Abstract
ABIN-1 (encoded by the gene Tnip1) is a ubiquitin-binding protein that can interact with ubiquitin-editing enzyme A20 (encoded by the gene TNFAIP3) to restrain the activation of necroptosis and NF-κB activation. Genetic variants in the genes Tnip1 and TNFAIP3 are both strongly associated with susceptibility to autoimmune chronic inflammatory diseases such as psoriasis vulgaris and systemic lupus erythematosus (SLE) in humans. Here we investigated the mechanism by which ABIN-1 regulated innate immune responses. We show that ABIN-1 heterozygosity sensitizes cells to antiviral response by mediating NF-κB-dependent and RIPK1-independent expression of pattern recognition molecules, including TLR3, RIG-I, and MDA5, in MEFs. Furthermore, we demonstrate that increased interaction of ABIN-1 and A20 with prolonged poly(I:C) stimulation of WT cells leads to A20-dependent reduction of ABIN-1 protein. Finally, we show that ABIN-1 heterozygosity sensitizes innate immune response of Abin-1+/− mice in vivo by promoting the production of proinflammatory cytokines, which can be blocked upon inhibition of RIPK1 kinase. Inhibition of RIPK1 kinase activity in vivo partially reduces the expression of MDA5, RIG-I, and caspase-11 in Abin-1+/− mice but not in WT mice. Thus, we conclude that ABIN-1 is a suppressor of innate immune response and the interaction of ABIN-1 with A20 controls innate immunity response through the NF-κB pathway and in both RIPK1 kinase activity-independent and dependent manner.
Subject terms: Cell biology, Microbiology
Introduction
The gene Tnip1 encodes ABIN-1 (A20 binding and inhibitor of nuclear factor (NF)-κB-1), a ubiquitin-binding protein with a UBAN domain, while the gene TNFAIP3 encodes A20, a ubiquitin-editing enzyme [1, 2]. ABIN-1 is recruited into the tumor necrosis factor receptor 1 (TNFR1) signaling complexes in a linear ubiquitination-dependent manner to promote the recruitment of A20 [3]. Cells deficient for either ABIN-1 or A20 are strongly sensitized to the activation of RIPK1 to promote necroptosis [3, 4]. Suppression of necroptosis by Ripk3 deficiency or inactivation of RIPK1 kinase rescues the embryonic lethality of Abin-1−/− mice. On the other hand, both ABIN-1 and A20 have also been implicated in modulating NF-κB activation [5, 6]. A20 can function as a negative-feedback regulator of NF-κB activation in response to multiple stimuli, including TNF-α, interleukin (IL)-1, Toll-like receptor (TLR) and NLR [NOD (nucleotide-binding oligomerization domain)-like receptor] ligands [7]. ABIN-1 has been proposed to physically link A20 to NF-κB essential modulator (NEMO) and promote A20-mediated de-ubiquitination of NEMO to terminate NF-κB signaling [8]. Thus ABIN-1 and A20 can cooperate to restrain both necroptosis and NF-κB activation.
TLR3, retinoic acid-inducible gene (RIG-I), and melanoma differentiation-associated gene 5 (MDA5) are cellular viral receptors involved in mediating defense response against RNA viruses [9]. TLR3 is mainly located in endosomal membrane of endosome, while RIG-I and MDA5, two RNA helicases with a high degree of functional and structural homology, serve as cytoplasmic double-stranded RNA (dsRNA) receptors that can respond to different types of dsRNAs and RNA viruses [10]. Upon binding to dsRNAs, TLR3 recruits the adaptor TIR-domain-containing adapter-inducing interferon (IFN) that in turn interacts with E3 ubiquitin ligase TRAF3 to activate TBK1 and IKKε. TBK1 and IKKε further phosphorylate and activate the IFN regulatory factor (IRF)-3 and IRF-7 to transactivate the expression of type I IFNs, including IFN-α and IFN-β [11, 12]. In addition, upon binding to dsRNAs in the cytosol, RIG-I and MDA5 can also associate with the IFN promoter stimulated-1 (IPS-1) adaptor (also known as MAVS, VISA, and Cardif), which further recruits and activates TRAF3, TBK1, and IKKε, leading to phosphorylation of IRF-3 and IRF-7 and production of type I IFNs [13].
Caspase-11 is involved in mediating both inflammatory response and cell death [14–16]. Caspase-11 is highly inducible in response to proinflammatory stimuli such as bacterial endotoxin lipopolysaccharide (LPS) and polyinosinic:polycytidylic (poly(I:C)) [17, 18]. The induction of caspase-11 is at least in part mediated by NF-κB pathway [19]. Our previous studies revealed that caspase-11−/− mice are highly resistant to endotoxic shock induced by LPS [14]. Thus the induction of caspase-11 can serve as an excellent biomarker for the activation of innate immunity response.
In this study, we demonstrate that ABIN-1 is an important regulator of innate immunity that controls the expression of intracellular viral RNA receptors. We show that ABIN-1 heterozygosity sensitizes cells to antiviral responses by promoting the expression of key viral pattern recognition molecules including TLR3, RIG-I, and MDA5 in a NF-κB-dependent manner independent of RIPK1 kinase. Furthermore, we show that the increased interaction of ABIN-1 with A20 in viral-infected cells leads to the decline of ABIN-1 protein levels in an A20-dependent manner. Finally, we demonstrate that ABIN-1 heterozygosity sensitizes Abin-1+/− mice to poly(I:C)-induced innate immunity response in vivo. Blocking RIPK1 kinase activity inhibits the systemic production of proinflammatory cytokines and partially reduces the induction of RIG-I and MDA5 in Abin-1+/− mice stimulated by poly(I:C) but has no effect in that of wild-type (WT) mice. Thus we conclude that ABIN-1 heterozygosity promotes innate immune response in both RIPK1-dependent and RIPK1-independent manner.
Results
ABIN-1 deficiency sensitizes cells to innate immune signaling
To simulate the process of viral infection, we transfected synthetic dsRNA poly(I:C) into mouse embryonic fibroblasts (MEFs) to induce innate immune responses. Cytoplasmic poly(I:C) can initiate RIG-I-, MDA5-, and TLR3-mediated antiviral signaling pathways to promote the expression of proinflammatory cytokines [10, 20, 21]. Interestingly, poly(I:C) transfection led to transcriptional induction of IFN-β, IL-6, and TNF-α in Abin-1+/+, Abin-1+/− and Abin-1−/− MEFs, which was inversely related to the levels of ABIN-1 (Fig. 1a–c). Treatment with RIPK1 inhibitor Nec-1s did not block poly(I:C)-induced mRNA transcription of these cytokines (Fig. S1a and b).
Fig. 1.
