Abstract
Transmembrane protein 268 (TMEM268) encodes a novel human protein of previously unknown function. This study analyzed the biological activities and molecular mechanisms of TMEM268 in vivo and in vitro. We found that TMEM268 deletion decreases cell viability, proliferation, and cell adhesion as well as causing S-phase cell cycle arrest and disrupts cytoskeleton remolding. Xenograft tumor mouse model studies showed that TMEM268 deletion inhibits the tumorigenesis of BGC823 gastric cancer cells. In addition, TMEM268-deleted BGC823 cells failed to colonize the lungs after intravenous injection and to form metastatic engraftment in the peritoneum. Molecular mechanism studies showed a C-terminal interaction between TMEM268 and integrin subunit β4 (ITGB4). TMEM268 knockout promotes ITGB4 ubiquitin-mediated degradation, increasing the instability of ITGB4 and filamin A (FLNA). The reduced ITGB4 protein levels result in the disassociation of the ITGB4/PLEC complex and cytoskeleton remodeling. This study for the first time demonstrates that TMEM268 plays a positive role in the regulation of ITGB4 homeostasis. The above results may provide a new perspective that targeting the TMEM268/ITGB4 signaling axis for the treatment of gastric cancer, which deserves further investigation in the future.
Subject terms: Tumour-suppressor proteins, Integrins
Introduction
Integrins are transmembrane glycoproteins that form an important family of heterodimeric cell adhesion receptors [1]. They have been shown to be critical in controlling cell−cell and cell−matrix interactions, and in connecting the extracellular matrix (ECM) to the cell cytoskeleton. They exert diverse effects on the proliferation, migration, and invasion of cancer cells [2–5], and regulate differentiation, angiogenesis, epithelial-to-mesenchymal transition, and even therapeutic outcomes [6–8]. These properties make integrins attractive prognostic indicators and therapeutic targets for cancer [9, 10].
The integrin subunit β4 (ITGB4) heterodimerizes with the integrin subunit α6 (ITGA6) to form a receptor for laminin (LN). ITGB4 has an unusually long cytoplasmic domain that has distinctive cytoskeletal and signaling functions [11]. One fundamental function of integrin α6β4 in polarized epithelial cells is to form a stable attachment to the basal membrane through the formation of hemidesmosomes. It is linked to the actin cytoskeleton. In vitro and in vivo experiments indicate that ITGB4 promotes tumorigenesis and metastasis in different types of cancers [12, 13]. Increasing evidence indicates that the overexpression of ITGB4 is correlated with an aggressive phenotype and poor prognosis in breast cancer, lung cancer, pancreatic cancer, cervical cancer, and gastric cancer [14–18]. These observations suggest that expression of ITGB4 confers an evolutionary advantage during tumor progression and potential therapeutic target. The regulatory mechanism for the expression of ITGB4 protein remains to be clarified.
As part of a human genomics project, we cloned hundreds of functionally unknown human open-reading frames (ORFs) by searching the human RefSeq and expressed sequence tag databases in GenBank. Using a cell-based high-throughput assay, we identified several novel genes associated with cell viability, including transmembrane protein 268 (TMEM268), previously identified as C9orf91 [19]. Transcription of the human TMEM268 gene produces two experimental confirmed mRNAs (TMEM268-v1 and TMEM268-v2), both of which encompass nine exons and eight introns. The full length of human TMEM268-v1 cDNA is 4413 base pairs (bps) comprising an ORF encoding a predicted 37.6 kDa protein of 342 amino acids. This TMEM268-v1 has been chosen as the canonical sequence, usually abbreviated as TMEM268. The full-length of TMEM268-v2 cDNA is 4481 bps long, its ORF encodes a predicted 37.7 kDa protein of 343 amino acids. The difference between the amino acid sequences of TMEM268-V1 and TMEM268-V2 is that the latter has an extra Glutamine (Q) behind 71 Isoleucine (I) (71: I → IQ), and all the other amino acids are the same (https://www.uniprot.org/uniprot/Q5VZI3). Transmembrane analysis (www.cbs.dtu.dk/services/TMHMM-2.0/) suggests that TMEM268 has two conserved TM domains (amino acids 104–126 and 130–152) and a domain of unknown function (DUF4481, amino acids 38–328) [20]. To our knowledge, no functional studies have been performed on this protein.
In the present study, we demonstrate that TMEM268 deficiency in gastric cancer cells inhibits cell growth, adhesion, and causes cell cycle arrest. Mechanistically, TMEM268 interacts with ITGB4; TMEM268 deletion promotes ITGB4 degradation via the protease pathway. Additionally, deletion of TMEM268 facilitates the disintegration of ITGB4 and Plectin, impairs FLNA stability and the F-actin network, which eventually leads to cytoskeletal remolding in cancer cells.
Results
Inactivation of TMEM268 inhibits cell growth and reduces tumorigenesis in gastric cancers cells
Data from RT-PCR and western blotting proved that TMEM68 is expressed in many human cell lines (Fig. S1A and B). Immunofluorescence assay demonstrated that the TMEM268 protein was mainly present in the endoplasmic reticulum and plasma membrane (Fig. S2). These data are consistent with data reported in The Human Protein Atlas for TMEM268 (http://www.proteinatlas.org/ENSG00000157693-TMEM268).
To clarify the physiological role of TMEM268, we conducted a series of experiments in TMEM268-depleted cells. We first identified an effective shRNA against TMEM268 in BGC823 and SGC7901 cell lines (Fig. S3A). MTS assay showed that cell viability of shTMEM268-transfected cells is significantly lower than that in the shControl group (Fig. S3B and C). A 5-ethynyl-2′-deoxyuridine (EdU) incorporation assay demonstrated that shTMEM268-transfected cells contain lower percentage of EdU-positive cells than control cells (Fig. S3D and E). This antiproliferative effect was further demonstrated in a colony formation assay (Fig. S3F and G).
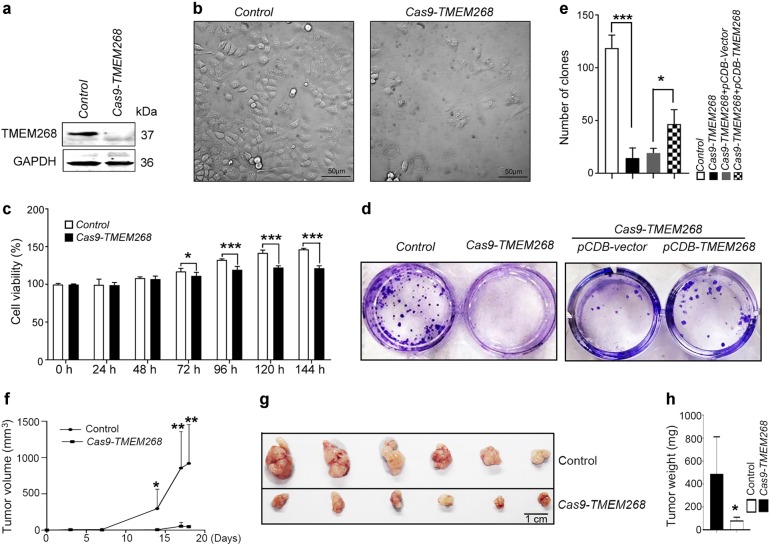
To further prove the function of TMEM268 in gastric cancer cells, we knocked out TMEM268 in BGC823 cells. Through a series of screenings, a Cas9-TMEM268/BGC823 clone was selected. Sequence analysis revealed that the selected clone contained a 4 bp deletion (ACAATG → TG) resulting in a frame shift which disrupts the TMEM268 ORF, leading to deletion of the TM domains and C-terminal (Fig. S4). Western blotting confirmed that the TMEM268 protein was not detectable in Cas9-TMEM268/BGC823 cells (Fig. 1a). It was found that Cas9-TMEM268/BGC823 cells displayed a slow cell growth, accompanied by cell flattening, as well as detachment from the culture dish (Fig. 1b). The growth arrest of Cas9-TMEM268/BGC823 cells was determined by MTS assay (Fig. 1c) and colony formation assay (Fig. 1d, e). The specific effects of TMEM268 knockout were assessed in a rescue experiment. As shown in Fig. 1d, e, overexpression of TMEM268 in Cas9-TMEM268/BGC823 cells significantly increases the number of clones compared to pCDB-vector transfected cells. These results further confirm that knockout of TMEM268 inhibits growth of gastric cancer cells.
