ABSTRACT
PLEKHG2 is a Gβγ- and Gαs-dependent guanine nucleotide exchange factor for Rac1 and Cdc42 small GTPases and has been shown to mediate signaling pathways such as those for actin cytoskeletal reorganization and serum response element (SRE)-dependent gene transcription. We have shown that the four-and-a-half LIM domains (FHL) 1 acts as a positive regulator of PLEKHG2. Here, we evaluated the other FHL family members and found that the FHL1A specifically regulate the PLEKHG2 activity. Moreover, FHL1A further enhanced Gβγ- and PLEKHG2-induced SRE-dependent gene transcription, whereas FHL1A partially restored the attenuated PLEKHG2-induced SRE-dependent gene transcription by Gαs. Our results suggest that FHL1A specifically interacts with PLEKHG2 to regulate a function of PLEKHG2 that is modified by the interaction of Gβγ and Gαs.
KEYWORDS: four-and-a-half LIM domain 1 (FHL1), guanine nucleotide exchange factor (GEF), protein-protein interaction, Rho family small GTPases (Rho)
Introduction
The cell morphological changes induced by actin cytoskeletal rearrangement are important for cell migration, cell division and cell differentiation. It is well known that Rho family small GTPases (Rho), including RhoA, Rac1 and Cdc42, regulate these morphological changes thorough the assembly and organization of F-actin in response to extracellular stimuli (i.e., receptor tyrosine kinases and heterotrimeric G protein-coupled receptors (GPCR)). Like other small GTPases, Rho is activated by their conversion from the GDP-bound state to the GTP-bound state, and this exchange is catalyzed by Rho-specific guanine nucleotide exchange factors (RhoGEFs).1
RhoGEFs comprise two families. One is the diffuse B-cell lymphoma (Dbl) protein family, whose members include a Dbl-homology (DH) domain followed by a pleckstrin-homology (PH) domain. The other is the dedicator of cytokinesis (DOCK) family.2 The DH domain plays a critical role in the conversion of GDP of Rho to GTP. We previously identified that PLEKHG2, one of the novel Dbl family RhoGEFs for Rac1 and Cdc42, was regulated by the heterotrimeric G proteins in response to GPCR stimuli. Heterotrimeric G protein Gβγ subunits bound to the N-terminal region of the DH domain of PLEKHG2 and enhanced the RhoGEF activity of PLEKHG2.3 On the other hand, the Gαs subunit has been shown to attenuate the function of PLEKHG2 by direct interaction.4
RhoGEFs are also regulated by various intracellular signals through an intramolecular multi-functional domain in RhoGEF.5 For example, two other Gβγ-dependent RhoGEFs, P-Rex1 and P-Rex2, are regulated via phosphorylation by several protein kinases, including p21-activated kinase 1, protein kinase A and protein kinase C.6-9 Recently we reported that the phosphorylation at Thr680 of PLEKHG2 by EGF/Ras/MAPK pathways regulated the PLEKHG2 functions related to cell morphological change.10 We also reported that the tyrosine phosphorylation at Tyr489 of PLEKHG2 by cSrc induced a tyrosine phosphorylation-dependent interaction between PLEKHG2 and PIK3R3 or ABL1.11 And we found that both β- and γ-actin acted as negative regulators of PLEKHG2 through direct interaction.12 Despite these discoveries, however, the regulatory mechanisms governing the functions of PLEKHG2 remain largely unknown.
In a previous study, we identified a zinc finger domain-containing protein, four-and-a-half LIM domains (FHL) 1, as a positive regulator of PLEKHG2.13 The FHL proteins are characterized by four cysteine-rich double-zinc-finger LIM domains with a half-LIM domain. LIM domains mediate protein-protein interactions and have been implicated in various cellular processes including cell adhesion. The family consists of FHL1 to -4 and the activator of CREM in testis (ACT, also known as FHL5).14 It is thought that FHL1, FHL2, and FHL3 are important in cancer development and progression.15 It has also been reported that the FHL1, -2, and -3 proteins inhibit human hepatoma cell growth both in vitro and in vivo through their physical and functional interaction with SMAD proteins. Previous studies have shown that FHL1 to -3 are located in both the nuclei and focal adhesions of C2C12 myoblasts. The overexpression of FHL1 to -3 has been shown to induce cell spreading and cell migration. Specifically, FHL1 inhibits integrin-mediated myoblast adhesion and promotes cell spreading and migration, resulting in the regulation of integrin-mediated cytoskeleton rearrangement.16 FHL2 has been shown to bind actin in order to modulate cytoskeleton dynamics and also functions as a lipid-triggered signaling molecule, which regulates migration and contraction during wound healing.17 FHL3 also binds actin and regulates α-actinin, resulting in increased cell spreading and the disruption of actin stress fibers, thereby promoting cell motility. The modulation of α-actinin levels by FHL3 could increase the motility and the tumorigenic properties of cells.18 Thus, it seems that not only FHL1 but also FHL2 and FHL3 may play roles in the Rho signaling pathway that is involved in cell morphological change. Here, we demonstrated that FHL1, but not FHL2 or FHL3, interacted with PLEKHG2 and influenced a function of PLEKHG2 that is regulated by heterotrimeric G protein signaling.