ABIN-1 deficiency sensitizes MEFs to poly(I:C) transfection-induced innate immune responses. a–c qPCR analysis of IFN-β (a), IL-6 (b), and TNF-α (c) expression upon 2 μg/ml poly(I:C) transfection for 1–3 h in Abin-1+/+, Abin-1+/−, and Abin-1−/− MEFs. d, e ELISA analysis of IFN-β (d) and IL-6 (e) production upon 2 μg/ml poly(I:C) transfection for 4 and 8 h in the presence or absence of 10 μM Nec-1s. f, g qPCR assay of IRF-1 (f) and IRF-7 (g) expression upon 2 μg/ml poly(I:C) transfection for 0–3 h. h Phosphorylation of IRF-3 and IκBα in response to 2 μg/ml poly(I:C) transfection for 0–16 h. i Nec-1s has no effect on poly(I:C) transfection-induced caspase-11 expression. Data are presented as the mean ± S.D. from at least three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001
We also determined the effect of ABIN-1 deficiency on the production of poly(I:C)-induced cytokine by enzyme-linked immunosorbent assay (ELISA). Heterozygosity of ABIN-1 enhanced the expression of IFN-β and IL-6 in cells stimulated by poly(I:C), which was not affected by the addition of RIPK1 inhibitor Nec-1s (Fig. 1d, e). In addition, ABIN-1 deficiency also sensitized MEFs to non-transfected poly(I:C)-induced IL-6 production (Fig. S1c). Thus cytokine production in cells directly stimulated by poly(I:C) is induced in RIPK1-kinase-independent manner.
To explore the mechanism that mediates the increased induction of pro-inflammatory cytokines in ABIN-1-deficient cells in response to poly(I:C), we measured the levels of IRF-1, IRF-2, and IRF-7, the IFN regulatory factors that regulate the production of these cytokines. We found that the levels of IRF-1 and IRF-7, but not IRF-2, were also increased dose-dependently in response to the reduction of ABIN-1 in Abin-1+/− and Abin-1−/− MEFs and the induction was not blocked by addition of Nec-1s (Fig. 1f, g, Fig. S1d and e).
Because the phosphorylation of IRF-3 and IκBα represent critical regulatory steps in antiviral responses, we next investigated whether the phosphorylation of IRF-3 and IκBα were affected by ABIN-1 deficiency. We found that phosphorylation of IRF-3 and IκBα was both activated earlier and stronger in Abin-1+/− and Abin-1−/− MEFs than that of WT MEFs (Fig. 1h). The addition of Nec-1s did not block the phosphorylation of IRF-3 and IκBα (Fig. S1f and g). In addition, the expression of caspase-11 was more strongly induced by poly(I:C) in Abin-1+/− and Abin-1−/− MEFs than that of WT MEFs, which was not affected by the addition of pan caspase inhibitor zVAD.fmk or Nec-1s (Fig. 1i; Fig. S1h). Thus the induction of phosphorylation of IRF-3 and IκBα was not affected by RIPK1 kinase activity.
Thus ABIN-1 deficiency sensitizes cells to the activation of host defense response by increasing the expression and phosphorylation of multiple transcriptional regulators, including IRFs and NF-κB pathways, in RIPK1-kinase-independent manner. We conclude that ABIN-1 is an important suppressor of innate immune response that can control multiple key innate immune signaling pathways.
ABIN-1 deficiency promotes antiviral responses to Sendai virus
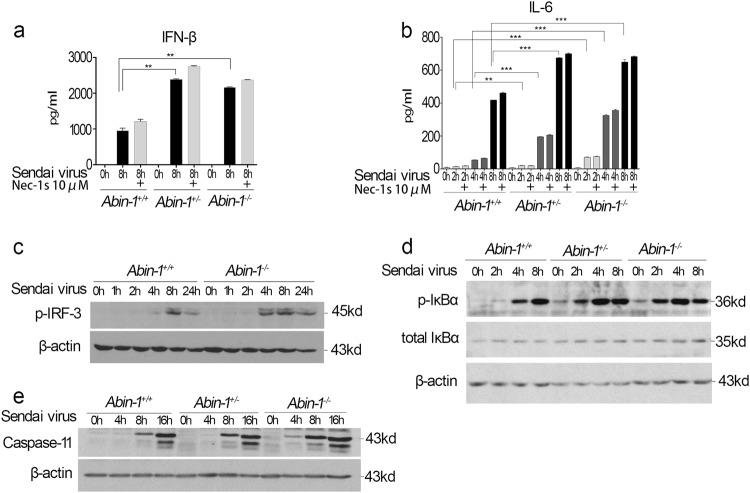
Next, we characterized the role of ABIN-1 in mediating host defense response upon infection with Sendai virus. We found that the production of IFN-β and IL-6 in Abin-1+/− and Abin-1−/− MEFs stimulated by Sendai virus was significantly elevated compared to that of WT MEFs; however, the addition of Nec-1s had no effect on the production of these cytokines (Fig. 2a, b). Consistent with the increased cytokine production, Sendai virus induced more rapid and sustained phosphorylation of IRF-3 and IκBα in ABIN-1-deficient MEFs than in WT MEFs (Fig. 2c, d). Addition of Nec-1s had no effect on phosphorylation of IRF-3 and IκBα (Fig. S2). In addition, Sendai virus also induced higher levels of caspase-11 expression in ABIN-1-deficient cells at earlier time point than that of WT (Fig. 2e). Taken together, these results demonstrate that ABIN-1 deficiency enhances antiviral responses independent of RIPK1 kinase activity.
Fig. 2.
