 Iridium based antimicrobial agents have shown efficacy against S. aureus, including MRSA, and appear to be safe in mice.
Iridium based antimicrobial agents have shown efficacy against S. aureus, including MRSA, and appear to be safe in mice.
Abstract
A new class of piano-stool iridium complexes with 1,2-diaminoethane ligands are shown to be effective and safe antimicrobials with activity against Staphylococcus aureus, including various isolates of methicillin-resistant strains (MRSA). Comparison to other piano stool complexes with activity against mycobacteria are made along with a discussion of structure–activity relationships. The structures of one the most active complexes with the ligand cis-1,2-diaminocyclohexane and one of the least active complexes with the ligand trans-1,2-diaminocyclohexane are compared and discussed with respect to their drastically different activities. In vitro toxicity studies for all of the complexes are described. In addition, a mouse study with one of the complexes, [(pentamethylcyclopentadienyl)(cis-1,2-diaminocyclohexane)(chloro)iridium]chloride, showed no ill effects on the mice at high doses.
1. Drug resistance: a serious threat
In 2010 the World Health Organization (WHO) determined that antimicrobial resistance is one of the three greatest threats facing humanity today. That same year, the Infectious Diseases Society of America put forth the “10 × '20 Initiative”1 stating that it will be necessary to develop 10 new antimicrobials by 2020 to address this issue. As of the writing of this article, that goal is nowhere near having been achieved. This initiative was developed in order to combat the growing number of multi-drug resistant strains of bacteria and the threat of an imminent infectious disease epidemic. The WHO has also reported that 2 billion people are infected with some form of Mycobacterium tuberculosis. The increased frequency of antibiotic-resistant mutants is adding to growing numbers of multi-drug resistant (MDR) strains of all tuberculosis developing each year.2 On top of the growing number of antibiotic-resistant strains of tuberculosis, treatment for these types of infections can take years to complete.3 The longer patients are prescribed a specific set of antimicrobials, the higher the risk for the drugs to select resistant mutant strains. These long term dosage regimes are a major concern when it comes to the dangers of side effects toxic to patients and often increase the mutation selection among these microbes.
While methicillin-resistant forms of S. aureus (MRSA) have been known since the 1960s, the occurrence, virulence, and further resistance to antibiotics has risen steadily.4–7 MRSA as a food-borne pathogen has also increased making it an “evolving threat”.8 As the number of MDR strains increase, current treatments are becoming more and more inadequate. The United States Center for Disease Control (CDC) made the alarming statement that “We are in a post-antibiotic era where doctors and patients are faced with untreatable infections”.9,10 Overall, there is no shortage of papers and reports extolling the crisis of antimicrobial resistance.11–15
There is no doubt that much of our current crisis has been fueled by misuse and overuse of antibiotics thus resulting in the evolution of pathogens resistant to the current library of antimicrobials.16–19 While education and better treatment practices will help to slow the growth of this crisis, the addition of new antimicrobials to our current arsenal is absolutely essential in going forward in the fight against dangerous, multi-drug resistant pathogens. On the natural product front and on the synthetic organic chemistry front, new discoveries are being made on a daily basis, yet we have still not developed much in the way of new antimicrobial agents that can address the crisis in untreatable infections.
While antimicrobial resistance is a complex issue that will not be solely resolved by finding new antimicrobials, one thesis of this paper is that the scientific community is not yet daring enough in its approaches and is still “stuck inside a box” of conventional thinking with respect to what kinds of molecules could act as antimicrobial agents. Interest in metal based antimicrobial agents is growing, though many of the recent reports feature complexes of metals with organic molecules that are already known to have antimicrobial properties.20–22 Indeed, a new edition of “Metals in Medicine” has a larger section on antimicrobials than did the previous edition, but there still, the bulk of the section deals with complexes of known drugs.23 Lately, we have reported on a class of what are known as “half-sandwich” metal complexes that can be tuned for activity against mycobacteria24,25 as well as against S. aureus including MRSA.26 This paper gives further details of our work on anti-staph complexes including an initial toxicity screening and makes the case that, as strange as this class of compounds may seem to many practitioners in the field, it is time to be bold in a much deeper and broader examination of metal compounds.27
2. Experimental
2.1. General synthetic techniques
The syntheses of the various diamine complexes discussed in this paper have been described before.26 All compounds are air-stable and water soluble, so no special handling techniques are needed. The best candidates of all of the diamine complexes tested were subject to a number of in vitro procedures to test for toxicity toward mammalian cells. The effect of our compounds on both vero28 and HEK cells were measured using ATP assays.29 One of the best candidates was the subject of a mouse toxicity safety study described here. All animal experiments were performed in accordance with the “Guide for the Care and Use of Laboratory Animals” published by the National Research Council, with protocols reviewed and approved by Virginia Tech's Institutional Animal Care and Use Committee (IACCUC).
2.2. X-ray crystallography
The crystal structures of the Cp* iridium complexes of both the cis- and trans-isomers of 1,2-diaminocyclohexane were determined. Data was collected on an Oxford diffraction Gemini_Mo_Eos diffractometer with crystals maintained at 99.9 K during data collection. Structures were solved and refined using SHELX30 software and figures and other crystallographic output were made using OLEX2.31 Full details of the experimental data for the crystal structures may be found in the supplemental material and the cif files may be downloaded from the Cambridge Crystallographic Data Centre, CCDC ; 1901126 for 1 and CCDC ; 1901125 for 2.32
2.3. Toxicology studies
Hemolysis of sheep red blood cells was carried out following the procedure described previously.33 The sheep red blood cells were obtained from VWR (Atlanta, GA) as a 10% suspension. In vitro cytotoxicity of the transition metal–diamine complexes were measured using Vero cells and the CellTiter 96 One Solution Cell Proliferation assay (Promega Corp., Madison, WI). Vero Cells were obtained from ATCC.34 Further in vitro toxicity testing was carried out by studying the effect of compound 1 on ATP concentration in human embryonic kidney cells.35 HEK-93 cells were obtained from Sigma-Aldrich (St. Louis, MO).
