Abstract
Skeletal muscle is divided into slow- and fast-type muscles, which possess distinct contractile and metabolic properties. Myogenic progenitors associated with each muscle fiber type are known to intrinsically commit to specific muscle fiber lineage during embryonic development. However, it is still unclear whether the functionality of postnatal adult myogenic cells is attributable to the muscle fiber in which they reside, and whether the characteristics of myogenic cells derived from slow- and fast-type fibers can be distinguished at the genetic level. In this study, we isolated adult satellite cells from slow- and fast-type muscle individually and observed that satellite cells from each type of muscle generated myotubes expressing myosin heavy chain isoforms similar to their original muscle, and showed different metabolic features. Notably, we discovered that slow muscle-derived cells had low potential to differentiate but high potential to self-renew compared with fast muscle-derived cells. Additionally, cell transplantation experiments of slow muscle-derived cells into fast-type muscle revealed that slow muscle-derived cells could better contribute to myofiber formation and satellite cell constitution than fast muscle-derived cells, suggesting that the recipient muscle fiber type may not affect the predetermined abilities of myogenic cells. Gene expression analyses identified T-box transcriptional factor Tbx1 as a highly expressed gene in fast muscle-derived myoblasts. Gain- and loss-of-function experiments revealed that Tbx1 modulated muscle fiber types and oxidative metabolism in myotubes, and that Tbx1 stimulated myoblast differentiation, but did not regulate myogenic cell self-renewal. Our data suggest that metabolic and myogenic properties of myogenic progenitor cells vary depending on the type of muscle from which they originate, and that Tbx1 expression partially explains the functional differences of myogenic cells derived from fast-type and slow-type muscles.
Subject terms: Cell biology, Molecular biology
Introduction
Skeletal muscle possesses a high capacity to regenerate following severe damage caused by exercise, toxins, or diseases. Muscle satellite cells, which are mononuclear myogenic stem cells, play a principal role during regeneration by proliferating and differentiate into myofibers [1, 2]. While the majority of myogenic cells undergo differentiation to form myofibers, a small population of myogenic cells exit their cell cycle, and eventually return to a quiescent state to maintain the satellite cell pool [3].
Maintaining the number as well as the functional attributes of muscle satellite cells is essential to ensure continued homeostasis and to achieve muscle regeneration. However, in certain circumstances, including aging [4, 5] and diseases such as Duchenne muscular dystrophy [6], satellite cell numbers and their proliferative potential significantly decline. Additionally, age-related reduction of satellite cell content is more severe in fast-type muscle fibers than in slow ones [5, 7]. Given in previous studies that the decrease of satellite cell number was due to a reduction of the capacity to self-renew [4, 8, 9], there are two possibilities: (1) slow-type muscle fibers secrete specific factors or possess niches to boost satellite cell self-renewal capacity, or (2) satellite cells, on slow-type muscle fibers, exhibit high potential to maintain their number.
Skeletal muscles are classified into four myosin heavy chain (MyHC) isoforms (types I, IIA, IID/X, and IIB). According to previous reports, satellite cells associated with each muscle fiber type are functionally different [3, 10–12]. For instance, a higher number of satellite cells were found in slow-type muscle than in fast-type muscle [3, 10], and the potential of cell proliferation or differentiation differed between them [10–12]. Additionally, adult slow- or fast-type muscle-derived myoblasts differentiated into specific muscle fibers [11, 12]. These observations indicated that the functionality of satellite cells is associated with the muscle fiber in which they reside.
Notably, in vivo and in vitro experiments have demonstrated that myogenic cells differentiate into muscle fibers with distinct identities [11, 12], implying that adult myogenic cells are intrinsically committed to differentiating into distinct muscle fibers. However, others demonstrated that engrafted adult myoblasts randomly formed all fiber types, concluding that the differentiation pattern of muscle fibers is independent of the cell lineage [13]. As such, no definitive conclusion has been reached on this issue, and it is still unclear whether the environmental conditions and niches surrounding satellite cells affect their differences or whether the characteristics of myogenic cells derived from slow- and fast-type fibers can be distinguished at the genetic level.
In the present study, we isolated satellite cells separately from slow-type-enriched soleus (SOL) and fast-type-enriched tibialis anterior (TA) and observed that SOL-derived myoblasts (SOL-MBs) generated slow-type myofibers while TA-derived myoblasts (TA-MBs) formed fast-type ones. Notably, SOL-MBs had low potential to differentiate but high potential to self-renew. Using gene expression analyses, we identified that T-box-containing gene Tbx1 was highly expressed in TA-MBs, and found that Tbx1 modulated muscle metabolism in myotubes and myogenic differentiation in myoblasts. These observations highlighted the functional differences of slow- and fast-type muscle-derived satellite cells and suggested Tbx1's involvement in regulating biological differences between TA- and SOL-MBs.
Results
Myogenic cell potentials are governed by the muscle fiber types from which satellite cells originate
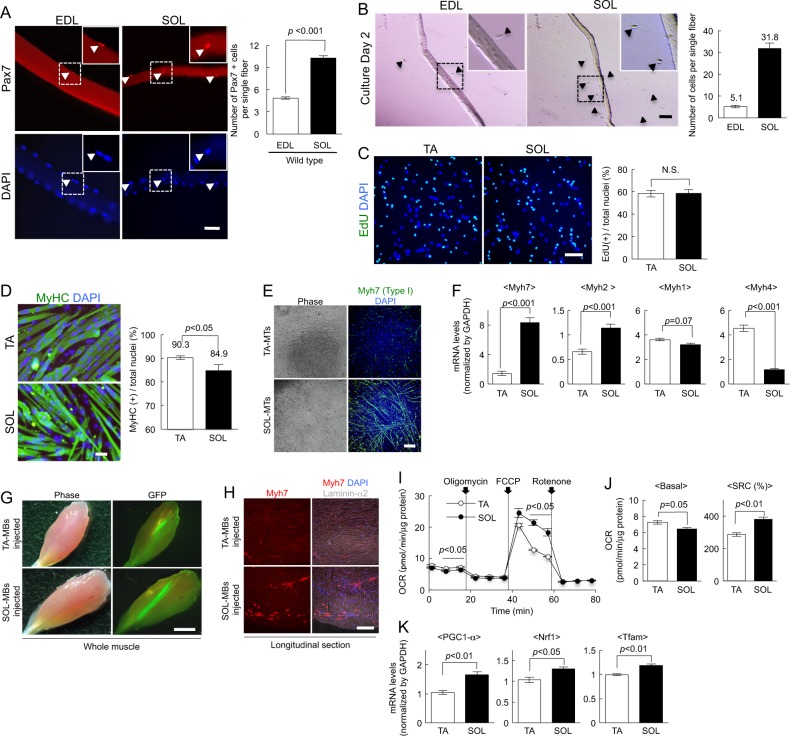
To investigate satellite cell functions between slow- and fast-type fibers, the cells were isolated from the SOL and fast-type fiber-enriched extensor digitorum longus (EDL). First, the cells were stained for Pax7 (Paired box 7), which is a marker for satellite cells, to count the number of satellite cells in single muscle fibers from both EDL and SOL. Single muscle fibers from SOL contained more Pax7(+) cells than fibers from EDL (Fig. 1a), as also previously reported [3, 10]. Additionally, initial activation of satellite cells occurred more rapidly in SOL than in EDL after 48 h of culture (Fig. 1b). Next, we conducted proliferation and differentiation assays. Since the proportion of type I fibers in TA was lower than in EDL (Fig. S1A) [14], TA was used to represent fast-type fiber. Myoblasts from TA and SOL were cultured with 5-ethynyl-2′-deoxyuridine (EdU) and then EdU(+) proliferating cells were counted. There was no difference in the number of EdU(+) cells (Fig. 1c). Five days after the differentiation, more cells differentiated into MyHC(+) myotubes in the culture of TA-MBs than for from SOL (Fig. 1d) [12], indicating that TA-MBs had higher potential to form myotubes.
Fig. 1.
