Abstract
LC3 is a protein that can associate with autophagosomes, autolysosomes, and phagosomes. Here, we show that LC3 can also redistribute toward the damaged Golgi apparatus where it clusters with SQSTM1/p62 and lysosomes. This organelle-specific relocation, which did not involve the generation of double-membraned autophagosomes, could be observed after Golgi damage was induced by various strategies, namely (i) laser-induced localized cellular damage, (ii) local expression of peroxidase and exposure to peroxide and diaminobenzidine, (iii) treatment with the Golgi-tropic photosensitizer redaporfin and light, (iv) or exposure to the Golgi-tropic anticancer peptidomimetic LTX-401. Mechanistic exploration led to the conclusion that both reactive oxygen species-dependent and -independent Golgi damage induces a similar phenotype that depended on ATG5 yet did not depend on phosphatidylinositol-3-kinase catalytic subunit type 3 and Beclin-1. Interestingly, knockout of ATG5 sensitized cells to Golgi damage-induced cell death, suggesting that the pathway culminating in the relocation of LC3 to the damaged Golgi may have a cytoprotective function.
Subject terms: Autophagy, Lysosomes
Introduction
The subcellular redistribution and lipidation of microtubule-associated proteins 1A/1B light chain 3B (MAP1LC3B, best known as LC3) is generally monitored in the context of autophagy research as an early marker of autophagy induction [1]. Indeed, LC3 is a member of the phylogenetically conserved ATG8 protein family that all redistribute from a usually diffuse cytoplasmic pattern toward so-called autophagic puncta [2] when they associate with the membranes of autophagosomes (which by definition are double-membraned) and autolysosomes (which result from the fusion of autophagosomes and lysosomes, causing the digestion of the inner membrane of the autophagosomes, hence yielding single-membraned vesicles) [3]. The C terminus of newly synthesized LC3 is hydrolyzed by a cysteine protease called ATG4B-exposing Gly120, termed LC3-I. Through a series of ubiquitin-like reactions involving the enzymes ATG7, ATG3 and ATG12-ATG5-ATG16, LC3-I is conjugated to the head group of the lipid phosphatidylethanolamine [4, 5]. This lipidation step, which yields LC3-II, favors the insertion of the C terminus of the protein into membranes. LC3 possesses a domain in its N terminus that interacts with proteins containing a LIR (LC3-interacting region). Hence, LC3 acts as an adaptor that facilitates the recruitment of specific cargo proteins with LIR motifs toward autophagosomes for their selective degradation [6]. One such cargo protein is sequestosome-1 (SQSTM1, generally known as p62) [7].
Autophagy assays often rely on the detection of this lipidation step which increases the electrophoretic mobility of LC3 (meaning that LC3-II migrates more quickly than LC3-I, yielding a band with an apparently lower molecular weight) [1]. Moreover, it is a common strategy to fuse the N terminus of LC3 with green fluorescent protein (GFP) to generate a chimeric protein, GFP-LC3, that can be stably transfected into cells [2, 8]. As the native LC3 protein, GFP-LC3 is mostly contained in the cytoplasm of cells, in a diffuse pattern, and relocates to GFP-LC3-positive puncta when autophagy is induced and/or when the final step of autophagy is blocked [2]. Similarly, autophagic flux can be monitored by assessing the abundance of intracellular LIR motif-containing proteins such as p62 (ref. [9]).
Although GFP-LC3 relocation to puncta is generally monitored in the context of autophagy, there are examples in which GFP-LC3 does not incorporate into double-membraned autophagosomes but into single-membraned structures. This has been documented for the so-called LC3-associated phagocytosis (LAP) in which LC3 incorporates into phagosome membranes [10]. Moreover, this occurs in the context of entosis, the engulfment of cells by others, in which LC3 derived from the cannibal cell associates with the membrane that contains the engulfed cell [11].
Here, we describe yet another case in which GFP-LC3 is recruited to single-membraned vesicles that are associated with, or derived from, the Golgi apparatus. This redistribution of GFP-LC3 occurs in response to a range of different stimuli that damage the Golgi apparatus.
Results and discussion
Direct Golgi damage induces local recruitment of LC3
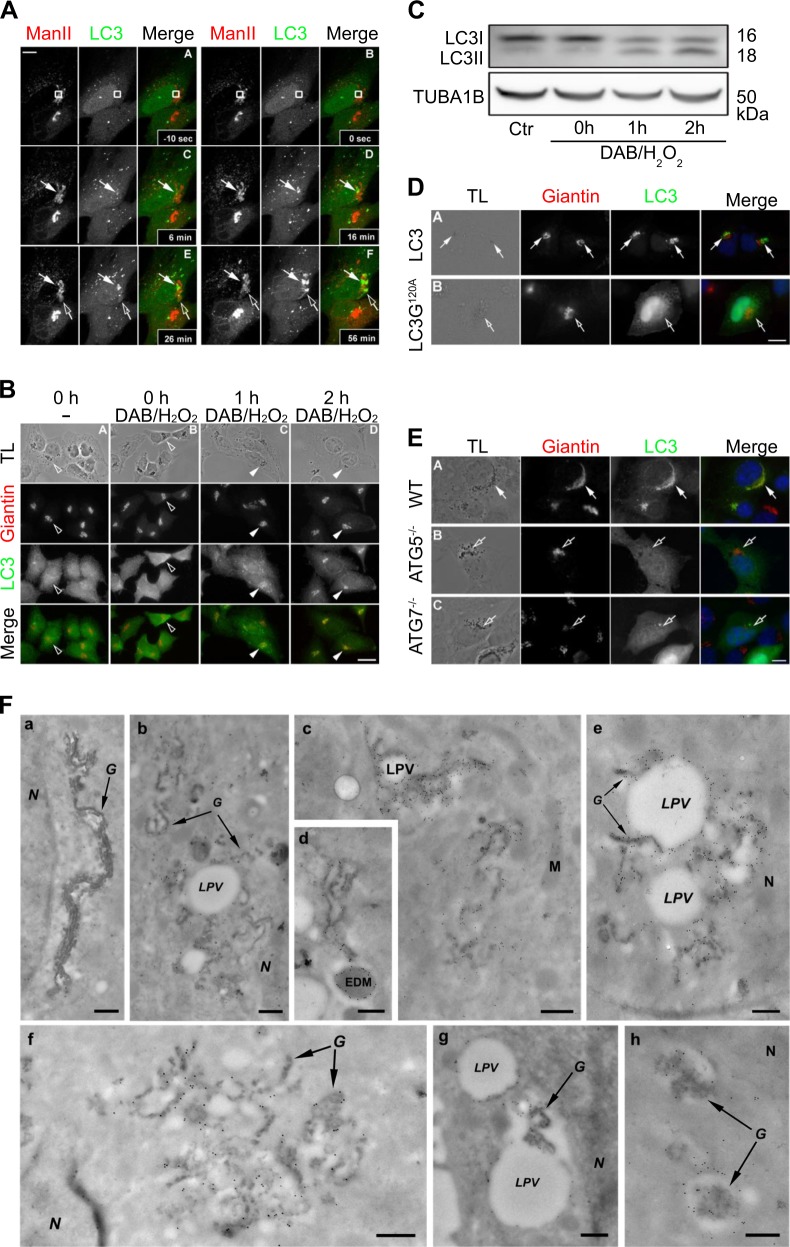
As a first approximation to the question whether local Golgi damage might result into local enrichment of LC3, we took advantage of cervical carcinoma HeLa cells stably transduced with GFP-LC3-expressing lentivirus and transfected with ManII−mCherry [12] to mark the Golgi apparatus. The Golgi complex in these cells was locally damaged by means of a short laser pulse using two-photon laser excitation. Local recruitment of GFP-LC3 to the damaged area of the cell was then observed as early as 6 min post damage. GFP-LC3 recruitment increased over time and, whereas it was only localized on the irradiated area soon after damage, it was later recruited to other areas of the Golgi, as indicated by colocalization of the green (GFP-dependent) and red (mCherry-dependent) fluorescence (Fig. 1a). Next, we damaged the Golgi complex using an independent system. GFP-LC3 was transduced into a cell line expressing the horseradish peroxidase (HRP) in the lumen of the Golgi apparatus through its fusion with ManII (ManII-HRP) [12]. When these ManII-HRP-expressing cells were transiently exposed to low amounts of hydrogen peroxide (H2O2) in the presence of diaminobenzidine (DAB), they locally produced polybenzimidazole in the Golgi, damaging the organelle [13]. Again, we observed that GFP-LC3 translocated to the damaged Golgi, as detected by immunofluorescence staining using an anti-giantin antibody to label the Golgi complex (Fig. 1b). LC3 recruitment was induced by membrane damage and not by perturbation of Golgi activity because if endosomes were crosslinked using a similar approach (endocytosis of HRP followed by DAB-dependent cross-link), damaged endosomes, but not Golgi membranes, were labeled by LC3 (Fig. S1). Recruitment of LC3 to damaged Golgi was accompanied by lipidation of endogenous LC3 protein (as indicated by an increase in the electrophoretic mobility of LC3, leading to the detection of LC3-II) (Fig. 1c). Accordingly, introduction of a mutation into GFP-LC3 (GFP-LC3 G120A) that precludes its lipidation [2] abolished its translocation to the Golgi (Fig. 2d). Moreover, knockout of the essential autophagy genes Atg5 or Atg7, which are known to contribute to LC3 lipidation [14], precluded the recruitment of LC3 to damaged Golgi complex (Fig. 1e). Although LC3 lipidation and its association with specific cellular membranes is typically observed in autophagy [15], transmission electron microscopy coupled to immunogold detection of GFP-LC3 failed to detect double-membraned autophagosomes. Rather, this ultrastructural analysis confirmed the translocation of GFP-LC3 on single membranes that were associated to DAB-positive as well as to electron-lucent large vesicles. (Fig. 1f).
Fig. 1.