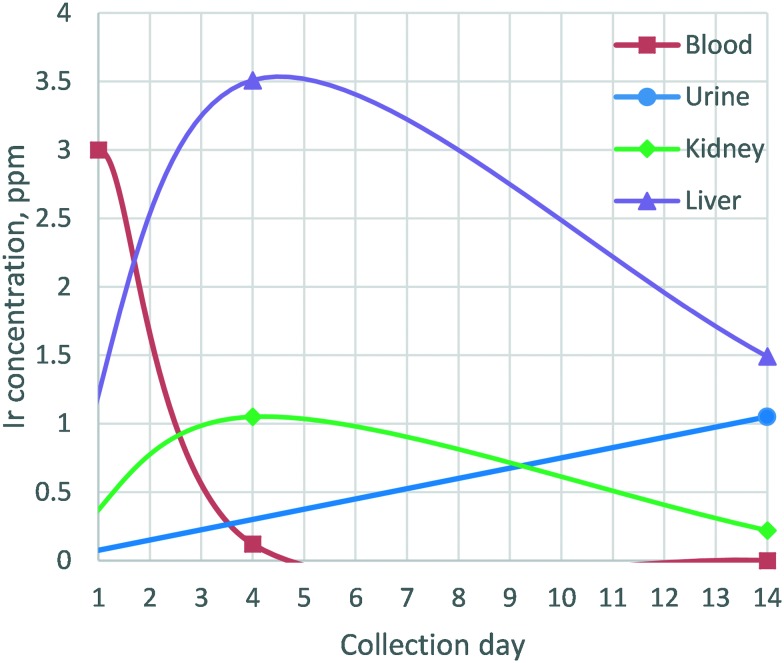
Insufficient resources were available to carry out complete pharmacokinetic and biodistribution studies for compound 1. However, a model safety experiment with the most active anti-MRSA iridium-containing complex was carried out. Compound 1 could be safely given intravenously (IV) to adult male outbred white mice (ICR strain) at a dose of 2.5 mg kg–1: a dose that provided blood concentrations >10-fold above the minimal inhibitory concentration (MIC) of S. aureus without causing neurobehavioral, pathologic, or histopathologic changes in treated mice.36 The same was true for mice given a double dose of 5.0 mg kg–1. The administered IV dose was similar to that used for aminoglycoside antibiotics such as tobramycin, the lipopeptide daptomycin, and the polymyxin colistimethate.37,38 Blood samples were drawn periodically during the two week study. Weights of mouse groups did not significantly change during the two week observation period. At the end of two weeks, all mice were sacrificed, a necropsy was performed, and the blood, organs, and urine (not quantitatively, simply for analysis) were collected. The collected urine, intermediate blood samples, and final organ digestions were analyzed for Ir content by ICP-OES. Fig. 8 shows a comparison between the levels of iridium found in mouse blood, urine, and organ samples taken one, four, and fourteen days post administration of an iridium metal complex antimicrobial (Ir-MCA). This representative data readily demonstrates that a significant portion of the iridium is eliminated from the blood over a relatively short period of time. The mouse studies were conducted in approved facilities with an approved protocol, IACUC #12-CVM.
Fig. 8. Detection of iridium in blood and urine (ppm) over the course of the 2 week study. Iridium content was determined via ICP-OES.
2.4. Calculations
The X-ray crystal structure of complex 1 showed two conformations of the 1,2-diaminocyclohexane group in the independent unit. In solution, as measured by proton NMR spectroscopy, only one species of complex 1 was observed, suggesting that a rapid interconversion of conformers is taking place in solution. To obtain some support for this hypothesis, Gaussian calculations to determine the energy of the conformers as well as the activation barrier for interconversion were carried out. Calculations were performed using Gaussian 0939,40 at the B3LYP level of theory41,42 using the lanl2dz basis set.43 The relative energy levels of the conformers shown in Fig. 5 and 6 were 0 kcal mol–1 assigned for the conformer in Fig. 5 and +0.47 kcal mol–1 higher for the conformer in Fig. 6. A calculation of the optimized transition state geometry showed the barrier to the conformational change was less than 10 kcal mol–1.