Differences in the capacity of myogenic cells from slow- and fast-type muscles. a Freshly isolated intact myofibers from EDL or SOL muscle of wild-type mice were stained against Pax7 (red) and with DAPI (blue). Typical Pax7(+) and DAPI(+) cells are shown (arrowheads). The number of Pax7(+) satellite cells per fiber was quantified. Values are presented as mean ± SE (n = 3). Scale bar = 50 µm. b Myofibers isolated from SOL or EDL muscle were cultured under growth conditions for 48 h. The activated satellite cells that migrated from each muscle fiber (arrowheads) were counted. Values are presented as mean ± SE (n = 3). Scale bar = 100 µm. c Myoblasts from SOL or TA muscle were cultured in growth medium with EdU. After staining with EdU (green) and DAPI (blue), the proportion of EdU(+) cells among the total nuclei was counted. Values are presented as mean ± SE (n = 3). Scale bar = 100 µm. d Myoblasts from SOL or TA muscle were cultured in differentiation medium for 4 days, and then myotubes and nuclei were stained against MyHC (green) and with DAPI (blue). The proportion of MyHC(+) cells among the total nuclei was quantified. Values are presented as mean ± SE (n = 3). Scale bar = 50 µm. e Myotubes of TA- and SOL-derived myoblasts were stained against Myh7 (MyHC type I slow; green) and with DAPI (blue). Scale bar = 200 µm. f The expression levels of Myh7, Myh2, Myh1, and Myh4 in myotubes were quantified by qPCR. The expression values were normalized to GAPDH expression and are presented as mean ± SE (n = 4). g TA- or SOL-MBs derived from GFP mouse muscles were injected into TA muscles of NOD/scid mice. Typical phase image (left) and GFP expression (right) are presented. Scale bar = 2 mm. h Longitudinal sections of engrafted TA muscles stained against slow-type myosin heavy chain (red) and laminin-α2 (gray), and with DAPI (blue). Scale bar = 500 µm. i OCR in SOL- or TA-derived myotubes was measured after treatment with oligomycin and FCCP. Values are presented as mean ± SE (n = 3). j Basal OCR and SRC of myotubes were quantified. Values are presented as mean ± SE (n = 3). k The expression levels of PGC-1α, Nrf1, and Tfam in TA- and SOL-MTs were quantified by qPCR. Values are presented as mean ± SE (n = 3)
To investigate the distribution of MyHC isoforms in myotubes formed from SOL- and TA-derived cells, myotubes were stained with anti-Myh7 (MyHC type-I) antibody. Slow-type Myh7 was expressed in myotubes from SOL-MBs (SOL-MTs), but not in myotubes from TA-MBs (TA-MTs: Fig. 1e). Additionally, quantitative PCR (qPCR) experiments revealed that SOL-MTs highly expressed Myh7, while Myh4 (MyHC type IIB) expression was low (Fig. 1f). Conversely, TA-MTs highly expressed Myh4, whereas Myh7 expression was low (Fig. 1f). To further confirm this in vivo, myoblasts from wild-type SOL or TA were labeled using a GFP-expressing retrovirus vector, and then transplanted into injured TA muscles of NOD/Scid immunodeficient mice. Three weeks later, we confirmed the GFP expression in TA-MB- or SOL-MB-engrafted TA muscles (Fig. 1g). After Myh7 staining, TA muscle transplanted with SOL-MBs exhibited some Myh7(+) fibers, while TA-MB-injected muscles did not (Fig. 1h). These suggested that each myogenic cell was predetermined to differentiate into a specific fiber type, and that the recipient muscle fibers may not affect the fate of injected cells.
Muscle fibers can use both oxidative phosphorylation (OXPHOS) and aerobic glycolysis for energy [15], and myosin types are well known to be related to muscle fiber metabolism. To assess the functional difference between TA-MTs and SOL-MTs in vitro, we evaluated oxygen consumption rate (OCR), an indicator of OXPHOS in myotubes, and observed that OCR in SOL-MTs was lower than in TA-MTs under basal conditions (Fig. 1i). When carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP) was added to measure the maximal flux of OCR, OCR was higher in SOL-MTs than in TA-MTs (Fig. 1i). Additionally, SOL-MTs possessed high spare respiratory capacity (SRC), which is defined as the quantitative difference between the maximal and basal OCRs (Fig. S1B) [16], in comparison to TA-MTs (Fig. 1i, j). According to the previous studies [17, 18], the steady state rate OCR in slow-type muscles is lower than, or comparable to, fast-type muscle. Additionally, the maximum OCR after isometric twitch stimulation is higher in slow-type than in fast-type muscle [17, 18]. These observations are consistent with our in vitro experiments (Fig. 1i, j). Therefore, the in vitro metabolic features of SOL- and TA-MTs may reflect those of slow- and fast-type muscles in vivo.
We also assessed the expression of the mitochondrial biogenesis markers peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α), nuclear respiratory factor 1 (Nrf1), and mitochondrial transcription factor A (Tfam) [19, 20] by qPCR. These gene expressions were significantly higher in SOL-MTs than in TA-MTs (Fig. 1k). Together, these findings suggested functional metabolic differences between TA- and SOL-MTs.
Slow-type muscle fiber-derived myogenic cells have a high potential for self-renewal
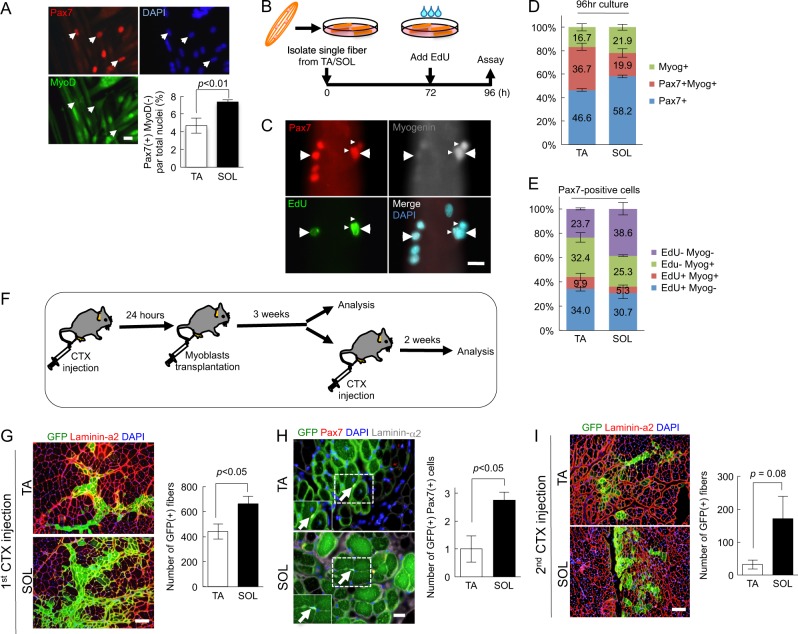
The capacity for myoblast differentiation in SOL-MTs was low (Fig. 1). To clarify the differences in myoblast self-renewal potency between SOL-MBs and TA-MBs, both cell types were cultured in differentiation medium for 5 days to produce reserve cells, which are equivalent to self-renewing satellite cells in vitro [3, 21–23]. These cells were stained for Pax7 and MyoD, which are the master myogenic transcriptional factors. The number of Pax7(+)MyoD(−) reserve cells were significantly higher in SOL-MBs than in TA-MBs (Fig. 2a). To confirm our observations, we conducted single-muscle-fiber culture experiments. Satellite cells attached to muscle fibers can enter the cell cycle within the first 72 h and some proportion of cells undergo either differentiation or self-renewal after 72 h [23]. Therefore, 24 h before harvest, EdU was added to the culture medium (Fig. 2b), and this was followed by immunostaining for EdU, Pax7, and Myogenin, which are the major myogenic differentiation markers (Fig. 2c). SOL fibers contained a higher number of Pax7(+) cells and a lower number of Myogenin(+) differentiated cells than TA muscle fibers, and the number of Pax7(+)EdU(−)Myogenin(−) self-renewing cells were higher in SOL fibers than in TA (Fig. 2d, e), indicating that satellite cells associated with SOL had a higher ability to self-renew.
Fig. 2.