ABIN-1 deficiency increases antiviral responses of MEFs to Sendai virus. a, b ELISA analysis of IFN-β (a) and IL-6 (b) production upon Sendai virus infection at the indicated time points in the presence or absence of 10 μM Nec-1s in Abin-1+/+, Abin-1+/−, and Abin-1−/− MEFs. Sendai virus strain: PI-1. Multiplicity of infection (MOI), 10. c Phosphorylation of IRF-3 upon Sendai virus infection for 0–24 h in Abin-1+/+ and Abin-1−/− MEFs. d Phosphorylation of IκBα upon Sendai virus infection for 0–8 h in MEFs. e Western blot analysis of caspase-11 expression upon Sendai virus infection for 0–16 h. Data are presented as the mean ± S.D. from at least three independent experiments. **P < 0.01, ***P < 0.001
ABIN-1 deficiency promotes the expression of TLR3, RIG-I, and MDA5
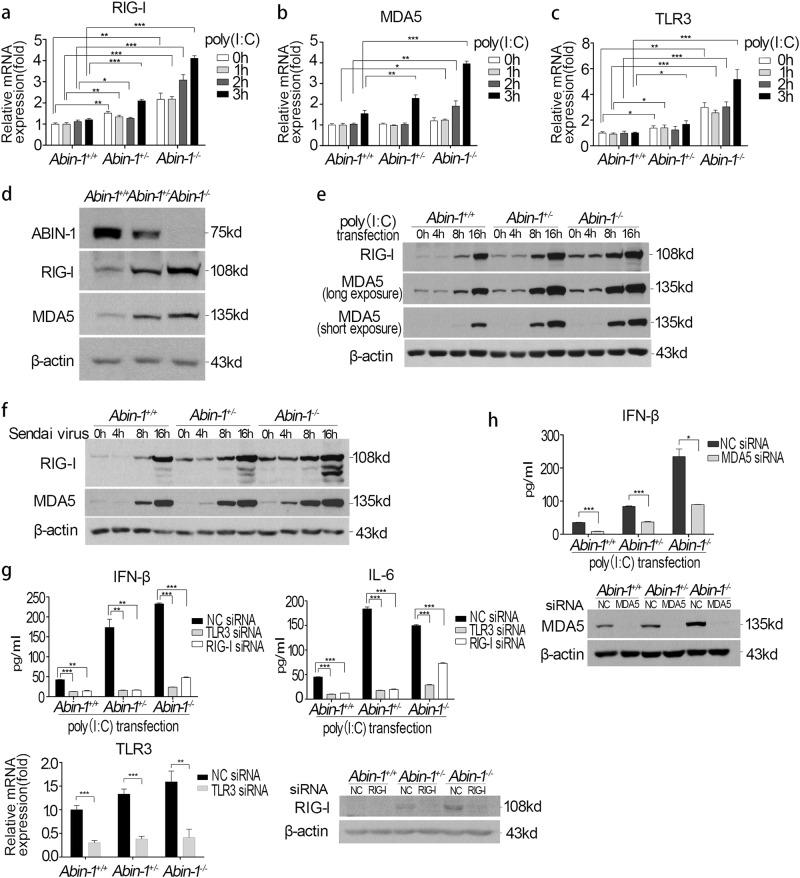
We next characterized the involvement of viral pattern recognition molecules, including TLR3, RIG-I, and MDA5, in the increased sensitivity of ABIN-1-deficient cells to virus. We found that the mRNA levels of RIG-I, MDA5, and TLR3 were increased in an inverse correlation manner to the reduction in ABIN-1 levels in Abin-1+/− and Abin-1−/− MEFs compared to WT MEFs (Fig. 3a–c). On the other hand, the expression levels of TLR7 and IPS-1 were not significantly different among Abin-1+/+, Abin-1+/−, and Abin-1−/− MEFs (Fig. S3a and b). The increased levels of RIG-I and MDA5 proteins were found in un-stimulated, poly(I:C)-transfected and Sendai-virus-infected Abin-1+/− and Abin-1−/− MEFs compared to WT MEFs (Fig. 3d, f). The protein levels of TLR3 were not examined owing to the lack of a specific antibody. Because the levels of K48 ubiquitinated RIG-I and MDA5 showed no difference in Abin-1+/+, Abin-1+/−, and Abin-1−/− MEFs (Fig. S3c), the increased levels of RIG-I and MDA5 in ABIN-1-deficient cells were not likely due to difference in the rate of protein degradation.
Fig. 3.
ABIN-1 deficiency increases basal and poly (I:C)/virus-induced TLR3, RIG-I, and MDA5 expression. a–c qPCR assay of RIG-I (a), MDA5 (b), and TLR3 (c) expression upon 2 μg/ml poly(I:C) transfection for 0–3 h in Abin-1+/+, Abin-1+/−, and Abin-1−/− MEFs. d Western blot analysis of the basal levels of ABIN-1, RIG-I, and MDA5 in Abin-1+/+, Abin-1+/−, and Abin-1−/− MEFs. e Western blot analysis of poly(I:C) transfection-induced RIG-I and MDA5 expression at the indicated time points (0, 4, 8, and 16 h). Poly(I:C), 2 μg/ml. f Western blot analysis of Sendai virus-induced RIG-I and MDA5 expression at different time points (0-16 h). g Effect of TLR3 or RIG-I knockdown on poly(I:C)-induced IFN-β and IL-6 production. MEFs were transfected with 50 nM negative control (NC)/RIG-I/TLR3 siRNA for 48 h and then transfected with 2 μg/ml poly(I:C) for 4.5 h, followed by collection of supernatants for ELISA. The lower two graphs are knockdown efficiency of TLR3 and RIG-I. h Effect of MDA5 knockdown on poly(I:C)-induced IFN-β production. Procedures are the same as described in g. The lower graph is the MDA5 knockdown efficiency. Data are presented as the mean ± S.D. from at least three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001
To explore the role of pattern recognition molecules in the increased sensitivity of ABIN-1-deficient cells to poly(I:C), we used small interfering RNAs (siRNAs) to knockdown TLR3, RIG-I, and MDA5. Similar to WT cells, knockdown of TLR3, RIG-I, and MDA5 blocked the poly(I:C)-transfection-induced IFN-β and IL-6 production in ABIN-1-deficient MEFs (Fig. 3g, h). These results indicate that the increased antiviral responses in ABIN-1-deficient cells are due to elevated transcription of three viral dsRNA receptors, TLR3, RIG-I, and MDA5.
ABIN-1 deficiency promotes NF-κB-mediated transcription of RIG-I and MDA5 genes
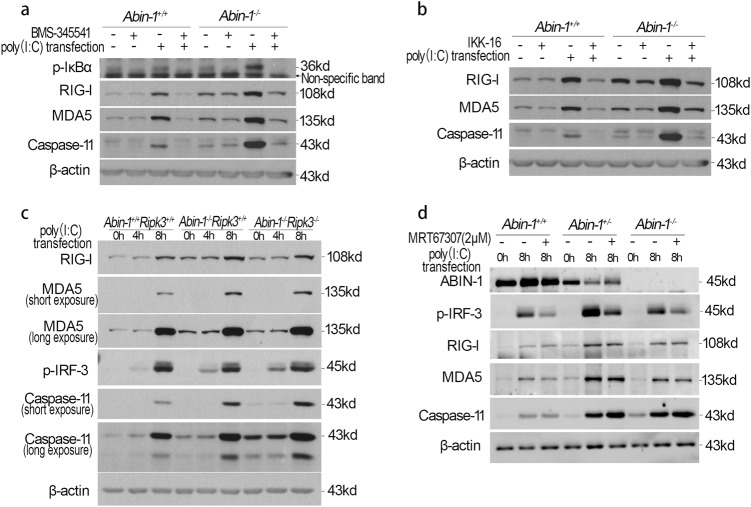
We further investigated the mechanism that promoted the expression of RIG-I and MDA5 in ABIN-1-deficient cells. Consistent with the increased phosphorylation of IκBα in ABIN-1-deficient MEFs (Figs. 1h and 2d), the presence of NF-κB inhibitors BMS-345541 or IKK-16 inhibited the elevated expression of RIG-I, MDA5, and caspase-11 (Fig. 4a, b). However, deletion of Ripk3 had no effect on the increased expression of MDA5, RIG-I, and caspase-11 in ABIN-1-deficient MEFs (Fig. 4c). The addition of TBK1 inhibitor MRT67307 blocked the phosphorylation of IRF3 as expected [22], but had no effect on the expression of MDA5, RIG-I, and caspase-11 in WT or ABIN-1-deficient MEFs (Fig. 4d). These results suggest that increased activation of NF-κB, but not IRF3, is involved in mediating the elevated expression of virus dsRNA receptors RIG-I and MDA5 in ABIN-1-deficient cells.