Results
There are three isoforms in FHL1: FHL1A, FHL1B and FHL1C. FHL1A interacts more strongly with PLEKHG2 and more potently enhances PLEKHG2-induced serum response element (SRE)-dependent gene transcription as compared to FHL1B. On the other hand, FHL2 and FHL3 also have two isoforms each (Fig. 1). As with FHL1A, the isoforms with four-and-a-half LIM domains structures are FHL2 isoform a (FHL2a) and FHL3 isoform 1 (FHL3-1). The total amino acid sequence of the FHL1A protein shows relatively high levels of amino acid sequence homology to FHL2a (48%) and FHL3-1 (44%), respectively. To examine whether FHL2a and FHL3-1 show the same effect as FHL1A on PLEKHG2-induced SRE-dependent gene transcription, which is known to be induced by Rho family activation, expression plasmids of Myc-tagged PLEKHG2 and Flag-tagged FHL1A, FHL2a or FHL3-1 were co-transfected into HEK293 cells. PLEKHG2-induced SRE-dependent gene transcription was significantly elevated in cells co-expressing FHL1A and PLEKHG2, whereas no increase was observed in cells co-expressing FHL2a or FHL3-1 (Fig. 2A). Since the enhancement of PLEKHG2-induced SRE-dependent gene transcription by FHL1A is due to the interaction of FHL1A and PLEKHG2, we examined whether PLEKHG2 interacted with FHL2a or FHL3-1. Myc-tagged PLEKHG2 and Flag-tagged FHL1A, FHL2a or FHL3-1 were overexpressed in HEK293 cells, and PLEKHG2 was immunoprecipitated with specific antibody against the Myc epitope tag. FHL1A was co-immunoprecipitated with PLEKHG2, whereas FHL2a and FHL3-1 were not co-immunoprecipitated (Fig. 2B). These results suggest that the interaction of PLEKHG2 and FHL1A is specific, and PLEKHG2 does not interact with other highly homologous FHL proteins, including FHL2a or FHL3-1. In terms of SRE-dependent gene transcription, the function of PLEKHG2 may be regulated by the interaction with FHL1A.
Figure 1.
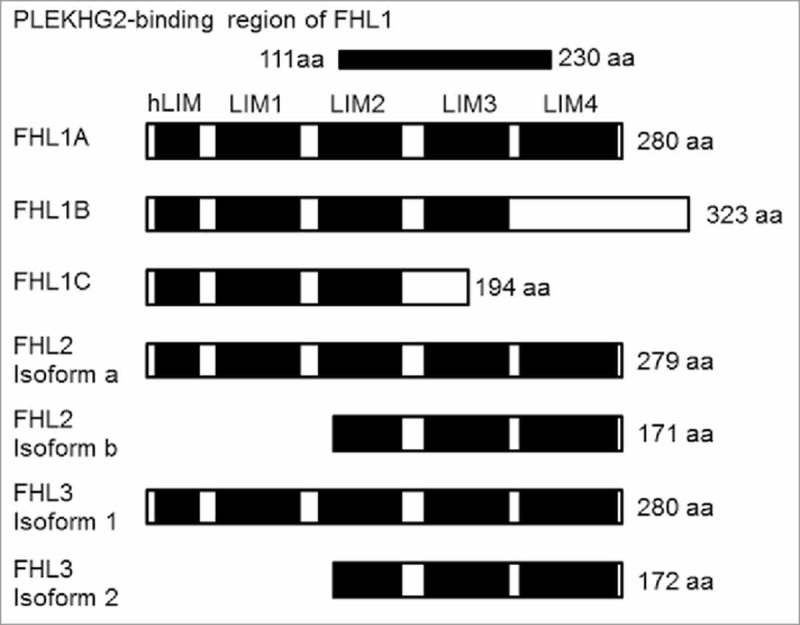
Schematic representation of the protein structure of the FHL family. FHL1A, FHL2a and FHL3-1 comprise an N-terminal half LIM domain (single zinc finger) followed by four complete LIM domains. aa, amino acids; hLIM, half LIM domain; LIM, LIM domain.
Figure 2.
