Abstract
Human nasopharyngeal carcinoma (NPC) has the highest metastatic rate in head and neck. However, the mechanisms underlying NPC metastasis remain unclear. Here using propensity-score-matched miRNA microarray analysis, miR-142-3p is identified to be the most correlated with distant-metastasis-free survival and downregulated in paraffin-embedded NPC with distant metastasis, which is validated in both internal cohort and external GEO dataset from Canada. miR-142 locus hypermethylation was observed and found to be associated with miR-142-3p downregulation in metastatic NPC. Furthermore, miR-142-3p was epigenetically silenced by EZH2-recruited DNMT1 and suppressed NPC cell metastasis and EMT. Intersecting PCR array gene profiling with bioinformatic prediction, we identify ZEB2 as a direct and functional target of miR-142-3p in NPC. Reversal of miR-142-3p silencing efficiently suppresses NPC cell invasion and metastasis. Moreover, epigenetic miR-142 hypermethylation is correlated with unfavorable prognosis in both training and validation cohorts. This study identifies miR-142-3p as a key suppressive regulator in NPC metastasis and reveals a DNMT1-mediated epigenetic mechanism for miR-142-3p silencing, providing a potential prognostic marker and therapeutic target to combat NPC metastasis.
Introduction
Nasopharyngeal carcinoma (NPC) is the most common head and neck cancer in Southeast Asia, especially in Southern China with 60,600 incidences and 34,100 deaths in 2015 [1]. NPC is characterized by high invasion and early metastasis, and patients with NPC often present with advanced diseases at first diagnosis [2]. Although local-regional control has been significantly improved by development in radiotherapy and chemotherapy, metastasis remains the major reason of treatment failure and cancer-related death in NPC [3, 4]. The outcome for patients with metastatic NPC is very poor, with progression-free survival of only 6 months [5]. However, it still lacks effective prediction patterns to stratify patients with high risk of distant metastasis for individual treatment. Therefore, understanding the molecular mechanisms underlying metastasis is of crucial importance for developing novel therapeutic strategies to treat NPC.
MicroRNAs (miRNAs) are a group of small (20–23 nucleotides) noncoding RNAs that repress gene expression by promoting target mRNA degradation or inhibiting mRNA translation [6, 7]. Functionally, miRNAs play crucial roles in numerous biological processes, including cell differentiation, proliferation, development, and motility [8, 9]. Accumulating studies have shown that miRNAs are aberrantly expressed and involved in the initiation and progression of various human malignancies, including NPC [10]. To date, several miRNAs have been identified to be downregulated and affect cell death, proliferation, migration, and invasion in NPC [11–14]. However, these studies mainly focus on the differentiated miRNAs in NPC tissues compared with noncancer nasopharyngeal tissues, and the expression patterns remain unclear between patients with and without distant metastasis. Therefore, the roles and mechanisms of miRNAs underlying NPC metastasis remain largely unknown, and identifying differentially expressed miRNAs in distant-metastatic NPC may help to achieve further knowledge about NPC metastasis.
miR-142-3p was initially described as a hematopoietic-specific miRNA [15]. Emerging data indicate that miR-142-3p is expressed in embryonic and adult hematopoietic tissues and required for the differentiation and development of hematopoietic stem cells and mature immune cells [16–19]. Recently, miR-142-3p has also been shown to be dysregulated and implicated in biological functions in cancer cells [20–23]. However, the functional roles and molecular mechanisms of miR-142-3p have not been clearly elucidated in NPC metastasis.
Here based on propensity-score-matched miRNA profiling, we identify and validate miR-142-3p as the specific miRNA downregulated in NPC with distant metastasis. The silencing of miR-142-3p is caused by DNA methylation mediated by EZH2-recruited DNMT1. miR-142-3p functions as a tumor suppressor in NPC cell metastasis and epithelial-to-mesenchymal transition (EMT) through directly targeting ZEB2. Moreover, miR-142 hypermethylation was correlated with poor distant metastasis-free survival (DMFS) and identified as an independent prognostic factor in NPC patients. Thus, this newly established DNMT1/miR-142-3p/ZEB2 axis plays a key role in NPC metastasis and represents a novel prognostic and therapeutic strategy to treat NPC metastasis.
Results
miR-142-3p is downregulated in NPC patients with distant metastasis
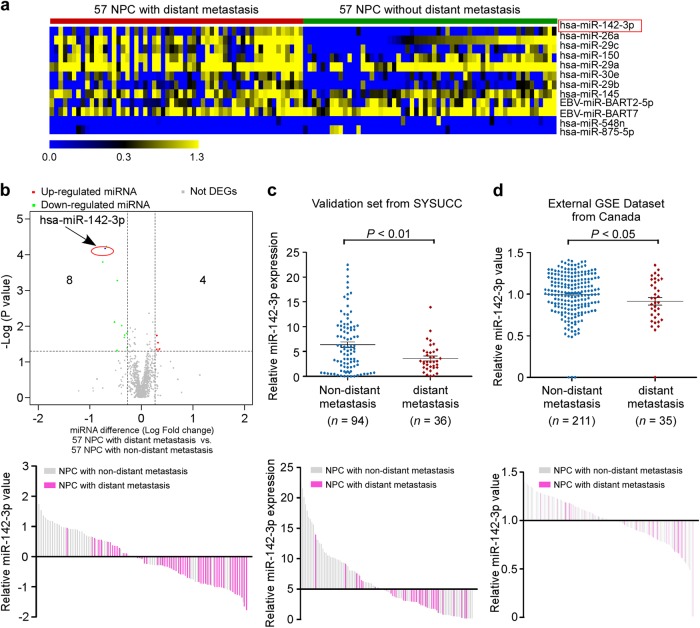
To identify distant-metastasis-specific miRNAs, we conducted two equally clinical-pathological matched NPC cohorts with (n = 57) vs. without (n = 57) distant metastasis from our previous miRNA microarray study (NCBI/GEO/GSE32960, ref. 11). After analyzing the miRNA profiles, 12 miRNAs were identified to be significantly differentially expressed in distant-metastatic NPC compared with nondistant metastatic NPC (fold change > 1.25, P value < 0.05; Fig. 1a). Among which, miR-142-3p was the most correlated with overall survival (OS) and DMFS in the 114 propensity-matched NPC patients (Supplementary Table S1), and significantly downregulated in specimens with distant metastasis (Fig. 1b). To estimate the reproducibility and validity of miR-142-3p expression, we performed internal validation by quantitative RT-PCR using dataset comprising 94 nondistant metastatic and 36 distant metastatic NPC patients from Sun Yat-sen University Cancer Center, and found that miR-142-3p was notably downregulated in NPC patients with distant metastasis (Fig. 1c). Furthermore, in an external GEO dataset from Toronto, Canada (GSE70970, ref. 12), miR-142-3p expression was also validated to be significantly reduced in distant metastatic NPC group using NanoString (n = 246, Fig. 1d). Taken together, these results suggest that miR-142-3p was downregulated in NPC patients with distant metastasis.
Fig. 1.
miR-142-3p is the specific miRNA downregulated in NPC with distant metastasis. a Heatmap clustering of differentially expressed miRNAs with a threshold of >1.25-fold change and P value < 0.05 symmetrically in NPC with distant metastasis (n = 57) and NPC without distant metastasis (n = 57). Columns: individual samples; Rows: miRNAs; Blue: low expression; yellow: high expression. b Volcano plot for the visualization of notably differentially expressed miRNAs. c Real-time PCR analysis of miR-142-3p expression in an internal set of 94 nondistant metastatic NPC and 36 distant metastatic NPC. P value was determined by one-tailed t test. d Published NanoString analysis of miR-142-3p expression in an independent set of 211 nondistant metastatic NPC and 35 distant metastatic NPC from Toronto, Canada (GSE70970). P value was determined by one-tailed t test
miR-142-3p downregulation is associated with DNA hypermethylation in NPC with distant metastasis
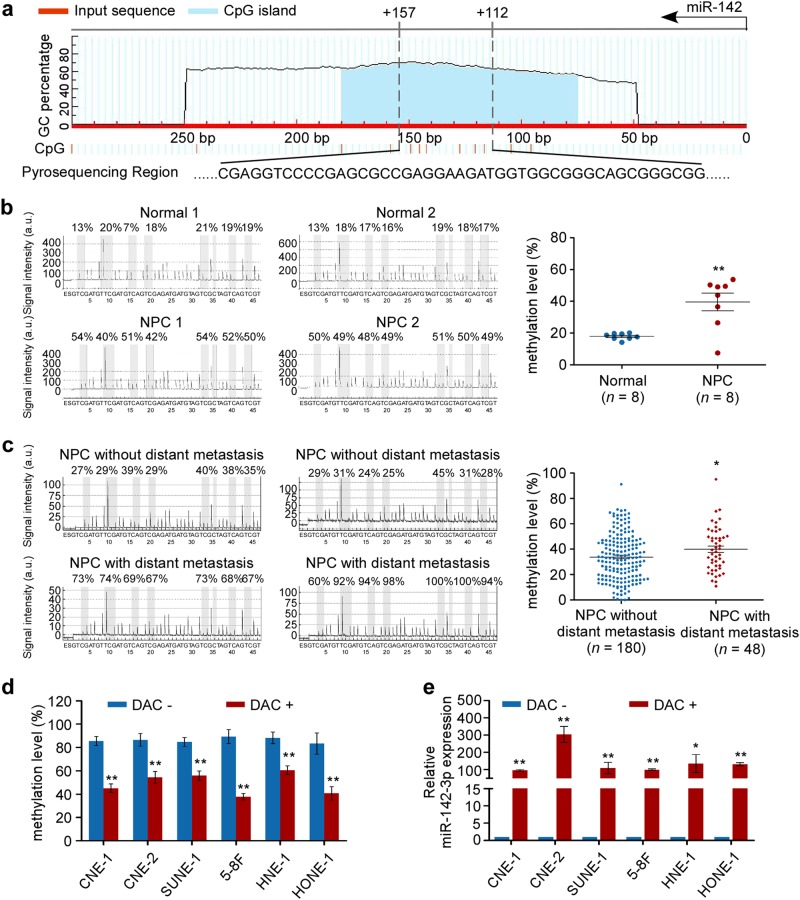
We next sought to elucidate the mechanism responsible for the downregulation of miR-142-3p in NPC. We found CpG islands located in the miR-142-coding region, raising the possibility that DNA methylation might be involved in the regulation of miR-142 expression (Fig. 2a). Bisulfite pyrosequencing was then performed to assess the status of CpG methylation within the miR-142 locus in fresh-frozen (n = 8) and paraffin-embedded NPC specimens (n = 228). The results showed that NPC tissues exhibited a significantly higher level of methylation within the miR-142 locus than the normal nasopharyngeal epitheliums (Fig. 2b, P < 0.01). Moreover, the methylation levels of the miR-142 locus was significantly upregulated in distant-metastatic NPC (Fig. 2c, P < 0.05). To further investigate the association between miR-142 downregulation and methylation, NPC cell lines were treated with or without 5-aza-2′-deoxycytidine (DAC, an inhibitor of DNA methyltransferase) for 72 h. Following DAC treatment, the methylation levels within the miR-142 locus were remarkably reduced in NPC cells (Fig. 2d, and Supplementary Fig. S1, P < 0.05). Meanwhile, miR-142-3p expression was conversely increased in NPC cells after DAC treatment (Fig. 4e, P < 0.05). Altogether, these findings suggest that hypermethylation in the miR-142 locus contributes to the downregulation of miR-142-3p in NPC.
