 Staphylococcus aureus (S. aureus) is an asymptomatic colonizer of 30% of all human beings. It is also the most dangerous of all Staphylococcal bacteria.
Staphylococcus aureus (S. aureus) is an asymptomatic colonizer of 30% of all human beings. It is also the most dangerous of all Staphylococcal bacteria.
Abstract
Staphylococcus aureus (S. aureus) is an asymptomatic colonizer of 30% of all human beings. While generally benign, antibiotic resistance contributes to the success of S. aureus as a human pathogen. Resistance is rapidly evolved through a wide portfolio of mechanisms including horizontal gene transfer and chromosomal mutation. In addition to traditional resistance mechanisms, a special feature of S. aureus pathogenesis is its ability to survive on both biotic and abiotic surfaces in the biofilm state. Due to this characteristic, S. aureus is a leading cause of human infection. Methicillin-resistant S. aureus (MRSA) in particular has emerged as a widespread cause of both community- and hospital-acquired infections. Currently, MRSA is responsible for 10-fold more infections than all multi-drug resistant (MDR) Gram-negative pathogens combined. Recently, MRSA was classified by the World Health Organization (WHO) as one of twelve priority pathogens that threaten human health. In this targeted mini-review, we discuss MRSA biofilm production, the relationship of biofilm production to antibiotic resistance, and front-line techniques to defeat the biofilm-resistance system.
I. Introduction
Nosocomial infections are a major global health concern.1–7 While significant progress has been made preventing transmission, on any given day, approximately 5% of patients in developed countries and 10% of patients in developing countries will acquire a hospital-associated infection (HAI).8–11 Higher rates of HAI are seen in developing countries due to limited resources.12,13 Furthermore, HAI rates can rise to around 50% for patients in intensive care units (ICUs).14
Staphylococcus aureus (S. aureus) is a common cause of nosocomial infection.15–17 S. aureus is a Gram-positive commensal that persistently colonizes the skin and mucosae of approximately 30% of the human population.18 Another 60% of people are transiently colonized.19 While the nose is the most frequent carriage site, the skin, axillae, perineum, and pharynx are also common sites of colonization.20
While S. aureus appears as an innocuous commensal, it is responsible for a major infectious disease burden.18 As an adaptable pathogen, S. aureus can cause a wide range of illnesses after an open wound or “entry point” is inoculated.21 For example, the most common type of staph infection in adults is the boil, a pocket of pus that develops in a hair follicle or an oil gland. In children, the most common infection is impetigo, a highly contagious skin infection that appears as red sores on the face near the mouth and nose. Other clinical manifestations of staph infection include endocarditis, osteoarticular infection, pneumonia, toxic shock syndrome, and prosthetic device and catheter infections.22
Staphylococcal infections occur when host defense mechanisms are low as a result of debilitating illness, open wounds, or treatment with steroids or other drugs that compromise immunity (Fig. 1). Indeed, S. aureus infection rates in ICUs are of particular concern, and the risk of infection increases with the duration of a patient's stay in these units.14,23,24 This characteristic of Staphylococcal infections is largely attributable to the fact that S. aureus is an opportunistic pathogen that possesses an extensive arsenal of virulence factors that enable the organism to take advantage of a compromised host.25,26 Moreover, a number of strains possess a battery of resistance mechanisms against conventional antibiotics.27 To compound the problem, S. aureus can live in the biofilm state. Biofilms are organized populations of bacteria encapsulated in a self-produced extracellular polymeric matrix that adheres to biotic and abiotic surfaces.28,29 Importantly, biofilms provide protection from antibiotics and the host immune system. Additionally, bacteria in the biofilm state display increased resistance to stress compared to those in the planktonic state. Given the ability of biofilms to shield bacteria from harsh host environments, biofilm adds an additional level of complexity to the problem of antimicrobial resistance.
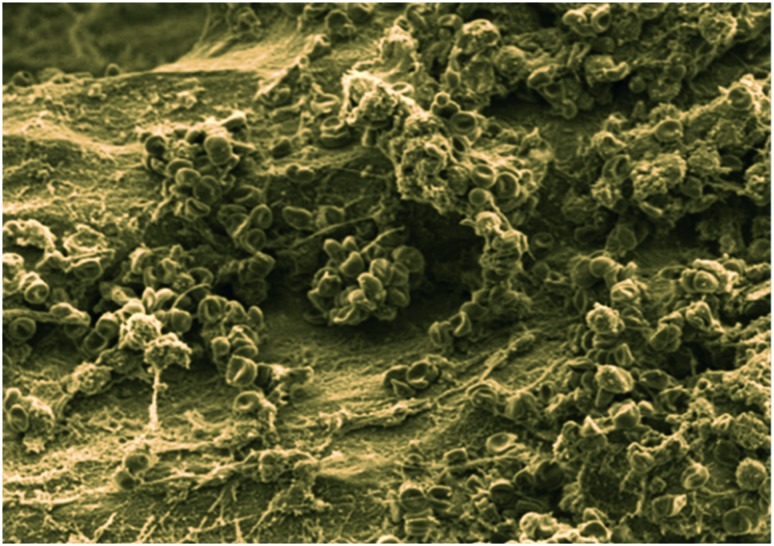
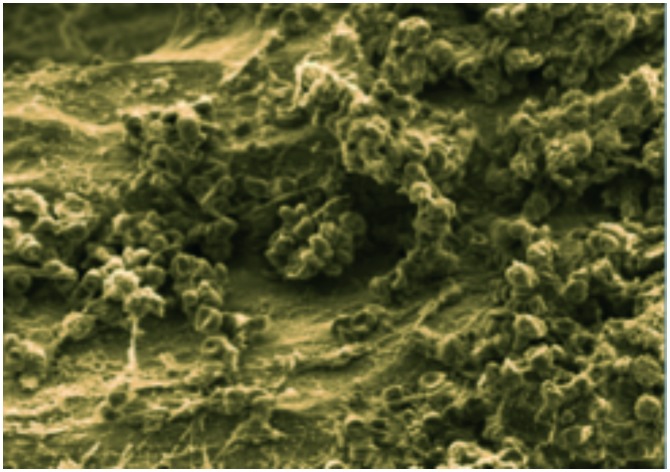
Fig. 1. Scanning electron microscope (SEM) image of Staphylococcus aureus-infected bone (image courtesy of Dr. Jennifer Gaddy at Vanderbilt University).
II. Methicillin-resistant S. aureus (MRSA)
S. aureus is an adaptable organism with the ability to evolve resistance to an array of antibiotics. Resistance development and subsequent dissemination are consequences of horizontal gene transfer (HGT), i.e. the lateral movement of genetic information between organisms. Notably, HGT enables new, antibiotic-resistant variants to arise without the need for genetic mutation.30–32 This mode of action is often encountered in hospitals where selective pressure for resistance is enhanced. Inevitably, hospital-associated resistant strains enter and spread throughout the community.
