W, Zhou J, Sun M, Qin X, Qiu Y, Zhu Z, et al. Low body mass index can be predictive of bracing failure in patients with adolescent idiopathic scoliosis: a retrospective study. Eur Spine J. 2017;26:1665–9. doi: 10.1007/s00586-016-4839-z. [DOI] [PubMed] [Google Scholar]
- 5.Bartley CE, Yaszay B, Bastrom TP, Shah SA, Lonner BS, Asghar J, et al. Perioperative and delayed major complications following surgical treatment of adolescent idiopathic scoliosis. J Bone Jt Surg Am. 2017;99:1206–12. doi: 10.2106/JBJS.16.01331. [DOI] [PubMed] [Google Scholar]
- 6.Pourabbas Tahvildari B, Erfani MA, Nouraei H, Sadeghian M. Evaluation of bone mineral status in adolescent idiopathic scoliosis. Clin Orthop Surg. 2014;6:180–4. doi: 10.4055/cios.2014.6.2.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yim AP, Yeung HY, Hung VW, Lee KM, Lam TP, Ng BK, et al. Abnormal skeletal growth patterns in adolescent idiopathic scoliosis--a longitudinal study until skeletal maturity. Spine. 2012;37:E1148–1154. doi: 10.1097/BRS.0b013e31825c036d. [DOI] [PubMed] [Google Scholar]
- 8.Burner WL, 3rd, Badger VM, Sherman FC. Osteoporosis and acquired back deformities. J Pediatr Orthop. 1982;2:383–5. doi: 10.1097/01241398-198210000-00006. [DOI] [PubMed] [Google Scholar]
- 9.Ishida K, Aota Y, Mitsugi N, Kono M, Higashi T, Kawai T, et al. Relationship between bone density and bone metabolism in adolescent idiopathic scoliosis. Scoliosis. 2015;10:19. doi: 10.1186/s13013-015-0043-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Li XF, Li H, Liu ZD, Dai LY. Low bone mineral status in adolescent idiopathic scoliosis. Eur Spine J. 2008;17:1431–40. doi: 10.1007/s00586-008-0757-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yip BH, Yu FW, Wang Z, Hung VW, Lam TP, Ng BK, et al. Prognostic value of bone mineral density on curve progression: a longitudinal cohort study of 513 girls with adolescent idiopathic scoliosis. Sci Rep. 2016;6:39220. doi: 10.1038/srep39220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng JC, Hung VW, Lee WT, Yeung HY, Lam TP, Ng BK, et al. Persistent osteopenia in adolescent idiopathic scoliosis--longitudinal monitoring of bone mineral density until skeletal maturity. Stud Health Technol Inform. 2006;123:47–51. [PubMed] [Google Scholar]
- 13.Garg P, Mazur MM, Buck AC, Wandtke ME, Liu J, Ebraheim NA. Prospective review of mesenchymal stem cells differentiation into osteoblasts. Orthop Surg. 2017;9:13–19. doi: 10.1111/os.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibon E, Lu L, Goodman SBAging. inflammation, stem cells, and bone healing. Stem Cell Res & Ther. 2016;7:44. doi: 10.1186/s13287-016-0300-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, Wang Y, Gou W, Lu Q, Peng J, Lu S. Role of mesenchymal stem cells in bone regeneration and fracture repair: a review. Int Orthop. 2013;37:2491–8. doi: 10.1007/s00264-013-2059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin Y, Guan J, Zhang C. Mesenchymal stem cells: mechanisms and role in bone regeneration. Postgrad Med J. 2014;90:643–7. doi: 10.1136/postgradmedj-2013-132387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhuang Q, Li J, Wu Z, Zhang J, Sun W, Li T, et al. Differential proteome analysis of bone marrow mesenchymal stem cells from adolescent idiopathic scoliosis patients. PLoS ONE. 2011;6:0018834. doi: 10.1371/journal.pone.0018834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park WW, Suh KT, Kim JI, Kim SJ, Lee JS. Decreased osteogenic differentiation of mesenchymal stem cells and reduced bone mineral density in patients with adolescent idiopathic scoliosis. Eur Spine J. 2009;18:1920–6. doi: 10.1007/s00586-009-1129-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, He L, Du Y, Zhu P, Huang G, Luo J, et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 2015;16:413–25. doi: 10.1016/j.stem.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 22.Zhu P, Wang Y, Huang G, Ye B, Liu B, Wu J, et al. lnc-beta-Catm elicits EZH2-dependent beta-catenin stabilization and sustains liver CSC self-renewal. Nat Struct Mol Biol. 2016;23:631–9. doi: 10.1038/nsmb.3235. [DOI] [PubMed] [Google Scholar]