Fig. 2.
miR-142 locus hypermethylation is associated with miR-142-3p downregulation in NPC with distant metastasis. a Schematic location of CpG islands (blue area) within the miR-142 locus. The transcriptional start site is indicated with an arrow. The regions analyzed by bisulfite pyrosequencing are listed (bottom panel). b, c Bisulfite pyrosequencing analysis and quantitative methylation levels of fresh-frozen normal nasopharyngeal epithelial tissues (n = 8) and NPC (n = 8) (b) and paraffin-embedded NPC specimens (n = 228) (c). Student’s t test, *P < 0.05, **P < 0.01. d, e Methylation levels (d) and miR-142-3p expression (e) in NPC cell lines with or without 5-aza-2′-deoxycytidine (DAC) treatment. Mean (n = 3) ± s.d. Student’s t test, *P < 0.05, **P < 0.01
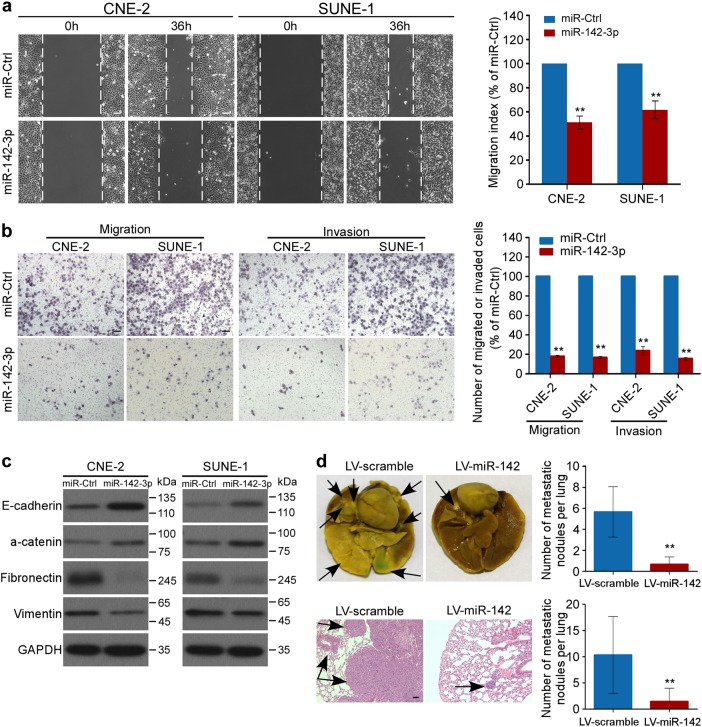
Fig. 4.
miR-142-3p attenuates NPC cell metastasis and EMT. a, b Representative images and quantification of wound-healing (a) and transwell (b) assays in NPC cells transfected with miR-142-3p mimics or control. Mean (n = 3) ± s.d. Student’s t test, *P < 0.05, **P < 0.01. c Western blot of E-cadherin, a-catenin, Fibronectin, and Vimentin. d A total of 1×106 SUNE-1 cells were intravenously injected via the tail vein, and the formation of lung metastatic colonization was evaluated after 8 weeks. Representative images and quantification of macroscopic and microscopic metastatic nodules in lungs of mice with or without stable miR-142 overexpression. The arrows indicated tumors. Mean (n = 9) ± s.d. Student’s t test, **P < 0.01. Scale bar, 100 μm
Epigenetic silencing of miR-142-3p is modulated by EZH2-recruited DNMT1 in NPC cells
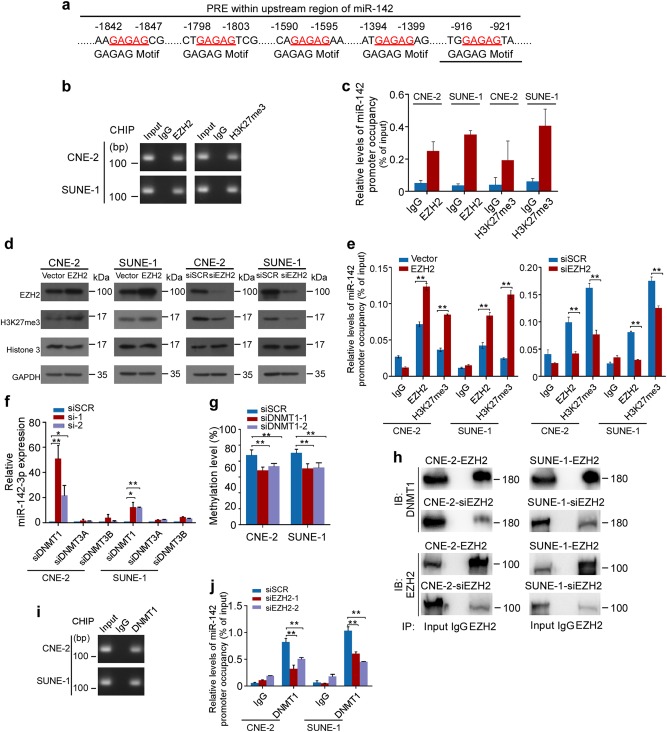
To investigate the mechanism regulating the miR-142 locus methylation abnormality, we analyzed the upstream and coding region of miR-142 and identified consensus polycomb response elements on the upstream region (Fig. 3a). As the polycomb repressive complex 2 (PRC2) complex has been shown to recruit DNA methyltransferases (DNMTs) to methylate target genes and thereby causes transcription repression [24, 25], this result suggests a possible involvement of PCR2 in the repression of miR-142-3p. Moreover, Enhancer of zeste 2 polycomb repressive complex 2 subunit, EZH2, is the core unit of PRC2 to catalyze the trimethylation of histone H3 at lysine 27 (H3K27me3) [24]. We found that EZH2 bound to the upstream region of miR-142, while high level of the repressive histone methylation marker H3K27me3 was also observed (Fig. 3b, c and Supplementary Fig. S2). In addition, EZH2 overexpression increased the levels of H3K27me3 and the occupancies of EZH2 in miR-142 upstream region, while EZH2 downregulation decreased H3K27 trimethylation and the bindings of EZH2 to the upstream region of the miR-142 locus (Fig. 3d, e and Supplementary Fig. S2, P < 0.01).
Fig. 3.
miR-142-3p is repressed by hypermethylation mediated by EZH2-recruited DNMT1. a Schematic location of the core DNA motif (red) of the polycomb response element (PRE). b, c PCR (b) and CHIP-qPCR (c) assay for anti-EZH2 and H3K27me3 in NPC cells. d Western blot analysis of EZH2, H3K27me3, and Histone3 in NPC cells with EZH2 overexpression or downregulation or control. e CHIP-qPCR assays for NPC cells after EZH2 modification. f, g Real-time PCR (f) and methylation level (g) assays for NPC cells after DNMTs downregulation. h Physical interactions between EZH2 and DNMT1 were examined via a Co-IP assay in NPC cells with EZH2 overexpression or downregulation. i PCR assay for DNMT1 occupancy in NPC cells. j CHIP-qPCR assays for anti-DNMT1 in NPC cells after EZH2 downregulation. Mean (n = 3) ± s.d. Student’s t test, *P < 0.05, **P < 0.01
We next performed quantitative PCR assays to test whether DNA methyltransferases are required for miR-142-3p downregulation. The results showed that decrease of DNA methyltransferase 1 (DNMT1) significantly restored miR-142-3p expression, but not DNMT3A or DNMT3B (Fig. 3f, P < 0.05). Meanwhile, the methylation status within the miR-142 locus was remarkably reduced after DNMT1 downregulation in NPC cells (Fig. 3g and Supplementary Fig. S3, P < 0.01). Furthermore, we evaluated whether EZH2 is responsible for DNMT1 binding to the miR-142 upstream region. Co-immunoprecipitation (co-IP) assays showed that EZH2 physically recruited DNMT1 in NPC cells, whereas the recruitment was remarkably decreased following EZH2 downregulation (Fig. 3h and Supplementary Fig. S4). CHIP assays confirmed the binding of DNMT1 (Fig. 3i and Supplementary Fig. S2) and showed that EZH2 inhibition resulted in a significant reduction of DNMT1 occupancies in the upstream region of miR-142 (Fig. 3j and Supplementary Fig. S2, P < 0.05). These data suggest that EZH2-recruited DNMT1 mediates the hypermethylation and thereby silencing of miR-142-3p.
