Abstract
Introduction:
The present study aimed to report the prevalent HIV-1 drug-resistant mutations in patients with HIV-1 alone and tuberculosis (TB) coinfection alone to improve our understanding of the mutation patterns and aid treatment decisions.
Methods:
Patients with HIV-1 and HIV-TB on treatment for more than 1 year with suspected failure were recruited. Sequencing of protease and two-thirds of the region of reverse transcriptase gene was done for drug-resistant mutations.
Results:
In the HIV-TB group (n = 25), 88%, 92%, and 12% had mutations to nucleoside reverse transcriptase inhibitors (NRTIs), non-nucleoside reverse transcriptase inhibitors (NNRTIs), and protease inhibitors (PIs), respectively. In the HIV-alone group (n = 25), 84%, 100%, and 4% had mutations to NRTIs, NNRTIs, and PIs, respectively. M184V, M41L, D67N, G190A, A98G, and K103N were the most common mutations seen.
Conclusion:
There is a high prevalence of drug-resistant mutations in HIV and HIV-TB coinfected patients.
Keywords: antiviral resistance, HIV-tuberculosis, HIV, treatment failure
What Do We Already Know About This Topic?
With increasing availability of antiretroviral therapy, there is emerging data to suggest an increase in HIV drug-resistant mutations.
How Does Your Research Contribute to the Field?
As tuberculosis and HIV are co-endemic diseases, we propose that the tuberculosis or its treatment may be driving these mutations.
What Are Your Research’s Implications Toward Theory, Practice, or Policy?
Universal viral load and drug susceptibility testing will help identify patients harboring drug-resistant mutations, thereby decreasing risk of opportunistic infections and spread.
Introduction
HIV has been a public health challenge for over 3 decades. According to the Joint United Nations Program on HIV/AIDS (UNAIDS) 2016 fact sheet, there were 36.7 million people living with HIV/AIDS at the end of 2015, with approximately 2 million new HIV infections in the same year.1 Since the introduction of effective antiretroviral therapy (ART) in the 1990s, there has been a marked decline in HIV-related morbidity and mortality. However, as the virus has adapted itself to the chemotherapy, drug-resistant mutations (DRMs) have emerged.2 In the struggle to control the HIV epidemic by 2030, the threat of increasing DRMs looms large.3
According to the UNAIDS 2016 fact sheet, the prevalence of HIV in Indian adults aged 15 to 49 years is 0.3%.4 Of the 2.1 million people living with HIV, only 49% are on ART. The current first-line ART regimen in India includes 2 nucleoside reverse transcriptase inhibitors (NRTIs) and 1 non-nucleoside reverse transcriptase inhibitor (NNRTI).5,6 The most common regimen consists of tenofovir disoproxil fumarate (TDF), lamivudine (3TC), and efavirenz (EFV). If the first-line treatment fails, the second-line treatment consists of 2 NRTIs and a protease inhibitor (PI; usually atazanavir with ritonavir boost [ATV/r]). Integrase inhibitor-based therapy and baseline viral load testing for all are currently not available in the national program. HIV drug susceptibility testing leads to improved survival.7,8 Due to cost constraints, universal viral load or drug susceptibility testing is not done in India.
There are 2 types of HIV drug resistance: primary and secondary. Primary drug resistance (PDR) is the resistance when an individual is infected by a strain of HIV which was already resistant to one or more drugs; secondary drug resistance (SDR) develops during ART. According to the World Health Organization (WHO) data for 2017, the prevalence of PDR against NNRTIs in HIV-positive patients is >10%.3 The prevalence of SDR in patients with first-line ART failure in Asia is around 87% to 95%, with 50% to 64% harboring DRMs of 2 classes of drugs.9,10 A study from north India reported an SDR prevalence of 93.9% in patients with ART failure.11
Since tuberculosis (TB) is a common opportunistic infection in people living with HIV12 and causes high mortality, we assessed the prevalence and pattern of HIV DRMs in HIV-TB coinfected patients and in patients with HIV alone. This knowledge will help design treatment plans as mandatory genotyping of HIV becomes the rule in future.