Fig. 1.
TMEM268 knockout inhibits cell growth and reduces tumorigenicity. a Western blot analysis of TMEM268 expression in control cells (TMEM268 WT) and Cas9-TMEM268/BGC823 cells. b Control and Cas9-TMEM268/BGC823 cells were seeded in six-well plates (1×105 cells/well). Seventy-two hours later, representative images were obtained by optical microscopy. c Control and Cas9-TMEM268/BGC823 cells were seeded in 96-well plates (3×103 cells/well; five replicates), serum-starved for 18 h and then pulsed with 10% FCS for different lengths of time. Cell viability was detected by MTS assay. Data represent the mean ± SD of three independent experiments. d Representative images of colony formation by indicated cells. e Number of clones counted in three independent experiments. Data are expressed as mean ± SD. *P < 0.05, ***P < 0.001. f The Control or Cas9-TMEM268/BGC823 cells were injected subcutaneously in BALB/c nude mice. Development of tumors (mean volume ± SD) was monitored using calipers. g, h Excised xenograft tumors were imaged and weighed on day 19. *P < 0.05, **P < 0.01, ***P < 0.001
The in vivo effects of TMEM268 were evaluated using a gastric cancer xenograft model established in BALB/C nude mice. These mice were subcutaneously injected with TMEM268 wild-type BGC823 cells or Cas9-TMEM268/BGC823 cells. The tumor growth curves are shown in Fig. 1f. The Control group developed grossly visible tumors at the site of injection. By comparison, the Cas9-TMEM268 group displayed smaller tumors. The tumor weights in the Cas9-TMEM268 group are markedly lighter than those of the Control group (Fig. 1g, h). Collectively, these data indicate that the inactivation of TMEM268 inhibits cell proliferation in gastric cancer cells.
TMEM268 knockout causes S-phase cell cycle arrest
We next analyze whether the growth arrest induced by TMEM268 loss is mediated by apoptosis. Data from flow cytometry analysis indicated that the apoptotic cells were not significantly different between Control and Cas9-TMEM268/BGC823 cells (Fig. S5), suggesting that the decreased viability of Cas9-TMEM268/BGC823 cells is not related to apoptosis.
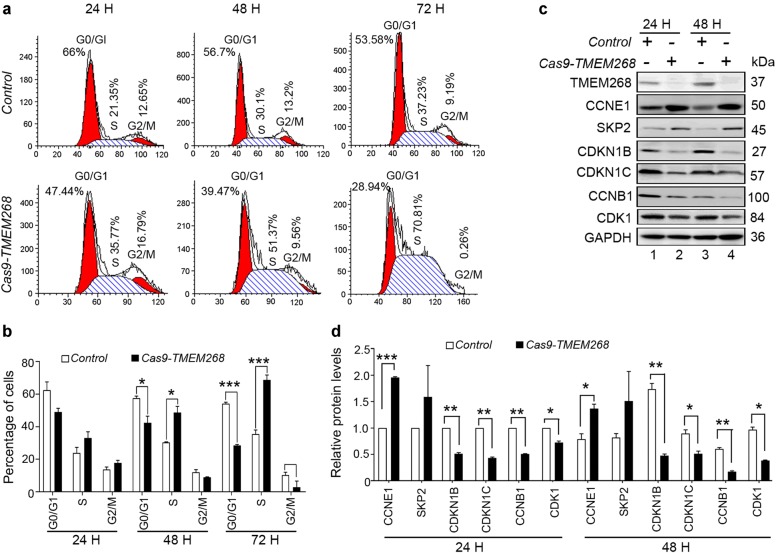
We further explored the effect of TMEM268 on cell cycle progression. As shown in Fig. 2a, b, a much higher percentage of cells in S phase is observed in Cas9-TMEM268/BGC823 cells than in the Control group. In each case, there is a concomitant reduction in the proportion of cells in the G0/G1 and G2/M phases.
Fig. 2.
TMEM268 knockout causes S-phase cell cycle arrest. a Control and CAS9-TMEM268/BGC823 cells were serum-starved for 24 h and then pulsed with 10% FCS for different amounts of time. Cell cycle distribution was assayed by flow cytometry. b Percentages of G0/G1, S, and G2/M phase cells. Each bar represents the mean ± SD of three independent experiments. c Cells were treated as described in (a), and protein levels detected by western blotting. d Quantification of the levels of the indicated proteins relative to GAPDH in cells treated as described in (a). Average value for control cells (24 h) was normalized to 1. Data shown represent the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001
Cell cycle progression depends on the sequential expression of stage-specific cyclins and the activity of their corresponding cyclin-dependent kinases (CDKs) [21]. G1/S transition is required for the activity of the cyclin E1 (CCNE1)/CDK2 complex. CCNE1 protein accumulates at the G1/S-phase boundary and is degraded as cells progress through S phase, triggering G1 → S transition [22]. CDK inhibitor 1B (CDKN1B) and CDK inhibitor 1C (CCKN1C) bind to and prevent the activation of the CCNE1/CDK2 complex, thus controlling cell cycle progression at G1 [23]. S-phase kinase-associated protein 2 (SKP2) regulates the S-phase entry of cells by inducing the degradation of the CDK inhibitors CDKN1B and CDKN1C [24]. The cyclin B1 (CCNB1)/CDK1 complex is essential for S/G2 and G2/M phase cell cycle transitions [25]. In accordance with the observed S-phase arrest, we found that knockout of TMEM268 increased the expression of CCNE1 and SKP2 and decreased the levels of CDKN1B and CCKN1C (Fig. 2c, d), allowing the cells to progress from G1 to the S phase. The levels of CCNB1 and CDK1 were also downregulated in Cas9-TMEM268/BGC823 cells (Fig. 2c, d), thus inhibiting S/G2 and G2/M transitions. Collectively, these data suggest that TMEM268 knockout induces S-phase arrest resulting from enhanced initiation and progression from G1 to S phase and the concomitant inhibition of the S to G2/M checkpoint through inactivation of the CCNB1/CDK1 complex.
TMEM268 knockout impairs cell adhesion
To examine the effects of endogenous TMEM268 on cell migration, a Transwell assay was performed in Cas9-TMEM268/BGC823 and Control cells. The results revealed that the number of cells adhered to the membrane is significantly decreased in the Cas9-TMEM268 group compared with the Control group (Fig. S6A and B). However, many cells were observed at the well bottom and in suspension in Cas9-TMEM268 group, few cells were observed in the Control group. Statistical analysis did not show any differences in total cell number between Cas9-TMEM268 group and Control group, which includes membrane-adhered cells and the shedding cells (Fig. S6C). These data suggest that inactivation of TMEM268 may not affect cell migration in vitro, it reflects a decrease of cell adhesion.
