Abstract
Numerous nascent proteins undergo folding and maturation within the luminal and membrane compartments of the endoplasmic reticulum (ER). Despite the presence of various factors in the ER that promote protein folding, many proteins fail to properly fold and assemble and are subsequently degraded. Regulatory proteins in the ER also undergo degradation in a way that is responsive to stimuli or the changing needs of the cell. As in most cellular compartments, the ubiquitin-proteasome system (UPS) is responsible for the majority of the degradation at the ER—in a process termed ER-associated degradation (ERAD). Autophagic processes utilizing ubiquitin-like protein-conjugating systems also play roles in protein degradation at the ER. The ER is continuous with the nuclear envelope (NE), which consists of the outer nuclear membrane (ONM) and inner nuclear membrane (INM). While ERAD is known also to occur at the NE, only some of the ERAD ubiquitin-ligation pathways function at the INM. Protein degradation machineries in the ER/NE target a wide variety of substrates in multiple cellular compartments, including the cytoplasm, nucleoplasm, ER lumen, ER membrane, and the NE. Here, we review the protein degradation machineries of the ER and NE and the underlying mechanisms dictating recognition and processing of substrates by these machineries.
Keywords: Protein degradation, ER-associated degradation, proteasome, ubiquitin, retrotranslocation, endoplasmic reticulum
1. Introduction
The endoplasmic reticulum (ER) is a massive intracellular organelle composed of a continuous membrane system that includes the peripheral ER and nuclear membranes. The nuclear envelope (NE) is a double lipid bilayer consisting of an outer nuclear membrane (ONM) and inner nuclear membrane (INM) that encapsulates the nucleus. While the ONM is generally viewed as an extension of the peripheral ER, the INM contains distinct physical characteristics along with a specific subset of membrane-residing proteins, albeit with some overlap [1]. The ER has many functions in cellular regulation [2]; perhaps the most prominent and conserved role of the ER is as the major site for protein synthesis and subsequent maturation of membrane and secreted proteins. Numerous factors present at the ER guide the proper folding and modification of nascent proteins [3]; nevertheless, many proteins fail to mature properly and need to be extracted from the ER and degraded. Additionally, protein levels must be carefully coordinated in response to environmental cues, such as nutrient availability and proteotoxic stress. Protein modification by covalent ubiquitin addition and subsequent degradation by the proteasome, as well as autophagic processes that utilize ubiquitin-like protein (Ubl)-conjugating systems are key to controlling protein levels both in the ER and elsewhere in the cell [4].
2. The Ubiquitin-Proteasome System
Protein degradation is a closely regulated process that serves to eliminate short-lived regulatory proteins as well as misfolded proteins [5]. Failure to discard misfolded proteins often leads to their accumulation as protein aggregates, a common theme in the development of many degenerative diseases [6]. The ubiquitin-proteasome system (UPS) is responsible for most selective protein degradation in eukaryotes. In this system, proteins are marked for proteasomal degradation by the covalent attachment of ubiquitin to a protein, a process termed ubiquitylation [7, 8]. Ubiquitylation begins with an ATP-dependent reaction that is catalyzed by an E1 ubiquitin-activating enzyme, leading to a high-energy thioester linkage between the ubiquitin C-terminus (Gly76) and an active-site cysteine residue of the E1. Ubiquitin is then transferred from the E1 to an active-site cysteine of a ubiquitin-conjugating (E2) enzyme. Finally, in the most common mechanism, an E3 ubiquitin ligase mediates the interaction between the E2~ubiquitin and target protein, stimulating ubiquitin transfer to the target protein, usually to a lysine side chain(s) [9]. The E3s are the largest class of enzymes in the ubiquitylation cascade, consisting of approximately 100 E3s in budding yeast and at least 600 in humans [10, 11]. Consistent with their vast number, the E3s are the primary UPS components responsible for substrate recognition and the remarkable specificity exhibited by the UPS.
Proteins marked for proteasome-mediated degradation typically contain at least one poly-ubiquitin chain or multiple mono-ubiquitin additions [12]. Poly-ubiquitin chains are formed when the C-terminus of a donor ubiquitin is attached to one of the seven lysine residues (K6, K11, K27, K29, K33, K48, and K63) or the alpha-amino group of the first methionine (M1) of an acceptor ubiquitin. Ubiquitin modifications are dynamic and can be removed through the action of deubiquitylating enzymes (DUBs) [13]. Proteins tagged with ubiquitin, most commonly in K48-linked ubiquitin chains, are recognized by the 26S proteasome and degraded in an ATP-dependent manner into peptides [14].
3. Protein Degradation at the ER
Due to the massive biosynthetic influx of proteins at the ER, an array of factors at the ER promote the proper folding of nascent proteins [3, 15]. Proteins that fail to fold properly or to be correctly modified are degraded through a branch of the UPS termed ER-Associated Degradation (ERAD), the process by which proteins are ubiquitylated at the ER membrane and subsequently degraded. ERAD has an essential role in eliminating proteins that could otherwise prove toxic. These protein quality control substrates include aberrantly folded proteins as well as protein complex subunits present in excess of their proper stoichiometry. Furthermore, ERAD regulates protein levels in response to cellular demands [16].
In the budding yeast Saccharomyces cerevisiae, there are three ER/NE membrane-resident E3 ligases responsible for the majority of the protein degradation initiated at these membranes. These include the two canonical ERAD ligase complexes centered on the Doa10 and Hrd1 E3s as well as the more recently characterized Asi complex. While Doa10 and Hrd1 have clear homologs in humans, no human orthologs for any Asi subunits have been defined [17, 18]. In addition to the mammalian Doa10 ortholog TEB4 (MARCH6) and the mammalian Hrd1 orthologs HRD1 and gp78, E3 ligases involved in mammalian ERAD include TRC8, TMEM129, Rfp2, RNF5/RMA1, RNF145, RNF170, and RNF185 [19–25]. There are likely additional ER-resident E3 ligases involved in mammalian ERAD that remain to be characterized [26]. The ERAD machinery is capable of degrading a diverse subset of proteins including those located within the ER lumen, ER membrane, NE, cytoplasm, and nucleoplasm. The present review will focus on studies that have illuminated our understanding of ERAD in yeast and mammals.
3.1. ERAD-L and the Hrd1 ubiquitin ligase complex
ERAD substrates with degradation signals (degrons) located within the ER lumen are termed ERAD-L substrates (Fig. 1). The degradation of these substrates depends on the UPS machinery, which resides primarily within the cytoplasm and nucleoplasm. All the major ERAD E3 ligases expose their catalytic RING domains on the cytoplasmic or nucleoplasmic side of the ER membrane. Therefore, ERAD-L substrates must be recognized within the ER lumen and transported across the membrane to undergo ubiquitylation at the ER surface. These substrates must then be extracted into the cytosol for proteasome-mediated degradation. Collectively, protein transport across the ER membrane and subsequent membrane extraction is termed retrotranslocation and will be discussed in Section 6.
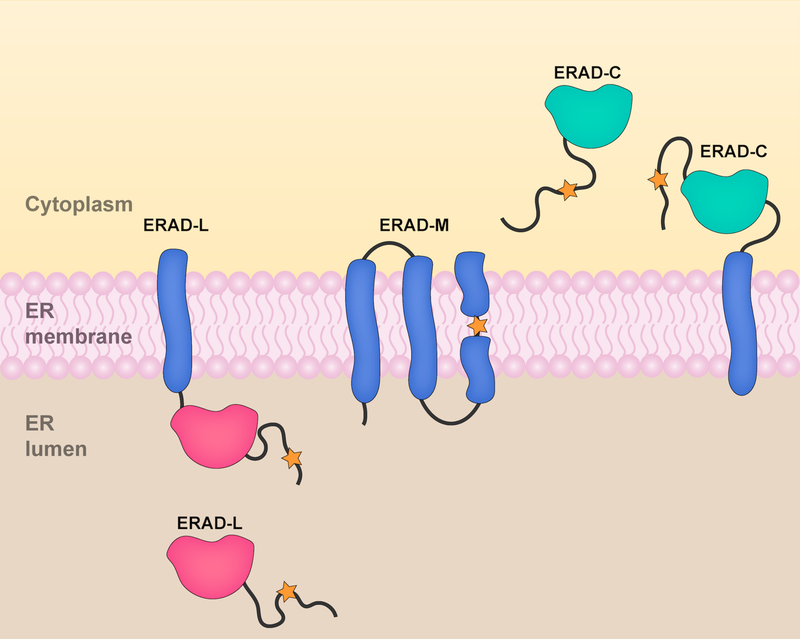
Figure 1. ERAD substrate classes.
Proteins targeted by ERAD are classified into three substrate classes based on the location of their misfolded domains or degrons (designated by an orange star). The different substrate classes bear degrons located in the ER lumen (ERAD-L), ER membrane (ERAD-M), or cytoplasm/nucleoplasm (ERAD-C). ERAD-L and ERAD-C substrates include integral membrane proteins as well as soluble proteins that are targeted for degradation at the ER membrane.
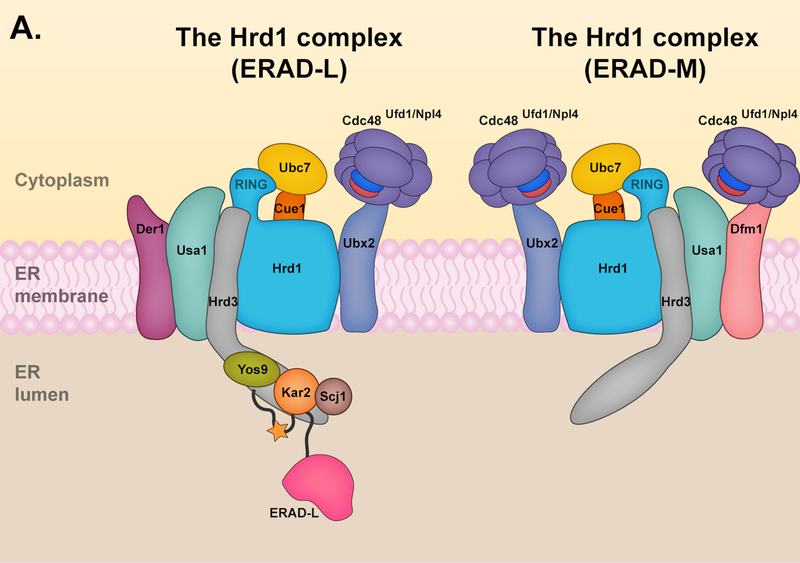
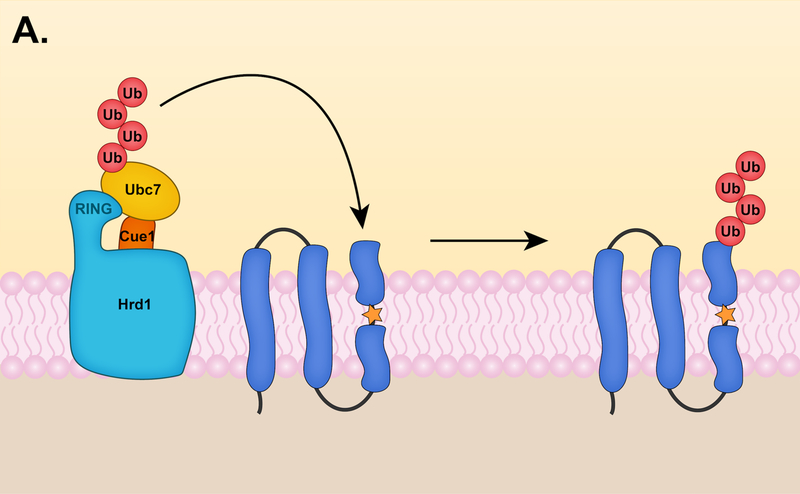
In budding yeast, the Hrd1 E3 ligase complex appears to be the only E3 responsible for targeting ERAD-L substrates (Fig. 2A). The central component of the Hrd1 complex is Hrd1, a protein with eight transmembrane helices (TMs) [27] and a catalytic RING domain [28]. The RING interacts with the E2 enzyme Ubc7 to promote substrate ubiquitylation. A transmembrane protein, Cue1, helps to tether Ubc7 to the ER membrane and is a Ubc7 activator [29]. In addition, the Hrd1 complex includes the membrane proteins Der1, Hrd3, and Usa1, all of which are involved in the degradation of ERAD-L substrates [30]. Hrd3 (SEL1L in mammals) is particularly important for the structural integrity of the Hrd1 complex and when absent, Hrd1 undergoes auto-ubiquitylation and rapid degradation [31]. Additionally, Hrd3 directly interacts with the luminal protein Yos9 (OS9 and XTP3-B in mammals). Yos9 is a lectin that recognizes misfolded glycosylated proteins in the ER lumen and delivers them to the Hrd1 complex through its interactions with Hrd3 [30, 32]. Misfolded luminal proteins can also be distinguished independently of N-glycan signals and recruited to the Hrd1 complex by the luminal Hsp70 chaperone Kar2 (BiP or Grp78 in mammals) [32].