Material and Methods
All patients, aged ≥18 years and on ART for at least 1 year with suspected clinical or immunological failure, were screened for eligibility during the period from July 2012 to January 2016. A detailed treatment history was taken from all patients and spouses (in case they were on ART). Exclusion criteria included a viral load <1000 RNA copies/mL; serological evidence of acute hepatitis A, B, C, or E; HBsAg positive; anti-hepatitis C virus antibody positive; or pregnancy. Baseline demographic data for every patient were recorded in a predesigned performa.
Ethical Approval
Our study was approved by the All India Institute of Medical Sciences ethics committee (approval number IESC/T-88/1.2.2013), and written informed consent was taken from all patients prior to participation.
Twenty-five patients were recruited in each group, where group 1 had HIV-positive patients with TB coinfection and group 2 had HIV-positive patients without TB coinfection.
Specimen Collection
Ten milliliters of whole blood sample from each patient was collected in K3-EDTA vacutainer tubes (Becton Dickinson, San Jose, California). Three milliliters was used for CD4 count estimation, and the remaining was centrifuged within 6 hours of collection at 400g for 10 minutes to separate plasma, and aliquots of 1 mL were stored at −80°C. These were further used for HIV viral load and HIV-1 genotyping as per the WHO HIV ResNet Laboratory Working Group guidelines.8
Viral Load Testing and CD4 T-Cell Estimation
Viral load testing was performed using the standard protocol of AMPLICOR HIV-1 Monitor Test, version 1.5 (Roche Molecular Systems Inc, Branchburg, New Jersey). CD4/CD8 T-cell counts were determined by flow cytometry using BD FACS CALIBUR (BD Biosciences, San Jose, California). The lower and higher limits of detection were 40 HIV-1 RNA copies/mL and 10 million HIV-1 RNA copies/mL, respectively.
HIV-1 Genotyping and Drug Resistance
HIV-1 genotyping and mutation analysis was performed using the ViroSeq HIV-1 Genotyping Systems (Abbott Diagnostics, Wiesbaden, Germany).13
Analysis was done with HIV-1 RNA isolation, reverse transcription (RT) with Moloney murine leukemia virus RT, and a single 40-cycle polymerase chain reaction (PCR) with AmpliTaq Gold (Abbott Diagnostics). Polymerase chain reaction yields a 1.3-kb DNA product. Polymerase chain reaction amplifications were performed with a uracil N-glycosylase contamination control system to reduce the risk of contamination of the PCR mixtures with products from previous amplification reactions. Polymerase chain reaction product bands with minimum DNA >20 ng were selected for DNA sequencing. DNA sequence analysis was performed with premixed BigDye sequencing reagents with 7 different primers (Abbott Diagnostics). BigDye terminator chemistry version 1.1 with Dye set E was used and provided 98% accuracy at 550 bases for the ABI 3130xl Genetic Analyzer (Applied Biosystems, Foster City, California), which was used for DNA analyses. Data collection software version 3.0 and sequence analysis software version 5.3 were used for obtaining data from the DNA sequencer. ViroSeq HIV-1 Genotyping System Software version 2.8 was used to assemble sequence data from different primers into a single project and generate a contiguous sequence spanning the entire protease and up to codon 335 of the RT gene. This consensus is compared to a known reference strain, HXB-2, to identify points of variance. An HIV drug resistance clinical report was exported and printed. The mutations picked up by the ViroSeq software were also compared to the HIVdb online Stanford database for updated known resistance mutations and for subtype/circulating recombinant forms and phylogenetic analysis.14 Sequencing was indigenously carried out at our laboratory on the 16-capillary automated ABI PRISM 3130xl Genetic Analyzer.
ViroSeq mutations and sequences were submitted to Stanford University HIV Drug Resistance Database14 to determine the drug resistance profile and subtype of each sample. Therefore, an updated genotypic–clinical correlation was obtained.
Clade Typing and Phylogenetic Tree
1.3 kb DNA sequences of the PI and RT regions from HIV-1 obtained were analyzed and aligned with reference sequences from different subtypes. Phylogenetic analysis was done using MEGA 7 software for bigger data sets.15 For phylogenetic study, nucleotide sequences were aligned using software and the ClustalW program.16 DNA distance and neighbor-joining tree were generated using the program within the MEGA 7 software.