LC3 recruitment to wounded or intoxicated Golgi apparatus (GA). a Human cervix carcinoma HeLa cells stably transduced with GFP-LC3 (LC3) were transfected with pManII-mCherry (ManII). Photodamage was applied on a confocal microscope equipped with a 2-Photon Laser. Pictures show cells 10 seconds before photodamage A, after photodamage with the rectangle indicating the photodamaged zone B and representative images of cells at several time points after photodamage C–F. White arrows follow the photodamaged zone showing early LC3 recruitment. Empty arrows indicate appearance of late LC3 recruitment elsewhere in the GA. B HeLa cells stably co-expressing ManII-HRP and GFP-LC3 were fixed directly A or were treated with DAB-H2O2 mix for 30 min at 4 °C then either fixed B or incubated in fresh complete medium for 1 or 2 h before fixation C, D. Cells were subsequently immunostained with anti-Giantin antibodies (Giantin). White arrowheads indicated recruitment of LC3 to the damaged GA. Empty arrowheads indicate absence of LC3 in the Golgi apparatus surroundings. Transmitted light (TL) images were taken to detect the DAB precipitates. Scale bars equal 10 μm. c HeLa ManII-HRP cells without prior treatment or upon treatment with DAB-H2O2 mix for 30 min at 4 °C then were either lysed directly or incubated in fresh complete medium for different time points prior to lysis. Lysates were separated by SDS-PAGE, and then electrotransferred onto a nitrocellulose membrane for immunodetection with anti-LC3 and anti-tubulin antibodies. d HeLa cells expressing ManII-HRP were transiently transfected with GFP-LC3 wt a or GFP-LC3 G120A mutant plasmids b. Cells were treated with DAB-H2O2, fixed 2 h after wash-out, and immunostained with anti-Giantin antibodies. e Wildtype, ATG5−/− or ATG7−/− mouse embryonic fibroblasts (MEFs) were co-transfected with pManII-HRP and pEGFP-LC3 wt plasmids 24 h before the experiment. Cells were treated with DAB-H2O2, fixed 2 h after wash-out and stained with anti-Giantin antibodies. White arrows indicate colocalization of LC3 with Giantin and DAB precipitate. Empty arrows indicate absence of LC3 at the GA surrounding. Transmitted light (TL) images were taken to detect the DAB precipitates. Scale bars equal 10 μm. f HeLa ManII-HRP GFP-LC3 were treated with DAB-H2O2 for 30 min at 4 °C then incubated in fresh complete medium for 0, 2, or 4 h before processing for immuno-EM. GFP-LC3 was detected using antibodies against the GFP tag and secondary antibodies labeled with 10 nm gold particles. Representative images are depicted for 0 a, 2 b–d, and 4 h after DAB-H2O2 treatment e–h. LPV = LC3-positive vacuoles; G = Golgi; N = nucleus; M = mitochondria. Scale bars equal 300 nm
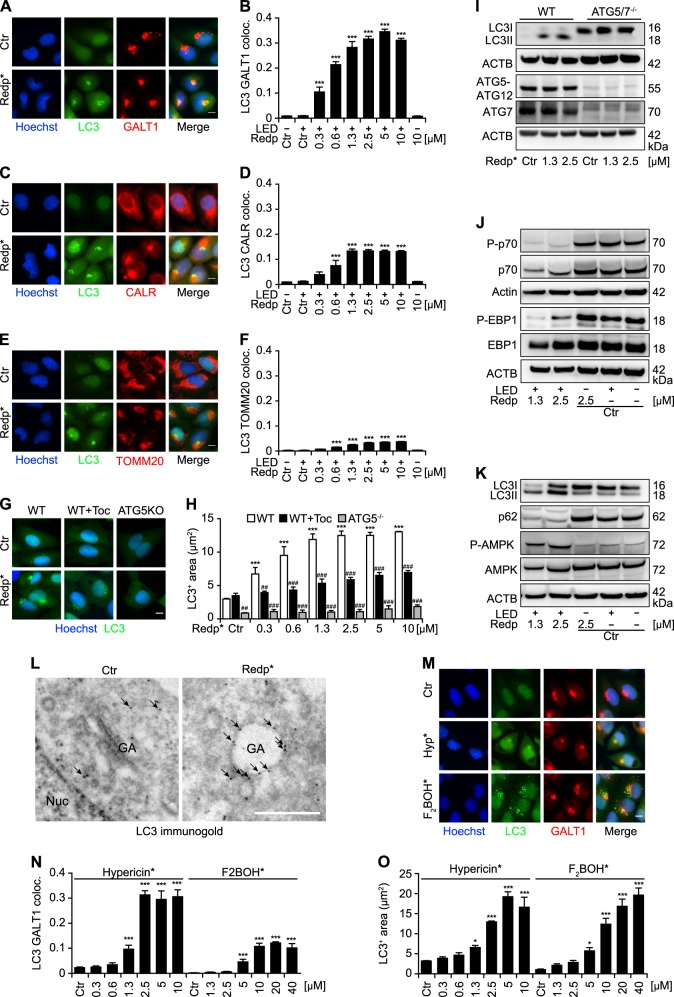
Fig. 2.
Redaporfin-PDT (redp*) induces LC3 aggregation at the Golgi apparatus (GA) and the mTOR-dependent autophagic pathway. a–f Representative images of LC3 aggregation (GFP-LC3+) and its colocalization with GA; ER; or mitochondria are shown in a, b; c, d; and e, f, respectively. Human osteosarcoma U2OS cells stably expressing GFP-LC3 were incubated with redaporfin (redp), at the indicated concentrations, for 20 h followed by irradiation (*) at 750 nm. Six hours later, immunostaining was performed for the GA marker GALT1 a, the ER marker calreticulin c, CALR, and the mitochondrial marker Tomm20 e. Data are presented as means ± SD of triplicates of one representative experiment out of two repeats. (One-way ANOVA, ***p < 0.001 versus untreated cells). Size bar equals 10 µm. g, h Representative images of LC3 aggregation in the presence of the antioxidant tocopherol (Toc) or in ATG5 KO cells. Quantitative analysis represents the average area of GFP-LC3 puncta/cell (µm2). U2OS WT and ATG5−/− cells stably expressing GFP-LC3 were incubated with redp as mentioned above. Toc (500 µM) was added to U2OS cells stably expressing GFP-LC3 4 h before irradiation and cells were fixed 6 h post irradiation. Data are presented as means ± SD of triplicates of one representative experiment out of 2–4 repeats. (Two-way ANOVA, ***p < 0.001 versus untreated cells; ##p < 0.01, ###p < 0.001 versus the presence of Toc or absence of ATG5). Size bar equals 10 µm. i–k Activation of the mTOR signaling pathway and involvement of ATG5/7 proteins in the LC3 lipidation induced by redaporfin-photodynamic therapy (PDT). Mouse embryonic fibroblasts (MEFs) WT or ATG5−/− i, and human osteosarcoma U2OS WT cells (j, k) cells were treated with redp*. Six hours post irradiation, proteins were collected and analyzed by immunoblotting for LC3 lipidation, p62 degradation, AMPK, p70, and EBP1 phosphorylation. Representative immunoblots are depicted. l Representative images of transmission electron microscopy showing vacuolization and LC3 accumulation at the GA upon redp*. Six hours after photoactivation of redp (2.5 µM) in U2OS cells expressing GFP-LC3, GFP-LC3 was detected by immunogold and analyzed by transmission electron microcopy. m–o Hypericin-PDT induces LC3 recruitment into the GA in contrast to F2BOH-mediated PDT. U2OS cells stably expressing GFP-LC3 were incubated with hypericin or F2BOH, at the indicated concentrations, for 20 h, followed by irradiation (*) and immunostaining for GALT1 at 6 h post irradiation. Representative images are shown in m, the average area of GFP-LC3+ dots per cell and the colocalization between GFP-LC3+ dots and GALT1+ structures are shown in n, o. Data indicate means ± SD of triplicates of one representative experiment out of 2–4 repeats. (one-way ANOVA, *p < 0.05, ***p < 0.001 versus untreated cells). Size bar equals 10 µm
To further study LC3 recruitment on damaged Golgi membranes, we decided to use an alternative strategy for Golgi damage that acts at the population level and that can be fine-tuned using photodynamic therapy combining Golgi-targeted photosensitizers and appropriate light sources.
Photodynamic therapy with redaporfin or hypericin induces GFP-LC3 translocation to the Golgi
GFP-LC3-expressing U2OS osteosarcoma cells were exposed to various doses of the photosensitizer redaporfin, which accumulates in the Golgi apparatus and in the endoplasmic reticulum [16], and either left untreated or exposed to infrared light (at 750 nm). The localization of GFP-LC3 was then analyzed in fixed cells counterstained to visualize the Golgi apparatus (Fig. 2a, b), the endoplasmic reticulum (Fig. 2c, d) or mitochondria (Fig. 2e, f). Again, a preferential relocation of GFP-LC3 toward the Golgi was observed (Fig. 2a–f). GFP-LC3 puncta induced by photodynamic therapy (PDT) with redaporfin were not only observed in U2OS cells but also in HeLa cervix carcinoma and H4 neuroblastoma cells (Fig. S2). Induction of general autophagy by the mTORC1 inhibitors rapamycin or torin failed to cause the recruitment of GFP-LC3 to Golgi membranes positive for GALT1 (Fig. S3). The formation of GFP-LC3 puncta induced by PDT required ATG5 and was suppressed by the lipophilic antioxidant tocopherol (Fig. 2g, h). Immunoblot analyses confirmed the lipidation of LC3 induced by PDT and the requirement of Atg5/7 for this lipidation (Fig. 2i). The activating phosphorylation of AMP-dependent kinase (AMPK) was also detected as well as the inhibition of the kinase activity of mechanistic target of rapamycin (MTOR), as suggested by the dephosphorylation of its substrates p70S6K and EBP1 (Fig. 2j, k). As above, immunogold staining of EM preparations confirmed GFP-LC3 localization on single-membrane organelles, without the formation of double-membraned autophagosomes (Fig. 2l). Of note, PDT with hypericin (which also targets the endoplasmic reticulum and Golgi) [17], but not PDT with F2BOH (which targets lysosomes, not the Golgi) [16], also stimulated the relocation of GFP-LC3 to the Golgi (Fig. 2m–o). PDT induced the relocation of endogenous LC3A, LC3B and GABARAP-L1 (but not LC3C and GABARAP) to the Golgi, as determined by immunofluorescence staining (Fig. S4), meaning that several proteins of the ATG8 (LC3/GABARAP) family can translocate to damaged Golgi membranes. The accumulation of GFP-LC3 toward discrete areas of the cells was not inhibited by cycloheximide (although it did reduce the overall abundance of GFP-LC3) (Fig. S5), indicating that pre-existing LC3 can move to the Golgi.
