Abstract
Cervical cancer (CC) is the third most common carcinoma and the fourth leading cause of cancer-associated mortality in women. Here, we report that MDM2-DHX9 interaction mediates CC motility and angiogenesis in a long noncoding RNA-dependent fashion. A long noncoding RNA, named lnc-CCDST, is significantly downregulated in CC tissues, and binds to pro-oncogenic DHX9. DHX9 is upregulated in CC tissue, and promotes CC cell motility and angiogenesis. The lnc-CCDST and DHX9 interaction promotes DHX9 degradation through the ubiquitin proteasome pathway. Furthermore, DHX9 bound to E3 ubiquitin ligase MDM2, and this interaction is enhanced by lnc-CCDST. Thus, lnc-CCDST promotes DHX9 degradation by serving as a scaffold to facilitate the formation of MDM2 and DHX9 complexes. Moreover, HPV oncogenes E6 and E7 abolish the expression of lnc-CCDST resulting in the increase of DHX9. Our results have revealed a novel mechanism by which high-risk HPVs promote motility and angiogenesis of CC by inhibiting expression of lnc-CCDST to disrupt MDM2 and DHX9 interaction, and DHX9 degradation, and identified a potential therapeutic target for CC.
Subject terms: Tumour angiogenesis, Metastasis, Pathogenesis
Introduction
Although the incidence and mortality rate of cervical cancer (CC) have continued to decline in recent years, CC remains as the third most common carcinoma and the fourth leading cause of cancer-associated mortality in females worldwide. According to the WHO datum, 528,000 new cases of CC were diagnosed and 266,000 patients died of the cancer in 2012. CC may be completely free of symptoms in the early stages but may be present in the abdomen, lungs, or elsewhere in advanced disease, particularly in metastatic stage [1]. Although early screening programs have reduced the mortality rates of CC patients, tumor metastasis continues to occur and negatively impact survival even for some diagnosed in early stage [2]. While many studies have focused on CC, the molecular mechanisms underlying the progression of CC remain unclear.
Human papillomavirus (HPV) infection causes more than 90% of CC cases [3]. Globally, high-risk HPV 16 and 18 are the causes of 75% of CC cases with the remaining cases associated with other high-risk HPVs [3]. HPV oncoproteins E6 and E7 are directly linked to the process of malignant progression [4, 5]. E6 and E7 have evolved numerous strategies to alter cell cycle and apoptosis control, and evade host immuno-surveillance while maintain viral persistence.
Long noncoding RNAs (lncRNAs) are non-protein coding transcripts, which are longer than 200 nucleotides. They have been implicated in human cancers, including CC [6]. Mounting evidence show that lncRNAs can regulate cell functions in different aspects, for example, the imprinted X-chromosome in female placental mammals is regulated by one of the best characterized lncRNA, Xist [7]. An earlier study has found that a lncRNA in the antisense direction of the Apolipoprotein A1 (APOA1) regulates the transcription of APOA1 through epigenetic modifications [8]. In addition to regulating gene transcription, lncRNAs control many aspects of post-transcriptional processes, such as RNA splicing [9], mRNA translation [10], lncRNA-microRNA axis [11], and so on. Additionally, lncRNAs can function as scaffolds to regulate protein–protein interactions [12].
Although the lnc-CC3 promotes epithelial–mesenchymal transition [13] and lncRNA RSU1P2 serves as a competitive endogenous RNA (ceRNA) to counter let-7a in CC [14], the specific molecular mechanism on how lncRNAs regulate cervix cell phenotypes remains largely unknown. Because HPV infection is critical for cancer development and progression, it could be interesting to further investigate on how lncRNAs mediate HPV-induced carcinogenesis. The mechanism by which lncRNAs mediate E6- and E7-induced carcinogenesis remains to be further explored.
In this study, we report an important role of a lncRNA named cervical cancer DExH-box helicase 9 (DHX9) suppressive transcript (abbreviated as CCDST), which is significantly downregulated in CC tissues. We have examined the negative effect of lnc-CCDST on cell migration, invasion, anchorage-independent growth, and angiogenesis in HPV-positive cell lines. Mechanistically, lnc-CCDST binds to DHX9 and causes its degradation through the ubiquitin proteasome pathway by recruiting E3 ubiquitin ligase MDM2. The HPV oncoproteins E6 and E7 decrease the expression of lnc-CCDST resulting in increased DHX9 expression. These results provide insights into the role of a cellular lncRNA in the pathogenesis of CC and identify DHX9-lncRNA-MDM2 as an important functional interaction, which could serve as a potential therapeutic target for HPV-associated malignancies.
Results
Lnc-CCDST is downregulated in CC tissues
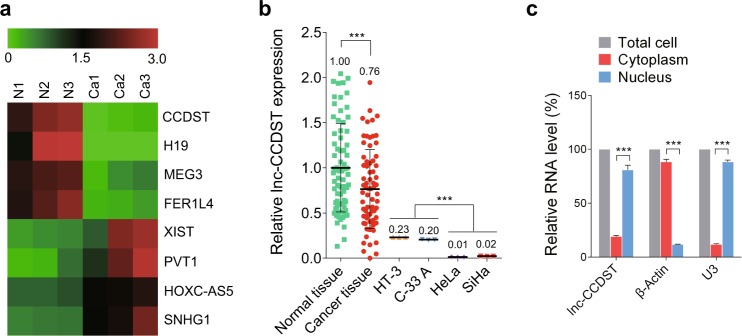
To acquire the transcriptional profile for lncRNAs in CC, three HPV + CC tissues (n = 3) and three paired normal tissues (n = 3) were analyzed using ArrayStar lncRNA microarray. When the criteria P < 0.05 and fold change > 2.0 were accepted, we found similar numbers of lncRNAs that were significantly upregulated or downregulated in CC tissues compared with the matched normal tissues. The screening identified eight candidate lncRNAs (Fig. 1a) that differed significantly between cancer and paired normal tissues. Some of these lncRNAs have previously been investigated in other types of cancer, however, there is no report in cancer on lncRNA ENST00000429352, which we named lnc-CCDST. We examined lnc-CCDST expression in 72 paired clinical CC and normal tissues. The results showed downregulation of lnc-CCDST in cancer tissues compared with matched normal tissues (Fig. 1b). To compare the lnc-CCDST level between the established cell lines of CC and normal tissues, the lnc-CCDST expression was examined in HT-3, C-33 A, HeLa, and SiHa cells. The lnc-CCDST exhibited a decreased expression in the established cell lines compared with the normal tissues (Fig. 1b). Interestingly, the lnc-CCDST levels in HT-3 and C-33 A (both are HPV-negative cells) were higher than those in HeLa and SiHa (both are HPV-positive cells) (Fig. 1b). Nuclear and cytoplasm fraction was isolated from HeLa cells, as shown in Fig. 1c, the isolated nuclear fraction displayed a higher level of lnc-CCDST than the cytoplasmic fraction.
Fig. 1.