Statistical Analysis
Mean and standard deviation were computed for data following parametric distribution. Median with range was computed for data following nonparametric distribution. The unpaired t test was used to find significant differences in normal data, while the Wilcoxon rank sum test was used to find differences in CD4 count and viral load. A sample size of convenience was taken because of financial constraints. A P value of <.05 was considered significant.
Results
There were 25 patients in each of the 2 groups (ie, HIV-TB and HIV alone). Baseline demographic and laboratory values of the patients in both groups are summarized in Tables 1 and 2, respectively. Patients with HIV-TB coinfection were younger and had a lower body mass index (P < .05). There was no difference in the CD4 count or the viral load.
Table 1.
Demographics and Treatment Details.a
| Variable | HIV and TB (n = 25) | HIV Alone (n = 25) |
|---|---|---|
| Ageb (years) | 37 (33-40) | 42 (36-48) |
| BMIb (kg/m2) | 17.6 (1.8) | 19.8 (2.4) |
| Male | 18 (72) | 21 (84) |
| Heterosexual mode of transmission | 24 (96) | 23 (92) |
| Site of TB | ||
| Pulmonary | 9 (36) | - |
| Extrapulmonary | 13 (52) | - |
| Disseminated | 3 (12) | - |
| ART duration (in months) | 60 (27-119) | 50 (19-96) |
| ART regimen, n (%) | ||
| D4T + 3TC + NVP | 1 (4) | 9 (36) |
| D4T + 3TC + EFV | 11 (40) | 0 |
| ZDV + 3TC + NVP | 0 | 16 (64) |
| ZDV + 3TC + EFV | 13 (52) | 0 |
| Change in ART regimen | ||
| No change | 7 (28) | 14 (56) |
| Once | 12 (48) | 9 (36) |
| Twice | 6 (24) | 2 (8) |
Abbreviations: ART, antiretroviral therapy; BMI, body mass index; D4T, stavudine; EFV, efavirenz; EPTB, extrapulmonary tuberculosis; NVP, nevirapine; TB, tuberculosis; 3TC, lamivudine; ZDV, zidovudine.
aData are presented as mean (standard deviation); median (interquartile range) or frequency (percentages).
b P value < .05; unpaired t test.
Table 2.
Laboratory Values at the Time of ART Failure.a
| Variable | HIV and TB, n = 25 | HIV Alone, n = 25 |
|---|---|---|
| CD4 count (cells/μL) | 75 (40-154) | 92 (58-137) |
| Log10 viral load (copies/mL) | 5.7 (4.9-5.8) | 5.3 (4.9-5.6) |
| Hemoglobin (g/dL) | 10 (8.4-11.1) | 10.1 (9.4-10.8) |
| TLC (cells/μL) | 4700 (3870-6400) | 6400 (4700-7300) |
| ESRb (mm/1 hour) | 48 (40-55) | 15 (10-25) |
| Total bilirubinb (mg/dL) | 0.8 (0.61-0.9) | 0.5 (0.5-0.7) |
| ASTb (IU/L) | 49 (35-66) | 32 (29-43) |
| ALTb (IU/L) | 41 (33-48) | 30 (21-38) |
| Total protein (g/dL) | 7.1 (6.7-7.8) | 8.0 (7.6-8.5) |
| Albuminb (g/dL) | 3.4 (3-4.4) | 4.4 (4-4.6) |
| Ureab (mg/dL) | 22 (19-28) | 18 (16-21) |
| Creatinine (mg/dL) | 0.8 (0.7-0.9) | 0.7 (0.6-0.8) |
Abbreviations: ALT, alanine transaminase; AST, aspartate transaminase; CD, cluster of differentiation; ESR, erythrocyte sedimentation rate; TLC, total leukocyte count.
aData are presented as median (interquartile range).
b P value <.05, Mann-Whitney U test.
Both groups were on first-line ART as per guidelines. There was no significant difference in the median duration of ART in the 2 groups. The ART regimen and changes made in the regimen are summarized in Table 1.