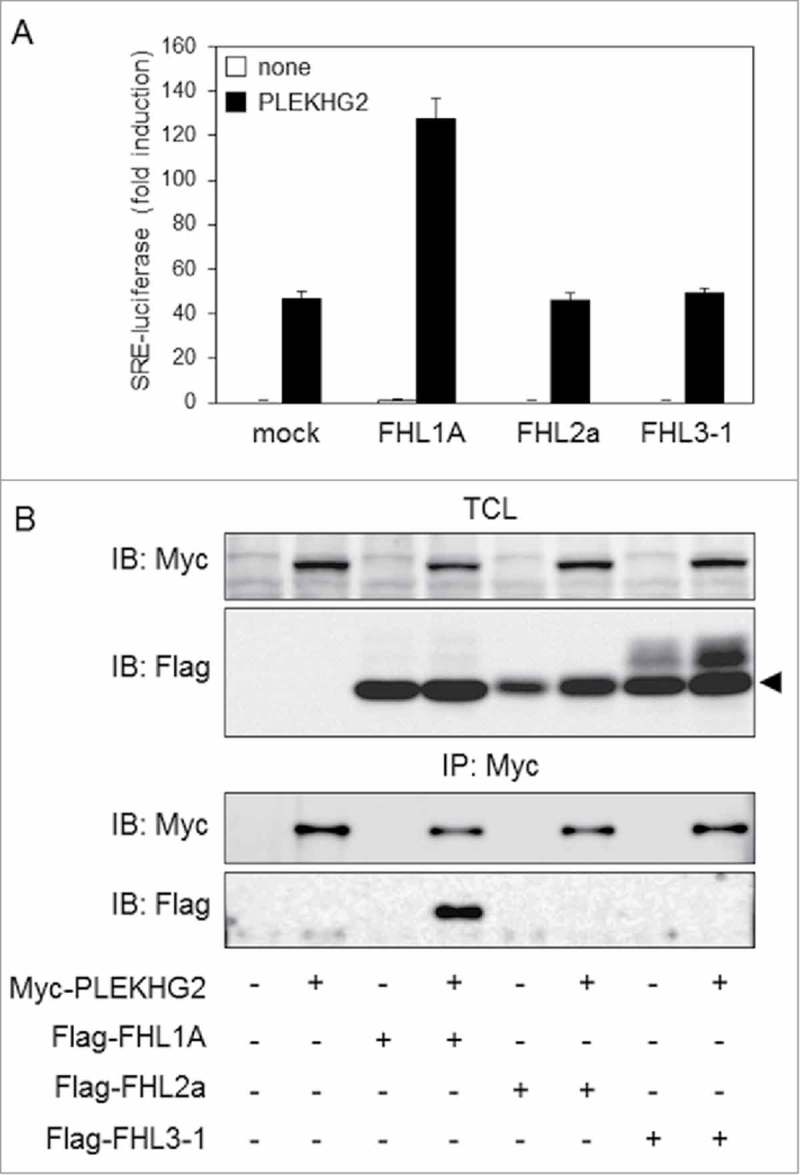
Effect of the FHL family on PLEKHG2-induced SRE-dependent gene transcription and the interaction with PLEKHG2. (A) HEK293 cells were co-transfected with pSRE.L-luciferase, pRL-SV40, Myc-tagged PLEKHG2 and Flag-tagged FHL1A, FHL2a or FHL3-1, as indicated. Transfected cells were lysed 24 h after transfection, and the effects of FHL1A, FHL2a and FHL3-1 on PLEKHG2-induced transcription were analyzed by a luciferase reporter gene assay. The experiment was performed in triplicate, and the values are the means ± SD (error bars). The data shown are representative of three independent experiments. (B) HEK293 cells were co-transfected with Myc-tagged PLEKHG2 and Flag-tagged FHL1A, FHL2a or FHL3-1, as indicated. Transfected cells were lysed 24 h after transfection, and PLEKHG2 was immunoprecipitated with anti-Myc antibody. Precipitated proteins were separated by SDS-PAGE and immunoblotted with anti-Myc (for PLEKHG2) antibody and anti-FLAG antibody (for FHL1A, FHL2a and FHL3-1). TCL, total cell lysate; IP, immunoprecipitation; IB, immunoblotting. The data shown are representative of three independent experiments.