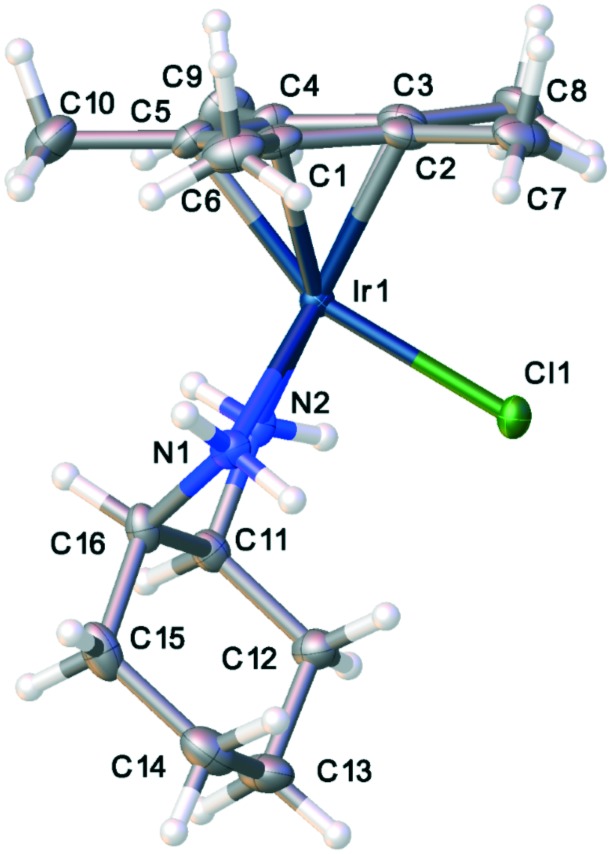
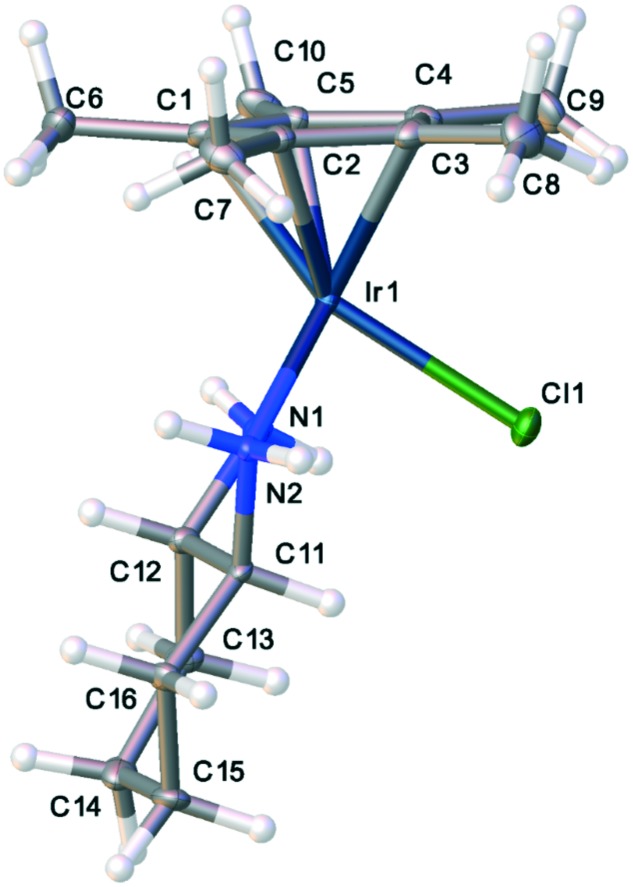
Fig. 5. Thermal ellipsoid plot of one conformer of the cis-diaminocyclohexyl compound, 1.
Fig. 6. Thermal ellipsoid plot of a second conformer of the cis-diaminocyclohexyl compound, 1.
3. Results and discussion
3.1. A brief summary of previous work with piano stool complexes
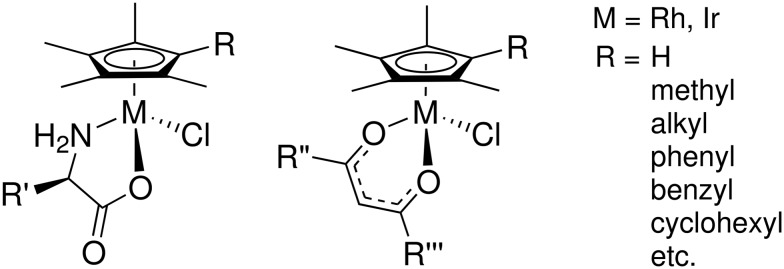
In a previous paper, we showed that piano stool compounds of cobalt, rhodium and iridium with Cp*R ligands and chelating amino acids, (Fig. 1, left), were effective antimicrobial agents against mycobacteria, including tuberculosis (TB).24 We also reported on an extensive series of β-diketonate Cp* rhodium and iridium complexes (Fig. 1, right) with antimycobacterial activity.25
Fig. 1. Examples of (Cp*R)M complexes found to be active against mycobacteria. Additional details regarding the substituents are given in ref. 24 and 25.
In both of these series, similar structure activity relationships (SARs) were observed: a) longer alkyl chains (R) on the Cp*R ligand increased activity up to the point at which the longer chains significantly decreased water solubility; b) hydrophobic groups on the amino acid or β-diketonate ligands also increased activity. For the amino acid complexes, one of the more subtle relationships is that l-amino acid complexes were active, but a direct comparison to the corresponding d-amino acid complex showed significantly decreased activity. The fact that there are strong relationships between antimicrobial activity and the identity of the chelating ligand especially shows that activity depends, in part, on those ligands and is not the result of decomposition of the complex to some indiscriminately poisonous metal-containing fragment.
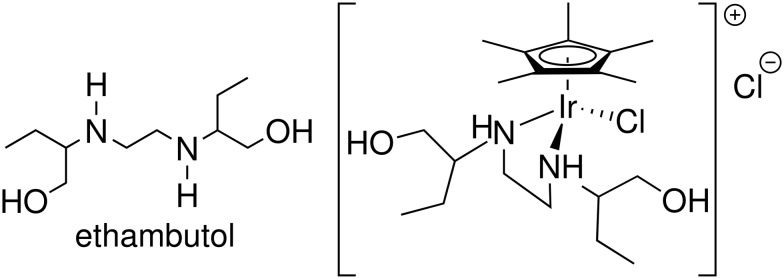
As mentioned above, many of the metal-based antimicrobial agents that have been investigated in the literature feature chelating ligands that are already known to have antimicrobial properties, including biguanide21 and sulfadoxine.22 While the majority of ligands discussed in this article are not known to have antimicrobial activity, we noted that the known tuberculosis drug, ethambutol,44,45 is a 1,2-diamine and we thought to make a metal complex of it (Fig. 2) to determine if the metal complex would retain its potency against Mycobacterium tuberculosis.