miR-142-3p suppresses NPC cell metastasis
To evaluate the potential role of miR-142-3p in NPC metastasis, we ectopically introduced miR-142-3p mimics in CNE-2 and SUNE-1 cells to further explore the biological function of miR-142-3p in NPC cell motility and invasion ability (Supplementary Fig. S5a). In wound-healing assays, NPC cells with miR-142-3p overexpression migrated more slowly than control cells (Fig. 4a, P < 0.01). In transwell assays, forced expression of miR-142-3p significantly inhibited NPC cell migration and invasion (Fig. 4b, P < 0.01). We also investigated the effects of miR-142-3p on cell proliferation. The MTT (3- (4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) and colony formation assays demonstrated that miR-142-3p overexpression had minimal effects on cell viability and colonization (Supplementary Fig. S6, P > 0.05). Moreover, ectopic miR-142-3p expression markedly increased the protein levels of epithelial markers (E-cadherin and a-catenin) and decreased the protein levels of mesenchymal markers (Vimentin and Fibronectin), indicating that miR-142-3p repressed the EMT in NPC cells (Fig. 4c and Supplementary Fig. S7). Altogether, these data suggest that miR-142-3p inhibits NPC cell migration, invasion, and EMT in vitro.
To further investigate the effect of miR-142-3p in NPC metastasis in vivo, we employed a lung metastasis model in mice. SUNE-1 cells with or without ectopic miR-142-3p expression were injected into nude mice via the tail vein (Supplementary Fig. S5b). miR-142-3p overexpression significantly attenuated lung metastasis and histological analysis confirmed that mice with miR-142-3p overexpression exhibited fewer and smaller metastatic tumors in the lung (Fig. 4d and Supplementary Table S2, P < 0.01). Taken together, these findings indicate that miR-142-3p suppresses metastasis in NPC.
ZEB2 is a direct target responsible for miR-142-3p-mediated NPC cell metastasis
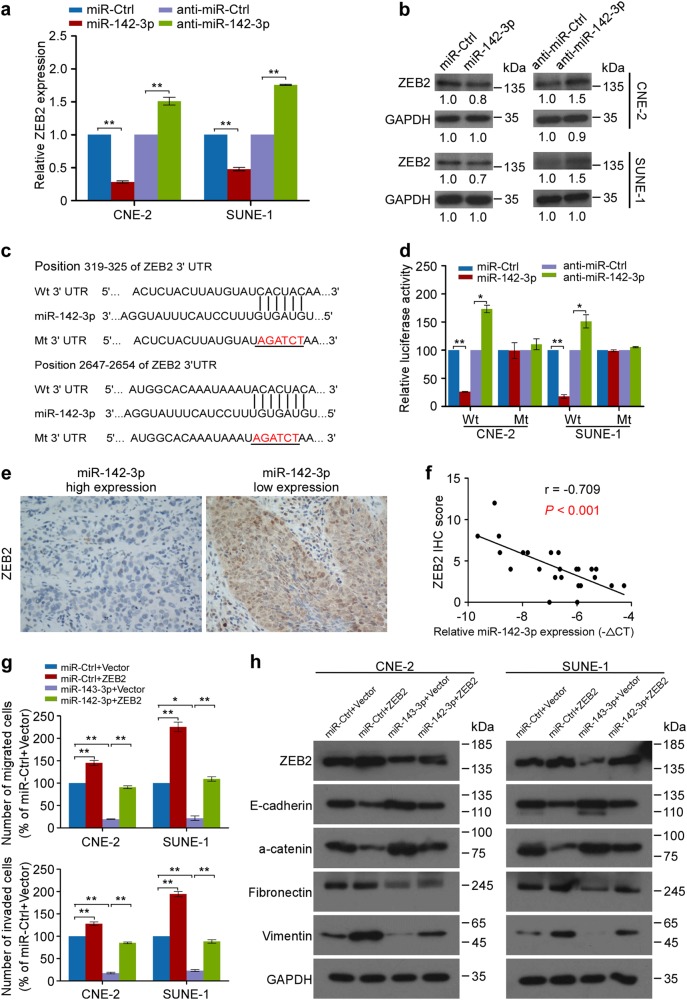
To explore the mechanisms of miR-142-3p in NPC cell metastasis and EMT, we evaluated the gene expression profiles of NPC cells with or without miR-142-3p overexpression using a human Epithelial to Mesenchymal Transition (EMT) RT2 Profiler PCR Array containing 84 genes associated with EMT and metastasis. As a result, nine downregulated and one upregulated candidate genes with more than a two-fold change in mRNA level were identified in CNE-2 and SUNE-1 cells after miR-142-3p overexpression (Supplementary Table S3 and Supplementary Fig. S8, P < 0.01). Using bioinformatic integration with TargetScan, miRanda and miRWalk and expression validation, only ZEB2 mRNA level was significantly reduced after miR-142-3p overexpression, while knocking down miR-142-3p elevated ZEB2 expression with over a 1.5-fold change (Fig. 5a and Supplementary Fig. S9). Similar changes in ZEB2 protein expression were examined by western blotting in CNE-2 and SUNE-1 cells (Fig. 5b). Furthermore, miR-142-3p overexpression or attenuation significantly suppressed or enhanced the luciferase activity of the wild-type ZEB2 3′ untranslated region (3′ UTR) reporter gene, respectively, but had no effect on the mutant one (Fig. 5c, d, P < 0.05). Clinically, ZEB2 expression was inversely correlated with miR-142-3p levels in NPC tissues (Fig. 5e, f; r = −0.709, P < 0.01). Collectively, these results suggest that ZEB2 might be a direct downstream target of miR-142-3p in NPC metastasis.
Fig. 5.
ZEB2 is a direct and functional target responsible for miR-142-3p-induced NPC cell metastasis and EMT. a, b Real-time PCR (a) and Western blot (b) of ZEB2 expression in NPC cells with miR-142-3p overexpression or inhibition. Mean (n = 3) ± s.d. Student’s t test, **P < 0.01. c Predicted and mutant binding sites of miR-142-3p on ZEB2 3′ UTR. d Luciferase reported assay after cotransfection with wild-type or mutant reporter genes and miR-142-3p mimics, inhibitor or control. Mean (n = 3) ± s.d. Student’s t test, *P < 0.05, **P < 0.01. e, f Representative images (e) and Spearman’s correlation analysis of ZEB2 protein expression and miR-142-3p miRNA expression in fresh-frozen NPC tissues (n = 24). f U6 was used as normalization control and −ΔCT equation was calculated by miR-142-3p CT value subtracted from U6 CT value. g Transwell assays of NPC cells cotransfected with ZEB2 or Vector and miR-142-3p mimics or control. Mean (n = 3) ± s.d. Student’s t test, *P < 0.05, **P < 0.01. h Western blot of E-cadherin, a-catenin, Fibronectin, and Vimentin
We next investigated whether ZEB2 was required for the miR-142-3p-induced inhibitory effects on NPC cell invasiveness and EMT. ZEB2 was introduced into CNE-2 and SUNE-1 cells with miR-142-3p overexpression. The wound-healing and transwell assays showed that ZEB2 restoration significantly abrogated the migratory and invasive inhibition induced by miR-142-3p in NPC cells (Fig. 5g, and Supplementary Fig. S10, P < 0.01). In addition, enforced ZEB2 overexpression reversed the inhibited EMT induced by miR-142-3p, as evidenced by the impaired expression of epithelial markers (E-cadherin and a-catenin) and the increased expression of mesenchymal markers (Vimentin and Fibronectin) (Fig. 5h). These studies provide evidence that ZEB2 is a direct and functional target of miR-142-3p during cell migration, invasion, and EMT in NPC.
DNMT1 regulates ZEB2 through repressing miR-142-3p
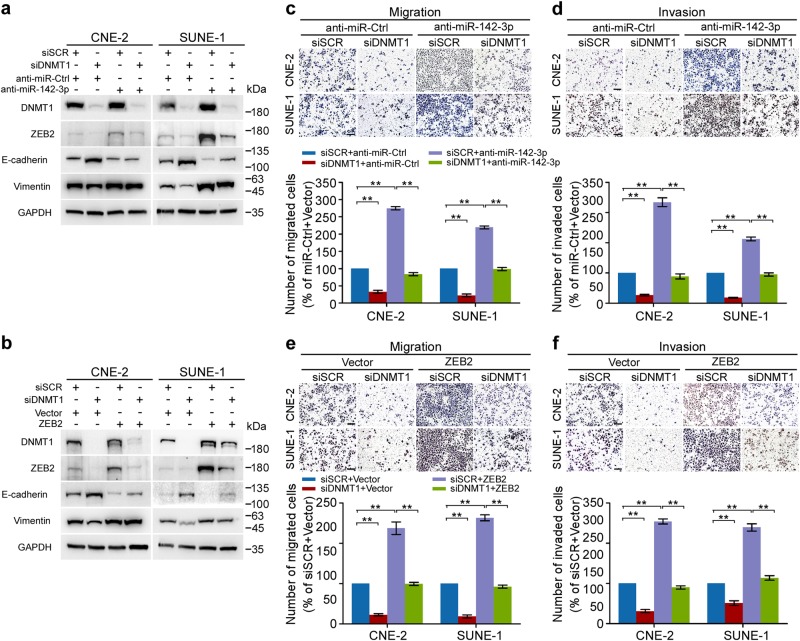
These new findings suggested that DNMT1 might be involved in the regulation of the miR-142-3p-ZEB2 axis during NPC metastasis. As expected, DNMT1 knockdown resulted in decreased expression of ZEB2 and Vimentin, but increased expression of E-cadherin (Fig. 6a, b). Conversely, miR-142-3p inhibition and exogenous ZEB2 manipulation promoted cell EMT and markedly restored the inhibitory effects induced by DNMT1 downregulation in NPC cells (Fig. 6a, b). Functionally, downregulation of DNMT1 exhibited repressed migration and invasion, which were significantly abrogated by inhibitory miR-142-3p expression and ectopic ZEB2 expression (Fig. 6c–f, P < 0.01). In addition, DAC treatment significantly inhibited the NPC cell migration and invasion, while miR-142-3p downregulation remarkably suppressed the sensitivity of NPC cells to DAC treatment (Supplementary Fig. S11, P < 0.05). Together, these results establish that DNMT1-mediated miR-142-3p repression leads to ZEB2 activation and promoted NPC cell metastasis and EMT.