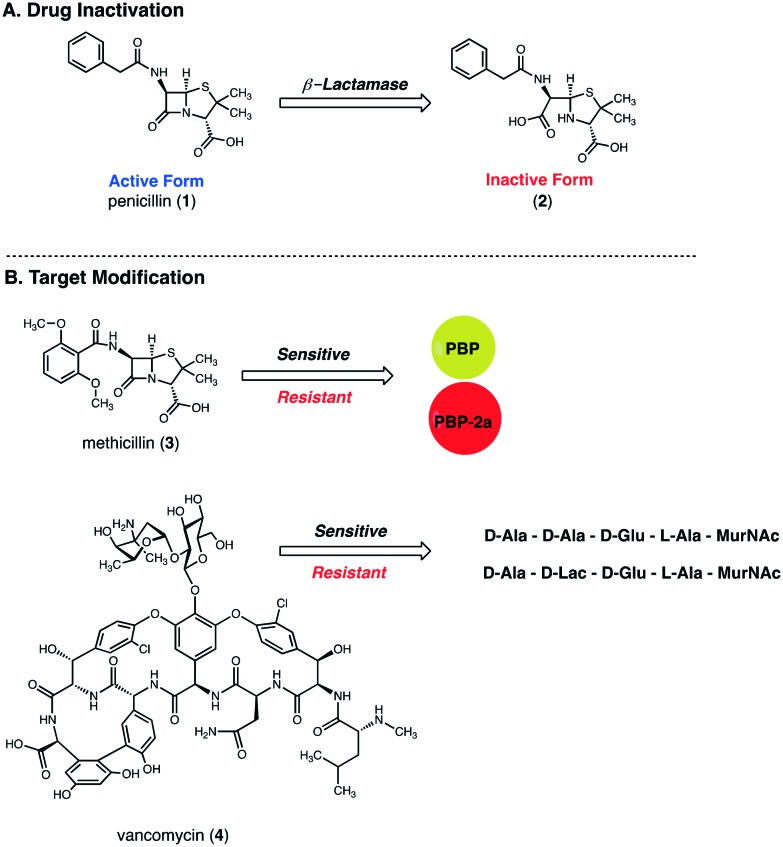
Antibiotic resistance in S. aureus was first observed in the 1940s when infections caused by penicillin-resistant S. aureus (PRSA) emerged in hospitals.33,34 These strains produce a plasmid-encoded lactamase (penicillinase) capable of hydrolyzing the β-lactam ring of penicillin (1). As this ring is the antimicrobial warhead of penicillin, its hydrolysis renders the drug inactive (2) (Fig. 2A). Within a few years after its appearance in hospitals, PRSA had spread to the community. By the 1950s and 1960s, penicillin-resistant strains in the community had reached pandemic levels.33 Today, more than 90% of Staphylococcal isolates produce penicillinase and are consequently resistant to penicillin.35
Fig. 2. Mechanisms of Staphylococcus aureus resistance to penicillin (1), methicillin (3), and vancomycin (4). (A) Penicillin is inactivated by bacterial β-lactamases that hydrolyze the β-lactam ring, which forms an inactive penicilloic acid. (B) Resistance to methicillin, a modified-penicillin scaffold featuring a larger aryl side chain that is resistant to β-lactamase action, is driven by the expression of the alternative transpeptidase, PBP2a, which has a lower affinity for methicillin. Resistance to vancomycin results from modification of the terminal dipeptide of cell wall peptidoglycan chains, which reduces the affinity of the dipeptide for vancomycin.
In an attempt to combat penicillin resistance, methicillin (3) was introduced in 1959.33,34 Methicillin features a larger aryl moiety near the β-lactam ring which reduces its affinity for Staphylococcal β-lactamases.36 Unfortunately, the first reports of methicillin resistance were observed in 1961, just 2 years after methicillin's introduction. Contrary to penicillin resistance, methicillin resistance is not a result of drug inactivation, i.e. hydrolysis of the β-lactam ring, but rather a result of drug target modification (Fig. 2B). Methicillin-resistant S. aureus (MRSA) strains express an additional penicillin-binding protein (PBP), known as PBP2a, which has been hypothesized to have originated from Staphylococcus sciuri.36
PBPs are membrane-bound enzymes that catalyze the cross-linking or transpeptidation reactions that link peptidoglycan chains in the bacterial cell wall.35 In the absence of resistance mechanisms, β-lactams inhibit the transpeptidase domain of PBPs. This results in inhibition of the cross-linking reactions which are integral to formation of a stable peptidoglycan layer. Without a structurally sound peptidoglycan layer, bacterial cell walls become weak and lack the ability to contain the cytoplasmic contents of the cell.36 While PBP2a shares the structural features associated with penicillin binding that are common to other PBPs, PBP2a has a low affinity for all β-lactams. Indeed, the PBP2a active site is able to block the binding of β-lactams while simultaneously allowing cross-linking to proceed.35 Importantly, while β-lactamase-mediated resistance is a narrow-spectrum mechanism, i.e. only penicillin is inactivated by the enzyme, methicillin resistance due to PBP2a expression is a broad-spectrum resistance mechanism. All β-lactams, including penicillins, cephalosporins, and carbapenems, are inactive against bacterial strains expressing PBP2a.
The inability of β-lactams to combat staph infections has led to an increased use of vancomycin (4) and the inevitable evolution of vancomycin-resistant S. aureus (VRSA) strains.37 Similar to methicillin resistance, vancomycin resistant S. aureus strains derive their resistance from structural modification of the target. Modification of the terminal dipeptide of cell wall peptidoglycan chains from d-alanyl-d-alanine (d-Ala-d-Ala) to d-alanyl-d-lactate (d-Ala-d-Lac) reduces the affinity of the dipeptide for vancomycin, thus preventing disruption of peptidoglycan cross-linking (Fig. 2B).38
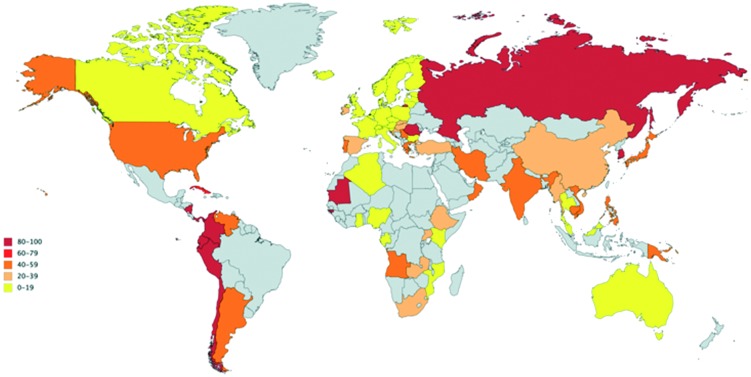
Today, MRSA is pandemic. The rise to pandemic status started with hospital-acquired MRSA clones in the 1960s. This then fostered community-acquired MRSA clones in the 1990s and finally livestock-associated MRSA clones in the 2000s. The evolution of MRSA from initial reports to widespread dissemination parallels the trajectory of PRSA in the 1940s. Unsurprisingly, MRSA is highly prevalent in hospitals (Fig. 3). The highest rates of MRSA (>50%) are reported in North and South America, Asia, and Malta. Intermediate rates (25–50%) are reported in China, Australia, Africa, and several European countries [e.g. Portugal (49%), Greece (40%), Italy (37%) and Romania (34%)]. Most European countries have low prevalence rates (e.g. Netherlands and Scandinavia).39–41
Fig. 3. Global prevalence of hospital-acquired MRSA.
III. The biofilm state
Implantable medical devices have revolutionized modern healthcare. Unfortunately, attachment to indwelling devices by surface-adhering bacteria increases patient morbidity and mortality. Biofilms formed by Staphylococci are the most common cause of biofilm-associated infections with S. aureus being among the most common cause of device related infections (DRI).42–44 All implanted medical devices are susceptible to colonization by Staphylococci. As a result, biofilm-associated infections have been associated with devices such as implanted catheters, prosthetic heart valves, cardiac pacemakers, contact lenses, cerebrospinal fluid shunts, joint replacements, and intravascular lines. To exacerbate the problem, infections associated with biofilms are particularly difficult to treat as bacteria within the matrix are more resistant to antimicrobial agents and the host immune response than planktonic bacteria. This increased resistance is attributable both to the protection afforded by the biofilm matrix as well as the unique phenotypic characteristics of bacteria within the matrix.