In conclusion, several distinct protocols designed to inflict physical or oxidative damage to the Golgi compartment uniformly induced the recruitment of GFP-LC3 to the GA and activated biochemical changes usually linked to autophagy induction (LC3 lipidation, AMPK activation, MTOR inhibition), yet failed to induce bona fide signs of autophagy, including the formation of double-membraned autophagosomes detectable by transmission electron microscopy.
Golgi recruitment of core components of the autophagic machinery
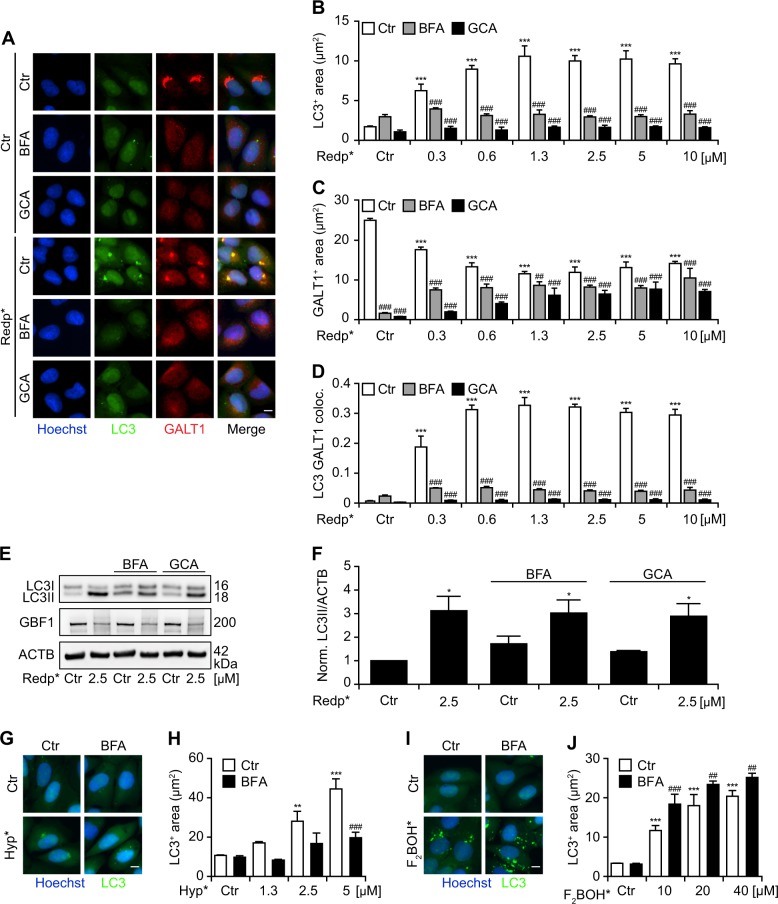
Dispersion of the Golgi apparatus by treatment with brefeldin A (which prevents the association of the COP-I coat to the Golgi membrane) [18] or golgicide A (which inhibits the Golgi brefeldin A resistant guanine nucleotide exchange factor 1, GBF1) [19] largely inhibited the redaporfin-PDT induced accumulation of GFP-LC3 in cytoplasmic puncta, supporting the idea that the Golgi is indeed the source of the primary target for GFP-LC3 relocation upon PDT (Fig. 3a–d). Although GFP-LC3 relocation to puncta was strongly inhibited, the lipidation of endogenous LC3 was only partially inhibited, meaning that the ratio between LC3-II (lipidated) and LC3-I (unlipidated) increased in response to photodynamic treatment with redaporfin even in the presence of brefeldin A and golgicide A (Fig. 3e, f). Of note, two distinct highly potent and specific cell-permeable inhibitors of lysosomal V-ATPases, concanamycin A, and bafilomycin A1, both caused dispersion of the Golgi and also prevented the punctuate redistribution of GFP-LC3 in response to phototoxic damage inflicted by the combination of redaporfin and light (Fig. S6). In agreement with our previous data, brefeldin A prevented LC3 aggregation after PDT of cells treated with hypericin but not with F2BOH (Fig. 3g–j).
Fig. 3.
Requirement of the Golgi apparatus (GA) structure for the aggregation of LC3 in response to Redaporfin-PDT (redp*). a–d Impact of Brefeldin A (BFA) and golgicide (GCA) on the LC3 aggregation and its colocalization with the GA marker, GALT1. Human osteosarcoma U2OS cells expressing GFP-LC3 were incubated with Redp, at the indicated concentrations, for 20 h followed by addition of BFA (5 µg/mL) or GCA (5 µM). Four hours later, cells were irradiated (*) and immunostaining was performed 6 h post irradiation for the GA marker, GALT1. Representative images are shown in a and the quantitative analysis that reflects the average area of GFP-LC3+ dots and GALT1+ Golgi structures per cell are shown in b and c. The level of colocalization (co-ocurrence) between GFP-LC3 dots and GALT1+ structures is depicted in d. (Two-way ANOVA, ***p < 0.001 versus untreated cells; ###p < 0.001 versus the presence of BFA or GCA). Size bar equals 10 µm. e, f Representative immunoblot and densitometry (means ± SEM of three independent experiments) for LC3 lipidation in U2OS cells submitted to redp* in the presence of BFA or GCA. g–j Impact of BFA on the LC3 aggregation triggered by hypericin or F2BOH-mediated PDT. U2OS cells stably expressing GFP-LC3 were incubated with hypericin or F2BOH, at the indicated concentrations for 20 h followed by addition of BFA (5 µg/mL). Four hours later, cells were irradiated and at 6 h post irradiation, cells were fixed with PFA and the nuclei were counterstained with Hoechst 33342. Representative images and the quantitative analysis that reflects the average area of GFP-LC3+ dots are shown for hypericin in g, h and for F2BOH in i, j. (Two-way ANOVA, **p < 0.01, ***p < 0.001 versus untreated cells; ##p < 0.01, ###p < 0.001 versus the presence of BFA or GCA). Size bar equals 10 µm
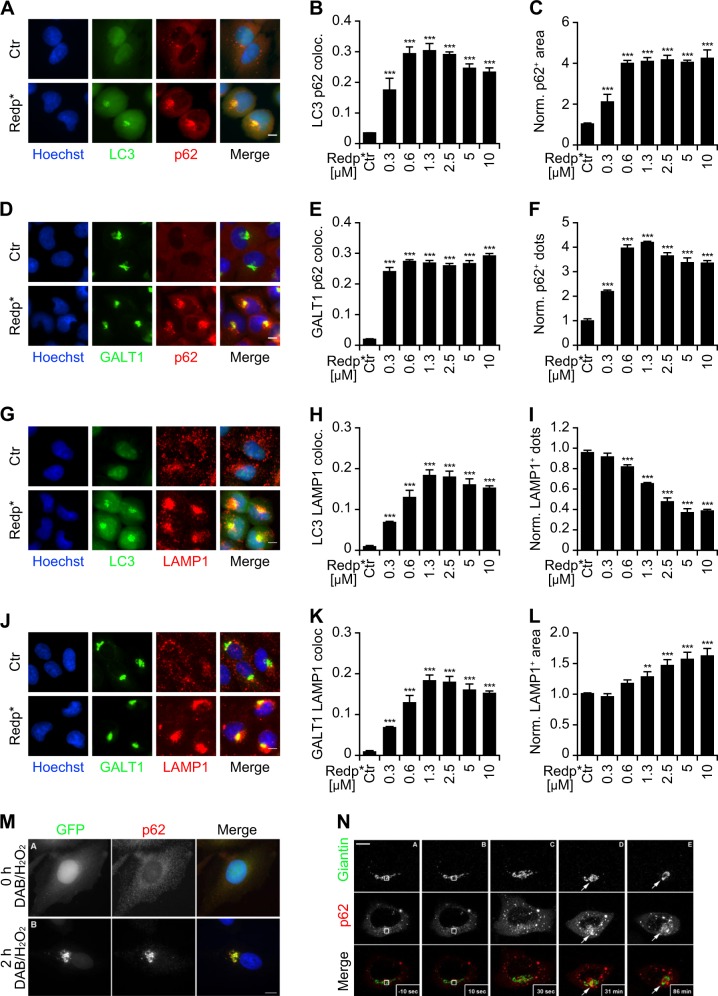
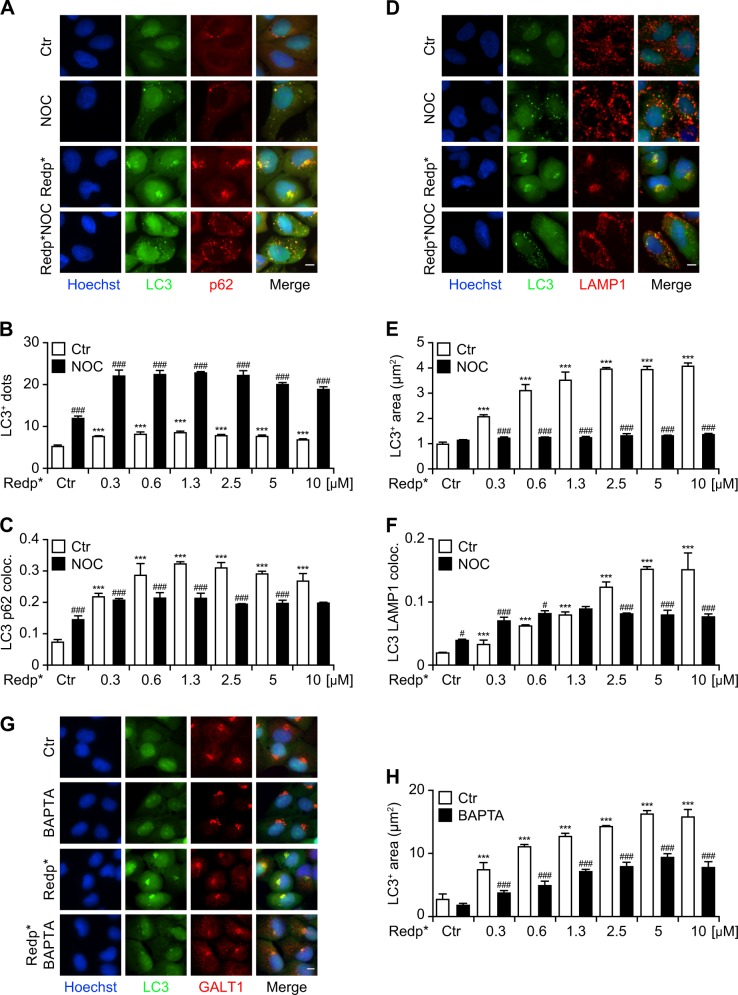
In response to redaporfin-PDT, a Golgi-centric redistribution was also noted for the autophagic adaptor protein sequestosome-1 (SQTM1, best known as p62), which colocalized with both LC3 (Fig. 4a–c) and the Golgi marker GALT1 (Fig. 4d–f). Lysosomes detectable by LAMP1 immunostaining also aggregated close to LC3-positive puncta (Fig. 4g–i) and GALT1 (Fig. 4j–l) after PDT. Expectedly, mTOR was also found on the LC3/GALT1/LAMP1-positive cluster (Fig. S7). Very similar recruitment of p62 to the damaged Golgi was observed in cells expressing Golgi-located peroxidase treated with H2O2 and DAB (Fig. 4m) or in response to localized laser damage (Fig. 4n). The clustering of GFP-LC3, p62 and LAMP1 in the same area of the cytoplasm was prevented by inhibiting microtubule polymerization using nocodazole, which caused dispersion of the Golgi complex and reduced the generation of GFP-LC3 puncta (Fig. 5a–f) Similarly, the relocation of LC3 to sites of the redaporfin-PDT damaged Golgi depends on cytosolic calcium as it was reduced in the presence of the intracellular calcium chelator BAPTA-AM (Fig. 5g, h)
Fig. 4.