Slow-type muscle-derived myogenic cells exhibit high potential for self-renewal. a Myoblasts from SOL or TA muscle were cultured in differentiation medium for 5 days, and then stained against Pax7 (red) and MyoD (green), and with DAPI (blue). The number of Pax7(+)MyoD(−) reserve cells were quantified. Values are presented as mean ± SE (n = 3). Scale bar = 50 µm. b Scheme for single muscle fiber culture experiments. Single muscle fibers of SOL or TA from the wild-type mice were isolated and cultured. EdU was added to the culture medium 24 h prior to harvest. c–e After 96 h of culture, the muscle fibers were stained against Pax7, EdU, and Myogenin (c), and then the ratio was quantified (d, e). Values are presented as mean ± SE (n = 3). Scale bar = 20 µm. f Scheme for myoblast transplantation experiments. GFP-positive myoblasts (2 × 105 cells) from TA- or SOL-derived myoblasts were transplanted into CTX-injected TA muscles of NOD/scid mice. Three weeks after transplantation, the engrafted muscles were analyzed or injected with CTX to re-induce muscle regeneration. Two weeks after the second CTX injection, the transplanted muscles were analyzed. g Three weeks after transplantation, cross-sections of transplanted TA muscles were stained against GFP (green) and laminin-α2 (red), and the number of GFP(+) fibers was quantified. Values are presented as mean ± SE (n = 4–6). Scale bar = 100 µm. h Cross-sections of transplanted TA muscles stained against GFP (green), Pax7, (red), and laminin-α2 (orange) antibodies. Values are presented as mean ± SE (n = 4–6). Scale bar = 20 µm. i Two weeks after the second CTX injection, the transplanted muscles were dissected and cross-sections were stained against GFP (green) and laminin-α2 (red) antibodies. Values are presented as mean ± SE (n = 4–6). Scale bar = 100 µm
Next, we performed myoblast transplantation to examine whether SOL-MBs preferentially contribute to satellite cell constitution in vivo (Fig. 2f). SOL- and TA-MBs, which were labeled with GFP-expressing retrovirus vector, were transplanted into the injured TA muscle. After the transplantation, SOL-MBs produced a higher number of GFP(+) myofibers (Fig. 2g). Additionally, SOL-MBs yielded a higher number of Pax7(+) satellite cells than TA-MBs (Fig. 2h), indicating that SOL-MBs had a high potential of self-renewal. To further confirm the capacity of SOL- and TA-MBs to self-renew, engrafted TA muscles were re-injured by the second injection of cardiotoxin (CTX) (Fig. 2f) [24]. After the second CTX injection, the number of GFP(+) fibers was significantly higher in SOL-MB-transplanted muscle than in TA-MB-transplanted muscle (Fig. 2i). Our findings suggested that SOL-MBs had high potential to contribute to muscle regeneration by forming myofibers and generating satellite cells.
Tbx1 regulates muscle fiber type and metabolism
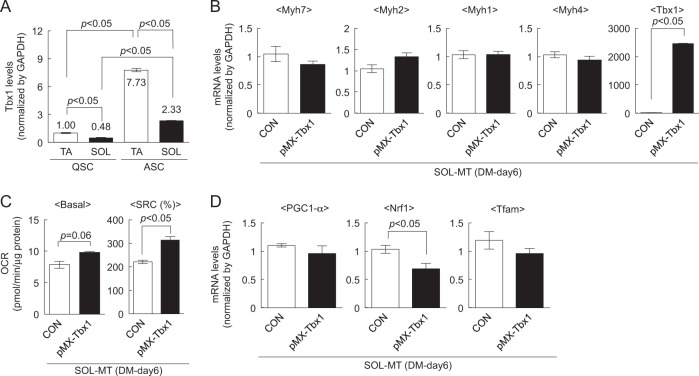
To reveal the biological differences between TA- and SOL-MBs, a microarray analysis was conducted. Gene expression analysis revealed that a transcription factor, T-box 1 (Tbx1), was highly expressed in TA-MBs (Table 1), which was confirmed by qPCR (Fig. 3a). Additionally, we isolated quiescent (QSC) and activated (ASC) satellite cells from TA and SOL, and then assessed Tbx1 expression. Tbx1 expression in QSC from TA was higher than in those from SOL. Additionally, Tbx1 expression increased in ASC and continued to be higher in TA-MBs (Fig. 3a), suggesting that Tbx1 is intrinsically high in fast-type muscle-derived satellite cells, and that some upstream factors may modulate Tbx1 expression upon satellite cell activation.
Table 1.
Relative expression levels in TA-MBs compared with those in SOL-MBs
| RefSeq_id | Symbol | Description | Ratio (vs SOL) |
|---|---|---|---|
| (A) | |||
| NM_153786 | Vgll2 | Vestigial like 2 homolog (Drosophila) | 3.06 |
| NM_011532 | Tbx1 | T-box 1 | 2.87 |
| NM_021550 | C1galt1c1 | C1GALT1-specific chaperone 1 | 2.59 |
| NM_025824 | Bzw1 | Basic leucine zipper and W2 domains 1 | 2.40 |
| NM_009033 | Rbmxrt | RNA binding motif protein, X chromosome retrogene | 2.26 |
| (B) | |||
| NM_013496 | Crabp1 | Cellular retinoic acid binding protein I | 0.20 |
| NM_021459 | Isl1 | ISL1 transcription factor, LIM/homeodomain | 0.21 |
| NM_011535 | Tbx3 | T-box 3 | 0.36 |
| NM_007555 | Bmp5 | Bone morphogenetic protein 5 | 0.43 |
| NM_009462 | Usp10 | Ubiquitin specific peptidase 10 | 0.49 |
Data are for the five genes that were most highly expressed in TA-MBs (A) or in SOL-MBs (B)
Fig. 3.
Tbx1 induces fast fiber-like properties in slow-type muscle-derived myotubes. a The expression levels of Tbx1 in QSC and ASC from TA and SOL muscles were quantified by qPCR. Values are presented as mean ± SE (n = 3). b The expression levels of Myh7, Myh2, Myh1, Myh4, and Tbx1 in Tbx1-expressing SOL-MTs were quantified by qPCR. Values are presented as mean ± SE (n = 4). c Basal OCR and SRC of myotubes were calculated. Values are presented as mean ± SE (n = 3). d The expression levels of PGC-1α, Nrf1, and Tfam in Tbx1-expressing SOL-MTs were quantified by qPCR. Values are presented as mean ± SE (n = 4)
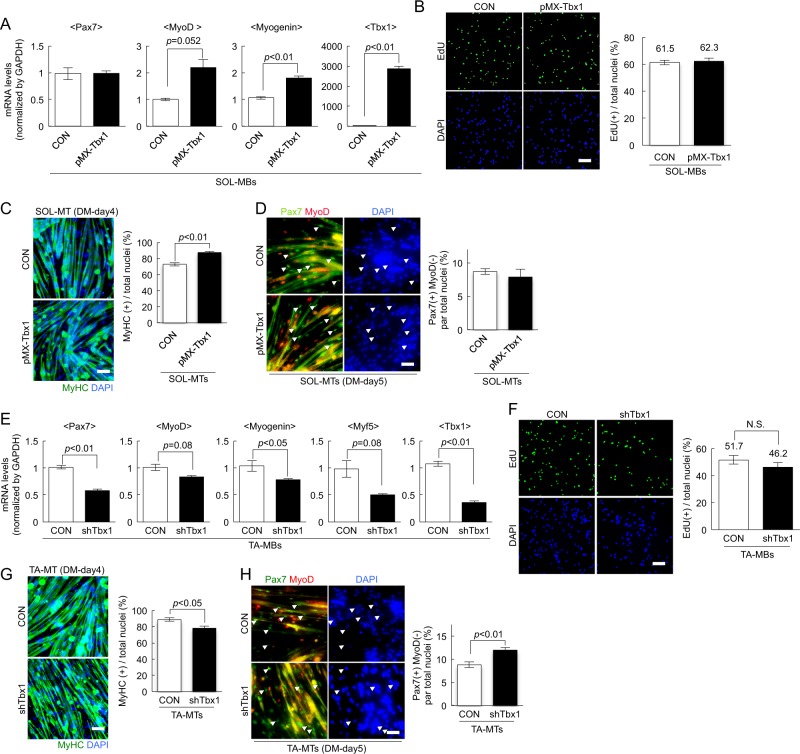
Since Tbx1 was highly expressed in TA-MBs, we hypothesized that Tbx1 plays a role in generating fast-type muscle fibers. To clarify this hypothesis, Tbx1 was overexpressed in SOL-MBs (Fig. 3b), and then Tbx1-overexpressing SOL-MBs were differentiated into myotubes. After qPCR analysis, Tbx1 slightly attenuated Myh7 expression in SOL-MTs (Fig. 3b). We also observed increases of the basal OCR and SRC (Fig. 3c) in Tbx1-expressing SOL-MTs compared with those in the control. Additionally, Nrf1 was significantly reduced while that of PGC-1α and Tfam was slightly attenuated in Tbx1-expressing SOL-MTs (Fig. 3d). The trends of decreases in Myh7 and mitochondrial biogenesis-related gene expression and increase of basal OCR induced by Tbx1 in SOL-MTs are reminiscent of characteristics of fast-like myotubes generated by TA-MBs (Fig. 1).