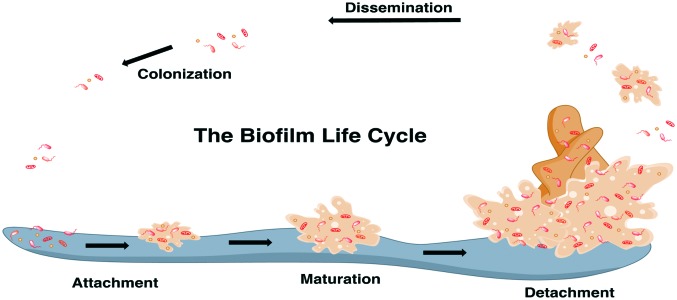
The first stage of biofilm formation is the attachment of a bacterial cell to a living (biotic) or non-living (abiotic) surface (Fig. 4).45 Following attachment, bacteria in the biofilm state progress through a growth and maturation phase.46 At the molecular level, the biofilm matrix is composed of an extracellular polymeric substance (EPS) composed primarily of oligosaccharides, DNA, and proteins.47 The primary oligosaccharide in S. aureus biofilm matrices is a polymer of N-acetyl-β-(1-6)-glucosamine (polysaccharide intercellular adhesin or PIA), while the accumulation-associated protein (Aap) is a common biofilm-associated protein. Teichoic acids are also common biofilm components. At the end of the biofilm cycle, cell clusters detach from the larger biofilm structure. Detachment is facilitated by expression of surfactant-like peptides, which are also critical to biofilm integrity and three-dimensional structure. Once detached, cell clusters can start new biofilm colonies on other surfaces.
Fig. 4. The biofilm life cycle.
S. aureus pathogenesis and biofilm development is controlled by cell-to-cell communication using a ubiquitous regulatory system called quorum sensing.48–52 During its growth and maturation phase, S. aureus produces an autoinducing peptide (AIP) that accumulates in the extracellular environment. Once AIP levels reach a specific concentration, the signal binds to a bacterial surface receptor and activates a regulatory cascade. The outcome is an increased expression of invasive factors such as toxins, hemolysins, proteases, and other tissue-degrading enzymes. Interestingly, these factors alter the metabolic status of the bacteria which subsequently changes their biofilm-forming capacity. Unfortunately, the relationship between environmental stress and pathogenesis remains poorly understood.
IV. Biofilm-mediated antimicrobial resistance
It has long been recognized that biofilms increase resistance to antimicrobial action from both external agents, such as antibiotics, and internal agents of the innate immune system, such as antimicrobial peptides (AMPs).53 Broadly speaking, two mechanisms are responsible for biofilm-mediated resistance. The first is prevention of chemotherapeutics from reaching their target due to limited diffusion or repulsion caused by the biofilm matrix itself.28,54 The second mechanism involves alteration of the physiology of biofilm-dwelling bacteria compared to planktonic bacteria.
Cells within the biofilm, particularly those deep within the matrix, are generally thought to exist in a slow-growing state; these slow-growing cells are referred to as dormant or persister cells. Persister cells are a small fraction of exponentially growing cells, but are ca. 1% of bacteria in both the stationary phase and in biofilms. The decreased growth rate of persister cells can limit the efficacy of antibiotics, especially those that target active cell processes, without the need for genetic alteration.55–57 For example, this type of cell would be immune to β-lactams that target cell wall formation in actively dividing cells.28,29,54 The ability of dormant cells to survive numerous rounds of antibiotic treatment also makes them key contributors to the restoration of biofilm communities.54
V. Strategies to combat MRSA biofilms
The development of strategies to prevent, remove, or disperse biofilms are as critical to treating staph infections as the development of new antibiotics.58–63 A frontier approach in the battle against S. aureus is to develop anti-biofilm strategies that can be combined with conventional antibiotics as a means to restore antibiotic efficacy to levels observed when treating planktonic bacteria. In this section, we will discuss several approaches used to eradicate MRSA biofilms. These strategies can be broken down broadly into two categories: prevention of biofilm formation (antibiotic chemotherapy, anti-adhesive coatings/surfaces) and elimination of established S. aureus biofilms.
A. Antibiotic therapy
The best method for treating a biofilm-related infection is by preventing initial infection altogether. Unfortunately, the facile evolution of antibiotic resistance by S. aureus poses a significant challenge to this approach. Biofilms compound this issue by significantly increasing antibiotic minimum inhibitory concentrations (MICs) compared to cells in the planktonic state.64 For example, the MIC for vancomycin, the most commonly administered drug for S. aureus biofilm-associated infections, is 10-times higher for biofilm-bound cells than for planktonic, free-floating cells (planktonic cell MIC = ca. 2 μg ml–1, biofilm bound cell MIC = ca. 20 μg ml–1).65
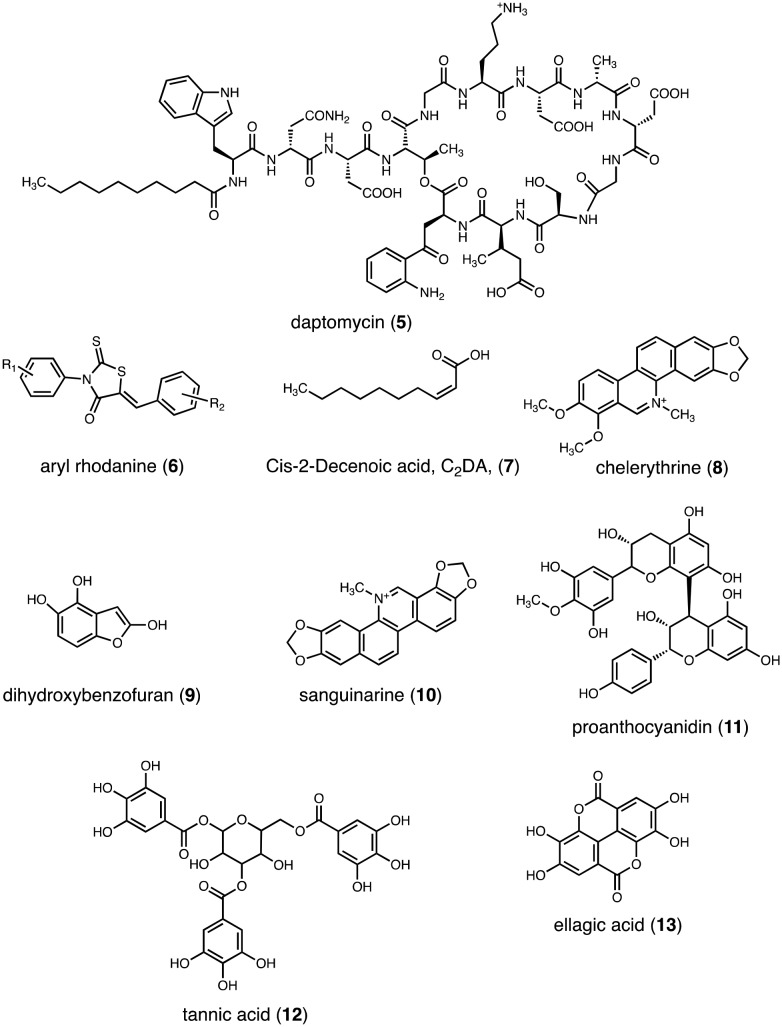
Despite growing resistance levels, there do exist antibiotics, such as daptomycin (5) that are effective at treating even VRSA biofilm-related infections (Fig. 5). Daptomycin, a cyclic lipopeptide molecule, is a novel antibiotic that disrupts the cytoplasmic membrane via rapid depolarization and interruption of DNA, RNA, and protein synthesis. Importantly, daptomycin is one of the most effective antibiotics at clearing S. aureus from an existing biofilm.66 Moreover, because the mode of action for daptomycin does not require cells to be in a metabolically active state, it is a particularly useful agent in the fight against persister cells embedded deep within the biofilm matrix.