Fig. 2. The known anti-tuberculosis drug ethambutol (left) and [Cp*IrCl(ethambutol)]Cl (right), found to be active against staphylococcus.
Interestingly, we found that the ethambutol complex of Cp*Ir was inactive against mycobacteria, but did have some activity against S. aureus, including MRSA (Table 1). That led us to begin an investigation into the activity of chelating diamine piano-stool compounds of Rh and Ir. Before moving on to the discussion of S. aureus, it is important to note another significance of this total lack of activity against mycobacteria: side by side MIC comparisons of ethambutol and an ethambutol complex showed that there can be no decomposition of the ethambutol complex leading to free ethambutol in the solutions. Had this decomposition taken place, even to a small degree, enough ethambutol would have been released to show some antimycobacterial activity at the higher concentrations of the complex tested.
Table 1. MICs of MCAs against S. aureus and MRSA, μg mL–1 (μM). More details are provided in ref. 26.
| Compound | S. aureus | MRSA |
| Cp*IrCl(L-phengly) | >64 (>125) | >64 (>125) |
| [Cp*IrCl(ethylenediamine)]Cl | 35 (76.3) | 33 (72.0) |
| [Cp*IrCl(N,N′-dimethylethylenediamine)]Cl | 40 (82.2) | 40 (82.2) |
| [Cp*IrCl(N-benzylethylenediamine)]Cl | 7.5 (13.7) | 7.5 (13.7) |
| [Cp*IrCl(cis-1,2-diaminocyclohexane)]Cl | 5 (9.8) | 7.5 (14.6) |
| [Cp*IrCl(trans-1,2-diaminocyclohexane)]Cl | >64 (>125) | >64 (>125) |
| [Cp*IrCl(ethambutol)]Cl | 35 (58.1) | 40 (66.4) |
| [Cp*IrCl2]2 | >64 (>80.3) | >64 (>80.3) |
| Ethylenediamine | >64 (>1065) | >64 (>1065) |
| cis-1,2-Diaminocyclohexane | >64 (>560) | >64 (>560) |
| trans-1,2-Diaminocyclohexane | >64 (>560) | >64 (>560) |
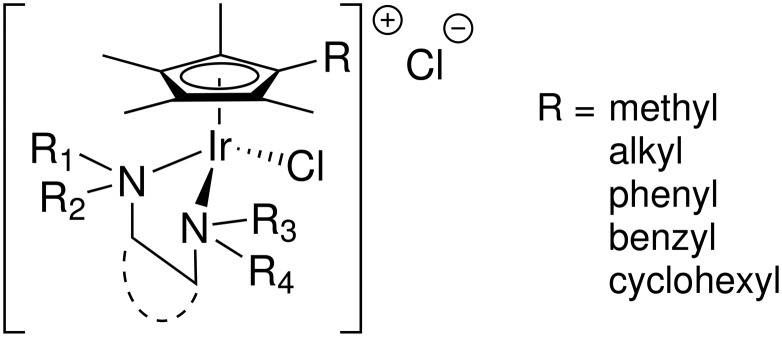
3.2. 1,2-Diamino alkane complexes – effective against MRSA
The experience with ethambutol led us to investigate more complexes of 1,2-diamines (Fig. 3) from the simplest 1,2-diaminoethane (ethylenediamine, en) to more complicated and substituted diamines. The structure–activity relationships for (Cp*R)Ir(1,2-diamine)Cl cations were reported previously,26 but the overall theme would suggest some of the most important characteristics were: a) a longer hydrophobic chain on the Cp*R ring; b) increased hydrophobicity of the substituents on the 1,2-diaminoalkane, either on the nitrogen or the alkyl backbone; and, c) the presence of hydrogen-bond donors (N–H) on the diamine ligand all correlated with higher antimicrobial activity. However, there was one subtlety that will be discussed in greater detail below: a large difference in activity was found between cis-1,2-diaminocyclohexane and trans-1,2-diaminocyclohexane complexes. While both complexes could be synthesized readily and are quite stable, the complex of the cis isomer was significantly more active than the trans. Moreover, neither diamine was active as the free ligand (Table 1), thus none of the activity could be attributable to a decomposition of the diamine complexes leading to release of the free ligand. Indeed, this was already seen with the ethambutol complex – no decomposition was observed as evidenced by no activity against mycobacteria.
Fig. 3. Examples of (Cp*R)Ir complexes found to be active against staphylococcus. Additional details regarding the substituents are given in ref. 26.
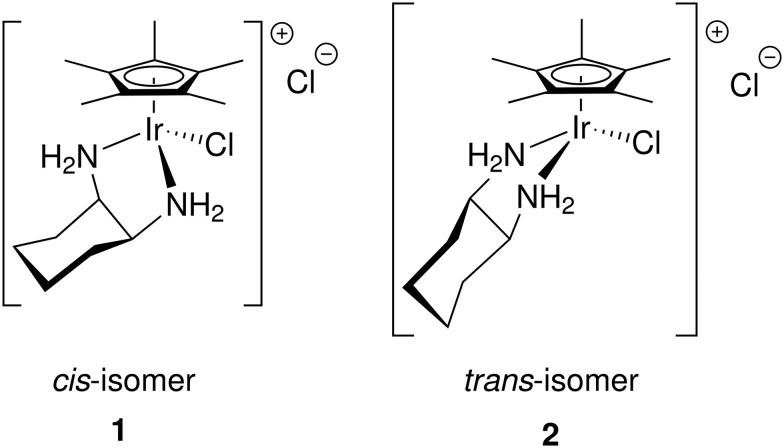
3.3. (Cp*R)IrCl complexes of 1,2-diaminocyclohexane: comparison between the R,S (cis) and the S,S (+) (trans)isomers
One very distinct difference between the antimycobacterial compounds (Cp*R)MCl(acac) and (Cp*R)MCl(aa) (Fig. 1), and the anti-MRSA complexes (Fig. 3) described herein is that the former are neutral and the latter are cationic, with a non-coordinating chloride as the counter-anion. This surely must affect how these two classes of compounds cross cellular membranes as well as with which sites they may interact to provide their antimicrobial activity. Hydrolysis studies may help elucidate differences between these compounds, but there is no spectroscopic evidence for chloride dissociation for these already cationic complexes. Further, complex 1 was given in isotonic saline solution which, as well as chloride concentration in blood, should further inhibit any chloride dissociation. Studies to help identify the site of action of both classes of molecules are ongoing.