Recently we reported that the Gαs subunit attenuated PLEKHG2-induced SRE-dependent gene transcription through its direct interaction with PLEKHG2. A constitutively active mutant of the Gαs subunit (GαsQ213L) also attenuated Gβγ-enhanced PLEKHG2-induced SRE-dependent gene transcription. On the other hand, we also reported that FHL1A further enhanced Gβγ-enhanced PLEKHG2-induced SRE-dependent gene transcription through its interaction with PLEKHG2. To investigate whether FHL1A has an effect on Gαs subunit-attenuated PLEKHG2-induced SRE-dependent gene transcription, Myc-tagged PLEKHG2 and Flag-tagged FHL1A with or without Gβγ or a GαsQ213L were overexpressed in HEK293 cells. Co-expression of FHL1A significantly enhanced Gβγ-enhanced PLEKHG2-induced SRE-dependent gene transcription as previously reported. On the other hand, co-expression of FHL1A could not restore Gαs-attenuated PLEKHG2-induced SRE-dependent gene transcription to the control levels, but slightly enhanced SRE-dependent gene transcription (Fig. 3A). To examine how co-expression of FHL1A affects the interaction between Gαs and PLEKHG2, Myc-tagged PLEKHG2 and Flag-tagged FHL1A with or without GαsQ213L were overexpressed in HEK293 cells. Co-expression of FHL1A slightly attenuated the interaction of PLEKHG2 and GαsQ213L (Fig. 3B). Furthermore, as shown in Fig. 3C, we performed quantification analysis. We confirmed the interaction between PLEKHG2 and GαsQ213L were reduced by the expression of Flag tagged FHL1A. These results suggested that the attenuation of the interaction slightly enhanced PLEKHG2-induced SRE-dependent gene transcription. Moreover, it appeared that FHL1A could not substantially weaken the inhibitory effect on PLELHG2-induced gene transcription of Gαs, because there is a possibility that FHL1A interacted with an amino acid region of PLEKHG2 that was different from the Gαs-binding region.
Figure 3.
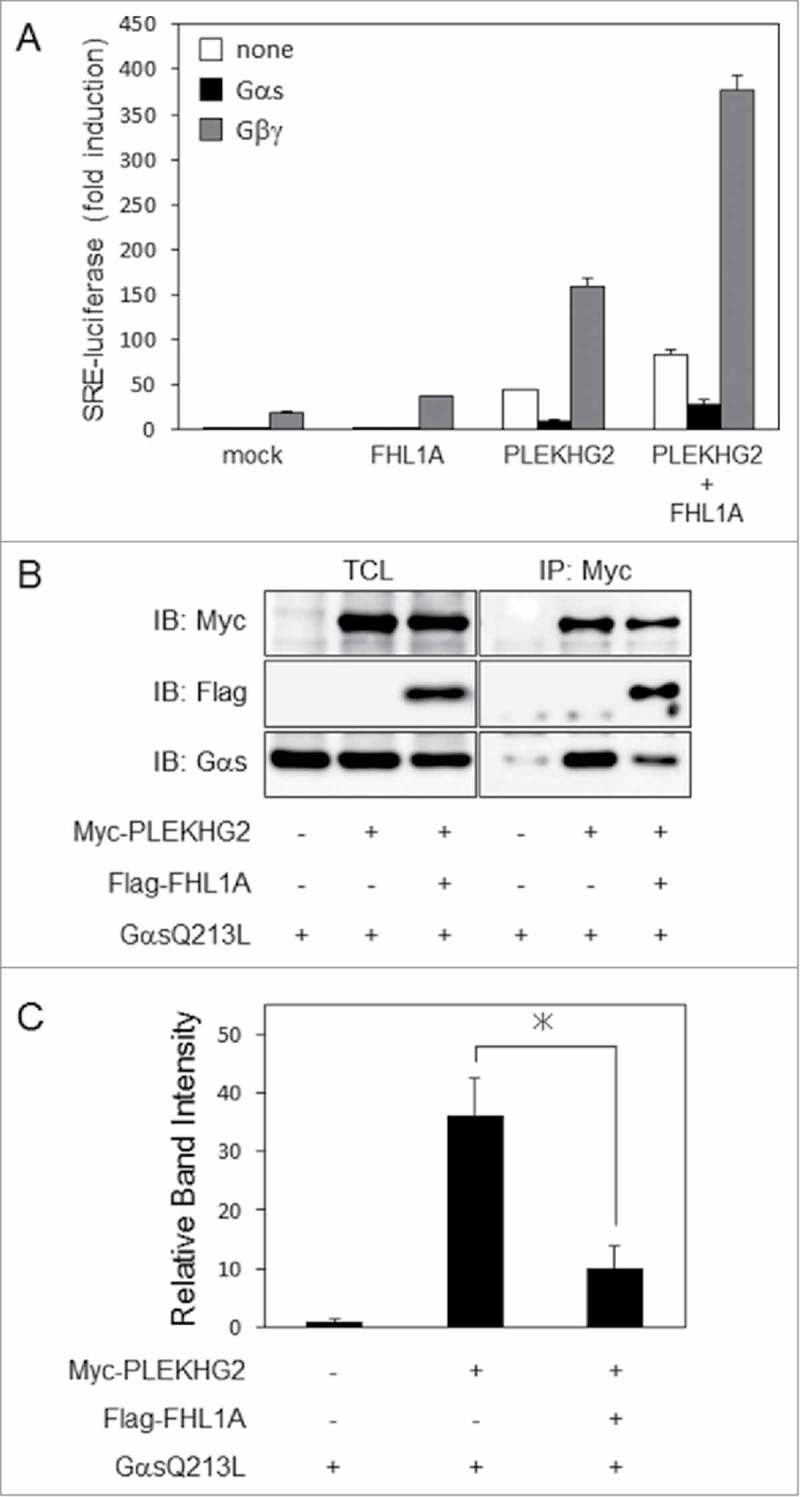
Effect of FHL1 on PLEKHG2-induced SRE-dependent gene transcription and interaction-regulated Gβγ or Gαs. (A) HEK293 cells were co-transfected with pSRE.L-luciferase, pRL-SV40, Myc-tagged PLEKHG2 and Flag-tagged FHL1A, GαsQ213L or Gβγ as indicated. Transfected cells were lysed 24 h after transfection, and the effects of GαsQ213L on FHL1A- and PLEKHG2-induced transcription were analyzed by a luciferase reporter gene assay. The effect of Gβγ on FHL1A- and PLEKHG2-induced transcription is also shown a positive control. The experiment was performed in triplicate, and the values are the means ± SD (error bars). The data shown are