Lnc-CCDST is downregulated in CC tissues. a The heatmap of the representative result of lncRNAs in CC tissues analyzed by microarray. The three HPV + CC tissues and paired adjacent normal cervical tissues were subjected to the lncRNAs microarray analysis and pseudo-color represented the intensity scale of cancer tissue group vs. normal cervical tissue group. The color “green” and “red” denote low and high expression, respectively. b Lnc-CCDST is decreased in CC tissues and CC cell lines. The lnc-CCDST expression levels were examined in 72 paired CC and adjacent normal cervical tissues by RT-qPCR, data are presented as mean ± SD. Every point in the normal or cancer tissue group contains three technical replicates, only the average lnc-CCDST level of each tissue was used to analyze the lnc-CCDST expression difference between the two groups; two tailed paired Student’s t test for lnc-CCDST analysis between the tissues was used, ***P < 0.001. The lnc-CCDST expression levels were also examined in HPV-negative cells (HT-3 and C-33A) and HPV-positive cells (HeLa and SiHa) by RT-qPCR. β-actin was used as an internal control for normalization. Every point in the CC cell group indicates one technical replicate, and data are presented as mean. Two-tailed unpaired Student’s t test for the lnc-CCDST analysis was used. ***P < 0.001. c Lnc-CCDST is enriched in HeLa nuclear fraction. Levels of lnc-CCDST, β-actin, and U3 RNA in purified HeLa nuclear and cytoplasm fractions were detected by RT-qPCR. Data are presented as mean ± SEM, determined from three independent experiments (n = 3), two-tailed unpaired Student’s t test. ***P < 0.001
Lnc-CCDST inhibits cell migration, invasion, anchorage-independent growth, and angiogenesis
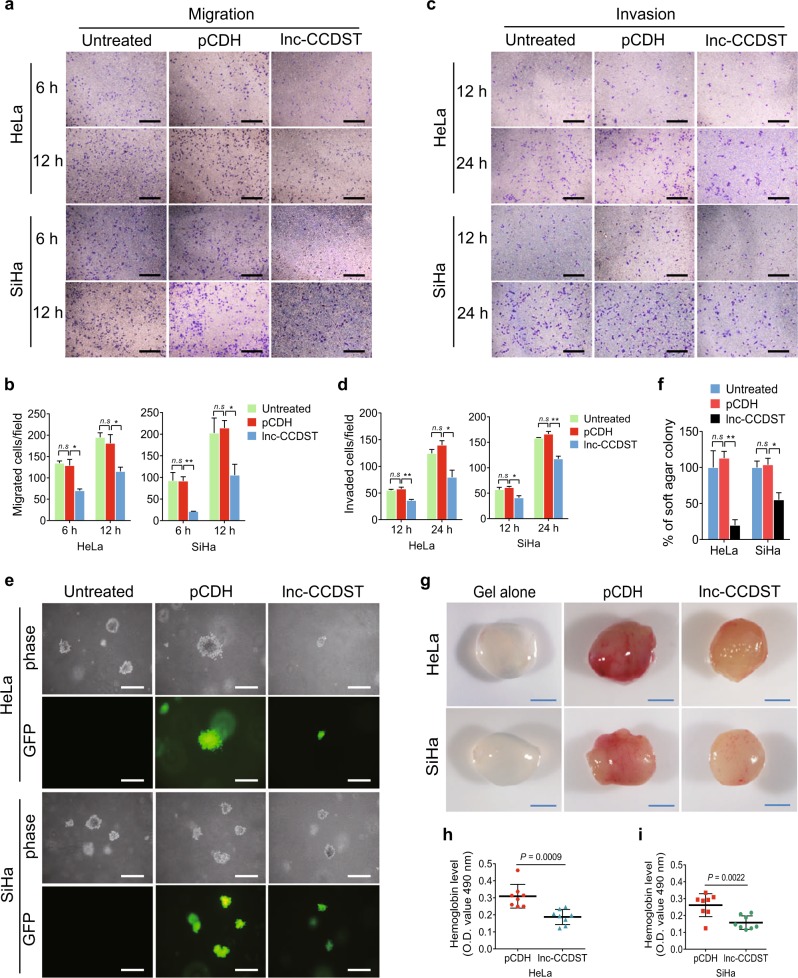
To determine the function of lnc-CCDST, we transduced CC cells with different multiplicities of infection (MOIs) of a lentiviral lnc-CCDST. The RT-qPCR results revealed that lnc-CCDST was successfully overexpressed in CC cells (Supplementary Fig. 1a). To obtain a physiological lnc-CCDST expression level, we chose the indicated MOIs based on the differences between the cell lines and normal tissues for the subsequent experiments (Fig. 1b; Supplementary Fig. 1a). Compared with cells transduced with the vector control (pCDH), HeLa cells transduced with lnc-CCDST had no effect on proliferation (Supplementary Fig. 1b). However, expression of lnc-CCDST inhibited HeLa and SiHa cell migration (Fig. 2a, b) and invasion (Fig. 2c, d). lnc-CCDST also inhibited the motility of C-33 A cells (Supplementary Fig. 2a). We further evaluated the role of this lncRNA in in vitro oncogenicity. The colony formation assay in soft agar showed that lnc-CCDST significantly reduced the sizes of the colonies of CC cells (Fig. 2e, f, Supplementary Fig. 2b). To determine if lnc-CCDST might regulate cancer angiogenesis, we incubated human umbilical vein endothelial cells (HUVECs) with conditioned medium collected from HeLa or SiHa cells expressed either lnc-CCDST or the vector control, and examined capillary tube formation. Compared with the vector control, conditioned medium from HeLa or SiHa cells overexpressing lnc-CCDST significantly inhibited capillary tube formation (Supplementary Figs. 3a, b).
Fig. 2.
Lnc-CCDST inhibits cell migration, invasion, anchorage-independent cell growth, and angiogenesis. a Transwell migration assay reveals that lnc-CCDST inhibits cells migration. Transwell migration assays were performed in HeLa/SiHa transduced with lnc-CCDST or the empty vector control cells. The representative images were captured at 6 and 12 h after seeding 104 cells in the upper chamber as described in the “Materials and methods” section. Scar bars, 40 μm. b Quantification of the results in a. Data are mean ± SEM determined from three independent experiments (n = 3), with each experiment containing six technical replicates. Two-tailed unpaired Student’s t test was used. *P < 0.05, **P < 0.01. c. Transwell invasion assay demonstrates that lnc-CCDST inhibits cell invasion. Matrigel invasion assays were performed in HeLa/SiHa cells transduced with lnc-CCDST or the empty vector control cells. The representative images were captured at 12 and 24 h after seeding 5 × 104 cells in the upper chamber as described in the “Materials and methods” section. Scar bars, 40 μm. d Quantification of the results in c. Data are presented as mean ± SEM determined from three independent experiments (n = 3), with each experiment containing six technical replicates, and two-tailed unpaired Student’s t test was used. *P < 0.05, **P < 0.01. e Soft agar colony formation assay shows that lnc-CCDST inhibits anchorage-independent cell growth. HeLa/SiHa were transduced with lnc-CCDST or the empty vector; and the representative images were captured at day 14 after seeding 1 × 104 cells per well into a 6-well plate as described in the “Materials and methods” section. Scar bars, 40 μm. f Quantification of the results in e. The areas of the colonies were scanned by the Image J, with the untreated group considered as 100%. Data are presented as mean ± SEM determined from three independent experiments (n = 3); and each experiment contains six technical replicate, and two-tailed unpaired Student’s t test was used. *P < 0.05, **P < 0.01. g Representative photographs showing lnc-CCDST inhibition of angiogenesis in the Matrigel Plug assay. HeLa or SiHa cells were examined for their proangiogenic effects in the Matrigel plug assay in nude mice as described in the “Materials and methods” section. Representative photographs showing extent of angiogenesis are presented. Scar bars, 1 cm. h, i. Statistical analysis of hemoglobin protein levels in different plugs in g. Data are mean ± SD, and each group contains eight tumors (n = 8). Two independent experiments were performed and similar results were obtained. Two-tailed unpaired Student’s t test was used
We further evaluated the angiogenesis effect of lnc-CCDST in vivo. Overexpression of lnc-CCDST significantly inhibited angiogenesis induced by HeLa and SiHa cells in a chicken chorioallantoic membrane (CAM) assay (Supplementary Figs. 3c–e). We then employed a mouse Matrigel plug angiogenesis assay. Mice were subcutaneously injected with Matrigel plugs containing HeLa or SiHa cells expressed either lnc-CCDST or vector control. As expected, overexpression of lnc-CCDST significantly reduced the angiogenic activity of HeLa or SiHa cells compared with the vector control cells, as shown by the redness of the plugs (Fig. 2g) and the hemoglobin content (Fig. 2h, i). H&E staining showed that there were less extensive dense neovascularization and hemorrhagic necrotic foci in plugs containing HeLa or SiHa cells expressing lnc-CCDST than the vector control (Supplementary Fig. 3f). Taken together, these results indicated that lnc-CCDST inhibited cell migration, invasion, anchorage-independent growth, and angiogenesis of CC cells.
Lnc-CCDST interacts with DHX9 and inhibits its expression
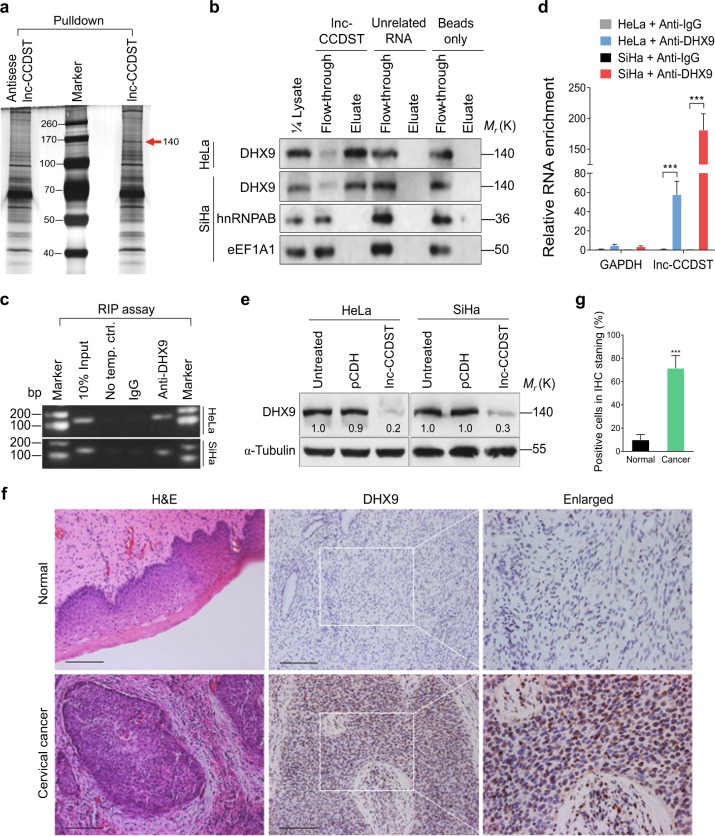
To investigate the mechanism of lnc-CCDST’s inhibitory functions, we hypothesized that lnc-CCDST might bind to a protein that mediates these effects. To identify such a protein, we first incubated in vitro-transcribed biotinylated lnc-CCDST sense or antisense RNA with cell lysate, and then performed pulldown with streptavidin beads. Silver staining revealed that there were more lnc-CCDST pulldown proteins than those of antisense RNA (Fig. 3a). The isolated proteins were subjected to mass spectrometry. We found that DHX9 (also known as RHA), hnRNPAB, and eEF1A1 were enriched in the lnc-CCDST pulldown proteins. To confirm that lnc-CCDST indeed interacted with DHX9, we incubated lnc-CCDST and an unrelated RNA with cell lysates of HeLa and SiHa cells, and then performed pulldown with streptavidin beads. The pulldown proteins were subjected to western blotting. As shown in Fig. 3b, DHX9 rather than hnRNPAB or eEF1A1 interacts with lnc-CCDST. DHX9 did not interact with the unrelated RNA or the beads alone. Next, we performed RNA immunoprecipitation (RIP) with a DHX9 antibody. The DHX9 antibody instead of the control antibody successfully pulled down lnc-CCDST as detected by polymerase chain reaction (PCR) (Fig. 3c) and quantitative PCR (qPCR) (Fig. 3d).