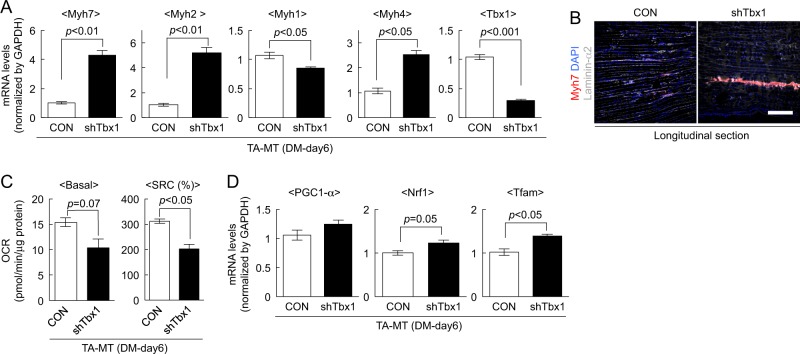
To further confirm the effect of Tbx1 in myotubes, Tbx1 was suppressed by shTbx1 in TA-MTs (Fig. 4a). ShTbx1 significantly increased Myh7, Myh2, and Myh4 expression, while Myh1 was apparently attenuated by shTbx1 in TA-MTs (Fig. 4a). We also performed myoblast transplantation experiments in vivo, and found that TA muscles injected with shTbx1-infected myoblasts exhibited clusters of Myh7(+) fibers (Fig. 4b). Additionally, we observed decreases of basal OCR and SRC (Fig. 4c) and increases of Nrf1 and Tfam expression (Fig. 4d), in shTbx1-treated TA-MTs compared with the levels in the control. These observations indicated that Tbx1 suppression may induce features associated with a predisposition to generate slow fibers. Taken together, the inherited metabolic and fiber-type properties of myogenic cells are regulated at least in part by Tbx1.
Fig. 4.
shTbx1 induces slow fiber-like properties in fast-type muscle-derived myotubes. a The expression levels of Myh7, Myh2, Myh1, Myh4, and Tbx1 in shTbx1-infected TA-MTs were quantified by qPCR. Values are presented as mean ± SE (n = 4). b shTbx1-infected TA-MBs were injected into TA muscles of NOD/Scid mice. Longitudinal sections of engrafted TA muscles stained against slow-type myosin heavy chain (red), laminin-α2 (gray), and DAPI (blue). Scale bar = 500 µm. c Basal OCR and SRC of myotubes were calculated. Values are presented as mean ± SE (n = 3). d The expression levels of PGC-1α, Nrf1, and Tfam in shTbx1-treated TA-MTs were quantified by qPCR. Values are presented as mean ± SE (n = 4)
Tbx1 modulates myogenic cell differentiation
Another topic of interest in this study is the effect of Tbx1 on the myogenic cells since the potential for myogenesis differed between TA- and SOL-MBs (Figs. 1 and 2). Quantitative PCR results demonstrated increases of MyoD and Myogenin expression without alteration of Pax7 expression in Tbx1-overexpressing SOL-MBs compared with the levels in control cells (Fig. 5a). Proliferation assay demonstrated no difference in the number of EdU(+) cells between control and Tbx1-expressing cells (Fig. 5b). Five days after myoblast differentiation, the number of MyHC(+) cells were significantly increased in Tbx1-expressing SOL-MBs compared with that in the control (Fig. 5c). The number of Pax7(+)MyoD(−) reserve cells were slightly reduced, albeit not significantly, in Tbx1-expressing myoblasts compared with that in control cells (Fig. 5d). These observations indicate that Tbx1 induces myogenic differentiation.
Fig. 5.
Tbx1 regulates myogenesis in SOL- and TA-muscle-derived myoblasts in vitro. a, e The expression of Pax7, MyoD, Myogenin, Myf5, and Tbx1 in Tbx1-overexpressing (a) or Tbx1-suppressed (e) myoblasts as quantified. Values are presented as mean ± SE (n = 4). b, f Tbx1-overexpressing (b) or Tbx1-suppressed (f) MBs were cultured in growth medium with EdU. The number of EdU(+) cells were counted. Values are presented as mean ± SE (n = 4). Scale bar = 100 µm. c, g Tbx1-overexpressing (c) or Tbx1-suppressed (g) TA-MBs were cultured in differentiation medium for 4 days, and then stained against MyHC (green) and with DAPI (blue). The proportion of MyHC(+) cells among total nuclei was quantified. Values are presented as mean ± SE (n = 3). Scale bar = 100 µm. d, h Tbx1-overexpressing (d) or Tbx1-suppressed (h) TA-MBs were cultured with differentiation medium for 5 days, and then stained for Pax7 (red) and MyoD (green). The number of Pax7(+)MyoD(−) reserve cells were quantified. Values are presented as mean ± SE (n = 3). Scale bar = 100 µm
In contrast, qPCR experiments revealed significant decreases in the expression of Pax7, MyoD, Myogenin, and Myf5, which regulate myogenic differentiation, in Tbx1-inhibited TA-MBs compared with the control cells (Fig. 5e). A proliferation assay revealed that the number of EdU(+) cells were slightly, but not significantly, decreased in shTbx1-treated TA-MBs compared with that in control cells (Fig. 5f). After the induction of differentiation, the number of MyHC(+) cells were significantly decreased (Fig. 5g), while the number of Pax7(+)MyoD(−) reserve cells were significantly increased in Tbx1-inhibited TA-MBs compared with the levels in control cells (Fig. 5h).
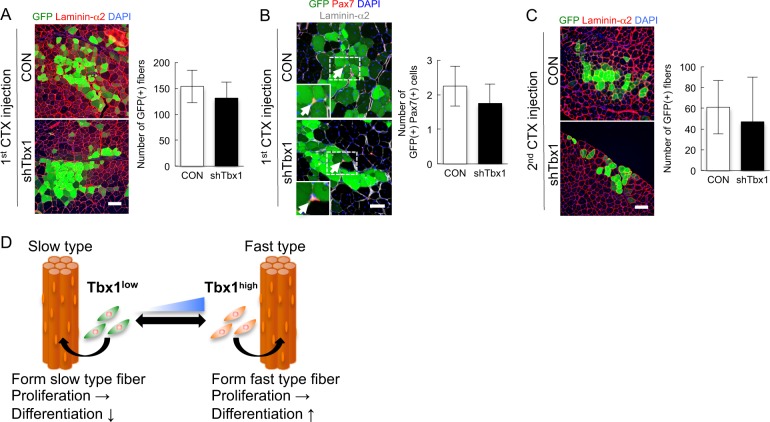
To examine whether the inhibition of Tbx1 in TA-MBs contributes to the formation of muscle fiber and satellite cells in vivo, shTbx1-treated TA-MBs derived from GFP-transgenic mice were transplanted into injured TA muscle. ShTbx1-treated TA-MBs produced a low number of GFP(+) myofibers (Fig. 6a) compared with control TA-MBs, while Tbx1 inhibition did not affect the production of GFP(+) Pax7(+) satellite cells (Fig. 6b). Additionally, 2 weeks after the second CTX injection, the number of GFP(+) fibers was decreased in muscle into which shTbx1-treated TA-MBs had been injected, compared with that in muscle into which control TA-MBs had been injected (Fig. 6c). Our findings suggest that Tbx1 is one of the modulators of myogenic cell differentiation, but cannot regulate myogenic cell self-renewal on its own.