Fig. 5. Select antibiofilm small molecules.
B. Physical methods for biofilm removal
Second to preventing initial infection and, by extension, initial formation of a biofilm matrix, the next simplest method to treat an S. aureus biofilm-mediated infection is through surgical removal of the biofilm abcess.67 Removal can occur through debridement of wounds or surgical implants. Irrigation and pulsed lavage are also strategies that are commonly employed. Unfortunately, techniques that apply purely physical tools have limited success. For example, pulse lavage irrigation is ineffective at eliminating S. aureus biofilms present on indwelling devices.68
C. Attachment prevention
Attachment of bacteria to abiotic surfaces is mediated by a number of factors such as adhesion surface proteins, fimbriae or pili, and exopolysaccharides.69,70 Adhesion occurs most readily on surfaces that are coarse or hydrophobic. As hospitals are rich with these types of surfaces, hospitals are a major source of device-associated infections. In a similar vein, indwelling medical devices often feature coarse or hydrophobic surfaces and thus present another potential colonization surface. Due to the prevalence of device-related infections, there has been increased interest in developing anti-infective strategies to prevent colonization.71–74
While adhesion to abiotic surfaces, such as metal and plastics, proceeds through nonspecific mechanisms, adherence to biotic surfaces is dependent on surface proteins that are anchored to the cell wall peptidoglycan.75,76 Indeed, cell surface proteins, which are designed to recognize host surfaces, are critical for S. aureus adherence to host tissues as well as subsequent tissue colonization and ultimately the survival of MRSA infections. Surface proteins known to play important roles in biofilm formation include Bap, clumping factors (ClfB), FnBPs, SasC, SasG, and protein A. ClfB, FnBPs and protein A are widely distributed.77–81 To target these proteins, and thus disrupt attachment, the Clubb group used an array of small molecules to inhibit MRSA transpeptidase sortase A; MRSA transpeptidase sortase A is a protein that anchors surface proteins to the cell wall.82,83 In theory, cell surface proteins are a novel therapeutic target to disrupt adhesion or adherence and mitigate biofilm formation.
Whether dealing with biotic or abiotic surfaces, the frontier challenge in attachment prevention methods remains understanding how bacteria coordinate the expression of different effectors and how various surfaces, particularly cellular surfaces, react to these effectors. If this communication system can be deciphered, one can develop strategies to eradicate biofilms by blocking initial adherence of the microbe. In the proceeding sections, several coatings that prevent bacterial attachment to and growth on surfaces are described.
C1. Small molecules
Aryl rhodanines (6) are 5-membered ring heterocycles that are known to inhibit biofilm formation in several Gram-positive models, including Staphylococcal and Enterococcal species (Fig. 5).84 Aryl rhodanines function by inhibiting attachment of bacterial cells through a mechanism that likely involves complexation of the rhodanines to one or more adhesins located on the microbial cell surface. Interestingly, aryl rhodanines are inactive against Gram-negative microbes. Importantly, while rhodanines possess anti-biofilm activity, they do not possess antimicrobial activity and are not cytotoxic against human cells. From a therapeutic perspective, rhodanines have the potential to be important tools in the battle against MRSA as their lack of antimicrobial activity reduces selective pressure. In other words, this class of small molecule is less likely to produce resistant strains or to induce high levels of biofilm production as a means to protect against a strong antimicrobial substance.
C2. Abiotic surface coating
Catheters coated with tetracyclines and ansamycins, both of which are bacteriostatic as opposed to bactericidal antibiotics, have been shown to decrease the frequency of MRSA central line-associated bloodstream infection (CLABSI) in ICUs.85,86 This result suggests that alteration of the surface properties of an indwelling device by coating the surface with bacteriostatic agents can prevent biofilm-associated infections.
A number of metals have also been used to coat abiotic surfaces, such as catheters, in an effort to prevent biofilm formation.87 The most well-known example is silver in the form of elemental silver, silver ions, and/or silver nanoparticles.88–90 Silver is effective at preventing biofilm formation against both Gram-positive and Gram-negative microbes, including MRSA. Interestingly, although silver coatings are frequently used, the mechanism of action behind silver-mediated biofilm production prevention remains unknown. However, changes to bacterial cell morphology have hinted at several mechanisms. For example, silver nanoparticles have been shown to attach to the bacterial membrane and penetrate the cell. After gaining entrance, the nanoparticles engage sulfide-containing proteins and DNA. This resultantly inhibits DNA replication and transcription. Thus, it is thought that silver prevents biofilm production by serving as an antimicrobial agent.
While silver-coating is common, there are cytotoxicity concerns with this method. Silver accelerates thrombin formation and platelet activation which subsequently places patients at higher risk for thrombosis. To avoid this issue, stainless steel and titanium have also been used to coat implant materials.91–93 Interestingly, a number of medical devices have also been coated with vancomycin to prevent MRSA adherence.
D. Treatment or dispersal of established biofilms
D1. Small molecules
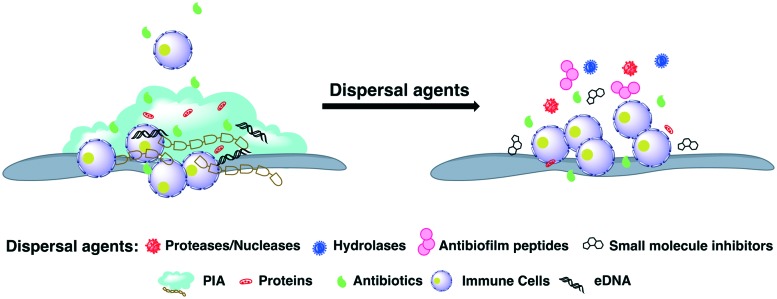
Cis-2-Decenoic acid (C2DA, 7) is a medium-chain fatty acid produced by Pseudomonas aeruginosa that has been shown not only to possess the ability to disperse established MRSA biofilms but also to completely inhibit MRSA biofilm formation (Fig. 5).94,95 In addition to this lipid, it has been shown that d-amino acids disperse established biofilms in S. aureus. Incorporation of d-amino acids into the peptidoglycan layer results in the release of amyloid fibers, a component of the extracellular matrix that connects cells in the biofilm matrix.96–99 Kolodkin-Gal demonstrated that d-amino acids disperse Bacillus subtilis biofilms by affecting the function of these amyloid fibers.100 Mechanistically, when noncanonical amino acids are incorporated into the peptidoglycan layer, they interfere with the normal anchoring that helps maintain biofilm architecture integrity. Moreover, d-amino acids compete with canonical amino acids for positions in the peptidoglycan layer which interferes with transpeptidation and transglycosylation. Importantly, this disruption of bacterial cell wall composition caused by d-amino acid incorporation interferes with biofilm formation (Fig. 6).
Fig. 6. General methods for biofilm dispersal.