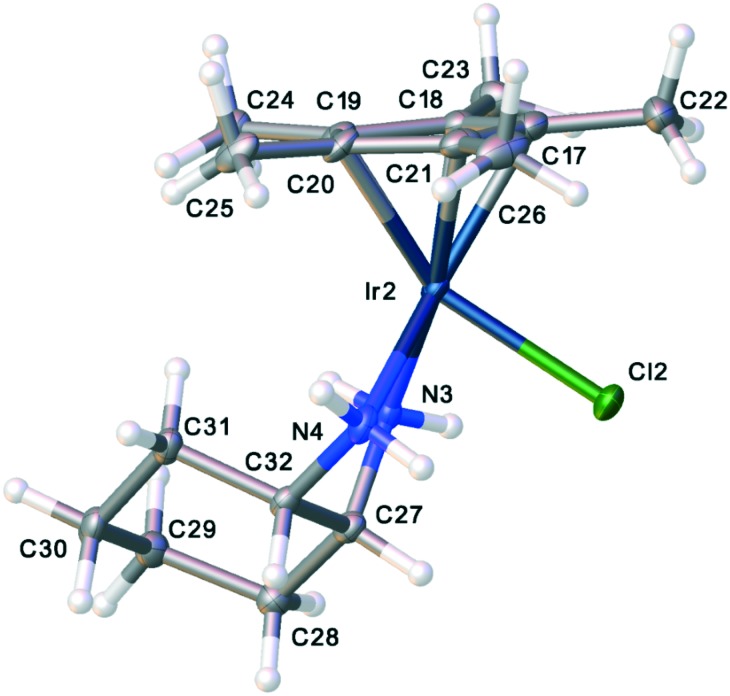
One of the most remarkable activity differences was found in direct comparison between [Cp*IrCl(cis-1,2-diaminocyclohexane)]Cl and [Cp*IrCl(trans(+)-1,2-diaminocyclohexane)]Cl (Fig. 4). While the complex of the cis isomer was very active, the complex of the trans isomer showed very little activity. This led us to conduct single crystal X-ray diffraction analysis of complexes of both isomers, complexes 1 and 2. Though Cp*R complexes with R = C6H13 were more active, crystals of Cp* were more easily obtained and should still provide relevant information on the key differences between the cis- and trans-complexes. The full details and tables of the two structures may be found in the ESI† as well as from the Cambridge Crystallographic Data Centre (CCDC) (1: CCDC ; 1901126; 2: CCDC ; 1901125). Fig. 5 and 6 show the thermal ellipsoid plot of the cationic [Cp*IrCl(cis-1,2-diaminocyclohexane)]+ and Fig. 7 shows the thermal ellipsoid plot of a cation of [Cp*IrCl(trans-1,2-diaminocyclohexane)]+. Figures showing the full asymmetric units for both structures may be found in the ESI.†
Fig. 4. Labeling notation for the (Cp*R)Ir complexes primarily discussed in this article.
Fig. 7. Thermal ellipsoid plot of the trans-diaminocyclohexyl compound, 2.
The most striking and, perhaps, the only significant difference between the two structures is the conformation of the cyclohexyl rings. The inactive complex with the trans-1,2-diaminocyclohexane shows a conformation with the cy ring laying fairly flat and projecting away from the Ir atom. The distance between Ir and the distal H atom of the cy ring in the trans complex is 6.31 Å. The same distances for all of the molecules in the asymmetric unit (4 unique molecules) is 5.7 Å. Another indicator of the conformational difference between the complexes of the two isomers is the difference in the measurements of the “bounding box” around the two molecules. The “bounding box” is the box into which the molecule just “fits” using the van der Waals radii of the atoms. For the complex of the trans isomer, the bounding box has dimensions of 9.027 × 8.542 × 11.845 with a volume of 913.37 Å3. The more folded structure of the complex of the cis isomer has a bounding box of 8.389 × 8.539 × 11.423 with a volume of 818.35 Å3. One supposition is that the complex of the cis isomer fits into a pocket of the target of activity while that of the trans isomer does not.
In the crystal lattice, there are two conformers of the complex of the cis isomer of 1,2-diaminocyclohexane, one with the ring tucked under the metal and pointing toward the chlorine atom (Fig. 5) and one with the ring flipped and pointing away from the chlorine atom (Fig. 6). The interconversion between these two conformers is similar to the conformational flip observed in (cis)-decalin, which shows a barrier to ring inversion of about 8 kcal mol–1. It is likely that the barrier to this conformational flip is similar to that of cis-decalin and that there is interconversion between conformations at room temperature in solution.46 Indeed, the proton NMR spectrum for complex 1 shows evidence for only one species in solution. Further support for a low barrier to conformer interconversion comes from theoretical calculations showing the barrier is less than 10 kcal mol–1.