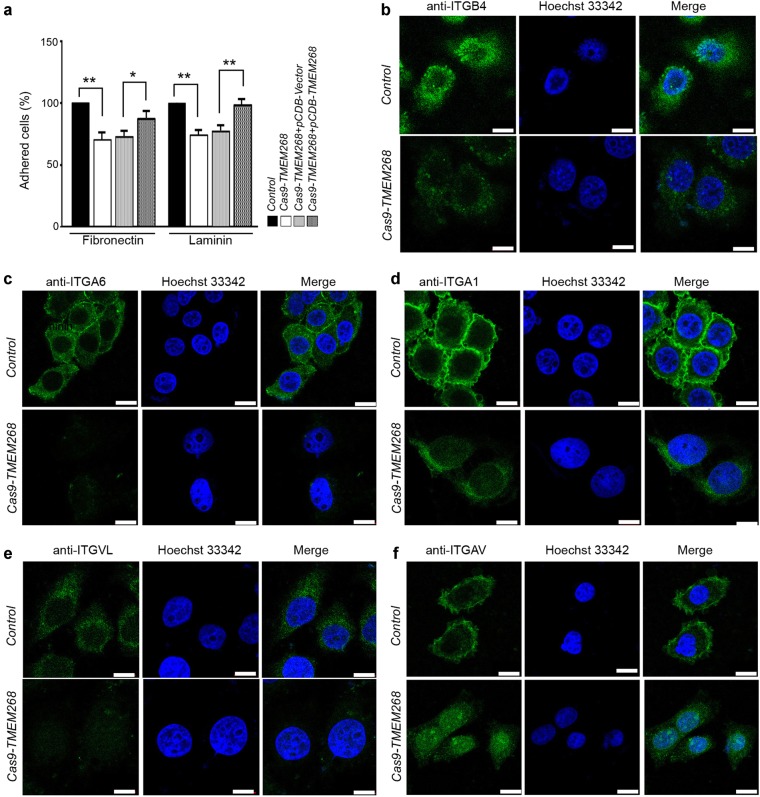
We next explored the effect of TMEM268 deficiency on cell adhesion ability. As shown in Fig. 3a, compared with the control group, the ability of Cas9-TMEM268/BGC823 cells to adhere to laminin (LN) and fibronectin (FN) is decreased, indicating a decrease in cell−ECM adhesion. Data from rescue experiments show that overexpression of TMEM268 in Cas9-TMEM268/BGC823 cells significantly increases cell−LN and cell–FN adhesion compared with the pCDB-Vector group, demonstrating the rescue of cell−ECM adhesion.
Fig. 3.
TMEM268 knockout decreases cell adhesion. a Control and Cas9-TMEM268/BGC823 cells were allowed to attach to 96-well plates precoated with fibronectin or laminin. The unadhered cells were removed at 30 min, and the number of adhered cells was determined using an MTS assay (*P<0.05, **P<0.01). b−f Control and Cas9-TMEM268/BGC823 cells were plated on glass slides for 24 h, incubated with the indicated FITC-conjugated antibodies. Nuclei were stained with Hoechst 33342. Representative images obtained from confocal microscopy are shown. Scale bars, 10 μm
We further analyzed adhesion molecules on the cell membrane associated with cell−matrix interactions. It was observed that in Cas9-TMEM268/BGC823 cells, the ITGB4 fluorescence signal on the surface of the cell membrane was significantly downregulated compared to Control cells (Fig. 3b), while the fluorescence signal of ITGA6 can hardly be detected (Fig. 3c). The membrane fluorescence of integrin subunit alpha 1 (ITGA1) and integrin subunit alpha L (ITGAL) was also decreased in Cas9-TMEM268/BGC823 cells (Figs. 3d, e). The fluorescence of integrin subunit alpha V (ITGAV) could not be detected on the cell surface in Cas9-TMEM268/BGC823 cells (Fig. 3f), but intracellular fluorescence signal was observed, suggesting that the internalization of ITGAV might occur. Collectively, our data suggest that TMEM268 inactivation may inhibit cell adhesion ability by downregulating adhesion molecules on the cell surface.
TMEM268 associates with ITGB4 via their C-terminals, and TMEM268 deletion promotes ITGB4 ubiquitination and degradation
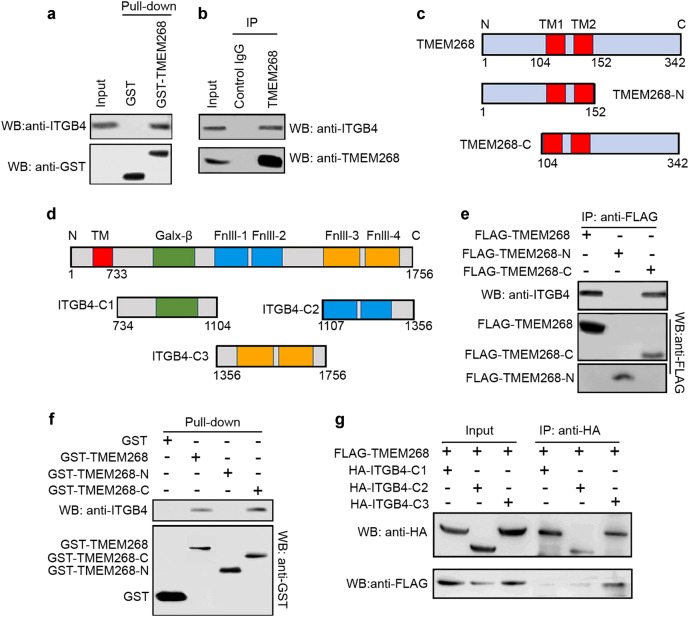
To identify TMEM268 binding partner, we performed a pull-down in BGC823 cell lysates using GST-tagged full-length TMEM268 protein. We found that GST-TMEM268 fusion protein pulled down endogenous ITGB4 (Fig. 4a). The endogenous co-immunoprecipitation (Co-IP) assay revealed that TMEM268 coprecipitated with ITGB4 (Fig. 4b), indicating TMEM268 and ITGB4 may associate together as a complex.
Fig. 4.
The TMEM268/ITGB4 interaction occurs at their C-terminals. a GST or GST-TMEM268 fusion protein were immobilized on glutathione-Sepharose beads and incubated with BGC823 cell lysates at 4 °C for 4 h. ITGB4 and GST were detected in the washed beads by western blotting. b Total BGC823 cell extracts were subjected to IP using either an anti-TMEM268 antibody or an IgG isotype control. ITGB4 was detected in the immunoprecipitates by western blotting. c, d Construction of truncated TMEM268 and ITGB4 plasmids. e BGC823 cells were transfected with FLAG-TMEM268, FLAG-TMEM268-N, or FLAG-TMEM268-C for 24 h. Total cell extracts were subjected to IP using either an anti-FLAG antibody or IgG isotype control. ITGB4 and FLAG were detected in the immunoprecipitates by western blotting. f GST and GST-TMEM268, GST-TMEM268-N or GST-TMEM268-C fusion protein immobilized on glutathione-Sepharose beads were incubated with BGC823 cell lysates. ITGB4 and GST were detected in the washed beads by western blotting. g 293T cells were cotransfected with FLAG-TMEM268 and either HA-ITGB4-C1, HA-ITGB4-C2 or HA-ITGB4-C3 for 24 h. Total cell extracts were subjected to IP using an anti-HA antibody. ITGB4 mutants and FLAG-TMEM268 were detected in the immunoprecipitates by western blotting
We next constructed different TMEM268 and ITGB4 mutants (Fig. 4c, d) and examined which domains were responsible for the TMEM268−ITGB4 interaction. Data from the Co-IP experiments suggest that the C-terminal of TMEM268 (TMEM268104–342) mediates the TMEM268−ITGB4 association (Fig. 4e). GST pull-down further confirmed the interaction between ITGB4 and GST-TMEM268104–342 (Fig. 4f). Further Co-IP assays indicated that the C-terminal of ITGB4 (ITGB41356–1756), which contains Fnlll3 and Fnlll4, interacts with TMEM268.