D2. Matrix degrading enzymes
Disruption of biofilm matrix structural integrity is an attractive approach to limit the protective effects the matrix affords cells enclosed within it. This method is the reason for the addition of exogenous enzymes, such as dispersion B or DNAase, to S. aureus biofilms.101–104 DNAase works by degrading the extracellular DNA in the biofilm matrix EPS, while dispersin B targets the polysaccharide EPS component. As biofilm matrices consist largely of extracellular DNA and polysaccharides, the actions of dispersin B and DNase serve to destabilize the matrix. It is important to note, however, that the use of exogenous enzymes like as dispersin B and DNase to disrupt S. aureus biofilm formation does have its shortcomings. For example, the susceptibility of S. aureus to dispersin B differs significantly among strains. Moreover, a number of clinically relevant MRSA strains produce biofilms that contain little polysaccharide which serves to limit the influence of dispersin B treatment on biofilm production.
D3. Plant-derived natural compounds
Natural products are critical to the discovery and development of new anti-infective agents against MRSA.59–62 For example, extracts from the broths of Krameria, Aesculus hippocastanum, and Conopodium majus each contains four compounds that have all been shown to inhibit S. aureus biofilm formation: chelerythrine (8), dihydroxybenzofuran (9), sanguinarine (10), and proanthocyanidin (11) (Fig. 5).105 American cranberry extracts, which contain proanthocyanidins (PAC), have also been shown to inhibit S. aureus biofilm formation as well as S. aureus growth.106,107 Moreover, polyphenolic compounds found in plant tissues, such as tannic acid (12), are known to inhibit S. aureus biofilm formation (Fig. 5).108,109
Tea-tree oil, an essential oil extracted from the leaves of Melaleuca alternifolia, eradicates biofilm production by S. aureus, including MRSA, by damaging the extracellular matrix.110,111 This damage initiates subsequent removal of the biofilm from biotic surfaces. Ellagic acid (13) derivatives from Rubus ulmifolius limit S. aureus biofilm production and also enhance the susceptibility of S. aureus to the antibiotics daptomycin, clindamycin, and oxacillin without contaminant cytotoxicity to mammalian cells (Fig. 5).112,113
Although the agents discussed in the section are effective at combatting biofilms, their modes of action remain unclear.
Conclusion and future outlook
Rising MRSA infection rates pose a significant threat to human health. While increasing antibiotic resistance is a well-appreciated contributing factor, a lesser appreciated but equally important factor is the ability of S. aureus to form biofilms. Biofilms serve to protect S. aureus from host defenses and antibiotics alike and are consequently integral to S. aureus pathogenesis. Indeed, biofilm-dwelling bacteria are generally able to tolerate much higher antibiotic concentrations than their planktonic counterparts. The increased resistance of biofilm-associated bacteria against antimicrobial action is attributable to the physical barrier between bacteria and antimicrobial afforded by the biofilm matrix as well as the phenotypic shift bacteria embedded in the matrix undergo. As a result, biofilm-associated infections are notoriously difficult to eradicate.
Indicative of the benefit of biofilm production for S. aureus survival, most chronic MRSA infections leverage the biofilm state in their pathogenesis. This is especially true for those associated with indwelling medical devices. As most therapeutic strategies are only effective at treating planktonic cells or acute infections, there is an urgent need to develop new therapeutic strategies capable of targeting S. aureus in the biofilm state. Unfortunately, despite much effort, the development of useful biofilm inhibitors and/or dispersal agents for Staphylococcal biofilms is in its infancy. While many innovative approaches to eradicate S. aureus biofilms have been achieved over the past two decades such as small molecules that prevent biofilm formation, enzymes that weaken biofilm matrix structural integrity, and antibodies and vaccines that target specific biofilm life cycle stages, these approaches lack clinical validation.
One potential future source of antibiofilm compounds are cationic small molecules. Indeed, several recent studies have showcased the ability of positively-charged molecules to disrupt biofilm matrices and inhibit biofilm formation by a number of pathogens.114–119 However, the antibiofilm activity of this class of molecule is generally accompanied by antimicrobial activity. Although this may seem beneficial, the antimicrobial activity is likely to induce selective pressure and promote the evolution of resistant bacteria. Additionally, care must be taken with cationic molecules to limit cytotoxicity to mammalian cells. Given these concerns, identifying cationic small molecules with exclusive antibiofilm activity represents an exciting research avenue.
Another approach, one which our lab has begun to investigate, is to use host defense mechanisms as a source of molecular inspiration. We recently demonstrated that human milk oligosaccharides (HMOs), non-conjugated oligosaccharides abundant in human milk, modulate growth and biofilm production for several bacterial pathogens, including MRSA.120 However, we have yet to identify the mechanism of action behind the antibiofilm activity observed. In a parallel study, we discovered that conversion of the ubiquitous HMO 2′-fucosyllactose (2′-FL) to an anomeric, amino-variant gave a compound with impressive antibiofilm activity against Group B Streptococcus.121 Once again, the mechanism behind this antibiofilm activity remains unknown. Thus, future studies are directed at elucidating a mechanism of action as well as investigating if this result translates to an S. aureus model.
In addition to these approaches, as previously mentioned, further elucidation of how bacteria coordinate the expression of various effectors and how surfaces react with these effectors will be paramount to the development of antibiofilm compounds. Indeed, a greater understanding of this communication system has the potential to identify unique bacterial targets that can be engaged to target biofilm production selectively without accompanying antimicrobial activity.
Conflicts of interest
There are no conflicts to declare.
Acknowledgments
Financial support is provided by Vanderbilt University, the Vanderbilt Microbiome Initiative (VMI), the Vanderbilt Pre3 Initiative, a Deans Faculty Fellowship, Glycosyn, FrieslandCampina, and the National Science Foundation (CAREER Award to SDT: CHE-1847804). K. M. C. is supported by the Vanderbilt Chemical Biology Interface (CBI) training program (T32 GM065086), the Vanderbilt Pre3 Initiative (travel grant), and the Mitchum E. Warren, Jr. Graduate Research Fellowship. Johny M. Nguyen acknowledges the Gates Millennium Scholars (GMS) Program for a graduate research fellowship. Lawrence J. Berg acknowledges the Beckman Foundation for a Beckman Scholars Program Fellowship.
References
- Mo Y., Low I., Tambyah S. K., Tambyah P. A. J. Hosp. Infect. 2018 doi: 10.1016/j.jhin.2018.08.013. [DOI] [PubMed] [Google Scholar]
- Choi J. Y., Kwak Y. G., Yoo H., Lee S. O., Kim H. B., Han S. H. J. Hosp. Infect. 2016;92(4):363–371. doi: 10.1016/j.jhin.2015.12.012. [DOI] [PubMed] [Google Scholar]
- Alai N. Curr. Med. Res. Opin. 2016;32(10):1757–1758. doi: 10.1080/03007995.2016.1208642. [DOI] [PubMed] [Google Scholar]
- Lackner P., Mueller C., Beer R., Broessner G., Fischer M., Helbok R. Curr. Drug Targets. 2017;18(12):1417–1423. doi: 10.2174/1389450117666160401124426. [DOI] [PubMed] [Google Scholar]
- Sutcu M., Salman N., Akturk H., Dalgic N., Turel O., Kuzdan C. Am. J. Infect. Control. 2016;44(10):1139–1143. doi: 10.1016/j.ajic.2016.03.056. [DOI] [PubMed] [Google Scholar]
- Heister T., Kaier K., Wolkewitz M. Am. J. Infect. Control. 2017;45(1):94–95. doi: 10.1016/j.ajic.2016.07.030. [DOI] [PubMed] [Google Scholar]
- Tsitsopoulos P. P., Iosifidis E., Antachopoulos C., Anestis D. M., Karantani E., Karyoti A. Acta Neurochir. 2016;158(9):1647–1654. doi: 10.1007/s00701-016-2890-5. [DOI] [PubMed] [Google Scholar]
- Stone P. W., Braccia D., Larson E. Am. J. Infect. Control. 2005;33(9):501–509. doi: 10.1016/j.ajic.2005.04.246. [DOI] [PubMed] [Google Scholar]
- Zingg W., Hopkins S., Gayet-Ageron A., Holmes A., Sharland M., Suetens C. Lancet Infect. Dis. 2017;17(4):381–389. doi: 10.1016/S1473-3099(16)30517-5. [DOI] [PubMed] [Google Scholar]
- Kariya N., Sakon N., Komano J., Tomono K., Iso H. J. Infect. Chemother. 2018;24(5):347–352. doi: 10.1016/j.jiac.2017.12.004. [DOI] [PubMed] [Google Scholar]
- Wong S. S., Huang C. H., Yang C. C., Hsieh Y. P., Kuo C. N., Chen Y. R. Antimicrob. Resist. Infect. Control. 2018;7:34. doi: 10.1186/s13756-018-0320-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagheri Nejad S., Allegranzi B., Syed S. B., Ellis B., Pittet D. Bull. W. H. O. 2011;89(10):757–765. doi: 10.2471/BLT.11.088179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO, Health care-associated infections FACT SHEET.