Fig. 3.
Lnc-CCDST interacts with and inhibits DHX9. a Silver staining of gel showing lnc-CCDST and the control pulldown proteins. Lnc-CCDST was transcribed in vitro and labeled with biotin. The biotin-labeled lnc-CCDST and the controls were subjected to pulldown using HeLa cell lysate. The pulldown solutions were then subjected to silver staining. b. Lnc-CCDST interacts with DHX9. Western blotting validated that DHX9, rather than hnRNPAB or eEF1A1, interacted with lnc-CCDST in the pulldown assay as described in the “Materials and methods” section. c, d. PCR (c) and qPCR (d) results showing that lnc-CCDST is precipitated by DHX9 in RIP assay as described in the “Materials and methods” section. The HeLa or SiHa cell lysate was subjected to the anti-DHX9 RNA immunoprecipitation, and the precipitated RNAs were examined by PCR or RT-qPCR. GAPDH served as a control to validate DHX9-lnc-CCDST interaction. Data are presented as mean ± SEM determined from three independent experiments (n = 3), and two-tailed unpaired Student’s t test was used. ***P < 0.001. e Western blotting analysis revealed that DHX9 was decreased in lnc-CCDST overexpressing HeLa and SiHa cells. The cells overexpressing lnc-CCDST of the control vector were subjected to western blotting to examine the DHX9 expression using the indicated antibodies. f DHX9 is upregulated in CC tissues. Hematoxylin and eosin staining (H&E) analysis of histologic features (left panel) and IHC staining analysis of the expression of DHX9 (middle and right panel) in CC and normal tissues were shown. Scar bars, 100 μm. g Quantification of the results in f. Positive IHC staining cells were counted and subjected to statistical analysis. Data are mean ± SD, n = 10, and two-tailed unpaired Student’s t test was used. ***P < 0.001
To identify the consequence of lnc-CCDST and DHX9 interaction, we measured the DHX9 protein expression in lnc-CCDST overexpressed cells. We found that the DHX9 protein level was significantly decreased after lnc-CCDST overexpression (Fig. 3e). Because lnc-CCDST was downregulated in CC tissues, we examined the expression of DHX9 in CC tissues. We found that there were more DHX9-positive cells in CC tissues than in normal tissues (Fig. 3f, g, Supplementary Fig. 4). These results were in line with those reported in lung cancer [15].
Lnc-CCDST promotes DHX9 degradation through the ubiquitin proteasome pathway by enhancing MDM2-DHX9 interaction
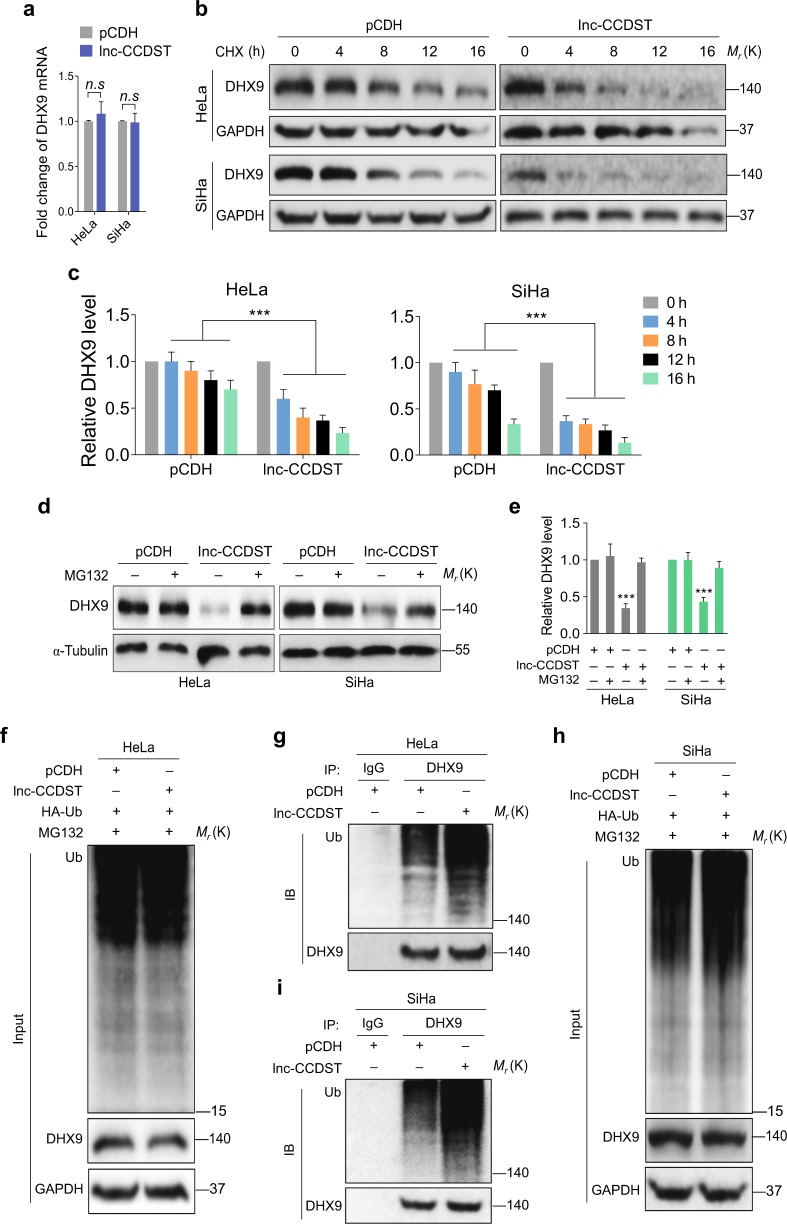
Since lnc-CCDST reduced the DHX9 protein level in HeLa and SiHa cells, we determined whether overexpression of lnc-CCDST could affect the DHX9 mRNA level. The results of qPCR indicated that overexpression of lnc-CCDST did not alter the level of DHX9 mRNA (Fig. 4a). Examination of a database (The Cancer Genome Atlas, TCGA, http://www.cbioportal.org/) revealed that there was no significant dysregulation of DHX9 mRNA in 307 CC cases.
Fig. 4.
Lnc-CCDST destabilizes DHX9 via ubiquitin proteasome pathway. a Lnc-CCDST has no effect on DHX9 mRNA level. DHX9 mRNA expression was detected by RT-qPCR in lnc-CCDST-transduced HeLa and SiHa cells. The values of RNA expression levels following normalization to GAPDH were shown. Data are mean ± SEM determined from three independent experiments (n = 3), and two-tailed unpaired Student’s t test was used. b Lnc-CCDST promotes DHX9 degradation. Lnc-CCDST transduced HeLa and SiHa or control cells were treated with CHX at 20 μg ml−1 for 0, 4, 8, 12, and 16 h, and then the DHX9 expression level was examined by western blotting using the indicated antibodies. c Quantification of the results in b. Data are mean ± SEM, n = 3, and two-tailed unpaired Student’s t test was used. ***P < 0.001. d Lnc-CCDST mediates degradation through the ubiquitin proteasome pathway. Lnc-CCDST transduced HeLa and SiHa or control cells were treated with proteasome inhibitor MG132 at 5 μM for 4 h, and then the DHX9 expression level was examined by western blotting. e Quantification of the results in d. Data are mean ± SD, n = 3, and two-tailed unpaired Student’s t test was used. ***P < 0.001. f The pCDH- and lnc-CCDST-transduced HeLa cells were transfected with HA-Ub plasmid and then treated with 5 μM MG132 for 4 h, then the cell lysates were examined by western blotting. g Increased DHX9 ubiquitination is observed after overexpression of lnc-CCDST. For denaturing, the cell lysates in f were heated at 95 °C for 5 min in the presence of 1% SDS, followed by 10-fold dilution with lysis buffer (to 0.1% SDS) and sonication (the same denaturing treatment was used in other ubiquitination assays). Then the cell lysates were subjected to co-immunoprecipitation with the antibody against DHX9 (IP: DHX9) followed by Western blotting using the indicated antibodies. h Cell lysates of pCDH- and lnc-CCDST-transduced SiHa cells transfected with HA-Ub plasmid and treated with 5 μM MG132 for 4 h were examined by western blotting. i Increased DHX9 ubiquitination is observed after overexpression of lnc-CCDST. The cell lysates in h were subjected to co-immunoprecipitation with the antibody against DHX9 (IP: DHX9) followed by western blotting using the indicated antibodies
We further determined whether lnc-CCDST might reduce DHX9 protein level by promoting its degradation. HeLa and SiHa cells were overexpressed with either lnc-CCDST or the vector control and then treated with cycloheximide (CHX) to inhibit de novo protein synthesis. We found that compared with the vector control, lnc-CCDST reduced the stability of DHX9 protein in both HeLa and SiHa cells (Fig. 4b, c). However, lnc-CCDST-mediated degradation of DHX9 was completely inhibited by the proteasome inhibitor MG132 (Fig. 4d, e) but not NH4Cl and 3-methyladenine (3-MA), which are inhibitors for the lysosome and autophagosome pathways, respectively (Supplementary Fig. 5). Furthermore, we observed increased DHX9 ubiquitination following lnc-CCDST overexpression in both HeLa and SiHa cells compared with the vector control (Fig. 4f–i). Collectively, these results suggested that lnc-CCDST might destabilize DHX9 protein by promoting its proteasome-mediated ubiquitination and degradation.