representative of three independent experiments. (B) HEK293 cells were co-transfected with Myc-tagged PLEKHG2 and Flag-tagged FHL1A with or without a GαsQ213L, as indicated. Transfected cells were lysed 24 h after transfection, and PLEKHG2 was immunoprecipitated with anti-Myc antibody. Precipitated proteins were separated by SDS-PAGE and immunoblotted with anti-Myc (for PLEKHG2) antibody and anti-FLAG antibody (for FHL1A, FHL2a and FHL3-1). TCL, total cell lysate; IP, immunoprecipitation; IB, immunoblotting. The data shown are representative of three independent experiments. (C) Quantification analysis of the precipitated GαsQ213L. The band of the GαsQ213L that interacted with Myc-tagged PLEKHG2 were quantified by ImageJ software. Each intensity of the bands was normalized by the total GαsQ213L and fold intensity was showed. *, P < 0.05.
Discussion
In this study, we show that only FHL1A, not FHL2a or FHL3-1, interacted with PLEKHG2 to enhance PLEKHG2-induced SRE-dependent gene transcription. In addition, we demonstrate that FHL1 modified heterotrimeric G protein signaling-regulated PLEKHG2-induced SRE-dependent gene transcription through its interaction with PLEKHG2 (Fig. 4).
Figure 4.
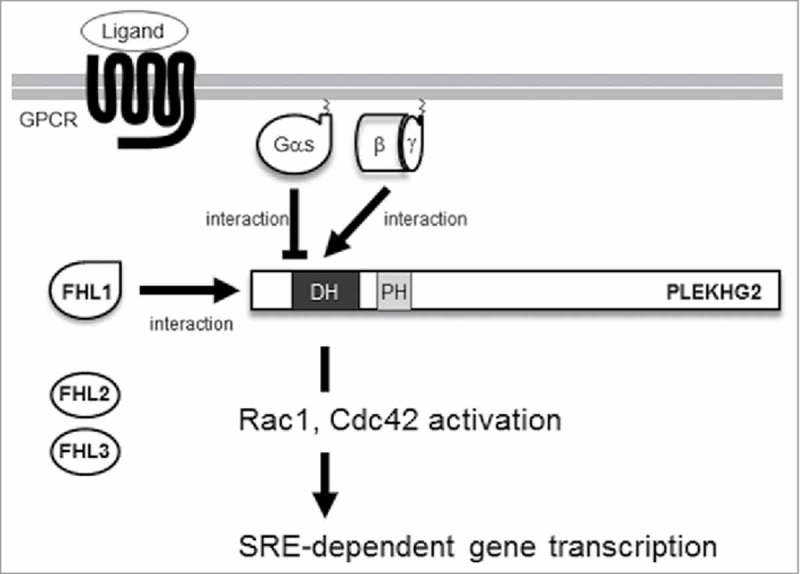
Proposed regulatory mechanisms of PLEKHG2 by FHL1. After GPCR was activated by its specific ligands, the heterotrimeric G proteins resolved into Gαs/olf and Gβγ. Gβγ interacts with PLEKHG2 and mediates actin cytoskeleton remodeling. Gαs/olf binds to the 1–310 a.a. region of PLEKHG2 and attenuates its activity. FHL1 binds to the 58–150 a.a. region of PLEKHG2 and enhances its activity. DH, Dbl homology domain; PH, pleckstrin homology domain.
We previously reported that PLEKHG2 interacted with the 111–230 amino acid region of FHL1A based on the results of a yeast two-hybrid system and immunoprecipitation.13 The 111–230 amino acid region of FHL1A contains a LIM3 domain and portions of a LIM2 and LIM4 domain (Fig. 1). The 111–230 amino acid region of FHL1A has relatively high homologies of 56% with the FHL2a and 50% with the FHL3-1. However, even with these relatively high homologies, FHL2a and FHL3-1 could not bind to PLEKHG2 in this study. It is well known that protein-protein interactions are often determined by the sequence of a few amino acids. And indeed, in this comparison of FHL1A with FHL2a or FHL3-1, some of the amino acid sequences in FHL1A were different from their counterparts in FHL2a and FHL3-1. In the future, these amino acid sequences may be found to be important for the interaction with PLEKHG2.