Clustering of LC3, Golgi apparatus (GA), lysosomes and p62 after Redaporfin-PDT (redp*) and DAB-H2O2 treatments. a–f Recruitment of p62 at the GA after redp*. Human osteosarcoma U2OS cells expressing GFP-LC3 or the GA marker GALT1-GFP were submitted to Redp-PDT. Six hours later, cells were immunostained for p62. Representative images of p62 aggregation at sites of LC3 aggregation and at GA structures are shown in a and d, respectively, alongside with the level of colocalization in b and e. The average number and area of p62+ structures (normalized to untreated controls) are depicted in c and f. Bars indicate means ± SD of triplicates of one representative experiment out of two repeats. (One-way ANOVA, ***p < 0.001 versus untreated cells). Size bar equals 10 µm. g–l Recruitment of lysosomes at GA after redp*. U2OS cell expressing GFP-LC3 or the GA marker GALT1-GFP were submitted to redaporfin-PDT. Six hours later, cells were immunostained for LAMP1. Representative images of LAMP1 aggregation at sites of LC3 aggregation and at GA structures are shown in g and j alongside with the level of colocalization in h and k. The average number and area of LAMP1+ structures (normalized to untreated controls) are depicted in i and l. Data indicate means ± SD of triplicates of one representative experiment out of two repeats. (One-way ANOVA, **p < 0.1, ***p < 0.001 versus untreated cells). Size bar equals 10 µm. m, n Human cervix carcinoma HeLa cells co-expressing the Golgi-targeted peroxidase ManII-HRP and GFP-LC3 cells were treated with DAB-H2O2 then either fixed or incubated in fresh complete medium for 2 h before fixation. Cells were then immunostained with anti-p62 antibody and analyzed by fluorescence microscopy m. HeLa cells expressing both GFP-Giantin and mCherry-p62 were subjected to Golgi-targeted photodamage performed by 10 seconds of irradiation with a confocal microscope equipped with a 2-photon laser. Representative images are depicted for 10 sec before photodamage A, 10 sec, B 30 sec, C 31 min D, and 86 min E after photodamage. White box indicates the photodamaged zone. White arrows show p62 recruitment to the photodamaged Golgi apparatus. Scale bar equals 10 µm
Fig. 5.
The clustering of LC3, Golgi apparatus (GA), lysosomes, and p62 is dependent on microtubule dynamics. a–f Impact of nocodazole (Noc) on the formation of the LC3, p62, and lysosome clustering after redaporfin-PDT. Human osteosarcoma U2OS cells expressing GFP-LC3 were incubated with redaporfin (Redp), at the indicated concentrations, for 20 h, followed by addition of Noc (2.5 µM). Two hours later, cells were irradiated (*) and immunostaining was performed, 6 h post irradiation, for p62 a or LAMP1 proteins d. Representative images are shown in a for p62 and in d for LAMP1. The quantitative analysis that reflects the number and average area of GFP-LC3+ dots is shown in b, e and the colocalization between GFP-LC3+ puncta and p62+ or LAMP1+ structures is shown in c, f. U2OS cell expressing GFP-LC3 were submitted to redaporfin-PDT in the presence or the absence of the intracellular calcium chelator BAPTA-AM. Following cells were immunostained for GALT1. Representative images of LC3 aggregation at sites of GALT1 stained GA structures are shown in g and quantitively assessed in h. Data are presented as means ± SD of triplicates of one representative experiment out of 2–4 repeats. (Two-way ANOVA, ***p < 0.001 versus untreated cells; #p < 0.05, ###p < 0.001 versus the presence of Noc). Size bar equals 10 µm
Altogether, these data suggest that several different components of the autophagolysosomal system are recruited to the damaged Golgi apparatus.
Non-oxidative Golgi damage can cause LC3 recruitment
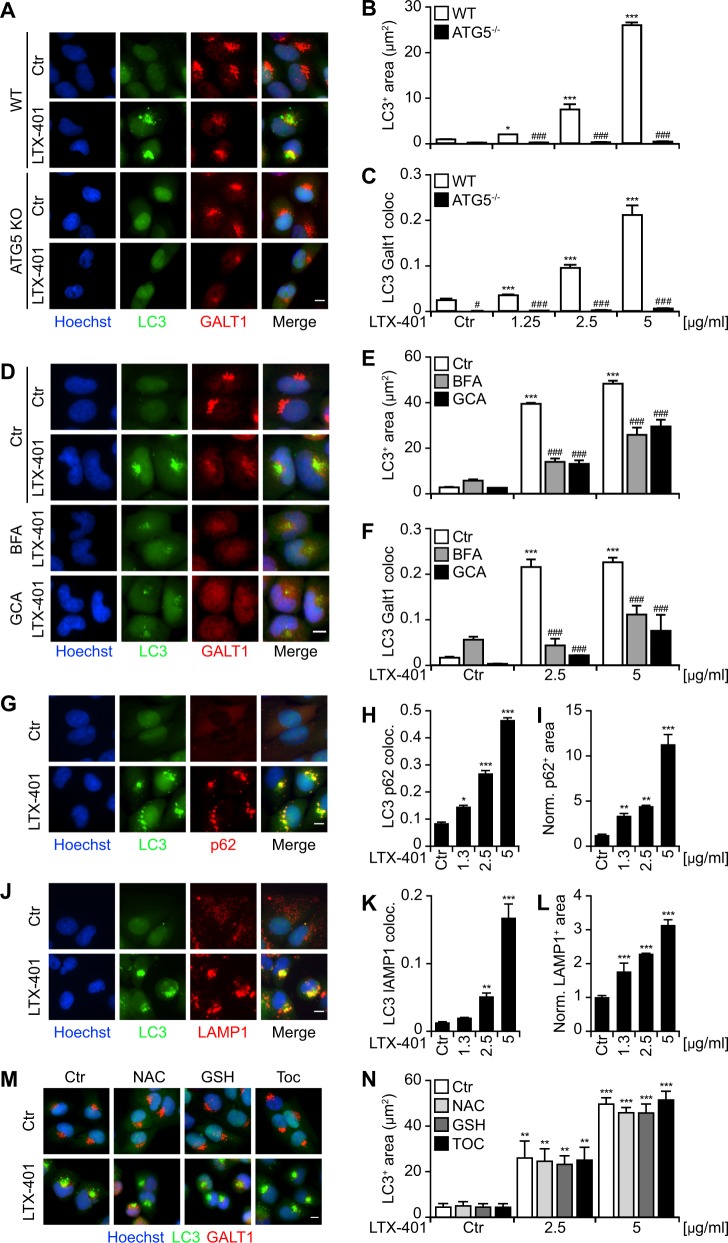
All the methods that were used above to damage Golgi membranes rely on light or peroxidase and may thus generate free radicals. To test whether production of free radicals and/or oxidation was essential to induce the recruitment of the autophagic machinery to damaged Golgi membranes, we explored an independent method to damage the Golgi complex. LTX-401 is an amphipathic peptide derivative that selectively targets the Golgi, disrupting its integrity, thereby triggering a lethal pathway that culminates in caspase-independent cell death [20]. We observed that Golgi damage using LTX-401 led to GFP-LC3 recruitment to this organelle (Fig. 6a). Electron microscopic analysis confirmed the recruitment of GFP-LC3 on Golgi damaged by LTX-401 but, again, no double membrane was detected (Fig. S8). As above, GFP-LC3 recruitment was ATG5-dependent (Fig. 6a–c), GFP-LC3 puncta formation was repressed in the presence of brefeldin A, golgicide A (Fig. 6d–f), concanamycin A or bafilomycin A1 (Fig. S8) and p62 and lysosomes were recruited toward GFP-LC3/GALT1-positive structures (Fig. 6g–l). As observed above, cycloheximide did not prevent recruitment of LC3 on damaged Golgi (Fig. S5). Interestingly, antioxidants such as N-acetylcysteine, reduced glutathione and tocopherol were unable to prevent GFP-LC3 puncta formation in response to LTX-401 (Fig. 6m, n), supporting the notion that this compound mediates direct membrane damage without oxidative stress and that oxidative stress is not necessary to promote LC3 recruitment to damaged Golgi complex.