Fig. 6.
DNMT1 regulates ZEB2 expression and function through miR-142-3p. a, b CNE-2 and SUNE-1 cells were cotransfected with either siDNMT1 or NC control and an miR-142-3p inhibitor or miR-Ctrl (a). CNE-2 and SUNE-1 cells were cotransfected with either siDNMT1 or NC control and the empty vector (Vector) or plasmid overexpressing DNMT1. b Western blot analysis of EZH2, ZEB2, E-cadherin, and Vimentin. c–f Transwell migration (left) or invasion (right). Representative images (upper panel) and quantification (lower panel) of transwell. Mean (n = 3) ± s.d. Student’s t test, **P < 0.01. Scale bar, 100 μm
Reversal of miR-142-3p silencing suppresses NPC cell invasion and metastasis in vivo
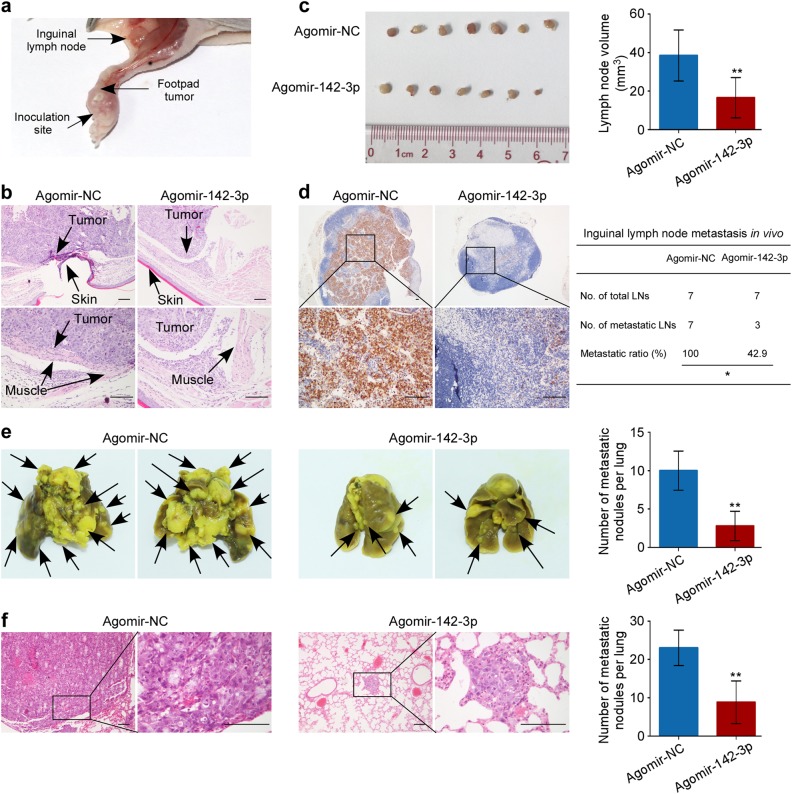
Based on these findings, we considered the therapeutic efficacy of miR-142-3p in NPC metastasis. To test this notion, we firstly constructed inguinal lymph node metastasis models through inoculating SUNE-1 cells into mice foot-pads and injected intratumoral agomir to overexpressing 142-3p or agomir NC control after 2 weeks growth (n = 7/group, Fig. 7a). Histological results showed that food-pad tumors formed in group injected with agomir-142-3p displayed sharp edges expanding as spheroids and less aggressive phenotypes invading towards skin and muscle than the control group (Fig. 7b). In addition, ectopic miR-142-3p overexpression significantly reduced the volumes and the ratios of inguinal metastatic lymph nodes, and also the numbers of pan-cytokeratin-positive metastatic tumor cells (Fig. 7c, d, P < 0.05). Collectively, these findings imply that miR-142-3p suppresses NPC cell invasion and lymph node metastasis in vivo.
Fig. 7.
Administration of miR-142-3p suppresses NPC cell aggression in mouse model. a SUNE-1 cells (2×105 cells in 30 µl PBS) were inoculated into mice foot-pads. After 2 weeks’ growth, intratumoral injection of agomir to overexpressing 142-3p or agomir NC control were injected into foot-pad tumors twice a week for 2 weeks. b Representative images of microscopy foot-pad tumors. Scale bar, 100 μm. c Representative images and quantification of inguinal lymph nodes. Mean (n = 7) ± s.d. Student’s t test, **P < 0.01. d Representative images of pan-cytokeratin-positive tumor cells and metastatic ratios of inguinal lymph nodes. *P < 0.05, scale bar, 100 μm. e, f Representative images and quantification of macroscopic (e) and microscopic metastatic nodules (f) in the lungs of mice after systemic administration of agomir-miR-142-3p or agomir control. Mean (n = 5) ± s.d. Student’s t test, **P < 0.01. Scale bar, 100 μm
Secondly, we established a lung metastatic colonization model to determine whether systemic reversal of miR-142-3p affected NPC tumor progression. Two weeks after model construction, agomir-142-3p or agomir control was administered to mice by tail vein injection twice a week for 4 weeks. As shown in Fig. 7e, systemic administration of miR-142-3p resulted in a significant reduction in the numbers of metastatic colonies compared with the control (P < 0.01). Meanwhile, these results were confirmed by hematoxylin and eosin staining (Fig. 7f, P < 0.01).Furthermore, we evaluate whether ZEB2 is involved in the above inhibitory effects of miR-142-3p on lung metastatic colonization. The results showed that ectopic ZEB2 overexpression significantly restored the lung metastasis suppressed by miR-142-3p systemic delivery (Supplementary Fig. S12). Moreover, the ZEB2 expression levels were significantly repressed by miR-142-3p overexpression (Supplementary Fig. S12c, P < 0.05). Taken together, these data imply that delivery of miR-142-3p expression inhibits NPC cell metastasis through ZEB2 and may serve as a potential therapeutic strategy for NPC metastasis.
MiR-142 hypermethylation is associated with poor survival in NPC
Collectively, these data suggest that DNMT1-mediated epigenetic silencing of miR-142-3p contributes to metastasis and EMT in NPC. Therefore, we next examined the CpG methylation status of miR-142 locus in 392 NPC specimens obtained from two centers. Supplementary Table S4 lists the pathoclinical features of patients with NPC in the training and validation cohorts. Bisulfite pyrosequencing analysis revealed that miR-142 locus was differentially methylated in NPC specimens. In the training cohort (n = 228), the cutoff value was determined using receiver operating characteristic (ROC) analysis.
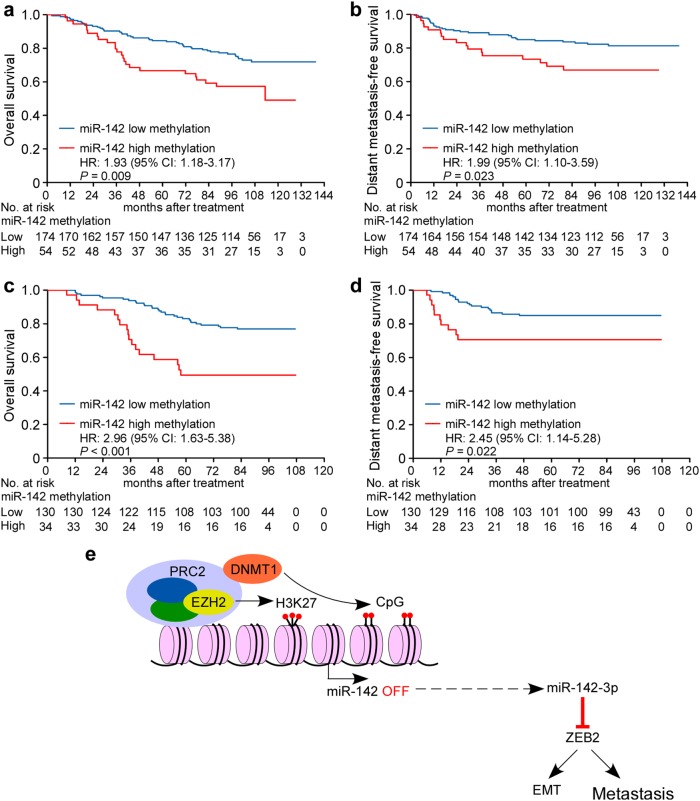
With this cutoff, patients in the training cohort were divided into miR-142 high methylation (methylation level ≥ 48.29) or low methylation groups (methylation level < 48.29%). As shown in Supplementary Table S4, no significant correlation was found between miR-142 methylation status and patient age, sex, pathological type, viral capsid antigen immunoglobulin A (VCA-IgA), early antigen immunoglobulin A (EA-IgA) and clinical stage. However, the methylation status of miR-142 locus was significantly correlated with death (P = 0.012) and distant metastasis (P = 0.031). Patients with high miR-142 methylation levels showed significantly lower OS (5-year OS; 66.7 vs. 84.5%, P = 0.009) and DMFS rates (5-year DMFS; 73.3 vs. 84.9%, P = 0.023; Fig. 8a, b) than those with low miR-142 methylation levels.