- 23.Zhu P, Wang Y, Wu J, Huang G, Liu B, Ye B, et al. LncBRM initiates YAP1 signalling activation to drive self-renewal of liver cancer stem cells. Nat Commun. 2016;7:13608. doi: 10.1038/ncomms13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo S, Lu JY, Liu L, Yin Y, Chen C, Han X, et al. Divergent lncRNAs regulate gene expression and lineage differentiation in pluripotent cells. Cell Stem Cell. 2016;18:637–52. doi: 10.1016/j.stem.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 25.Anderson KM, Anderson DM, McAnally JR, Shelton JM, Bassel-Duby R, Olson EN. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature. 2016;539:433–6. doi: 10.1038/nature20128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang W, Huang H, Ding L, Zhu P, Saiyin H, Ji G, et al. Regulation of cell cycle of hepatocellular carcinoma by NF90 through modulation of cyclin E1 mRNA stability. Oncogene. 2015;34:4460–70. doi: 10.1038/onc.2014.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song D, Huang H, Wang J, Zhao Y, Hu X, He F, et al. NF90 regulates PARP1 mRNA stability in hepatocellular carcinoma. Biochem Biophys Res Commun. 2017;488:211–7. doi: 10.1016/j.bbrc.2017.05.037. [DOI] [PubMed] [Google Scholar]
- 28.Thiagarajan L, Abu-Awwad HAM, Dixon JE. Osteogenic Programming of Human Mesenchymal Stem Cells with Highly Efficient Intracellular Delivery of RUNX2. Stem Cells Transl Med. 2017;6:2146–59. doi: 10.1002/sctm.17-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhuang Q, Mao W, Xu P, Li H, Sun Z, Li S, et al. Identification of Differential Genes Expression Profiles and Pathways of Bone Marrow Mesenchymal Stem Cells of Adolescent Idiopathic Scoliosis Patients by Microarray and Integrated Gene Network Analysis. Spine. 2016;41:840–55. doi: 10.1097/BRS.0000000000001394. [DOI] [PubMed] [Google Scholar]
- 30.Chen C, Xu C, Zhou T, Gao B, Zhou H, Chen C, et al. Abnormal osteogenic and chondrogenic differentiation of human mesenchymal stem cells from patients with adolescent idiopathic scoliosis in response to melatonin. Mol Med Rep. 2016;14:1201–9. doi: 10.3892/mmr.2016.5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song Xiao-Xing, Jin Lin-Yu, Li Xin-Feng, Qian Lie, Shen Hong-Xing, Liu Zu-De, Yu Bu-Wei. Effects of Low Bone Mineral Status on Biomechanical Characteristics in Idiopathic Scoliotic Spinal Deformity. World Neurosurgery. 2018;110:e321–e329. doi: 10.1016/j.wneu.2017.10.177. [DOI] [PubMed] [Google Scholar]
- 32.Akazawa T, Kotani T, Sakuma T, Katogi T, Minami S, Niki H, et al. Bone mineral density and physical performance of female patients 27 years or longer after surgery for adolescent idiopathicscoliosis. Asian Spine J. 2017;11:780–6. doi: 10.4184/asj.2017.11.5.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hung VW, Qin L, Cheung CS, Lam TP, Ng BK, Tse YK, et al. Osteopenia: a new prognostic factor of curve progression in adolescent idiopathic scoliosis. J Bone Jt Surg Am. 2005;87:2709–16. doi: 10.2106/JBJS.D.02782. [DOI] [PubMed] [Google Scholar]
- 34.Noshchenko A, Hoffecker L, Lindley EM, Burger EL, Cain CM, Patel VV, et al. Predictors of spine deformity progression in adolescent idiopathic scoliosis: a systematic review with meta-analysis. World J Orthop. 2015;6:537–58. doi: 10.5312/wjo.v6.i7.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flynn RA, Chang HY. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell. 2014;14:752–61. doi: 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu XY, Wang L, Yu B, Zhuang QY, Wang YP. Expression signatures of long noncoding RNAs in adolescent idiopathic Scoliosis. Biomed Res Int. 2015;2015:276049. doi: 10.1155/2015/276049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–66. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kao PN, Chen L, Brock G, Ng J, Kenny J, Smith AJ, et al. Cloning and expression of cyclosporin A- and FK506-sensitive nuclear factor of activated T-cells: NF45 and NF90. J Biol Chem. 1994;269:20691–9. [PubMed] [Google Scholar]
- 40.Shi L, Godfrey WR, Lin J, Zhao G, Kao PN. NF90 regulates inducible IL-2 gene expression in T cells. J Exp Med. 2007;204:971–7. doi: 10.1084/jem.20052078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reichman TW, Parrott AM, Fierro-Monti I, Caron DJ, Kao PN, Lee CG, et al. Selective regulation of gene expression by nuclear factor 110, a member of the NF90 family of double-stranded RNA-binding proteins. J Mol Biol. 2003;332:85–98. doi: 10.1016/S0022-2836(03)00885-4. [DOI] [PubMed] [Google Scholar]