Fig. 6.
LTX-401, a Golgi apparatus (GA)-targeting oncolytic agent, induces the recruitment of the autophagic machinery to the GA. a–c Treatment with the oncolytic compound, LTX-401 induces LC3 aggregation (GFP-LC3+) at the site of the GA that requires ATG5. WT and ATG5−/− human osteosarcoma U2OS cells expressing GFP-LC3 cells were treated with LTX-401 at different concentrations, for 6 h, in complete medium. The presence of serum partially decreased the cytotoxicity of LTX-401, allowing the study of its effects at the cellular level. Representative images of GALT1 immunostaining are shown in a, whereas the average area of GFP-LC3+ puncta and the level of colocalization between GFP-LC3+ and GALT1+ structures are depicted in b, c. Data indicate means ± SD of triplicates of one representative experiment out of 2–4 repeats. (Two-way ANOVA, *p < 0.5, ***p < 0.001 versus untreated cells; #p < 0.05,###p < 0.001 versus ATG5 knockout). Size bar equals 10 µm. d–f The GA-disrupting agents golgicide A (GCA) and brefeldin A (BFA) inhibited the aggregation of LC3 induced by LTX-401. U2OS cells expressing GFP-LC3 were treated with LTX-401 in the presence of BFA (5 µg/mL) or in the presence of GCA (5 µM) for 6 h followed by GALT1 immunostaining. Representative images are depicted in d and quantitative analysis showing average area of GFP-LC3+ puncta or GALT1+ structures, as well as the level of colocalization between GFP-LC3+ and GALT1+ structures, are presented in e, f. Data are presented as means ± SD of triplicates of one representative experiment out of 2–4 repeats. (Two-way ANOVA, ***p < 0.001 versus untreated cells; ###p < 0.001 versus the presence of BFA or GCA). Size bar equals 10 µm. g–l LTX-401 induces GFP-LC3+ puncta, Golgi apparatus, p62, and lysosomes clustering. U2OS-GFP-LC3 cells were treated with LTX-401 for 6 h followed by immunostaining for p62 g or LAMP1 j proteins. Representative images are shown in g for p62 and in j for LAMP1. The quantitative analysis that reflects the colocalization of GFP-LC3+ puncta with p62+ or LAMP1+ structures are shown in h, k and the changes in the average area of p62+ or LAMP1+ structures after LTX-401 treatment are shown in i, l. Data are represented as means ± SD of triplicates of one representative experiment out of 2–4 repeats. (One-way ANOVA, *p < 0.5, **p < 0.01, ***p < 0.001 versus untreated cells). Size bar equals 10 µm. m, n Impact of different antioxidants on the LC3 aggregation mediated by LTX-401. U2OS cells expressing GFP-LC3 were incubated with the antioxidants n-acetyl-cysteine (NAC, 10 mM), glutathione (GSH, 5 mM), or tocopherol (TOC, 500 µM) for 3 h, followed by co-incubation with LTX-401 in the presence of the same antioxidants. Representative images are shown in m and the quantitative analysis that reflects the average area of GFP-LC3+ dots per cell are shown in n. Data are represented as means ± SD of triplicates of one representative experiment out of 2–4 repeats. (Two-way ANOVA, **p < 0.01, ***p < 0.001 versus ctr cells; ns versus the presence of antioxidants). Size bar equals 10 µm
LC3 recruitment to the Golgi does not involve the canonical autophagic pathway yet might be cytoprotective
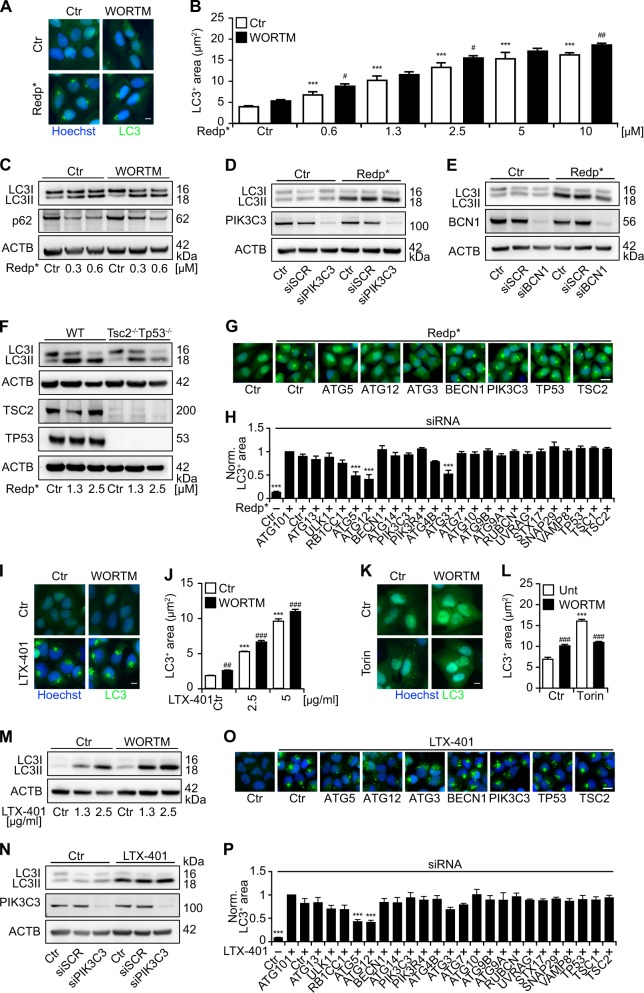
Canonical autophagy relies on the generation of phosphatidylinositol-3-phosphate by phosphatidylinositol-3-kinase catalytic subunit type 3 (PIK3C3, commonly known as VPS34) and the PIK3C3-activatory complex organized around Beclin-1 (BECN1) [21]. Recently, we demonstrated that the redistribution of LC3 to the Golgi complex induced by unsaturated fatty acids (such as oleate) depends on ATG5, yet occurs without, and independently of, PIK3C3 and BECN1 [22]. Driven by these considerations, we determined whether inhibition of PIK3C3 by wortmannin would abolish the translocation of LC3 to the Golgi complex. Wortmannin failed to prevent the Golgi enrichment of GFP-LC3 induced by PDT with redaporfin (Fig. 7a–c) or by treatment with LTX-401 (Fig. 7i, j, m). As a control, we confirmed that Wortmannin used in the same conditions prevented torin-induced GFP-LC3 puncta formation (Fig. 7k, l). Similarly, knockdown of either PIK3C3 or BECN1 failed to inhibit the lipidation of LC3 induced by redaporfin-based PDT (Fig. 7d–e) or LTX-401 (Fig. 7n). Moreover, knockdown of the negative mTORC1 regulators TSC1 or TSC2, as well as the knockout of TSC2, failed to interfere with PDT-induced LC3 lipidation or relocation (Fig. 7f–h). We found that knockdown of ATG3, 5 and 12 could inhibit the redaporfin-induced LC3 relocation (Fig. 7g, h) process, and the knockdown of ATG5 and 12 inhibited the LTX-401-induced LC3 relocation (Fig. 7o, p). In contrast, genes/proteins involved in the ULK1 complex (ULK1, RBCC1, ATG14), in Beclin-1 complexes (BCN1, UVRAG, RUBCN, PIK3C), the ATG9 pathway (ATG9A, AtG9B) and the lysosomal-autophagosome fusion process (STX17, SNAP29, VAMP8) are not involved in this process. Moreover, knockdown of negative regulators of the mTORC1 complex (TSC1, TSC2) is unable to interfere with the relocation of LC3 to the damaged Golgi (Fig. 7g, h, o, p). Altogether, these results support the notion that the Golgi-specific redistribution of LC3 coupled to its lipidation occurs in an ATG5/12-dependent fashion, yet does not require other major autophagy-related protein complexes.
Fig. 7.