Fig. 4.
Poly(I:C) transfection-induced RIG-I, MDA-5, and caspase-11 expression in MEFs is NF-κB dependent. a Effect of NF-κB inhibitor BMS-345541 on poly(I:C) transfection-induced RIG-I, MDA-5, and caspase-11 expression at the indicated times in Abin-1+/+ and Abin-1−/− MEFs. BMS-345541, 8 μM, pretreatment for 30 min; poly(I:C), 2 μg/ml. b Effect of NF-κB inhibitor IKK-16 on poly(I:C) transfection-induced RIG-I, MDA-5, and caspase-11 expression at the indicated times. IKK-16, 5 μM, pretreatment for 30 min; poly(I:C), 2 μg/ml. c Effect of Abin-1 knockout and Abin-1/Ripk3 double knockout on RIG-I, MDA-5, p-IRF-3, and caspase-11 expression upon poly(I:C) transfection for 4 and 8 h in primary MEFs. Poly(I:C), 2 μg/ml. d Effect of TBK1 inhibitor MRT67307 on poly(I:C) transfection-induced RIG-I, MDA-5, and caspase-11 expression at the indicated times. MRT67307 (MR), 2 μM; poly(I:C), 2 μg/ml. Results shown are representative of at least three independent experiments
A20-dependent decline of ABIN-1 protein levels with sustained poly(I:C) stimulation
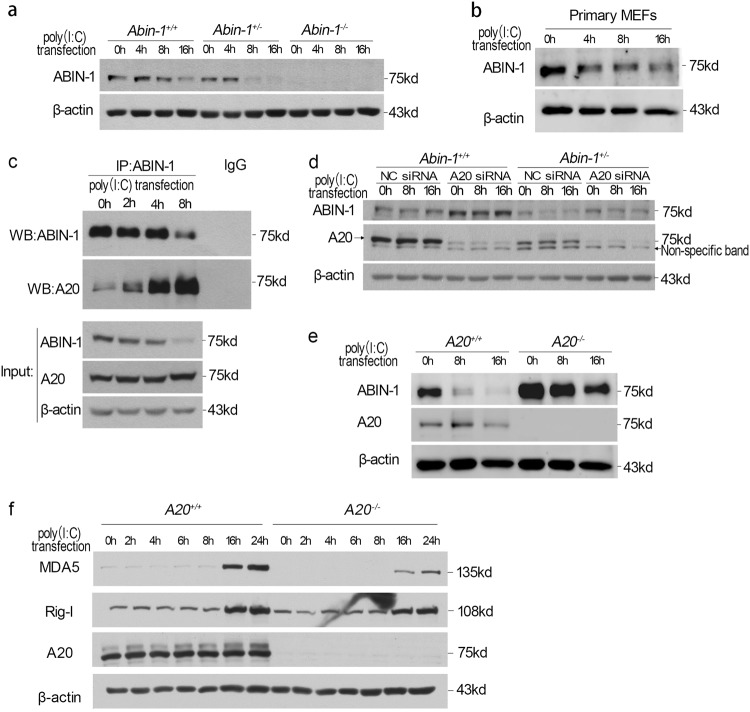
The increased sensitivity of Abin-1+/− MEFs to poly(I:C) led us to explore whether changes in ABIN-1 levels might be involved in mediating dynamics of innate immune response in WT cells. Interestingly, we found that sustained stimulation of poly(I:C) led to the decline of ABIN-1 protein levels in both immortalized WT and Abin-1+/− MEFs (Fig. 5a). Similarly, poly(I:C) transfection also led to the decline of ABIN-1 levels in primary MEFs (Fig. 5b). We found that sustained poly(I:C) stimulation increased the binding of ABIN-1 and A20 (Fig. 5c).
Fig. 5.
Poly(I:C) transfection promotes the decline of ABIN-1 in an A20-dependent manner. a Decline of ABIN-1 protein in response to 2 μg/ml poly(I:C) transfection for 0–16 h in Abin-1+/+ and Abin-1+/− immortalized MEFs. b Decline of ABIN-1 protein in response to 2 μg/ml poly(I:C) transfection for 0–16 h in primary MEFs. c Immunoprecipitation analysis for the interaction between ABIN-1 and A20 upon 2 μg/ml poly (I:C) transfection for 0–8 h in wild-type MEFs. d Effect of A20 knockdown on ABIN-1 levels in Abin-1+/+ and Abin-1+/− MEFs. Cells were transfected with 50 nM control NC siRNA or A20 siRNA for 36 h, followed by transfection with 2 μg/ml poly(I:C) for 8 and 16 h. e Effect of A20 knockout on ABIN-1 level. A20+/+ and A20−/− immortalized MEFs were transfected with 2 μg/ml poly(I:C) for 0–16 h. f RIG-I and MDA5 expression upon 2 μg/ml poly(I:C) transfection for 0–24 h in A20+/+ and A20−/− immortalized MEFs. Results shown are representative of at least three independent experiments
To characterize the role of A20 in mediating the decline of ABIN-1 levels after prolonged innate signaling, we knocked down A20 with siRNAs and observed that ABIN-1 levels were significantly increased in A20-deficient cells (Fig. 5d). The protein levels of ABIN-1 were considerably higher in A20 knockout cells with or without poly(I:C) stimulation (Fig. 5e). On the other hand, the presence of pan caspase inhibitor zVAD.fmk had no effect on the decline of ABIN-1 protein (Fig. S4a), suggesting that caspases are not involved in regulating the ABIN-1 levels. The addition of autophagy inhibitor 3-methyladenine (3-MA) had a partial effect in reducing the loss of ABIN-1 at 8 h time point (Figure S4b). Because A20 can mediate K48-linked polyubiquitination [5] and binds to ABIN-1 avidly, and the levels of A20 inversely correlated with that of ABIN-1, these results suggest that A20 may mediate the degradation of ABIN-1 in viral-infected cells. Consistent with the role of A20 in negatively regulating ABIN-1, the induction of MDA5 and RIG-I by poly(I:C) was reduced in A20−/− cells compared to that of WT (Fig. 5f). Taken together, these results suggest that A20 negatively controls ABIN-1 in modulating innate immune response.