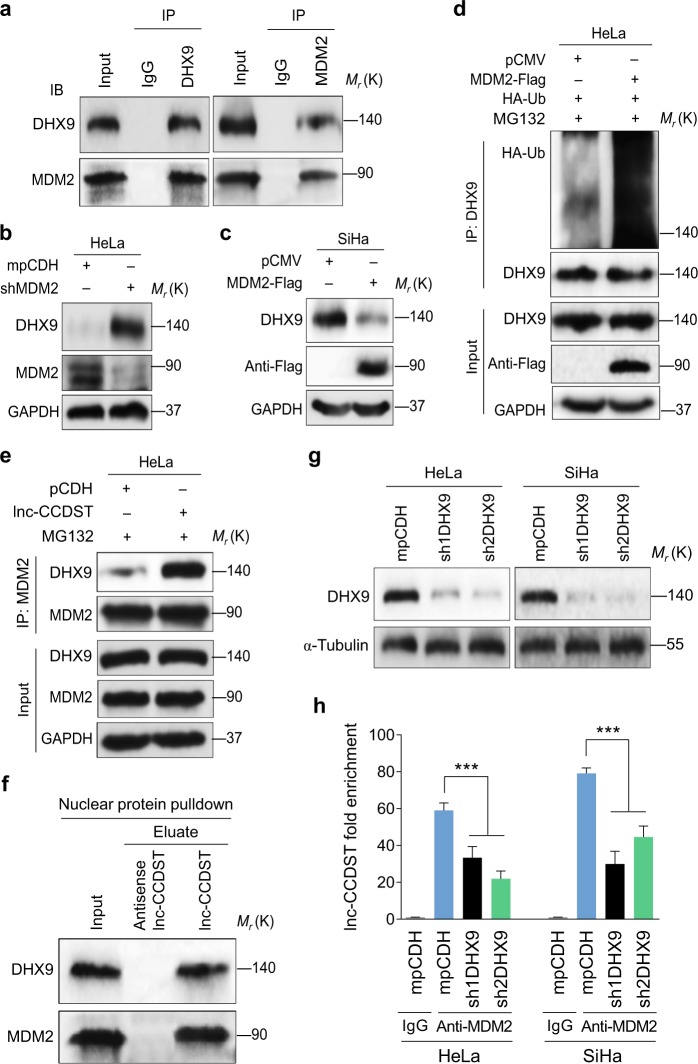
To identify the ubiquitin ligase that might target DHX9 for degradation in CC cells, we searched candidate DHX9 ligases in the Human Protein Reference Database (www.hprd.org). The MDM2 LC/MS/MS results were uploaded in the Supplementary data, which identified DHX9 as a candidate MDM2 interaction protein [16]. Indeed, we detected interaction of endogenous DHX9 and MDM2 in HeLa cells by immunoprecipitation (Fig. 5a). Importantly, knockdown of MDM2 with shRNAs increased the DHX9 protein level (Fig. 5b, Supplementary Fig. 6), while overexpression of MDM2 reduced the DHX9 protein level (Fig. 5c). As expected, increased DHX9 ubiquitination was observed following MDM2 overexpression (Fig. 5d). We further examined the role of lnc-CCDST in MDM2-mediated DHX9 degradation. Overexpression of lnc-CCDST enhanced the interaction of DHX9 and MDM2 (Fig. 5e). To validate the lnc-CCDST–DHX9-MDM2 interaction in the nucleus, the HeLa nucleus fraction was extracted and subjected to lnc-CCDST RNA pulldown assay. Both DHX9 and MDM2 interacted with lnc-CCDST in the cell nucleus (Fig. 5f). To determine whether DHX9 could affect lnc-CCDST–MDM2 interaction, the DHX9 was knocked down with two shRNAs in HeLa and SiHa cells (Fig. 5g). The cells were then subjected to anti-MDM2 RIP assay. qPCR results indicated that knockdown of DHX9 significantly impaired the lnc-CCDST–MDM2 interaction (Fig. 5h). Taken together, these results revealed that lnc-CCDST promoted DHX9 degradation by serving as a scaffold to bring MDM2 and DHX9 together.
Fig. 5.
Lnc-CCDST enhances binding of E3 ubiquitin ligase MDM2 to DHX9. a HeLa is subjected to co-immunoprecipitation with an antibody against immunoglobulin G (IgG), or DHX9 or MDM2 followed by western blotting using the indicated antibodies. Experiments were independently repeated three times with similar results and results from a representative experiment were shown. b, c. Results of knockdown (b), or overexpression MDM2 (c) showing that MDM2 negatively regulates DHX9 expression. DHX9 and MDM2 were examined by western blotting in HeLa or SiHa transduced with shMDM2 or transfected with MDM2-Flag plasmid. d MDM2 promotes DHX9 ubiquitination. HeLa cells were transfected with MDM2-Flag or its control pCMV, after treated with 5 μM MG132 for 4 h, the cells were subjected to co-immunoprecipitation with the antibody against DHX9 (IP: DHX9) followed by western blotting using the indicated antibodies. e Lnc-CCDST promotes MDM2 binding to DHX9. Cell lysates from HeLa cells transduced with lentiviral lnc-CCDST or its control pCDH and treated with 5 μM MG132 for 4 h were subjected to co-immunoprecipitation with the antibody against MDM2 (IP: MDM2) followed by Western blotting using the indicated antibodies. f Lnc-CCDST binds to nuclear MDM2 and DHX9. The nuclear proteins from HeLa cells were subjected to the lnc-CCDST pulldown assay as described in the “Materials and methods” section. The lnc-CCDST-MDM2-DHX9 interaction was examined by western blotting using the indicated antibodies. g. Western blotting reveals that DHX9 is successfully knocked down by the two shRNAs. HeLa and SiHa cells transduced with DHX9 shRNAs or its control mpCDH were examined for the knockdown efficiencies by Western blotting. h Knockdown of DHX9 impairs the lnc-CCDST-MDM2 interaction. HeLa or SiHa cells were transduced with DHX9 shRNAs or the empty vector. Then the cells were subjected to the anti-MDM2 RIP assay as described in the “Materials and methods” section. Data are mean ± SEM determined from three independent experiments (n = 3), and two-tailed unpaired Student’s t test was used. ***P < 0.001
Lnc-CCDST inhibits DHX9 promotion of cell migration, invasion, and anchorage-independent growth
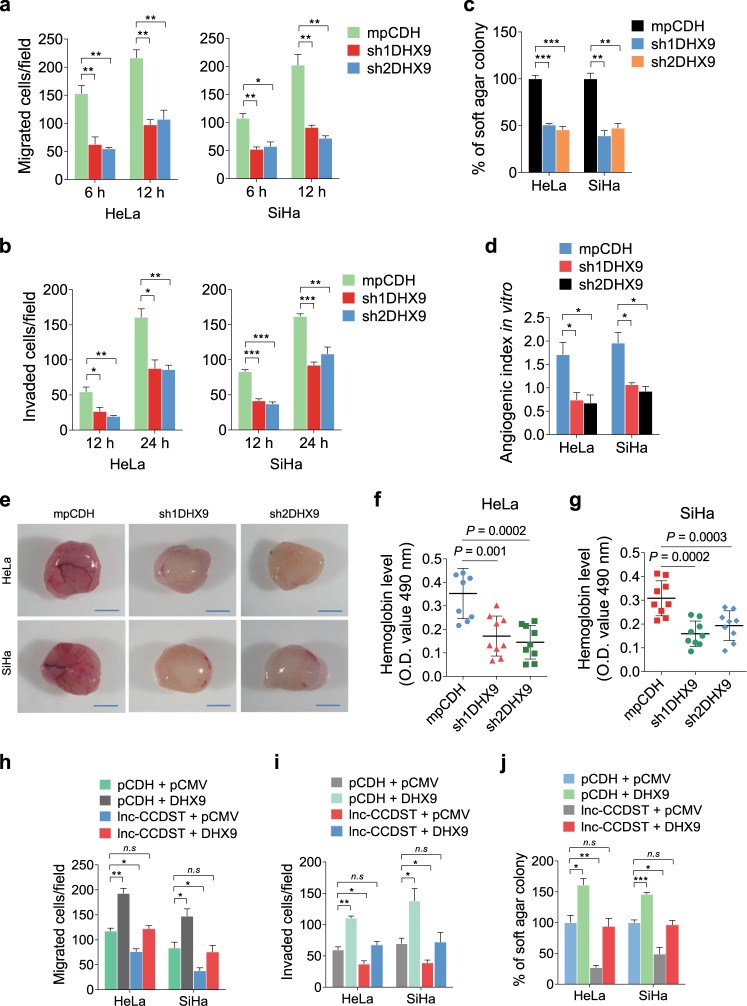
DHX9 has been reported to promote cell survival in different human cells [17]. However, the role of DHX9 in CC is still unclear. We found that knockdown of DHX9 expression with shRNAs in HeLa and SiHa cells significantly inhibited cell migration (Fig. 6a, Supplementary Fig. 7a). Knockdown of DHX9 also inhibited cell invasion (Fig. 6b, Supplementary Fig. 7b), anchorage-independent growth (Fig. 6c, Supplementary Fig. 7c), and in vitro (Fig. 6d) and in vivo angiogenic activities (Fig. 6e–g). Similar results were also observed in C-33A cells (Supplementary Fig. 8). Since cell migration, invasion, anchorage-independent growth, and angiogenesis were regulated by both lnc-CCDST and DHX9, and lnc-CCDST decreased the level of DHX9, we examined whether DHX9-mediated lnc-CCDST regulated inhibition of these phenotypes. We expressed DHX9 in lnc-CCDST overexpressing HeLa and SiHa cells (Supplementary Fig. 9). We found that overexpression of DHX9 significantly rescued lnc-CCDST-inhibited cell migration (Fig. 6h), invasion (Fig. 6i), and anchorage-independent growth (Fig. 6j), suggesting that lnc-CCDST-mediated inhibition of DHX9 expression was an important mechanism by which lnc-CCDST exerted its inhibitory effect on these biological processes.
