Abstract
The emergence of high-throughput DNA sequencing technologies sparked an immediate revolution in the field of genomics that has rippled into many branches of the life and physical sciences. The remarkable sensitivity, specificity, throughput, and multiplexing capacity that are inherent to massively parallel DNA sequencing have since motivated its use as a broad-spectrum molecular counter in small molecule and peptide-based inhibitor discovery, high-throughput biochemistry, protein and cellular detection and diagnostics, and even materials science. A key aspect of extrapolating DNA sequencing to ‘non-traditional’ applications is the underlying need to append nucleic acid barcodes to entities of interest. In this review, we describe the chemical and biochemical approaches that have enabled facile nucleic acid barcoding of proteinaceous and non-proteinaceous materials and provide exciting examples of downstream technologies that have been made possible by DNA-encoded molecules. Considering that commercially available high-throughput sequencers were first released fewer than 15 years ago, we believe related applications will continue to mature for years to come and close by proposing potential new frontiers to support this assertion.
High-throughput DNA sequencing: revolutionizing genomics and beyond
High-throughput DNA sequencing technologies have revolutionized our understanding of genomics, transcriptomics, epigenetics, and many other nuclear and cytosolic processes. Since it was first demonstrated in 2005[1], massively parallel DNA sequencing has been in a constant state of evolution and is currently able to simultaneously generate DNA sequence information for over a billion surface-immobilized DNA templates in just a few days[2]. In addition to increased throughput, these advances have also driven the cost of sequencing the human genome (~3 billion base pairs) from $2.7 billion dollars in the early 2000s (Human Genome Project[3]) to around $1,000 today[4]. Moreover, the most common ‘sequencing-by-synthesis’ platforms (hereto referred to as ‘next-generation sequencing’ or ‘NGS’)[5] require just picograms of DNA starting material for high quality library generation and exhibit a quantitative readout with an error rate of ~0.1%[6]. While nearly all NGS strategies are dependent upon clonal amplification of spatially distinct immobilized DNA strands, recent years have seen great strides in single molecule sequencing approaches, including zero-mode waveguides[7] and nanopore sequencing[8] (commonly referred to as ‘de novo’ or ‘third generation’ sequencing), that offer much longer read lengths (10–100kb) and have the ability to directly gather sequence information as well as other chemical features of native nucleic acid molecules. Not surprisingly, the ever-improving cost, throughput, sensitivity, and multiplexing capacity of DNA sequencing have morphed what was initially developed as a relatively monolithic technology by Frederick Sanger over 40 years ago[9] into a broadly utilized ‘molecular counter’ for biologists and chemists alike.
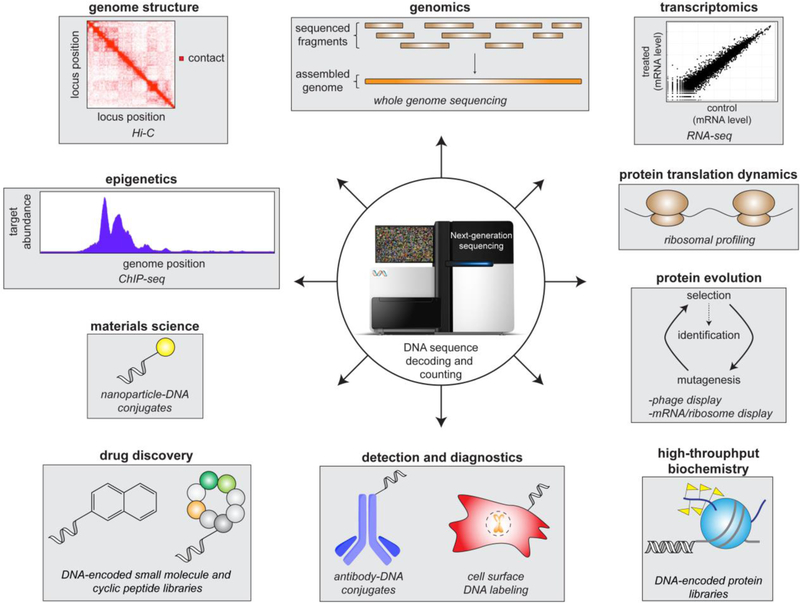
Beyond the genome and transcriptome sequencing efforts that took center stage at the onset of the high-throughput DNA sequencing era, many ‘non-traditional’ methods that exploit DNA sequencing have been developed (Figure 1). Such methods have enabled high resolution mapping of chromatin-associated proteins and chromatin modifications across the genome (ChIP-seq[10]), determination of genome structure (DNase-seq[11] and Hi-C[12]), comprehensive analysis of protein translation activity (ribosome profiling[13]), and genome-wide analysis of many other nucleic acid-templated processes[14]. Furthermore, common in vitro selection technologies, including phage display[15], yeast-2-hybrid screening[16], and other cell surface display technologies[17], experienced vast improvements in throughput and cost[18]. These methods, which utilize different biological organisms to express libraries of user-defined or randomized peptides and proteins, previously required DNA sequencing of each individually selected clone to identify molecules with desirable properties. By adapting NGS readouts, entire selected populations can now be quantitatively analyzed in a single sequencing run, thus vastly improving overall coverage of positive adaptations and useful protein sequence space[18]. Similarly, NGS has facilitated the development of laboratory techniques to evolve biomolecules that exhibit unnatural function[19], engineer cellular populations with unique fitness attributes[20], and trace cell lineages throughout organism development[21]. Notably, all of the technologies described above are cell-based approaches wherein the DNA that is ultimately analyzed is generated in and extracted from a cellular environment. To further expand the utility of high-throughput DNA sequencing as a molecular identifier and counter, many innovative chemical and biochemical strategies have been employed to tether unique, synthetic DNA sequences (‘barcodes’) to conceivably any moiety of interest. This review is meant to bridge the gap between biologists and chemists interested in such DNA barcoding approaches by describing methods that have been developed to assemble DNA-conjugated materials and highlighting the exciting DNA-encoded library technologies that have resulted from these efforts. In doing so, we hope to inspire new DNA barcoding methodologies and NGS-based applications in biochemistry, cell biology, and nanotechnology.
Figure 1.
Examples of high-throughput sequencing–based methods and DNA-encoded molecules that span many scientific disciplines.
Chemical approaches for generating DNA-encoded synthetic molecules
Even prior to the invention of NGS, the idea of ‘encoded combinatorial chemical libraries, in which each chemical sequence is labeled by an appended genetic tag’ had been discussed as a potentially powerful and versatile method for drug screening[22]. Since this original proposal by Brenner and Lerner, many creative methods have been developed that enable massively parallel DNA-encoded small molecule synthesis and subsequent ligand screening. Library synthesis approaches fall into two major classes: 1.) ‘DNA-recorded chemistry’ wherein each chemical transformation step is followed by attachment of a unique DNA sequence to the resulting molecule, resulting in a ‘record’ of synthesis history, and 2.) ‘DNA-templated chemistry’ wherein a programmable DNA strand is used to ‘template’ chemical reactions. Importantly, in both of these approaches, the chemical identity of the final molecule can be ‘decoded’ from the corresponding DNA sequence, thus making NGS the ideal ligand screening assay readout.
DNA-recorded chemistry
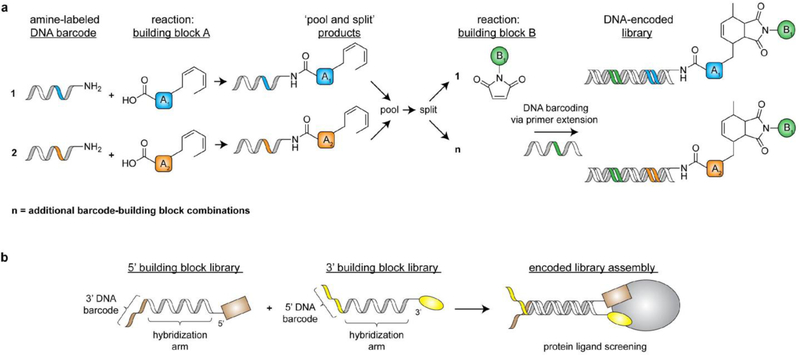
Most DNA-recorded chemistry approaches follow a similar workflow wherein a functionalized building block is conjugated to a 5’-functionalized oligonucleotide (typically with a commercially available amine or thiol moiety) to form an initial DNA-tagged chemical scaffold. This scaffold is then ‘split’ into multiple reaction vessels and subjected to different chemical transformations, each of which is ‘recorded’ by extending the DNA tag with a unique DNA sequence (Figure 2a). This ‘reaction barcoding’ can be accomplished after completion of each chemical transformation via ligase-catalyzed DNA conjugation[23] or polymerase-catalyzed primer extension[24] of a reaction-specific DNA strand. Once barcoded, all reaction products can be ‘pooled’, split again, and subjected to another DNA-encoded synthesis step. This ‘split-and-pool’ approach can be iterated to generate large (hundreds of millions of compounds), diverse libraries for ligand identification efforts. Following library incubation with a biomolecule of interest, high-affinity compounds can be eluted and identified via high-throughput DNA sequencing.
Figure 2.
Common strategies to synthesize DNA-recorded small molecule libraries. a) A split-and-pool-based method to generate a library of DNA-encoded bicyclic molecules via Diels-Alder chemistry as carried out by the Neri lab. b) A workflow for encoded self-assembling chemical (ESAC) library assembly and screening.
In the past 20 years, many chemical synthesis strategies have been implemented to diversify molecular functionalities and topologies in DNA-recorded small molecule libraries. Pioneering studies out of the Neri laboratory demonstrated the general applicability of diverse coupling strategies, including amide bond formation[25] and Diels-Alder cycloadditions[24], to generate 4,000 member libraries in two split-and-pool steps. More complex scaffolds that enable stepwise synthesis of highly branched structures (ex. triazine scaffolds) have also been implemented to explore diverse molecular geometries and greatly expand library size to millions of compounds[23]. Complementary library synthesis strategies have also been developed, including ‘encoded self-assembling chemical (ESAC) libraries’, in which single-stranded DNA-fused molecules are brought in close special proximity via hybridization of complementary DNA sequences (Figure 2b)[26]. In doing so, ESAC technologies enable display of unique molecular combinations, and hits can be identified via various code transfer methods (ex. proximity-based DNA ligation of hybridized barcodes) and subsequent NGS[27]. Over recent years DNA-encoded library sizes have continued to grow, with academic and pharmaceutical laboratories now reporting libraries with billions or even trillions of unique compounds[28]. Today, the use of DNA-recorded chemistry in academia and industry has become commonplace and has resulted in lead compound identification for a wide range of high profile biomedical targets including GPCRs[29], kinases[30], proteases[31], and many others[32].