LC3 aggregation is independent of PIK3C3 and Beclin-1 but dependent on ATG5, ATG12 and ATG3. a–h Human osteosarcoma U2OS wildtype or stably GFP-LC3 expressing cells were pre-treated with wortmannin (WORTM; 1 µM) or submitted to the RNA interference-mediated silencing of PIK3C3 and BCN1 followed by photodynamic therapy (PDT) with redaporfin (Redp*) a–e LTX-401 i, j, and m, n and torin k, l. Six hours later, LC3 aggregation was evaluated by microscopy in GFP-LC3 expressing cells whereas WT cells were processed for immunoblotting for the assessment of LC3 lipidation. MEFs WT or MEFs DKO for TSC2 and TP53 were also submitted to redp-PDT followed by the assessment of LC3 lipidation by means of immunoblotting f. Alongside, U2OS cells expressing GFP-LC3 cells were submitted to RNA interference-mediated silencing of 24 different autophagy-related genes followed by redp* g, h or LTX-401 o, p treatment. Six hours later, GFP-LC3+ puncta were evaluated by fluorescence microscopy. Representative images of GFP-LC3+ puncta are depicted for redp* a, g; LTX-401 i, o; or torin k and the quantitative analysis that shows the average area of GFP-LC3+ puncta is presented in b, h; j, p; and l, respectively. Representative western blots of LC3 lipidation are depicted in c–f for redp* and in m, n for LTX-401. Data are presented as means ± SEM of at least two independent experiments. (Two-way ANOVA, ***p < 0.001 versus untreated cells; #p < 0.5, ##p < 0.01, ###p < 0.001 versus the presence of wortm or PIK3C3 or BCN1 silencing. For h, p, two-way ANOVA, ***p < 0.001 versus treated cells in the presence of the siCTR). Size bar equals 10 µm
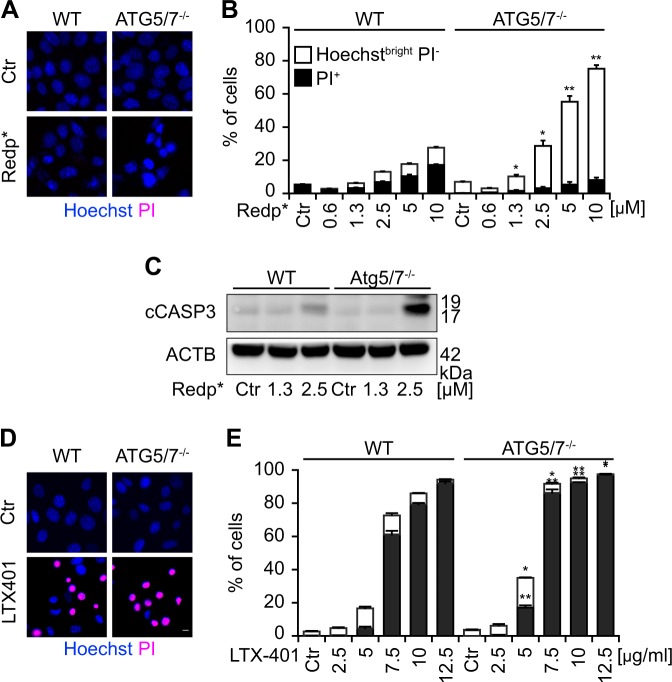
Autophagy is mostly considered as a cytoprotective process that reduces the propensity of cell to succumb to stress [23]. Although Golgi recruitment of LC3 cannot be considered as a canonical autophagy-like event (because it fails to yield double-membraned vesicles and because it does not rely on PIK3C3 and BECN1, see above), we asked whether the pathway involved in the recruitment of LC3 to the Golgi would be cytoprotective. For this, we took advantage of MEF cells lacking Atg5/7 that do not allow the translocation of LC3 to the Golgi (see above). As compared with WT cells, the knockout of Atg5/7 sensitized the cells to PDT- and LTX-401-induced cell death (measured by assessment of Hoechst 33342-detectable pyknosis, propidium iodide-detectable plasma membrane permeabilization) and caspase-3 activation (measured by immunoblot) (Fig. 8a–e). Similarly, GFP-LC3-expressing U2OS cells carrying a knockout of ATG5 (which then fail to recruit GFP-LC3 to the Golgi, see above Fig. 6a–c) became more sensitive to PDT-induced loss of clonogenicity and cleavage of poly-(adenosyl ribose) polymerase-1 (Fig. S9A–C), as well as to LTX-401-mediated killing (Fig. S9D, E). These results suggest that the non-canonical pathway leading to the redistribution of LC3 to the Golgi compartment might have a cytoprotective function.
Fig. 8.
ATG5/ATG7 knockout (KO) sensitizes cells to redaporfin-PDT (redp*) and oncolysis by LTX-401. a–e Mouse embryonic fibroblasts (MEFs) wildtype (WT) as well as KO for Atg5/7 (Atg5/7−/−) were treated with redp* or with the oncolytic compound LTX-401 at the indicated concentrations. Six hours later, cell viability was evaluated by double-staining with PI and Hoechst 33342. Representative images are presented in a, d and the corresponding quantitative results regarding dying cancer cells (Hoechst bright and PI−) and dead cells (PI+ cells) in b, e. c Cleaved caspase-3 (cCASP3) was evaluated by immunoblotting at 6 h post irradiation in MEFs WT as well as in MEFs KO for Atg5/7 (Atg5/7−/−). Data are presented as means ± SD of triplicates of one representative experiment out of three repeats. (*p < 0.5, **p < 0.01 versus untreated cells). Size bar equals 10 µm
Concluding remarks
In this work, we present evidence that damage of the Golgi complex using various independent approaches lead to the translocation of LC3 to the organelle. This was observed upon physical damage induced by localized laser irradiation (in the absence of sensitizing chemical agents), by local luminal precipitation of DAB driven by a Golgi-localized peroxidase or by photodynamic therapy with redaporfin or hypericin. Although these damaging strategies may all be related to local oxidation, non-oxidative damage by another Golgi-targeted agent, LTX-401 [20], as well as unsaturated cis-fatty acids [22], also stimulated the redistribution of LC3 to this organelle together with the LC3 interactor p62. This phenomenon was accompanied by the local accumulation of lysosomes at or close to Golgi markers.
Importantly, Golgi-directed relocation of LC3 occurred in the absence of bona fide signs of autophagy and in particular the absence of double-membraned autophagosomes and a lack of implication of the PIK3C3/BECN1 complex. Similar observations were made before for oleate-induced LC3 translocation to the Golgi [22] and are here confirmed for mechanical, oxidative or LTX-401-mediated Golgi damage. The relocation of LC3 to single-membraned organelles of the endolysosomal system, has been described before, following a range of macroendocytic engulfment events such as entosis (live-cell cannibalism) [11] and LAP, during which lipidated LC3 is recruited to phagosomes housing microbial pathogens or dead cells, respectively [10]. However, to the best of our knowledge, the autophagy-independent recruitment of LC3 to a specific intracellular organelle in response to different damages has not been reported before. There is an ample literature showing that mitochondrial damage leading to the permeabilization of the inner membrane (with consequent dissipation of the mitochondrial transmembrane potential) or permeabilization of the outer membrane (with exposure of proteins such as prohibitin-2) leads to the recruitment of LC3 and mitophagy, i.e., the selective removal of damaged mitochondria by autophagy [24, 25]. Similarly, it has been reported that oxidative damage of the endoplasmic reticulum leads to the recruitment of LC3 and ER-specific autophagy, reticulophagy [26]. However, these instance of damage-induced organelle-specific autophagy involve the formation of double-membraned autophagosomes, contrasting with the single-membrane association of LC3 observed after Golgi damage.
An intriguing observation about the LC3 translocation to damaged Golgi complex is its independence from PIK3C3/BECN1. On the one hand, the phenomenon of organelle-specific LC3 recruitment strongly correlates with LC3 lipidation (and is suppressed when LC3 cannot be lipidated due the G120A mutation) and actually depends on ATG5, an enzyme that are involved in an ubiquitinylation-like conjugation system required for LC3 lipidation [4]. On the other hand, knockdown of PIK3C3 or BECN1, as well as enzymatic inhibition PIK3C3 fails to interfere with the Golgi damage induced local LC3 recruitment. Similarly, knockout of proteins involved in the ULK1 complex failed to interfere with the Golgi recruitment of LC3. This applies to all the Golgi-damaging agents characterized in this respect including reactive oxygen species (ROS)-dependent damage by PDT and ROS-independent stimuli such as LTX-401. We previously reported that the Golgi recruitment of LC3 following exposure to cis-unsaturated fatty acids occurred in an ATG5/7-dependent but PIK3C3/BECN1 and ULK1 independent fashion [22]. Thus, translocation of LC3 to the Golgi appears to be generally independent of PIK3C3/BECN1 and ULK1.
Even though Golgi recruitment of LC3 appears to occur without the formation of autophagosomes and without the involvement of PIK3C3/BECN1 and ULK1, this pathway is potentially cytoprotective. Indeed, knockout of ATG5 (alone or together with ATG7), that prevented LC3 recruitment, sensitized mouse or human cells to killing by redaporfin-mediated PDT or LTX-401. At this point, it is unclear through which precise mechanism, ATG5 and LC3 may contribute to cytoprotection. Prior work suggested that lysosomes contribute to the repair of plasma membranes [27]. It is tempting to speculate, yet remains to be investigated in further detail that the recruitment of molecules from the autophagolysosomal system to the damaged Golgi may help avoiding cell death as well.
Material and methods
Cell culture and reagents
Human osteosarcoma U2OS wildtype, or expressing GALT1-GFP, GFP-LC3 or knockout (KO) for ATG5, human cervix carcinoma HeLa cells expressing GFP-LC3 or HRP-ManII, human neuroglioma H4 cells expressing GFP-LC3, MEFs wildtype or double KO for ATG5 and 7 cells were cultured in Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Gibco® Thermo Fisher Scientific), 1% nonessential amino acids (Thermo Fisher Scientific) and 1% penicillin/streptomycin (Thermo Fisher Scientific) in a humidified incubator with 5% CO2 at 37 °C. The medium for HeLa ManII-HRP and HeLa GFP-LC3 cells was further supplemented with 0.2 mg/ml geneticin (Thermo Fisher Scientific). DAB-H2O2-based organelle cross-linking was performed as described previously (Brachet et al., 1999; Muller et al., 1983; Jollivet et al., 2007). Redaporfin (LUZ11) and F2BOH (LUZ10) were provided by Luzitin SA (Coimbra, Portugal) and LTX-401 was from Lytix Biopharma (Tromsø, Norway). Brefeldin A (B6542), Nocodazole (SML1665), Wortmannin (W3144), DAB (D5905), H2O2 (H1009), Tocopherol (T3251), n-acetyl-cysteine (NAC; A9165), glutathion (GSH; G6013), and ATG5 (A2859) and ATG7 (A2856) antibodies were purchased from Sigma-Aldrich (St Louis, MO, USA). Golgicide A (3584), bafilomycin A1 (1334), and concanamycin A (2656/10U) were from Tocris Bioscience (Bristol, UK). Primary antibodies against CALR (ab ab2907), TOMM20 (ab78547), GBF1 (ab 86071), actin-HRP (ab 49900), LAMP1 (ab25630), LC3A (ab62720) and GABARAP-L1 (ab86497) came from Abcam (Cambridge, UK). Antibodies for LC3 (2775), P-AMPK (2535), AMPK (2603), P-p70 (9205), p70(2708), P-4EBP1 (sc18091), 4EBP1 (sc9577), mTOR (2983), cleaved caspase-3 (9661), and GABARAP (9137330) were purchased from Cell Signaling Technology (Danvers, MA, USA), the antibody against p62 (H00008878-M01) and B4GALT1 (PAB20512) were from Abnova (Taipei, Taiwan) and the antibody for LC3C was purchased from Thermo Fisher Scientific, Carlsbad, CA, USA). The recombinant antibodies anti-Giantin (TA10) and anti-alpha tubulin (F2C) were previously described (Nizak et al., 2003; Moutel et al., 2009). We used here the hFc forms of these antibodies. Anti-GM130 antibody was received from Transduction Laboratories (Becton Dickinson, Francklin Lakes, NJ, USA). Mouse anti-GFP antibody was purchased from Roche Diagnostics (Roche Diagnostics, Risch-Rotkreuz, Switzerland). Secondary AlexaFluor®488 or 647-labeled antibodies and Hoechst 33342 were from Thermo Fisher Scientific. Lipofectamine 2000 reagent (Thermo Fisher Scientific) was used to transiently transfect HeLa cells. Lipofectamine LTX and Plus Reagent (Thermo Fisher Scientific) were used to transfect MEFs. Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific) was used to transfect cells with siRNA.