Fig. 6.
DHX9 promotes cell migration, invasion, angiogenesis, and anchorage-independent cell growth, and is involved in the lnc-CCDST-mediated phenotypes. a Transwell migration assay for HeLa and SiHa transduced with two DHX9 shRNAs or empty vector. The images were captured at 6 and 12 h post seeding at 104 cells in the upper chamber as described in the “Materials and methods” section. Data are mean ± SEM determined from three independent experiments (n = 3), each containing six technical replicates. Two-tailed unpaired Student’s t test was used. *P < 0.05, **P < 0.01. b Transwell invasion assay for HeLa and SiHa cells transduced with two DHX9 shRNAs or empty vector. The images were captured at 12 and 24 h following seeding of 5 × 104 cells in the upper chamber as described in the “Materials and methods” section. Data are mean ± SEM determined from three independent experiments (n = 3), each contains six technical replicates. Two-tailed unpaired Student’s t test was used. *P < 0.05, **P < 0.01. c Quantification of the results of soft agar colony formation assay for HeLa and SiHa transduced with two DHX9 shRNAs or an empty vector mpCDH. The representative images were captured at day 14 after seeding a total number of 1 × 104 cells into a 6-well plate as described in the “Materials and methods” section. Data are represented with mean ± SEM determined from three independent experiments (n = 3), each with six technical replicates. Two-tailed unpaired Student’s t test was used. **P < 0.01, ***P < 0.001. d Quantification of the results of tubule formation assay for HeLa and SiHa transduced with two DHX9 shRNAs or empty vector. Data are mean ± SEM determined from three independent experiments (n = 3), each with five technical replicates. Two-tailed unpaired Student’s t test was used. *P < 0.05. e. Representative photographs showing angiogenesis in the Matrigel Plug assay of cells with knockdown of DHX9. HeLa or SiHa cells were examined for their proangiogenic effects in a Matrigel plug assay in nude mice as described in the “Materials and methods” section. Representative photographs showing extent of angiogenesis are presented. Scar bars, 1 cm. f, g The statistical analysis of hemoglobin protein levels in two shDHX9 groups and control group. Data are mean ± SD, each with eight or nine tumors (n = 9). Two independent experiments were performed and similar results were obtained. Two-tailed unpaired Student’s t test was used. h Transwell migration assay for HeLa and SiHa transduced with lentivirus-mediated lnc-CCDST or empty vector followed by co-transfection with DHX9 or its control pCMV plasmids, respectively. The images were captured at 6 and 12 h post seeding 104 cells in the upper chamber as described in the “Materials and methods” section. Data are mean ± SEM determined from three independent experiments (n = 3), each with six technical replicates. Two-tailed unpaired Student’s t test was used. *P < 0.05, **P < 0.01. i Transwell invasion assay for cells treated as in h. The images were captured at 12 and 24 h post seeding of 5 × 104 cells per well in the upper chamber as described in the “Materials and methods” section. Data are mean ± SEM determined from three independent experiments (n = 3), each with six technical replicates. Two-tailed unpaired Student’s t test was used. *P < 0.05, **P < 0.01. j Soft agar colony formation assay for cells treated as described in h. Cells were seeded at 1 × 104 into a 6-well plate as described in the “Materials and methods” section and the images were captured at day 14. Data are mean ± SEM determined from three independent experiments (n = 3), each with six technical replicates. Two-tailed unpaired Student’s t test was used. *P < 0.05, **P < 0.01
HPV regulates lnc-CCDST and DHX9 expression
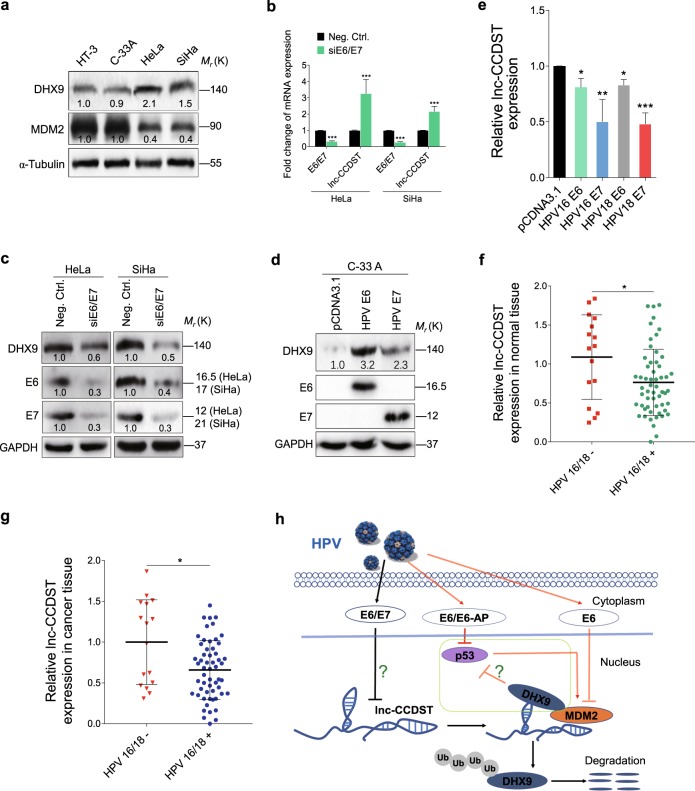
Because lnc-CCDST was decreased in HPV-positive cells (HeLa and SiHa) compared with HPV-negative cells (HT-3 and C-33A) (Fig. 1b), and lnc-CCDST negatively regulated DHX9 expression (Fig. 3e), we examined DHX9 and MDM2 expression level in these cells. As expected, DHX9 was increased and MDM2 was decreased in HeLa and SiHa cells when compared with HT-3 and C-33 A cells (Fig. 7a). To determine whether HPV oncoproteins E6 and E7 might downregulate lnc-CCDST, we performed knockdown of E6 and E7 with a pool of siRNAs. Significantly, knockdown of E6 and E7 dramatically increased the level of lnc-CCDST in HeLa and SiHa cells (Fig. 7b). More importantly, the DHX9 protein level was reduced following knockdown of E6 and E7 proteins (Fig. 7c). Similarly, DHX9 was induced and lnc-CCDST was decreased when we overexpressed HPV E6 and E7 proteins in HPV-negative C-33 A CC cells (Fig. 7d, e). We then examined the incidence of HPV infection in 72 CC patients. A total of 56 (78%) patients were identified as HPV16/18 positive. We evaluated the lnc-CCDST expression difference between HPV-positive and HPV-negative groups. The results showed that lnc-CCDST was significantly decreased in the HPV-positive group compared with the HPV-negative in both normal and cancer tissues (Fig. 7f, g).
Fig. 7.
HPV E6/E7 decreases lnc-CCDST level and elevates DHX9 expression. a DHX9 protein level is increased while MDM2 protein level is decreased in HPV-positive cells. DHX9 and MDM2 in HPV-negative cells (HT-3 and C-33A) and HPV-positive cells (HeLa and SiHa) were examined by Western blotting using the indicated antibodies. b Lnc-CCDST level is upregulated after E6/E7 knockdown. Lnc-CCDST was detected by RT-qPCR in HeLa and SiHa cells transfected with an E6/E7 siRNA pool. GAPDH served as internal control for normalization. Data are mean ± SEM determined from three independent experiments (n = 3), each with four technical replicates. Two-tailed unpaired Student’s t test was used. ***P < 0.001. c DHX9 protein level was decreased after E6/E7 knockdown. Western blotting analysis of DHX9 protein was performed in HeLa and SiHa transfected with an E6/E7 siRNA pool. d DHX9 protein level was increased in C-33 A cells transducing with E6/E7. HPV-negative C-33 A cells were transfected with HPV E6 and E7 plasmids. DHX9, E6, and E7 proteins were examined by Western blotting using the indicated antibodies. e Lnc-CCDST level was decreased in E6/E7 transducing C-33A cells. lnc-CCDST in C-33A cells transfected with E6 and E7 plasmids was detected by RT-qPCR. GAPDH was used as an internal control for normalization. Data are mean ± SEM determined from three independent experiments (n = 3), each with four technical replicates. Two-tailed unpaired Student’s t test was used. *P < 0.05, **P < 0.01, ***P < 0.001. f In normal tissues, the level of lnc-CCDST is significantly lower in HPV16/18-positive group than in HPV16/18-negative group. The lnc-CCDST expression levels were examined in HPV16/18-positive or -negative cervical normal tissues. β-actin was used as an internal control for normalization. Data are mean ± SD, and two-tailed unpaired Student’s t test was used. *P < 0.05. g In cancer tissues, lnc-CCDST is significantly lower in HPV16/18-positive group than in HPV16/18-negative group. The lnc-CCDST expression levels were examined in HPV16/18-positive or -negative CC tissues. β-actin was used as an internal control for normalization. Data are mean ± SD, and two-tailed unpaired Student’s t test was used. *P < 0.05. h A schematic model of this work. Our results (the black arrows) indicate that HPV E6/E7 decreases lnc-CCDST level by an unknown mechanism in cervical cells, which stabilizing DHX9 by reducing the MDM2/DHX9 interaction. We extend the HPV knowledge in the literature (the red arrows) that HPV E6 degrades p53 through E6-AP rather than MDM2, and p53 possesses the ability to transcriptionally activate MDM2, which might explain why E6 also inhibits MDM2 expression in HPV-infected cells. p53 is also inhibited by DHX9 in CC cells but the mechanism remains unclear
Discussion
The lncRNA was initially described as the “dark matter” of the genome and transcriptional noise. It is now apparent that a subset of these lncRNAs is functional. The functions of lncRNAs include signaling of protein metabolism, recruitment of chromatin modifiers, and regulation of cell cycle control, and cellular functions [18]. In this study, we have reported that lnc-CCDST (ENST00000429352), a 397 nt intergenic non coding RNA located at chromosome 1: 152,205,858–152,207,057, is downregulated in most CC patients. Overexpression of lnc-CCDST inhibits cell migration, invasion, anchorage-independent growth, and angiogenesis, implying that this lncRNA might play an important role in the development and progression of CC.