DNA-templated chemistry
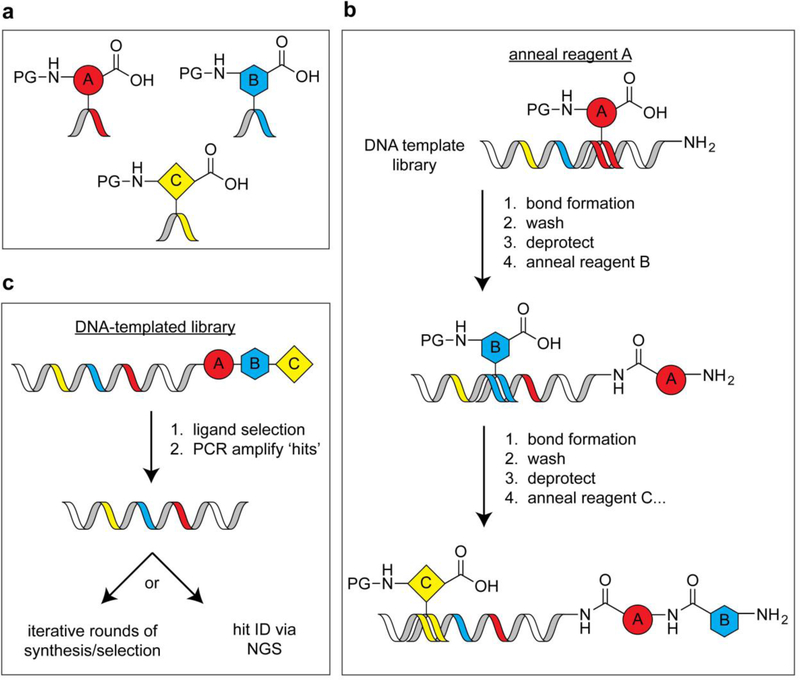
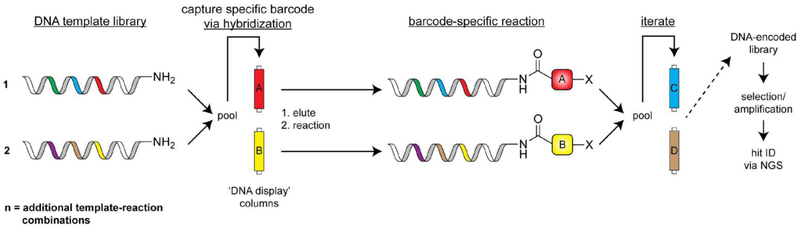
DNA-templated synthesis was inspired by the desire to apply iterative in vitro selection and amplification principles to synthetic molecules. Such an approach requires the ability to translate DNA sequences into man-made chemical entities, which can be accomplished by using DNA sequences to direct specific chemical synthesis steps and encode molecular identity. Major breakthroughs in this field came in the early 2000s, when a series of papers from the Liu[33] and Harbury[34] labs reported complementary strategies for DNA-directed chemical synthesis. The Liu approach exploits hybridization between a functionalized DNA template strand and a DNA-linked chemical building block to increase the effective molarity between these two molecules, thus ‘templating’ a chemical reaction (Figure 3). By iterating this process and including unique template sequences and building blocks, very large DNA-templated small molecule libraries can be achieved. While this approach was initially used direct amine acylation reactions using trinucleotide-labeled building blocks that yielded a 65-member macrocycle library[33c], second-generation libraries have reached 256,000 compounds[35]. The Harbury approach is fundamentally different in that DNA-conjugated building blocks are not required (Figure 4). To accomplish this, template DNA strands comprising a reactive handle and multiple unique DNA barcodes are assembled. These templates are then captured on a column that displays a DNA sequence that is complementary to one of the pre-installed template barcodes. After barcode-specific isolation of template strands, corresponding chemical transformations are performed. The products are then pooled, and the process repeated for other barcodes that reside within a given template. In a proof-of-concept study, a library of one million compounds was assembled by performing multiple, iterative synthesis steps on each template in parallel[34b]. The most crucial conceptual advancement from the DNA-templated methods described here is the ability to translate a DNA sequence into a specific synthetic molecule. Consequently, iterative cycles of library selection followed by synthetic ligand amplification can be performed to enrich for hits in a given screen, which can be deconvoluted and quantitatively profiled in a single NGS run[35].
Figure 3.
Design of DNA-fused chemical building blocks and the corresponding DNA-templated chemistry workflow. a) ‘Anticodon’ building blocks for DNA-templated library synthesis (PG = protecting group). b) A library synthesis workflow wherein hybridization of anticodon building blocks with a template strand is followed by a bond formation step. c) A library screening approach that enables selection and amplification of small molecule libraries.
Figure 4.
DNA-templated synthesis of small molecule libraries as facilitated by DNA display columns. Template DNA strands are captured and isolated via immobilization on DNA display columns to enable barcode-specific reactions. This technology affords a split-and-pool approach to DNA-directed synthesis of small molecules.
Other notable approaches for DNA-templated synthesis include the ‘YoctoReactor’[36] and ‘densely functionalized nucleic acid polymers’[37]. Much like the Liu approach described above, the YoctoReactor exploits DNA hybridization to direct chemical reactions through modulating effective molarity. This method is unique in that the DNA-encoded building blocks are programmed to position their respective reactive chemical moieties at the center of three-way DNA junctions to achieve an unprecedented reaction volume (1 yoctoliter (10−24 L))[36]. Densely functionalized nucleic acid polymers are DNA polymers in which every third nucleotide contains a functionalized base moiety. To assemble such libraries in a single-pot, functionalized trinucleotides (or ‘codons’) are prepared and hybridized with a library of single-stranded DNA templates that present unique codon reading frames. After synthetic codon annealing, DNA ligase is used to ligate the codons and create the functionalized nucleic acid polymer. Excellent reviews further describing these and other DNA-encoded small molecule library technologies and their success in drug discovery can be found elsewhere[38].
Hijacking the ribosome: co-translational nucleic acid-barcoding of proteins in vitro
A vast majority of proteins and peptide macrocycles are out of reach of the synthetic chemist, and thus not amenable to the chemical approaches described for DNA-encoded small molecule library assembly. Therefore, unique methodologies are required to create DNA-encoded biomolecular libraries, which are powerful tools for high-throughput analysis of protein function, interaction networks, and ligand discovery and characterization. Traditionally, library-scale analysis of proteins is performed via yeast-2-hybrid screening, a process in which protein-coding DNA libraries are expressed and analyzed in live cells[39]. Various shortcomings that can limit yeast-two-hybrid utility include the requirement for a cell-based screening assay, the fact that library size is limited by DNA uptake of the target organism, and the need to switch back and forth between in vivo screening and in vitro amplification during iterative selection protocols (a laborious process). The process of in vitro translation (IVT), which was first reported by Nirenberg and Matthaei in 1961, enables cell-free ribosomal synthesis of proteins by utilizing cellular lysates[40] or purified protein translation components[41]. To circumvent the limitations imposed by yeast-two-hybrid approaches, several IVT-based technologies have been developed in which ribosome-synthesized protein products become fused to their corresponding mRNA construct during cell-free protein synthesis. Such co-translational nucleic acid barcoding of proteins facilitates massively parallel, one-pot synthesis of protein and peptide macrocycle libraries in vitro, which have become instrumental in the characterization of biological signaling pathways and the discovery of biological drug candidates. Here, we discuss the two major strategies for co-translational DNA barcoding, ribosome display and mRNA display, and highlight key downstream applications.
Ribosome display and high-throughput identification of protein-protein interactions
Ribosome display represents the first method to physically link protein genotype to phenotype in vitro, and was initially demonstrated on polysomes by Matteakis et al. in 1994[42] and on individual ribosomes by the Pluckthun laboratory in 1997[43]. This strategy is based on ribosome stalling, which can be accomplished in several ways, including the omission of a stop codon from the gene(s) of interest, omission of release factors from an IVT mixture[41], or addition of ribosome stalling agents (ex. chloramphenicol or a ribosome stalling mRNA segment). As a result, a stable peptidyl tRNA linkage is formed and the final protein product remains tethered to the ribosome along with the corresponding mRNA molecule (Figure 5a). Following an assay of interest, ‘selected’ mRNAs are isolated and reverse transcribed to generate DNA for hit analysis or iterative rounds of library synthesis and selection. The resulting library sizes are therefore limited only by the number of ribosomes in solution and can reach >1014 members[44], several order of magnitude higher than transformation efficiency-limited techniques like phage display and yeast-2-hybrid screening (library sizes 107-1010 members)[45].
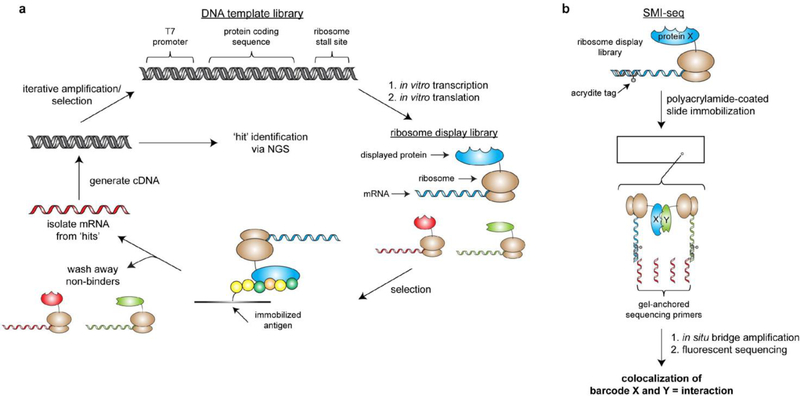
Figure 5.
Ribosome display library synthesis and downstream assays for high-throughput identification of protein ligands and interaction partners. a) Workflow for co-translational nucleic acid-barcoding of proteins via ribosome display and subsequent screening for protein-binding partners of an immobilized antigen of interest. Isolated mRNA from ‘hits’ can be identified via NGS or subjected to additional rounds of library generation and screening. b) The single molecule interaction-sequencing (‘SMI-seq’) method developed by the Church lab.
Since its inception, ribosome display has remained a commonly applied technology for directed evolution of protein-binding module specificities and stabilities (including antibodies[46], nanobodies[47], and designed ankyrin repeat proteins[48]), enzyme activity[49], and peptide-based ligands[50], all of which are useful in biological research, diagnostics, and disease therapy. A typical workflow for ligand selection via ribosome display begins by incubating the library (user-defined collection of ribosome-displayed natural, mutated, and/or randomized polypeptide sequences) with an immobilized peptide or protein of interest (Figure 5a). After washing away binding-incompetent members, mRNA constructs corresponding to ‘hits’ are isolated and reverse transcribed to generate cDNA templates. These cDNA templates can then be amplified via PCR for iterative rounds of selection or identified by DNA sequencing. To enable directed evolution and affinity maturation, error-prone PCR or other mutagenic approaches can be used in the cDNA amplification step prior to additional rounds of selection[51]. Initially, ribosome display selection analysis was dependent upon Sanger sequencing, and 102-103 clones were typically isolated and identified[52]. The advent of NGS and the associated bioinformatics tools have led to a >10,000-fold increase in the number of sequences (>107) that could be analyzed while reducing costs and obviating the need for clonal isolation. This significant boost in experimental analysis has led to more effective design of second-generation libraries and enables deep mutational scanning of selection results[53], both of which vastly increase the power of ribosome display.