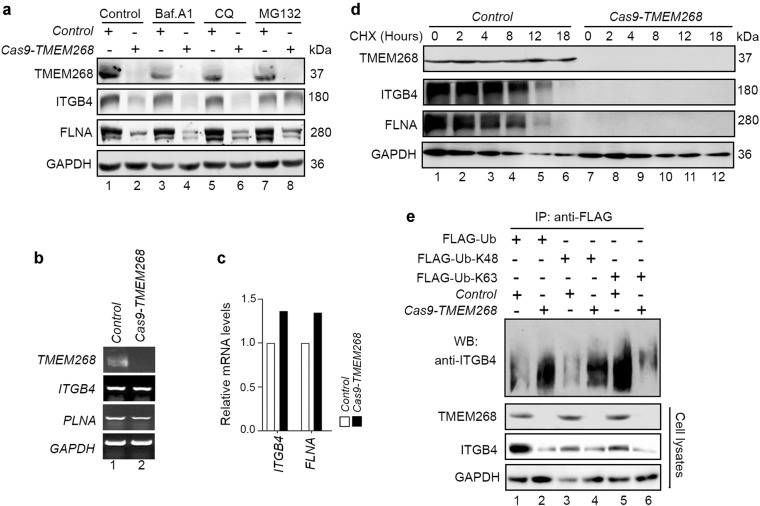
We next explored the biochemical consequence of the TMEM268−ITGB4 interaction. In Cas9-TMEM268/BGC823 cells, the levels of ITGB4 are significantly decreased (Fig. 5a, lane 2 vs. lane 1), consistent with the results of the immunofluorescence assay (Fig. 3b). These data indicate that TMEM268 may mediate the stability of ITGB4. TMEM268 deletion also downregulates the expression of FLNA (Fig. 5a, lane 2 vs. lane 1). Notably, the mRNA levels of ITGB4 and FLNA obtained from RT-PCR and quantitative RT-PCR assays remain unchanged in TMEM268-deleted cells (Fig. 5b, c), suggesting that TMEM268 may regulate the post-translational modification of ITGB4.
Fig. 5.
TMEM268 knockout promotes ITGB4 ubiquitination and degradation. a Control and Cas9-TMEM268/BGC823 cells were treated with different small molecules for different time (10 nM of Baf.A1 or 25 μM of CQ for 4 h; 10 mM MG-132 for 6 h). The cell lysates were then probed with anti-ITGB4 and anti-FLNA antibodies. GAPDH was used as the loading control. b Control and Cas9-TMEM268/BGC823 cells were treated with cycloheximide (CHX, 10 nM) for the indicated times. The levels of endogenous ITGB4 and FLNA were measured by western blotting. c, d RT-PCR and quantitative real-time RT-PCR were performed to assess mRNA levels of ITGB4 and FLNA in Cas9-TMEM268/BGC823 cells. e Control and Cas9-TMEM268/BGC823 cells were transfected with the indicated plasmids for 24 h. The cell lysates were then immunoprecipitated with an anti-FLAG antibody and probed with an anti-ITGB4 antibody
We next investigated whether ITGB4 protein stability is related to proteasomal or lysosomal degradation. We added the proteasome inhibitor MG-132 (10 μM), or the autophagic inhibitors chloroquine (CQ) or bafilomycin A1 (Baf.A1) which block autophagosome fusion with lysosomes, to Cas9-TMEM268/BGC823 or Control cells. The results show that ITGB4 degradation in Cas9-TMEM268/BGC823 cells is significantly blocked after treatment with MG-132 (Fig. 5a, lane 8 vs. lane 7), but not CQ or Baf.A1 (Fig. 5a, lane 6 vs. lane 5, lane 4 vs. lane 3), demonstrating that deletion of TMEM268 leads to increased proteasomal degradation of ITGB4. We next measured the half-life of ITGB4 using the protein translation inhibitor cycloheximide (CHX) in CAS9-TMEM268/BGC823 cells. As shown in Fig. 5d, inactivation of TMEM268 significantly promotes the decay of ITGB4, indicating that TMEM268 may be a positive regulator of ITGB4.
We next investigated whether TMEM268 regulates ITGB4 stability by modulating its ubiquitylation. The results were shown in Fig. 5e (lane 2 vs. lane 1), total polyubiquitylated ITGB4 protein is significantly increased in Cas9-TMEM268/BGC823 cells. Interestingly, TMEM268-deleted cells exhibit an increase in K48-linked (Fig. 5e, lane 4 vs. lane 3) and a decrease in K63-linked ubiquitylation (Fig. 5e, lane 6 vs. lane 5) in the ITGB4 immunoprecipitates. This result suggests that TMEM268 may prevent K48-linked but facilitate K63-linked polyubiquitination of ITGB4. Combined with the results of Fig. 3b, that showed decreased ITGB4 fluorescent signaling in Cas9-TMEM268/BGC823 cells, we conclude that TMEM268 may play an important role in maintaining the homeostasis of the ITGB4 protein.
TMEM268 deficiency causes cytoskeleton remodeling and cytoskeletal network damage
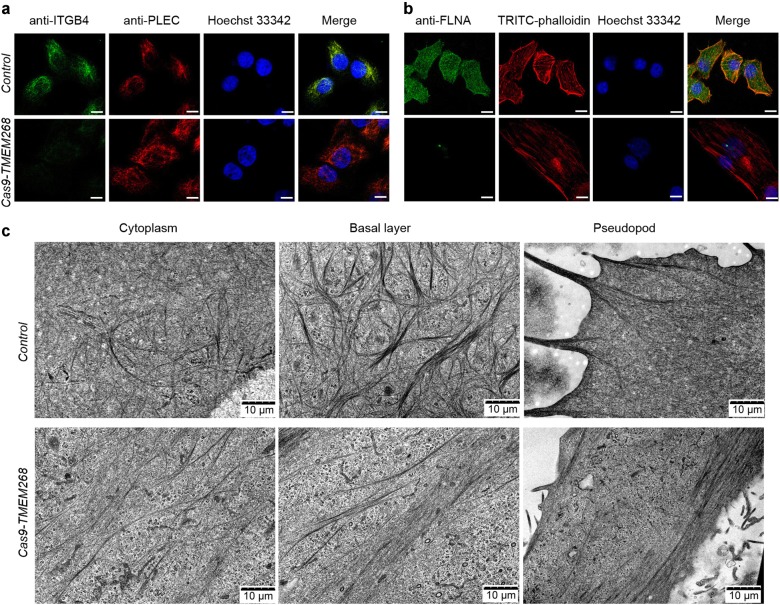
It is widely speculated that the association of integrin with the cytoskeleton is necessary to stabilize adhesion to the ECM [26]. PLEC binds ITGB4 and mediates the hemidesmosomal cytoskeleton, which plays an important role in the integrity of the cytoskeleton [27]. In control BGC823 cells, the fluorescence signals of ITGB4 and PLEC showed cluster distribution and complete colocalization (Fig. 6a, upper panel). In Cas9-TMEM268/BGC823 cells, the fluorescence signal of ITGB4 was almost undetectable, while PLEC displayed a diffuse filamentous distribution (Fig. 6a, lower panel).
Fig. 6.
TMEM268 deficiency causes cytoskeleton remodeling and cytoskeletal network damage. a, b Control and Cas9-TMEM268/BGC823 cells were plated on glass slides for 24 h, then an immunofluorescence assay performed with the indicated antibodies. Nuclei were stained with Hoechst 33342. Representative images obtained from confocal microscopy are shown. Scale bars, 10 μm. c Changes in the microfilament network structure in control and Cas9-TMEM268/BGC823 cells were observed by transmission electron microscopy. Representative images obtained are shown
It has been shown that when ITGB4 expression is reduced, dissociated PLEC can bind to intracellular F-actin to maintain the homeostasis of PLEC [28]. Using TRITC-phalloidin staining, we analyzed the distribution of F-actin and the colocalization between PLEC and F-actin in Cas9-TMEM268/BGC823 cells. The F-actin microfilament monomer is polymerized into a stable intersecting structure, with no obvious colocalization between PLEC and F-actin in Control cells (Fig. S7, upper panel). In Cas9-TMEM268/BGC823 cells, F-actin shows a long and sparsely filamentous structure, and the colocalization of PLEC/F-actin significantly increases (Fig. S7, lower panel). The results imply that the ITGB4 degradation induced by TMEM268 inactivation may directly cause the disintegration and structural damage of hemidesmosomes. Excess PLEC combines with F-actin, and the cytoskeleton is remodeled.