We previously reported a 58–150 amino acid region of PLEKHG2 that functions as an FHL1A-binding region.13 We also recently reported that a Gαs subunit interacted with the 1–310 amino acid region of PLEKHG2.4 These reports suggest that the FHL1A-binding region of PLEKHG2 is contained in the Gαs-binding region of PLEKHG2. In this study, co-expression of FHL1A had little effect on the attenuation of PLEKHG2-induced SRE-dependent gene transcription by a constitutively active form of Gαs. From the above, the following two possibilities are considered. The affinity of Gαs for PLEKHG2 may be stronger than that of FHL1A, because the Gαs-binding region and FHL1-binding region on PLEKHG2 are identical. An alternative explanation is that the Gαs-binding region and FHL1-binding region on PLEKHG2 are different, and the influence of the interaction of Gαs with PLEKHG-2 is greater than the interaction of FHL1A with PLEKHG2. In the future, structural analyses will be needed to elucidate the molecular mechanisms underlying the complex formation of PLEKHG2 and FHL1 or Gβγ and Gαs.
Previously, we showed that the overexpression of PLEKHG2, Gβγ and FHL1 influences for cell morphology.13 Since interaction of the FHL1 and PLEKHG2 were found by the yeast two-hybrid with using the human fetal brain cDNA library as bait, we speculate this system has function in a neural system. On the other hand, PLEKHG2 also activate in the lymphoid cells such as T lymphocytes.19 Furthermore, another Gβγ dependent RhoGEF, P-Rex1 stimulated by the one of the GPCR, CXCR4 darling the barest caner metastasis.20 These observations suggest that PLEKHG2 function not only the neural system but also other cell types.
As we mentioned above, FHL family protein also involved in the cell spreading, migration and actin rearrangement.17-18 The effect toward the cell morphology or actin polymerization are dependent on the cell type. Several studies have reported that FHL2 are involved in the sphingosine1-phosphate (S1P) receptor (S1PR). In fibroblasts, S1P signaling activates RhoA to induce translocation of FHL2 into the nucleus.21 In the immune system, immature dendritic cells, signaling via S1PR2 induce Rho activation to induce translocation of FHL2 into the nucleus.22 Considering S1PR1 and S1PR2 are one of the GPCR, the interaction between PLEKHG2 and FHL1A also might play in the S1P signaling. Although we could not investigate in this report, by clarifying the spatiotemporal control of the binding within the cell, together with the details mechanisms of the interaction, the physiological role may be clarified.
Materials and methods
Plasmids
Human FHL2a and FHL3-1 were cloned by PCR amplification from human brain cDNA (QUICK-Clone™ cDNA; Clontech, Mountain View, CA) in the same manner as FHL1 in our previous report.2 Amplified fragments were digested by an SgfI/PmeI Flexi Enzyme Mix (Promega, Madison, WI) and ligated into pF5A-FLAG-CMVneo vector. The pFN21A-Myc-PLEKHG2, pF4A-CMV-neo-GαsQ213L, pF4A-CMV-neo-Gβ1, pF4A-CMV-neo-Gγ2, pSRE.L-luciferase reporter plasmid and pRL-SV40 were described previously.3
Cell culture and transfection
HEK293 cells were grown in DMEM supplemented with 10% fetal bovine serum at 37°C. Transient transfection was performed using polyethylenimine (PolySciences Inc., Warrington, PA).23 Cells were transfected with DNA for 6 h and then washed with serum-free DMEM and incubated 18 h in DMEM.
Assay of SRE-dependent gene transcription
Cells seeded in 24-well plates were co-transfected with the indicated expression plasmids. After transfection, cells were washed once with ice-cold PBS and lysed with passive lysis buffer. Luciferase activities were determined by using a Dual-Luciferase Reporter assay system (Promega). The activity of the experimental reporter was normalized against the activity of the control vector.