Fig. 8.
miR-142 hypermethylation is associated with metastasis in NPC patients. a, b Kaplan−Meier analysis of overall survival (OS, a) and distant metastasis-free survival (DMFS, b) according to the miR-142 methylation status in 228 NPC patients in the training cohort. The P value was determined using the log-rank test. c, d Kaplan−Meier analysis of OS (c) and DMFS (d) in 164 NPC patients in the validation cohort. The P value was determined using the log-rank test. e Proposed model for miR-142-3p downregulation in NPC. DNMT1-mediated epigenetic silencing of miR-142-3p determines ZEB2 activation and leads to EMT and metastasis in NPC
Furthermore, we assessed the miR-142 methylation level, clinical characteristics, and survival status in the validation set. Patients (n = 164) were classified into high or low miR-142 methylation groups with the same cutoff identified in the training set. As expected, the miR-142 methylation level was strongly correlated with death and distant metastasis (P < 0.05), while pathological and clinical features did not differ between these two groups (Supplementary Table S4). Patients with high miR-142 methylation showed significantly shorter OS (5-year OS; 49.4 vs. 83.1%, P = 0.009) and DMFS (5-year DMFS; 43.9 vs. 73.8%, P = 0.023; Fig. 8c, d) than those with low miR-142 methylation.
Finally, multivariate analysis was carried out to reveal prognostic factors in both patient cohorts. In the training cohort, miR-142 methylation status (OS, hazard ratio (HR), 1.76; 95% confidence interval (CI), 1.07–2.88; P = 0.026; DMFS, HR, 1.83; 95% CI, 1.01–3.31; P = 0.046) and TNM stage (OS, HR, 2.28; 95% CI, 1.16–4.46; P = 0.017; DMFS, HR, 3.13; 95% CI, 1.24–7.91; P = 0.016) were identified as independent prognostic factors for OS and DMFS survival (Supplementary Table S5). Similarly, we verified miR-142 methylation and TNM stage as independent prognostic factors for OS and DMFS in the validation cohort (Supplementary Table S5). Taken together, these new findings confirm that epigenetic suppression of miR-142-3p might depend on hypermethylation mediated by EZH2-recruited DNMT1 and contributes to metastasis and poor clinical outcomes in NPC through targeting ZEB2 (Fig. 8e).
Discussion
Our current study provides novel evidence that epigenetic silencing of miR-142-3p is the specific event leading to metastasis in NPC. First, miR-142-3p was identified to be downregulated in NPC with distant metastasis, which was validated in internal set and external GEO dataset in Canada. Second, downregulation of miR-142-3p was associated with miR-142 locus hypermethylation in metastatic NPC. Third, miR-142-3p was epigenetically silenced by EZH2-recruited DNMT1 and suppressed NPC cell metastasis and EMT in a ZEB2-dependent manner. Restoration of miR-142-3p silencing repressed NPC cell invasion and metastasis in vivo. Finally, miR-142 locus methylation was an independent prognostic factor and associated with unfavorable metastasis-free survival in NPC patients. These new findings reveal a critical DNMT1/miR-142-3p/ZEB2 cascade for the promotion of NPC metastasis, suggesting the application of miRNA as a new prognostic marker and therapeutic strategy for patients with, or at risk of, metastasis.
Metastasis, a hallmark of cancer, is a complex process characterized by tumor cell invasion into surrounding sites, dissemination throughout the body circulation and establishment of secondary tumors at distant organs [26, 27]. Genetic and epigenetic alterations acquired by tumor cells likely contribute to the formation of metastasis [28, 29]. Recently, it has emerged that miRNAs play important roles in cancer initiation, development, drug resistance, and metastasis [30]. Here, our propensity-score-matched miRNA microarray data identified miR-142-3p to be downregulated in NPC with distant metastasis. The roles and mechanisms of miR-142-3p in NPC metastasis remain undefined. We demonstrated that miR-142-3p functioned as a suppressor in NPC metastasis, which is consistent with the inhibitory effects of miR-142-3p in other cancer types [31–33]. However, previous study showed a conversely promoted phenotype of miR-142-3p in NPC cell proliferation [34]. It has speculated that cancer cells can undergo dynamic cycles of each cell process, including proliferation, motility, and dissemination, instead of both at the same time in a mechanism called phenotype-switching [35, 36]. Therefore, miR-142-3p may influence regulators on proliferation and motility transitions and ultimately results in suppressive tumor metastasis.
To date, the mechanism for the regulation of miR-142-3p has not been clearly demonstrated. Previous studies showed that miR-142-3p expression could be modulated by the transcriptional regulator Spi-1 proto-oncogene (SPI1) or vav guanine nucleotide exchange factor 1 (VAV1) in hematopoietic malignancies, and H4 histone acetylation in lupus CD4+ T cells [37–39]. Here, we demonstrated that DNA methylation within the miR-142 locus contributed to the reduction of miR-142-3p expression in metastatic NPC. Moreover, miR-142 methylation was associated with DMFS in NPC patients, indicating the regulation role and prognostic value of miR-142 hypermethylation in NPC metastasis.
DNA methylation is a reversible enzyme-mediated modification involved in embryonic and stem cell development and cancer progression [40]. DNMT1, DNMT3A, and DNMT3B are three catalytic active DNA methyltransferases for the maintenance of DNA methylation in mammals [41]. EZH2, a catalytic subunit of the PCR2 complex, serves as a recruitment platform for DNMTs [25, 42]. In the current study, we reported that EZH2-recruited DNMT1 occupied the upstream region of miR-142 and inhibited miR-142-3p expression through DNA methylation. Previous studies showed that DNMT1 is overexpressed and plays a role in the metastasis of NPC [43, 44]. Consistently, we found that DNMT1 promoted NPC cell migration and invasion by regulating miR-142-3p and subsequent ZEB2 expression. Our study provides new insight into the mechanism of DNMT1 regulating miR-142-3p expression and its contribution to NPC cell metastasis.
It is well known that miRNAs negatively regulate gene expression by interacting with 3′ UTR of target gene mRNA [7]. After intersecting gene expression profile PCR array analysis with bioinformatic algorithms, we identified that ZEB2 served as a direct target of miR-142-3p. Further analysis confirmed that miR-142-3p overexpression inhibited ZEB2 expression at both the mRNA and protein levels. As reported, ZEB2 is a member of the ZEB family of transcription factors, which are described as transcriptional repressors of E-cadherin to induce the EMT process [45]. EMT and its intermediate dynamic states are critical drivers of embryonic development and tumor progression, metastasis and chemotherapy resistance [46, 47]. Emerging evidence indicates that ZEB2 promotes tumor invasion and metastasis by inducing an EMT program [48, 49]. Our study verified that enforced ZEB2 expression restored the inhibitory effects of miR-142-3p on NPC cell migration, invasion, and EMT, evidenced by attenuated expression of E-cadherin and enhanced expression of Vimentin, thus supporting the contribution of miR-142-3p-regulated ZEB2 to NPC metastasis and EMT.
Recently, miRNAs have emerged as therapeutic targets due to their important roles in multiple cancer processes [10]. Given that some suppressive miRNAs are epigenetically silenced, the reversal of their expression may be an effective therapeutic strategy for cancer treatment. Accumulating studies have confirmed that ectopic miRNA expression via synthetic mimics displays antitumor effects [50]. Similarly, we found that restoration of miR-142-3p repressed cell invasion and lung metastasis in a mouse model. These findings indicate a new therapeutic strategy for NPC metastasis through modifying miRNA expression.
In conclusion, we identify miR-142-3p as a metastasis-specific suppressor in NPC and reveal a novel pathway in which DNMT1 epigenetically mediates miR-142-3p repression with resultant ZEB2 activation, leading to tumor metastasis in NPC. Our study also highlights the prognostic value of miR-142 methylation and the therapeutic effect of miR-142-3p in NPC metastasis, thereby facilitating the development of novel therapeutic strategies against NPC metastasis.
Materials and methods
MiRNA microarray and expression analysis
MiRNA microarray and NanoString expression data of human NPC tissues were extracted and re-analyzed to identify miRNA signature between NPC patients with and without distant metastasis. Propensity score matching (PSM) analysis was used to adjust patient selection bias and create two equally matched cohorts with vs. without distant metastasis from our previous 312-patient group that conducted miRNA microarrays (NCBI/GEO/GSE32960, ref. 11). The propensity score was calculated with host and tumor factors, including age, gender, T stage, N stage, WHO pathological type, Epstein−Barr virus antigen immunoglobulin (VCA-IgA and EA-IgA), radiotherapy methods, and chemotherapy. We conducted one-to-one PSM using the caliper algorithm with 0.05 with set to yield sufficient power and similarity. After adjusting, miRNA microarray data from 57 nondistant-metastatic NPC patients and 57 distant-metastatic NPC patients were background subtracted, normalized and analyzed using Biometric Research Branch-Array Tools version 3.8.0. Comparison was calculated with unpaired t test to identify differentially expressed miRNAs between nondistant-metastatic and metastatic groups with a threshold of 1.25-fold change and P value less than 0.05. Target miRNA was validated in the NanoString datasets GSE70970 [12] of 211 NPC patients without distant metastasis and 35 NPC patients with distant metastasis collected from Princess Margaret Cancer Center, Toronto, Canada. Unpaired one-tailed t test was used to calculate P value with the Stata Statistical Computer Package (STATA 10, StataCorp LP, Texas, USA).