Recent studies have indicated that lncRNAs interact with proteins to modulate protein function, regulate protein–protein interactions, or directly localize within cellular compartments to regulate cellular functions [19]. Our results have shown that lnc-CCDST decreases DHX9 protein level via direct interaction. DHX9, a member of the DEAH-containing family of RNA helicases, has many regulatory roles in a broad range of cellular processes. As a multi-domain protein, DHX9 is capable of unwinding DNA, RNA, and complex polynucleotide structures [20]. Its functions include regulation of DNA replication [21], transcription [22], RNA transport [23], translation [24], microRNA and circular RNA processing [25], and genome maintenance [26]. There are increasing evidence that DHX9 might be involved in various human cancers. Intriguingly, DHX9 is hijacked by numerous viruses, including HIV-1 [27], HCV [28], HEV [29], HCMV [30], and influenza A virus [31] to enhance infection efficiency. To our knowledge, this is the first report of DHX9 regulation through ubiquitination post-translational modification. More importantly, we have shown that DHX9 ubiquitination is regulated by a cellular lncRNA and HPV oncoproteins E6 and E7.
MDM2 has oncogenic properties, and is able to immortalize mouse NIH3T3 and primary mouse embryonic fibroblasts cells, and accelerate the growth of human fibroblasts [32]. Several in vivo models have revealed MDM2’s oncogenic roles. For instance, MDM2 overexpression could induce sarcoma or lymphomas, and carcinoma [33]. In CC, MDM2 T309G polymorphism has been suggested to be a risk factor for carcinogenesis [34]. However, IHC staining did not detect significant difference of MDM2 expression between CC and normal tissues [35], suggesting alternative mechanism might regulate its function. Nevertheless, MDM2 promotes cervix cell proliferation [36] and migration [37, 38]. In this study, we have shown that lnc-CCDST can act as a scaffold structure to allow MDM2 to bind and destabilize with DHX9.
Infection by high-risk HPVs is the risk factor for CC. HPV16 and HPV18 cause more than two thirds of the world’s CC burden while other high-risk HPVs, such as HPV31, HPV33, HPV45, and HPV58 are predominantly associated with the remaining CC. HPV encodes two oncoproteins, E6 and E7, which directly contribute to the development and progression of HPV-induced carcinogenesis. HPV E6 and E7 jointly regulate various cellular functions, including cell cycle and apoptosis, leading to cell immortalization and malignant proliferation. The combined action of HPV E6 and E7 is important for HPV-induced malignancies. However, how lncRNAs mediate E6- and E7-induced carcinogenesis remains unclear. We showed that knockdown of E6 and E7 led to an increase of lnc-CCDST expression and a decrease of DHX9. Hence, E6 and E7 inhibit the expression of lnc-CCDST to increase the level of pro-oncogenic DHX9. Although E6 strongly induces DHX9 than E7 (Fig. 7d), lnc-CCDST is preferentially decreased by E7 (Fig. 7e). These paradoxical results imply that E6/E7 may regulate DHX9 expression by other unknown manners, such as by modulating promoter activity, mRNA translation efficiency, protein degradation, and signal transduction. Moreover, how E6 and/or E7 regulate lnc-CCDST is unclear and it is worth further investigations. It is possible that E6 and/or E7 might reduce the activities of transcription factors bound to the lnc-CCDST’s promoter, or accelerate the degradation of lnc-CCDST.
Our results have revealed that lnc-CCDST might have a tumor suppressor function. In the normal cervix tissues, it is likely that high levels of lnc-CCDST serve as scaffolds to recruit MDM2 to DHX9, hence increasing DHX9 degradation, and suppressing the oncogenic function of DHX9. Upon infection by high-risk HPVs, E6 and E7 decreased the level of lnc-CCDST relieving the binding of MDM2 to DHX9, hence leading to a higher level of DHX9 and oncogenicity. DHX9 was reported to decrease p53 level in the mouse and human diploid fibroblasts model [39, 40], and we also observed similar results when we knocked down DHX9 in HeLa cells (data not shown). However, we found that overexpression of lnc-CCDST had no effect on p53 protein level (data not shown). We assume that other p53 regulators may also be controlled by lnc-CCDST to offset the negative effect of DHX9 on p53 expression. It is well established that p53 is degraded by MDM2, and MDM2 is transcriptional activated by p53 [41–43]. However, MDM2 is not responsible for p53 degradation in HPV-positive cells [44], though p53 may still have the ability to promote MDM2 transcription. These results are consistent with the observations that both p53 and MDM2 were increased after HPV E6 knockdown [35, 45], as well as our results that DHX9 was upregulated (Fig. 7a), and p53 [46, 47] and MDM2 (Fig. 7a) were downregulated in HPV-positive cells compared with the HPV-negative cells. However, the mechanism of MDM2 downregulation in HPV-positive cell lines remains unclear. Therefore, our working model is that HPV E6/E7 inhibits lnc-CCDST by an unknown mechanism, which reduces the effect of MDM2 targeting DHX9 for degradation, hence promoting malignant cell proliferation and tumorigenesis in HPV-infected cells (Fig. 7h). Nevertheless, the mechanism of HPV and, more specifically, E6/E7 induce deregulation of lnc-CCDST and DHX9, and other possible relationship between lnc-CCDST and DHX9 remain to be further investigated. Our findings have identified novel potential therapeutic targets for CC. Future studies should examine the mechanism by which HPV oncoproteins regulate lnc-CCDST expression and the specific lysine(s) of DHX9 protein ubiquitinated by MDM2.
Materials and methods
Clinical samples
Fresh frozen CC tissues and their adjacent normal tissues were obtained from the First Affiliated Hospital of Nanjing Medical University and Affiliated Gynecology and Obstetrics Hospital of Nanjing Medical University. The samples were taken within 10 min after tumor excision, and immediately stored at −80 °C until application in the experiments. Written informed consents were acquired from all patients and this study was approved by the Ethics Committee of the hospitals.
Microarray analysis
Total RNA of three CC tissues and paired adjacent normal tissues were extracted, and the expression profile of 30,584 human lncRNAs was conducted with the Arraystar Human LncRNA Microarray V3.0. Volcano plot filtering, set at a threshold of ≥ 2.0 folds, was used to screen for lncRNAs that exhibited significantly different (P < 0.05; unpaired t test) expression levels in the two study groups.
Preparation of nuclear and cytoplasmic extracts
The nuclear and cytoplasmic fraction of cells was extracted using a RNA Subcellular Isolation Kit (Active motif, 25501). Cells were washed three times with PBS on ice followed by centrifugation at 300 g for 5 min. Cell pellets were resuspended in cell fraction buffer from the RNA Subcellular Isolation Kit, incubated on ice for 10 min, and then centrifuged at 500 g for 5 min at 4 °C. Nuclear and cytoplasmic pellets were homogenized with the cell disruption buffer from the RNA Subcellular Isolation Kit.
Cell culture, treatment, plasmids, transfection, and lentiviral transduction
HeLa, SiHa, C-33 A, HT-3, HUVECs, and HEK 293 T cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), and 1% penicillin/streptomycin (pen/strep). All cell lines were cultured in an atmosphere of 5% CO2, 99% relative humidity at 37 °C.
For cell treatment, 20 μg ml−1 CHX (Sigma-Aldrich) were incubated with cells for the indicated periods of time. Cells were treated with proteasome inhibitor MG132 (Selleckchem) at 5 μM for 4 h. Plasmid and siRNA transfections were performed using Lipofectamine 2000 (Life Technologies). Lnc-CCDST was cloned into green fluorescent protein (GFP) based lentiviral transferring plasmid pCDH-CMV-MCS-EF1-copGFP (abbreviated as pCDH) to generate recombinant pCDH-Lnc-CCDST. The shRNA plasmids (shDHX9, shMDM2) were constructed in the modified pCDH vector (mpCDH), which contains miR-30 precursor stem-loops sequence. The pCDH/mpCDH plasmids were co-transfection with psPAX2 and pMD2.G to generate indicated lentiviruses. The pCMV3-HA-DHX9, pCMV3-Flag-MDM2 plasmids were purchased from Sino Biological Inc. (Beijing, China). siRNA oligonucleotides targeting HPVs E6/E7 and non-targeting siRNA were purchased from GenePharma. The short hairpin RNA (shRNA) expressing lentiviral vector was constructed as previously described [48]. The sequences of siRNAs and shRNAs used in this study are summarized in Supplementary Table 1.
RNA extraction and RT-qPCR analysis
Total RNA from the cervical tissue specimens and cell lines used in this study was extracted with TRIzol reagent (Life Technologies). We synthesized cDNA by using the HiScript II Q RT SuperMix for qPCR (Vazyme Biotech). Relative RNA levels determined by RT-qPCR were measured on a Roche lightcycler 96 by using the ChamQ SYBR qPCR Master Mix (Vazyme Biotech); the sequences for the gene-specific primers used are listed in Supplementary Table 2. GAPDH or actin was employed as an internal control for quantification of lnc-CCDST and the mRNA levels of other genes. The relative expression of RNAs was calculated using the comparative Ct method.