ABIN-1 deficiency enhances antiviral responses in vivo
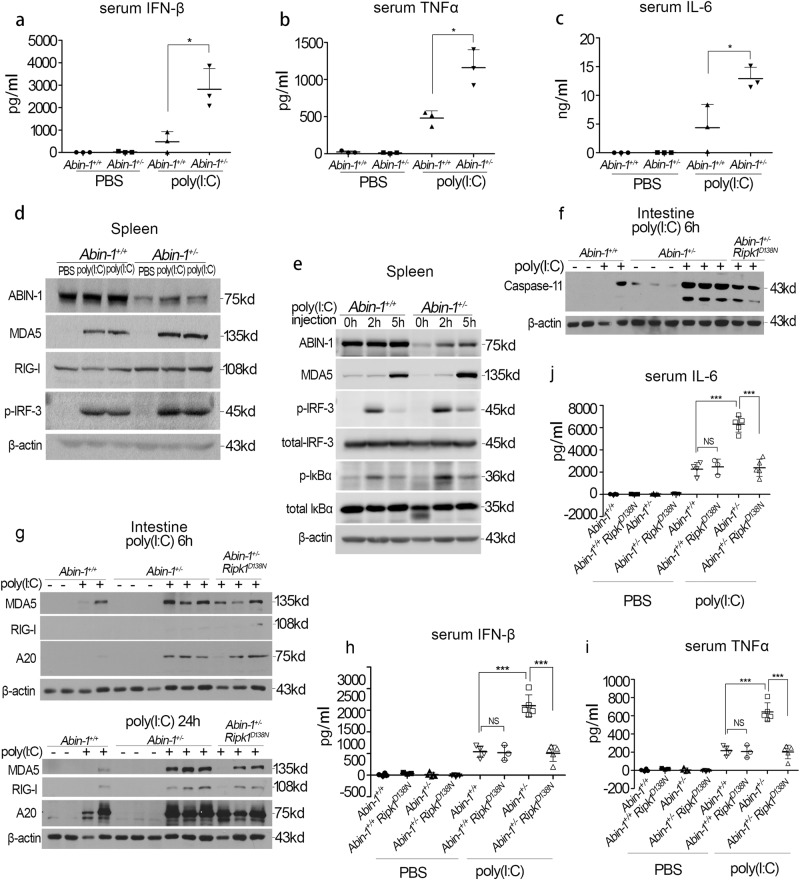
To further characterize the role of ABIN-1 in regulating the antiviral responses in vivo, WT mice and Abin-1+/− mice were injected intraperitoneally with phosphate-buffered saline (PBS) buffer or poly(I:C) and analyzed for their responses. The levels of IFN-β, IL-6, and TNF-α were much higher in the serum of poly(I:C)-injected Abin-1+/− mice compared to that of WT mice (Fig. 6a–c). Consistent with the in vitro results, we observed that increased levels of MDA5 and RIG-I and phosphorylation of IRF-3 and IκBα in the spleens of Abin-1+/− mice compared to that of WT mice (Fig. 6d, e). Furthermore, the levels of MDA5, RIG-I, and caspase-11 were also higher in the small intestine of Abin-1+/− mice compared to that of WT mice (Fig. 6f, g). Blocking RIPK1 kinase activity by Ripk1-D138N mutation in Abin-1+/− mice partially reduced the induction of RIG-I, MDA5, and caspase-11 induced by poly(I:C) (Fig. 6f, g; Fig. S5a-d). Furthermore, inhibition of RIPK1 in Abin-1+/− mice blocked poly(I:C)-induced systemic increases in the serum levels of inflammatory cytokines, including TNF-α, IL-6, and IFN-β (Fig. 6h–j). However, inactivation of RIPK1 kinase in Abin-1+/+ (WT) mice did not significantly affect the levels of poly(I:C)-induced TNF-α, IL-6, and IFN-β secretion or that of MDA5 and RIG-I (Fig. 6h–j; Fig. S5e).
Fig. 6.
ABIN-1 deficiency enhances the poly(I:C)-induced antiviral responses in vivo. a–c ELISA analysis of serum IFN-β (a), TNF-α (b), and IL-6 (c). Abin-1+/+ and Abin-1+/− mice were i.p. injected with 5 mg/kg body weight poly(I:C) and sacrificed 1 h later. n = 3. d RIG-I, MDA5, p-IRF-3, and ABIN-1 expression in the spleen of Abin-1+/+ and Abin-1+/− mice i.p. injected with PBS or poly(I:C) (5 mg/kg body weight). Mice were sacrificed 3.5 h after drugs' injection, and the spleens were homogenized using RIPA buffer and subjected for western blot analysis. e MDA5, p-IRF-3, total IRF-3, p-IκBα, total IκBα, and ABIN-1 expression in the spleen of Abin-1+/+ and Abin-1+/− mice i.p. injected with PBS or poly(I:C) (5 mg/kg body weight). Mice were sacrificed 2 and 5 h after drugs' injection, and the spleens were homogenized using RIPA buffer and subjected for western blot analysis. f Caspase-11 expression in the small intestine of Abin-1+/+, Abin-1+/−, and Abin-1+/− Ripk1D138N mice i.p. injected with PBS or poly(I:C) (5 mg/kg body weight). Mice were sacrificed 6 h after drugs' injection, and the small intestines were homogenized using RIPA buffer and subjected for western blot analysis. g RIG-I, MDA5, and A20 expression in the small intestine of Abin-1+/+, Abin-1+/−, and Abin-1+/− Ripk1D138N mice i.p. injected with PBS or poly(I:C) (5 mg/kg body weight). Mice were sacrificed either 6 or 24 h after drugs' injection, and the small intestines were homogenized using RIPA buffer and subjected for western blot analysis. h–j ELISA assay of serum IFN-β (h), TNF-α (i), and IL-6 (j). Abin-1+/+, Abin-1+/− Ripk1D138N, Abin-1+/−, and Abin-1+/− Ripk1D138N mice were i.p. injected with 5 mg/kg body weight poly(I:C) and sacrificed 6 h later. Data are presented as the mean ± S.D. from at least three independent experiments. *P < 0.05, ***P < 0.001, NS no significant difference
Thus the kinase activity of RIPK1 is not involved in regulating the antiviral response in WT ABIN-1 background; however, increased innate immunity response in ABIN-1 heterozygosity in vivo at least in part involves RIPK1 kinase activity.