Cross-linking reaction
Organelle cross-linking was performed as previously described [13]. Cells expressing HRP in Golgi cisternae, or pulsed with soluble-HRP to target endosomes, were washed in phosphate-buffered saline (PBS) and incubated with DAB (Sigma-Aldrich) in the presence of 0.003% H2O2 (Sigma-Aldrich) for 30 min at 0 °C. Cells were then washed and fixed or further incubated in normal medium for imaging, fixation or lysis.
Plasmids and lentiviruses
The pManII-HRP plasmid was previously described [13]. pEGFP-Giantin was constructed by introducing the Giantin coding sequence amplified by PCR into a pEGFP vector (TaKaRa Bio, Mountain view, CA, USA).
Retroviral infections and generation of stable cell lines
HeLa ManII-HRP cells were generated as described previously [13]. GFP-LC3 expressing cell lines were generated using a lentiviral expression system. pExp-Dh1-GFP-LC3-Ef1-Puro, psPAX2, and pVSV-G vectors were used to transfect HEK 293 T cells to generate recombinant lentiviruses. HeLa cells were infected with recombinant lentiviruses and selected by FACS for medium expression levels. All infections were performed in a security level 3 facility.
Photodamage and time-lapse imaging
Cells were plated in a glass-bottom dish and maintained under standard culture conditions followed by photodamage and associated time-lapse acquisitions performed with a Zeiss LSM 710 NLO (Jena, Germany) equipped with a Mai-Tai HP two-photon laser (Spectra Physics, Santa Clara, CA, USA) for microablation. The acquisition was performed with a 63 ×/1.4 NA objective. This microscope was steered with ZEN (Zeiss) and resulting images were processed with ZEN, Metamorph (Molecular Devices, Sunnyvale, CA, USA), Image J (freely available at https://imagej.nih.gov/ij) and Adobe Photoshop (Adobe, San Jose, CA, USA). Spinning-disc confocal microscopy was carried out with a Yokogawa CSU-22 spinning-disc head (Tokyo, Japan) on a Nikon TE-2000U microscope (Tokyo, Japan) equipped with a Coolsnap HQ2 camera, a NanoScanZ piezo focusing stage (Prior Scientific, Cambridge, UK) a motorized scanning stage (Märzhäuser, Wetzlar, Germany) and a × 63/1.4 NA objective, operated with Metamorph. Images were processed with Metamorph.
Treatment with photodynamic therapy
Cells were seeded in black 96-well plates (Greiner Bio-One, Kremsmünster, Austria). After 24 h, photosensitizers (redaporfin, F2BOH or hypericin) were added to cells and incubated for 20 h. After washing cells to remove non-internalized photozensitizer, photoactivation of redaporfin, and F2BOH was carried out at 750 nm with a light-emitting diode (LED) from Marubeni (model L740-66-60-550, Tokyo, Japan). A light dose (LD) of 0.075 J/cm2 or 0.6 J/cm2 was delivered respectively to redaporfin and F2BOH, with the exception of viability studies in which a LD of 0.2 J/cm2 was applied. Hypericin photoactivation was carried out with a GU10 LED, 1.5 W 220–240 V, covered with a filter that results in a peak at 583 nm. A LD of 0.3 J/cm2 was delivered. Untreated cells, cells incubated with the photosensitizer (at the highest tested concentration) without photoactivation and cells submitted to light (in the absence of the photosensitizer) were included in all experiments as negative controls. All inhibitors such as tocopherol (Toc, 500 µM), brefeldin A (BFA, 5 µg/mL), golgicide A (GCA, 5 µM), were added to the cells 4 h before irradiation and then let present until the end of the assay.
Immunofluorescence staining
Six hours after PDT treatment, cells were fixed in paraformaldehyde 4 % (w/v) in PBS containing 2 µM Hoechst 33342, for 45 min at room temperature (RT). Cells were then permeabilized using 0.1% Triton X-100 (Sigma-Aldrich) in PBS for 5 min at RT. For LC3B and GABARAP staining, cells were fixed by means of ice-cold 100% methanol for 15 min at − 20 °C followed by three washing steps with PBS and permeabilization with 0.3% Triton X-100. Unspecific binding was reduced by incubating cells with 2.5% bovine serum albumin (BSA) in PBS for 30 min at RT followed by incubation with the primary antibody, overnight at 4 °C. Afterwards, cells were washed thrice with PBS and further incubated with AlexaFluor®-coupled secondary antibody (AF488 or AF647) for 1.5 h at RT. After washing three times with PBS, images were acquired using an ImageXpress micro XL automated bioimager (Molecular Devices) equipped with a PlanApo 20X/0.75 NA objective (Nikon). A minimum of four images was taken per well. Alternatively, cells were grown on coverslips, fixed, and stained as described above. Coverslips were mounted in Mowiol (Merck Millipore, Darmstadt, Germany) supplemented with 4′,6-diamidino-2-phenylindole to stain DNA and then imaged using a Leica DMRA (Leica, Wetzlar, Germany) equipped with a CoolSnap HQ camera (Photometrics) and equipped with 40 ×/1.25 NA, 63 ×/1.4 NA and 100 ×/1.4 NA objectives, a Leica DM6000 B equipped with a CoolSnap HQ2 camera and equipped with 40 ×/1.25 NA, 63 ×/1.4 NA and 100 ×/1.4 NA objectives, or a Nikon Eclipse 90i equipped with a CoolSnap HQ2 camera and a 100 ×/1.4 NA objective. All microscopes were operated with Metamorph and images were processed with Metamorph and Adobe Photoshop 2.0.0.975.
Western blot analysis
Cells were harvested and the obtained pellet was resuspended in radioimmunoprecipitation buffer (89900, Thermo Fisher Scientific) supplemented with phosphatase and protease inhibitors (88669, Thermo Fisher Scientific). Protein concentration was measured by DCTM Protein Assay kit (5000112, BioRad, Hercules, CA, USA) and 20 µg of protein was mixed with Nupage®LDS sample buffer 4 × and Nupage® sample reducing agent 10 × (NP0008 and NP 0009, Life Technologies) followed by denaturation at 100 °C for 10 min. Proteins were then separated by polyacrylamide gel electrophoresis using 4–12% Bis-Tris pre-casted gels (Life Technologies) in MES running buffer (Life Technologies) and then transferred to polyvinylidene difluoride membrane (Merck Millipore, Darmstadt, Germany) using a Tris-glycine buffer (Life Technologies) with 20% of ethanol. In order to reduce unspecific binding, membranes were incubated for 1 h with a mixture of Tris-buffered saline (Life Technologies) and Tween 20 (0.1%) (Sigma-Aldrich) supplemented with 5% of BSA (Euromedex, Souffelweyersheim, France). Following membranes were incubated with the primary antibody overnight at 4 °C followed and with the appropriate secondary antibody conjugated to horseradish peroxidase (Southern Biotech, Birmingham, AL, USA) for 1 h at room temperature. After washing, proteins were revealed with ECL (GE Healthcare Life Sciences, Little Chalfont, UK). Chemiluminescence images were acquired with an ImageQuant LAS 4000 imaging system (GE Healthcare Life Science). Beta-actin was used for loading control.
Immunogold labeling and transmission electron microscopy
Cells were seeded in petri dishes (Greiner Bio-One) and let to adapt for 24 h. Following the cells were treated with redaporfin (2.5 µM) plus photoactivation or LTX-401 (5 µg/ml). Six hours later, cells were fixed in 4% formaldehyde at 4 °C for 1 h. Cell pellets were then dehydrated in methanol and embedded in lowicryl K4M at − 20 °C in an AFS2 Freeze Substitution Processor apparatus (Leica Microsystems). Polymerization under UV light was carried out for 2 days at − 20 °C, followed by 2 days at 20 °C. Ultrathin sections were blocked and incubated with primary antibody for GFP (Santa Cruz, sc-9996 or Abcam, ab290), for 1 h at room temperature, and then with secondary antibodies conjugated to 10- or 15-nm gold particles (BBI International, Cardiff, UK). Images were acquired with a Tecnai 12 electron microscope (FEI, Eindhoven, the Netherlands). Alternatively, cells were fixed with 2% paraformaldehyde and 0.2% glutaraldehyde in 0.1 M sodium phosphate buffer, pH 7.4. After washing in PBS/Glycin 0.02 M, cells were pelleted by centrifugation, embedded in 12% gelatin, cooled in ice and cut into 5 mm3 blocks. The blocks were infused overnight with 2.3 M sucrose at 4 °C, frozen in liquid nitrogen and stored until cryo-ultramicrotomy. Sections of 70 nm were cut with a diamond knife (Diatome) at − 112 °C using a Leica EM-UC7. Ultrathin sections were picked up in a mix of 1.8% methyl cellulose and 2.3 m sucrose (1:1) according to Liou et al. [28], and transferred to formvar carbon-coated copper grids. Double immunolabeling was performed using optimal combination of gold particle sizes and sequence of antibodies. Cryosections were incubated with rabbit polyclonal antibodies (anti-GFP antibody, Thermo Fisher Scientific) followed by protein A gold. A rabbit anti–mouse immunoglobulin antibody was used as a bridging antibody when monoclonal antibodies were used (anti-GM130 antibody, Becton dickinson). After labeling, the sections were treated with 1% glutaraldehyde, counterstained with uranylacetate, and embedded in methyl cellulose uranylacetate [29]. Grids were observed on a Tecnai G2 spirit (FEI) equipped with a 4-k Quemesa CCD camera (Olympus, Tokyo, Japan).