While directed evolution of biomolecules and ligand selection have been achieved by a number of different in vitro selection techniques, there are several methods for high-throughput analysis of protein function that are specific to ribosome display. In 2014, the Elledge lab developed PLATO (parallel analysis of translated ORFs), which takes advantage of a rapid, highly parallel Gateway cloning strategy to display a large subset of the human ORFeome (14,582 unique cDNAs)[54]. The authors used this library to identify novel binding partners for the LYN tyrosine kinase as well as targets for antibodies from patients with autoimmune diseases. Importantly, this approach offers advantages over protein microarray technologies, including increased detection sensitivity and the fact that molecules do not need to be immobilized and spatially segregated. Another protein interaction study from the Church lab (so-called ‘single molecular interaction-sequencing’ or ‘SMI-seq’[55]) integrates acrydite-labeled DNA-mRNA hybrids into ribosome display complexes to immobilize entire ribosome display libraries on a polyacrylamide film (Figure 5b). Subsequent solid-phase PCR can then be performed with two gel-anchored primers by following an isothermal bridge amplification protocol similar to that used in the Illumina NGS platform[6]. Because the amplification process is performed in a flow cell with fluorescent probes, all barcodes can be spatially resolved and quantified (greater than one million barcodes per square millimeter of film). Protein interactions can therefore be inferred from co-localized barcodes and, because all bound and unbound molecules are quantified, this method enables high-throughput calculation of protein-protein interaction dissociation constants. The effectiveness of SMI-seq was initially demonstrated by identifying binding preferences for antibodies and the GTPase H-Ras, and accompanying mathematical models suggest that thousands of interactions can be quantified in a single assay. It is important to note that one shortcoming of these ribosome display technologies is that they require assays that are compatible with ribosome-fused proteins in ribosome-stable buffers. When these factors preclude the use of ribosome display, complementary mRNA display-based methods can be pursued.
mRNA display and DNA-encoded macrocyclic peptide libraries
Similarly to ribosome display, mRNA display can be used to tether proteins to their transcripts during IVT. However, this approach utilizes puromycin-fused mRNA templates that result in a covalently linked polypeptide-nucleic acid product, which can be isolated from ribosomes for downstream library applications (Figure 6a). In 1997, the Szostak[56] and Yanagawa[57] groups independently reported similar methods to fuse puromycin (an antibiotic that mimics the tRNA aminoacyl moiety) to the 3’ end of an mRNA construct via a DNA linker. The mRNA segment of the resulting hybrid nucleic acid can be translated into the corresponding polypeptide by the ribosome, which stalls once it reaches the 3’ DNA segment. Upon translation stalling, the 3’ puromycin molecule enters the A-site of the ribosome and accepts the growing polypeptide via amide bond formation, thus resulting in release of an mRNA-DNA-peptide conjugate from the ribosome. From this point, mRNA display is conceptually equivalent to ribosome display and has been used for many similar applications[58]. Key mRNA display-based studies include high-throughput characterization of transcription factors[59] and cellular apoptosis[60], protein interaction networks, whole organism proteome library generation and analysis[61], and directed evolution of functional proteins[62], enzymes[63] and antibodies[64]. Because mRNA display is most effective for polypeptides under 300 amino acids in length[61], ribosome display remains the method of choice for analysis of large proteins. However, the stability and minimally invasive nature (relative to ribosomal tethering) of puromycin-linked nucleic acid-polypeptide conjugates make mRNA display ideal for studying short peptide constructs. Building off these principles, mRNA display has been combined with recent advances in genetic code reprogramming, peptide cyclization chemistry, reconstituted translation systems, and NGS to revolutionize the development of encoded peptide macrocycle libraries for drug discovery (described below).
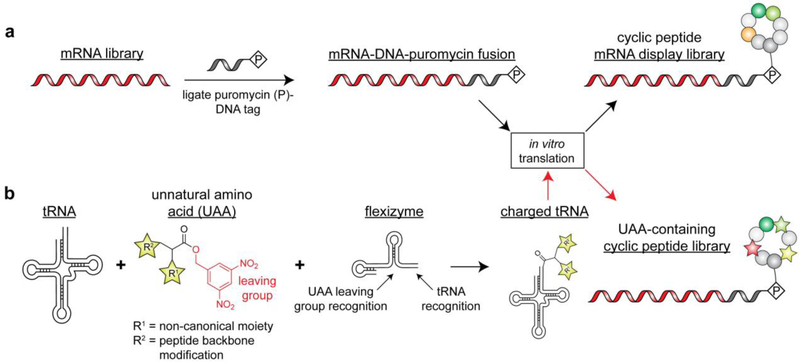
Figure 6.
Generation of cyclic peptide libraries via mRNA display. a) Workflow for generating cyclic peptide libraries containing natural amino acids. Cyclization can be induced in a variety of ways including cysteine side chain disulfide formation or addition of side chain-reactive crosslinkers. b) Workflow for generating cyclic peptide libraries containing unnatural amino acids via the Flexizyme technology as pioneered by the Suga lab.
Macrocyclic natural products, including those that emerge from polyketide synthase[65] and nonribosomal peptide synthase[66] biosynthetic pathways, represent a diverse chemical space that is unique from that of classic small molecule drugs. Interestingly, analysis of 1,071 known nonribosomal peptides revealed greater than 500 unique monomer building blocks, which extend far beyond the 20 naturally occurring amino acids to include those with non-canonical side chains, d-stereochemistry, backbone N-methylation/alkylation, β-amino acids, and many other modifications[67]. The size and chemical diversity of these molecules, along with the constrained structural entropy imposed by cyclization, make them particularly useful for targeting features beyond classically ‘druggable’ hydrophobic pockets, such as protein-protein interfaces[68]. Currently, over 40 naturally occurring and de novo peptidic macrocycles are in clinical use in a vast array of therapeutic areas, including antibiotics (ex. vancomycin), immunosuppressants (ex. cyclosporine), chemotherapeutics (ex. lanreotide), and others[69]. Indeed, these successes have demonstrated the potential of peptide macrocycles as therapeutics and a recent increase in FDA-approvals suggest that these molecules have only just begun to claim their place in drug discovery[69].
Similar to the small molecule drug screening platforms described above, much effort has gone into creating large and diverse libraries of nucleic acid-barcoded peptide macrocycles. Fully synthetic approaches that follow a split-and-pool workflow have been employed wherein coupling of each monomer is followed by attachment of a unique DNA barcode. Cyclization can then be initiated by activating complementary reactive handles that are installed during synthesis, such as azide-alkyne groups[70]. This strategy was recently used to generate a library of >1012 macrocycles consisting of natural and unnatural amino acid building blocks 4–20 units in length. One shortcoming related to this approach is that the selection process cannot be iterated to enrich for specific binders. To circumvent this problem, the Liu laboratory has extended DNA templated synthesis to include DNA-conjugated building blocks that may be cyclized through Wittig olefination after the final building block coupling step[35]. This approach enables iterative rounds of amplification and selection; however, the library size is relatively modest (256,000 members) when compared to peptide libraries that can be compiled via peptide display methods. In order to generate cyclic peptide libraries following ribosomal synthesis of peptides (ex. for phage display), cyclization is typically accomplished via disulfide crosslinking of cysteine residues[71]. Cyclization can also be accomplished via post-translational addition of crosslinkers that specifically react with amino acid side chain functionalities, such as disuccinimidyl glutarate (DSG) for amine-amine crosslinking[72]. Notably, unlike disulfide crosslinking, this approach results in more stable, non-reducible macrocycle bridges. Bicyclic molecules can also be generated using similar methods, as is the case when 1,3,5-tris(bromomethyl)benzene (TBMB) is added to display peptides that contain three cysteine residues to form multiple thioether bonds[73].
Unnatural amino acid (UAA) incorporation technologies have been successfully implemented to install handles for stable peptide cyclization[74], such as initiation with an N-chloroacetyl amino acid to form thioether bonds with cysteine residues[75]. Other groups have utilized split-intein fusion constructs to accomplish rapid and spontaneous circular ligation of randomized peptides (so-called ‘SICLOPPS’) in cells[76], which require two-hybrid type cell based assays as a readout. One major limitation of these display technologies is the need to incorporate a wide array of UAAs to access the plethora of unique monomer building blocks found in naturally occurring macrocycles. Indeed, UAA incorporation technologies have been successfully implemented in cell- and phage-based library approaches, but the number of unnatural units per peptide remains limited by UAA incorporation efficiency[68]. Therefore, IVT-based display technologies are superior because they enable user-defined protein synthesis cocktails to be used for library expression[74]. Such IVT systems make genetic code reprogramming (i.e. reassignment of redundant codons to UAAs) remarkably effective because redundant, naturally occurring tRNA sythetases and/or corresponding tRNA molecules can be omitted from a reaction[77]. This eliminates competition between natural and unnatural codon assignments and permits multiple unique UAAs to be installed in a single peptide[78]. However, a major limitation associated with genetic code reprogramming that mRNA display cannot overcome is the need to generate UAA-charged tRNAs. This is typically accomplished enzymatically by utilizing directed evolution to create aminoacyl tRNA synthetases for a UAA of interest[79] or chemically by either ligating modified amino acid building blocks to tRNA molecules[80] or modifying the amino acid moiety of natural aminoacylated tRNAs[81]. While enzymatic tRNA charging can be performed during the IVT reaction, the chemical approach requires pre-synthesized aminoacylated tRNAs to be added to the IVT reaction for ribosomal incorporation into peptides. Notably, these strategies require unique chemical approaches or evolved tRNA synthetases for each UAA of interest. To this end, the Suga lab has developed a suite of tRNA acylation ribozymes, which are able to charge tRNA molecules in an indiscriminant fashion with respect to the identity of the UAA[82]. These ‘flexizymes’ recognize the universal 3’ sequence of tRNA molecules and a bulky leaving group that can be chemically installed on any unnatural amino acid, thus inducing proximity between these two molecules catalyzing the aminoacylation reaction (Figure 6b). Currently, the flexizyme dFx (which recognizes 3,5-dinitrobenzyl ester leaving groups) is the most generic and used for a wide array of tRNA acylation reactions while eFx (which recognizes 4-chloro-benzyl thioester leaving groups) is useful for sterically demanding UAA side chains[83]. This system has been used in combination with mRNA display to create macrocyclic peptide libraries exceeding 1012 members with a highly diverse composition, include building blocks with non-canonical side chains, d-stereochemistry, N-alkylated or acylated α-amino groups, β-amino acids, α-hydroxy acids, and even peptide foldamers[84]. Such nucleic acid-encoded libraries can be synthesized and screened in just days to weeks when combined with NGS and have been used to identify lead compounds for an array of high priority drug targets, including bacterial transporters, mammalian cell surface receptors, and intercellular proteins and enzymes[84]. It is important to also note that a vast majority of macrocyclic peptide inhibitors are unable to penetrate cells, and new advances in drug delivery and/or medicinal chemistry will be necessary to fully realize the potential and versatility of these molecules[85].
Bioorthogonal chemistry for DNA barcoding of proteins and other biomaterials
Despite the aforementioned advances in nucleic acid barcoding via ribosome and mRNA display, these technologies are incompatible with proteins and protein complexes that cannot be reconstituted using IVT. In these cases, proteins must be individually barcoded using post-translational bioorthogonal labeling strategies. Although this is a relatively low throughput approach that drastically reduces the potential library size, the remarkable detection sensitivity, specificity, and multiplexing capacity afforded by NGS, as well as the relatively inert presence of DNA tags in biochemical and biological assays, have motivated the development of myriad labeling strategies and downstream applications. Furthermore, many of these approaches can be extended to non-proteinaceous materials including carbohydrates, cell surfaces and tissues, and even non-biological materials.