Clinical specimens
For miR-142-3p expression analysis, 130 paraffin-embedded NPC specimens were collected from Sun Yat-sen University Cancer Center (Guangzhou, China). For methylation level analysis, a total of 392 paraffin-embedded NPC specimens were obtained from Sun Yat-sen University Cancer Center (n = 228) and Zhejiang Cancer Hospital (Hangzhou, China; n = 164) between January 2004 and April 2007. This study was approved by the Institutional Ethical Review Boards of Sun Yat-sen University Cancer Center and Zhejiang Cancer Hospital. Written informed consent was received from participants prior to inclusion in the study. All specimens were pathologically reassessed by two pathologists (J. Yun and J. Zeng), and each NPC sample contained more than 70% tumor cells. No patient had received radiotherapy or chemotherapy before biopsy, and all patients were diagnosed with nonmetastatic NPC. The TNM stage was reclassified according to the seventh edition of the AJCC Cancer Staging Manual. The median follow-up period was 95.1 months (range, 2.4−139.5 months). All patients received radiotherapy, and patients with stage III-IV NPC received concurrent platinum-based chemotherapy [4, 51]. The details of the clinicopathological characteristics are shown in Supplementary Table S4.
Cell culture
The human immortalized nasopharyngeal epithelial cell line (NP69) and human NPC cell lines (CNE-1, CNE-2, SUNE-1, 5–8F, HNE-1, and HONE-1) were obtained from Professor Musheng Zeng within 6 months (Sun Yat-sen University, Guangzhou, China). The NP69 cell line was maintained in keratinocyte/serum-free medium (Invitrogen, Grand Island, NY, USA) supplemented with bovine pituitary extract (BD Biosciences, San Diego, CA, USA). NPC cell lines were cultured in RPMI 1640 (Invitrogen) supplemented with 10% FBS (fetal bovine serum) (Gibco, Grand Island, NY, USA); 293FT cells were obtained from ATCC and grown in DMEM (Dulbeccoʼs Modified Eagle Media) (Invitrogen) supplemented with 10% FBS. All the cells were authenticated using short-tandem repeat profiling and tested for mycoplasma contamination.
RNA extraction, real-time PCR, and the Human Epithelial to Mesenchymal Transition (EMT) RT2 Profiler PCR Array
Total RNA from NPC specimens and cultured cells was extracted and real-time PCR was performed as previously described [14]. Reverse transcription was performed using reverse transcriptase (Promega, Madison, WI, USA) with random primers (Promega) for genes or Bulge-Loop miRNA-specific RT-primers (RiboBio, Guangzhou, China) for miR-142-3p. Real-time PCR reactions were performed with SYBR Green qPCR SuperMix-UDG reagents (Invitrogen) on a CFX96 Touch sequence detection system (Bio-Rad, Hercules, CA, USA). GAPDH and U6 were used as internal controls for genes and miR-142-3p, respectively. The Human Epithelial to Mesenchymal Transition (EMT) RT2 Profiler PCR Array (Qiagen, Hilden, Germany) was performed on a LightCycler 480 system (Roche, Basel, Switzerland), according to the manufacturer’s instructions. The relative expression was calculated with the 2−ΔΔCT equation [52].
Oligonucleotides, plasmids, and stable cell line generations
Mimics and inhibitors of miR-142-3p and small-interfering RNAs targeting EZH2 were purchased from GenePharma (Suzhou, China). The sequences were as follows: miR-142-3p mimics: 5′-UGUAGUGUUUCCUACUUUAUGGA-3′; miR-142-3p inhibitor: 5′-UCCAUAAAGUAGGAAACACUACA-3′; siEZH2-1: 5′-GGAUCACCGAGAUGAUAAATT-3′; siEZH2-2: 5′-CGGCUUCCCAAUAACAGUATT-3′; siDNMT1-1: 5′-GCAGGCGGCUCAAAGAUUUTT-3′; siDNMT1-2: 5′-GGAUGAGUCCAUCAAGAAATT-3′. The pENTER-vetcor and pENTER-ZEB2 plasmids were purchased from Vigene Biosciences (Jinan, China). The sequences of pri-miR-142 and human EZH2 gene were synthesized and cloned into the lentiviral plasmid pSin-EF2-puromycin (Addgene, Cambridge, MA). The primers used for amplification were as follows: miR-142-Forward: 5′-CCGGAATTCGGGGTTCACAGAACTGAAGG-3′, miR-142-Reverse: 5′-TATCATATGGCTGCATCAGGGTTCCACAT-3′; EZH2-Forward: 5′-CGGGATCCATGGGCCAGACTGGGAAGA-3′; EZH2-Reverse: 5′-GGACTAGTTCAAGGGATTTCCATTTCTCT-3′; DNMT1-Forward: TGGTGATGGTGTGTACCTGC; DNMT1-Reverse: TTCCGGTAGTGCTCTGGGTA. SUNE-1 and CNE-2 cells were transfected with oligonucleotides (100 nM) or plasmids (2 μg) using Lipofectamine 2000 reagent or Lipofectamine 3000 reagent (Invitrogen), and then cells were harvested for assays at 48 h after transfection. Stable SUNE-1 cell lines expressing miR-142 or EZH2 were established after infection with lentiviruses generated by 293FT cells and selected using 0.5 µg ml−1 puromycin.
Western blotting
Cells were lysed on ice in RIPA buffer containing protease inhibitor cocktail (Fdbio Science, Hangzhou, China). Equal amounts of cell lysis material were separated by 10–14% SDS-PAGE gels and then transferred to polyvinylidene fluoride membranes (Merck Millipore, Billerica, MA, USA) for antibody blotting. Antibodies included those targeting E-cadherin (1:2000; BD Biosciences, San Diego, CA; 610181), a-catenin (1:500; BD Biosciences, 610193), Fibronectin (1:500; Proteintech, Rosemont, IL, USA; 156131-AP), Vimentin (1:1,000; Proteintech, 10366-1-AP), ZEB2 (1:500; Abcam, Cambridge, MA, USA, ab138222), EZH2 (1:5000; Cell Signaling Technology, Beverly, MA; 07-449), H3K27me3 (1:5000; Millipore, 4905), histone 3 (1:1000; Cell Signaling Technology, 5246), GAPDH (1:5000; Abcam, ab128915), and a-tubulin (1:500; Sigma-Aldrich, Ronkonkoma, NY, USA, T3526). Horseradish peroxidase-conjugated antibody was used as the secondary antibody (anti-mouse or anti-rabbit; 1:5,000; Cell Signaling Technology, 7076 or 7074). Proteins were visualized with the ECL detection system (ThermoFisher Scientific, Waltham, MA).
5-aza-2′-deoxycytidine treatment
For DAC (Sigma-Aldrich) treatment, cells were cultured with or without DAC (10 μM) for 72 h, replacing the medium containing DAC every 24 h. After treatment, cells were harvested for DNA, RNA or protein extraction.
DNA extraction and bisulfite pyrosequencing methylation analysis
DNA was isolated from NPC fresh-frozen samples, FFPE specimens or cells using the AllPrep DNA/RNA Mini Kit (Qiagen, Hilden, Germany), QIAamp DNA FFPE Tissue Kit (Qiagen) or EZ1 DNA tissue Kit (Qiagen), respectively, according to the manufacturer’s instructions. Bisulfite modification was performed with 1 μg of genomic DNA using the EpiTect Bisulfite Kit (Qiagen). Bisulfite-treated DNA was amplified with primers designed by PyroMark Assay Design Software 2.0 (Qiagen). The primer sequences were as follows: Forward: 5′-GGAATTAGGAAGGGTAGGAAAGTTA-3′, Reverse: 5′-CTTCCAAAATCACCCCACTACTAC-3′, and sequencing: 5′-GTAGTAAGGTGTTTATTTGTTATA-3′. Bisulfite sequencing reactions and quantification of methylation were carried out using the PyroMark Q96 system (Qiagen). The average methylation from individual CpG sites was calculated as the percentage of methylation. For methylation analysis of FFPE NPC specimens, ROC curve analysis was used to determine cutoff values, and classification was performed as follows: low methylation with average methylation < 48.29%, and high methylation ≥ 48.29 [53, 54].
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were performed using an EZ-Magna ChIP kit (Millipore), according to the manufacturer’s instructions. Cells were fixed with 1% formaldehyde to crosslink; then, nucleoprotein complexes were sheared to 200–500 bases in length with sonication and then immunoprecipitated with anti-EZH2 (1:100; Cell Signaling Technology, 07-449), or anti-H3K27me3 (1:100; Millipore, 07-449) antibodies overnight at 4 °C. PCR and real-time PCR were used to detect the enrichment of DNA fragments in the putative binding sites of the miR-142 upstream coding region. The primers used to detect the miR-142 upstream sequence were as follows: P1 Forward: 5′-GACCTGATCCTCACCCAGCTCTG-3′, Reverse: 5′-TGGCACTAGGCTAGGTGTTCACG-3′; P2 Forward: 5′-CGTGAACACCTAGCCTAGTGCCATC-3′, Reverse: 5′-ACCCCACAAAGCCACCCCTCCTG-3′; P3 Forward: 5′-CTGAGTGTTGGGAAGGGAAGGGAG-3′, Reverse: 5′-CCCAAAACGGGGAGTCCAGAATT-3′; P4 Forward: 5′-CTTCTGGGGTCACCCTCAATGCC-3′, Reverse: 5′-GGGAGGACAGACTGACAGGAAACAG-3′; and P5 Forward: 5′-CGGGGAAAGAGGTGTCCAAG-3′, Reverse: 5′-CTTCTCAGCCTCTGCGTACC-3′. For the real-time PCR analysis, the percentage of bound DNA enrichment was quantified related to input.
Co-immunoprecipitation (co-IP) assays
For co-IP assay, cells were lysed with RIPA lysis buffer and protein extracts were subsequently incubated using anti-EZH2 (1:50; Cell Signaling Technology, 4905) and anti-H3K27me3 (2µ, Millipore, 07–449) antibodies, followed by precipitation with protein A agarose (Invitrogen) and immobilized on protein G Sepharose beads. The precipitated proteins were detected by western blotting. Finally, the blots were visualized using a chemiluminescence system.