The actin-binding protein FLNA can crosslink F-actin filaments to form a stable cytoskeleton. Western blotting results indicate that the FLNA levels are significantly lower in Cas9-TMEM268/BGC823 cells than in Control cells (Fig. 5a). Similar to ITGB4, TMEM268 knockout also promotes the decay of FLNA (Fig. 5d). Results from immunofluorescence assays show that the fluorescence signal of FLNA is almost quenched in Cas9-TMEM268/BGC823 cells (Fig. 6b). The colocalization between F-actin and FLNA, therefore, failed to occur (Fig. 6b), indicating that TMEM268 may regulate the stability of FLNA. These results provide further evidence that TMEM268 knockout affects the reconstruction of the cytoskeleton.
Data from transmission electron microscope analysis demonstrated that actin filaments show cross-linked architecture in the cytoplasm and pyknotic network structure in the basal layer in Control cells (Fig. 6c, upper panel). These three-dimensional structures possess high strength, which can support cell pseudopodium extension. In Cas9-TMEM268/BGC823 cells, the three-dimensional reticular architecture of actin filaments is destroyed, and instead the filaments are constructed into a compact bundle presenting a long and straight filamentous structure. These parallel microfilaments affect the formation and extension of pseudopodia (Fig. 6c, lower panel), resulting in the destruction of the cytoskeleton. Collectively, we have demonstrated that TMEM268 binds to ITGB4, and may act as an adaptor between ITGB4 and microfilament skeleton remodeling.
TMEM268-deleted cells failed to colonize the lungs after intravenous injection and to form metastatic engraftment in the peritoneum
Since TMEM268 inactivation impairs the adhesion and growth of gastric cancer cells, we therefore assessed whether TMEM268-deleted BGC823 cells lose the ability for distant metastasis.
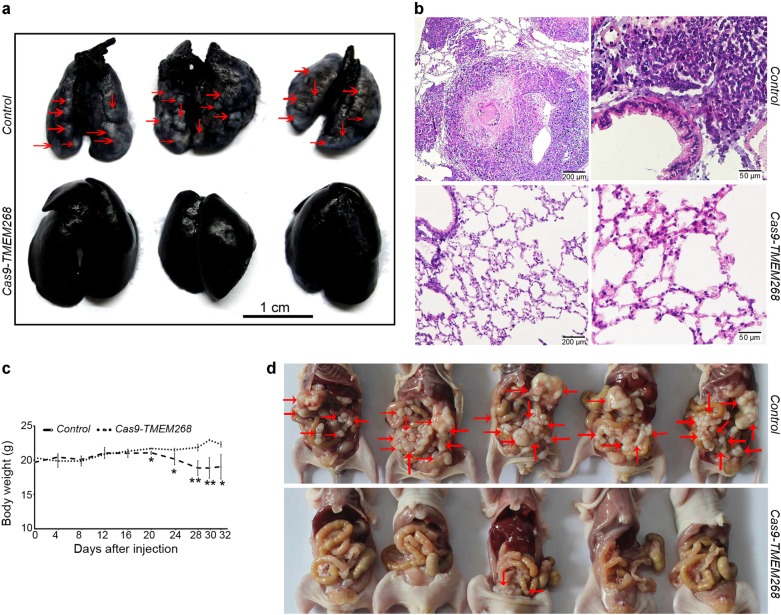
Cas9-TMEM268/BGC823 cells and Control cells were injected into nude mice via the tail vein. On day 54, the mice from each group were sacrificed. The lungs were filled with India ink via the upper trachea and fixed. Metastatic nodules on the black lung surface were observed and stained with hematoxylin and eosin (H&E). As shown in Fig. 7a, gross examination revealed that many metastatic tumors appear as white nodules on the black lung surfaces in the Control group, while no metastatic nodules are observed in the Cas9-TMEM268 group. Data obtained from histological analysis demonstrates that, in the Control group, there are many tumor cell masses in the alveoli, the Cas9-TMEM268 group displays the normal tissue structure of the lungs (Fig. 7b). These results suggest that TMEM268-deleted cells failed to colonize the lungs after intravenous injection.
Fig. 7.
TMEM268 deletion prevents the metastasis of BGC823 cells. a Control or Cas9-TMEM268/BGC823 cells (1×106 cells/mouse) were injected into BALB/c nude mice (n = 6) via the tail vein. On day 54, the mice from each group were sacrificed, and the lungs filled with India ink via the upper trachea and fixed. Metastatic lesions (red arrows) on the black lung surface were observed and photographed. b H&E staining of lung tissues. c Control or Cas9-TMEM268/BGC823 cells (8×106 cells/mouse) were injected into the abdominal cavity of mouse (n = 6). The changes in body weight were monitored every 4 days after injection (*P < 0.05; **P < 0.01). d Images of the peritoneal metastatic nodules were obtained at day 32. Arrows indicate the disseminated tumor nodules.
To further confirm the effects of TMEM268 on tumor metastasis, an animal model of peritoneal metastasis was established by the intraperitoneal injection of Control or Cas9-TMEM268/BGC823 cells into mice. The weight curve revealed that the body weight of the Control mice decreased after 20 days, while Cas9-TMEM268 group was maintained at normal level or slightly upregulated (Fig. 7c). All mice were sacrificed at day 32 after injection. As shown in Fig. 7d, a large number of metastatic nodules were found in the peritoneal cavity in Control mice. By contrast, the Cas9-TMEM268 group had little or no tumor nodules. These data suggested that inactivation of TMEM268 might prevent peritoneal homing and growth of metastatic nodules in gastric cancer cells.
Discussion
TMEM268, a novel human gene associated with cell viability was first identified by our group. It is evolutionarily conserved and widely expressed in many cell lines and tissues. Our studies have demonstrated that inactivation of TMEM268 can inhibit cell growth, reduce cell adhesion, induce cell cycle arrest, and disrupt cytoskeleton reorganization. Subsequently as a result, the abilities of tumorigenicity and metastasis were significantly decreased (Fig. S8).
Integrin is a complex composed of α and β subunits that associate by noncovalent binding. The different heterodimeric forms can bind different ligands and mediate a variety of cellular activities [29]. The subunits ITGB4 and ITGA6 can bind specifically and form the LN receptor. This complex is involved in cell−ECM and cell−cell adhesion. By integrating the intracellular domain of ITGB4 and its downstream signaling molecules, the cells can react efficiently and accurately to external stimuli. ITGB4 and PLEC form the main structure of hemidesmosomes, which are important structures for epithelial cell adherence to the ECM and for protection of cells from external mechanical stress [30, 31]. Our study found that the loss of TMEM268 caused ITGB4 polyubiquitination and degradation, as well as reduced ITGA6 levels. Downregulation or disappearance of the ITGA6-ITGB4 complex directly decreases the adhesion ability of TMEM268 knockout cells and LN. Simultaneously, downregulated ITGA1, ITGAL1, and ITGAV adhesion molecules were also found in TMEM268 knockout cells, which resulted in a decrease in cell−cell and cell−ECM adhesion. Subsequently, this leads to cells detaching from the bottom of culture plates. Our results suggest that TMEM268 may be a new membrane stability molecule, which plays an important regulatory role in the homeostasis and quality control of adhesion molecules.