Chemical methods for appending DNA to natural and unnatural functional groups
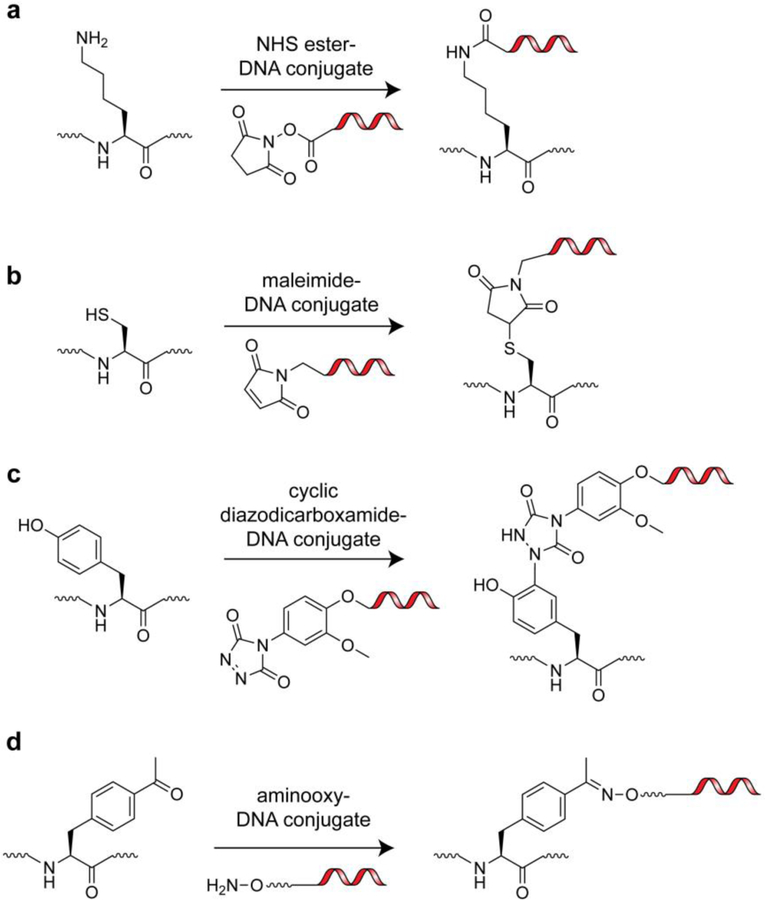
There are many rapid and efficient strategies for appending DNA tags to isolated proteins in mild, aqueous buffers that take advantage of naturally occurring protein functionalities[86]. The use of Michael acceptors (ex. maleimides) for cysteine side chain thiol targeting and activated esters (ex. N-Hydroxysuccinimide (NHS)-esters) for lysine side chain ε-amine targeting represent the most common amino acid-specific conjugation approaches (Figure 7a,b). Importantly, maleimide and NHS-ester homobifunctional and heterobifunctional linkers are commercially available, as are pre-functionalized DNA primers, all of which have greatly facilitated the widespread implementation of cysteine and lysine labeling. Of these strategies, cysteine labeling is particularly attractive as it is a low abundance amino acid in proteins. This makes engineering a single, non-perturbative reactive site in a protein of interest relatively straightforward[87]. The use of diazocarboxamides for tyrosine phenol targeting has also been reported as an efficient means to conjugate DNA to proteins (Figure 7c); however, this approach has not been widely adapted, likely because it still requires the user to synthesize diazocarboxamide-containing heterobifunctional linkers[88]. Another promising strategy for targeting a single site in proteins is N-terminal (α-amine) protein labeling via 2-pyridinecarboxyaldehydes as initially reported by the Francis lab[89]. This technique has the potential to enable facile synthesis of DNA-protein conjugates from native proteins and aldehyde-tagged DNA fragments. Other methods for targeting native functional groups such as carboxylates (Asp, Glu, C-terminus), guanidinium (Arg), thioether (Met), and imidazole (His) are available but not ideal due to issues with specificity, efficiency, and reaction conditions that are not compatible with certain folded proteins[90].
Figure 7.
Chemical functionalities for labeling of natural and unnatural amino acid side chains with DNA barcodes. a) Lysine side chain labeling via an NHS-ester-DNA conjugate. b) Cysteine side chain labeling via a maleimide-DNA conjugate. c) Tyrosine side chain labeling via a cyclic diazodicarboxamide-DNA conjugate. d) p-acetylphenylalanine side chain labeling via an aminooxy-DNA conjugate.
Genetic code reprogramming can also be exploited to introduce reactive handles into proteins for downstream DNA tethering (Figure 7d)[91]. These approaches grant access to robust bioorthogonal reactions including copper-catalyzed and strain promoted azide-alkyne cycloadditions, the inverse electron demand Diels-Alder reaction, the Staudinger ligation, and oxime/hydrazine ligations[92]. Importantly, the Schultz laboratory and others have placed a great deal of effort on developing and distributing an array of UAA-tRNA synthetase pairs to enable facile incorporation of a variety of these ‘clickable’ functionalities into a protein of interest, and many of the complementary reactive groups can be purchased pre-conjugated to commercially available, custom DNA primers.
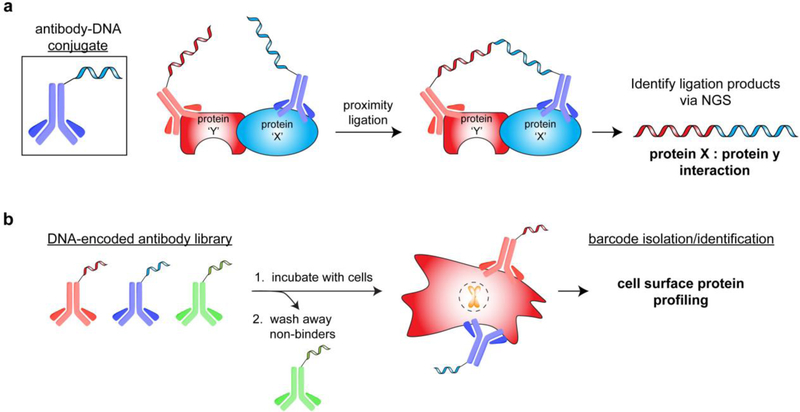
Antibodies represent one of the most common classes of proteins to which amino acid side chain-DNA conjugation methods have been applied[91]. In 1992, the Cantor lab showed that antibody-DNA conjugates could be used for immuno-PCR, an ELISA-based method that utilizes PCR to detect antigen-antibody interactions[93]. The 10–1,000-fold increase in detection sensitivity for PCR over traditional ELISA signal agents has inspired development of many complementary methods, including DNA-encoded antibody libraries for use in protein diagnostics, biomarker detection, cell sorting, and protein imaging in cells (via rolling circular amplification[94])[95]. One such example is the use of a DNA-barcoded 90 antibody library (designed to detect hallmark cancer antigens) to profile cancer cells (Figure 8a)[96]. After incubation of the antibody library with a cell line of interest, the DNA barcodes that remained bound to cells were identified to determine potential pathway dependencies in patient tumor samples. Notably, the Wells lab recently reported a similar workflow using a library of preselected, phage-displayed antibodies for high-throughput identification of cell surface proteins via NGS[97]. Another common DNA-antibody conjugate technique for detection of proteins and protein complexes is proximity ligation[98], which occurs when antigen recognition by two independent antibodies induces proximity between the corresponding barcodes (Figure 8b). The high effective molarity of the two localized DNA strands can be exploited to promote barcode ligation, and ligated sequences can be identified in a high-throughput format by NGS to determine protein-protein complex identities[99]. This approach greatly reduces background, enhances specificity, and can be used to detect zeptomole (10−21) amounts of material[98].
Figure 8.
Antibody-DNA conjugate applications. a) Identification of protein-protein interactions via proximity ligation of antibody-fused DNA strands. b) High-throughput profiling of cell surface proteins via a library of DNA-encoded antibodies.
Using unique protein features to guide site-specific DNA conjugation
One major caveat in targeting naturally occurring side chain moieties for DNA conjugation is that it is often difficult to achieve homogenous labeling of a single site. To overcome this, several methods have been developed that exploit unique protein features to guide site-specific labeling. One such method developed by the Niemeyer laboratory utilizes a DNA-conjugated heme to replace the natural cofactor in myoglobin, an approach that has been extended to other co-factors and proteins[100]. Another strategy is to conjugate DNA to known protein ligands to create a protein-targeting vehicle[101]. Once this targeting vehicle is bound to the protein of interest, a complementary DNA strand bearing an otherwise promiscuous reactive moiety (ex. 5’ maleimide, NHS-ester, or photoactivatable crosslinker) can be added to the solution in stoichiometric amounts at a low concentration. Hybridization with the ligand-DNA fusion induces a high effective molarity between the reactive strand and the protein surface, resulting in quantitative labeling of a single local residue. This general workflow can also be achieved by employing protein-binding DNA aptamers to template the labeling reaction[102].
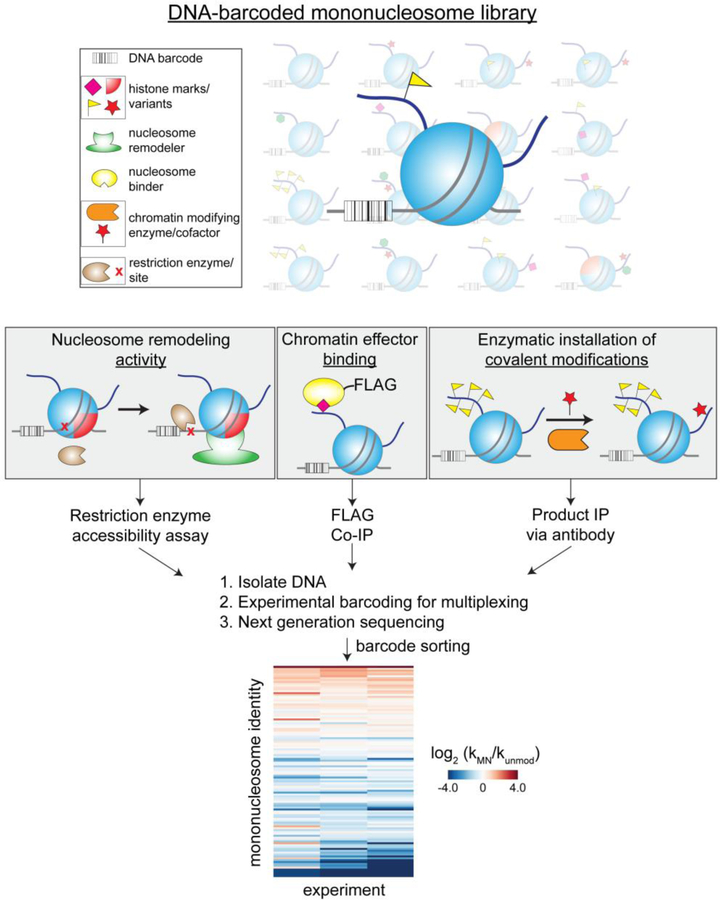
Naturally occurring nucleoprotein complexes also offer a convenient mechanism for homogenous DNA barcoding wherein the nucleic acid component can be tagged with a unique sequence identifier. Our lab has exploited this concept to generate a DNA-barcoded library of homogenously modified mononucleosomes, the fundamental repeating unit of chromatin[103]. Mononucleosomes are composed of histone proteins and DNA, both of which exhibit extensive modification landscapes that serve to regulate DNA accessibility and gene transcription. To analyze the role that these modifications play in chromatin effector activity, we synthesized 115 homogenously modified mononucleosomes via a variety of protein chemistry approaches and appended unique DNA sequences to the nucleic acid component of each library member[104]. Once barcoded, the mononucleosomes were pooled, and the resulting library was used in protein interaction and enzymatic assays (Figure 9). Following an assay of interest, all products (or efficient binding partners) can be isolated using pull-down or immunoprecipitation protocols, and the corresponding DNA sequences identified and quantified by NGS. This workflow has enabled high-throughput substrate characterization of a variety of epigenetic regulators, including chromatin remodelers, nucleosome binding proteins, and histone-modifying enzymes[105]. Importantly, these studies demonstrate the benefits of using NGS as an assay readout even with modest library sizes (102 members). Specifically, all modified nucleosome substrates are analyzed in a single, competition-based format to yield a highly sensitive spectrum of activities, and the PCR amplification-dependent signal greatly reduces the amount of substrate nucleosomes and effector protein required relative to nearly all other detection platforms. Notably, the Ruthenburg lab has also utilized DNA-barcoded, homogenously modified mononucleosomes as internal standards to calibrate ChIP-seq data[106].