Figure 2. The three known ER/NE-resident ERAD E3 ligases in S. cerevisiae.
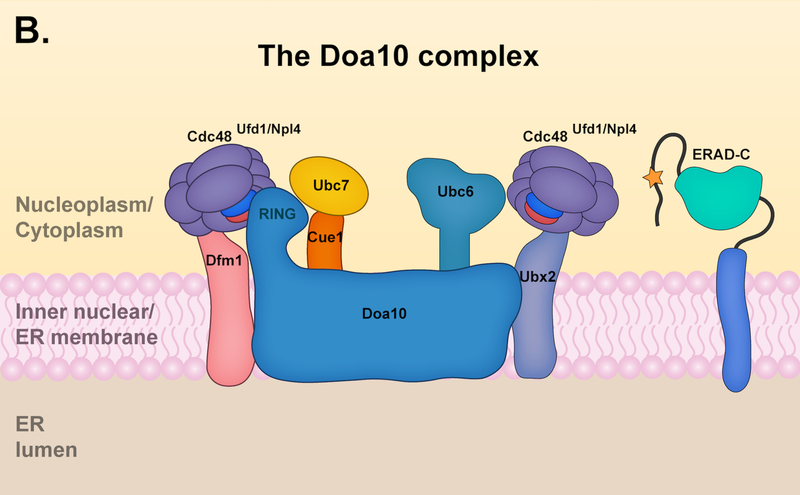
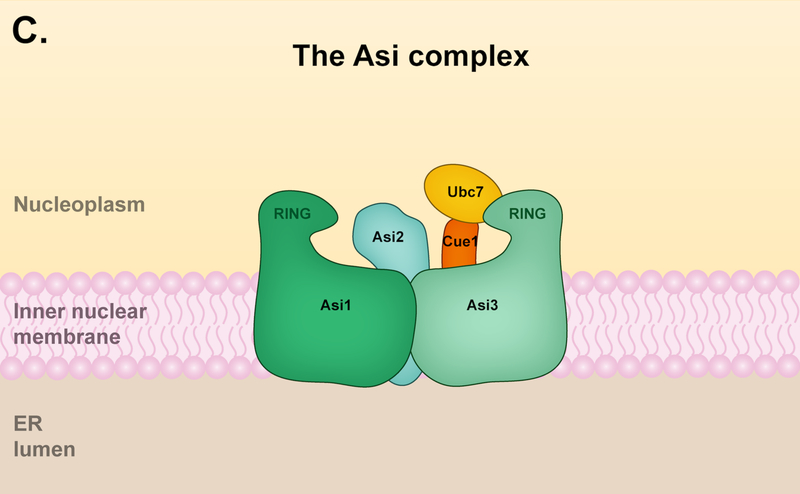
(A) The Hrd1 complex. The Hrd1 complex resides in the ER but is limited in its ability to reach the inner nuclear membrane (INM); it is capable of targeting both ERAD-L and ERAD-M substrates. The components of the Hrd1 complex involved in degrading ERAD-M and ERAD-L have some distinctions. The core components of the Hrd1 complex – Hrd1p, Hrd3 and Usa1 – are involved in both ERAD-M and ERAD-L pathways. Hrd1 functions primarily with the E2 enzyme Ubc7, which is activated and recruited to the ER membrane by the cofactor Cue1. Hrd3 is important for the structural integrity of the Hrd1 complex and substrate recruitment, while Usa1 promotes Hrd1 oligomerization. The degradation of ERAD-L substrates (left panel) requires the luminal components Yos9, Kar2, and Scj1 as well as the Derlin Der1. The luminal components and Der1 are dispensable for targeting of ERAD-M substrates (right panel) by the Hrd1 complex, which instead requires the Derlin Dfm1. Ubx2 or Dfm1 recruits Cdc48 and its associated cofactors Ufd1 and Npl4 to the ER membrane. The Cdc48 ATPase complex mediates the extraction of Hrd1 substrates into the cytosol prior to proteasomal degradation. (B) The Doa10 complex. The Doa10 complex localizes throughout the ER, including the INM. It primarily targets ERAD-C substrates, although a few ERAD-M substrates have been characterized. Doa10 function usually requires the presence of two E2 enzymes, Ubc6 and Ubc7. The Cdc48 ATPase complex contributes to the extraction of Doa10 membrane substrates into the cytosol for proteasomal degradation. The Derlin Dfm1 might also be required for the extraction of Doa10 membrane substrates. (C) The Asi complex. The Asi complex resides in the INM. Substrate features recognized by the Asi complex are less defined, but the complex might target both ERAD-C and ERAD-M substrates. Asi1 and Asi3 are essential components of the Asi complex while Asi2 is only required for the degradation of certain substrates. The Asi complex functions with the Ubc7 E2, and early characterization also suggests Ubc6 and Ubc4 involvement (not shown). The Cdc48 ATPase complex (not shown) is required for the extraction of Asi membrane substrates, but it is unclear if Ubx2 or Dfm1 is also required for this process.
Usa1 (Herp in mammals) promotes the oligomerization of the Hrd1 complex and mediates interactions between Der1 and Hrd1 [32, 33]. Der1 is required specifically for the degradation of ERAD-L substrates, yet its exact role remains unclear [32]; it may be involved in substrate recognition or the retrotranslocation of ERAD-L substrates (or both) [32, 34]. Der1 is the founding member of a group of proteins called Derlins, which are catalytically inactive members of the rhomboid protease family [35]. A Der1 paralog, Dfm1, is also present in yeast. While Dfm1 probably shares a similar topology with Der1 [36, 37], the two yeast Derlins have distinct roles in ERAD. In contrast to Der1, Dfm1 is specifically required for the degradation of Hrd1 membrane substrates and probably those of Doa10 as well (see Section 3.2) [38]. Both Derlins have been implicated in the retrotranslocation process, but for different classes of substrates. The mammalian orthologs of Der1, Derlin-1, −2, and −3, have also been implicated in the retrotranslocation of ERAD substrates (discussed in Section 6) [35, 39, 40]. In addition to their role in retrotranslocation, the Derlins are components of a complex, which includes Sec61α and HRD1, involved in degrading specific ER proteins under ER stress conditions in mammalian cells [41, 42]. In a pathway termed ER stress-induced pre-emptive quality control (ERpQC), Derlin-1 facilitates the re-routing of ER-targeted proteins from the translocation pathway to the cytosol for HRD1-mediated degradation, thereby reducing the protein folding load at the ER [41, 42].
The number of mammalian ERAD E3 ligases has expanded significantly relative to yeast. The involvement of the various mammalian E3 ligases in the degradation of luminal proteins is less defined; however, the major components of the ERAD-L machinery in S. cerevisiae are found in mammals and ERAD-L substrates are likely recognized and processed in a similar manner in yeast and mammals.
3.2. The ERAD-M and ERAD-C pathways
Other than those that are ERAD-L substrates, integral membrane proteins targeted by ERAD are typically categorized into two other subclasses based on the location of their misfolded domains or degrons. Membrane proteins containing a degron within their membrane-spanning region are termed ERAD-M substrates while those containing a degron in the cytoplasm or nucleoplasm are termed ERAD-C substrates (Fig. 1). Following their ubiquitylation, membrane substrates are retrotranslocated into the cytoplasm or nucleoplasm and degraded by the proteasome. The ERAD-C pathway also targets soluble cytoplasmic or nuclear proteins; however, soluble ERAD-C substrates do not require retrotranslocation for proteasome-mediated degradation.
The yeast Hrd1 complex is also involved in degrading ERAD-M substrates. The components of the Hrd1 complex involved in degrading ERAD-M substrates have some distinctions from those involved in ERAD-L (Fig. 2A). The core components of the Hrd1 complex – Hrd1, Hrd3 and Usa1 – are involved in both ERAD-M and ERAD-L pathways [30, 33]. By contrast, recent data suggest Dfm1 is a component of the ERAD-M pathway and not ERAD-L, which instead requires the Der1 paralog [38]. Yos9, Kar2, and Der1, which have central roles in ERAD-L, are dispensable for ERAD-M, suggesting Hrd1 detects ERAD-M substrates by a distinct mechanism.
Yeast Doa10 (degradation of alpha2) was originally identified in a screen for mutants with defects in the degradation of a soluble nuclear substrate bearing the Deg1 degron; Deg1 is a degron from the transcription factor MATalpha2 [43]. While the first Doa10 substrate identified was a soluble substrate, numerous membrane substrates were identified subsequently [44–46]. Membrane proteins containing a cytoplasmic degron (ERAD-C) are primarily targeted for degradation by the Doa10 complex in S. cerevisiae (Fig. 2B). The main component of the Doa10 complex is Doa10, a large E3 ubiquitin ligase with 14 TMs and an N-terminal RING domain [43]. Doa10 is conserved throughout eukaryotes and the human Doa10 ortholog TEB4/MARCH6 appears to contain a similar membrane topology and localization [47, 48].
As noted above, Doa10 functions with two E2s, Ubc6 and Ubc7 [49]; both have mammalian orthologs. The Ubc7 ortholog, Ube2g2, associates with TEB4 to promote K48 chain formation in vitro [50]. Ubc6 is a tail-anchored membrane protein that contains two mammalian orthologs, Ube2j1 and Ube2j2, which were originally thought not to function with TEB4 [51]; however, a recent study determined that Ube2j2 works with TEB4 to degrade substrates containing the cytosolic CL1 degron, a Doa10-dependent degron in yeast [52, 53]. Moreover, depletion of Ube2j2 increases protein levels of another TEB4 substrate, squalene monooxygenase (SM), as occurs following proteasomal inhibition or TEB4 depletion, suggesting Ube2j2 also functions with TEB4 to degrade SM. While depletion of the E2 enzyme Ube2D3 was required for complete stabilization of mCherry-CL1 through the TEB4 pathway, Ube2D3 depletion did not significantly affect SM protein levels [52]. More work is needed to fully understand which E2s operate with TEB4 and how these may differ depending on the substrate.
Until recently, Doa10 and Hrd1 were thought to be the only ER-resident E3 ligases participating in ERAD in budding yeast; however, recent studies have identified the transmembrane Asi complex as part of a previously uncharacterized ERAD pathway operating exclusively at the inner membrane of the NE (Fig. 2C) [17, 18]. Because of this spatial restriction, it might be useful to call it NE-associated degradation (NEAD). The components of the Asi complex were originally identified as amino acid sensor-independent (ASI) genes that negatively regulate an amino acid-sensing pathway by degrading the transcription factors Stp1 and Stp2 [54, 55]. The Asi complex comprises Asi1, Asi2, and Asi3, all integral membrane proteins that localize to the INM [56, 57]. Asi2 has two transmembrane segments, while the paralogous Asi1 and Asi3 each have five apparent TMs and a C-terminal RING domain [54, 57]. The RING domains interact with the E2s Ubc7, Ubc6, and, to a lesser extent, Ubc4 [18]. Notably, deletion of UBC7 only partially blocks the degradation of the Asi substrate Erg11, suggesting the Asi complex works with multiple E2 enzymes [17]. Although Asi2 is required for Erg11 degradation, Asi2 is dispensable for other substrates, implying a regulatory or structural role specific to certain substrates [17, 18]. The Asi complex has no clear mammalian orthologs [17, 18]; however, E3 ligases dedicated to maintaining protein quality control at the INM are likely also present in mammals.
3.3. Other ERAD pathways
In addition to the canonical ERAD-L, -C, and -M pathways, recent studies have provided insight into more specialized ERAD pathways mediating protein quality control at the ER. One of these newly characterized pathways involves the degradation of ribosome-associated polypeptide chains at the ER membrane, which has been termed ERAD of ribosome-associated proteins (ERAD-RA)[58]. ER-targeted polypeptides within stalled ribosome complexes are marked for proteasomal degradation by the E3 ligase Ltn1/Rkr1 in budding yeast [58]. Ltn1 interacts with ribosomes and has a general role in mediating ribosome-associated protein quality control [59, 60]. The ERAD-RA pathway is likely conserved throughout eukaryotes as the Ltn1 mammalian ortholog Listerin also appears to mediate ERAD-RA [61].
ERAD-T (translocon-associated) is another ERAD pathway that mediates the degradation of proteins associated with the translocon, which may arise from aberrant interactions as well as abnormalities in the translocation process. The Hrd1 complex is capable of targeting ERAD-T substrates and likely has a general role in clearing proteins aberrantly associated with the translocon [62]. The intramembrane protease Ste24 is also involved in clearing proteins that aberrantly interact with and obstruct the translocon due to defects in signal recognition particle (SRP)-independent translocation [63]. Ste24 interacts with obstructed translocons and cleaves “clogging” substrates, generating fragments that are ultimately degraded by the proteasome [63]. Hrd1 and Ste24 might work in separate pathways to eliminate translocon-associated substrates with distinct characteristics. Many of the molecular mechanisms governing the recognition and processing of ERAD-T substrates remain to be determined.