Discussion
Genetic variants in the genes Tnip1 and TNFAIP3 in humans are both strongly associated with susceptibility to autoimmune chronic inflammatory diseases, such as psoriasis vulgaris and systemic lupus erythematosus (SLE) [23, 24]. Thus understanding the mechanism by which ABIN-1 and A20 regulates inflammation and cell death may have direct relevance to these important human diseases. Here we show that the interaction of ABIN-1 and A20 provide differential controls on innate responses and inflammation mediated by NF-κB activation and RIPK1 kinase activation. Mimicking viral stimulation by poly(I:C) transfection promotes the interaction of ABIN-1 and A20 and degradation of ABIN-1 in an A20-dependent manner. Thus ABIN-1 is an important “gate-keeper” that modulates this innate immune response. Although both ABIN-1 and A20 have been identified as negative regulators of both NF-κB pathway and RIPK1 kinase activity and ABIN-1 can act to recruit A20 in TNFR1 signaling pathway [3–6], our result suggests that A20 may also provide feedback regulation on ABIN-1, which may provide additional layer of regulation in the control of innate immune response. In addition, since the levels of ABIN-1 may reduce after prolonged innate immune response, heterozygosity of ABIN-1 may mimic cells with prolonged viral infection to lead to sustained antiviral responses that may promote tissue damage.
Regulating the expression of multiple viral pattern recognition molecules including RIG-I, MDA5, and TLR3 as well as phosphorylation of IRF3 and IκBα by ABIN-1 and A20 makes a concerted effort to coordinate important intracellular antiviral responses. Because A20 is a polyubiquitin-editing enzyme that adds K48-linked polyubiquitin and removes K63-linked polyubiquitin to specific proteins [5, 25], our results suggest that ABIN-1 is a substrate of A20 and interaction of ABIN-1 with A20 may promote its degradation. However, because MG-132 and Velcade are both highly toxic to cells in combination of poly(I:C) transfection, we cannot directly examine the effect of proteasome in the degradation of ABIN-1. The ability of A20 deficiency to stabilize the protein levels of ABIN-1 suggests that the proinflammatory phenotype of A20−/− mice can be partially compensated by elevated levels of ABIN-1. While the embryonic lethality of Abin-1−/− mice and proinflammatory phenotype of A20−/− mice can both be inhibited upon inactivation of RIPK1 [3, 26], A20−/− mice can survive significantly longer than that of Abin-1−/− mice. The ability of A20−/− mice to survive past embryonic development may be in part due to the compensatory expression of ABIN-1 in A20-deficient background. Consistent with this possibility, the loss of either A20 or ABIN-1 alone from intestinal epithelial cells (IECs) leads to negligible pathology, whereas double deletion of both A20 and ABIN-1 leads to rapid IEC death and mouse lethality [27]. Inhibition of RIPK1 kinase is sufficient to block the activation of RIPK1-dependent apoptosis and necroptosis in A20−/−; Abin-1−/− cells [3]. Thus ABIN-1 and A20 may cooperate to suppress the activation of RIPK1.
While RIPK1 kinase activity does not directly regulate the antiviral response mediated by ABIN-1/A20, the interaction of ABIN-1 with A20 regulates systemic inflammatory response in RIPK1-dependent manner in ABIN-1-deficient background. Sustained viral infection in ABIN-1 heterozygosity individuals may facilitate the activation of RIPK1 to promote secondary inflammation. Thus the coordinated action of ABIN-1/A20 is important to restrain the activation of RIPK1 and necroptosis. Sustained innate immune response in ABIN-1-deficient individuals may lead to tissue damage and inflammatory diseases mediated by RIPK1 kinase. Interestingly, at least in our experimental paradigm, inhibition of RIPK1 did not change the induction of viral receptors or proinflammatory cytokines in WT background, suggesting the specificity of RIPK1 kinase activation under disease conditions. The contribution of TNF-α pathway to pathogenesis of psoriasis has been well recognized [28]. Our results suggest that pharmacological inhibition of RIPK1 might provide an important mechanism to modulate the TNF-α pathway for the treatment of psoriasis and SLE by blocking systemic inflammation under disease condition.
Materials and methods
Reagents
zVAD.fmk (Z) (Sigma, Cat# V116) was dissolved in dimethyl sulfoxide (DMSO) at a concentration of 20 mM and used at a final concentration of 20 μM. R-7-Cl-O-Nec-1 (Nec-1s) was made by custom synthesis, dissolved in DMSO at a concentration of 10 mM, and used at a final concentration of 10 μM. ELISA kit for mouse IFN-β was from PBL Assay Science (Cat# 42400), and ELISA kits for mouse TNF-α and IL-6 were from BioLegend (Cat# 431304 and Cat# 430904). The following reagents were used: Poly(I:C) (InvivoGen, Cat# tlrl-pic-5), BMS-345541 (Sigma-Aldrich, Cat# B9935), IKK-16 dihydrochloride (Sigma-Aldrich, Cat# SML1138), MRT67307 HCl (Selleckchem, Cat# S7948), and 3-MA (Selleckchem, Cat# S2767).
Animals
Abin-1+/− (Tnip1+/−) mice were kindly provided by Dr. Averil Ma of UCSF. Ripk1D138N mice were kindly provided by Dr. Manolis Pasparakis of U. Cologne, Germany and Dr. Michelle Kelliher of U. Mass. Ripk3−/− mice were kindly provided by Dr. Vishva Dixit of Genentech. In animal studies, female mice (6–10 weeks) were used for poly(I:C) injection. All animals were cultured in a pathogen-free environment, and experiments on mice were conducted according to the protocols approved by the Harvard Medical School Animal Care Committee.
Mouse genotyping
Genotyping used the following primers:
For Abin-1 (5’–3’): ATGGGTGGGTAGGCATAGGGATAG (common), CCTCAAACAGCAGAAGAGGAAAGC (WT), and TTGATTCCCCTTCGCCCATTCCAGC (KO). For Ripk3 (5’–3’): CGCTTTAGAAGCCTTCAGGTTGAC (common), GCAGGCTCTGGTGACAAGATTCATGG (WT), and CCAGAGGCCACTTGTGTAGCG (KO).
For Ripk1D138N (5’–3’):
TACCTTCTAACAAAGCTTTCC (common), AATGGAACCACAGCATTGGC (WT), and CCCTCGAAGAGGTTCACTAG (KD).
Generation and immortalization of MEFs
Male and female mice with different genotypes were crossed and pregnancy was terminated at E11–E13 stage. Embryos were homogenized individually and treated with trypsin/EDTA, sieved through 70-micron filter, and primary MEFs were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 15% fetal bovine serum (FBS) and antibiotics. At passages 4–6, primary MEFs were immortalized by transfection with SV40 small+large T antigen-expressing plasmid (Addgene, Cat# 22298), using Lipofectamine 2000 or Lentivirus system.
Sendai virus infection
The Sendai virus PI-1 strain was obtained from Charles River. Virus infection was carried out as described before [29]. Briefly, the cells were infected with Sendai virus in virus infection medium (DMEM supplemented with 2% FBS) at multiplicity of infection of 10. Cells were incubated with virus for 1 h with gentle agitation every 10 min. After 1 h of incubation, the virus was removed and the cells were washed twice with DMEM. The cells were then placed in complete medium and harvested at the times indicated.