- Vincent J. L., Rello J., Marshall J., Silva E., Anzueto A., Martin C. D. JAMA, J. Am. Med. Assoc. 2009;302(21):2323–2329. doi: 10.1001/jama.2009.1754. [DOI] [PubMed] [Google Scholar]
- Choo E. J. Infect. Chemother. 2017;49(2):158–159. doi: 10.3947/ic.2017.49.2.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valaperta R., Tejada M. R., Frigerio M., Moroni A., Ciulla E., Cioffi S. New Microbiol. 2010;33(3):223–232. [PubMed] [Google Scholar]
- Alvarez C. A., Yomayusa N., Leal A. L., Moreno J., Mendez-Alvarez S., Ibanez M. Am. J. Infect. Control. 2010;38(4):315–318. doi: 10.1016/j.ajic.2009.05.013. [DOI] [PubMed] [Google Scholar]
- Tong S. Y., Davis J. S., Eichenberger E., Holland T. L., Fowler Jr. V. G. Clin. Microbiol. Rev. 2015;28(3):603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D. N., Feazel L. M., Bessesen M. T., Price C. S., Janoff E. N., Pace N. R. PLoS One. 2010;5(5):e10598. doi: 10.1371/journal.pone.0010598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollid J. U., Furberg A. S., Hanssen A. M., Johannessen M. Infect., Genet. Evol. 2014;21:531–541. doi: 10.1016/j.meegid.2013.03.020. [DOI] [PubMed] [Google Scholar]
- Kobayashi S. D., Malachowa N., DeLeo F. R. Am. J. Pathol. 2015;185(6):1518–1527. doi: 10.1016/j.ajpath.2014.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creech C. B., Al-Zubeidi D. N., Fritz S. A. Infect. Dis. Clin. North Am. 2015;29(3):429–464. doi: 10.1016/j.idc.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oztoprak N., Cevik M. A., Akinci E., Korkmaz M., Erbay A., Eren S. S. Am. J. Infect. Control. 2006;34(1):1–5. doi: 10.1016/j.ajic.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Hardy K. J., Hawkey P. M., Gao F., Oppenheim B. A. Br. J. Anaesth. 2004;92(1):121–130. doi: 10.1093/bja/aeh008. [DOI] [PubMed] [Google Scholar]
- Gallardo-Garcia M. M., Sanchez-Espin G., Ivanova-Georgieva R., Ruiz-Morales J., Rodriguez-Bailon I., Vinuela Gonzalez V. Eur. J. Clin. Microbiol. Infect. Dis. 2016;35(5):821–828. doi: 10.1007/s10096-016-2603-2. [DOI] [PubMed] [Google Scholar]
- Lacey K. A., Geoghegan J. A., McLoughlin R. M. Pathogens. 2016;5(1):22. doi: 10.3390/pathogens5010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock S. J., Paterson G. K. Annu. Rev. Biochem. 2015;84:577–601. doi: 10.1146/annurev-biochem-060614-034516. [DOI] [PubMed] [Google Scholar]
- Costerton J. W., Stewart P. S., Greenberg E. P. Science. 1999;284(5418):1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Davies D. Nat. Rev. Drug Discovery. 2003;2(2):114–122. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- Haug M. C., Tanner S. A., Lacroix C., Stevens M. J., Meile L. FEMS Microbiol. Ecol. 2011;78(2):210–219. doi: 10.1111/j.1574-6941.2011.01149.x. [DOI] [PubMed] [Google Scholar]
- Andam C. P., Fournier G. P., Gogarten J. P. FEMS Microbiol. Rev. 2011;35(5):756–767. doi: 10.1111/j.1574-6976.2011.00274.x. [DOI] [PubMed] [Google Scholar]
- Summers A. O. Anim. Biotechnol. 2006;17(2):125–135. doi: 10.1080/10495390600957217. [DOI] [PubMed] [Google Scholar]
- Chambers H. F., Deleo F. R. Nat. Rev. Microbiol. 2009;7(9):629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks C. W., Blatnik J. A., Krpata D. M., Novitsky Y. W., Rosen M. J. Hernia. 2014;18(1):65–70. doi: 10.1007/s10029-012-1035-x. [DOI] [PubMed] [Google Scholar]
- Lowy F. D. J. Clin. Invest. 2003;111(9):1265–1273. doi: 10.1172/JCI18535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton P. D., Taylor P. W. Sci. Prog. 2002;85:57–72. doi: 10.3184/003685002783238870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuinness W. A., Malachowa N., DeLeo F. R. Yale J. Biol. Med. 2017;90(2):269–281. [PMC free article] [PubMed] [Google Scholar]
- Okano A., Isley N. A., Boger D. L. Chem. Rev. 2017;117(18):11952–11993. doi: 10.1021/acs.chemrev.6b00820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassoun A., Linden P. K., Friedman B. Crit. Care. 2017;21(1):211. doi: 10.1186/s13054-017-1801-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igrejas G., Correia S., Silva V., Hebraud M., Canica M., Torres C. Front. Microbiol. 2018;9:2964. doi: 10.3389/fmicb.2018.02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu H., Yamashita K., Kunisawa S., Fushimi K., Imanaka Y. Am. J. Infect. Control. 2016;44(12):1628–1633. doi: 10.1016/j.ajic.2016.05.008. [DOI] [PubMed] [Google Scholar]
- Vinh D. C., Embil J. M. J. Long-Term Eff. Med. Implants. 2005;15(5):467–488. doi: 10.1615/jlongtermeffmedimplants.v15.i5.20. [DOI] [PubMed] [Google Scholar]
- Duran L. W. Med. Device Technol. 2000;11(6):14–17. [PubMed] [Google Scholar]
- Lin T. L., Lu F. M., Conroy S., Sheu M. S., Su S. H., Tang L. Med. Device Technol. 2001;12(8):26–30. [PubMed] [Google Scholar]
- Schachter B. Nat. Biotechnol. 2003;21(4):361–365. doi: 10.1038/nbt0403-361. [DOI] [PubMed] [Google Scholar]
- Gregor R., David S., Meijler M. M. Chem. Soc. Rev. 2018;47(5):1761–1772. doi: 10.1039/c7cs00606c. [DOI] [PubMed] [Google Scholar]
- Flemming H.-C., Wingender J. Nat. Rev. Microbiol. 2010;8(9):623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- Welsh M. A., Blackwell H. E. FEMS Microbiol. Rev. 2016;40(5):774–794. doi: 10.1093/femsre/fuw009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praneenararat T., Palmer A. G., Blackwell H. E. Org. Biomol. Chem. 2012;10(41):8189–8199. doi: 10.1039/c2ob26353j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarwood J. M., Schlievert P. M. J. Clin. Invest. 2003;112(11):1620–1625. doi: 10.1172/JCI20442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarwood J. M., Bartels D. J., Volper E. M., Greenberg E. P. J. Bacteriol. 2004;186(6):1838–1850. doi: 10.1128/JB.186.6.1838-1850.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong K. F., Vuong C., Otto M. Int. J. Med. Microbiol. 2006;296(2–3):133–139. doi: 10.1016/j.ijmm.2006.01.042. [DOI] [PubMed] [Google Scholar]