The marked difference in MIC between the complexes of the two ligand isomers is further indication that these complexes are stable under physiological conditions. Had these complexes decomposed to some other species of the Cp*Ir fragment, they would have shown similar activity to each other. This point is further bolstered by the lack of activity of ligand-free [(Cp*)IrCl2]2 (Table 1). Thus, neither the ligand nor the ligand-free iridium complex show activity. While it appears that this argument has been overly repeated in this paper, this is the most important point of this paper: to indicate that at least some of these half-sandwich compounds are safe for administration to mammals and to overcome a widely-held view that metal compounds are only appropriate for anti-cancer studies.
3.4. Safety
A common theme from reviewers of both papers and proposals dealing with the antimicrobial activity of transition metal compounds is that these complexes could never be used as antibiotics because they will surely be toxic to mammalian cells. This misunderstanding is easy to rationalize. Following the discovery of the anticancer activity of cisplatin, nearly all studies of the biological activity of transition metal complexes have been directed toward their anticancer properties.47 Thus, they always seem to be viewed as toxic to mammalian cells. We were prompted to carry out both in vitro and in vivo studies to show that, at levels therapeutic for bacteria, in this case S. aureus, our most active complex was not a problem for mammalian cells.
3.4.1. In vitro studies
All compounds in our studies at 50% toxicity concentration showed no hemolytic activity with sheep red blood cells. In addition, testing cytotoxicity with an ATP concentration assay using vero cells and also with human embryonic kidney (HEK) cells showed no toxicity at concentrations at least 50 times higher than MICs.
3.4.2. In vivo study
In a study involving the [Cp*IrCl(cis-1,2-diaminocyclohexane)]Cl complex 1, a toxicity screening was carried out using n = 10 per group of adult outbred white mice (ICR strain) obtained at weights of 25–30 g and dosed when they had an average weight of 40 g. In addition to a control group given vehicle alone (saline), one group of mice was given a single IV dose of 1 at a level of 2.5 mg kg–1 while a second group was dosed at 5 mg kg–1. Mice were observed initially and then periodically over a two week period. Mice were examined for a series of behavioral indices using a modified functional observational battery at 1, 4, and 6 h, and 1, 2, 7, 9, 12, and 14 days post-dosing.48 Body weight was measured and recorded at the time of each assessment. Blood and tissue samples were collected at 24 h, 4 days, and 14 days for clinical pathology and histopathological assessment. Of the animals receiving 1, 5/19 animals showed immediate signs of distress following dosing. Signs of distress ceased within one minute and animals had normal appearance, activity, and locomotion within 5 minutes. No subsequent evidence of neurobehavioral, clinical or histopathological detriments was noted. There were no observations of any untoward behavioral changes during the two weeks – mice had expected weight gains and displayed normal behavior. Further blood test information indicating no significant effects on various blood parameters may be found in the ESI.†
ICP-OES analysis of those samples for iridium showed the following (Fig. 8): 1. the concentration of Ir in the blood on day 1 was what was expected for the given dose and the expected blood volume in the mouse; 2. the concentration of iridium in the blood samples at day 4 dropped precipitously and liver and kidney showed significant presence of iridium; 3. there was no Ir detectable in blood samples taken following sacrifice on day 14 and the amount of iridium in the organs studied dropped. Individual urine samples were not obtained, but rather urine was collected at the end of the 14 day period (unfortunately, not quantitatively) and ICP-OES showed significant Ir content in the urine. The blood, urine, and organ analysis together show that mice have a mechanism to distribute the complex for uptake into organs, and for elimination from the organs and body overall. It is important to note that, in this study, the identity of the eliminated iridium species was not determined since we only quantified the presence of total elemental iridium content.
In addition to behavioral observation during the two week trial, following sacrifice, necropsies were performed and there were no lesions, tumors, discoloration, or malformations of any of the internal organs. Thus, as much as one can conclude from this limited toxicity study, these compounds are safe in mammals.
4. Conclusions
A class of transition metal piano-stool complexes has been shown to exhibit potent activity against Staphylococcus aureus, including against several strains of methicillin-resistant S. aureus (MRSA). In vitro and in vivo toxicity studies show that, to the extent knowable at this time, complex 1 is both potent against one of the most problematic drug resistant bacterial strain and well tolerated by mammalian cells, including in mice. This provides strong evidence that, in order to address the crisis of drug resistance across the globe, it is time to truly think “outside the box” of purely organic/natural product compounds for antimicrobial agents and continue to study the alternatives of transition metal complexes in greater depth and breadth. Nearly all of the studies advocating approaches to fighting the growing concern over drug resistance in pathogens make two strong recommendations: 1) better education so that antimicrobials are not abused with such abuse leading to development of resistance; and, 2) that new “weapons in our arsenal” to combat drug resistance must be developed. This paper shows one pathway to combat drug resistance that we must continue to explore.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
The Virginia Tech Foundation is gratefully acknowledged for funding the mouse safety study described in this paper. The Virginia Tech Center for Drug Discovery is also acknowledged for funds that allowed the purchase of some materials and supplies for this project. The remainder of funds were provided by private donors to the Virginia Tech Foundation's Fund for Research in the Merola group and, for their philanthropy, we are very grateful. The authors would also like to thank Dr. David Morris for help with crystallographical analyses and Professor Diego Troya for discussions on Gaussian calculations.