In vitro cellular assays
The in vitro cell migration and invasion assays were performed using Transwell chambers (Corning, Corning, NY) without or with Matrigel (BD Biosciences). Briefly, 5×104 or 1×105 transfected cells were suspended in serum-free medium and plated in the upper chamber for the migration or invasion assays, respectively. Medium in the lower chamber was supplemented with 10% FBS. After 16 or 24 h of incubation, migrated or invaded cells were fixed, stained, and counted using an inverted microscope. For the wound-healing assays, transfected cells were seeded in six-well plates and subjected to serum-free medium for 24 h. An artificial wound was scratched on the confluent cell monolayer, and images were taken at 0 and 36 h after serum starvation.
In vivo lung metastasis model
For in vivo lung metastasis assays, 4- to 6-week-old female BALB/c nude mice were purchased from the Medical Experimental Animal Center of Guangdong Province (Guangzhou, China). SUNE-1 cells (1×106 cells in 200 µl phosphate buffer saline (PBS)) with or without stable miR-142 expression were injected into mice via the tail vein. After 8 weeks, the mice were sacrificed, and lung metastatic colonization was validated and quantified by histopathological analysis.
Dual luciferase reporter assay
Two binding sites on the ZEB2 3′ UTR were predicted to be regulated by miR-142-3p using TargetScan 6.2 and miRanda (position 1: 319–325, position 2: 2647–2654), and these sites were mutated via site-directed mutagenesis. The mutated (Mt) and wild-type (Wt) sequences of the ZEB2 3′ UTR were constructed and cloned into the psiCHECK-2 luciferase reporter vectors (Promega). For the reporter assays, Wt or Mt or the control psiCHECK-2 vector (2 μg) was cotransfected with miR-142-3p mimics (100 nM) or miR-142-3p inhibitor (100 nM) or miRNA negative control (miR-Ctrl, 100 nM) using Lipofectamine 3000 reagent (Invitrogen). Luciferase activity was measured 24 h after transfection using the Dual-Luciferase Reporter Assay System (Promega).
Immunofluorescence staining
For immunofluorescence staining assays, cells grown on coverslips were fixed in 2% paraformaldehyde for 30 min, permeabilized in 0.5% Triton X-100 in PBS for 15 min at room temperature and followed by incubation with primary anti-E-cadherin (1:100; BD Biosciences, 610181) and anti-Vimentin (1:100; Proteintech, 10366-1-AP) antibodies overnight at 4 °C. The coverslips were incubated with species-matched Alexa Fluor 594 goat IgG secondary antibody (1:1,000; Life Technologies, Carlsbad, CA, USA; A-11008 or A-11001), followed by counterstaining with 4′, 6-diamidino-2-phenylindole. Images were viewed with a confocal laser-scanning microscope (Olympus FV1000, Tokyo, Japan).
Delivery of miR-142-3p to lymph node metastasis and lung metastatic colonization model
Four- to 6-week-old female BALB/c nude mice were purchased from the Medical Experimental Animal Center of Guangdong Province (Guangzhou, China). For in vivo lymph node metastasis, SUNE-1 cells (2 × 105 cells in 30 µl PBS) were inoculated into mice foot-pads. After 2 weeks’ growth, intratumoral injection of agomir to overexpressing 142-3p or agomir NC control (10 nM, RiboBio) were injected into foot-pad tumors twice a week for 2 weeks. The foot-pad tumors and inguinal lymph nodes were excised for analysis. For lung metastasis model construction, 1 × 106 SUNE-1 cells were injected into mice via the tail vein. Two weeks later, agomir-miR-142-3p or agomir NC control was administered via tail vein injection at a dose of 5 nM, twice per week for 4 weeks. The animals’ health status was observed, including body weight, activity, food intake, and abnormalities such as dehydration and diarrhea. Eight weeks after model construction, the mice were sacrificed and lung metastatic colonization was validated and quantified by histopathological analysis.
Immunohistochemical staining
Immunohistochemistry assays were performed on paraffin-embedded sections prepared from in vivo experiments to detect the expression levels of ZEB2. The antibody used was anti-ZEB2 (1:100; Abcam, ab138222). The degree of staining was examined separately by two pathologists. The staining intensity of stained cells was graded as follows: no staining (score 0); weak staining, light yellow (score 1); moderate staining, yellow brown (score 2); and strong staining, brown (score 3). The extent of staining was scored according to the percentage of positively stained cells: <10% cells (score 1); 10–35% (score 2); 35–70% (score 3); and >70% (score 4), as previously described [55]. The staining index was calculated by multiplying the staining intensity and percentage of positive cells.
Statistical analysis
Animal experiments were performed using five mice in each group. Data are presented as the mean ± s.d. resulting from at least three independent experiments. Two-tailed Student’s t tests were used to compare groups. Comparisons among categorical variables were performed with χ2 and Fisher’s exact tests. The Kaplan−Meier method was conducted to calculate survival curves, and the log-rank test was used to compare the differences. Independent prognostic factors were assessed by multivariate analysis using a Cox proportional hazards regression model. P values less than 0.05 were considered significant.
Electronic supplementary material
Acknowledgements
We would like to thank Hanqi Yin (Guangdong Longsee Biomedical Corporation, China) for bioinformatics and statistical consultation and Professor Musheng Zeng for cell lines.
Funding
This study was supported by grants from the National Natural Science Foundation of China (81402516, 81773229), Guangdong Special Support Program (2017TQ04R754), Natural Science Foundation of Guangdong Province (2018B030306045, 2017A030312003), Health & Medical Collaborative Innovation Project of Guangzhou City, China (201803040003), Innovation Team Development Plan of the Ministry of Education (No. IRT_17R110), and Overseas Expertise Introduction Project for Discipline Innovation (111 Project, B14035).
Author contributions
YL, JM and NL designed the experiments. YL, QH, XW, XH and XY conducted experiments and acquiring data. YL, XH, XY, XT, PZ, YL and YS analyzed the data. QH, YW and JZ provided reagents. YL, JM and NL wrote the manuscript. All authors read and approved the final manuscript.
Data availability
The microarray datasets used in this paper have been deposited at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under the series accession number GSE32960 and GSE70970. The authenticity of this article has been validated by uploading the key raw data onto the Research Data Deposit public platform (http://www.researchdata.org.cn), with the approval RDD number as RDDB2018000218.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
All animal studies were approved by the Institutional Animal Care and Use Ethics Committee of Sun Yat-sen University Cancer Center (approval number: L102012015090I). The experimental methods comply with the Helsinki Declaration.
Footnotes
Edited by Richard A. Knight
These authors contributed equally: Yingqin Li, Qingmei He, Xin Wen and Xiaohong Hong
Contributor Information
Jun Ma, Email: majun2@mail.sysu.edu.cn.
Na Liu, Email: liun1@sysucc.org.cn.
Electronic supplementary material
The online version of this article (10.1038/s41418-018-0208-2) contains supplementary material, which is available to authorized users.