Fig. 6.
Tbx1 inhibition did not affect myogenic cell self-renewal in TA-muscle-derived myoblasts in vivo. a Three weeks after transplantation (1 × 105 cells/mouse), cross-sections of engrafted TA muscles were stained against GFP and laminin-α2, and the number of GFP(+) fibers was quantified. Values are presented as mean ± SE (n = 4). Scale bar = 100 µm. b Cross-sections of transplanted TA muscles stained against GFP, Pax7, and laminin-α2. Values are presented as mean ± SE (n = 4). Scale bar = 50 µm. c Two weeks after the second CTX injection, the transplanted muscles were dissected and cross-sections were stained against GFP and laminin-α2. Values are presented as mean ± SE (n = 4). Scale bar = 100 µm. d A model for the regulation of muscle fiber type and myogenic cell potential involving Tbx1
Discussion
In this study, we found that the initial activation of satellite cells occurs more rapidly in slow-type muscle than in fast-type muscle fibers (Fig. 1b), and that fast-type muscle-derived cells have a high potential to form myotubes (Figs. 1 and 2). According to a previous study, the initiation of muscle regeneration occurs earlier in SOL than in fast-type EDL [25]. However, muscle regeneration is completed earlier in EDL than in SOL [25]. These differences in muscle regeneration between the two types may be explained by differences between the fast- and slow-type muscle-derived cells.
We also confirmed that myogenic cells are committed to specific muscle fiber types (Figs. 1 and 2). Notably, we could separately culture slow- or fast-type fibers in vitro without innervation. These observations are in agreement with previous results [11, 12, 26]. It was generally considered that muscle fiber-type components are predominantly influenced by innervation or exogenous stimuli [27, 28] since muscle fiber types are transitioned between fast and slow type muscle in several situations, including immobilization, denervation, starvation, or aging, which are considered to be unrelated to satellite cell activation. However, our results suggested that the predetermined property of satellite cells is also one of the key factors influencing muscle fiber types.
In this context, a question arises: Why are myogenic cells predetermined to commit to specific fibers? One possible answer is related to the potential role of myogenic cells in the maintenance of fiber-type composition. The overall composition and relative ratios of muscle fiber types are conserved throughout life [29–31], except in specific situations including muscle wasting and diseases [32]. Even after denervation and injury, the expression of slow MyHC was found to be maintained in SOL [33]. These observations suggest that myogenic cells are intrinsically dedicated to a specific muscle fiber type. However, it is still unclear whether the muscle fiber shift leads to a change of myogenic potential in satellite cells, or whether the transition of muscle fiber types occurs before or after the alteration of myogenic cell abilities. These questions should be answered by future studies.
We also found that SOL-MBs have higher potential to self-renew than TA-MBs (Fig. 1). Our results are supported by previous studies demonstrating that in comparison to TA fibers, SOL muscle fibers make a major contribution toward muscle fiber formation and to the reconstitution of self-renewing satellite cells after muscle fiber transplantation [3]. These observations suggest that satellite cells, which exist in slow-type muscle fibers, have a strong capacity to maintain their number. However, there remains the possibility that the environmental conditions or niche surrounding slow-type muscle fibers affect satellite cell self-renewal. Previous reports demonstrated that aged muscle fiber-derived fibroblast growth factor 2 (Fgf2) signals led to satellite cell activation, resulting in the depletion of satellite cells [34]. Several groups also demonstrated that satellite cell self-renewal is regulated by several extracellular factors, including Ang1/Tie2, FGF, MEGF10, collagen VI, and fibronectin [35], suggesting that factors from muscle fibers, the niche, or extracellular components might regulate satellite cell self-renewal. However, our results have proved that, after the transplantation of SOL-MBs into fast-type TA muscle, SOL-MBs could better contribute to myofiber formation and satellite cell constitution than TA-MBs (Fig. 2), indicating that the recipient muscle fiber type may not affect the abilities of myogenic cells. These findings suggest the possibility that the individual satellite cell fate is already determined prior to myogenic differentiation.
Tbx1 is expressed in pharyngeal mesoderm, which gives rise to the branchiomeric muscles, and is the major candidate gene for DiGeorge syndrome [36–38]. According to previous reports, Tbx1 is also expressed in adult skeletal muscle [39, 40]. However, few reports explained the function of Tbx1. In our study, we have shown that Tbx1 was highly expressed in TA-MBs, and that Tbx1-inhibited myoblasts contribute to the formation of slow-type muscle fibers in vivo (Fig. 4b), strongly indicating that Tbx1 is a modulator of muscle fiber types. Though we cannot fully explain why the Myh4 gene was upregulated by Tbx1-inhibition (Fig. 4a), future experiments may clarify whether or how Tbx1 directly or indirectly regulates the Myh4 gene.
Previous microarray experiments demonstrated that Tbx1 was highly expressed in glycolytic muscles compared with slow-type muscles [40, 41]. Additionally, Tbx1 expression was low in brown and white adipocytes, but enriched in beige cells, which have low UCP-1 levels and a high capacity to turn on respiration and increase energy expenditure [42]. In relation to Tbx1, Tbx15, a member of the Tbx1 family that is highly expressed in TA-MBs (data not shown), was also detected at a high level in glycolytic muscle [40, 43], and ablation of Tbx15 resulted in a decrease in the number of glycolytic fibers and in a modest increase in oxidative fibers [43]. Given these observations, Tbx1 is a possible regulator of muscle fiber type and/or metabolism.
Intriguingly, Tbx1 modulated Nrf1 expression and the OCR of cultured myotubes without the alteration of PGC-1α, which is a regulator of mitochondrial biogenesis [19] (Figs. 3d and 4d). Nrf1 is a key factor that regulates metabolism-related genes required for mitochondrial biogenesis and functions [44]. A previous report revealed that Nrf1 is an important cofactor of PGC-1α, and is required for PGC-1α-dependent mitochondrial biogenesis. Additionally, Nrf1-knockdown reduces Sirt6 activity, resulting in induction of glycolysis [45], whereas Nrf1-overexpression in muscle promotes an increase in the levels of mitochondrial proteins without affecting PGC-1α [46]. These reports, together with our results, suggest that the modulation of Nrf1 by Tbx1 independently of PGC-1α can explain the mechanism whereby Tbx1 alters the metabolic features in muscle. Whether Tbx1 directly or indirectly regulates metabolism-related genes, including Nrf1, will be clarified in future studies.
We also found that Tbx1 could modulate the differentiation in myogenic cells (Figs. 5 and 6). As previously reported, the activation of myogenic regulatory factors, Myf5 and MyoD, failed to occur in pharyngeal mesoderm of Tbx1-knockout mice [47, 48]. Additionally, in Tbx1/Myf5 double-knockout mice, Tbx1 affected MyoD expression, resulting in the defect of first branchial arch-derived muscles [49]. Consistent with these results observed during embryogenesis, Tbx1 regulated myogenic regulatory factors in adult myogenic cells (Figs. 5 and 6), suggesting that Tbx1 is a critical factor for myogenesis.
However, Tbx1 expression was not sufficient to completely explain the functional differences in TA- and SOL-MBs. SOL-MBs contributed to myofiber formation and satellite cell constitution (Fig. 2), whereas the inhibition of Tbx1 in TA-MBs did not (Fig. 6). One of the possible reasons for this is a reduction of myogenic-related gene expressions by Tbx1 inhibition. Pax7 and MyoD were suppressed by Tbx1 inhibition (Fig. 5), but slightly increased in SOL-MBs, compared with the levels in TA-MBs (data not shown). Given that Pax7, MyoD, and Myf5 play essential roles in muscle regeneration and satellite cell maintenance [50–53], Tbx1 could be a regulator of myogenic cell differentiation, but not of self-renewal. The differences in self-renewal ability in TA- and SOL-MBs could be regulated by other factors.