The main structure of hemidesmosomes is composed of PLEC and ITGB4. PLEC is an important part of the cytoskeleton, and it is widely expressed in various types of cells [32]. PLEC contains a conserved actin-binding domain, which interacts with F-actin and respond to external stimuli [28]. In a normal physiological state, TMEM268 binds with the ITGB4-PLEC complex and may maintain hemidesmosome formation into three-dimensional filamentous structures. In cells lacking TMEM268, the reduced levels of ITGB4 protein results in the disassociation of the ITGB4/PLEC complex. Dissociated PLEC binds to F-actin and forms a long bunchy structure. Additionally, reduced FLNA makes actin F-actin unable to form a vertical branching structure. Such effects can lead to the disorder of the three-dimensional structure of the cytoskeleton microfilaments and damage the cytoskeleton network structure [33–35]. The detailed mechanism of TMEM268 affecting cytoskeleton remodeling needs further investigation.
Metastasis is a complicated biological cascade that starts with local invasion by tumor cells and continues with migration to distant tissues and ultimately colonization [36]. Among the cancer cells that arrive at a metastatic target organ, only the cells that have acquired the capacities of adhesion, migration, invasion, and proliferation can form a new tumor focus. Increasing evidence demonstrates that extracellular matrix protein 1 promotes cell metastasis by interacting with ITGB4 and inducing ITGB4/FAK signal pathway [17]. ITGB4-mediated invasion and migration of gastric cancer cells is also regulated by metallopanstimulin-1 [18]. Data from in vitro and animal model experiments suggest that TMEM268 interacts with ITGB4, and its levels are greatly depressed in TMEM268-deleted cells via proteasome degradation pathway. Such effect thereby leads to a decrease in cell adhesion, survival, and proliferation of TMEM268-inactivated cells. Obviously, these circulating tumor cells in the blood may lose the ability to adhere to the vascular endothelium, fail to extravasate into the new tissue (such as lung) and eventually deprives metastatic colonization. The above results provide a new perspective that targeting the TMEM268/ITGB4 signaling axis may be effective in the treatment of metastasis in gastric cancer, which requires further investigation.
In conclusion, we demonstrate that TMEM268 is a positive regulator of ITGB4. TMEM268 deficiency in gastric cancer cells inhibits adhesion, decreases cell proliferation, regulates cytoskeleton remodeling, and finally depresses tumorigenesis and metastasis. Further studies will focus on collaborating with the clinical laboratory to explore the relationship between TMEM268 levels and clinicopathological parameters of gastric cancer, which will offer evidence for estimating the possibility of TMEM268 as a potential target to prevent/treat gastric cancer.
Materials and methods
Antibodies and reagents
Polyclonal or monoclonal antibodies were as follows: anti-TMEM268(Sigma-Aldrich, HPA017199), anti-PLEC (Abcam, ab11220); anti-ITGB4 (14803), anti-FLNA (4762), and anti-ubiquitin (3936) from Cell Signaling Technology; anti-GAPDH (KM9002), anti-GST (KM8005), anti-FLAG (KM8002L), anti-GFP (KM8009L) from Tianjin Sungene. These antibodies conjugated with FITC were purchased from Biolegend: anti-ITGA1 (328307), anti-ITGA6 (313605), anti-ITGAL (301206), anti-ITGAV (327907), and anti-ITGB4 (327805). Following antibodies were from Santa Cruz Biotechnology: anti-CCNE1 (sc-377100), anti-CDKN1B (sc-1641), anti-CCKN1C (sc-56341), anti-SKP2 (sc-74477), anti-CCNB1 (sc-166210), anti-CDK1 (sc-54). Secondary antibodies included DyLight 800/DyLight 680-conjugated IgG against mouse (Rockland, 610-145-002/610-144-002) or rabbit (Rockland, 611-145-002/611-144-002) and FITC/RBITC-conjugated IgG against mouse (Bioss Inc., bs-0296GFITC/bs-0296G-RBITC) or rabbit (Bioss Inc., bs-0295G-FITC/bs-0295G-RBITC). Other reagents used in this study were from Sigma-Aldrich: bafilomycin A1 (BafA1, B1793), MG-132 (C2211), Hoechst 33342 (14533), TRITC-phalloidin (P1951). Cycloheximide (CHX, 239763) was from Calbiochem.
Plasmid construction
The TMEM268 cDNA was amplified from BGC823 cells by PCR using the forward primer (5′-CCGGAATTCATGGCCTgTgAACCACAGGTG-3′) and reverse primer (5′- CGGGATCCGCGCTCACCTCGCCAGGAACGG-3′). The insert was released by EcoRI and BamHI, then subcloned into the EcoRI site of pcDNA.3.1/myc-His(-)B (Life Technologies, V85520) to construct the pcDB-TMEM268 plasmid. Based on this plasmid, we constructed the following plasmids in succession: GFP-TMEM268, Flag-TMEM268, GST-tagged TMEM268 and mutants (Fig. 4c). Different GFP-ITGB4 mutants (Fig. 4d) were also constructed in our laboratory. All constructs were confirmed by DNA sequencing. Specific shRNA mediating TMEM268 gene knockdown with the targeting sequence, 5′- CACCGCCAGTTGTGTGTTGTCATTTCAAGAGAATGACAACACACAACTGGCTTTTTTG-3′, and nonsilencing shRNA vector were constructed by ORIGEN Corporation. The shRNA vector sequence with no sequence homology to any known human gene was used as the control.
Cell culture, transfections, and treatments
BGC823, SGC7901, and HEK293 cell lines were cultured in Dulbecco's Modification of Eagle's Medium (DMEM) (Invitrogen, 12800-017) supplemented with 10% Ausbian fetal calf serum (FCS) (Shanghai VIAN-SAGA Biotech. Ltd, Shanghai, China). Cells were transfected with plasmids using NEOFECT Reagent (Neofect, Beijing, TF201201), according to the manufacturer’s instruction, while shRNA transfection was performed using Lipofectamine 3000 reagent (Life Technologies, L3000-015).
Knockout of the TMEM268 gene by CRISPR/Cas9-mediated genome editing
TMEM268 knockout cell line by CRISPR/Cas9-mediated genome editing was established in BGC823 cells. Target sequences for CRISPR interference were designed by Shanghai Biomodel Organism Science & Technology Development Co., Ltd (China). The target sequences for human TMEM268 are CAATGGCCAGGTCCTCACTG (Exon 2). The TMEM268 DNA was amplified by PCR using the forward primer (5′- TCTCACATTGTGGTCCTAGGA-3′) and reverse primer (5′- TACGACAGCTGGTACAGA-3′). Sequence analysis revealed that the mutation (ACAATG → TG) resulted in frameshifts with premature termination by introducing stop codon, removal of translation initiation codon and deletion of functional domain such as TM domain of TMEM268.
Reverse transcription PCR and quantitative real-time PCR assays
Total RNA samples were extracted from cells with the TRIzol reagent (Invitrogen, 15596-026). RT-PCR was performed using the ThermoScript RT-PCR System (Invitrogen, 11146-024). Quantitative real-time PCR was performed using SYBR Premix Ex Taq (TaKaRa, Japan). The primers against the indicated genes used in this study were listed in Supplementary Table 1.
Immunofluorescence, fluorescence, and confocal microscopy assays
Cells were cultured in confocal dishes and treated as indicated, fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100. The dishes were then incubated with fetal bovine serum overnight and exposed to primary antibodies for 1 h at 4 °C. After washing three times with phosphate-buffered saline (PBS), the dishes were covered with FITC/TRITC-conjugated secondary antibodies. Nuclei were stained with Hoechst 33342. Morphological alterations in the cells were observed and documented using an Olympus FluoViewTM FV1000 Confocal Microscope (Olympus, Melville, NY, USA).