Immunoprecipitation
Transfected cells seeded in 6-cm dishes were washed once with ice-cold PBS and lysed with lysis buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.1 mM EDTA, 1 mM Na3VO4, 0.5% Nonidet P-40, phosphatase inhibitor solution (Roche Diagnostics GmbH, Mannheim, Germany), and protease inhibitor solution (Roche Diagnostics)). The lysates were centrifuged to remove residue (16,100 xg for 10 min). Clear lysates were incubated with 1.0 μg of anti-Myc IgG for 2 h at 4°C and then mixed with protein G-agarose (EMD Milipore, Billerica, MA) for 1 h at 4°C. The beads were washed three times with washing buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.1 mM EDTA, 1 mM Na3VO4, 0.1% Nonidet P-40, phosphatase inhibitor solution, and protease inhibitor solution). Bound proteins were eluted with sample buffer and resolved by SDS-PAGE. The transferred PVDF membrane was blocked by PVDF Blocking Reagent for Can Get Signal (Toyobo Co., Ltd., Osaka, Japan). For the detection of Myc-tag and Flag-tag, we used HRP-conjugated mouse anti-Myc IgG (Wako, Osaka, Japan) and mouse anti-Flag IgG (Wako), respectively. Visualization of HRP-labeled proteins was performed using enzyme-linked chemiluminescence (PerkinElmer Life Science, Waltham, MA) and an LAS-4000 luminescent image analyzer (GE Healthcare, Buckinghamshire, UK).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This study was supported by JSPS KAKENHI Grants (nos. 23590073 and 15K07927). The DNA sequence analysis was supported by the Division of Genomic Research in the Life Science Research Center at Gifu University.
References
- [1].Raftopoulou M, Hall A. Cell migration: Rho GTPases lead the way. Dev Biol 2004; 265:23-32; PMID:14697350; http://doi.org/ 10.1016/j.ydbio.2003.06.003 [DOI] [PubMed] [Google Scholar]
- [2].Lazer G, Katzav S. Guanine nucleotide exchange factors for RhoGTPases: good therapeutic targets for cancer therapy? Cell Signal 2011; 23:969-79; PMID:21044680; http://doi.org/ 10.1016/j.cellsig.2010.10.022 [DOI] [PubMed] [Google Scholar]
- [3].Ueda H, Nagae R, Kozawa M, Morishita R, Kimura S, Nagase T, Ohara O, Yoshida S, Asano T. Heterotrimeric G protein betagamma subunits stimulate FLJ00018, a guanine nucleotide exchange factor for Rac1 and Cdc42. J Biol Chem 2008; 283:1946-53; PMID:18045877; http://doi.org/ 10.1074/jbc.M707037200 [DOI] [PubMed] [Google Scholar]
- [4].Sugiyama K, Tago K, Matsushita S, Nishikawa M, Sato K, Muto Y, Nagase T, Ueda H. Heterotrimeric G protein Gαs subunit attenuates PLEKHG2, a Rho family-specific guanine nucleotide exchange factor, by direct interaction. Cell Signal 2017; 32:115-23; PMID:28108261; http://doi.org/ 10.1016/j.cellsig.2017.01.022 [DOI] [PubMed] [Google Scholar]
- [5].Schmidt A, Hall A. Guanine nucleotide exchange factors for Rho GTPases:turning on the switch. Genes Dev 2002; 16:1587-609; PMID:12101119; http://doi.org/ 10.1101/gad.1003302 [DOI] [PubMed] [Google Scholar]
- [6].Barrows D, He JZ, Parsons R. PREX1 protein function is negatively regulated downstream of receptor tyrosine kinase activation by p21-activated Kinases (PAKs). J Biol Chem 2016; 291:20042-54; PMID:27481946; http://doi.org/ 10.1074/jbc.M116.723882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mayeenuddin LH, Garrison JC. Phosphorylation of P-Rex1 by the cyclic AMP-dependent protein kinase inhibits the phosphatidylinositiol(3,4,5)-trisphosphate and Gbetagamma-mediated regulation of its activity. J Biol Chem 2006; 281:1921-8; PMID:16301320; http://doi.org/ 10.1074/jbc.M506035200 [DOI] [PubMed] [Google Scholar]
- [8].Urano D, Nakata A, Mizuno N, Tago K, Itoh H. Domain-domain interaction ofP-Rex1 is essential for the activation and inhibition by G protein betagamma subunits and PKA. Cell Signal 2008; 20:1545-54; PMID:18514484; http://doi.org/ 10.1016/j.cellsig.2008.04.009 [DOI] [PubMed] [Google Scholar]