We have not clarified the mechanisms mediated by Tbx1 in adult myogenic cells. In cardiovascular development, Tbx1 interacts with several factors including Smad1 [54] and Serum Response Factor [55], and regulates downstream genes including Hes1 [56], Mef2c [57], and Pitx2 [58]. These genes may synergistically act with Tbx1 for the regulation of myogenic differentiation. To fully clarify the biological differences between TA- and SOL-MBs, the Tbx1-related mechanisms should be elucidated in the future.
In conclusion, we revealed that adult myogenic cells are intrinsically committed to specific muscle fibers, and that Tbx1 plays roles in muscle fiber type determination, metabolism, and myogenic cell function. Identification of the regulator of Tbx1 may help clarifying the metabolic system and myogenic differentiation, resulting in the development of novel therapies for muscle atrophy and for a range of muscular diseases.
Methods and materials
Animals
All animal procedures were approved by the Animal Care and Use Committee of the Tokyo Metropolitan Geriatric Hospital and Institute of Gerontology. C57BL/6 mice were purchased from Japan SLC (Hamamatsu, Japan) and NOD/Scid immunodeficient mice were purchased from Charles River. GFP-transgenic mice were kindly provided by Dr. M. Okabe (Osaka University, Osaka, Japan) [59]. All mice were housed and bred in accordance with standard procedures.
Isolation of muscle satellite cells
Mononuclear cells were prepared from C57BL/6 mouse muscles and GFP-transgenic mice (8–12 weeks old), as previously described [60]. Cells were suspended with 2% fetal bovine serum (FBS; Invitrogen, Burlington, CA, USA) in Dulbecco’s modified Eagle’s medium (DMEM; Wako Pure Chemical Industries, Ltd., Osaka, Japan), and stained with anti-CD31-PE antibody (1:200, clone 390; eBioscience, San Diego, CA, USA), anti-CD45-PE (1:200, clone 30-F11; eBioscience), anti-Sca1-PE (1:200, clone D7; eBioscience), anti-Integrin α7 (1:200, clone 3C12; MBL International, Woburn, MA, USA), anti-PE MicroBeads (1:20; Miltenyi Biotec Inc., Bergisch Gladbach, Germany), and anti-mouse IgG MicroBeads (1:20; Miltenyi Biotec Inc.). After staining, cells were separated with Mini & MidiMACS Starting Kit (Miltenyi Biotec Inc.). Isolated cells were cultured in growth medium consisting of DMEM with 20% FBS, 2.5 ng/ml basic fibroblast growth factor (Invitrogen), and penicillin (100 U/ml)–streptomycin (100 mg/ml) on culture dishes coated with Matrigel (BD Biosciences, Franklin Lakes, NJ) at 37 °C. To induce myoblast differentiation and reserve cell formation, the medium was replaced with DMEM supplemented with 5% horse serum (Invitrogen) and penicillin–streptomycin.
RNA extraction and SYBR green-based qPCR
Total RNAs were extracted from myoblasts and myotubes using an RNeasy RNA isolation kit (Qiagen, Hilden, Germany). First-strand cDNA was produced using a Go-script Reverse Transcription Kit (Promega, Madison, WI) and mixed with GoTaq® qPCR Master Mix (Promega). The specific primers for mRNA expression used for PCR are listed in the Supplementary Table 1. The expression levels of mRNA were quantified on an ABI StepOneTM real-time PCR machine (Applied Biosystems, Foster City, CA), following the manufacturer’s instructions. Individual mRNA expression was normalized against the expression of GAPDH. The expression levels of mRNA were quantified by the comparative Ct (ΔΔCt) method [61]. Gene expression profiling was performed using a 3D-Gene Mouse Oligo Chip 24 k (Toray Industries, Kamakura, Japan).
Measurement of OCR and extracellular acidification rate
OCR was measured as previously described [16]. Briefly, cells were plated on XFp cell culture plates (Seahorse Bioscience, North Billerica, MA) and cultured for 1 day, and then induced to undergo differentiation for 6 days. Prior to the assay, the medium in the well was replaced with the appropriate assay medium. The sensor cartridge was calibrated and the cell plate was incubated at 37 °C without CO2 for 1 h. The OCR measurement was performed simultaneously using a Seahorse XFp Extracellular Flux Analyzer (Seahorse Bioscience), following the manufacturer’s instructions. Mitochondrial function was assessed using an XF Cell Mito Stress Test Kit (Seahorse Bioscience), following the manufacturer’s instructions. The following compounds, oligomycin (6.25 μM), FCCP, (1 μM), and rotenone/antimycin A (1 μM), were injected for a mitostress test. After analysis, protein was extracted from the myotubes in each well using cell lysis buffer (1% NP-40 in TBS). The protein content was measured using a PierceTM BCA protein assay kit (Thermo Fisher Scientific, Waltham, MA).
Isolation and culture of single muscle fibers
The EDL, TA, and SOL muscles isolated from mice were digested with 2 mg/ml collagenase type I (Worthington Biochemical), as previously described [11], and the isolated muscle fibers were suspended and cultured in growth medium (DMEM containing 15% horse serum [Invitrogen] with 0.5% chick embryo extract [Gemini Bio-Products, West Sacramento, CA] and penicillin–streptomycin) at 37 °C [23]. EdU (Invitrogen) was added to the culture medium at a final concentration of 10 μM and cultured for 24 h.
Myoblast transplantation and muscle dissection
Twenty-four hours before myoblast transplantation, 50 μl of cardiotoxin (CTX, 10 μM; Sigma-Aldrich, St. Louis, MO) was injected into TA muscles of NOD/Scid mice to induce muscle regeneration. Myoblasts were injected into the regenerating TA muscles. Three weeks after transplantation, the TA muscles were harvested. For re-injury experiments, 3 weeks after myoblast transplantation, 50 μl of CTX was injected into the TA muscles, and the muscles were harvested 2 weeks after the second CTX injection. The dissected muscles were fixed in 4% paraformaldehyde (PFA) for 30 min, immersed in 10% sucrose/phosphate-buffered saline (PBS) and then in 20% sucrose/PBS, and subsequently frozen in cooled 2-methylbutane (Sigma) with liquid nitrogen. The frozen muscles were sliced into 6–8-µm cross and longitudinal sections for immunohistochemistry.
Retrovirus production
A retrovirus-based expression plasmid (pMX) containing GFP was purchased from Cell Biolabs Inc. Tbx1 cDNA (GenScript Corp., Piscataway, NJ) was cloned into pMX vectors. Viral particles were prepared by introducing the pMX vectors into PLAT-E retrovirus packaging cells, and the filtered supernatant was added to the cultured myoblasts
RNA silencing
Tbx1 and control short hairpin RNA (shRNA) lentiviral plasmids were purchased from Sigma. Lentiviral vectors, along with packaging plasmids (MDL/RRE, Rev, and VSV-G), were transfected into HEK293T cells using Lipofectamine 2000 (Invitrogen). Three days after transfection, the viral supernatants were collected, mixed with Lenti-X lentivirus concentrator (Clontech, Mountain View, CA), and incubated overnight at 4 °C. The following day, the virus was concentrated by centrifugation at 1500×g for 60 min at 4 °C. The concentrated viruses were added to the myoblasts in culture to suppress Tbx1 expression. Seventy-two hours after induction, the selection of Tbx1-inhibited myoblasts was performed using puromycin.
Immunohistochemistry and cytochemistry
Cultured cells, fibers, and muscle sections were fixed with acetone or 2% PFA and blocked with 1% BSA in PBS containing 5% goat serum. After blocking, they were stained with the following primary antibodies: anti-Pax7 (R&D Systems, Minneapolis, MN), polyclonal anti-MyoD (Santa Cruz Biotechnology, Santa Cruz, CA), polyclonal anti-Myogenin (Santa Cruz Biotechnology), anti-MyHC (clone: MF20; Developmental Studies Hybridoma Bank, Iowa City, IA), anti-Laminin-α2 (clone 4H8-2; Sigma), anti-GFP (EMD Millipore Corporation, Billerica, MA), anti-MyHC I (clone NOQ7.5.4D; Sigma), and anti-MyHC type IIB (clone BF-F3; Developmental Studies Hybridoma Bank). After staining, they were incubated with a secondary antibody conjugated with Alexa-488, -568, or -647 (Molecular Probes, Waltham, MA). EdU staining was performed using the Click-iT EdU imaging kit (Invitrogen) after primary and secondary staining, in accordance with the manufacturer’s instructions. Nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI).