4. Substrate Recognition
A fundamental task of the ERAD machinery is to distinguish its substrates from other proteins it will encounter in the cell. Substrates include misfolded proteins as well as regulatory proteins whose levels must be tightly controlled. Sequences sufficient to induce degradation when appended to normally stable proteins are called degradation signals or degrons [14]. Here we discuss general properties of characterized degrons from the different ERAD pathways and how they are thought to be recognized.
4.1. Targeting of ERAD-L Substrates
ERAD-L substrates are targeted for degradation through recognition mechanisms that rely on protein folding or glycosylation status. N-linked glycosylation is mediated by the multimeric oligosaccharyltransferase complex (OST), which attaches a branched glycan moiety (Gly3Man9GlcNAc2) onto nascent polypeptides bearing a N-linked glycosylation site (N-X-S/T) [64]. N-linked glycans are trimmed by a series of ER-resident glycosidases to generate glycan moieties that either promote protein folding or entry into ERAD [65]. Early-acting glycosidases generate N-linked glycans that promote protein folding through the action of lectin chaperones [65]. Conversely, glycoproteins retained in the ER lumen are processed by late-acting glycosidases to generate a glycan degradation signal (Man7GlcNAc2) [66, 67]. A complex consisting of the mannosidase Htm1 and the protein disulfide isomerase Pdi1 (Htm1-Pdi1) preferentially recognizes misfolded glycoproteins containing Man8GlcNAc2 and processes them into Man7GlcNAc2; the latter is recognized by the Hrd1 complex through the lectin Yos9 (OS9 and XTP3-B in mammals) [68–70]. Binding of Htm1 to Pdi1 is required for Htm1 mannosidase activity, and disruptions of this interaction impair ERAD of glycosylated substrates [69, 70]. The mammalian counterpart of Htm1, EDEM1, forms a complex with the disulfide reductase ERdj5, and EDEM1–3 mannosidase activity mediates ERAD of misfolded glycoproteins [71–73]. Most N-linked glycan structures found in yeast are also present in mammals [65], suggesting similar substrate recognition mechanisms for detecting misfolded glycoproteins.
An early example of an ERAD-L substrate influenced by N-linked glycosylation was a mutant form of the vacuolar protease carboxypeptidase Y (CPY*), which fails to fold properly in the ER lumen and is subsequently degraded by the Hrd1 pathway [74–76]. CPY* contains four N-glycosylation sites, and their glycosylation enhances CPY* degradation rates [77]. Like other ERAD-L substrates, CPY* can be degraded independently of N-glycosylation, albeit more slowly [77]. Yos9 and Hrd3 can both detect CPY* independently of N-linked glycosylation [32]. Additionally, the Hsp70 chaperone Kar2 (BiP or Grp78 in mammals) also detects misfolded proteins independently of N-linked glycans and is required for the degradation of ERAD-L substrates [32, 78, 79]. Kar2 interacts with Yos9/Hrd3 and may act to recruit terminally misfolded proteins to the Hrd1 complex for ubiquitylation [32]. Genetic analyses had suggested Kar2 works in conjunction with the Hsp40 co-chaperones Scj1 and Jem1 [78, 80], but a more recent study determined that Scj1, but not Jem1, is required for the degradation of ERAD-L substrates such as CPY* [81]. Interestingly, Scj1 binds Hrd3; in its absence, CPY* degradation is blocked and the interaction between CPY* and Hrd3 is stabilized. Scj1 is not required for substrate recognition and instead could mediate substrate release from Hrd3/Kar2 [81]. Der1 is situated near Hrd3 and might facilitate substrate delivery and/or Hrd1-mediated retrotranslocation following substrate release by Scj1 [82].
O-mannosylation is another post-translational modification that influences protein folding and substrate recognition by the ERAD machinery. This type of glycosylation is catalyzed by protein O-mannosyltransferases (PMTs), which attach a short oligomannose chain to the hydroxyl group of serine or threonine residues [83]. It has been suggested that O-mannosylation terminates repeated cycles of chaperone-assisted folding, thereby directing misfolded proteins to ERAD [84]. O-mannosylation contributes to the degradation of several luminal proteins [85–87]; however, it remains unclear how ERAD substrates are targeted for O-mannosylation and how the ERAD machinery recognizes these modified substrates. The Pmt1-Pmt2 complex interacts with the Hrd1 complex; potentially, O-mannosylation of misfolded substrates bound to Hrd3/Kar2 commits them to ERAD instead of refolding [85]. A full understanding of the role of O-mannosylation in ER homeostasis and substrate recognition by the ERAD machinery will require further investigation.
In summary, ERAD-L substrates are recognized based on the presence of misfolded luminal domains and specific glycan signals that are also conserved in mammals [65, 83]. While these central recognition features are likely conserved throughout eukaryotes, previous studies have suggested additional mechanisms underlying the detection of ERAD-L substrates in mammalian cells [88, 89]. The recognition of misfolded luminal proteins in yeast is mediated by components of the Hrd1 complex, including Hrd3, Yos9, Kar2, and possibly Der1. Following substrate recognition, ERAD-L substrates must be transported from the ER lumen to the cytosolic face of the ER for ubiquitylation. These processes will be discussed in Sections 5 (Substrate Ubiquitylation) and 6 (Retrotranslocation).
4.2. ERAD-M Substrates
ERAD-M substrates are targeted for degradation based on the presence of intramembrane lesions. ERAD-M degrons appear to be characterized by hydrophilic residues within intramembrane regions, whereas degrons immersed in aqueous compartments often contain exposed hydrophobic regions as key elements. An early example of an ERAD-M substrate was yeast HMG-CoA reductase 2 (Hmg2), a sterol biosynthetic enzyme whose degradation is mediated by the Hrd1 pathway in a sterol-dependent manner [90]. The membrane accumulation of downstream products of the mevalonate pathway leads to structural changes in Hmg2 that promote its recognition by the Hrd1 pathway [91].
Degradation of mammalian Hmg2 (HMGR) is similarly regulated in a sterol-dependent manner and apparently by multiple ERAD pathways, including the gp78, TRC8, and TEB4 pathways [92–94]. Sterol-stimulated degradation of HMGR requires Insig-1 and its paralog Insig-2 [95]. Insig1/2 associates with gp78 as well as TRC8 and mediates HMGR recruitment through interactions with the sterol-sensing domain of HMGR in the presence of sterols [92, 93]. RNF145 is a more recently characterized membrane-residing E3 that is involved in sterol-stimulated HMGR degradation and was also found to interact with Insig-1/2 [24, 96]. The involvement of additional E3 pathways in HMGR turnover, like RNF145, could explain previous results from a study in gp78 (−/−) mouse embryonic fibroblasts that argued against gp78 and TRC8 involvement in sterol-stimulated HMGR degradation [97]. The yeast ortholog of Insig-1, Nsg1, also binds Hmg2 in a sterol-dependent manner; however, these interactions block Hmg2 degradation instead of enhancing it [98].
Recent evidence suggests ERAD-M substrates can be generated by ER-localized intramembrane proteases. Signal peptide peptidase (SPP) is an intramembrane protease primarily known for its role in cleaving signal peptides of secreted proteins; however, SPP has been implicated in a variety of other processes, including ERAD [99]. SPP interacts with Derlin-1 and the E3 TRC8 to form a complex capable of targeting membrane proteins for degradation [100, 101]. In the case of the tail-anchored membrane protein heme oxygenase-1 (HO-1), SPP cleavage generates HO-1 fragments that are recognized for degradation by the TRC8 and TEB4 pathways [52, 101].
The involvement of intramembrane proteases in ERAD is likely conserved throughout eukaryotes. For instance, in yeast the intramembrane protease Ypf1 and the Doa10 complex mediate the degradation of the zinc transporter Zrt1 [102]. Notably, Dfm1 is required for maximal degradation of Zrt1, and human Derlin-1 has been implicated in SPP-mediated ERAD [100, 102]. The role of Dfm1 and Derlin-1 in these cases is unclear. The Derlins might be required for the retrotranslocation of substrate fragments following intramembrane cleavage and ubiquitylation. Alternatively, the Derlins could be required prior to SPP substrate cleavage, as has been suggested for a transmembrane form of XBP1u, a regulator of the ER unfolded protein response (UPR) [100]. Overall, these studies suggest intramembrane proteases can generate ERAD substrates by disrupting the membrane structure or topology of membrane proteins. The mechanisms governing substrate recognition by intramembrane proteases as well as the prevalence of intramembrane proteases in ERAD are active areas of investigation.
How are intramembrane lesions detected by membrane-resident E3 ligases? Unlike the Hrd1 ERAD-L pathway, the recognition of ERAD-M substrates does not require the presence of the luminal factors Kar2 and Yos9 [32]. Instead, the recognition of ERAD-M substrates is likely mediated through the transmembrane domain of Hrd1. Hrd1 contains numerous conserved hydrophilic residues within its TMs, and alterations of these residues lead to substrate-specific degradation defects in the ERAD-M pathway. This suggests the transmembrane segments of Hrd1 itself, rather than an adaptor, may directly bind to ERAD-M substrates [103].
The Hrd1 complex was thought to be responsible for the ubiquitylation of all ERAD-M substrates in yeast. Recent studies, however, suggest both the Doa10 and Asi complexes contribute to the degradation of certain ERAD-M substrates [17, 104]. A mutant subunit of the translocon, Sec61–2, is an ERAD-M substrate whose degradation is primarily mediated by the Hrd1 pathway; however, appending a nuclear localization signal to Sec61–2 results in degradation by both the Asi and Hrd1 pathways, suggesting the Asi complex is capable of detecting ERAD-M degrons in the INM [17]. The mutant Sec61–2 protein contains a single point mutation (G213D) affecting a cytosolic residue near a TM, and it has been suggested this point mutation disrupts the TMs within Sec61 to generate an ERAD-M substrate [45, 78]. It is possible that the Asi and Hrd1 complexes recognize distinct features within Sec61–2. Another Asi substrate is the transcription factor Stp1, which contains an N-terminal degron termed RI that is predicted to form an amphipathic helix [55]. Amphipathic helices are a common feature of ERAD-C degrons (see Section 4.3), suggesting the Asi complex might also recognize ERAD-C substrates. A more detailed analysis of additional Asi degrons is required for understanding the exact degron features recognized by this E3.
4.3. ERAD-C Substrates
Yeast ERAD-C substrates contain degrons exposed to the cytoplasm or nucleoplasm and are primarily degraded through the Doa10 pathway. Doa10 localizes throughout the ER membrane, including the INM, and the ability of Doa10 to localize to the INM is required for efficient degradation of nuclear substrates [105]. The best-studied Doa10-dependent degron is Deg1, from the short-lived transcription factor MATalpha2 [43, 106]. Deg1 includes a predicted amphipathic helix, and mutations on the hydrophobic face of this helix strongly impair Doa10-dependent degradation [107, 108]. N-terminal acetylation of Deg1 was also proposed to be a central feature of its recognition by Doa10 [109], but this result could not be reproduced [37].
Additional Doa10-dependent degrons have been identified that share similar characteristics to Deg1. The Doa10-dependent CL1 degron is a cytosolic degron also predicted to form an amphipathic helix at the C-terminus of proteins bearing this sequence [45, 110]. Similar to Deg1, mutagenesis of CL1 hydrophobic residues impairs proteasomal degradation in yeast as well as mammalian cells [52, 111]. Another Doa10-dependent degron is DegAB, which is derived from the yeast kinetochore protein Ndc10. DegAB is a nuclear degron consisting of two elements, DegA and DegB, both of which are necessary for Doa10-dependent degradation [112]. The Doa10 complex most likely recognizes DegA, an element composed of two amphipathic helices, while DegB is required at a post-ubiquitylation step, possibly the initiation of proteasomal degradation [113].
Characterization of these degrons has revealed common properties of many Doa10-dependent degrons. In particular, they all contain predicted amphipathic helices, and disruptions of the hydrophobic surfaces of these helices impair Doa10-dependent degradation. This suggests that exposed hydrophobicity in helical elements is a major determinant for Doa10 substrate recognition. The importance of hydrophobicity for substrate recognition by the Doa10 pathway was recently validated in an elegant high-throughput analysis of unstable reporters [114].
While Doa10 primarily targets ERAD substrates with degrons exposed to the cytoplasm or nucleoplasm, Doa10 can also recognize specific ERAD-M substrates [104, 115]. The first evidence suggesting Doa10 could target intramembrane degrons came from studies showing that the Doa10 cofactor Ubc6 is a short-lived protein and that its C-terminal membrane anchor is likely to be part of the relevant degron [46, 116]. Ubc6 is an unusual substrate in that it also functions as an E2 enzyme within the Doa10 complex, so it was unclear if Doa10 could recognize additional ERAD-M degrons. Notably, a recently identified Doa10 substrate, the tail-anchored membrane protein Sbh2, which is a subunit of the Ssh1 alternative translocon, was demonstrated to contain an intramembrane degron [104]. Doa10 also mediates the degradation of several lipid droplet proteins, including Pcg1, through recognition of a C-terminal hydrophobic hairpin that mediates association with the membrane [115].