Figure 9.
Accelerated chromatin effector profiling via a library of chemically distinct, DNA-encoded mononucleosomes substrates. This workflow is amenable to many common chromatin biochemistry assays, including nucleosome binding, enzymatic modification, and remodeling.
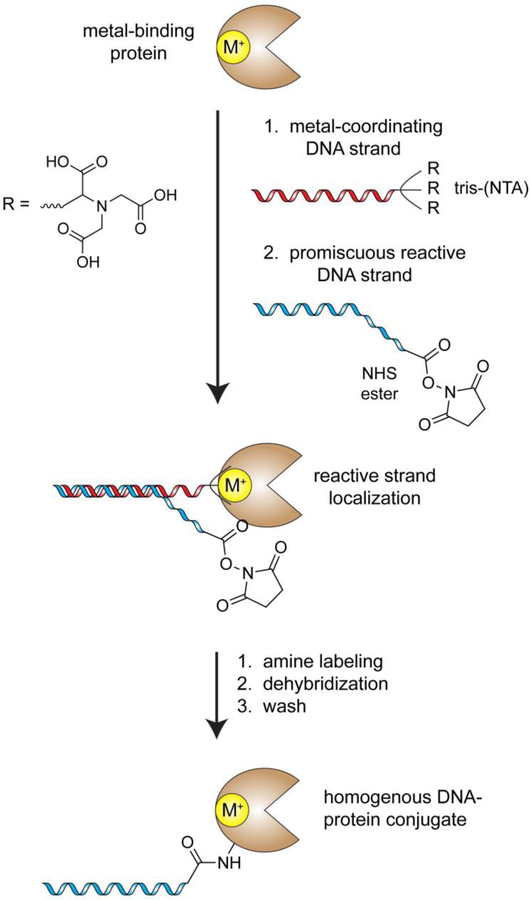
Despite their efficacy, the above DNA barcoding methods are limited to proteins with relatively specific structural features (co-factor or nucleic acid-binding site) or known small molecule ligands/DNA aptamers. In a much more generally applicable variation of these strategies, the Gothelf laboratory fused a DNA construct with tris-nitrilotriacetic acid (tris-NTA, a metal-binding functionality) to guide a template DNA strand to any protein that contains a metal binding site (note: an estimated one-third of all proteins contain a metal-binding site)[107]. As described above with other protein targeting vehicles, a complementary reactive DNA strand can then be added to achieve quantitative labeling of single local residue, and the metal binding template can be washed away to yield a homogenously modified functional protein (Figure 10). This approach has been successfully implemented with several metalloenzymes (ex. serotransferrin) and IgG antibodies, which possess histidine-rich clusters with metal-binding properties. While these and other conceptually similar methods are necessary for site-directed DNA tagging of native proteins, many genetically-encodable tags have been developed that enable facile and efficient DNA barcoding of recombinant proteins.
Figure 10.
DNA-templated protein conjugation as pioneered by the Gothelf laboratory. In this method, a metal-coordinating strand is used to localize the activity of an otherwise promiscuous reactive DNA strand, such as an NHS-ester-DNA conjugate. This method has proven effective with a broad spectrum of metal binding proteins. M+ = metal atom.
Genetically encodable tags for DNA conjugation
Although exogenous tags require genetic manipulation of the target protein(s), this is often acceptable when generating DNA-barcoded protein libraries for biochemical and cell-based assays (Figure 11). Among the most commonly applied tagging technologies for creating synthetic ligand-protein conjugates are the SNAP- (~19 kDa)[108], Halo- (~33 kDa)[109], and CLIP-tag (~20 kDa)[110], all of which utilize engineered enzymes that ultimately form a stable covalent bond with synthetic ligands. A variety of cloning vectors and easily functionalized synthetic ligand derivatives are now commercially available, making these orthogonal labeling approaches accessible to researchers across many fields of study. The Church lab recently employed the HaloTag technology (Figure 11a) to append DNA barcodes to nanodisc-embedded G-protein coupled receptors (GPCRs)[55]. This enabled parallel screening for GPCR agonist and antagonist specificity using their aforementioned SMI-seq platform. Much like the mononucleosome library technology described earlier, this work makes use of NGS as an assay readout with a relatively small DNA-encoded library (3 GPCRs) to take advantage of detection sensitivity, specificity, and multiplexing capacity. Indeed, these advantages in combination with decreasing costs and rapid turn-around time have made NGS an attractive alternative to many traditional assay readouts.
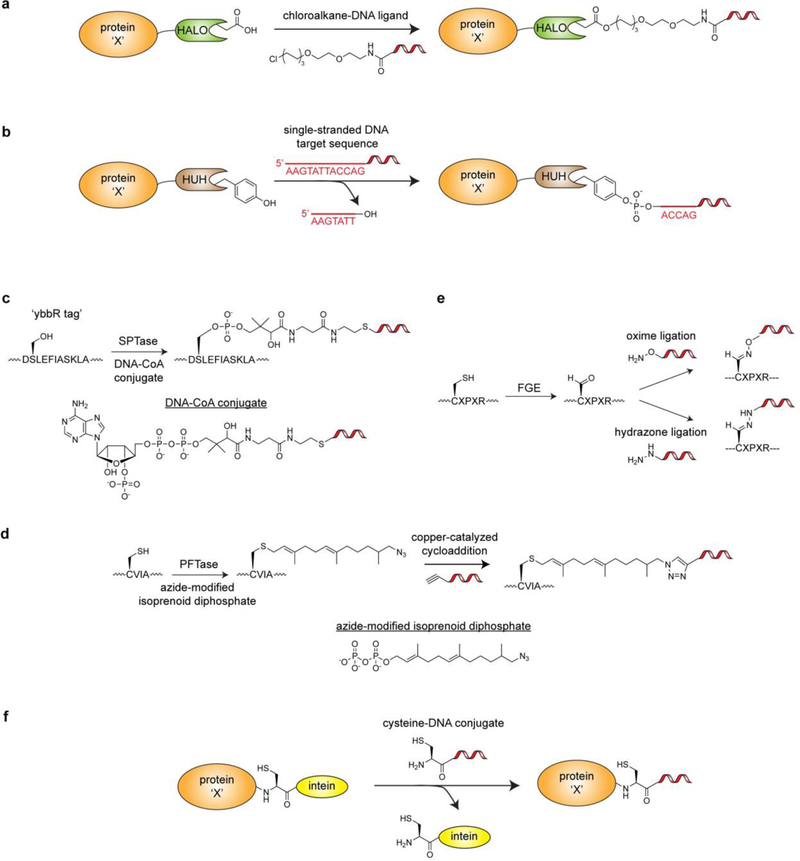
Figure 11.
Genetically encodable fusion tags that enable bioorthogonal labeling of proteins with DNA. a) Labeling of a protein-HALO tag fusion with a chloroalkane-DNA conjugate. b) Labeling of a protein-HUH domain fusion with a single stranded DNA fragment via a stable 5’-phosphotyrosine linkage. c) Sfp phosphopantetheinyl transferase (SPTase)-mediated labeling of the ybbR tag via a DNA-CoA conjugate. d) Protein farnesyltransferase (PFTase)-mediated labeling of the CVIA motif with an azide-modified isoprenoid diphosphate substrate. A subsequent copper-catalyzed cycloaddition reaction can be used to append an alkyne-functionalized DNA strand. e.) Formylglycine Generating Enzyme (FGE)-mediated labeling of the CXPXR motif. FGE is used to generate an aldehyde handle that can be used in subsequent oxime and hydrazone ligations with appropriately functionalized DNA strands. f) Labeling of a protein-intein fusion with a cysteine-DNA conjugate.
Another innovative DNA-protein conjugation strategy that was recently reported by the Gordon and de la Cruz labs involves the use of HUH-endonuclease domain (~10–30 kDa; named for their histidine-hydrophobic residue-histidine motif) fusions (Figure 11b). In nature, HUH domains cleave specific single-stranded DNA sequences to create a free 3’-OH group and a covalent 5’-phosphotyrosine intermediate[111]. In biochemical isolation, the phosphotyrosine linkage persists as a stable, covalent bond between the protein and DNA[112]. One major advantage of this DNA conjugation approach is that it does not require preparation of a DNA-linked synthetic ligand and can be carried out directly with unmodified DNA strands. While HUH-domain fusions have been used primarily to target DNA-protein conjugates to specific coordinates on DNA nanostructures (‘DNA origami structures’), they could be easily adapted to generate barcoded protein libraries. Additionally, there are many HUH-domains with unique DNA sequence specificities, which allows for orthogonal labeling of multiple proteins in one-pot if necessary.
One important consideration regarding the enzyme-based fusions described above is their potential to interfere with downstream assays. Attractive alternatives include peptide fusion-based tagging systems and self-excising inteins that leave a relatively minor adapter scar (Figure 11c–e). In one iteration of peptide-tagging, the peptide sequence serves as a target for transferase enzymes that can utilize DNA-appended co-factors. Examples of this include the Sfp phosphopantetheinyl transferase that can append single-stranded DNA (and other small molecules) to an eleven amino acid sequence (the ‘ybbR tag’) using a DNA-conjugated Coenzyme A co-factor[113] (Figure 11c) and a protein farnesyltransferase (PFTase) that can label a four amino acid sequence with an azide-modified isoprenoid diphosphate substrate[114] (Figure 11d). Like the HUH-domain fusions, both of these approaches have been used to enable assembly of protein nanostructures on DNA origami scaffolds. A complementary strategy is the aldehyde tag, a five amino acid sequence containing a cysteine that is modified to formylglycine in the presence of Formylglycine Generating Enzyme (FGE)[115] (Figure 11e). This conversion yields a bioorthogonal aldehyde handle that can be targeted by aminooxy or hydrazide reagents to yield oxime and hydrazone ligation products, respectively[116]. The Gartner lab has taken advantage of this approach to generate antibody-DNA conjugates for the assembly of DNA-templated heterodimeric and heterotrimeric antibody scaffolds[117]. These molecules enable exploration of antibody geometry, valency, and combinatorial binding capacity for applications such as cell surface recognition. Conceivably, this approach could be extended to create DNA-encoded libraries of oligomeric antibodies for the identification of novel cell-specific recognition biomolecules via NGS-based screening assays. Indeed, there are many alternative peptide-based tagging approaches that can be used to generate DNA-protein conjugates (e.g. the ‘SpyTag/SpyCatcher’[118]), and a comprehensive review of these technologies can be found elsewhere[119].
Inteins offer a bioorthogonal approach to generate DNA-protein conjugates with an extremely minimal scar (a single Cys, Ser, or Thr residue) at the ligation junction (Figure 11f). In expressed protein ligation, a backbone thioester is formed at the fusion junction between the protein of interest and the intein tag[120]. This thioester can be intercepted by a molecule bearing an N-terminal cysteine through a transthioesterification reaction, which spontaneously rearranges to an amide bond via an S/O-to-N acyl shift. To take advantage of intein tags for DNA conjugation (Figure 11f), facile methods have been developed to conjugate a cysteine residue to the 5’-end of DNA molecules[121]. Our laboratory has utilized the intein tagging strategy to append a poly dA:dT immune stimulant to antibodies that target dendritic cells, thus inducing an adaptive immune response for vaccine development purposes[122]. Inteins were also employed by the Niemeyer lab to conjugate peptide nucleic acids to proteins for hybridization-dependent microarray immobilization[123]. Notably, all of the protein conjugation strategies discussed in this section have pros and cons related to optimal reaction conditions, turn-over efficiency, ligation ‘scar’, and many other variables that will dictate which method is most suitable on a case-by-case basis[124].