Footnotes
†Electronic supplementary information (ESI) available: Images showing the full asymmetric units for complexes 1 and 2, toxicology report, ICP data, sample Gaussian calculations, and experimental data for the X-ray crystallographic determinations of complexes 1 and 2. CCDC 1901125 and 1901126. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c9md00140a
References
- Gilbert D. N., Guidos R. J., Boucher H. W., Talbot G. H., Spellberg B., Edwards Jr J. E., Scheld W. M., Bradley J. S., Bartlett J. G. Clin. Infect. Dis. 2010;50:1081–1083. doi: 10.1086/652237. [DOI] [PubMed] [Google Scholar]
- Fair R. J., Tor Y. Perspect. Med. Chem. 2014;6:25–64. doi: 10.4137/PMC.S14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raviglione M. C., Smith I. M. N. Engl. J. Med. 2007;356:656–659. doi: 10.1056/NEJMp068273. [DOI] [PubMed] [Google Scholar]
- Carretto E., Visiello R. and Nardini P., Pet-to-Man Travel. Staphylococci A World Prog., 2018, vol. 4, pp. 225–235. [Google Scholar]
- Turner N. A., Sharma-Kuinkel B. K., Maskarinec S. A., Eichenberger E. M., Shah P. P., Carugati M., Holland T. L., Fowler V. G. Nat. Rev. Microbiol. 2019;17(4):203–218. doi: 10.1038/s41579-018-0147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challagundla L., Luo X., Tickler I. A., Didelot X., Coleman D. C., Shore A. C., Coombs G. W., Sordelli D. O., Brown E. L., Skov R., Larsen A. R., Reyes J., Robledo I. E., Vazquez G. J., Rivera R., Fey P. D., Stevenson K., Wang S.-h., Kreiswirth B. N., Mediavilla J. R., Arias C. A., Planet P. J., Nolan R. L., Tenover F. C., Goering R. V., Robinson D. A. mBio. 2018;9:1–15. doi: 10.1128/mBio.02016-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copin R., Sause W. E., Balasubramanian D., Dyzenhaus S., Jamil M., Kumar K., Lees J., Stachel A., Jason C., Drlica K., Phillips M., Weiser J. N., Paul J., Uhlemann A.-c., Altman D. R., Bakel H. V., Lighter J., Torres V. J., Copin R., Sause W. E., Fulmer Y., Balasubramanian D., Dyzenhaus S., Ahmed J. M. Proc. Natl. Acad. Sci. U. S. A. 2019;116:4747–4747. doi: 10.1073/pnas.1814265116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro A., Silva J. and Teixeira P., Foodborne Dis., Elsevier, 2018, vol. 15, pp. 213–238. [Google Scholar]
- Molteni M. and Patel J., Wired, 2017.
- NCEZID, Antibiotic-Resistant Infections, 2019, https://www.cdc.gov/ncezid/what-we-do/our-topics/fighting-antibiotic-resistance.html.
- Khamash D. F., Milstone A. M., Carroll K. C., Gadala A., Klein E., Maragakis L. L., Cosgrove S. E., Fabre V. Infect. Control Hosp. Epidemiol. 2019:1–2. doi: 10.1017/ice.2019.4. [DOI] [PubMed] [Google Scholar]
- Kesecioglu J., Mer M., Paiva J.-A., Carlet J., Zahar J.-R., Antonelli M., Timsit J.-F., Garnacho-Montero J., De Waele J. J., Poljak M., Roberts J. A., Bassetti M., Canton R., Dimopoulos G., De Backer D., Lipman J., Rodriguez Bano J., Akova M. Intensive Care Med. 2017;44:189–196. doi: 10.1007/s00134-017-5036-1. [DOI] [PubMed] [Google Scholar]
- Aslam B., Wang W., Arshad M. I., Khurshid M., Muzammil S., Rasool M. H., Nisar M. A., Alvi R. F., Aslam M. A., Qamar M. U., Salamat M. K. F., Baloch Z. Infect. Drug Resist. 2018;11:1645–1658. doi: 10.2147/IDR.S173867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. Adv. Microbiol. 2018;08:65–76. [Google Scholar]
- Leeson N. and Hsueh P.-R., Antimicrobial resistance in the 21st century, 2015, vol. 10, pp. 297–298. [DOI] [PubMed] [Google Scholar]
- Levy S. B., Marshall B. Nat. Med. 2004;10:S122–S129. doi: 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- Davies J., Davies D. Microbiol. Mol. Biol. Rev. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K., Courvalin P., Dantas G., Davies J., Eisenstein B., Huovinen P., Jacoby G. A., Kishony R., Kreiswirth B. N., Kutter E., Lerner S. A., Levy S., Lewis K., Lomovskaya O., Miller J. H., Mobashery S., Piddock L. J. V., Projan S., Thomas C. M., Tomasz A., Tulkens P. M., Walsh T. R., Watson J. D., Witkowski J., Witte W., Wright G., Yeh P., Zgurskaya H. I. Nat. Rev. Microbiol. 2011;9:894–896. doi: 10.1038/nrmicro2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDDEP, The State of World's Antibiotics 2015, Center for disease dynamics, economics & policy technical report, 2015.