The identification of Doa10 substrates containing ERAD-M degrons suggests Doa10 can recognize a broader spectrum of substrates than previously thought. While cytosolic and membrane degrons are placed into separate categories, it is possible these degrons share certain features. It had previously been noted that a Doa10 degron containing an amphipathic helix could associate tightly with membranes [117]; it therefore might engage Doa10 in a manner similar to the aforementioned ERAD-M substrates. Similarly, the amphipathic CL1 degron associates with membranes in mammalian cells and mutations in hydrophobic residues that disrupt membrane association also impair degradation [52].
Although several shared characteristics of Doa10-dependent degrons have been defined, it remains unclear if Doa10 directly interacts with substrates. Doa10 is a large protein that contains 14 TMs, and this membrane topology is highly conserved. It is possible that Doa10 interacts with substrates through its transmembrane segments, possibly forming an internal channel [43], similar to what has been suggested for the E3 ligase Hrd1 [27]. Conserved regions within Doa10 that might have a role in substrate recognition include the TEB4-Doa10 (TD) domain, which includes three TMs, and the highly conserved 16-residue C-terminal element (CTE) [47, 48].
Another possibility is that Doa10 substrate recognition is mediated through adaptor proteins, such as molecular chaperones. Degradation of several Doa10 substrates requires Hsp40 and Hsp70 molecular chaperones [44, 53, 118]. While the Hsp40 co-chaperone Ydj1 is required for the degradation of Ste6* and CL1-containing substrates [44, 53], it is dispensable for the degradation of DegAB- and Deg1-bearing substrates, which instead require the Hsp40 Sis1 [118]. It is worth noting that the E3 ligase Ubr1, which contains some substrate overlap with Doa10 [115, 119], also requires Ydj1 and Sis1 for the degradation of substrates [120]. A recent study showed that Ubr1 concentrates in the nucleus, and the Ubr1 pathway requires Ydj1 and the Hsp110 Sse1 for nuclear import of substrates [121]. Sis1 also mediates the nuclear import of misfolded proteins, but appears to be required at an additional step following nuclear import for the Ubr1 and San1 degradation pathways [121, 122]. Nuclear export, in addition to nuclear import, has been shown to be important for the degradation of some UPS substrates in yeast and mammals, although no ERAD substrate has been reported to require such export [123–125]. It is unlikely that the Doa10 pathway requires chaperones solely for nuclear import as Doa10 can target substrates in both the nucleus and cytoplasm [105]. These chaperones might be required simply for maintaining substrate solubility, which could be necessary for their ubiquitylation by Doa10. It is also possible that these chaperones act as adaptor proteins that deliver substrates or mediate their binding to Doa10 [51, 118, 126].
5. Substrate Ubiquitylation
The ERAD machinery generally ubiquitylates substrates at the cytoplasmic/nuclear face of the ER/NE. The primary components catalyzing substrate-specific ubiquitylation are the membrane-residing E3 ligases along with their cognate E2 enzymes. Ubiquitin can be attached to substrates in a variety of ways, many of which are recognized by the proteasome. Attachment of a single ubiquitin (mono-ubiquitylation) to a protein is not usually sufficient for recognition by the proteasome and is typically involved in other regulatory processes; however, mono-ubiquitylation at multiple sites within a protein can target it for proteasomal degradation [127]. More commonly, ubiquitin is attached to proteins in the form of ubiquitin chains, which are formed when the C-terminus of a donor ubiquitin (G76) is attached to one of the seven lysine side chains (isopeptide linkage) or the alpha-amino group of the first methionine of an acceptor ubiquitin. Poly-ubiquitin chains can be homotypic or heterotypic: the former is composed of a single linkage type while the latter contains multiple linkage types and exhibits a branched topology [127]. E2 enzymes possess specific structural properties that dictate the ubiquitin-ubiquitin linkage type [128].
The primary yeast E2 enzymes involved in ERAD are Ubc6 and Ubc7 (with some contribution by Ubc1), which have distinct catalytic properties [12]. Ubc7 functions in every known ERAD pathway and like its mammalian ortholog, Ube2g2, forms K48-linked chains in vitro [129, 130]. These K48-linked chains can be assembled onto the active site cysteine of Ube2g2 and transferred to a lysine residue of a substrate en bloc (Fig. 3A) [129]. Ube2g2 forms dimers in vitro, and this dimerization is required for ERAD in vitro [131]. Yeast two-hybrid analysis suggests Ubc7 also can form dimers [49].
Figure 3. Models for ERAD substrate ubiquitylation.
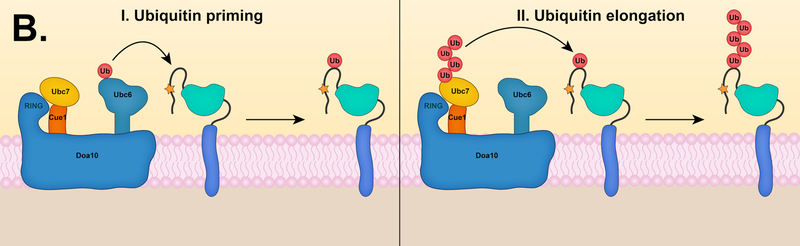
(A) Substrate ubiquitylation by Ubc7 and Hrd1. Ubc7 is thought to assemble K48-linked chains onto its active site cysteine residue, followed by en bloc chain transfer to a lysine residue of a substrate. Stepwise ubiquitin chain assembly on the substrate could also occur. (B) Sequential ubiquitylation mechanism by Ubc6 and Ubc7 in the Doa10 pathway. These E2 enzymes might target substrates by a sequential E2 mechanism, where Ubc6 and Ubc7 function at distinct steps. (I) In the first step, Ubc6 attaches a single ubiquitin molecule to a lysine, serine, or threonine residue of a substrate in a process called ubiquitin priming. Ubc6 is capable of forming K11-linked chains, and ubiquitin priming might involve the attachment of short K11-linked chains. (II) After substrates are modified with either a single ubiquitin or short K11-linked chain, Ubc7 transfers ubiquitin molecules stepwise or K48-linked chains en bloc to the initial ubiquitin (shown here).
Cue1 is a Ubc7 activator that tethers Ubc7 to the yeast ER membrane through interactions mediated by a C-terminal Ubc7-Binding Region (U7BR) [132]. Mammalian Ube2g2 is recruited to membranes through an analogous binding region, Ube2g2-Binding Region (G2BR), which can also be found in the E3 gp78 and the lipid droplet protein AUP1 [133, 134]. Structural analysis revealed the G2BR of gp78 binds the “backside” of Ube2g2, the face of the E2 opposite to its catalytic-site cysteine, and this interaction stimulates Ube2g2 activity in vitro [135]. Ubc7 interacts with the U7BR domain of Cue1 through a similar backside mechanism to stimulate ubiquitin transfer from Ubc7~ubiquitin to free or K48-linked ubiquitin [136]. Furthermore, the U7BR domain also increases Ubc7 binding to the RING domains of Doa10 and Hrd1 [136]. In addition, Cue1 bears a ubiquitin-binding CUE domain that preferentially binds K48-ubiquitin chains and promotes Ubc7-mediated K48-chain formation in vitro [137]. More recent work suggests the CUE domain specifically binds the ubiquitin moiety adjacent to the acceptor ubiquitin of a growing chain, which aligns Ubc7 with the distal end of the Ub chain and stimulates chain elongation [138]. Thus, the CUE domain promotes K48-chain formation and this function is required for the degradation of ERAD substrates [137]. While there is no apparent mammalian ortholog of Cue1, the E3 gp78 contains a CUE domain that promotes K48-chain formation in vitro [131].
The other major ERAD E2, Ubc6, is an integral component of the Doa10 pathway and early characterization of the Asi pathway also suggests Ubc6 involvement [17, 18]. Ubc6 and Ubc7 are both required for efficient degradation of most Doa10 substrates. This raises the question of why these ERAD pathways should require multiple E2s. These E2s have distinct catalytic properties. Ubc7 only synthesizes K48-linked chains, but this is not true for Ubc6. A stable isotope labeling with amino acids in cell culture (SILAC) experiment showed a ~40–50% global decrease in K11-chain formation in yeast strains lacking Ubc6 or Doa10 [12]. The degradation of Ubc6 was also significantly reduced in vivo in a K11R-ubiquitin mutant. These data suggest Doa10 acts with Ubc6 to assemble K11 chains [12]. K11 chains are rarely longer than dimers in yeast cells [139]; Ubc7 might attach K48 ubiquitin chains to Ubc6-catalyzed K11 dimers already linked to a substrate. Interestingly, the prevalence of K11 linkages increases in yeast and mammalian cells under ER stress conditions, suggesting K11-chain formation is an important component of ERAD [12, 140].
More recent work supports a sequential E2 model (Fig. 3B) based on in vitro ubiquitylation analysis with the Doa10 RING domain, Ubc7/Cue1ΔTM, and Ubc6ΔTM. Ubc7 only adds K48-chains onto the Doa10 RING domain if the RING is first mono-ubiquitylated by Ubc6 [141]. This model was also supported in vivo through degradation analysis of Sbh2 and Ubc6; attachment of a non-cleavable ubiquitin to the N-terminal of Sbh2 or Ubc6 leads to efficient degradation in the presence of Ubc7 alone [141]. These data suggest Ubc6 functions primarily in “ubiquitin priming,” or the initial ubiquitin attachment to a substrate. Interestingly, Ubc6 as well as its mammalian ortholog, Ube2j2, can attach ubiquitin not only to lysine amino groups but also to the hydroxyl side chains of serine and threonine residues, increasing the number of potential priming sites [141, 142]. E2 association sites within Doa10 are unknown, but yeast two-hybrid analysis implies that Ubc6 and Ubc7 function in the same Doa10 complex [49]. Finally, since interaction data suggest the Asi complex may also use multiple E2s [17, 18], it is possible that a sequential E2 mechanism might also operate in this pathway [141, 143].
6. Retrotranslocation
Nascent protein translocation involves the insertion of integral membrane proteins into the ER membrane or the transport of luminal and secretory proteins to the ER lumen [144]. Elimination of membrane or luminal proteins by ERAD requires the reverse of this process, known as retrotranslocation [145]. The molecular mechanisms regulating this process remain obscure. The Sec61 translocon has a protein-conducting channel that provides the route for translocation of nascent proteins into the ER, usually cotranslationally, and it has been postulated that a protein-conducting channel with similar properties would be required for retrotranslocation [144, 146]. Additionally, proteasomes have also been suggested to facilitate the retrotranslocation of certain ERAD substrates [147–149]. In this section, we discuss current ideas on how the different ERAD pathways might drive substrates through the ER membrane back into the cytosol for degradation by the proteasome [145, 150].
6.1. Retrotranslocation of ERAD-L substrates
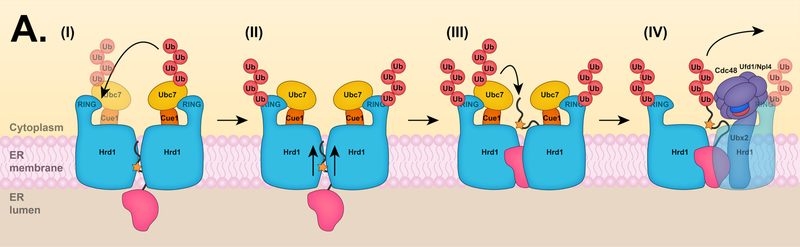
ERAD-L substrates must be able to dock on the luminal side of the ER membrane, initiate movement across the bilayer, and become decorated with the appropriate ubiquitin modifications on the cytoplasmic side of the ER for full extraction from the membrane and degradation by the proteasome. Probably the most mysterious of these steps has been the traversal of the lipid bilayer by an initially at least partially folded and often heavily glycosylated soluble protein. Over the years, several proteins have been proposed to serve as a retrotranslocation channel for ERAD-L substrates, including the Sec61 translocon, the Derlins, and Hrd1 itself [146]. Hrd1 is a promising candidate for this role. First, overexpression of Hrd1 bypasses the requirement of other Hrd1 complex membrane components, including Hrd3, Usa1, and Der1, for the degradation of ERAD-L substrates [151]. Moreover, Hrd1 interacts directly with ERAD-L substrates through its transmembrane domain [151, 152]. More recently, CPY* fused with a transmembrane domain (CPY*-TM) was reconstituted into proteoliposomes with Hrd1 [153]. Hrd1 alone catalyzes the retrotranslocation of CPY*-TM by a mechanism that requires auto-ubiquitylation of specific Hrd1 RING residues (Fig. 4A) [153].
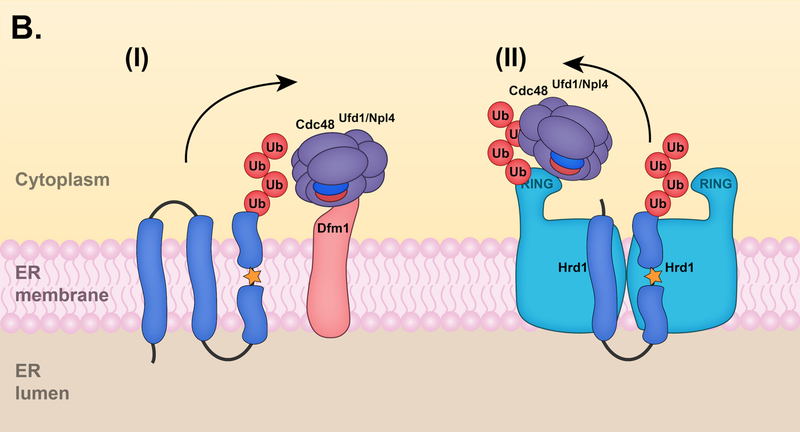
Figure 4. Hrd1 retrotranslocation models.