- 42.Kiesler P, Haynes PA, Shi L, Kao PN, Wysocki VH, Vercelli D. NF45 and NF90 regulate HS4-dependent interleukin-13 transcription in T cells. J Biol Chem. 2010;285:8256–67. doi: 10.1074/jbc.M109.041004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Castella S, Bernard R, Corno M, Fradin A, Larcher JC. Ilf3 and NF90 functions in RNA biology. Wiley Interdiscip Rev RNA. 2015;6:243–56. doi: 10.1002/wrna.1270. [DOI] [PubMed] [Google Scholar]
- 44.Kuwano Y, Kim HH, Abdelmohsen K, Pullmann R, Jr, Martindale JL, et al. MKP-1 mRNA stabilization and translational control by RNA-binding proteins HuR and NF90. Mol Cell Biol. 2008;28:4562–75. doi: 10.1128/MCB.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vumbaca F, Phoenix KN, Rodriguez-Pinto D, Han DK, Claffey KP. Double-stranded RNA-binding protein regulates vascular endothelial growth factor mRNA stability, translation, and breast cancer angiogenesis. Mol Cell Biol. 2008;28:772–83. doi: 10.1128/MCB.02078-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pei Y, Zhu P, Dang Y, Wu J, Yang X, Wan B, et al. Nuclear export of NF90 to stabilize IL-2 mRNA is mediated by AKT-dependent phosphorylation at Ser647 in response to CD28 costimulation. J Immunol. 2008;180:222–9. doi: 10.4049/jimmunol.180.1.222. [DOI] [PubMed] [Google Scholar]
- 47.Sakamoto S, Aoki K, Higuchi T, Todaka H, Morisawa K, Tamaki N, et al. The NF90-NF45 complex functions as a negative regulator in the microRNA processing pathway. Mol Cell Biol. 2009;29:3754–69. doi: 10.1128/MCB.01836-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 49.Wang KC, Helms JA, Chang HY. Regeneration, repair and remembering identity: the three Rs of Hox gene expression. Trends Cell Biol. 2009;19:268–75. doi: 10.1016/j.tcb.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klein D, Benchellal M, Kleff V, Jakob HG, Ergun S. Hox genes are involved in vascular wall-resident multipotent stem cell differentiation into smooth muscle cells. Sci Rep. 2013;3:2178. [DOI] [PMC free article] [PubMed]
- 51.Akam M. Hox genes: from master genes to micromanagers. Curr Biol. 1998;8:R676–8. doi: 10.1016/S0960-9822(98)70433-6. [DOI] [PubMed] [Google Scholar]
- 52.Ackema KB, Charite J. Mesenchymal stem cells from different organs are characterized by distinct topographic Hox codes. Stem Cells Dev. 2008;17:979–91. doi: 10.1089/scd.2007.0220. [DOI] [PubMed] [Google Scholar]
- 53.Chang HY, Chi JT, Dudoit S, Bondre C, van de Rijn M, Botstein D, et al. Diversity, topographic differentiation, and positional memory in human fibroblasts. Proc Natl Acad Sci USA. 2002;99:12877–82. doi: 10.1073/pnas.162488599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu YX, Xu B, Wu CL, Wu Y, Tong PJ, Xiao LW. Dynamic expression of DKK1 protein in the process whereby Epimedium-derived flavonoids up-regulate osteogenic and down-regulate adipogenic differentiation of bone marrow stromal cells in ovariectomized rats. Orthop Surg. 2011;3:119–26. doi: 10.1111/j.1757-7861.2011.00129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma XL, Liu ZP, Ma JX, Han C, Zang JC. Dynamic expression of Runx2, Osterix and AJ18 in the femoral head of steroid-induced osteonecrosis in rats. Orthop Surg. 2010;2:278–84. doi: 10.1111/j.1757-7861.2010.00100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–64. doi: 10.1016/S0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 57.Ducy P, Schinke T, Karsenty G. The osteoblast: a sophisticated fibroblast under central surveillance. Science. 2000;289:1501–4. doi: 10.1126/science.289.5484.1501. [DOI] [PubMed] [Google Scholar]
- 58.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–54. doi: 10.1016/S0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 59.Wang WJ, Sun C, Liu Z, Sun X, Zhu F, Zhu ZZ, et al. Transcription factor Runx2 in the low bone mineral density of girls with adolescent idiopathic scoliosis. Orthop Surg. 2014;6:8–14. doi: 10.1111/os.12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Han P, Li W, Lin CH, Yang J, Shang C, Nuernberg ST, et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature. 2014;514:102–6. doi: 10.1038/nature13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.