Poly(I:C) transfection
Poly(I:C) was transfected into MEFs using Lipofectamine™ 2000 Transfection Reagent (Cat# 11668019, Thermo Fisher Scientific). Briefly, 2 μg poly(I:C) and 2 μl Lipofectamine™ 2000 were individually diluted in 50 μl Opti-MEM I Reduced Serum Media (Cat# 31985062, Thermo Fisher Scientific). Diluted Poly(I:C) was transferred into diluted Lipofectamine 2000, followed by pipetting and incubation of the mixture for 30 min at room temperature. Transfection complex was added to 1 ml cell culture and the plate was shaken gently for a few seconds.
Cell lines, siRNA, and cell culture
A20−/− MEFs were kindly provided by Dr. Averil Ma of UCSF. Knockdowns were generated using Lipofectamine™ RNAiMAX (Thermo Fisher Scientific, Cat# 13778100) and the following siRNA sequences: TLR3: 5’-AAGGAUGUUUUCGGGCCGCCU-3’, RIG-I: 5’-CCGGACTTCGAACACGTTTAA-3’, MDA5: 5’-GAACGUAGACGACAUAUUA-3’, and A20: 5’-CAAAGCACUUAUUGACAGA-3’. MEFs were cultured in DMEM supplemented with 10% FBS, penicillin, and streptomycin. All cell lines were authenticated by quantitative PCR (qPCR) or western blot assay and tested for mycoplasma contamination.
qPCR analysis and primers
Total RNA was extracted from MEFs using an AxyPrep Multisource Total RNA Miniprep Kit (Axygen, Cat# AP-MN-MS-RNA-250). Reverse transcription was performed with a PrimeScript First Strand cDNA Synthesis Kit (Takara, Cat# 6110A). Real-time PCR primers were designed with the Primer 5.0 software, and the sequences were as follows (5’−3’): IFN-β: GGAAAGATTGACGTGGGAGA (F), CTGAGGCATCAACTGACAGG (R); IL-6: GAGGATACCACTCCCAACAGACC (F), AAGTGCATCATCGTTGTTCATACA (R); IRF-1: AGGCATCCTTGTTGATGTCC (F), AATTCCAACCAAATCCCAGG (R); IRF-7: AGCATTGCTGAGGCTCACTT (F), TGATCCGCATAAGGTGTACG (R); IRF-2: GGCATGGTACCCTCTCAATG (F), TTCAACTGACGGGCTTTCAT (R); RIG-I: CAGACAGATCCGAGACACTA (F), TGCAAGACCTTTGGCCAGTT (R); MDA5: CGATCCGAATGATTGATGCA (F), AGTTGGTCATTGCAACTGCT (R); TLR3: GAAGATGATGCAGTCTTTCCA (F), CCTGTATCATATTCTACTCCTTGC (R); TLR7: AAACCATCGAAACCCAAAGA (F), CCACCAGACCTCTTGATTCC (R); IPS-1: AGGGTGGGATGGACTGAGAT (F), CTAGGGGAGAATGAGGTCGG (R); and β-actin: AGAGGGAAATCGTGCGTGAC (F), CAATAGTGATGACCTGGCCGT (R). qPCR was performed on an Applied Biosystems 7900HT System using SYBR® Green Master Mix reagent (Applied Biosystems, Cat# 4309155).
Antibodies and immunoprecipitation (IP)
The following antibodies were used: ABIN-1 (Ubiquigent, Cat# 68-0001-100), K48 ubiquitin antibody (kindly provided by Genentech), A20 (Cell Signaling, Cat# 5630), RIG-I (Cell Signaling, Cat# 4200), MDA5 (Cell Signaling, Cat# 5321), Phospho-IRF-3 (Cell Signaling, Cat# 4947), IRF-3 (Cell Signaling, Cat# 4302; Proteintech Group, Cat. 11312-1-AP), Phospho-IκBα (Cell Signaling, Cat# 9246), IκBα (Cell Signaling, Cat# 4812), Caspase-11 (Lab-made rat monoclonal antibody) [17], and β-actin (Santa Cruz, Cat# sc-81178). For ABIN-1 IP, cells were harvested in RIPA buffer and incubated overnight with ABIN-1 antibody followed by 2 h incubation with Protein A/G ultra-link resin (Thermo Scientific, Cat# 53133). For K48 ubiquitin IP, cells were lysed with 6 M urea lysis buffer [6 M urea, 20 mM Tris-HCl, pH 7.5, 135 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 1% Triton X-100, Complete protease inhibitor cocktail (Roche), 20 mM NEM, 1 mM phenylmethanesulfonylfluoride, 5 mM NaPyrophoshate, 2 mM Na3VO4, 50 mM NaF, 5 mM Na-glycerophosphate] and centrifuged at full speed for 30 min in 4 C. Supernatants were diluted with lysis buffer (without urea) to bring urea concentration to 3 M, and 2–3 μg chain specific antibody was added and incubated at 4 °C overnight followed by 4 h incubation with Protein A agarose resin (Pierce, Cat# 20333). Beads were washed and proteins were eluted with 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer.
Statistics
Data were expressed as means ± standard deviation (S.D.) or means ± standard error of the mean (S.E.M) and analyzed by two-tailed t tests. Differences were considered statistically significant if *p < 0.05, **p < 0.01, or ***p < 0.001.
Electronic supplementary material
Acknowledgements
This work was supported in part by grants from the NIA (1R01AG047231 and RF1AG055521) (to JY) and R01 AI 106934 (to DMK) and the China National Natural Science Foundation (31670798) (to ZS). We thank Dr. Averil Ma of UCSF for providing Abin-1−/− mice and A20−/− MEFs, Dr. Vishva Dixit of Genentech for Ripk3−/− mice and for ab against K48 ubiquitin chains, Dr. Manolis Pasparakis of U. Cologne, Germany and Dr. Michelle A. Kelliher of U. Mass for providing Ripk1D138N mice. We thank Dr. Roderick Bronson for mouse histopathology analysis. We thank the members of Yuan laboratory for stimulating discussions.
Author contributions
JY conceived the concept, designed the experiments, and wrote the manuscript. ZS designed the experiments, executed majority of the experiments, prepared the figures, and drafted manuscript. SAD, DH, VJB, NB, WL, LQ, NJ, DO, AN, and DMK conducted or designed specific experiments. HZ did genotyping.
Compliance with ethical standards
Conflict of interest
JY is a consultant of Denali Therapeutics. The other authors declare that they have no conflict of interest.
Footnotes
Edited by H. Ichijo
Electronic supplementary material
The online version of this article (10.1038/s41418-018-0215-3) contains supplementary material, which is available to authorized users.