DNA labeling of carbohydrates
Carbohydrates represent an abundant class of biomolecules with a complex set of building blocks and oligosaccharide topologies[125]. As one of the most ubiquitous modifications in the cell, glycosylated proteins can mediate processes such as cell-to-cell communication and interactions, cancer metastasis, and the immune response. There are several chemical and enzymatic methods to produce diverse libraries of homogeneous and heterogeneous carbohydrate chains in order to greatly facilitate characterization of their biological function[126]. Interestingly, a DNA-encoded carbohydrate library was recently reported by the Flitsch lab, which combines chemical synthesis, enzymatic glycosylation, and split-and-pool workflow to conjugate unique carbohydrate chains to specific DNA sequences for downstream high-throughput analysis[127].
DNA labeling of cells
Cellular DNA barcodes are typically introduced by genome engineering techniques that generate a permanent genomic scar that persists through cell division. However, conjugation of single-stranded DNA molecules to live cell surfaces opens up new possibilities in cell biology and cellular/tissue engineering. Many of the methods that have been described throughout this review have been applied to cell surface labeling, including the use of NHS-ester-labeled DNA to modify exposed lysines[128], click chemistry to modify cell surface glycans containing metabolically incorporated carbohydrate-azide derivatives (ex. N‐α‐azidoacetylmannosamine)[129], oxime ligations to cell surface aldehyde groups that are generated by periodate cleavage of native surface glycans[130], and antibody-DNA conjugates that target cell surface proteins[95b]. Other labeling approaches include synthesis of DNA-lipid conjugates that can embed themselves into cellular membranes[131] and expression of cell surface-displayed DNA-binding proteins (ex. programmable zinc fingers) that can capture specific exogenous DNA strands[132]. DNA-labeled cells have been particularly useful for inducing user-specified self-assembly of cells and tunable adhesion to solid surfaces, and have even facilitated programmed synthesis of microtissue arrays[133]. While these applications do not require NGS, it is easy to envision how DNA barcode sequencing could inform related aspects, including synthetic tissue composition and biological consequences of different induced cell-to-cell contacts.
Nanoparticles
DNA labeling has also found use with various nanomaterials, including drug delivery vehicles[134], quantum dots (QDs)[135], and gold nanoparticles (AuNPs)[136]. One innovative approach from the Wang lab encapsulated DNA barcodes inside 30 unique drug delivery nanoparticles and administered them to mice[134]. Subsequently, different tissues were harvested, and NGS was used to determine the delivery efficiency of each vehicle to different cells types. To achieve DNA labeling of nanomaterials with valuable optical and physiochemical properties, such as QDs and AuNPs, commercially available functionalized (ex. thiol or amine-labeled) particles can be conjugated to single-stranded DNA molecules bearing complementary reactive moieties for downstream applications. Interesting applications include hybridization-induced surface immobilization, self-assembly of two- and three-dimensional nanoparticle structures, and homo- and hetero-oligomerization for sensing and imaging[137]. Overall, the concept of utilizing DNA to achieve programmable material assembly holds huge potential spanning from biology to material science (as comprehensively reviewed elsewhere[138]), and it will be exciting to watch as this field evolves.
Another emerging technology that combines nanomaterials and DNA barcoding is the use of silica to encapsulate and protect DNA molecules[139]. The DNA component of these ‘synthetic fossils’ is highly thermostable and resistant to a wide-range of otherwise detrimental chemical conditions, yet easily liberated via treatment with hydrogen fluoride. The Grass and Stark labs have demonstrated the potential of such nanoparticles for barcoding valuable and/or dangerous goods (ex. food products[140]) to protect against counterfeits. Future efforts in this area may lead to standardized barcoding of medicines, fuels, and cosmetics, and other materials as a way to rapidly and inexpensively confirm product originality through DNA sequencing.
Outlook and new frontiers
It is remarkable how quickly NGS has become integrated into the fields of biology, chemistry, and physics since the first commercial instrument became available in 2005. This interdisciplinary penetrance has been fueled by the development of many complementary, facile nucleic acid-barcoding technologies as well as improvements in NGS accessibility, cost, and turn-around time. Based on the current trajectory of these fields and the success stories that have been highlighted throughout this review, it is logical to predict that NGS-based methodologies will become even more commonplace in the near future. As the experimental feasibility of bioconjugation chemistry and high-throughput sequencing continue to become less prohibitive, it is tempting to speculate what new frontiers will be explored next via NGS.
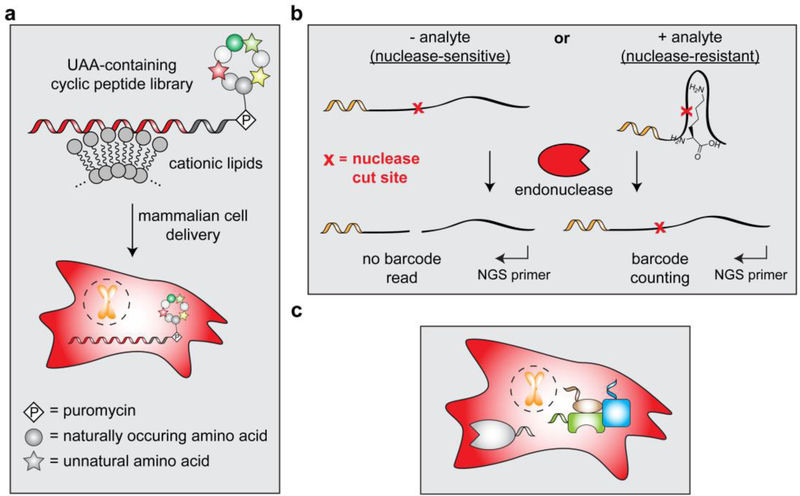
There are a number of immediately accessible advances that we can envision by simply combining barcoding with other nucleic acid-based technologies. One example of ‘multipurpose nucleic acid barcodes’ includes taking advantage of the RNA moiety on mRNA-displayed cyclic peptide libraries to deliver these molecules into mammalian cells via recently reported cationic lipid-mediated protein delivery methods[141]. In this case, the nucleic acid barcode would not only act as a molecular identifier but also serve as a handle for intracellular delivery of all library members, thus enabling high-throughput cell-based screening assays for macrocyclic peptides that contain a high density of unnatural building blocks (Figure 12a). Another potential development in the area of multipurpose nucleic acid barcodes may stem from nucleic acid aptamers and riboswitches. With the appropriate design elements, nucleic acid-based sensors can become nuclease-resistant in the presence of their target protein or small molecule to enable detection of sensor-analyte interactions via an exonuclease protection assay (as previously demonstrated[142]). Conceivably, this approach can be adapted to create an assay wherein nuclease activity removes a pre-installed barcode from the sensor after incubation in live cells or lysates (Figure 12b). Libraries of these ‘responsive barcode detection agents’ would enable protein and/or metabolite abundance determination in a one-pot (or single cell) fashion via NGS-based analysis.
Figure 12.
Potential future applications that take advantage of nucleic acid barcoding and NGS. a) mRNA-displayed cyclic peptide libraries can be delivered to cells using cationic lipids to enable cell-based screening assays. b) Barcoded nucleic acid-based sensors can be used in an endonuclease protection assay to facilitate detection and quantification of analytes. By implementing NGS as a readout, parallel analysis of multiple sensors is possible. c) A method for appending nucleic acid barcodes to proteins in live cells would allow for highly sensitive detection and reliable quantification of endogenous proteins.
There are many other frontiers that will require creativity and innovation beyond currently available methodologies but are worth mentioning here. One void in the field of DNA-encoded molecules is the lack of high-throughput methods to create libraries of recombinantly expressed proteins and protein complexes. These molecules still require individual purification and barcoding, thus severely limiting library size relative to ribosome display and mRNA display. Another exciting direction would be the development of systems that enable DNA barcoding of large subsets of proteins in a live cell (Figure 12c). This advance would have the potential to revolutionize the field of proteomics by increasing detection sensitivity and reducing detection bias relative to current mass spectrometry-based proteomics methods. Such an approach would likely require a reactive handle (ex. orthogonal HUH-domain fusions), and the functional effect of DNA barcoding would need to be considered on a case-by-case basis. DNA barcoding could also offer a possible solution for deconvoluting complex mixtures of proteins and/or nucleic acids that have been immobilized on a cryo-EM grid, thus enabling high-throughput structural characterization of macromolecules.
While specific details of our ideas are not offered here, we hope that the reader will appreciate the underlying principles and explore these and other exciting avenues. In a recent review, Shendure et al. predicted that, ‘…in the long view of history, the impact of DNA sequencing will be on a par with that of the microscope[2].’ Indeed, this technology has claimed a prominent role in many branches of science and will continue to thrive behind the strength of today’s highly collaborative and increasingly interdisciplinary style of research.
Acknowledgements
We thank Dr. Katharine Diehl, Dr. Robert Thompson, other current members of the Muir laboratory, and anonymous reviewers for suggestions and comments. Some of the work discussed herein was performed in the author’s laboratory and was supported by National Institutes of Health (NIH) Grants R37 GM086868, R01 GM107047, and P01 CA196539. G.P.L. was supported by an NIH Research Service Award (1F32GM110880).
References
- [1] a).Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, et al. , Nature 2005, 437, 376–380; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, et al. , Science 2005, 309, 1728–1732. [DOI] [PubMed] [Google Scholar]
- [2].Shendure J, Balasubramanian S, Church GM, Gilbert W, Rogers J, Schloss JA, et al. , Nature 2017, 550, 345–353. [DOI] [PubMed] [Google Scholar]
- [3] a).Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. , Nature 2001, 409, 860–921; [DOI] [PubMed] [Google Scholar]; b) Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. , Science 2001, 291, 1304–1351; [DOI] [PubMed] [Google Scholar]; c) http://www.genome.gov/27565109/ 2016.