Statistics
All quantitative data are presented as mean ± SE. The statistical analyses were performed using a two-tailed Student’s t-test to analyze differences between two groups, or a one-way analysis of variance followed by a Tukey’s test for multiple comparisons. A probability of less than 5% (P < 0.05) was considered statistically significant.
Electronic supplementary material
Acknowledgements
This work was supported by a Grant-in-Aid for a Japan Society for the Promotion of Science (JSPS) Fellowship for Research Abroad (NM), JSPS KAKENHI (NM: 15H05364), the Nakatomi Foundation (NM), the Uehara Memorial Foundation (NM), a General Insurance Association of Japan medical research grant on traffic accidents (AU), the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS; AA: R01 AR062142), and the Muscular Dystrophy Association (MDA; AA).
Author contributions
NM designed and performed the experiments, analyzed the data, and wrote the manuscript. AU designed the experiments and edited the manuscript. YS and ST contributed the mouse line. AA, MU, SM, YM, RT, and KS analyzed and interpreted the data.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by E. Baehrecke
Electronic supplementary material
The online version of this article (10.1038/s41418-018-0186-4) contains supplementary material, which is available to authorized users.
References
- 1.Mauro A. Satellite cell of skeletal muscle fibers. J Biophys Biochem Cytol. 1961;9:493–5. doi: 10.1083/jcb.9.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Charge SB, Rudnicki MA. Cellular and molecular regulation of muscle regeneration. Physiol Rev. 2004;84:209–38. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- 3.Collins CA, Olsen I, Zammit PS, Heslop L, Petrie A, Partridge TA, et al. Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell. 2005;122:289–301. doi: 10.1016/j.cell.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 4.Day K, Shefer G, Shearer A, Yablonka-Reuveni Z. The depletion of skeletal muscle satellite cells with age is concomitant with reduced capacity of single progenitors to produce reserve progeny. Dev Biol. 2010;340:330–43. doi: 10.1016/j.ydbio.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verdijk LB, Snijders T, Drost M, Delhaas T, Kadi F, van Loon LJ. Satellite cells in human skeletal muscle; from birth to old age. Age (Dordr) 2013;36:545–7. doi: 10.1007/s11357-013-9583-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Webster C, Blau HM. Accelerated age-related decline in replicative life-span of Duchenne muscular dystrophy myoblasts: implications for cell and gene therapy. Somat Cell Mol Genet. 1990;16:557–65. doi: 10.1007/BF01233096. [DOI] [PubMed] [Google Scholar]
- 7.Shefer G, Van de Mark DP, Richardson JB, Yablonka-Reuveni Z. Satellite-cell pool size does matter: defining the myogenic potency of aging skeletal muscle. Dev Biol. 2006;294:50–66. doi: 10.1016/j.ydbio.2006.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernet JD, Doles JD, Hall JK, Kelly-Tanaka K, Carter TA, Olwin BB. p38 MAPK signaling underlies a cell-autonomous loss of stem cell self-renewal in skeletal muscle of aged mice. Nat Med. 2014;20:265–71. doi: 10.1038/nm.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cosgrove BD, Gilbert PM, Porpiglia E, Mourkioti F, Lee SP, Corbel SY, et al. Rejuvenation of the muscle stem cell population restores strength to injured aged muscles. Nat Med. 2014;20:255–64. doi: 10.1038/nm.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ono Y, Boldrin L, Knopp P, Morgan JE, Zammit PS. Muscle satellite cells are a functionally heterogeneous population in both somite-derived and branchiomeric muscles. Dev Biol. 2010;337:29–41. doi: 10.1016/j.ydbio.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenblatt JD, Lunt AI, Parry DJ, Partridge TA. Culturing satellite cells from living single muscle fiber explants. In Vitro Cell Dev Biol Anim. 1995;31:773–9. doi: 10.1007/BF02634119. [DOI] [PubMed] [Google Scholar]
- 12.Lagord C, Soulet L, Bonavaud S, Bassaglia Y, Rey C, Barlovatz-Meimon G, et al. Differential myogenicity of satellite cells isolated from extensor digitorum longus (EDL) and soleus rat muscles revealed in vitro. Cell Tissue Res. 1998;291:455–68. doi: 10.1007/s004410051015. [DOI] [PubMed] [Google Scholar]
- 13.Hughes SM, Blau HM. Muscle fiber pattern is independent of cell lineage in postnatal rodent development. Cell. 1992;68:659–71. doi: 10.1016/0092-8674(92)90142-Y. [DOI] [PubMed] [Google Scholar]
- 14.Lexell J, Jarvis JC, Currie J, Downham DY, Salmons S. Fibre type composition of rabbit tibialis anterior and extensor digitorum longus muscles. J Anat. 1994;185:95–101. [PMC free article] [PubMed] [Google Scholar]
- 15.Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscles. Physiol Rev. 2011;91:1447–531. doi: 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- 16.Nicholls DG, Darley-Usmar VM, Wu M, Jensen PB, Rogers GW, Ferrick DA. Bioenergetic profile experiment using C2C12 myoblast cells. J Vis Exp. 2010;46:2511. [DOI] [PMC free article] [PubMed]
- 17.Bockman EL, McKenzie JE. Tissue adenosine content in active soleus and gracilis muscles of cats. Am J Physiol. 1983;244:H552–9. doi: 10.1152/ajpheart.1983.244.4.H552. [DOI] [PubMed] [Google Scholar]
- 18.Kushmerick MJ, Meyer RA, Brown TR. Regulation of oxygen consumption in fast- and slow-twitch muscle. Am J Physiol. 1992;263:C598–606. doi: 10.1152/ajpcell.1992.263.3.C598. [DOI] [PubMed] [Google Scholar]
- 19.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–24. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 20.Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 21.Baroffio A, Bochaton-Piallat ML, Gabbiani G, Bader CR. Heterogeneity in the progeny of single human muscle satellite cells. Differentiation. 1995;59:259–68. doi: 10.1046/j.1432-0436.1995.5940259.x. [DOI] [PubMed] [Google Scholar]
- 22.Yoshida N, Yoshida S, Koishi K, Masuda K, Nabeshima Y. Cell heterogeneity upon myogenic differentiation: down-regulation of MyoD and Myf-5 generates ‘reserve cells’. J Cell Sci. 1998;111:769–79. doi: 10.1242/jcs.111.6.769. [DOI] [PubMed] [Google Scholar]
- 23.Zammit PS, Golding JP, Nagata Y, Hudon V, Partridge TA, Beauchamp JR. Muscle satellite cells adopt divergent fates: a mechanism for self-renewal? J Cell Biol. 2004;166:347–57. doi: 10.1083/jcb.200312007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gayraud-Morel B, Chretien F, Jory A, Sambasivan R, Negroni E, Flamant P, et al. Myf5 haploinsufficiency reveals distinct cell fate potentials for adult skeletal muscle stem cells. J Cell Sci. 2012;125:1738–49. doi: 10.1242/jcs.097006. [DOI] [PubMed] [Google Scholar]
- 25.Bassaglia Y, Gautron J. Fast and slow rat muscles degenerate and regenerate differently after whole crush injury. J Muscle Res Cell Motil. 1995;16:420–9. doi: 10.1007/BF00114507. [DOI] [PubMed] [Google Scholar]
- 26.Rosenblatt JD, Parry DJ, Partridge TA. Phenotype of adult mouse muscle myoblasts reflects their fiber type of origin. Differentiation. 1996;60:39–45. doi: 10.1046/j.1432-0436.1996.6010039.x. [DOI] [PubMed] [Google Scholar]
- 27.Salmons S. Exercise, stimulation and type transformation of skeletal muscle. Int J Sports Med. 1994;15:136–41. doi: 10.1055/s-2007-1021035. [DOI] [PubMed] [Google Scholar]
- 28.Gambke B, Lyons GE, Haselgrove J, Kelly AM, Rubinstein NA. Thyroidal and neural control of myosin transitions during development of rat fast and slow muscles. FEBS Lett. 1983;156:335–9. doi: 10.1016/0014-5793(83)80524-9. [DOI] [PubMed] [Google Scholar]