- Bowler P. G. J. Wound Care. 2018;27(5):273–277. doi: 10.12968/jowc.2018.27.5.273. [DOI] [PubMed] [Google Scholar]
- Hall-Stoodley L., Costerton J. W., Stoodley P. Nat. Rev. Microbiol. 2004;2(2):95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- Miyaue S., Suzuki E., Komiyama Y., Kondo Y., Morikawa M., Maeda S. Front. Microbiol. 2018;9:1396. doi: 10.3389/fmicb.2018.01396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood T. K. Microb. Biotechnol. 2017;10(5):1054–1056. doi: 10.1111/1751-7915.12774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon B. P., Rowe S. E., Lewis K. Adv. Exp. Med. Biol. 2015;831:1–9. doi: 10.1007/978-3-319-09782-4_1. [DOI] [PubMed] [Google Scholar]
- Melander R. J., Melander C. Adv. Exp. Med. Biol. 2015;831:69–91. doi: 10.1007/978-3-319-09782-4_6. [DOI] [PubMed] [Google Scholar]
- Blackledge M. S., Worthington R. J., Melander C. Curr. Opin. Pharmacol. 2013;13(5):699–706. doi: 10.1016/j.coph.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington R. J., Richards J. J., Melander C. Org. Biomol. Chem. 2012;10(37):7457–7474. doi: 10.1039/c2ob25835h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowe S. D., Richards J. J., Tucker A. T., Thompson R., Melander C., Cavanagh J. Mar. Drugs. 2011;9(10):2010–2035. doi: 10.3390/md9102010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards J. J., Melander C. ChemBioChem. 2009;10(14):2287–2294. doi: 10.1002/cbic.200900317. [DOI] [PubMed] [Google Scholar]
- Richards J. J., Huigens Iii R. W., Ballard T. E., Basso A., Cavanagh J., Melander C. Chem. Commun. 2008;(14):1698–1700. doi: 10.1039/b719802g. [DOI] [PubMed] [Google Scholar]
- Melander R. J., Melander C. ACS Infect. Dis. 2017;3(8):559–563. doi: 10.1021/acsinfecdis.7b00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem A. H., Elkhatib W. F., Noreddin A. M. J. Pharm. Pharmacol. 2011;63(1):73–79. doi: 10.1111/j.2042-7158.2010.01183.x. [DOI] [PubMed] [Google Scholar]
- Smith K., Perez A., Ramage G., Gemmell C. G., Lang S. Int. J. Antimicrob. Agents. 2009;33(4):374–378. doi: 10.1016/j.ijantimicag.2008.08.029. [DOI] [PubMed] [Google Scholar]
- Bhattacharya M., Wozniak D. J., Stoodley P., Hall-Stoodley L. Expert Rev. Anti-infect. Ther. 2015;13(12):1499–1516. doi: 10.1586/14787210.2015.1100533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urish K. L., DeMuth P. W., Craft D. W., Haider H., Davis 3rd C. M. J. Arthroplasty. 2014;29(6):1128–1132. doi: 10.1016/j.arth.2013.12.012. [DOI] [PubMed] [Google Scholar]
- Arena M. P., Capozzi V., Spano G., Fiocco D. Appl. Microbiol. Biotechnol. 2017;101(7):2641–2657. doi: 10.1007/s00253-017-8182-z. [DOI] [PubMed] [Google Scholar]
- Berne C., Ducret A., Hardy G. G., Brun Y. V. Microbiol. Spectrum. 2015;3(4) doi: 10.1128/microbiolspec.MB-0018-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerli W., Sendi P. Antimicrob. Agents Chemother. 2018 doi: 10.1128/AAC.01746-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidelman J., Lewis S. S. Infect. Dis. Clin. North Am. 2018;32(4):861–876. doi: 10.1016/j.idc.2018.06.006. [DOI] [PubMed] [Google Scholar]
- Buhmann M. T., Stiefel P., Maniura-Weber K., Ren Q. Trends Biotechnol. 2016;34(12):945–948. doi: 10.1016/j.tibtech.2016.05.016. [DOI] [PubMed] [Google Scholar]
- Morgenstern M., Erichsen C., von Ruden C., Metsemakers W. J., Kates S. L., Moriarty T. F. Injury. 2016;47(7):1427–1434. doi: 10.1016/j.injury.2016.04.027. [DOI] [PubMed] [Google Scholar]
- Merghni A., Ben Nejma M., Dallel I., Tobji S., Ben Amor A., Janel S. Microb. Pathog. 2016;91:61–67. doi: 10.1016/j.micpath.2015.11.009. [DOI] [PubMed] [Google Scholar]
- Beloin C., Da Re S., Ghigo J. M. EcoSal Plus. 2005;1(2) doi: 10.1128/ecosalplus.8.3.1.3. [DOI] [PubMed] [Google Scholar]
- Kim S. J., Chang J., Rimal B., Yang H., Schaefer J. Biochim. Biophys. Acta, Biomembr. 2018;1860(3):749–756. doi: 10.1016/j.bbamem.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan J. A., Foster T. J. Curr. Top. Microbiol. Immunol. 2017;409:95–120. doi: 10.1007/82_2015_5002. [DOI] [PubMed] [Google Scholar]
- Foulston L., Elsholz A. K., DeFrancesco A. S., Losick R. mBio. 2014;5(5):e01667–14. doi: 10.1128/mBio.01667-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiecinski J., Jin T., Josefsson E. APMIS. 2014;122(12):1240–1250. doi: 10.1111/apm.12295. [DOI] [PubMed] [Google Scholar]
- Foster T. J., Geoghegan J. A., Ganesh V. K., Hook M. Nat. Rev. Microbiol. 2014;12(1):49–62. doi: 10.1038/nrmicro3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A. H., Wereszczynski J., Amer B. R., Yi S. W., Jung M. E., McCammon J. A. Chem. Biol. Drug Des. 2013;82(4):418–428. doi: 10.1111/cbdd.12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suree N., Yi S. W., Thieu W., Marohn M., Damoiseaux R., Chan A. Bioorg. Med. Chem. 2009;17(20):7174–7185. doi: 10.1016/j.bmc.2009.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opperman T. J., Kwasny S. M., Williams J. D., Khan A. R., Peet N. P., Moir D. T. Antimicrob. Agents Chemother. 2009;53(10):4357–4367. doi: 10.1128/AAC.00077-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull I. R., Buckman S. A., Horn C. B., Bochicchio G. V., Mazuski J. E. Surg. Infect.: Sel. Antibiot. Ther. 2018;19(1):40–47. doi: 10.1089/sur.2017.087. [DOI] [PubMed] [Google Scholar]
- Harron K., Mok Q., Hughes D., Muller-Pebody B., Parslow R., Ramnarayan P. PLoS One. 2016;11(3):e0151348. doi: 10.1371/journal.pone.0151348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczuk D., Ginalska G., Piersiak T., Miazga-Karska M. J. Biomed. Mater. Res., Part B. 2012;100(7):1874–1882. doi: 10.1002/jbm.b.32755. [DOI] [PubMed] [Google Scholar]