- [9].Montero JC, Seoane S, García-Alonso S, Pandiella A. Multisite phosphorylation of P-Rex1 by protein kinase C. Oncotarget 2016; 7:77937-49; PMID:27788493; http://doi.org/ 10.18632/oncotarget.12846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sato K, Sugiyama T, Nagase T, Kitade Y, Ueda H. Threonine 680 phosphorylation of FLJ00018/PLEKHG2, a Rho family-specific guanine nucleotide exchange factor, by epidermal growth factor receptor signaling regulates cell morphology of Neuro-2a cells. J Biol Chem 2014; 289:10045-56; PMID:24554703; http://doi.org/ 10.1074/jbc.M113.521880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sato K, Suzuki T, Yamaguchi Y, Kitade Y, Nagase T, Ueda H. PLEKHG2/FLJ00018, a Rho family-specific guanine nucleotide exchange factor, is tyrosine phosphorylated via the EphB2/cSrc signaling pathway. Cell Signal 2014; 26:691-6; PMID:24378532; http://doi.org/ 10.1016/j.cellsig.2013.12.006 [DOI] [PubMed] [Google Scholar]
- [12].Sato K, Handa H, Kimura M, Okano Y, Nagaoka H, Nagase T, Sugiyama T, Kitade Y, Ueda H. Identification of a Rho family specific guanine nucleotide exchange factor, FLJ00018, as a novel actin-binding protein. Cell Signal 2013; 25:41-9; PMID:23000341; http://doi.org/ 10.1016/j.cellsig.2012.09.015 [DOI] [PubMed] [Google Scholar]
- [13].Sato K, Kimura M, Sugiyama K, Nishikawa M, Okano Y, Nagaoka H, Nagase T, Kitade Y, Ueda H. Four-and-a-half LIM Domains 1 (FHL1) protein interacts with the Rho guanine nucleotide exchange factor PLEKHG2/FLJ00018 and regulates cell morphogenesis. J Biol Chem 2016; 291:25227-38; PMID:27765816; http://doi.org/ 10.1074/jbc.M116.759571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Shathasivam T, Kislinger T, Gramolini AO. Genes, proteins and complexes: the multifaceted nature of FHL family proteins in diverse tissues. J Cell Mol Med 2010; 14:2702-20; PMID:20874719; http://doi.org/ 10.1111/j.1582-4934.2010.01176.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Johannessen M, Møller S, Hansen T, Moens U, Van Ghelue M. The multifunctional roles of the four-and-a-half-LIM only protein FHL2. Cell Mol Life Sci 2006; 63:268-84; PMID:16389449; http://doi.org/ 10.1007/s00018-005-5438-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ding L, Wang Z, Yan J, Yang X, Liu A, Qiu W, Zhu J, Han J, Zhang H, Lin J, et al.. Human four-and-a-half LIM family members suppress tumor cell growth through a TGF-beta-like signaling pathway. J Clin Invest 2009; 119:349-61; PMID:19139564; http://doi.org/ 10.1172/JCI35930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Müller JM, Metzger E, Greschik H, Bosserhoff AK, Mercep L, Buettner R, Schüle R. The transcriptional coactivator FHL2 transmits Rho signals from the cell membrane into the nucleus. EMBO J 2002; 21:736-48; PMID:11847121; http://doi.org/ 10.1093/emboj/21.4.736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Coghill ID, Brown S, Cottle DL, McGrath MJ, Robinson PA, Nandurkar HH, Dyson JM, Mitchell CA. FHL3 is an actin-binding protein that regulates alpha-actinin-mediated actin bundling: FHL3 localizes to actin stress fibers and enhances cell spreading and stress fiber disassembly. J Biol Chem 2003; 278:24139-52; PMID:12704194; http://doi.org/ 10.1074/jbc.M213259200 [DOI] [PubMed] [Google Scholar]
- [19].Runne C, Chen S. PLEKHG2 promotes heterotrimeric G protein βγ-stimulated lymphocyte migration via Rac and Cdc42 activation and actin polymerization. Mol Cell Biol 2013; 33:4294-307; PMID:24001768; http://doi.org/ 10.1128/MCB.00879-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hynes NE, Gattelli A. P-Rex1, a guanine exchange factor that is overexpressed in breast cancer, is a convergence node for ErbB and CXCR4 signaling. Mol Cell 2011; 41:5-7; PMID:21211718; http://doi.org/ 10.1016/j.molcel.2010.12.024 [DOI] [PubMed] [Google Scholar]
- [21].Wixler V, Hirner S, Müller JM, Gullotti L, Will C, Kirfel J, Günther T, Schneider H, Bosserhoff A, Schorle H, et al.. Deficiency in the LIM-only protein Fhl2 impairs skin wound healing. J Cell Biol 2007; 177:163-72; PMID:17420295; http://doi.org/ 10.1083/jcb.200606043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].König K, Diehl L, Rommerscheidt-Fuss U, Golletz C, Quast T, Kahl P, Kolanus W, Knolle P, Buettner R, Heukamp LC. Four-and-a-half LIM domain protein 2 is a novel regulator of sphingosine 1-phosphate receptor 1 in CCL19-induced dendritic cell migration. J Immunol 2010; 185:1466-75; PMID:20592280; http://doi.org/ 10.4049/jimmunol.0903449 [DOI] [PubMed] [Google Scholar]
- [23].Godbey WT, Wu KK, Mikos AG. Poly(ethylenimine) and its role in gene delivery. J Control Release 1999; 60:149-60; PMID:10425321; http://doi.org/ 10.1016/S0168-3659(99)00090-5 [DOI] [PubMed] [Google Scholar]