- 29.Larsson L, Biral D, Campione M, Schiaffino S. An age-related type IIB to IIX myosin heavy chain switching in rat skeletal muscle. Acta Physiol Scand. 1993;147:227–34. doi: 10.1111/j.1748-1716.1993.tb09493.x. [DOI] [PubMed] [Google Scholar]
- 30.Holloszy JO, Chen M, Cartee GD, Young JC. Skeletal muscle atrophy in old rats: differential changes in the three fiber types. Mech Ageing Dev. 1991;60:199–213. doi: 10.1016/0047-6374(91)90131-I. [DOI] [PubMed] [Google Scholar]
- 31.Lexell J. Human aging, muscle mass, and fiber type composition. J Gerontol A Biol Sci Med Sci. 1995;50:11–6. doi: 10.1093/gerona/50a.special_issue.11. [DOI] [PubMed] [Google Scholar]
- 32.Ciciliot S, Rossi AC, Dyar KA, Blaauw B, Schiaffino S. Muscle type and fiber type specificity in muscle wasting. Int J Biochem Cell Biol. 2013;45:2191–9. doi: 10.1016/j.biocel.2013.05.016. [DOI] [PubMed] [Google Scholar]
- 33.Launay T, Noirez P, Butler-Browne G, Agbulut O. Expression of slow myosin heavy chain during muscle regeneration is not always dependent on muscle innervation and calcineurin phosphatase activity. Am J Physiol Regul Integr Comp Physiol. 2006;290:R1508–14. doi: 10.1152/ajpregu.00486.2005. [DOI] [PubMed] [Google Scholar]
- 34.Chakkalakal JV, Jones KM, Basson MA, Brack AS. The aged niche disrupts muscle stem cell quiescence. Nature. 2012;490:355–60. doi: 10.1038/nature11438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Motohashi N, Asakura A. Muscle satellite cell heterogeneity and self-renewal. Front Cell Dev Biol. 2014;2:1. doi: 10.3389/fcell.2014.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scambler PJ. The 22q11 deletion syndromes. Hum Mol Genet. 2000;9:2421–6. doi: 10.1093/hmg/9.16.2421. [DOI] [PubMed] [Google Scholar]
- 37.Lindsay EA, Vitelli F, Su H, Morishima M, Huynh T, Pramparo T, et al. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410:97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- 38.Yagi H, Furutani Y, Hamada H, Sasaki T, Asakawa S, Minoshima S, et al. Role of TBX1 in human del22q11.2 syndrome. Lancet. 2003;362:1366–73. doi: 10.1016/S0140-6736(03)14632-6. [DOI] [PubMed] [Google Scholar]
- 39.Chieffo C, Garvey N, Gong W, Roe B, Zhang G, Silver L, et al. Isolation and characterization of a gene from the DiGeorge chromosomal region homologous to the mouse Tbx1 gene. Genomics. 1997;43:267–77. doi: 10.1006/geno.1997.4829. [DOI] [PubMed] [Google Scholar]
- 40.de Wilde J, Hulshof MF, Boekschoten MV, de Groot P, Smit E, Mariman EC. The embryonic genes Dkk3, Hoxd8, Hoxd9 and Tbx1 identify muscle types in a diet-independent and fiber-type unrelated way. BMC Genomics. 2010;11:176. doi: 10.1186/1471-2164-11-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chemello F, Bean C, Cancellara P, Laveder P, Reggiani C, Lanfranchi G. Microgenomic analysis in skeletal muscle: expression signatures of individual fast and slow myofibers. PLoS One. 2011;6:e16807. doi: 10.1371/journal.pone.0016807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu J, Bostrom P, Sparks LM, Ye L, Choi JH, Giang AH, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–76. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee KY, Singh MK, Ussar S, Wetzel P, Hirshman MF, Goodyear LJ, et al. Tbx15 controls skeletal muscle fibre-type determination and muscle metabolism. Nat Commun. 2015;6:8054. doi: 10.1038/ncomms9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–38. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 45.Kim HS, Xiao C, Wang RH, Lahusen T, Xu X, Vassilopoulos A, et al. Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metab. 2010;12:224–36. doi: 10.1016/j.cmet.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baar K, Song Z, Semenkovich CF, Jones TE, Han DH, Nolte LA, et al. Skeletal muscle overexpression of nuclear respiratory factor 1 increases glucose transport capacity. FASEB J. 2003;17:1666–73. doi: 10.1096/fj.03-0049com. [DOI] [PubMed] [Google Scholar]
- 47.Kelly RG, Jerome-Majewska LA, Papaioannou VE. The del22q11.2 candidate gene Tbx1 regulates branchiomeric myogenesis. Hum Mol Genet. 2004;13:2829–40. doi: 10.1093/hmg/ddh304. [DOI] [PubMed] [Google Scholar]
- 48.Jerome LA, Papaioannou VE. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet. 2001;27:286–91. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- 49.Sambasivan R, Gayraud-Morel B, Dumas G, Cimper C, Paisant S, Kelly RG, et al. Distinct regulatory cascades govern extraocular and pharyngeal arch muscle progenitor cell fates. Dev Cell. 2009;16:810–21. doi: 10.1016/j.devcel.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 50.Oustanina S, Hause G, Braun T. Pax7 directs postnatal renewal and propagation of myogenic satellite cells but not their specification. EMBO J. 2004;23:3430–9. doi: 10.1038/sj.emboj.7600346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuang S, Charge SB, Seale P, Huh M, Rudnicki MA. Distinct roles for Pax7 and Pax3 in adult regenerative myogenesis. J Cell Biol. 2006;172:103–13. doi: 10.1083/jcb.200508001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rocheteau P, Gayraud-Morel B, Siegl-Cachedenier I, Blasco MA, Tajbakhsh S. A subpopulation of adult skeletal muscle stem cells retains all template DNA strands after cell division. Cell. 2012;148:112–25. doi: 10.1016/j.cell.2011.11.049. [DOI] [PubMed] [Google Scholar]
- 53.Yamamoto M, Legendre NP, Biswas AA, Lawton A, Yamamoto S, Tajbakhsh S, et al. Loss of MyoD and Myf5 in skeletal muscle stem cells results in altered myogenic programming and failed regeneration. Stem Cell Rep. 2018;10:956–69. doi: 10.1016/j.stemcr.2018.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fulcoli FG, Huynh T, Scambler PJ, Baldini A. Tbx1 regulates the BMP-Smad1 pathway in a transcription independent manner. PLoS One. 2009;4:e6049. doi: 10.1371/journal.pone.0006049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55.Chen L, Fulcoli FG, Tang S, Baldini A. Tbx1 regulates proliferation and differentiation of multipotent heart progenitors. Circ Res. 2009;105:842–51. doi: 10.1161/CIRCRESAHA.109.200295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Bueren KL, Papangeli I, Rochais F, Pearce K, Roberts C, Calmont A, et al. Hes1 expression is reduced in Tbx1 null cells and is required for the development of structures affected in 22q11 deletion syndrome. Dev Biol. 2010;340:369–80. doi: 10.1016/j.ydbio.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pane LS, Zhang Z, Ferrentino R, Huynh T, Cutillo L, Baldini A. Tbx1 is a negative modulator of Mef2c. Hum Mol Genet. 2012;21:2485–96. doi: 10.1093/hmg/dds063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nowotschin S, Liao J, Gage PJ, Epstein JA, Campione M, Morrow BE. Tbx1 affects asymmetric cardiac morphogenesis by regulating Pitx2 in the secondary heart field. Development. 2006;133:1565–73. doi: 10.1242/dev.02309. [DOI] [PubMed] [Google Scholar]
- 59.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. ‘Green mice’ as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–9. doi: 10.1016/S0014-5793(97)00313-X. [DOI] [PubMed] [Google Scholar]
- 60.Motohashi N, Asakura Y, Asakura A. Isolation, culture, and transplantation of muscle satellite cells. J Vis Exp. 2014. https://doi.org/10.3791/50846. [DOI] [PMC free article] [PubMed]
- 61.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.