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Wei WI, Sham JS. Nasopharyngeal carcinoma. Lancet. 2005;365:2041–54. doi: 10.1016/S0140-6736(05)66698-6. [DOI] [PubMed] [Google Scholar]
- 3.Chen L, Hu CS, Chen XZ, Hu GQ, Cheng ZB, Sun Y, et al. Concurrent chemoradiotherapy plus adjuvant chemotherapy versus concurrent chemoradiotherapy alone in patients with locoregionally advanced nasopharyngeal carcinoma: a phase 3 multicentre randomised controlled trial. Lancet Oncol. 2012;13:163–71. doi: 10.1016/S1470-2045(11)70320-5. [DOI] [PubMed] [Google Scholar]
- 4.Chen YP, Wang ZX, Chen L, Liu X, Tang LL, Mao YP, et al. A Bayesian network meta-analysis comparing concurrent chemoradiotherapy followed by adjuvant chemotherapy, concurrent chemoradiotherapy alone and radiotherapy alone in patients with locoregionally advanced nasopharyngeal carcinoma. Ann Oncol. 2014;26:205–11. doi: 10.1093/annonc/mdu507. [DOI] [PubMed] [Google Scholar]
- 5.Zhang L, Huang Y, Hong S, Yang Y, Yu G, Jia J, et al. Gemcitabine plus cisplatin versus fluorouracil plus cisplatin in recurrent or metastatic nasopharyngeal carcinoma: a multicentre, randomised, open-label, phase 3 trial. Lancet. 2016;388:1883–92. doi: 10.1016/S0140-6736(16)31388-5. [DOI] [PubMed] [Google Scholar]
- 6.Lee R, Feinbaum R, Ambros V. A short history of a short RNA. Cell. 2004;116:S89–92. doi: 10.1016/S0092-8674(04)00035-2. [DOI] [PubMed] [Google Scholar]
- 7.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 10.Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 11.Liu N, Chen NY, Cui RX, Li WF, Li Y, Wei RR, et al. Prognostic value of a microRNA signature in nasopharyngeal carcinoma: a microRNA expression analysis. Lancet Oncol. 2012;13:633–41. doi: 10.1016/S1470-2045(12)70102-X. [DOI] [PubMed] [Google Scholar]
- 12.Bruce JP, Hui AB, Shi W, Perez-Ordonez B, Weinreb I, Xu W, et al. Identification of a microRNA signature associated with risk of distant metastasis in nasopharyngeal carcinoma. Oncotarget. 2015;6:4537–50. doi: 10.18632/oncotarget.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu N, Jiang N, Guo R, Jiang W, He QM, Xu YF, et al. MiR-451 inhibits cell growth and invasion by targeting MIF and is associated with survival in nasopharyngeal carcinoma. Mol Cancer. 2013;12:123. doi: 10.1186/1476-4598-12-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li YQ, Ren XY, He QM, Xu YF, Tang XR, Sun Y, et al. MiR-34c suppresses tumor growth and metastasis in nasopharyngeal carcinoma by targeting MET. Cell Death Dis. 2015;6:e1618. doi: 10.1038/cddis.2014.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–6. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 16.Lu X, Li X, He Q, Gao J, Gao Y, Liu B, et al. miR-142-3p regulates the formation and differentiation of hematopoietic stem cells in vertebrates. Cell Res. 2013;23:1356–68. doi: 10.1038/cr.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao J, Cao Y, Lei Z, Yang Z, Zhang B, Huang B. Selective depletion of CD4 + CD25 + Foxp3 + regulatory T cells by low-dose cyclophosphamide is explained by reduced intracellular ATP levels. Cancer Res. 2010;70:4850–8. doi: 10.1158/0008-5472.CAN-10-0283. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Q, Haupt S, Prots I, Thummler K, Kremmer E, Lipsky PE, et al. miR-142-3p is involved in CD25 + CD4 T cell proliferation by targeting the expression of glycoprotein A repetitions predominant. J Immunol. 2013;190:6579–8. doi: 10.4049/jimmunol.1202993. [DOI] [PubMed] [Google Scholar]
- 19.Xu S, Wei J, Wang F, Kong LY, Ling XY, Nduom E, et al. Effect of miR-142-3p on the M2 macrophage and therapeutic efficacy against murine glioblastoma. J Natl Cancer Inst. 2014;106:dju162. doi: 10.1093/jnci/dju162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jia AY, Castillo-Martin M, Domingo-Domenech J, Bonal DM, Sanchez-Carbayo M, Silva JM, et al. A common MicroRNA signature consisting of miR-133a, miR-139-3p, and miR-142-3p clusters bladder carcinoma in situ with normal umbrella cells. Am J Pathol. 2013;182:1171–9. doi: 10.1016/j.ajpath.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin RJ, Xiao DW, Liao LD, Chen T, Xie ZF, Huang WZ, et al. MiR-142-3p as a potential prognostic biomarker for esophageal squamous cell carcinoma. J Surg Oncol. 2012;105:175–82. doi: 10.1002/jso.22066. [DOI] [PubMed] [Google Scholar]
- 22.Wang F, Wang XS, Yang GH, Zhai PF, Xiao Z, Xia LY, et al. miR-29a and miR-142-3p downregulation and diagnostic implication in human acute myeloid leukemia. Mol Biol Rep. 2012;39:2713–22. doi: 10.1007/s11033-011-1026-5. [DOI] [PubMed] [Google Scholar]
- 23.Dahlhaus M, Roolf C, Ruck S, Lange S, Freund M, Junghanss C. Expression and prognostic significance of hsa-miR-142-3p in acute leukemias. Neoplasma. 2013;60:432–8. doi: 10.4149/neo_2013_056. [DOI] [PubMed] [Google Scholar]
- 24.Piunti A, Shilatifard A. Epigenetic balance of gene expression by Polycomb and COMPASS families. Science. 2016;352:d9780. doi: 10.1126/science.aad9780. [DOI] [PubMed] [Google Scholar]
- 25.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 26.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–4. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 27.Cheung KJ, Ewald AJ. A collective route to metastasis: seeding by tumor cell clusters. Science. 2016;352:167–9. doi: 10.1126/science.aaf6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibson WJ, Hoivik EA, Halle MK, Taylor-Weiner A, Cherniack AD, Berg A, et al. The genomic landscape and evolution of endometrial carcinoma progression and abdominopelvic metastasis. Nat Genet. 2016;48:848–55. doi: 10.1038/ng.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown R, Curry E, Magnani L, Wilhelm-Benartzi CS, Borley J. Poised epigenetic states and acquired drug resistance in cancer. Nat Rev Cancer. 2014;14:747–53. doi: 10.1038/nrc3819. [DOI] [PubMed] [Google Scholar]
- 30.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 31.Mackenzie TN, Mujumdar N, Banerjee S, Sangwan V, Sarver A, Vickers S, et al. Triptolide induces the expression of miR-142-3p: a negative regulator of heat shock protein 70 and pancreatic cancer cell proliferation. Mol Cancer Ther. 2013;12:1266–75. doi: 10.1158/1535-7163.MCT-12-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu L, Cai C, Wang X, Liu M, Li X, Tang H. MicroRNA-142-3p, a new regulator of RAC1, suppresses the migration and invasion of hepatocellular carcinoma cells. FEBS Lett. 2011;585:1322–30. doi: 10.1016/j.febslet.2011.03.067. [DOI] [PubMed] [Google Scholar]
- 33.Berrout J, Kyriakopoulou E, Moparthi L, Hogea AS, Berrout L, Ivan C, et al. TRPA1-FGFR2 binding event is a regulatory oncogenic driver modulated by miRNA-142-3p. Nat Commun. 2017;8:947. doi: 10.1038/s41467-017-00983-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qi X, Li J, Zhou C, Lv C, Tian M. MiR-142-3p suppresses SOCS6 expression and promotes cell proliferation in nasopharyngeal carcinoma. Cel Physiol Biochem. 2015;36:1743–52. doi: 10.1159/000430147. [DOI] [PubMed] [Google Scholar]
- 35.Kemper K, de Goeje PL, Peeper DS, van Amerongen R. Phenotype switching: tumor cell plasticity as a resistance mechanism and target for therapy. Cancer Res. 2014;74:5937–41. doi: 10.1158/0008-5472.CAN-14-1174. [DOI] [PubMed] [Google Scholar]
- 36.Caino MC, Seo JH, Wang Y, Rivadeneira DB, Gabrilovich DI, Kim ET, et al. Syntaphilin controls a mitochondrial rheostat for proliferation-motility decisions in cancer. J Clin Invest. 2017;127:3755–69. doi: 10.1172/JCI93172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grassilli S, Nika E, Lambertini E, Brugnoli F, Piva R, Capitani S, et al. A network including PU.1, Vav1 and miR-142-3p sustains ATRA-induced differentiation of acute promyelocytic leukemia cells—a short report. Cell Oncol (Dordr) 2016;39:483–9. doi: 10.1007/s13402-016-0292-6. [DOI] [PubMed] [Google Scholar]
- 38.Sun Y, Sun J, Tomomi T, Nieves E, Mathewson N, Tamaki H, et al. PU.1-dependent transcriptional regulation of miR-142 contributes to its hematopoietic cell-specific expression and modulation of IL-6. J Immunol. 2013;190:4005–13. doi: 10.4049/jimmunol.1202911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang Q, Yang Y, Zhao M, Liang G, Wu H, Liu Q, et al. Mycophenolic acid upregulates miR-142-3P/5P and miR-146a in lupus CD4+ T cells. Lupus. 2015;24:935–42. doi: 10.1177/0961203315570685. [DOI] [PubMed] [Google Scholar]
- 40.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 41.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–57. doi: 10.1016/S0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 42.Sellers WR, Loda M. The EZH2 polycomb transcriptional repressor--a marker or mover of metastatic prostate cancer? Cancer Cell. 2002;2:349–50. doi: 10.1016/S1535-6108(02)00187-3. [DOI] [PubMed] [Google Scholar]
- 43.Tsai CL, Li HP, Lu YJ, Hsueh C, Liang Y, Chen CL, et al. Activation of DNA methyltransferase 1 by EBV LMP1 Involves c-Jun NH(2)-terminal kinase signaling. Cancer Res. 2006;66:11668–76. doi: 10.1158/0008-5472.CAN-06-2194. [DOI] [PubMed] [Google Scholar]
- 44.Zhang RL, Peng LX, Yang JP, Zheng LS, Xie P, Wang MY, et al. IL-8 suppresses E-cadherin expression in nasopharyngeal carcinoma cells by enhancing E-cadherin promoter DNA methylation. Int J Oncol. 2016;48:207–14. doi: 10.3892/ijo.2015.3226. [DOI] [PubMed] [Google Scholar]
- 45.Beltran M, Puig I, Pena C, Garcia JM, Alvarez AB, Pena R, et al. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Gene Dev. 2008;22:756–69. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 47.Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527:472–6. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xia L, Huang W, Tian D, Zhang L, Qi X, Chen Z, et al. Forkhead box Q1 promotes hepatocellular carcinoma metastasis by transactivating ZEB2 and VersicanV1 expression. Hepatology. 2014;59:958–73. doi: 10.1002/hep.26735. [DOI] [PubMed] [Google Scholar]
- 49.Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–29. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 50.Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12:847–65. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chua ML, Wee JT, Hui EP, Chan AT. Nasopharyngeal carcinoma. Lancet. 2016;387:1012–24. doi: 10.1016/S0140-6736(15)00055-0. [DOI] [PubMed] [Google Scholar]
- 52.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 53.Zweig MH, Campbell G. Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin Chem. 1993;39:561–77. [PubMed] [Google Scholar]
- 54.Jiang W, Liu N, Chen XZ, Sun Y, Li B, Ren XY, et al. Genome-wide identification of a methylation gene panel as a prognostic biomarker in nasopharyngeal carcinoma. Mol Cancer Ther. 2015;14:2864–73. doi: 10.1158/1535-7163.MCT-15-0260. [DOI] [PubMed] [Google Scholar]
- 55.Li Y, Tang X, He Q, Yang X, Ren X, Wen X, et al. Overexpression of mitochondria mediator gene TRIAP1 by miR-320b loss is associated with progression in nasopharyngeal carcinoma. PLoS Genet. 2016;12:e1006183. doi: 10.1371/journal.pgen.1006183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray datasets used in this paper have been deposited at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under the series accession number GSE32960 and GSE70970. The authenticity of this article has been validated by uploading the key raw data onto the Research Data Deposit public platform (http://www.researchdata.org.cn), with the approval RDD number as RDDB2018000218.