- Patra M., Gasser G., Metzler-Nolte N. Dalton Trans. 2012;41:6350–6358. doi: 10.1039/c2dt12460b. [DOI] [PubMed] [Google Scholar]
- Chen F., Moat J., McFeely D., Clarkson G., Hands-Portman I. J., Furner-Pardoe J. P., Harrison F., Dowson C. G., Sadler P. J. J. Med. Chem. 2018;61:7330–7344. doi: 10.1021/acs.jmedchem.8b00906. [DOI] [PubMed] [Google Scholar]
- Chellan P., Avery V. M., Duffy S., Triccas J. A., Nagalingam G., Tam C., Cheng L. W., Liu J., Land K. M., Clarkson G. J., Romero-Canelón I., Sadler P. J. Chem. – Eur. J. 2018;24:10078–10090. doi: 10.1002/chem.201801090. [DOI] [PubMed] [Google Scholar]
- Dabrowiak J. C., Metals in Medicine, John Wiley & Sons Ltd, Oxford, UK, 2017. [Google Scholar]
- Karpin G. W., Merola J. S., Falkinham J. O. Antimicrob. Agents Chemother. 2013;57:3434–3436. doi: 10.1128/AAC.00452-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuChane C. M., Brown L. C., Dozier V. S., Merola J. S. Organometallics. 2018;37:530–538. [Google Scholar]
- Karpin G. W., Morris D. M., Ngo M. T., Merola J. S., Falkinham J. O. MedChemComm. 2015;6:1471–1478. [Google Scholar]
- .
- Howard B. A., Rumore A. C., Lawrence C. B., Babiceanu M. C., Kita H. Front Microbiol. 2013;4:1–14. doi: 10.3389/fmicb.2013.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thermofisher, Molecular Probes. ATP Determination Kit (A22066), 2005, https://tools.thermofisher.com/content/sfs/manuals/mp22066.pdf.
- Sheldrick G. M. Acta Crystallogr., Sect. A: Found. Crystallogr. 2007;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- Dolomanov O. V., Bourhis L. J., Gildea R. J., Howard J. A. K., Puschmann H. J. Appl. Crystallogr. 2009;42:339–341. doi: 10.1107/S0021889811041161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groom C. R., Bruno I. J., Lightfoot M. P., Ward S. C. Acta Crystallogr., Sect. B: Struct. Sci., Cryst. Eng. Mater. 2016;72:171–179. doi: 10.1107/S2052520616003954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisuria B. B., Actis M. L., Hardrict S. N., Falkinham J. O., Cole M. F., Cihlar R. L., Peters S. M., MacRi R. V., Sugandhi E. W., Williams A. a., Poppe M. a., Esker A. R., Gandour R. D. Bioorg. Med. Chem. 2011;19:2918–2926. doi: 10.1016/j.bmc.2011.03.036. [DOI] [PubMed] [Google Scholar]
- ATCC, Vero (ATCC® CCL-81) Product information, https://www.atcc.org/Products/All/CCL-81.aspx.
- Leach F. R. and Webster J. J. B. T. M. i. E., Biolumin. Chemilumin. Part B, Academic Press, 1986, vol. 133, pp. 51–70. [Google Scholar]
- Ehrich M., Hinckley J., Jortner B. S., Boes K., Karpin G. W., Falkinham J. O., Merola J. S. Toxicology. 2015:135. [Google Scholar]
- Israel K. S., Welles J. S., Black H. R. J. Infect. Dis. 1976;134:S97–S103. doi: 10.1093/infdis/134.supplement_1.s97. [DOI] [PubMed] [Google Scholar]
- Wang Y., Burr D. H., Korthals G. J., Sugiyama H. Appl. Environ. Microbiol. 1984;48:951–955. doi: 10.1128/aem.48.5.951-955.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J. and Polik W., WebMO Enterprise, https://www.webmo.net/.
- Frisch M. J., Trucks G. W., Schlegel H. B., Scuseria G. E., Robb M. A., Cheeseman J. R., Scalmani G., Barone V., Petersson G. A., Nakatsuji H., Li X., Caricato M., Marenich A., Bloino J., Janesko B. G., Gomperts R., Mennucci B., Hratchian H. P., Ortiz J. V., Izmaylov A. F., Sonnenberg J. L., Williams-Young D., Ding F., Lipparini F., Egidi F., Goings J., Peng B., Petrone A., Henderson T., Ranasinghe D., Zakrzewski V. G., Gao J., Rega N., Zheng G., Liang W., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Vreven T., Throssell K., Montgomery J. J. A., Peralta J. E., Ogliaro F., Bearpark M., Heyd J. J., Brothers E., Kudin K. N., Staroverov V. N., Keith T., Kobayashi R., Normand J., Raghavachari K., Rendell A., Burant J. C., Iyengar S. S., Tomasi J., Cossi M., Millam J. M., Klene M., Adamo C., Cammi R., Ochterski J. W., Martin R. L., Morokuma K., Farkas O., Foresman J. B. and Fox D. J., Gaussian09, Gaussian, Inc., Wallingford CT, 2016.
- Becke A. D. J. Chem. Phys. 1993;98:5648–5652. [Google Scholar]
- Lee C., Yang W., Parr R. G. Phys. Rev. B: Condens. Matter Mater. Phys. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- Hay P. J., Wadt W. R. J. Chem. Phys. 1985;82:299–310. [Google Scholar]
- Place V. A., Thomas J. P. Am. Rev. Respir. Dis. 1963;87:901–904. doi: 10.1164/arrd.1963.87.6.901. [DOI] [PubMed] [Google Scholar]
- Telenti A., Stockbauer K. E., Kreiswirth B. N., Moghazeh S. L., Pan X., Sreevatsan S., Musser J. M., Jacobs W. R. Antimicrob. Agents Chemother. 2018;41:1677–1681. doi: 10.1128/aac.41.8.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerig J. T., Roberts J. D. J. Am. Chem. Soc. 1966;88:2791–2799. [Google Scholar]
- Dabrowiak J. C., Metals in Medicine, John Wiley & Sons Ltd., 2009, p. 334. [Google Scholar]
- Tegeris J. S., Balster R. L. Toxicol. Sci. 1994;22:240–250. doi: 10.1006/faat.1994.1028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.