(A) Retrotranslocation of ERAD-L substrates. (I) After an ERAD-L substrate is recognized by the Hrd1 complex, Hrd1 undergoes auto-ubiquitylation at specific RING residues. (II) Hrd1 auto-ubiquitylation promotes the movement of the substrate through the Hrd1 protein channel. (III) Hrd1 ubiquitylates the substrate on a cytosolically exposed region. (IV) At the final step of retrotranslocation, the Cdc48 ATPase complex is recruited to the ubiquitylated substrate and mediates substrate extraction into the cytosol, allowing for proteasomal degradation. Proteasomes may in some cases directly mediate substrate extraction. (B) Retrotranslocation of ERAD-M substrates. (I) Dfm1 can mediate the retrotranslocation of membrane proteins in the absence of Hrd1. Following E3-mediated ubiquitylation, Dfm1 recruits the Cdc48 ATPase complex to the ER membrane and facilitates the retrotranslocation of membrane proteins. (II) Alternatively, Hrd1 might also facilitate retrotranslocation of certain membrane proteins in the absence of Dfm1. Hrd1 auto-ubiquitylation probably recruits the Cdc48 complex to the ER membrane for ER extraction. The Cdc48 cofactor Ubx2 (not shown) is dispensable for the retrotranslocation of ERAD-M substrates in the absence of Dfm1. Other membrane-resident ERAD E3 ligases, such as Doa10, might also directly mediate retrotranslocation of their respective substrates.
A structure for Hrd1-Hrd3 was recently determined by electron cryomicroscopy (cryo-EM) methods to an average resolution of ~4 Å [27]. This structure revealed a Hrd1 dimer in which each protomer has eight TMs, six of which are arranged to form a pair of symmetry-related aqueous funnels that are sealed at the end nearer to the luminal side and lined with conserved hydrophilic residues [27]. The putative Hrd1 channels may permit entry of substrate elements with specific characteristics, such as hydrophilic residues that can transiently interact with hydrophilic residues in the transmembrane segments of the channel. The function of Hrd1 dimerization is unclear from the structure. ERAD-M substrates might enter the Hrd1 channel through a lateral gating mechanism, analogous to the movement of integral membrane proteins through the Sec61 lateral gate during protein translocation [27, 144]. Hrd1 contains conserved residues within its transmembrane domain that are found in the mammalian orthologs of Hrd1 (HRD1 and gp78) and other mammalian E3 ligases, such as RNF145 and TRC8, suggesting these membrane-resident E3 ligases could also form aqueous channels facilitating substrate retrotranslocation [27].
Additional components of the Hrd1 complex likely contribute to retrotranslocation. For instance, the polytopic membrane protein Der1 interacts with Hrd1 through the scaffold protein Usa1 [33, 81]. Der1 is required specifically for the degradation of ERAD-L substrates and its TMs interact directly with these substrates [32, 82]. Derlins might also be involved in mammalian ER retrotranslocation, and this function is likely conserved across eukaryotic Derlins, including yeast Der1 and Dfm1 (addressed below in Section 6.2).
6.2. Retrotranslocation of membrane substrates
While recent studies have provided strong support for a model in which Hrd1 acts as a retrotranslocation channel, other results involving a chimeric Hrd1 protein suggest otherwise, at least for ERAD-M substrates. A chimera composed of the Hrd1 RING domain fused to a stable ER membrane protein (Hmg1) undergoes auto-ubiquitylation, membrane extraction, and degradation in the absence of Hrd1 [154]. How then is this chimeric protein, also referred to as a self-ubiquitylating substrate (SUS), being retrotranslocated in the absence of the Hrd1 transmembrane domain? A recent genomic screen revealed that the Derlin Dfm1 is required for degradation of SUS [38]. Dfm1 is an integral membrane protein that binds the Cdc48 ATPase complex (discussed in Section 6.3) through two C-terminal SHP-box motifs [35].
The involvement of Dfm1 in ERAD has been controversial. While some groups could link Dfm1 to ERAD [102, 155], others observed no degradation defects in dfm1 null cells [47, 156]. In their recent study, Neal et. al determined that Dfm1 is required for the retrotranslocation of numerous integral membrane substrates, including several Hrd1 substrates as well as the ERAD-C substrate Ste6* [38]. Dfm1 function requires sequences conserved in the rhomboid and pseudorhomboid proteins as well as its SHP-box motifs [35, 38–40]. Furthermore, Dfm1 is important for Cdc48 association with the membrane, a function that was previously attributed to the Cdc48 cofactor Ubx2 [157, 158]. The exact mechanism of Dfm1-mediated retrotranslocation is still unclear; however, the Derlins have been proposed to aid in the recognition of misfolded substrates as well as promote their retrotranslocation through TM unwinding and local membrane thinning [159].
Conflicting results from earlier studies could be explained by the rapid phenotypic reversion that dfm1 null cells undergo upon ERAD-M substrate overexpression. Suppression of the dfm1 null phenotype occurs by increasing Hrd1 levels through chromosomal duplication, which also restores Cdc48 association with the membrane [38]. These results suggest Hrd1 may not normally be essential for the retrotranslocation of ERAD-M substrates, in contrast to ERAD-L substrates (Fig. 4B). Other integral membrane ERAD ligases may also form protein-conducting channels that facilitate ER extraction of their respective substrates, as previously suggested for Doa10 [43]. Whether Dfm1 is required for the retrotranslocation of all integral membrane substrates, including those targeted specifically by the Doa10 and Asi degradation pathways, remains to be determined. It is possible that different proteins promote retrotranslocation at different stages of the process, particularly for ERAD-L substrates. For instance, a substrate might be transferred between different translocation factors or channels may be formed from multiple components.
6.3. The Cdc48/p97 ATPase complex
Cdc48 (p97 or valosin-containing protein (VCP) in mammals) is a homohexameric AAA+ ATPase that separates polypeptides from within protein complexes or from membranes. This “segregase” activity is required for many cellular processes [160]. Cdc48 and its essential co-factors Ufd1 and Npl4 are a central component of the ERAD machinery required for the retrotranslocation of many luminal and membrane substrates. Cdc48Ufd1-Npl4 is a cytosolic protein complex recruited to the ER membrane for its role in ERAD. This recruitment is likely mediated by several receptors in the membrane, including the Ubx2 ER membrane protein, which contains an N-terminal ubiquitin-associated (UBA) domain as well as a C-terminal UBX domain that binds Cdc48 [158]; the Derlin Dfm1 with its Cdc48-binding SHP-boxes [38]; and the Hrd1 E3, given that its overexpression promotes Cdc48 membrane recruitment, potentially as a result of its auto-ubiquitylation [38, 152].
Recent structural and biochemical analysis using purified components has provided insight into some of the mechanistic details of Cdc48 activity. First, Cdc48Ufd1-Npl4 binds ubiquitylated substrates at the face of the Cdc48 ring complex containing the Cdc48 N-terminal domains. Cdc48 ATPase activity appears to drive substrate unfolding, which allows passage of the unfolded polypeptide through the Cdc48 central pore [161, 162]. This inference was supported by photocrosslinking experiments as well as the use of a Cdc48-FtsH protease chimera, which led to substrate proteolysis following polypeptide movement through the Cdc48 pore. Another Cdc48 cofactor, the deubiquitylating enzyme Otu1, is required for substrate release following substrate unfolding and initial polypeptide transfer through the Cdc48 pore [162]. Deletion of the OTU1 gene in yeast has no impact on ERAD; however, expression of a catalytically inactive Otu1 (C120S) stabilizes Cdc48-dependent substrates in vivo, suggesting functional redundancy among DUBs in this pathway [152]. Interestingly, the substrate that passed through the Cdc48 pore in the in vitro experiments still was conjugated to short ubiquitin oligomers, implying that the ubiquitin moieties were also unfolded in order to pass through the narrow pore, which cannot accommodate a folded ubiquitin molecule.
Another Cdc48 co-factor, Ufd2, is required for efficient degradation of several ERAD substrates [126, 149, 163, 164]. Ufd2 was originally identified as a ubiquitin chain-elongation enzyme (“E4”); however, a recent study suggests Ufd2 does not efficiently elongate ubiquitin chains and instead attaches a single ubiquitin moiety onto proximal ubiquitins to form K48-ubiquitin branches [165, 166]. Ufd2 may function following substrate release from Cdc48 to increase substrate affinity for the proteasome or the proteasome shuttle factors Rad23 and Dsk2 [167]. In yeast, Ufd2 interacts with the C-terminal tails of Cdc48 near where substrates are released; however, Ufd2 interacts with the N-terminal domain of p97 in humans, and it has yet to be determined whether Ufd2 functions before or after Cdc48/p97 action [168].
7. Other Ub-dependent degradation processes at the ER and NE
7.1. Autophagy at the ER and NE
Autophagy (macroautophagy) is the engulfment of a small volume of cell contents into a double membrane-bounded structure called an autophagosome, which eventually fuses with the lysosome (vacuole in yeast) where its contents are degraded by resident hydrolases [169]. While this review focuses on protein degradation mediated by the UPS, autophagic processes that use ubiquitin-like protein (UBL)-conjugating systems are also significant contributors to ER protein degradation, and these autophagic pathways can also specifically target ubiquitylated cargos [169]. At the center of autophagy are the UBLs Atg12 and Atg8, which have ubiquitin-like folds and are broadly conserved [170]. These UBLs undergo a series of reactions analogous to the ubiquitylation cascade to yield, on the one hand, an Atg12-Atg5 conjugate and on the other, an Atg8 conjugate to the lipid phosphatidylethanolamine (PE) [171]. The Atg12-Atg5 conjugate binds Atg16 to form an E3-like complex that stimulates Atg8-PE formation [172]. Atg8-PE conjugates are crucial for autophagosome formation [173, 174].
In selective autophagy, autophagy receptors bind both specific cargo and Atg8-PE, thereby bringing the cargo into developing autophagosomes [175]. Cargos are often ubiquitylated, and the autophagy receptors also have ubiquitin-binding domains. Selective autophagy pathways have been characterized for various cellular components, including the ER and nucleus, termed “ER-phagy” and “nucleophagy,” respectively [169]. Atg39 and Atg40 are yeast autophagy receptors that mediate the degradation of ER and INM components in response to nitrogen starvation [176]. Atg39 primarily localizes to the nuclear membrane, while Atg40 localizes to the cortical and cytoplasmic ER [176]. FAM134B is the functional counterpart of Atg40 in mammalian cells and has been implicated in sensory neuropathy in humans [177]. Atg39 does not contain any clear mammalian orthologs; however, nucleophagy has been reported in mammalian cells and a functional counterpart of Atg39 is likely present [178, 179]. Autophagosomes containing nuclear components have been described in mammalian cells encoding mutations in lamin A and emerin [178]. A more recent study determined that the nuclear lamina protein lamin B1 is degraded by nucleophagy in response to oncogene-induced senescence and other tumorigenic stresses [180]. Based on these studies, nucleophagy and ER-phagy represent important degradation processes that target ER and nuclear components in response to a variety of cellular stresses; however, many of the molecular mechanisms governing these processes have yet to be explored in detail.
7.2. Nuclear Pore Complex Quality Control
The ONM and INM fuse at nuclear pore complexes (NPCs). NPCs provide a selectivity filter that mediates the exchange of nuclear and cytoplasmic components and imposes a size barrier that limits passage of larger macromolecules, including transmembrane proteins with large extraluminal domains [1]. Passage of larger molecules requires active transport mechanisms mediated by nuclear transport receptors [1, 181]. NPCs are composed of ~30 different subunits, called nucleoporins (Nups), which are present in multiple copies, creating a structure of ~120 MDa in vertebrates [182]. The proper assembly of NPCs is vital for maintaining nuclear integrity and compartmentalization. Defective NPC assembly intermediates are recognized and cleared through a quality control mechanism involving the ESCRTIII machinery and the AAA+ ATPase Vps4 in budding yeast [183]. These misassembled NPCs are sequestered in distinct clusters, which are retained in the mother cells following mitosis [183]. Interestingly, perturbations of the nuclear assembly process leads to proteasomal degradation of Nup85 in an ESCRTIII/Vps4-dependent manner. It remains unclear which E3 pathway(s) targets Nup85 or if other misassembled NPC subunits are degraded by proteasomes. Other studies have indicated several additional Nups are ubiquitylated in yeast, and Nup96 is degraded by the UPS in a cell cycle-dependent manner in mammalian cells [184–186]. Overall, these studies suggest the UPS implements quality control mechanisms for misassembled NPC intermediates.