Transmission electron microscopy assays
Treated BGC823 cells were fixed in 0.1 M sodium phosphate buffer containing 2.5% glutaraldehyde (pH 7.4) for 1 h at 37 °C. Cells were embedded in Ultracut (LEICA ULTRACUT R, Bensheim, Germany) and sliced into 70-nm sections. Ultrathin sections were stained with uranyl acetate and lead citrate, and observed under a HT7700 transmission electron microscope (HITACHI-JPN, Ltd.).
Cell viability assay
Control and Cas9-TMEM268/BGC823 cells were treated with serum deprivation for 24 h. Then, cells were cultured with the complete culture medium for indicated times. Cell viability assays were performed using the CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega, G1111) according to the manufacturer’s instructions. The absorbance was measured on an EL-311SX ELISA Reader (Bio-Tec Instruments, USA) at 490 nm. Cell viability was calculated as follows: cell viability = absorbance of test group/absorbance of control cell group × 100%. Each experiment was performed in biological triplicate and independently repeated four times.
Clone formation assay
Cells were plated in triplicate at 200 cells/well in 24-well plates and cultured for 10−14 days. Cells were then fixed with 4% paraformaldehyde for 30 min and stained with crystal violet for 30 min. Colonies containing more than 50 cells were counted and photographed. All of the experiments were repeated three times, and the average values were reported.
Cell cycle analysis
Control and Cas9-TMEM268/BGC823 cells were treated with serum deprivation for 24 h. Then, cells were cultured with the complete culture medium for indicated times. The cells were fixed with 70% ethanol at −20 °C overnight, then treated with 100 μg/ml RNase A for 30 min at 37 °C, and stained using PI in 0.2% Triton X-100. Subsequently, the cells were collected on an FACS Calibur flow cytometer and the percentages of cells in the phases of the cell cycle (G0/G1, S and G2/M) were analyzed by ModFit software (BD Biosciences, Franklin Lake, NJ, USA).
Cell−matrix adhesion assays
Ninety-six-well plates were coated with 5 μg/ml plasma fibronectin (Sigma, F1141) or 5 μg/ml laminin (Corning, 354232) overnight at 4 °C. Subsequently, nonspecific binding sites were blocked with 1% bovine serum albumin (BSA) in PBS for 1 h, then plates were washed by PBS. The cells (4×104 cells/100 μl) diluted with DMEM were added to the coated 96-well plates and incubated at 37 °C for 30 min. Nonadherent cells were removed by washing with DMEM. Attached cells were analyzed using MTS according to the manufacturer’s instructions, and the optical density was measured at 490 nm. These experiments were performed in six duplicate and repeated three times.
GST affinity isolation assay
Recombinant GST, GST-TMEM268, or GST-TMEM268 mutants were expressed in Escherichia coli strain BL21 (DE3) and purified. Equal amounts of these proteins immobilized on glutathione-Sepharose TM 4B (GE Healthcare, 17-0756-01) were incubated with whole cell lysates extracted from transfected BGC823 cells at 4 °C for 4 h. The beads were then washed and resuspended in 2× sodium dodecyl sulfate (SDS) loading buffer and analyzed by western blot.
Immunoprecipitation and western blot analysis
For the IP analysis, treated cells were collected and disrupted in IP analysis buffer (300 mM NaCl, 50 mM Tris, pH 8.0, 0.4% NP-40, 10 mM MgCl2, 2.5 mM CaCl2) containing protease inhibitors (Roche Diagnostics, 04693116001). Total cell extracts (1 mg per sample) were mixed with precleared protein G SepharoseTM Fast Flow (GE Healthcare, 17-0618-01) and appropriate antibodies, followed by incubation for 4 h at 4 °C. The beads were collected by centrifugation, washed five times and resuspended in 2× SDS loading buffer and then subjected to western blotting. The protein bands were visualized using DyLight 800/DyLight 680-conjugated secondary antibodies, and the infrared fluorescence image was obtained using an Odyssey infrared imaging system (LI-CORBiosciences, Lincoln, NE, USA).
Tumorigenicity in nude mice
A nude mouse xenograft model was established using 6−8-week-old male BALB/c nude mice (Experimental Animal Center, Peking University Health Sciences Center, Beijing, China). Mice were housed and maintained in a pathogen-free facility, and all experimental procedures were approved by the Institutional Authority for Laboratory Animal Care of Peking University. Control or Cas9-TMEM268/BGC823 cells were subcutaneously injected in the right axilla of mice (4×106 cells/mouse, n = 6). Mice were sacrificed at 19 days after cell inoculation. Tumors were excised and photographed.
Long-term lung colonization assay in nude mice
Control or Cas9-TMEM268/BGC823 cells (1×106 cells/mouse) were injected into BALB/c nude mice (n = 6) through tail vein. At 53 days after inoculation, the mice were sacrificed. The lungs were filled with 15% India ink via the upper trachea and fixed with Fekete’s solution, then photographed. Some specimen were embedded in paraffin, sectioned, and subjected to H&E staining.
Peritoneal metastasis assay in nude mice
Control or Cas9-TMEM268/BGC823 cells (8×106 cells/mouse) were injected intra-peritoneum in BALB/c nude mice (n = 6). The changes in body weight were monitored every 4 days after injection. At 32 days after inoculation, the mice were sacrificed and the formation of metastatic nodules was observed and photographed.
Statistical analysis
Data are presented as the mean ± SD. Differences between groups were analyzed using the Student’s t test for continuous variables. Statistical significance in this study was set at P < 0.05. All analyses were performed using GraphPad Prism 7.
Electronic supplementary material
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (81522046, 31872827, and 81420108002), National Science and Technology Major Projects of China for “Major New Drugs Innovation and Development” (2018ZX09711003) and the Fundamental Research Funds for the Central Universities of China.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by JP Medema
These authors contributed equally: Dubeiqi Hong, Xuan Zhang
Contributor Information
Jian Lin, Email: linjian@pku.edu.cn.
Yingyu Chen, Email: yingyu_chen@bjmu.edu.cn.
Electronic supplementary material
The online version of this article (10.1038/s41418-018-0223-3) contains supplementary material, which is available to authorized users.