- [4].Wetterstrand K, http://www.genome.gov/sequencingcostsdata 2018. [Google Scholar]
- [5].Metzker ML, Nat. Rev. Genet 2010, 11, 31–46. [DOI] [PubMed] [Google Scholar]
- [6].Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, et al. , Nature 2008, 456, 53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, et al. , Science 2009, 323, 133–138. [DOI] [PubMed] [Google Scholar]
- [8].Manrao EA, Derrington IM, Laszlo AH, Langford KW, Hopper MK, Gillgren N, et al. , Nat. Biotechnol 2012, 30, 349–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sanger F, Nicklen S, Coulson AR, Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. , Nature 2007, 448, 553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, et al. , Cell 2008, 132, 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, et al. , Science 2009, 326, 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS, Science 2009, 324, 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Shendure J, Lieberman Aiden E, Nat. Biotechnol 2012, 30, 1084–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Fowler DM, Araya CL, Fleishman SJ, Kellogg EH, Stephany JJ, Baker D, et al. , Nat. Methods 2010, 7, 741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yu H, Tardivo L, Tam S, Weiner E, Gebreab F, Fan C, et al. , Nat. Methods 2011, 8, 478–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17] a).Lee SY, Choi JH, Xu Z, Trends Biotechnol 2003, 21, 45–52; [DOI] [PubMed] [Google Scholar]; b) Ho M, Pastan I, Methods Mol. Biol 2009, 525, 337–352, xiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18] a).Dias-Neto E, Nunes DN, Giordano RJ, Sun J, Botz GH, Yang K, et al. , PLoS One 2009, 4, e8338; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ravn U, Gueneau F, Baerlocher L, Osteras M, Desmurs M, Malinge P, et al. , Nucleic Acids Res 2010, 38, e193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Esvelt KM, Carlson JC, Liu DR, Nature 2011, 472, 499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, et al. , Nature 2009, 460, 894–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].McKenna A, Findlay GM, Gagnon JA, Horwitz MS, Schier AF, Shendure J, Science 2016, 353, aaf7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Brenner S, Lerner RA, Proc. Natl. Acad. Sci. USA 1992, 89, 5381–5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Clark MA, Acharya RA, Arico-Muendel CC, Belyanskaya SL, Benjamin DR, Carlson NR, et al. , Nat. Chem. Biol 2009, 5, 647–654. [DOI] [PubMed] [Google Scholar]
- [24].Buller F, Mannocci L, Zhang Y, Dumelin CE, Scheuermann J, Neri D, Bioorg. Med. Chem. Lett 2008, 18, 5926–5931. [DOI] [PubMed] [Google Scholar]
- [25].Mannocci L, Zhang Y, Scheuermann J, Leimbacher M, De Bellis G, Rizzi E, et al. , Proc. Natl. Acad. Sci. USA 2008, 105, 17670–17675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Melkko S, Scheuermann J, Dumelin CE, Neri D, Nat. Biotechnol 2004, 22, 568–574. [DOI] [PubMed] [Google Scholar]
- [27].Wichert M, Krall N, Decurtins W, Franzini RM, Pretto F, Schneider P, et al. , Nat. Chem 2015, 7, 241–249. [DOI] [PubMed] [Google Scholar]
- [28].Halford B, Chem. Eng. News 2017, 95, 28–33. [Google Scholar]
- [29] a).Ahn S, Kahsai AW, Pani B, Wang QT, Zhao S, Wall AL, et al. , Proc. Natl. Acad. Sci. USA 2017, 114, 1708–1713; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Cheng RKY, Fiez-Vandal C, Schlenker O, Edman K, Aggeler B, Brown DG, et al. , Nature 2017, 545, 112–115. [DOI] [PubMed] [Google Scholar]
- [30] a).Harris PA, Berger SB, Jeong JU, Nagilla R, Bandyopadhyay D, Campobasso N, et al. , J. Med. Chem 2017, 60, 1247–1261; [DOI] [PubMed] [Google Scholar]; b) Chan AI, McGregor LM, Jain T, Liu DR, J. Am. Chem. Soc 2017, 139, 10192–10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Mannocci L, Melkko S, Buller F, Molnar I, Bianke JP, Dumelin CE, et al. , Bioconjug. Chem 2010, 21, 1836–1841. [DOI] [PubMed] [Google Scholar]
- [32].Franzini RM, Neri D, Scheuermann J, Acc. Chem. Res 2014, 47, 1247–1255. [DOI] [PubMed] [Google Scholar]
- [33] a).Gartner ZJ, Liu DR, J. Am. Chem. Soc 2001, 123, 6961–6963; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Gartner ZJ, Kanan MW, Liu DR, J. Am. Chem. Soc 2002, 124, 10304–10306; [DOI] [PubMed] [Google Scholar]; c) Gartner ZJ, Tse BN, Grubina R, Doyon JB, Snyder TM, Liu DR, Science 2004, 305, 1601–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34] a).Halpin DR, Harbury PB, PLoS Biol 2004, 2, E173; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Halpin DR, Harbury PB, PLoS Biol 2004, 2, E174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Usanov DL, Chan AI, Maianti JP, Liu DR, Nat. Chem 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hansen MH, Blakskjaer P, Petersen LK, Hansen TH, Hojfeldt JW, Gothelf KV, et al. , J. Am. Chem. Soc 2009, 131, 1322–1327. [DOI] [PubMed] [Google Scholar]
- [37] a).Hili R, Niu J, Liu DR, J. Am. Chem. Soc 2013, 135, 98–101; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Chen Z, Lichtor PA, Berliner AP, Chen JC, Liu DR, Nat. Chem 2018, 10, 420–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38] a).Goodnow RA Jr., Dumelin CE, Keefe AD, Nat. Rev. Drug Discov 2017, 16, 131–147; [DOI] [PubMed] [Google Scholar]; b) Zimmermann G, Neri D, Drug Discov. Today 2016, 21, 1828–1834; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Kleiner RE, Dumelin CE, Liu DR, Chem. Soc. Rev 2011, 40, 5707–5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Fields S, Song O, Nature 1989, 340, 245–246. [DOI] [PubMed] [Google Scholar]
- [40].Nirenberg MW, Matthaei JH, Proc. Natl. Acad. Sci. USA 1961, 47, 1588–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, et al. , Nat. Biotechnol 2001, 19, 751–755. [DOI] [PubMed] [Google Scholar]
- [42].Mattheakis LC, Bhatt RR, Dower WJ, Proc. Natl. Acad. Sci. USA 1994, 91, 9022–9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Hanes J, Pluckthun A, Proc. Natl. Acad. Sci. USA 1997, 94, 4937–4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].He M, Taussig MJ, Nucleic Acids Res 1997, 25, 5132–5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Sidhu SS, Lowman HB, Cunningham BC, Wells JA, Methods Enzymol 2000, 328, 333–363. [DOI] [PubMed] [Google Scholar]
- [46].Yan X, Xu Z, Drug Discov. Today 2006, 11, 911–916. [DOI] [PubMed] [Google Scholar]
- [47].Helma J, Cardoso MC, Muyldermans S, Leonhardt H, J. Cell Biol 2015, 209, 633–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Pluckthun A, Annu. Rev. Pharmacol. Toxicol 2015, 55, 489–511. [DOI] [PubMed] [Google Scholar]
- [49] a).Amstutz P, Pelletier JN, Guggisberg A, Jermutus L, Cesaro-Tadic S, Zahnd C, et al. , J. Am. Chem. Soc 2002, 124, 9396–9403; [DOI] [PubMed] [Google Scholar]; b) Cesaro-Tadic S, Lagos D, Honegger A, Rickard JH, Partridge LJ, Blackburn GM, et al. , Nat. Biotechnol 2003, 21, 679–685. [DOI] [PubMed] [Google Scholar]
- [50] a).Gersuk GM, Corey MJ, Corey E, Stray JE, Kawasaki GH, Vessella RL, Biochem. Biophys. Res. Commun 1997, 232, 578–582; [DOI] [PubMed] [Google Scholar]; b) Weichhart T, Horky M, Sollner J, Gangl S, Henics T, Nagy E, et al. , Infect. Immun 2003, 71, 4633–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hanes J, Schaffitzel C, Knappik A, Pluckthun A, Nat. Biotechnol 2000, 18, 1287–1292. [DOI] [PubMed] [Google Scholar]
- [52].Rouet R, Jackson KJL, Langley DB, Christ D, Front. Immunol 2018, 9, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53] a).Fowler DM, Fields S, Nat. Methods 2014, 11, 801–807; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Araya CL, Fowler DM, Trends Biotechnol 2011, 29, 435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Larman HB, Liang AC, Elledge SJ, Zhu J, Nat. Protoc 2014, 9, 90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Gu L, Li C, Aach J, Hill DE, Vidal M, Church GM, Nature 2014, 515, 554–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Roberts RW, Szostak JW, Proc. Natl. Acad. Sci. USA 1997, 94, 12297–12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Nemoto N, Miyamoto-Sato E, Husimi Y, Yanagawa H, FEBS Lett 1997, 414, 405–408. [DOI] [PubMed] [Google Scholar]
- [58].Takahashi TT, Austin RJ, Roberts RW, Trends Biochem. Sci 2003, 28, 159–165. [DOI] [PubMed] [Google Scholar]
- [59].Miyamoto-Sato E, Fujimori S, Ishizaka M, Hirai N, Masuoka K, Saito R, et al. , PLoS One 2010, 5, e9289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Hammond PW, Alpin J, Rise CE, Wright M, Kreider BL, J. Biol. Chem 2001, 276, 20898–20906. [DOI] [PubMed] [Google Scholar]
- [61].Cotten SW, Zou J, Valencia CA, Liu R, Nat. Protoc 2011, 6, 1163–1182. [DOI] [PubMed] [Google Scholar]
- [62].Keefe AD, Szostak JW, Nature 2001, 410, 715–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Seelig B, Szostak JW, Nature 2007, 448, 828–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Fukuda I, Kojoh K, Tabata N, Doi N, Takashima H, Miyamoto-Sato E, et al. , Nucleic Acids Res 2006, 34, e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Hertweck C, Angew. Chem. Int. Ed 2009, 48, 4688–4716. [DOI] [PubMed] [Google Scholar]
- [66].Sussmuth RD, Mainz A, Angew. Chem. Int. Ed 2017, 56, 3770–3821. [DOI] [PubMed] [Google Scholar]
- [67].Caboche S, Leclere V, Pupin M, Kucherov G, Jacques P, Bacteriol J. 2010, 192, 5143–5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Josephson K, Ricardo A, Szostak JW, Drug Discov. Today 2014, 19, 388–399. [DOI] [PubMed] [Google Scholar]
- [69].Zorzi A, Deyle K, Heinis C, Curr. Opin. Chem. Biol 2017, 38, 24–29. [DOI] [PubMed] [Google Scholar]
- [70].Zhu Z, Shaginian A, Grady LC, O’Keeffe T, Shi XE, Davie CP, et al. , ACS Chem. Biol 2018, 13, 53–59. [DOI] [PubMed] [Google Scholar]
- [71].Deyle K, Kong XD, Heinis C, Acc. Chem. Res 2017, 50, 1866–1874. [DOI] [PubMed] [Google Scholar]
- [72].Millward SW, Fiacco S, Austin RJ, Roberts RW, ACS Chem. Biol 2007, 2, 625–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Heinis C, Rutherford T, Freund S, Winter G, Nat. Chem. Biol 2009, 5, 502–507. [DOI] [PubMed] [Google Scholar]