Western blotting and antibodies
Western blotting and immunoprecipitation were performed as previously described [49]. Briefly, protein extracts from cells or immunoprecipitation samples were prepared using detergent-containing lysis buffer. Total protein (60 μg) was subjected to SDS-PAGE and transferred to 0.45 μm PVDF membrane (Millipore). Antibodies against DHX9 (Abcam, ab26271), MDM2 (Abcam, ab16895), ubiquitin (Cell Signaling Technology, 3936), HPV16 E6 + HPV18 E6 (Abcam, ab70), HPV16 E7 (Santa Cruz Biotechnology, sc-6981), HPV18 E7 (Abcam, ab100953), Tubulin (Santa Cruz Biotechnology, sc-23948), and GAPDH (Santa Cruz Biotechnology, sc-137179) were used. Membranes were incubated overnight at 4 °C with primary antibody diluted 1:1, 000–1:10, 000 according to manufactures’ instructions, and proteins were detected with a peroxide lumiglo reagent (Cell Signaling Technology, 7003).
Analysis of cell migration, invasion, angiogenesis
Cell migration, invasion, and angiogenesis were performed as previously described [25, 50, 51]. In total, 1 × 104 cells were added to the upper 8-μm pore inserts (Millipore) in serum-free medium. We added DMEM containing 10% FBS to the lower chambers as a chemoattractant. After indicate time, cultured at 37 °C in an incubator at 5% CO2, cells that migrated through the inserts were fixed with methanol and stained with crystal violet. Cell numbers were counted in six random fields. The invasion assay was conducted in a similar fashion with coating the filters with 30 μg of Matrigel (BD Biosciences), and 5 × 104 cells were added to the upper inserts. After incubating for 48 h, the conditioned medium was collected from the same number of cells. Then the conditioned medium was added to 7 × 103 HUVECs, the capillary tubes were measured after 3 h incubation.
Soft agar colony formation assays
A bottom layer was prepared consisting of a 0.8% agarose (BD Biosciences) solution mixed with culture medium in six-well plates. HeLa or SiHa cells (1 × 104) suspended in 0.4% top agarose solution mixed with culture medium were placed on top of the bottom agarose. The plates were incubated at 37 °C with 5% CO2. Random five fields were chosen and photographed, and the colony areas were counted by NIH Image J software.
Cell proliferation assay
Cell Count Kit-8 was purchased from Dojindo Molecular Technologies (Tokyo, Japan) and used to examine cell proliferation according to the manufacturer’s instructions.
Immunohistochemistry (IHC)
The CC tissue specimens and the normal provided by different hospitals were applied for hematoxylin and eosin (H&E) and immunohistochemistry (IHC) staining. All the samples were formalin-fixed, paraffin-embedded, and immunostained with DHX9 antibodies. Three random fields were photographed under the microscope.
RNA pulldown and mass spectrometry analysis
In vitro transcription of lnc-CCDST was performed with primers containing the T7 promoter sequence. RNA pulldown assay was performed according to the manufacturer’s instructions. Briefly, a single biotinylated desthiobiotinylated cytidine was attached to RNA 3′ terminus by using T4 RNA ligase. The biotinylated lnc-CCDST was incubated with cellular protein extracts from HeLa or SiHa cells, and streptavidin beads were then added. Recovered proteins associated with lnc-CCDST or control were resolved by gel electrophoresis and silver staining. The eluted solutions were accomplished by mass spectrometry analysis (Shanghai Applied Protein Technology Co., Ltd) on Q Exactive mass spectrometer (Proxeon Biosystems, now Thermo Fisher Scientific).
RNA immunoprecipitation (RIP)
RIP experiments were performed using the Magna RIP RNA-Binding Protein Immunoprecipitation kit (Millipore, 17–700). HeLa and SiHa cells were lysed and the whole-cell lysates were incubated with protein A+G magnetic beads, which were conjugated using 5 μg of DHX9 antibodies (Abcam, ab26271) or control IgG at 4 °C for overnight. The immunoprecipitated RNA was purified and detected by RT-qPCR.
Chicken chorioallantoic membranes assay (CAM)
The CAM assay was performed as previously described [50]. Fertilized White Leghorn chicken eggs were incubated at 37 °C under conditions of constant humidity. A window was opened on the shell above the air sac on day 9. The opening on the eggs was sealed with paraffin. The cells were suspended in a medium containing 50% High Concentration Matrigel (BD Biosciences). Aliquots (0.2 ml) of the mixture were then applied onto the CAMs of 9-day-old embryos. The implanted Matrigel was photographed 4–5 days post-implantation, and the blood vessels were counted by two observers in a double-blind fashion.
The Matrigel plug assay in nude mice
All animal care and procedures were executed on the basis of standards established by Laboratory Animal Management Regulations approved by the State Council of People’s Republic of China. Nanjing Medical University Experimental Animal Welfare Ethics Committee approved the animal experiments (IACUC-1705037). The plug assay was performed as previously described [25, 51, 52]. Female BALB/c nude mice of 4-week-old were purchased from the Charles River Laboratories and maintained under pathogen-free conditions. The cells were resuspended in serum-free medium. In total, 0.2 ml cell aliquots were mixed with 0.2 ml of High Concentration Matrigel (BD Biosciences), and the mixture was immediately injected into the flanks of nude mice subcutaneously. Matrigel plugs were removed from the mice after the mice were killed 14 days postinjection. The hemoglobin level of the plugs was analyzed using Drabkin’s reagent kit (Sigma-Aldrich) according to the manufacturer’s instructions. The final hemoglobin concentration was detected at 540 nm and calculated from a standard calibration curve.
Statistical analysis
For the cell functional analyses, results are presented as mean ± SEM, unless otherwise stated; the comparison of means between two groups was conducted using Student’s t test, P < 0.05 is considered significant. All the experiments were repeated at least three times, unless otherwise stated.
Electronic supplementary material
Acknowledgements
We are grateful to Mi Zhang for collection of CC specimens. We also thank members from Dr. Lu laboratory for helpful discussion. This work was supported partially by grants from National Natural Science Foundation of China (81730062 and 81761128003), and Nanjing Medical University (KY101RC1710).
Author contributions
Conceived and designed the experiments: CL. Performed the experiments: XD, XJ, CW, and JX. Analyzed the data: XD, SJG, and CL. Wrote the paper: XD, SJG, and CL.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary material
The online version of this article (10.1038/s41418-018-0242-0) contains supplementary material, which is available to authorized users.
References
- 1.Canavan TP, Doshi NR. Cervical cancer. Am Fam Physician. 2000;61:1369–76. [PubMed] [Google Scholar]
- 2.Fleming ND, Frumovitz M, Schmeler KM, dos Reis R, Munsell MF, Eifel PJ, et al. Significance of lymph node ratio in defining risk category in node-positive early stage cervical cancer. Gynecol Oncol. 2015;136:48–53. doi: 10.1016/j.ygyno.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munoz N, Bosch FX, de Sanjose S, Herrero R, Castellsague X, Shah KV, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348:518–27. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- 4.Vande Pol SB, Klingelhutz AJ. Papillomavirus E6 oncoproteins. Virology. 2013;445:115–37. doi: 10.1016/j.virol.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roman A, Munger K. The papillomavirus E7 proteins. Virology. 2013;445:138–68. doi: 10.1016/j.virol.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hosseini ES, Meryet-Figuiere M, Sabzalipoor H, Kashani HH, Nikzad H, Asemi Z. Dysregulated expression of long noncoding RNAs in gynecologic cancers. Mol Cancer. 2017;16:107. doi: 10.1186/s12943-017-0671-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wutz A, Gribnau J. X inactivation Xplained. Curr Opin Genet Dev. 2007;17:387–93. doi: 10.1016/j.gde.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Halley P, Kadakkuzha BM, Faghihi MA, Magistri M, Zeier Z, Khorkova O, et al. Regulation of the apolipoprotein gene cluster by a long noncoding RNA. Cell Rep. 2014;6:222–30. doi: 10.1016/j.celrep.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan JH, Liu XN, Wang TT, Pan W, Tao QF, Zhou WP, et al. The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of lncRNA-PXN-AS1. Nat Cell Biol. 2017;19:820–32. doi: 10.1038/ncb3538. [DOI] [PubMed] [Google Scholar]
- 10.Centonze D, Rossi S, Napoli I, Mercaldo V, Lacoux C, Ferrari F, et al. The brain cytoplasmic RNA BC1 regulates dopamine D2 receptor-mediated transmission in the striatum. J Neurosci. 2007;27:8885–92. doi: 10.1523/JNEUROSCI.0548-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101–12. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoon JH, Abdelmohsen K, Kim J, Yang X, Martindale JL, Tominaga-Yamanaka K, et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun. 2013;4:2939. doi: 10.1038/ncomms3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang B, Sun R, Fang S, Qin C, Pan X, Peng L, et al. Lnc-CC3 increases metastasis in cervical cancer by increasing Slug expression. Oncotarget. 2016;7:41650–61. doi: 10.18632/oncotarget.9519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Q, Guo X, Que S, Yang X, Fan H, Liu M, et al. LncRNA RSU1P2 contributes to tumorigenesis by acting as a ceRNA against let-7a in cervical cancer cells. Oncotarget 2016;8:43768–81. [DOI] [PMC free article] [PubMed]