References
- 1.Case LB, Waterman CM. Integration of actin dynamics and cell adhesion by a three-dimensional, mechanosensitive molecular clutch. Nat Cell Biol. 2015;17:955–63. doi: 10.1038/ncb3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo W, Giancotti FG. Integrin signalling during tumour progression. Nat Rev Mol Cell Biol. 2004;5:816–26. doi: 10.1038/nrm1490. [DOI] [PubMed] [Google Scholar]
- 3.Ishizuka Y, Koshinaga T, Hirano T, Nagasaki-Maeoka E, Watanabe Y, Hoshi R, et al. NRP1 knockdown promotes the migration and invasion of human neuroblastoma-derived SK‑N‑AS cells via the activation of β1 integrin expression. Int J Oncol. 2018;53:159–166.2. doi: 10.3892/ijo.2018.4397. [DOI] [PubMed] [Google Scholar]
- 4.Siddharth S, Nayak A, Das S, Nayak D, Panda J, Wyatt MD, et al. The soluble nectin-4 ecto-domain promotes breast cancer induced angiogenesis via endothelial Integrin-β4. Int J Biochem Cell Biol. 2018;102:151–60. doi: 10.1016/j.biocel.2018.07.011. [DOI] [PubMed] [Google Scholar]
- 5.Knowles LM, Gurski LA, Engel C, Gnarra JR, Maranchie JK, Pilch J. Integrin αvβ3 and fibronectin upregulate Slug in cancer cells to promote clot invasion and metastasis. Cancer Res. 2013;73:6175–84. doi: 10.1158/0008-5472.CAN-13-0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Longmate W, DiPersio CM1. Beyond adhesion: emerging roles for integrins in control of the tumor microenvironment. F1000Res. 2017;6:1612. doi: 10.12688/f1000research.11877.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamidi Hellyeh, Ivaska Johanna. Every step of the way: integrins in cancer progression and metastasis. Nature Reviews Cancer. 2018;18(9):533–548. doi: 10.1038/s41568-018-0038-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seguin L, Desgrosellier JS, Weis SM, Cheresh DA. Integrins and cancer: regulators of cancer stemness, metastasis, and drug resistance. Trends Cell Biol. 2015;25:234–40. doi: 10.1016/j.tcb.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vicente-Manzanares M, Sánchez-Madrid F. Targeting the integrin interactome in human disease. Curr Opin Cell Biol. 2018;55:17–23. doi: 10.1016/j.ceb.2018.05.010. [DOI] [PubMed] [Google Scholar]
- 10.Raab-Westphal S, Marshall JF, Goodman SL. Integrins as therapeutic targets: successes and cancers. Cancers. 2017;9:E110. doi: 10.3390/cancers9090110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giancotti FG. Targeting integrin beta4 for cancer and anti-angiogenic therapy. Trends Pharmacol Sci. 2007;28:506–11. doi: 10.1016/j.tips.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 12.Wang H, Rana S, Giese N, Büchler MW, Zöller M. Tspan8, CD44v6 and alpha6beta4 are biomarkers of migrating pancreatic cancer-initiating cells. Int J Cancer. 2013;133:416–26. doi: 10.1002/ijc.28044. [DOI] [PubMed] [Google Scholar]
- 13.Nagata M, Noman AA, Suzuki K, Kurita H, Ohnishi M, Ohyama T, et al. TITGA3 and ITGB4 expression biomarkers estimate the risks of locoregional and hematogenous dissemination of oral squamous cell carcinoma. BMC Cancer. 2013;13:410. doi: 10.1186/1471-2407-13-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lipscomb EA, Simpson KJ, Lyle SR, Ring JE, Dugan AS, Mercurio AM. The alpha6beta4 integrin maintains the survival of human breast carcinoma cells in vivo. Cancer Res. 2005;65:10970–6. doi: 10.1158/0008-5472.CAN-05-2327. [DOI] [PubMed] [Google Scholar]
- 15.Stewart RL, West D, Wang C, Weiss HL, Gal T, Durbin EB, et al. Elevated integrin alpha6beta4 expression is associated with venous invasion and decreased overall survival in non-small cell lung cancer. Hum Pathol. 2016;54:174–83. doi: 10.1016/j.humpath.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stewart RL, O’Connor KL. Clinical significance of the integrin [alpha]6[beta]4 in human malignancies. Lab Invest. 2015;95:976–86. doi: 10.1038/labinvest.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gan L, Meng J, Xu M, Liu M, Qi Y, Tan C, et al. Extracellular matrix protein 1 promotes cell metastasis and glucose metabolism by inducing integrin β4/FAK/SOX2/HIF-1α signaling pathway in gastric cancer. Oncogene. 2018;37:744–55. doi: 10.1038/onc.2017.363. [DOI] [PubMed] [Google Scholar]
- 18.Yang ZY, Jiang H, Qu Y, Wei M, Yan M, Zhu ZG, et al. Metallopanstimulin-1 regulates invasion and migration of gastric cancer cells partially through integrin β4. Carcinogenesis. 2013;34:2851–60. doi: 10.1093/carcin/bgt226. [DOI] [PubMed] [Google Scholar]
- 19.Lorenz WW, McCann RO, Longiaru M, Cormier MJ. Isolation and expression of a cDNA encoding Renilla reniformis luciferase. Proc Natl Acad Sci USA. 1991;88:4438–42. doi: 10.1073/pnas.88.10.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–82. [PubMed] [Google Scholar]
- 21.Sherr CJ. The Pezcoller lecture: cancer cell cycles revisited. Cancer Res. 2000;60:3689–95. [PubMed] [Google Scholar]
- 22.Hwang HC, Clurman BE. Cyclin E in normal and neoplastic cell cycles. Oncogene. 2005;24:2776–86. doi: 10.1038/sj.onc.1208613. [DOI] [PubMed] [Google Scholar]
- 23.Bertoli C, Skotheim JM, de Bruin RA. Control of cell cycle transcription during G1 and S phases. Nat Rev Mol Cell Biol. 2013;14:518–28. doi: 10.1038/nrm3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Gao D, Fukushima H, Inuzuka H, Liu P, Wan L, et al. Skp2: a novel potential therapeutic target for prostate cancer. Biochim Biophys Acta. 2012;1825:11–17. doi: 10.1016/j.bbcan.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fisher D, Krasinska L, Coudreuse D, Novak B. Phosphorylation network dynamics in the control of cell cycle transitions. J Cell Sci. 2012;125:4703–11. doi: 10.1242/jcs.106351. [DOI] [PubMed] [Google Scholar]
- 26.Wu D, Xu Y, Ding T, Zu Y, Yang C, Yu L. Pairing of integrins with ECM proteins determines migrasome formation. Cell Res. 2017;27:1397–1400. doi: 10.1038/cr.2017.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Litjens SH, Koster J, Kuikman I, van Wilpe S, de Pereda JM, Sonnenberg A. Specificity of binding of the plectin actin-binding domain to beta4 integrin. Mol Biol Cell. 2003;14:4039–50. doi: 10.1091/mbc.e03-05-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geerts D, Fontao L, Nievers MG, Schaapveld RQ, Purkis PE, Wheeler GN, et al. Binding of integrin alpha6beta4 to plectin prevents plectin association with F-actin but does not interfere with intermediate filament binding. J Cell Biol. 1999;147:417–34. doi: 10.1083/jcb.147.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watt FM. Role of integrins in regulating epidermal adhesion, growth and differentiation. EMBO J. 2002;21:3919–26. doi: 10.1093/emboj/cdf399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raymond K, Kreft M, Song JY, Janssen H, Sonnenberg A. Dual role of alpha6beta4 integrin in epidermal tumor growth: tumor-suppressive versus tumor-promoting function. Mol Biol Cell. 2007;18:4210–21. doi: 10.1091/mbc.e06-08-0720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nievers MG, Kuikman I, Geerts D, Leigh IM, Sonnenberg A. Formation of hemidesmosome-like structures in the absence of ligand binding by the (alpha)6(beta)4 integrin requires binding of HD1/plectin to the cytoplasmic domain of the (beta)4 integrin subunit. J Cell Sci. 2000;113:963–73. doi: 10.1242/jcs.113.6.963. [DOI] [PubMed] [Google Scholar]
- 32.Steinbock FA, Wiche G. Plectin: a cytolinker by design. Biol Chem. 1999;380:151–8. doi: 10.1515/BC.1999.023. [DOI] [PubMed] [Google Scholar]
- 33.Bouameur JE, Favre B, Fontao L, Lingasamy P, Begre N, Borradori L. Interaction of plectin with keratins 5 and 14: dependence on several plectin domains and keratin quaternary structure. J Invest Dermatol. 2014;134:2776–83. doi: 10.1038/jid.2014.255. [DOI] [PubMed] [Google Scholar]
- 34.Koster J, van Wilpe S, Kuikman I, Litjens SH, Sonnenberg A. Role of binding of plectin to the integrin beta4 subunit in the assembly of hemidesmosomes. Mol Biol Cell. 2004;15:1211–23. doi: 10.1091/mbc.e03-09-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiche G. Role of plectin in cytoskeleton organization and dynamics. J Cell Sci. 1998;111:2477–86. doi: 10.1242/jcs.111.17.2477. [DOI] [PubMed] [Google Scholar]
- 36.Ganguly KK, Pal S, Moulik S, Chatterjee A. Integrins and metastasis. Cell Adh Migr. 2013;7:251–61. doi: 10.4161/cam.23840. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.