8. Conclusions
ERAD has been investigated for ~30 years, and most of the ERAD machinery has likely been identified. Recent studies have improved our understanding of the mechanisms underlying ERAD substrate recognition, ubiquitylation, and retrotranslocation. Still, many important mechanistic details remain unresolved. One of the most puzzling questions in the field has been centered on the retrotranslocation of substrates at the ER. Recent studies have provided insight into the components involved in retrotranslocation and have also suggested retrotranslocation can be achieved through multiple avenues. One of these is mediated by the membrane-resident E3 ligase Hrd1, which appears to form a protein-conducting channel. Additional ERAD E3 ligases may well also mediate the retrotranslocation of their respective substrates and might also form protein channels in the ER membrane. Many aspects of how the ERAD E3 ligases interact with their cognate E2 enzymes remain to be clarified; the same is true for understanding the mechanism and significance of different ubiquitin chain linkages in the degradation of ERAD substrates. The involvement of molecular chaperones in substrate recognition as well as the mechanisms enabling ERAD E3 ligases to recognize such a remarkably diverse array of substrates will require considerable additional investigation. With the help of recent advances in structural biology, most notably high-resolution cryo-EM methods, and the development of other novel approaches, future studies will undoubtedly provide many insights into these fundamental ERAD mechanisms in the coming years.
Acknowledgements:
We would like to thank Carolyn Allain and Christopher M. Hickey for their helpful comments on the manuscript.
Funding:
This work was supported by the National Institutes of Health Grant GM046904 and the National Institutes of Health Training Grant T32 GM7223–43.
9. References
- [1].Katta SS, Smoyer CJ, Jaspersen SL. Destination: inner nuclear membrane. Trends Cell Biol 2014;24:221–9. [DOI] [PubMed] [Google Scholar]
- [2].Schwarz DS, Blower MD. The endoplasmic reticulum: structure, function and response to cellular signaling. Cell Mol Life Sci 2016;73:79–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Braakman I, Hebert DN. Protein folding in the endoplasmic reticulum. Cold Spring Harb Perspect Biol 2013;5:a013201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lilienbaum A Relationship between the proteasomal system and autophagy. Int J Biochem Mol Biol 2013;4:1–26. [PMC free article] [PubMed] [Google Scholar]
- [5].Varshavsky A The ubiquitin system, an immense realm. Annu Rev Biochem 2012;81:167–76. [DOI] [PubMed] [Google Scholar]
- [6].Schwartz AL, Ciechanover A. Targeting proteins for destruction by the ubiquitin system: implications for human pathobiology. Annu Rev Pharmacol Toxicol 2009;49:73–96. [DOI] [PubMed] [Google Scholar]
- [7].Hochstrasser M Ubiquitin-dependent protein degradation. Annu Rev Genet 1996;30:405–39. [DOI] [PubMed] [Google Scholar]
- [8].Pickart CM. Ubiquitin in chains. Trends Biochem Sci 2000;25:544–8. [DOI] [PubMed] [Google Scholar]
- [9].Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem 2009;78:399–434. [DOI] [PubMed] [Google Scholar]
- [10].Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, et al. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS One 2008;3:e1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Finley D, Ulrich HD, Sommer T, Kaiser P. The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics 2012;192:319–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 2009;137:133–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Origin Hochstrasser M. and function of ubiquitin-like proteins. Nature 2009;458:422–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol 2008;9:679–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhang S, Xu C, Larrimore KE, Ng DTW. Slp1-Emp65: A Guardian Factor that Protects Folding Polypeptides from Promiscuous Degradation. Cell 2017;171:346–57 e12. [DOI] [PubMed] [Google Scholar]
- [16].Ruggiano A, Foresti O, Carvalho P. Quality control: ER-associated degradation: protein quality control and beyond. J Cell Biol 2014;204:869–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Foresti O, Rodriguez-Vaello V, Funaya C, Carvalho P. Quality control of inner nuclear membrane proteins by the Asi complex. Science 2014;346:751–5. [DOI] [PubMed] [Google Scholar]
- [18].Khmelinskii A, Blaszczak E, Pantazopoulou M, Fischer B, Omnus DJ, Le Dez G, et al. Protein quality control at the inner nuclear membrane. Nature 2014;516:410–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Stagg HR, Thomas M, van den Boomen D, Wiertz EJ, Drabkin HA, Gemmill RM, et al. The TRC8 E3 ligase ubiquitinates MHC class I molecules before dislocation from the ER. J Cell Biol 2009;186:685–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].van den Boomen DJ, Timms RT, Grice GL, Stagg HR, Skodt K, Dougan G, et al. TMEM129 is a Derlin-1 associated ERAD E3 ligase essential for virus-induced degradation of MHC-I. Proc Natl Acad Sci U S A 2014;111:11425–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Younger JM, Chen L, Ren HY, Rosser MF, Turnbull EL, Fan CY, et al. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell 2006;126:571–82. [DOI] [PubMed] [Google Scholar]
- [22].Lu JP, Wang Y, Sliter DA, Pearce MM, Wojcikiewicz RJ. RNF170 protein, an endoplasmic reticulum membrane ubiquitin ligase, mediates inositol 1,4,5-trisphosphate receptor ubiquitination and degradation. J Biol Chem 2011;286:24426–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].El Khouri E, Le Pavec G, Toledano MB, Delaunay-Moisan A. RNF185 is a novel E3 ligase of endoplasmic reticulum-associated degradation (ERAD) that targets cystic fibrosis transmembrane conductance regulator (CFTR). J Biol Chem 2013;288:31177–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Jiang LY, Jiang W, Tian N, Xiong YN, Liu J, Wei J, et al. Ring finger protein 145 (RNF145) is a ubiquitin ligase for sterol-induced degradation of HMG-CoA reductase. J Biol Chem 2018;293:4047–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lerner M, Corcoran M, Cepeda D, Nielsen ML, Zubarev R, Ponten F, et al. The RBCC gene RFP2 (Leu5) encodes a novel transmembrane E3 ubiquitin ligase involved in ERAD. Mol Biol Cell 2007;18:1670–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kaneko M, Iwase I, Yamasaki Y, Takai T, Wu Y, Kanemoto S, et al. Genome-wide identification and gene expression profiling of ubiquitin ligases for endoplasmic reticulum protein degradation. Sci Rep 2016;6:30955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Schoebel S, Mi W, Stein A, Ovchinnikov S, Pavlovicz R, DiMaio F, et al. Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3. Nature 2017;548:352–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gardner RG, Swarbrick GM, Bays NW, Cronin SR, Wilhovsky S, Seelig L, et al. Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. J Cell Biol 2000;151:69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Biederer T, Volkwein C, Sommer T. Role of Cue1p in ubiquitination and degradation at the ER surface. Science 1997;278:1806–9. [DOI] [PubMed] [Google Scholar]
- [30].Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell 2006;126:361–73. [DOI] [PubMed] [Google Scholar]
- [31].Plemper RK, Bordallo J, Deak PM, Taxis C, Hitt R, Wolf DH. Genetic interactions of Hrd3p and Der3p/Hrd1p with Sec61p suggest a retro-translocation complex mediating protein transport for ER degradation. J Cell Sci 1999;112 (Pt 22):4123–34. [DOI] [PubMed] [Google Scholar]
- [32].Denic V, Quan EM, Weissman JS. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell 2006;126:349–59. [DOI] [PubMed] [Google Scholar]
- [33].Horn SC, Hanna J, Hirsch C, Volkwein C, Schutz A, Heinemann U, et al. Usa1 functions as a scaffold of the HRD-ubiquitin ligase. Mol Cell 2009;36:782–93. [DOI] [PubMed] [Google Scholar]
- [34].Gauss R, Sommer T, Jarosch E. The Hrd1p ligase complex forms a linchpin between ER-lumenal substrate selection and Cdc48p recruitment. EMBO J 2006;25:1827–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Greenblatt EJ, Olzmann JA, Kopito RR. Derlin-1 is a rhomboid pseudoprotease required for the dislocation of mutant alpha-1 antitrypsin from the endoplasmic reticulum. Nat Struct Mol Biol 2011;18:1147–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hitt R, Wolf DH. Der1p, a protein required for degradation of malfolded soluble proteins of the endoplasmic reticulum: topology and Der1-like proteins. FEMS Yeast Res 2004;4:721–9. [DOI] [PubMed] [Google Scholar]
- [37].Zattas D, Adle DJ, Rubenstein EM, Hochstrasser M. N-terminal acetylation of the yeast Derlin Der1 is essential for Hrd1 ubiquitin-ligase activity toward luminal ER substrates. Mol Biol Cell 2013;24:890–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Neal S, Jaeger PA, Duttke SH, Benner C, C KG, Ideker T, et al. The Dfm1 Derlin Is Required for ERAD Retrotranslocation of Integral Membrane Proteins. Mol Cell 2018;69:306–20 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lilley BN, Ploegh HL. A membrane protein required for dislocation of misfolded proteins from the ER. Nature 2004;429:834–40. [DOI] [PubMed] [Google Scholar]
- [40].Oda Y, Okada T, Yoshida H, Kaufman RJ, Nagata K, Mori K. Derlin-2 and Derlin-3 are regulated by the mammalian unfolded protein response and are required for ER-associated degradation. J Cell Biol 2006;172:383–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kadowaki H, Nagai A, Maruyama T, Takami Y, Satrimafitrah P, Kato H, et al. Pre-emptive Quality Control Protects the ER from Protein Overload via the Proximity of ERAD Components and SRP. Cell Rep 2015;13:944–56. [DOI] [PubMed] [Google Scholar]
- [42].Kadowaki H, Satrimafitrah P, Takami Y, Nishitoh H. Molecular mechanism of ER stress-induced pre-emptive quality control involving association of the translocon, Derlin-1, and HRD1. Sci Rep 2018;8:7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Swanson R, Locher M, Hochstrasser M. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Mat’2 repressor degradation. GENES & DEVELOPMENT 2001;15:2660–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Huyer G, Piluek WF, Fansler Z, Kreft SG, Hochstrasser M, Brodsky JL, et al. Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J Biol Chem 2004;279:38369–78. [DOI] [PubMed] [Google Scholar]
- [45].Ravid T, Kreft SG, Hochstrasser M. Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. EMBO J 2006;25:533–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kreft SG, Hochstrasser M. An unusual transmembrane helix in the endoplasmic reticulum ubiquitin ligase Doa10 modulates degradation of its cognate E2 enzyme. J Biol Chem 2011;286:20163–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kreft SG, Wang L, Hochstrasser M. Membrane topology of the yeast endoplasmic reticulum-localized ubiquitin ligase Doa10 and comparison with its human ortholog TEB4 (MARCH-VI). J Biol Chem 2006;281:4646–53. [DOI] [PubMed] [Google Scholar]
- [48].Zattas D, Berk JM, Kreft SG, Hochstrasser M. A Conserved C-terminal Element in the Yeast Doa10 and Human MARCH6 Ubiquitin Ligases Required for Selective Substrate Degradation. J Biol Chem 2016;291:12105–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MAT alpha 2 repressor. Cell 1993;74:357–69. [DOI] [PubMed] [Google Scholar]
- [50].Hassink G, Kikkert M, van Voorden S, Lee SJ, Spaapen R, van Laar T, et al. TEB4 is a C4HC3 RING finger-containing ubiquitin ligase of the endoplasmic reticulum. Biochem J 2005;388:647–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Zattas D, Hochstrasser M. Ubiquitin-dependent protein degradation at the yeast endoplasmic reticulum and nuclear envelope. Crit Rev Biochem Mol Biol 2015;50:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Stefanovic-Barrett S, Dickson AS, Burr SP, Williamson JC, Lobb IT, van den Boomen DJ, et al. MARCH6 and TRC8 facilitate the quality control of cytosolic and tail-anchored proteins. EMBO Rep 2018;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Metzger MB, Maurer MJ, Dancy BM, Michaelis S. Degradation of a cytosolic protein requires endoplasmic reticulum-associated degradation machinery. J Biol Chem 2008;283:32302–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Forsberg H, Hammar M, Andreasson C, Moliner A, Ljungdahl PO. Suppressors of ssy1 and ptr3 null mutations define novel amino acid sensor-independent genes in Saccharomyces cerevisiae. Genetics 2001;158:973–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Omnus DJ, Ljungdahl PO. Latency of transcription factor Stp1 depends on a modular regulatory motif that functions as cytoplasmic retention determinant and nuclear degron. Mol Biol Cell 2014;25:3823–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Boban M, Zargari A, Andreasson C, Heessen S, Thyberg J, Ljungdahl PO. Asi1 is an inner nuclear membrane protein that restricts promoter access of two latent transcription factors. J Cell Biol 2006;173:695–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Zargari A, Boban M, Heessen S, Andreasson C, Thyberg J, Ljungdahl PO. Inner nuclear membrane proteins Asi1, Asi2, and Asi3 function in concert to maintain the latent properties of transcription factors Stp1 and Stp2. J Biol Chem 2007;282:594–605. [DOI] [PubMed] [Google Scholar]