- 15.Wei X, Pacyna-Gengelbach M, Schluns K, An Q, Gao Y, Cheng S, et al. Analysis of the RNA helicase A gene in human lung cancer. Oncol Rep. 2004;11:253–8. [PubMed] [Google Scholar]
- 16.Okoro DR, Arva N, Gao C, Polotskaia A, Puente C, Rosso M, et al. Endogenous human MDM2-C is highly expressed in human cancers and functions as a p53-independent growth activator. PLoS ONE. 2013;8:e77643. doi: 10.1371/journal.pone.0077643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee T, Paquet M, Larsson O, Pelletier J. Tumor cell survival dependence on the DHX9 DExH-box helicase. Oncogene. 2016;35:5093–105. doi: 10.1038/onc.2016.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–61. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 19.Chu HP, Cifuentes-Rojas C, Kesner B, Aeby E, Lee HG, Wei C, et al. TERRA RNA Antagonizes ATRX and Protects Telomeres. Cell. 2017;170:86–101 e116. doi: 10.1016/j.cell.2017.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jain A, Bacolla A, Chakraborty P, Grosse F, Vasquez KM. Human DHX9 helicase unwinds triple-helical DNA structures. Biochemistry. 2010;49:6992–9. doi: 10.1021/bi100795m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chakraborty P, Grosse F. WRN helicase unwinds Okazaki fragment-like hybrids in a reaction stimulated by the human DHX9 helicase. Nucleic Acids Res. 2010;38:4722–30. doi: 10.1093/nar/gkq240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson SF, Schlegel BP, Nakajima T, Wolpin ES, Parvin JD. BRCA1 protein is linked to the RNA polymerase II holoenzyme complex via RNA helicase A. Nat Genet. 1998;19:254–6. doi: 10.1038/930. [DOI] [PubMed] [Google Scholar]
- 23.Tang H, Gaietta GM, Fischer WH, Ellisman MH, Wong-Staal F. A cellular cofactor for the constitutive transport element of type D retrovirus. Science. 1997;276:1412–5. doi: 10.1126/science.276.5317.1412. [DOI] [PubMed] [Google Scholar]
- 24.Manojlovic Z, Stefanovic B. A novel role of RNA helicase A in regulation of translation of type I collagen mRNAs. RNA. 2012;18:321–34. doi: 10.1261/rna.030288.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aktas T, Avsar Ilik I, Maticzka D, Bhardwaj V, Pessoa Rodrigues C, Mittler G, et al. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 2017;544:115–9. doi: 10.1038/nature21715. [DOI] [PubMed] [Google Scholar]
- 26.Jain A, Bacolla A, Del Mundo IM, Zhao J, Wang G, Vasquez KM. DHX9 helicase is involved in preventing genomic instability induced by alternatively structured DNA in human cells. Nucleic Acids Res. 2013;41:10345–57. doi: 10.1093/nar/gkt804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Tang H, Mullen TM, Westberg C, Reddy TR, Rose DW, et al. A role for RNA helicase A in post-transcriptional regulation of HIV type 1. Proc Natl Acad Sci USA. 1999;96:709–14. doi: 10.1073/pnas.96.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He QS, Tang H, Zhang J, Truong K, Wong-Staal F, Zhou D. Comparisons of RNAi approaches for validation of human RNA helicase A as an essential factor in hepatitis C virus replication. J Virol Methods. 2008;154:216–9. doi: 10.1016/j.jviromet.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 29.Paingankar MS, Arankalle VA. Identification and characterization of cellular proteins interacting with Hepatitis E virus untranslated regions. Virus Res. 2015;208:98–109. doi: 10.1016/j.virusres.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 30.Lenarcic EM, Ziehr BJ, Moorman NJ. An unbiased proteomics approach to identify human cytomegalovirus RNA-associated proteins. Virology. 2015;481:13–23. doi: 10.1016/j.virol.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin L, Li Y, Pyo HM, Lu X, Raman SN, Liu Q, et al. Identification of RNA helicase A as a cellular factor that interacts with influenza A virus NS1 protein and its role in the virus life cycle. J Virol. 2012;86:1942–54. doi: 10.1128/JVI.06362-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lenos K, de Lange J, Teunisse AF, Lodder K, Verlaan-de Vries M, Wiercinska E, et al. Oncogenic functions of hMDMX in in vitro transformation of primary human fibroblasts and embryonic retinoblasts. Mol Cancer. 2011;10:111. doi: 10.1186/1476-4598-10-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lundgren K, Montes de Oca Luna R, McNeill YB, Emerick EP, Spencer B, Barfield CR, et al. Targeted expression of MDM2 uncouples S phase from mitosis and inhibits mammary gland development independent of p53. Genes Dev. 1997;11:714–25. doi: 10.1101/gad.11.6.714. [DOI] [PubMed] [Google Scholar]
- 34.Singhal P, Hussain S, Thakur N, Batra S, Salhan S, Bhambani S, et al. Association of MDM2 and p53 polymorphisms with the advancement of cervical carcinoma. DNA Cell Biol. 2013;32:19–27. doi: 10.1089/dna.2012.1718. [DOI] [PubMed] [Google Scholar]
- 35.Jin H, Lee K, Kim YH, Oh HK, Maeng YI, Kim TH, et al. Scaffold protein FHL2 facilitates MDM2-mediated degradation of IER3 to regulate proliferation of cervical cancer cells. Oncogene. 2016;35:5106–18. doi: 10.1038/onc.2016.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ren ZJ, Nong XY, Lv YR, Sun HH, An PP, Wang F, et al. Mir-509-5p joins the Mdm2/p53 feedback loop and regulates cancer cell growth. Cell Death Dis. 2014;5:e1387. doi: 10.1038/cddis.2014.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang AR, An HT, Ko J, Kang S. Ataxin-1 regulates epithelial-mesenchymal transition of cervical cancer cells. Oncotarget. 2017;8:18248–59. doi: 10.18632/oncotarget.15319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Q, Li M, Wang Y, Zhang Y, Jin S, Xie G, et al. RNA interference targeting CML66, a novel tumor antigen, inhibits proliferation, invasion and metastasis of HeLa cells. Cancer Lett. 2008;269:127–38. doi: 10.1016/j.canlet.2008.04.035. [DOI] [PubMed] [Google Scholar]
- 39.Mills JR, Malina A, Lee T, Di Paola D, Larsson O, Miething C, et al. RNAi screening uncovers Dhx9 as a modifier of ABT-737 resistance in an Emu-myc/Bcl-2 mouse model. Blood. 2013;121:3402–12. doi: 10.1182/blood-2012-06-434365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee T, Di Paola D, Malina A, Mills JR, Kreps A, Grosse F, et al. Suppression of the DHX9 helicase induces premature senescence in human diploid fibroblasts in a p53-dependent manner. J Biol Chem. 2014;289:22798–814. doi: 10.1074/jbc.M114.568535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–9. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 42.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 43.Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461–8. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Traidej M, Chen L, Yu D, Agrawal S, Chen J. The roles of E6-AP and MDM2 in p53 regulation in human papillomavirus-positive cervical cancer cells. Antisense Nucleic Acid Drug Dev. 2000;10:17–27. doi: 10.1089/oli.1.2000.10.17. [DOI] [PubMed] [Google Scholar]
- 45.Koivusalo R, Mialon A, Pitkanen H, Westermarck J, Hietanen S. Activation of p53 in cervical cancer cells by human papillomavirus E6 RNA interference is transient, but can be sustained by inhibiting endogenous nuclear export-dependent p53 antagonists. Cancer Res. 2006;66:11817–24. doi: 10.1158/0008-5472.CAN-06-2185. [DOI] [PubMed] [Google Scholar]
- 46.Srivastava S, Tong YA, Devadas K, Zou ZQ, Chen Y, Pirollo KF, et al. The status of the p53 gene in human papilloma virus positive or negative cervical carcinoma cell lines. Carcinogenesis. 1992;13:1273–5. doi: 10.1093/carcin/13.7.1273. [DOI] [PubMed] [Google Scholar]
- 47.Scheffner M, Munger K, Byrne JC, Howley PM. The state of the p53 and retinoblastoma genes in human cervical carcinoma cell lines. Proc Natl Acad Sci USA. 1991;88:5523–7. doi: 10.1073/pnas.88.13.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu M, Wang C, Li W, Lu W, Bai Z, Qin D, et al. A KSHV microRNA Directly Targets G Protein-Coupled Receptor Kinase 2 to Promote the Migration and Invasion of Endothelial Cells by Inducing CXCR2 and Activating AKT Signaling. PLoS Pathog. 2015;11:e1005171. doi: 10.1371/journal.ppat.1005171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li W, Yan Q, Ding X, Shen C, Hu M, Zhu Y, et al. The SH3BGR/STAT3 pathway regulates cell migration and angiogenesis induced by a gammaherpesvirus MicroRNA. PLoS Pathog. 2016;12:e1005605. doi: 10.1371/journal.ppat.1005605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu X, Guo Y, Yao S, Yan Q, Xue M, Hao T, et al. Synergy between Kaposi’s sarcoma-associated herpesvirus (KSHV) vIL-6 and HIV-1 Nef protein in promotion of angiogenesis and oncogenesis: role of the AKT signaling pathway. Oncogene. 2014;33:1986–96. doi: 10.1038/onc.2013.136. [DOI] [PubMed] [Google Scholar]
- 51.Yao S, Hu M, Hao T, Li W, Xue X, Xue M, et al. MiRNA-891a-5p mediates HIV-1 Tat and KSHV Orf-K1 synergistic induction of angiogenesis by activating NF-kappaB signaling. Nucleic Acids Res. 2015;43:9362–78. doi: 10.1093/nar/gkv988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xue M, Yao S, Hu M, Li W, Hao T, Zhou F, et al. HIV-1 Nef and KSHV oncogene K1 synergistically promote angiogenesis by inducing cellular miR-718 to regulate the PTEN/AKT/mTOR signaling pathway. Nucleic Acids Res. 2014;42:9862–79. doi: 10.1093/nar/gku583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.