References
- 1.Wagner S, Carpentier I, Rogov V, Kreike M, Ikeda F, Löhr F, et al. Ubiquitin binding mediates the NF-κ B inhibitory potential of ABIN proteins. Oncogene. 2008;27:3739–45. doi: 10.1038/sj.onc.1211042. [DOI] [PubMed] [Google Scholar]
- 2.Opipari A, Boguski M, Dixit V. The A20 cDNA induced by tumor necrosis factor alpha encodes a novel type of zinc finger protein. J Biol Chem. 1990;265:14705–8. [PubMed] [Google Scholar]
- 3.Dziedzic SA, Su Z, Jean Barrett V, Najafov A, Mookhtiar AK, Amin P, et al. ABIN-1 regulates RIPK1 activation by linking Met1 ubiquitylation with Lys63 deubiquitylation in TNF-RSC. Nat Cell Biol. 2018;20:58–68. doi: 10.1038/s41556-017-0003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Onizawa M, Oshima S, Schulze-Topphoff U, Oses-Prieto JA, Lu T, Tavares R, et al. The ubiquitin-modifying enzyme A20 restricts ubiquitination of the kinase RIPK3 and protects cells from necroptosis. Nat Immunol. 2015;16:618–27. doi: 10.1038/ni.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wertz IE, O’rourke KM, Zhou H, Eby M. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature. 2004;430:694–9. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 6.Oshima S, Turer EE, Callahan JA, Chai S, Advincula R, Barrera J, et al. ABIN-1 is a ubiquitin sensor that restricts cell death and sustains embryonic development. Nature. 2009;457:906–9. doi: 10.1038/nature07575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das T, Chen Z, Hendriks RW, Kool M. A20/tumor necrosis factor alpha-induced protein 3 in immune cells controls development of autoinflammation and autoimmunity: lessons from mouse models. Front Immunol. 2018;9:104. doi: 10.3389/fimmu.2018.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mauro C, Pacifico F, Lavorgna A, Mellone S, Iannetti A, Acquaviva R, et al. ABIN-1 binds to NEMO/IKKγ and co-operates with A20 in inhibiting NF-κB. J Biol Chem. 2006;281:18482–8. doi: 10.1074/jbc.M601502200. [DOI] [PubMed] [Google Scholar]
- 9.Slater L, Bartlett NW, Haas JJ, Zhu J, Walton RP, Sykes A, et al. Co-ordinated role of TLR3, RIG-I and MDA5 in the innate response to rhinovirus in bronchial epithelium. PLoS Path. 2010;6:e1001178. doi: 10.1371/journal.ppat.1001178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–5. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 11.Nasirudeen A, Wong HH, Thien P, Xu S, Lam KP, Liu DX. RIG-I, MDA5 and TLR3 synergistically play an important role in restriction of dengue virus infection. PLoS Negl Trop Dis. 2011;5:e926. doi: 10.1371/journal.pntd.0000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honda K, Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol. 2006;6:644–58. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 13.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, et al. IPS-1, an adaptor triggering RIG-I-and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–8. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 14.Wang S, Miura M, Jung YK, Zhu H, Li E, Yuan J. Murine caspase-11, an ICE-interacting protease, is essential for the activation of ICE. Cell. 1998;92:501–9. doi: 10.1016/S0092-8674(00)80943-5. [DOI] [PubMed] [Google Scholar]
- 15.Kang S, Wang S, Kuida K, Yuan J. Distinct downstream pathways of caspase-11 in regulating apoptosis and cytokine maturation during septic shock response. Cell Death Differ. 2002;9:1115–25. doi: 10.1038/sj.cdd.4401087. [DOI] [PubMed] [Google Scholar]
- 16.Yuan J, Najafov A, Py BF. Roles of caspases in necrotic cell death. Cell. 2016;167:1693–704. doi: 10.1016/j.cell.2016.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S, Miura M, Jung Y-k, Zhu H, Gagliardini V, Shi L, et al. Identification and characterization of Ich-3, a member of the interleukin-1β converting enzyme (ICE)/Ced-3 family and an upstream regulator of ICE. J Biol Chem. 1996;271:20580–7. doi: 10.1074/jbc.271.34.20580. [DOI] [PubMed] [Google Scholar]
- 18.Hagar JA, Powell DA, Aachoui Y, Ernst RK, Miao EA. Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science. 2013;341:1250–3. doi: 10.1126/science.1240988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobori M, Yang Z, Gong D, Heissmeyer V, Zhu H, Jung Y, et al. Wedelolactone suppresses LPS-induced caspase-11 expression by directly inhibiting the IKK complex. Cell Death Differ. 2004;11:123–30. doi: 10.1038/sj.cdd.4401325. [DOI] [PubMed] [Google Scholar]
- 20.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–7. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 21.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature. 2001;413:732–8. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 22.Clark K, Peggie M, Plater L, Sorcek RJ, Young ER, Madwed JB, et al. Novel cross-talk within the IKK family controls innate immunity. Biochem J. 2011;434:93–104. doi: 10.1042/BJ20101701. [DOI] [PubMed] [Google Scholar]
- 23.Caster DJ, Korte EA, Nanda SK, McLeish KR, Oliver RK, G’Sell RT, et al. ABIN1 dysfunction as a genetic basis for lupus nephritis. J Am Soc Nephrol. 2013;24:1743–54. doi: 10.1681/ASN.2013020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Musone SL, Taylor KE, Lu TT, Nititham J, Ferreira RC, Ortmann W, et al. Multiple polymorphisms in the TNFAIP3 region are independently associated with systemic lupus erythematosus. Nat Genet. 2008;40:1062–4. doi: 10.1038/ng.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hymowitz SG, Wertz IE. A20: from ubiquitin editing to tumour suppression. Nat Rev Cancer. 2010;10:332–41. doi: 10.1038/nrc2775. [DOI] [PubMed] [Google Scholar]
- 26.Newton K, Dugger D, Maltzman A, Greve J, Hedehus M, Martin-McNulty B, et al. RIPK3 deficiency or catalytically inactive RIPK1 provides greater benefit than MLKL deficiency in mouse models of inflammation and tissue injury. Cell Death Differ. 2016;23:1565–76. doi: 10.1038/cdd.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kattah MG, Shao L, Rosli YY, Shimizu H, Whang MI, Advincula R, et al. A20 and ABIN-1 synergistically preserve intestinal epithelial cell survival. J Exp Med. 2018;215:1839–52. [DOI] [PMC free article] [PubMed]
- 28.Conrad C, Gilliet M. Psoriasis: from pathogenesis to targeted therapies. Clin Rev Allergy Immunol. 2018;54:102–13. doi: 10.1007/s12016-018-8668-1. [DOI] [PubMed] [Google Scholar]
- 29.Peters K, Chattopadhyay S, Sen GC. IRF-3 activation by Sendai virus infection is required for cellular apoptosis and avoidance of persistence. J Virol. 2008;82:3500–8. doi: 10.1128/JVI.02536-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.