- Wang X., Wu J., Li P., Wang L., Zhou J., Zhang G. ACS Appl. Mater. Interfaces. 2018 doi: 10.1021/acsami.8b10972. [DOI] [Google Scholar]
- Pompilio A., Geminiani C., Bosco D., Rana R., Aceto A., Bucciarelli T. Front. Microbiol. 2018;9:1349. doi: 10.3389/fmicb.2018.01349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson H. N., Iveson S., Catherall P., Hardman M. J. Front. Microbiol. 2018;9:1450. doi: 10.3389/fmicb.2018.01450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias H. B., Bernardi M. I. B., Bauab T. M., Hernandes A. C., de Souza Rastelli A. N. Dent. Mater. 2019;35(2):e36–e46. doi: 10.1016/j.dental.2018.11.002. [DOI] [PubMed] [Google Scholar]
- Akens M. K., Chien C., Katchky R. N., Kreder H. J., Finkelstein J., Whyne C. M. BMC Musculoskeletal Disord. 2018;19(1):260. doi: 10.1186/s12891-018-2199-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauss M., Graf S., Gersbach S., Hintermann B., Ilchmann T., Knupp M. Clin. Orthop. Relat. Res. 2013;471(7):2312–2317. doi: 10.1007/s11999-013-2919-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amari D. T., Marques C. N., Davies D. G. J. Bacteriol. 2013;195(20):4600–4610. doi: 10.1128/JB.00707-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings J. A., Courtney H. S., Haggard W. O. Clin. Orthop. Relat. Res. 2012;470(10):2663–2670. doi: 10.1007/s11999-012-2388-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abenojar E. C., Wickramasinghe S., Ju M., Uppaluri S., Klika A., George J. ACS Infect. Dis. 2018;4(8):1246–1256. doi: 10.1021/acsinfecdis.8b00076. [DOI] [PubMed] [Google Scholar]
- Jia R., Li Y., Al-Mahamedh H. H., Gu T. Front. Microbiol. 2017;8:1538. doi: 10.3389/fmicb.2017.01538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmata A. J., Ma Y., Sanchez C. J., Zienkiewicz K. J., Elefteriou F., Wenke J. C. Clin. Orthop. Relat. Res. 2015;473(12):3951–3961. doi: 10.1007/s11999-015-4465-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausbacher D., Fallarero A., Kujala J., Maattanen A., Peltonen J., Strom M. B. Biofouling. 2014;30(1):81–93. doi: 10.1080/08927014.2013.847924. [DOI] [PubMed] [Google Scholar]
- Kolodkin-Gal I., Romero D., Cao S., Clardy J., Kolter R., Losick R. Science. 2010;328(5978):627–629. doi: 10.1126/science.1188628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddinger R. M., Luke-Marshall N. R., Sauberan S. L., Hakansson A. P., Campagnari A. A. mBio. 2018;9(1) doi: 10.1128/mBio.02089-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddinger R. M., Luke-Marshall N. R., Hakansson A. P., Campagnari A. A. mBio. 2016;7(4) doi: 10.1128/mBio.01235-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano C., Echeverz M., Lasa I. Curr. Opin. Microbiol. 2014;18:96–104. doi: 10.1016/j.mib.2014.02.008. [DOI] [PubMed] [Google Scholar]
- Rogers S. A., Krayer M., Lindsey J. S., Melander C. Org. Biomol. Chem. 2009;7(3):603–606. doi: 10.1039/b817923a. [DOI] [PubMed] [Google Scholar]
- Chung P. Y., Toh Y. S. Pathog. Dis. 2014;70(3):231–239. doi: 10.1111/2049-632X.12141. [DOI] [PubMed] [Google Scholar]
- Ulrey R. K., Barksdale S. M., Zhou W., van Hoek M. L. BMC Complementary Altern. Med. 2014;14:499. doi: 10.1186/1472-6882-14-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M., Grenier D. J. Appl. Microbiol. 2012;113(2):438–447. doi: 10.1111/j.1365-2672.2012.05329.x. [DOI] [PubMed] [Google Scholar]
- Dong G., Liu H., Yu X., Zhang X., Lu H., Zhou T. Nat. Prod. Res. 2018;32(18):2225–2228. doi: 10.1080/14786419.2017.1366485. [DOI] [PubMed] [Google Scholar]
- Lee J. H., Park J. H., Cho H. S., Joo S. W., Cho M. H., Lee J. Biofouling. 2013;29(5):491–499. doi: 10.1080/08927014.2013.788692. [DOI] [PubMed] [Google Scholar]
- Kwiecinski J., Eick S., Wojcik K. Int. J. Antimicrob. Agents. 2009;33(4):343–347. doi: 10.1016/j.ijantimicag.2008.08.028. [DOI] [PubMed] [Google Scholar]
- Raman A., Weir U., Bloomfield S. F. Lett. Appl. Microbiol. 1995;21(4):242–245. doi: 10.1111/j.1472-765x.1995.tb01051.x. [DOI] [PubMed] [Google Scholar]
- Sivasankar C., Maruthupandiyan S., Balamurugan K., James P. B., Krishnan V., Pandian S. K. Biofouling. 2016;32(4):397–410. doi: 10.1080/08927014.2016.1148141. [DOI] [PubMed] [Google Scholar]
- Quave C. L., Estevez-Carmona M., Compadre C. M., Hobby G., Hendrickson H., Beenken K. E. PLoS One. 2012;7(1):e28737. doi: 10.1371/journal.pone.0028737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque J., Konai M. M., Sequeira S. S., Samaddar S., Haldar J. J. Med. Chem. 2016;59(23):10750–10762. doi: 10.1021/acs.jmedchem.6b01435. [DOI] [PubMed] [Google Scholar]
- Carmona-Ribeiro A. M., de Melo Carrasco L. D. Int. J. Mol. Sci. 2013;14(5):9906–9946. doi: 10.3390/ijms14059906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque J., Akkapeddi P., Yarlagadda V., Uppu D. S., Kumar P., Haldar J. Langmuir. 2012;28(33):12225–12234. doi: 10.1021/la302303d. [DOI] [PubMed] [Google Scholar]
- Algburi A., Zhang Y., Weeks R., Comito N., Zehm S., Pinto J. Antimicrob. Agents Chemother. 2017;61(12) doi: 10.1128/AAC.00650-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente-Nunez C., Korolik V., Bains M., Nguyen U., Breidenstein E. B., Horsman S. Antimicrob. Agents Chemother. 2012;56(5):2696–2704. doi: 10.1128/AAC.00064-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donelli G., Francolini I., Piozzi A., Di Rosa R., Marconi W. Aust. J. Chem. 2002;14(5):501–507. doi: 10.1179/joc.2002.14.5.501. [DOI] [PubMed] [Google Scholar]
- Ackerman D. L., Craft K. M., Doster R. S., Weitkamp J. H., Aronoff D. M., Gaddy J. A. ACS Infect. Dis. 2018;4(3):315–324. doi: 10.1021/acsinfecdis.7b00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craft K. M., Townsend S. D. J. Antibiot. 2019 doi: 10.1038/s41429-019-0151-6. [DOI] [PubMed] [Google Scholar]