RNA interference
SMARTpool siGENOME siRNAs for different autophagy-related genes were obtained from GE Dharmacon (Chicago, IL, USA). Transfection was conducted in 96-well plates using the DharmaFECTTM transfection reagent and a final siRNA concentration of 25 nM following the manufacture’s protocol. Then cells were subjected to the standard protocol of redaporfin-PDT or treatment with LTX-401. Six hours after irradiation or incubation with LTX-401, cells were fixed and images were acquired as previously described.
Clonogenic assay
U2OS wildtype (WT) and U2OS ATG5−/− stably expressing GFP-LC3 cells were submitted to redaporfin-PDT or LTX-401 treatment in 96-well plates as previously described. Then, cells were detached using 100 µl of trypsin and 5 µl (~ 1000 cells) of each condition were transferred to six-well plates. Colonies growing from dilutions of cells were fixed after 7 days in normal culture conditions, by means of crystal violet staining solution according to standard procedures before enumeration of colonies using the freely available software Image J (https://imagej.nih.gov/ij/).
Image and statistical analyses
Image segmentation and analysis were performed with the MetaXpress® software (Molecular Devices). Co-occurrence analysis was determined by comparing the common area between the two target fluorophores with the area likely to be occupied by both fluorophores. Dead cells and debris were excluded from the analysis by using R software. Graphs were built with GraphPad Prism and data are presented as mean ± SD. Statistical analyses were performed using one- or two-way ANOVA with GraphPad Prism.
Electronic supplementary material
Acknowledgements
The plasmid pEGFP-LC3 wt was a gift from Dr Noboru Mizushima (The Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan). The pGalT-EGFP plasmid was a kind gift from J. White, EMBL, Heidelberg, Germany. The pDest-mCherry-p62 plasmid was a kind gift from Terje Johansen. The pExp-Dh1-GFP-LC3-Ef1-Puro for lentiviral production was a kind gift of François-Xavier Gobert and Philippe Benaroch. Luzitin SA provided redaporfin and F2BOH. Sabrina Forveille for her help on assessing colonies numbers. LCGdS is supported by Portuguese Science Foundation (ref. SFRH/BPD/93562/2013). GK is supported by the Ligue contre le Cancer (équipe labelisée); Agence National de la Recherche (ANR)—Projets blancs; ANR under the frame of E-Rare-2, the ERA-Net for Research on Rare Diseases; Association pour la recherche sur le cancer (ARC); Cancéropôle Ile-de-France; Institut National du Cancer (INCa); Institut Universitaire de France; Fondation pour la Recherche Médicale (FDM20140630126 and FDM 40739); the European Commission (ArtForce); the European Research Council (ERC); the LeDucq Foundation; the LabEx Immuno-Oncology; the RHU Torino Lumière, the SIRIC Stratified Oncology Cell DNA Repair and Tumor Immune Elimination (SOCRATE); the SIRIC Cancer Research and Personalized Medicine (CARPEM); and the Paris Alliance of Cancer Research Institutes (PACRI). FP is supported by the Centre National de la Recherche Scientifique (CNRS), the Institut Curie and the LabEx CelTisPhyBio (ANR-10-LBX-0038 part of the IDEX PSL no ANR-10-IDEX-0001–02).
Author contribution
L.C.G.d.S. performed the experiments with redaporfin-PDT and LTX-401 and A.J.J. performed the experiments of Golgi damage using laser photodamage and HRP-ManII-expressing cells. A.S. developed R scripts used for analysis. S.S., S.D., and M.S. performed electron microscopy, Ø.R. and B.S. provided LTX-401. L.A., O.K., G.K., and F.P. designed the study. G.K. and F.P. wrote the paper.
Compliance with ethical standards
Disclosure
BS and ØR are full-time employees of Lytix Biopharma and this study was partially funded by Lytix Biopharma.
Footnotes
Edited by H.-U. Simon
Contributor Information
Oliver Kepp, Email: captain.olsen@gmail.com.
Guido Kroemer, Email: kroemer@orange.fr.
Franck Perez, Email: franck.perez@curie.fr.
Electronic supplementary material
The online version of this article (10.1038/s41418-018-0221-5) contains supplementary material, which is available to authorized users.
References
- 1.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marino G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol. 2014;15:81–94. doi: 10.1038/nrm3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–2100. doi: 10.1091/mbc.e07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol. 2004;36:2503–18. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birgisdottir AB, Lamark T, Johansen T. The LIR motif - crucial for selective autophagy. J Cell Sci. 2013;126(Pt 15):3237–47. doi: 10.1242/jcs.126128. [DOI] [PubMed] [Google Scholar]
- 7.Ichimura Y, Kumanomidou T, Sou YS, Mizushima T, Ezaki J, Ueno T, et al. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem. 2008;283:22847–57. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- 8.Kepp O, Galluzzi L, Lipinski M, Yuan J, Kroemer G. Cell death assays for drug discovery. Nat Rev Drug Discov. 2011;10:221–37. doi: 10.1038/nrd3373. [DOI] [PubMed] [Google Scholar]
- 9.BenYounes A, Tajeddine N, Tailler M, Malik SA, Shen S, Metivier D, et al. A fluorescence-microscopic and cytofluorometric system for monitoring the turnover of the autophagic substrate p62/SQSTM1. Autophagy. 2011;7:883–91. doi: 10.4161/auto.7.8.15538. [DOI] [PubMed] [Google Scholar]
- 10.Martinez J, Malireddi RK, Lu Q, Cunha LD, Pelletier S, Gingras S, et al. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat Cell Biol. 2015;17:893–906. doi: 10.1038/ncb3192. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Overholtzer M, Mailleux AA, Mouneimne G, Normand G, Schnitt SJ, King RW, et al. A nonapoptotic cell death process, entosis, that occurs by cell-in-cell invasion. Cell. 2007;131:966–79. doi: 10.1016/j.cell.2007.10.040. [DOI] [PubMed] [Google Scholar]
- 12.Boncompain G, Divoux S, Gareil N, de Forges H, Lescure A, Latreche L, et al. Synchronization of secretory protein traffic in populations of cells. Nat Methods. 2012;9:493–8. doi: 10.1038/nmeth.1928. [DOI] [PubMed] [Google Scholar]
- 13.Jollivet F, Raposo G, Dimitrov A, Sougrat R, Goud B, Perez F. Analysis of de novo Golgi complex formation after enzyme-based inactivation. Mol Biol Cell. 2007;18:4637–47. doi: 10.1091/mbc.e07-08-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev. 2010;90:1383–435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 15.Tasdemir E, Galluzzi L, Maiuri MC, Criollo A, Vitale I, Hangen E, et al. Methods for assessing autophagy and autophagic cell death. Methods Mol Biol. 2008;445:29–76. doi: 10.1007/978-1-59745-157-4_3. [DOI] [PubMed] [Google Scholar]
- 16.Gomes-da-Silva LC, Zhao L, Bezu L, Zhou H, Sauvat A, Liu P, et al. Photodynamic therapy with redaporfin targets the endoplasmic reticulum and Golgi apparatus. EMBO J. 2018; in press. [DOI] [PMC free article] [PubMed]
- 17.Garg AD, Krysko DV, Verfaillie T, Kaczmarek A, Ferreira GB, Marysael T, et al. A novel pathway combining calreticulin exposure and ATP secretion in immunogenic cancer cell death. EMBO J. 2012;31:1062–79. doi: 10.1038/emboj.2011.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helms JB, Rothman JE. Inhibition by brefeldin A of a Golgi membrane enzyme that catalyses exchange of guanine nucleotide bound to ARF. Nature. 1992;360:352–4. doi: 10.1038/360352a0. [DOI] [PubMed] [Google Scholar]
- 19.Saenz JB, Sun WJ, Chang JW, Li J, Bursulaya B, Gray NS, et al. Golgicide A reveals essential roles for GBF1 in Golgi assembly and function. Nat Chem Biol. 2009;5:157–65. doi: 10.1038/nchembio.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou H, Sauvat A, Gomes-da-Silva LC, Durand S, Forveille S, Iribarren K, et al. The oncolytic compound LTX-401 targets the Golgi apparatus. Cell Death Differ. 2016;23:2031–41. doi: 10.1038/cdd.2016.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng X, Overmeyer JH, Maltese WA. Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking. J Cell Sci. 2006;119:259–70. doi: 10.1242/jcs.02735. [DOI] [PubMed] [Google Scholar]
- 22.Niso-Santano M, Malik SA, Pietrocola F, Bravo-San Pedro JM, Marino G, Cianfanelli V, et al. Unsaturated fatty acids induce non-canonical autophagy. EMBO J. 2015;34:1025–41. doi: 10.15252/embj.201489363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen S, Kepp O, Kroemer G. The end of autophagic cell death? Autophagy. 2012;8:1–3. doi: 10.4161/auto.8.1.16618. [DOI] [PubMed] [Google Scholar]
- 24.Wei Y, Chiang WC, Sumpter R, Jr, Mishra P, Levine B. Prohibitin 2 is an inner mitochondrial membrane mitophagy receptor. Cell. 2017;168:224–38. doi: 10.1016/j.cell.2016.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pickles S, Vigie P, Youle RJ. Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr Biol. 2018;28:R170–R185. doi: 10.1016/j.cub.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rubio N, Coupienne I, Di Valentin E, Heirman I, Grooten J, Piette J, et al. Spatiotemporal autophagic degradation of oxidatively damaged organelles after photodynamic stress is amplified by mitochondrial reactive oxygen species. Autophagy. 2012;8:1312–24. doi: 10.4161/auto.20763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reddy A, Caler EV, Andrews NW. Plasma membrane repair is mediated by Ca(2+)-regulated exocytosis of lysosomes. Cell. 2001;106:157–69. doi: 10.1016/S0092-8674(01)00421-4. [DOI] [PubMed] [Google Scholar]
- 28.Liou W, Geuze HJ, Slot JW. Improving structural integrity of cryosections for immunogold labeling. Histochem Cell Biol. 1996;106:41–58. doi: 10.1007/BF02473201. [DOI] [PubMed] [Google Scholar]
- 29.Slot JW, Geuze HJ, Gigengack S, Lienhard GE, James DE. Immuno-localization of the insulin regulatable glucose transporter in brown adipose tissue of the rat. J Cell Biol. 1991;113:123–35. doi: 10.1083/jcb.113.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.