- [58].Crowder JJ, Geigges M, Gibson RT, Fults ES, Buchanan BW, Sachs N, et al. Rkr1/Ltn1 Ubiquitin Ligase-mediated Degradation of Translationally Stalled Endoplasmic Reticulum Proteins. J Biol Chem 2015;290:18454–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Bengtson MH, Joazeiro CA. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature 2010;467:470–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Brandman O, Hegde RS. Ribosome-associated protein quality control. Nat Struct Mol Biol 2016;23:7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].von der Malsburg K, Shao S, Hegde RS. The ribosome quality control pathway can access nascent polypeptides stalled at the Sec61 translocon. Mol Biol Cell 2015;26:2168–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Rubenstein EM, Kreft SG, Greenblatt W, Swanson R, Hochstrasser M. Aberrant substrate engagement of the ER translocon triggers degradation by the Hrd1 ubiquitin ligase. J Cell Biol 2012;197:761–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Ast T, Michaelis S, Schuldiner M. The Protease Ste24 Clears Clogged Translocons. Cell 2016;164:103–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Mohorko E, Glockshuber R, Aebi M. Oligosaccharyltransferase: the central enzyme of N-linked protein glycosylation. J Inherit Metab Dis 2011;34:869–78. [DOI] [PubMed] [Google Scholar]
- [65].Aebi M, Bernasconi R, Clerc S, Molinari M. N-glycan structures: recognition and processing in the ER. Trends Biochem Sci 2010;35:74–82. [DOI] [PubMed] [Google Scholar]
- [66].Quan EM, Kamiya Y, Kamiya D, Denic V, Weibezahn J, Kato K, et al. Defining the glycan destruction signal for endoplasmic reticulum-associated degradation. Mol Cell 2008;32:870–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Clerc S, Hirsch C, Oggier DM, Deprez P, Jakob C, Sommer T, et al. Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J Cell Biol 2009;184:159–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Gauss R, Kanehara K, Carvalho P, Ng DT, Aebi M. A complex of Pdi1p and the mannosidase Htm1p initiates clearance of unfolded glycoproteins from the endoplasmic reticulum. Mol Cell 2011;42:782–93. [DOI] [PubMed] [Google Scholar]
- [69].Liu YC, Fujimori DG, Weissman JS. Htm1p-Pdi1p is a folding-sensitive mannosidase that marks N-glycoproteins for ER-associated protein degradation. Proc Natl Acad Sci U S A 2016;113:E4015–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Pfeiffer A, Stephanowitz H, Krause E, Volkwein C, Hirsch C, Jarosch E, et al. A Complex of Htm1 and the Oxidoreductase Pdi1 Accelerates Degradation of Misfolded Glycoproteins. J Biol Chem 2016;291:12195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Ushioda R, Hoseki J, Araki K, Jansen G, Thomas DY, Nagata K. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science 2008;321:569–72. [DOI] [PubMed] [Google Scholar]
- [72].Hosokawa N, Wada I, Hasegawa K, Yorihuzi T, Tremblay LO, Herscovics A, et al. A novel ER alpha-mannosidase-like protein accelerates ER-associated degradation. EMBO Rep 2001;2:415–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Ninagawa S, Okada T, Sumitomo Y, Kamiya Y, Kato K, Horimoto S, et al. EDEM2 initiates mammalian glycoprotein ERAD by catalyzing the first mannose trimming step. J Cell Biol 2014;206:347–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Finger A, Knop M, Wolf DH. Analysis of two mutated vacuolar proteins reveals a degradation pathway in the endoplasmic reticulum or a related compartment of yeast. Eur J Biochem 1993;218:565–74. [DOI] [PubMed] [Google Scholar]
- [75].Hiller MM, Finger A, Schweiger M, Wolf DH. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science 1996;273:1725–8. [DOI] [PubMed] [Google Scholar]
- [76].Bordallo J, Plemper RK, Finger A, Wolf DH. Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol Biol Cell 1998;9:209–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Kostova Z, Wolf DH. Importance of carbohydrate positioning in the recognition of mutated CPY for ER-associated degradation. J Cell Sci 2005;118:1485–92. [DOI] [PubMed] [Google Scholar]
- [78].Nishikawa SI, Fewell SW, Kato Y, Brodsky JL, Endo T. Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J Cell Biol 2001;153:1061–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Plemper RK, Bohmler S, Bordallo J, Sommer T, Wolf DH. Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature 1997;388:891–5. [DOI] [PubMed] [Google Scholar]
- [80].Buck TM, Kolb AR, Boyd CR, Kleyman TR, Brodsky JL. The endoplasmic reticulum-associated degradation of the epithelial sodium channel requires a unique complement of molecular chaperones. Mol Biol Cell 2010;21:1047–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Mehnert M, Sommermeyer F, Berger M, Kumar Lakshmipathy S, Gauss R, Aebi M, et al. The interplay of Hrd3 and the molecular chaperone system ensures efficient degradation of malfolded secretory proteins. Mol Biol Cell 2015;26:185–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Mehnert M, Sommer T, Jarosch E. Der1 promotes movement of misfolded proteins through the endoplasmic reticulum membrane. Nat Cell Biol 2014;16:77–86. [DOI] [PubMed] [Google Scholar]
- [83].Loibl M, Strahl S. Protein O-mannosylation: what we have learned from baker’s yeast. Biochim Biophys Acta 2013;1833:2438–46. [DOI] [PubMed] [Google Scholar]
- [84].Xu C, Wang S, Thibault G, Ng DT. Futile protein folding cycles in the ER are terminated by the unfolded protein O-mannosylation pathway. Science 2013;340:978–81. [DOI] [PubMed] [Google Scholar]
- [85].Goder V, Melero A. Protein O-mannosyltransferases participate in ER protein quality control. J Cell Sci 2011;124:144–53. [DOI] [PubMed] [Google Scholar]
- [86].Vashist S, Kim W, Belden WJ, Spear ED, Barlowe C, Ng DT. Distinct retrieval and retention mechanisms are required for the quality control of endoplasmic reticulum protein folding. J Cell Biol 2001;155:355–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Nakatsukasa K, Okada S, Umebayashi K, Fukuda R, Nishikawa S, Endo T. Roles of O-mannosylation of aberrant proteins in reduction of the load for endoplasmic reticulum chaperones in yeast. J Biol Chem 2004;279:49762–72. [DOI] [PubMed] [Google Scholar]
- [88].Bernasconi R, Galli C, Calanca V, Nakajima T, Molinari M. Stringent requirement for HRD1, SEL1L, and OS-9/XTP3-B for disposal of ERAD-LS substrates. J Cell Biol 2010;188:223–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Horimoto S, Ninagawa S, Okada T, Koba H, Sugimoto T, Kamiya Y, et al. The unfolded protein response transducer ATF6 represents a novel transmembrane-type endoplasmic reticulum-associated degradation substrate requiring both mannose trimming and SEL1L protein. J Biol Chem 2013;288:31517–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Hampton RY, Gardner RG, Rine J. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol Biol Cell 1996;7:2029–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Gardner RG, Shearer AG, Hampton RY. In vivo action of the HRD ubiquitin ligase complex: mechanisms of endoplasmic reticulum quality control and sterol regulation. Mol Cell Biol 2001;21:4276–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Song BL, Sever N, DeBose-Boyd RA. Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol Cell 2005;19:829–40. [DOI] [PubMed] [Google Scholar]
- [93].Jo Y, Lee PC, Sguigna PV, DeBose-Boyd RA. Sterol-induced degradation of HMG CoA reductase depends on interplay of two Insigs and two ubiquitin ligases, gp78 and Trc8. Proc Natl Acad Sci U S A 2011;108:20503–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Zelcer N, Sharpe LJ, Loregger A, Kristiana I, Cook EC, Phan L, et al. The E3 ubiquitin ligase MARCH6 degrades squalene monooxygenase and affects 3-hydroxy-3-methyl-glutaryl coenzyme A reductase and the cholesterol synthesis pathway. Mol Cell Biol 2014;34:1262–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Sever N, Yang T, Brown MS, Goldstein JL, DeBose-Boyd RA. Accelerated degradation of HMG CoA reductase mediated by binding of insig-1 to its sterol-sensing domain. Mol Cell 2003;11:25–33. [DOI] [PubMed] [Google Scholar]
- [96].Cook EC, Nelson JK, Sorrentino V, Koenis D, Moeton M, Scheij S, et al. Identification of the ER-resident E3 ubiquitin ligase RNF145 as a novel LXR-regulated gene. PLoS One 2017;12:e0172721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Tsai YC, Leichner GS, Pearce MM, Wilson GL, Wojcikiewicz RJ, Roitelman J, et al. Differential regulation of HMG-CoA reductase and Insig-1 by enzymes of the ubiquitin-proteasome system. Mol Biol Cell 2012;23:4484–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Theesfeld CL, Hampton RY. Insulin-induced gene protein (INSIG)-dependent sterol regulation of Hmg2 endoplasmic reticulum-associated degradation (ERAD) in yeast. J Biol Chem 2013;288:8519–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Voss M, Schroder B, Fluhrer R. Mechanism, specificity, and physiology of signal peptide peptidase (SPP) and SPP-like proteases. Biochim Biophys Acta 2013;1828:2828–39. [DOI] [PubMed] [Google Scholar]
- [100].Chen CY, Malchus NS, Hehn B, Stelzer W, Avci D, Langosch D, et al. Signal peptide peptidase functions in ERAD to cleave the unfolded protein response regulator XBP1u. EMBO J 2014;33:2492–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Boname JM, Bloor S, Wandel MP, Nathan JA, Antrobus R, Dingwell KS, et al. Cleavage by signal peptide peptidase is required for the degradation of selected tail-anchored proteins. J Cell Biol 2014;205:847–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Avci D, Fuchs S, Schrul B, Fukumori A, Breker M, Frumkin I, et al. The yeast ER-intramembrane protease Ypf1 refines nutrient sensing by regulating transporter abundance. Mol Cell 2014;56:630–40. [DOI] [PubMed] [Google Scholar]
- [103].Sato BK, Schulz D, Do PH, Hampton RY. Misfolded membrane proteins are specifically recognized by the transmembrane domain of the Hrd1p ubiquitin ligase. Mol Cell 2009;34:212–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Habeck G, Ebner FA, Shimada-Kreft H, Kreft SG. The yeast ERAD-C ubiquitin ligase Doa10 recognizes an intramembrane degron. J Cell Biol 2015;209:261–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Deng M, Hochstrasser M. Spatially regulated ubiquitin ligation by an ER/nuclear membrane ligase. Nature 2006;443:827–31. [DOI] [PubMed] [Google Scholar]
- [106].Hickey CM, Hochstrasser M. STUbL-mediated degradation of the transcription factor MATalpha2 requires degradation elements that coincide with corepressor binding sites. Mol Biol Cell 2015;26:3401–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Johnson PR, Swanson R, Rakhilina L, Hochstrasser M. Degradation signal masking by heterodimerization of MATalpha2 and MATa1 blocks their mutual destruction by the ubiquitin-proteasome pathway. Cell 1998;94:217–27. [DOI] [PubMed] [Google Scholar]
- [108].Kim I, Miller CR, Young DL, Fields S. High-throughput analysis of in vivo protein stability. Mol Cell Proteomics 2013;12:3370–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Hwang CS, Shemorry A, Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science 2010;327:973–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Gilon T, Chomsky O, Kulka RG. Degradation signals for ubiquitin system proteolysis in Saccharomyces cerevisiae. EMBO J 1998;17:2759–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Gilon T, Chomsky O, Kulka RG. Degradation signals recognized by the Ubc6p-Ubc7p ubiquitin-conjugating enzyme pair. Mol Cell Biol 2000;20:7214–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Furth N, Gertman O, Shiber A, Alfassy OS, Cohen I, Rosenberg MM, et al. Exposure of bipartite hydrophobic signal triggers nuclear quality control of Ndc10 at the endoplasmic reticulum/nuclear envelope. Mol Biol Cell 2011;22:4726–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Alfassy OS, Cohen I, Reiss Y, Tirosh B, Ravid T. Placing a disrupted degradation motif at the C terminus of proteasome substrates attenuates degradation without impairing ubiquitylation. J Biol Chem 2013;288:12645–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Kats I, Khmelinskii A, Kschonsak M, Huber F, Kniess RA, Bartosik A, et al. Mapping Degradation Signals and Pathways in a Eukaryotic N-terminome. Mol Cell 2018;70:488–501 e5. [DOI] [PubMed] [Google Scholar]