- [74].Bashiruddin NK, Suga H, Curr. Opin. Chem. Biol 2015, 24, 131–138. [DOI] [PubMed] [Google Scholar]
- [75].Goto Y, Ohta A, Sako Y, Yamagishi Y, Murakami H, Suga H, ACS Chem. Biol 2008, 3, 120–129. [DOI] [PubMed] [Google Scholar]
- [76].Tavassoli A, Benkovic SJ, Nat. Protoc 2007, 2, 1126–1133. [DOI] [PubMed] [Google Scholar]
- [77].Forster AC, Tan Z, Nalam MN, Lin H, Qu H, Cornish VW, et al. , Proc. Natl. Acad. Sci. USA 2003, 100, 6353–6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78] a).Josephson K, Hartman MC, Szostak JW, J. Am. Chem. Soc 2005, 127, 11727–11735; [DOI] [PubMed] [Google Scholar]; b) Iwane Y, Hitomi A, Murakami H, Katoh T, Goto Y, Suga H, Nat. Chem 2016, 8, 317–325. [DOI] [PubMed] [Google Scholar]
- [79].O’Donoghue P, Ling J, Wang YS, Soll D, Nat. Chem. Biol 2013, 9, 594–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Ellman J, Mendel D, Anthony-Cahill S, Noren CJ, Schultz PG, Methods Enzymol 1991, 202, 301–336. [DOI] [PubMed] [Google Scholar]
- [81].Merryman C, Green R, Chem. Biol 2004, 11, 575–582. [DOI] [PubMed] [Google Scholar]
- [82].Morimoto J, Hayashi Y, Iwasaki K, Suga H, Acc. Chem. Res 2011, 44, 1359–1368. [DOI] [PubMed] [Google Scholar]
- [83].Murakami H, Ohta A, Ashigai H, Suga H, Nat. Methods 2006, 3, 357–359. [DOI] [PubMed] [Google Scholar]
- [84].Passioura T, Suga H, Chem. Commun 2017, 53, 1931–1940. [DOI] [PubMed] [Google Scholar]
- [85].Walport LJ, Obexer R, Suga H, Curr. Opin. Biotechnol 2017, 48, 242–250. [DOI] [PubMed] [Google Scholar]
- [86] a).Trads JB, Torring T, Gothelf KV, Acc. Chem. Res 2017, 50, 1367–1374; [DOI] [PubMed] [Google Scholar]; b) Niemeyer CM, Angew. Chem. Int. Ed 2010, 49, 1200–1216. [DOI] [PubMed] [Google Scholar]
- [87].Boutureira O, Bernardes GJ, Chem. Rev 2015, 115, 2174–2195. [DOI] [PubMed] [Google Scholar]
- [88].Bauer DM, Ahmed I, Vigovskaya A, Fruk L, Bioconjug. Chem 2013, 24, 1094–1101. [DOI] [PubMed] [Google Scholar]
- [89].MacDonald JI, Munch HK, Moore T, Francis MB, Nat. Chem. Biol 2015, 11, 326–331. [DOI] [PubMed] [Google Scholar]
- [90].Basle E, Joubert N, Pucheault M, Chem. Biol 2010, 17, 213–227. [DOI] [PubMed] [Google Scholar]
- [91].Kazane SA, Sok D, Cho EH, Uson ML, Kuhn P, Schultz PG, et al. , Proc. Natl. Acad. Sci. USA 2012, 109, 3731–3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Kim CH, Axup JY, Schultz PG, Curr. Opin. Chem. Biol 2013, 17, 412–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Sano T, Smith CL, Cantor CR, Science 1992, 258, 120–122. [DOI] [PubMed] [Google Scholar]
- [94].Schweitzer B, Wiltshire S, Lambert J, O’Malley S, Kukanskis K, Zhu Z, et al. , Proc. Natl. Acad. Sci. USA 2000, 97, 10113–10119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95] a).Niemeyer CM, Adler M, Wacker R, Trends Biotechnol 2005, 23, 208–216; [DOI] [PubMed] [Google Scholar]; b) Bailey RC, Kwong GA, Radu CG, Witte ON, Heath JR, J. Am. Chem. Soc 2007, 129, 1959–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Ullal AV, Peterson V, Agasti SS, Tuang S, Juric D, Castro CM, et al. , Sci. Transl. Med 2014, 6, 219ra219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Pollock SB, Hu A, Mou Y, Martinko AJ, Julien O, Hornsby M, et al. , Proc. Natl. Acad. Sci. USA 2018, 115, 2836–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gustafsdottir SM, et al. , Nat. Biotechnol 2002, 20, 473–477. [DOI] [PubMed] [Google Scholar]
- [99].Nong RY, Wu D, Yan J, Hammond M, Gu GJ, Kamali-Moghaddam M, et al. , Nat. Protoc 2013, 8, 1234–1248. [DOI] [PubMed] [Google Scholar]
- [100] a).Fruk L, Niemeyer CM, Angew. Chem. Int. Ed 2005, 44, 2603–2606; [DOI] [PubMed] [Google Scholar]; b) Cosnier S, Gondran C, Dueymes C, Simon P, Fontecave M, Decout JL, Chem. Commun 2004, 1624–1625. [DOI] [PubMed] [Google Scholar]
- [101].Li G, Liu Y, Liu Y, Chen L, Wu S, Liu Y, et al. , Angew. Chem. Int. Ed 2013, 52, 9544–9549. [DOI] [PubMed] [Google Scholar]
- [102].Vinkenborg JL, Mayer G, Famulok M, Angew. Chem. Int. Ed 2012, 51, 9176–9180. [DOI] [PubMed] [Google Scholar]
- [103].Nguyen UT, Bittova L, Muller MM, Fierz B, David Y, Houck-Loomis B, et al. , Nat. Methods 2014, 11, 834–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Dann GP, Liszczak GP, Bagert JD, Muller MM, Nguyen UTT, Wojcik F, et al. , Nature 2017, 548, 607–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105] a).Liszczak G, Diehl KL, Dann GP, Muir TW, Nat. Chem. Biol 2018, 14, 837–840; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wojcik F, Dann GP, Beh LY, Debelouchina GT, Hofmann R, Muir TW, Nat. Commun 2018, 9, 1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Grzybowski AT, Chen Z, Ruthenburg AJ, Mol. Cell 2015, 58, 886–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107] a).Rosen CB, Kodal AL, Nielsen JS, Schaffert DH, Scavenius C, Okholm AH, et al. , Nat. Chem 2014, 6, 804–809; [DOI] [PubMed] [Google Scholar]; b) Waldron KJ, Rutherford JC, Ford D, Robinson NJ, Nature 2009, 460, 823–830. [DOI] [PubMed] [Google Scholar]
- [108].Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K, Nat. Biotechnol 2003, 21, 86–89. [DOI] [PubMed] [Google Scholar]
- [109].Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, et al. , ACS Chem. Biol 2008, 3, 373–382. [DOI] [PubMed] [Google Scholar]
- [110].Gautier A, Juillerat A, Heinis C, Correa IR Jr., Kindermann M, Beaufils F, et al. , Chem. Biol 2008, 15, 128–136. [DOI] [PubMed] [Google Scholar]
- [111].Chandler M, de la Cruz F, Dyda F, Hickman AB, Moncalian G, Ton-Hoang B, Nat. Rev. Microbiol 2013, 11, 525–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112] a).Sagredo S, Pirzer T, Aghebat Rafat A, Goetzfried MA, Moncalian G, Simmel FC, et al. , Angew. Chem. Int. Ed 2016, 55, 4348–4352; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lovendahl KN, Hayward AN, Gordon WR, J. Am. Chem. Soc 2017, 139, 7030–7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113] a).Yin J, Straight PD, McLoughlin SM, Zhou Z, Lin AJ, Golan DE, et al. , Proc. Natl. Acad. Sci. USA 2005, 102, 15815–15820; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Pippig DA, Baumann F, Strackharn M, Aschenbrenner D, Gaub HE, ACS Nano 2014, 8, 6551–6555. [DOI] [PubMed] [Google Scholar]
- [114].Duckworth BP, Chen Y, Wollack JW, Sham Y, Mueller JD, Taton TA, et al. , Angew. Chem. Int. Ed 2007, 46, 8819–8822. [DOI] [PubMed] [Google Scholar]
- [115].Appel MJ, Bertozzi CR, ACS Chem. Biol 2015, 10, 72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Kolmel DK, Kool ET, Chem. Rev 2017, 117, 10358–10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Liang SI, McFarland JM, Rabuka D, Gartner ZJ, J. Am. Chem. Soc 2014, 136, 10850–10853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Min D, Arbing MA, Jefferson RE, Bowie JU, Protein Sci 2016, 25, 1535–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Lotze J, Reinhardt U, Seitz O, Beck-Sickinger AG, Mol. Biosyst 2016, 12, 1731–1745. [DOI] [PubMed] [Google Scholar]
- [120].Muir TW, Sondhi D, Cole PA, Proc. Natl. Acad. Sci. USA 1998, 95, 6705–6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Lovrinovic M, Niemeyer CM, Biochem. Biophys. Res. Commun 2005, 335, 943–948. [DOI] [PubMed] [Google Scholar]
- [122].Barbuto S, Idoyaga J, Vila-Perello M, Longhi MP, Breton G, Steinman RM, et al. , Nat. Chem. Biol 2013, 9, 250–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Lovrinovic M, Seidel R, Wacker R, Schroeder H, Seitz O, Engelhard M, et al. , Chem. Commun 2003, 822–823. [DOI] [PubMed] [Google Scholar]
- [124].Stephanopoulos N, Francis MB, Nat. Chem. Biol 2011, 7, 876–884. [DOI] [PubMed] [Google Scholar]
- [125].Bertozzi CR, Kiessling LL, Science 2001, 291, 2357–2364. [DOI] [PubMed] [Google Scholar]
- [126].Liang R, Yan L, Loebach J, Ge M, Uozumi Y, Sekanina K, et al. , Science 1996, 274, 1520–1522. [DOI] [PubMed] [Google Scholar]
- [127].Thomas B, Lu X, Birmingham WR, Huang K, Both P, Reyes Martinez JE, et al. , ChemBioChem 2017, 18, 858–863. [DOI] [PubMed] [Google Scholar]
- [128].Hsiao SC, Shum BJ, Onoe H, Douglas ES, Gartner ZJ, Mathies RA, et al. , Langmuir 2009, 25, 6985–6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Chandra RA, Douglas ES, Mathies RA, Bertozzi CR, Francis MB, Angew. Chem. Int. Ed 2006, 45, 896–901. [DOI] [PubMed] [Google Scholar]
- [130].Vogel K, Glettenberg M, Schroeder H, Niemeyer CM, Small 2013, 9, 255–262. [DOI] [PubMed] [Google Scholar]
- [131].Selden NS, Todhunter ME, Jee NY, Liu JS, Broaders KE, Gartner ZJ, J. Am. Chem. Soc 2012, 134, 765–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Mali P, Aach J, Lee JH, Levner D, Nip L, Church GM, Nat. Methods 2013, 10, 403–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Todhunter ME, Jee NY, Hughes AJ, Coyle MC, Cerchiari A, Farlow J, et al. , Nat. Methods 2015, 12, 975–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Dahlman JE, Kauffman KJ, Xing Y, Shaw TE, Mir FF, Dlott CC, et al. , Proc. Natl. Acad. Sci. USA 2017, 114, 2060–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Wang G, Li Z, Ma N, ACS Chem. Biol 2018. [DOI] [PubMed] [Google Scholar]
- [136].Niemeyer CM, Ceyhan B, Hazarika P, Angew. Chem. Int. Ed 2003, 42, 5766–5770. [DOI] [PubMed] [Google Scholar]
- [137] a).Wang CC, Wu SM, Li HW, Chang HT, ChemBioChem 2016, 17, 1052–1062; [DOI] [PubMed] [Google Scholar]; b) Banerjee A, Pons T, Lequeux N, Dubertret B, Interface Focus 2016, 6, 20160064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Jones MR, Seeman NC, Mirkin CA, Science 2015, 347, 1260901. [DOI] [PubMed] [Google Scholar]
- [139].Paunescu D, Fuhrer R, Grass RN, Angew. Chem. Int. Ed 2013, 52, 4269–4272. [DOI] [PubMed] [Google Scholar]
- [140].Puddu M, Paunescu D, Stark WJ, Grass RN, ACS Nano 2014, 8, 2677–2685. [DOI] [PubMed] [Google Scholar]
- [141].Zuris JA, Thompson DB, Shu Y, Guilinger JP, Bessen JL, Hu JH, et al. , Nat. Biotechnol 2015, 33, 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Wang XL, Li F, Su YH, Sun X, Li XB, Schluesener HJ, et al. , Anal. Chem 2004, 76, 5605–5610. [DOI] [PubMed] [Google Scholar]