- [115].Ruggiano A, Mora G, Buxo L, Carvalho P. Spatial control of lipid droplet proteins by the ERAD ubiquitin ligase Doa10. EMBO J 2016;35:1644–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Walter J, Urban J, Volkwein C, Sommer T. Sec61p-independent degradation of the tail-anchored ER membrane protein Ubc6p. EMBO J 2001;20:3124–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Arteaga MF, Wang L, Ravid T, Hochstrasser M, Canessa CM. An amphipathic helix targets serum and glucocorticoid-induced kinase 1 to the endoplasmic reticulum-associated ubiquitin-conjugation machinery. Proc Natl Acad Sci U S A 2006;103:11178–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Shiber A, Breuer W, Brandeis M, Ravid T. Ubiquitin conjugation triggers misfolded protein sequestration into quality control foci when Hsp70 chaperone levels are limiting. Mol Biol Cell 2013;24:2076–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Stolz A, Besser S, Hottmann H, Wolf DH. Previously unknown role for the ubiquitin ligase Ubr1 in endoplasmic reticulum-associated protein degradation. Proc Natl Acad Sci U S A 2013;110:15271–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Summers DW, Wolfe KJ, Ren HY, Cyr DM. The Type II Hsp40 Sis1 cooperates with Hsp70 and the E3 ligase Ubr1 to promote degradation of terminally misfolded cytosolic protein. PLoS One 2013;8:e52099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Prasad R, Xu C, Ng DTW. Hsp40/70/110 chaperones adapt nuclear protein quality control to serve cytosolic clients. J Cell Biol 2018;217:2019–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Park SH, Kukushkin Y, Gupta R, Chen T, Konagai A, Hipp MS, et al. PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell 2013;154:134–45. [DOI] [PubMed] [Google Scholar]
- [123].Chen L, Madura K. Degradation of specific nuclear proteins occurs in the cytoplasm in Saccharomyces cerevisiae. Genetics 2014;197:193–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Dang FW, Chen L, Madura K. Catalytically Active Proteasomes Function Predominantly in the Cytosol. J Biol Chem 2016;291:18765–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Freedman DA, Levine AJ. Nuclear export is required for degradation of endogenous p53 by MDM2 and human papillomavirus E6. Mol Cell Biol 1998;18:7288–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Nakatsukasa K, Huyer G, Michaelis S, Brodsky JL. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell 2008;132:101–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Yau R, Rape M. The increasing complexity of the ubiquitin code. Nat Cell Biol 2016;18:579–86. [DOI] [PubMed] [Google Scholar]
- [128].Stewart MD, Ritterhoff T, Klevit RE, Brzovic PS. E2 enzymes: more than just middle men. Cell Res 2016;26:423–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Li W, Tu D, Brunger AT, Ye Y. A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate. Nature 2007;446:333–7. [DOI] [PubMed] [Google Scholar]
- [130].Bazirgan OA, Hampton RY. Cue1p is an activator of Ubc7p E2 activity in vitro and in vivo. J Biol Chem 2008;283:12797–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Liu W, Shang Y, Zeng Y, Liu C, Li Y, Zhai L, et al. Dimeric Ube2g2 simultaneously engages donor and acceptor ubiquitins to form Lys48-linked ubiquitin chains. EMBO J 2014;33:46–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Kostova Z, Mariano J, Scholz S, Koenig C, Weissman AM. A Ubc7p-binding domain in Cue1p activates ER-associated protein degradation. J Cell Sci 2009;122:1374–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Chen B, Mariano J, Tsai YC, Chan AH, Cohen M, Weissman AM. The activity of a human endoplasmic reticulum-associated degradation E3, gp78, requires its Cue domain, RING finger, and an E2-binding site. Proc Natl Acad Sci U S A 2006;103:341–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Spandl J, Lohmann D, Kuerschner L, Moessinger C, Thiele C. Ancient ubiquitous protein 1 (AUP1) localizes to lipid droplets and binds the E2 ubiquitin conjugase G2 (Ube2g2) via its G2 binding region. J Biol Chem 2011;286:5599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Das R, Mariano J, Tsai YC, Kalathur RC, Kostova Z, Li J, et al. Allosteric activation of E2-RING finger-mediated ubiquitylation by a structurally defined specific E2-binding region of gp78. Mol Cell 2009;34:674–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Metzger MB, Liang YH, Das R, Mariano J, Li S, Li J, et al. A structurally unique E2-binding domain activates ubiquitination by the ERAD E2, Ubc7p, through multiple mechanisms. Mol Cell 2013;50:516–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Bagola K, von Delbruck M, Dittmar G, Scheffner M, Ziv I, Glickman MH, et al. Ubiquitin binding by a CUE domain regulates ubiquitin chain formation by ERAD E3 ligases. Mol Cell 2013;50:528–39. [DOI] [PubMed] [Google Scholar]
- [138].von Delbruck M, Kniss A, Rogov VV, Pluska L, Bagola K, Lohr F, et al. The CUE Domain of Cue1 Aligns Growing Ubiquitin Chains with Ubc7 for Rapid Elongation. Mol Cell 2016;62:918–28. [DOI] [PubMed] [Google Scholar]
- [139].Tsuchiya H, Burana D, Ohtake F, Arai N, Kaiho A, Komada M, et al. Ub-ProT reveals global length and composition of protein ubiquitylation in cells. Nat Commun 2018;9:524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Yau RG, Doerner K, Castellanos ER, Haakonsen DL, Werner A, Wang N, et al. Assembly and Function of Heterotypic Ubiquitin Chains in Cell-Cycle and Protein Quality Control. Cell 2017;171:918–33 e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Weber A, Cohen I, Popp O, Dittmar G, Reiss Y, Sommer T, et al. Sequential Poly-ubiquitylation by Specialized Conjugating Enzymes Expands the Versatility of a Quality Control Ubiquitin Ligase. Mol Cell 2016;63:827–39. [DOI] [PubMed] [Google Scholar]
- [142].Wang X, Herr RA, Rabelink M, Hoeben RC, Wiertz EJ, Hansen TH. Ube2j2 ubiquitinates hydroxylated amino acids on ER-associated degradation substrates. J Cell Biol 2009;187:655–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Rodrigo-Brenni MC, Morgan DO. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell 2007;130:127–39. [DOI] [PubMed] [Google Scholar]
- [144].Rapoport TA. Protein transport across the endoplasmic reticulum membrane. FEBS J 2008;275:4471–8. [DOI] [PubMed] [Google Scholar]
- [145].Tsai B, Ye Y, Rapoport TA. Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat Rev Mol Cell Biol 2002;3:246–55. [DOI] [PubMed] [Google Scholar]
- [146].Romisch K A Case for Sec61 Channel Involvement in ERAD. Trends Biochem Sci 2017;42:171–9. [DOI] [PubMed] [Google Scholar]
- [147].Lipson C, Alalouf G, Bajorek M, Rabinovich E, Atir-Lande A, Glickman M, et al. A proteasomal ATPase contributes to dislocation of endoplasmic reticulum-associated degradation (ERAD) substrates. J Biol Chem 2008;283:7166–75. [DOI] [PubMed] [Google Scholar]
- [148].Bar-Nun S, Glickman MH. Proteasomal AAA-ATPases: structure and function. Biochim Biophys Acta 2012;1823:67–82. [DOI] [PubMed] [Google Scholar]
- [149].Smith N, Adle DJ, Zhao M, Qin X, Kim H, Lee J. Endoplasmic Reticulum-associated Degradation of Pca1p, a Polytopic Protein, via Interaction with the Proteasome at the Membrane. J Biol Chem 2016;291:15082–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Wolf DH, Stolz A. The Cdc48 machine in endoplasmic reticulum associated protein degradation. Biochim Biophys Acta 2012;1823:117–24. [DOI] [PubMed] [Google Scholar]
- [151].Carvalho P, Stanley AM, Rapoport TA. Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell 2010;143:579–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Stein A, Ruggiano A, Carvalho P, Rapoport TA. Key steps in ERAD of luminal ER proteins reconstituted with purified components. Cell 2014;158:1375–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Baldridge RD, Rapoport TA. Autoubiquitination of the Hrd1 Ligase Triggers Protein Retrotranslocation in ERAD. Cell 2016;166:394–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Garza RM, Sato BK, Hampton RY. In vitro analysis of Hrd1p-mediated retrotranslocation of its multispanning membrane substrate 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase. J Biol Chem 2009;284:14710–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Stolz A, Schweizer RS, Schafer A, Wolf DH. Dfm1 forms distinct complexes with Cdc48 and the ER ubiquitin ligases and is required for ERAD. Traffic 2010;11:1363–9. [DOI] [PubMed] [Google Scholar]
- [156].Sato BK, Hampton RY. Yeast Derlin Dfm1 interacts with Cdc48 and functions in ER homeostasis. Yeast 2006;23:1053–64. [DOI] [PubMed] [Google Scholar]
- [157].Schuberth C, Buchberger A. Membrane-bound Ubx2 recruits Cdc48 to ubiquitin ligases and their substrates to ensure efficient ER-associated protein degradation. Nat Cell Biol 2005;7:999–1006. [DOI] [PubMed] [Google Scholar]
- [158].Neuber O, Jarosch E, Volkwein C, Walter J, Sommer T. Ubx2 links the Cdc48 complex to ER-associated protein degradation. Nat Cell Biol 2005;7:993–8. [DOI] [PubMed] [Google Scholar]
- [159].Greenblatt EJ, Olzmann JA, Kopito RR. Making the cut: intramembrane cleavage by a rhomboid protease promotes ERAD. Nat Struct Mol Biol 2012;19:979–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Xia D, Tang WK, Ye Y. Structure and function of the AAA+ ATPase p97/Cdc48p. Gene 2016;583:64–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Ye Y, Meyer HH, Rapoport TA. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J Cell Biol 2003;162:71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Bodnar NO, Rapoport TA. Molecular Mechanism of Substrate Processing by the Cdc48 ATPase Complex. Cell 2017;169:722–35 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Liu C, van Dyk D, Xu P, Choe V, Pan H, Peng J, et al. Ubiquitin chain elongation enzyme Ufd2 regulates a subset of Doa10 substrates. J Biol Chem 2010;285:10265–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Richly H, Rape M, Braun S, Rumpf S, Hoege C, Jentsch S. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell 2005;120:73–84. [DOI] [PubMed] [Google Scholar]
- [165].Liu C, Liu W, Ye Y, Li W. Ufd2p synthesizes branched ubiquitin chains to promote the degradation of substrates modified with atypical chains. Nat Commun 2017;8:14274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 1999;96:635–44. [DOI] [PubMed] [Google Scholar]
- [167].Bodnar N, Rapoport T. Toward an understanding of the Cdc48/p97 ATPase. F1000Res 2017;6:1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Bohm S, Lamberti G, Fernandez-Saiz V, Stapf C, Buchberger A. Cellular functions of Ufd2 and Ufd3 in proteasomal protein degradation depend on Cdc48 binding. Mol Cell Biol 2011;31:1528–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Khaminets A, Behl C, Dikic I. Ubiquitin-Dependent And Independent Signals In Selective Autophagy. Trends Cell Biol 2016;26:6–16. [DOI] [PubMed] [Google Scholar]
- [170].Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep 2008;9:859–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Nakatogawa H Two ubiquitin-like conjugation systems that mediate membrane formation during autophagy. Essays Biochem 2013;55:39–50. [DOI] [PubMed] [Google Scholar]
- [172].Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem 2007;282:37298–302. [DOI] [PubMed] [Google Scholar]
- [173].Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 2007;130:165–78. [DOI] [PubMed] [Google Scholar]
- [174].He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 2009;43:67–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Zaffagnini G, Martens S. Mechanisms of Selective Autophagy. J Mol Biol 2016;428:1714–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Mochida K, Oikawa Y, Kimura Y, Kirisako H, Hirano H, Ohsumi Y, et al. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature 2015;522:359–62. [DOI] [PubMed] [Google Scholar]
- [177].Khaminets A, Heinrich T, Mari M, Grumati P, Huebner AK, Akutsu M, et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature 2015;522:354–8. [DOI] [PubMed] [Google Scholar]
- [178].Park YE, Hayashi YK, Bonne G, Arimura T, Noguchi S, Nonaka I, et al. Autophagic degradation of nuclear components in mammalian cells. Autophagy 2009;5:795–804. [DOI] [PubMed] [Google Scholar]
- [179].Dou Z, Ivanov A, Adams PD, Berger SL. Mammalian autophagy degrades nuclear constituents in response to tumorigenic stress. Autophagy 2016;12:1416–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Dou Z, Xu C, Donahue G, Shimi T, Pan JA, Zhu J, et al. Autophagy mediates degradation of nuclear lamina. Nature 2015;527:105–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Stewart M Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol 2007;8:195–208. [DOI] [PubMed] [Google Scholar]
- [182].Webster BM, Lusk CP. Border Safety: Quality Control at the Nuclear Envelope. Trends Cell Biol 2016;26:29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Webster BM, Colombi P, Jager J, Lusk CP. Surveillance of nuclear pore complex assembly by ESCRT-III/Vps4. Cell 2014;159:388–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Chakraborty P, Wang Y, Wei JH, van Deursen J, Yu H, Malureanu L, et al. Nucleoporin levels regulate cell cycle progression and phase-specific gene expression. Dev Cell 2008;15:657–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Hayakawa A, Babour A, Sengmanivong L, Dargemont C. Ubiquitylation of the nuclear pore complex controls nuclear migration during mitosis in S. cerevisiae. J Cell Biol 2012;196:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, et al. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol 2003;21:921–6. [DOI] [PubMed] [Google Scholar]