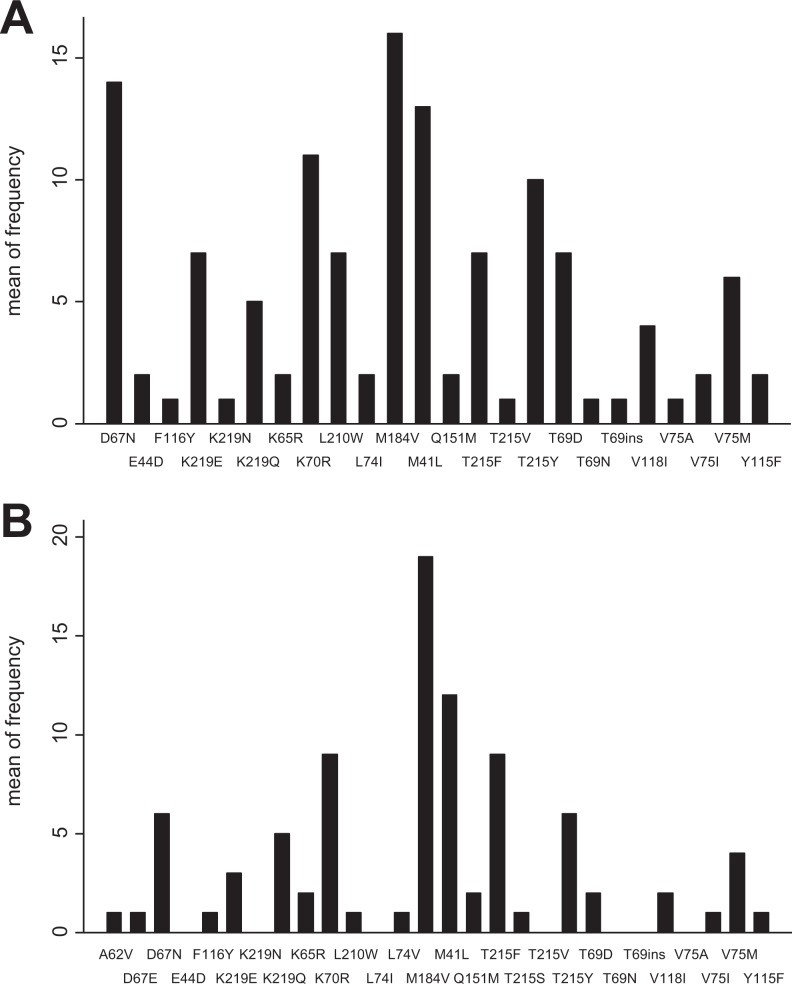
In patients with HIV-TB (n = 25), 24 (96%) had DRMs in at least one of the ART drug classes. Twenty-two (88%) patients had both NRTIs and NNRTIs DRMs. Twenty-two patients had DRMS to NRTIs, while 23 patients had DRMs to NNRTIs. Only 3 PI DRMs were found (1 minor and 2 major). The DRMs are summarized in Table 3 and Figures 1A and 2A. Thymidine analog mutations were found in 22 (88%) patients, with more than 3 mutations in 52% (13/25) of the patients.
Table 3.
Secondary Drug Resistance Mutations in Patient with HIV and TB.a
| Mutations (n = 25) | |||
|---|---|---|---|
| NRTI DRMs | 22 (88) | ||
| M41L | 13 (52) | Y115F | 2 (8) |
| E44D | 2 (8) | F116Y | 1 (4) |
| K65R | 2 (8) | V118I | 4 (16) |
| D67N | 14 (56) | Q151M | 2 (8) |
| T69D | 7 (28) | M184V | 16 (64) |
| T69N | 1 (4) | L210W | 7 (28) |
| T69ins | 1 (4) | T215F | 7 (28) |
| K70R | 11 (44) | T215V | 1 (4) |
| L74I | 2 (8) | T215Y | 10 (40) |
| V75M | 6 (24) | K219E | 7 (28) |
| V75I | 2 (8) | K219N | 1 (4) |
| V75A | 1 (4) | K219Q | 5 (20) |
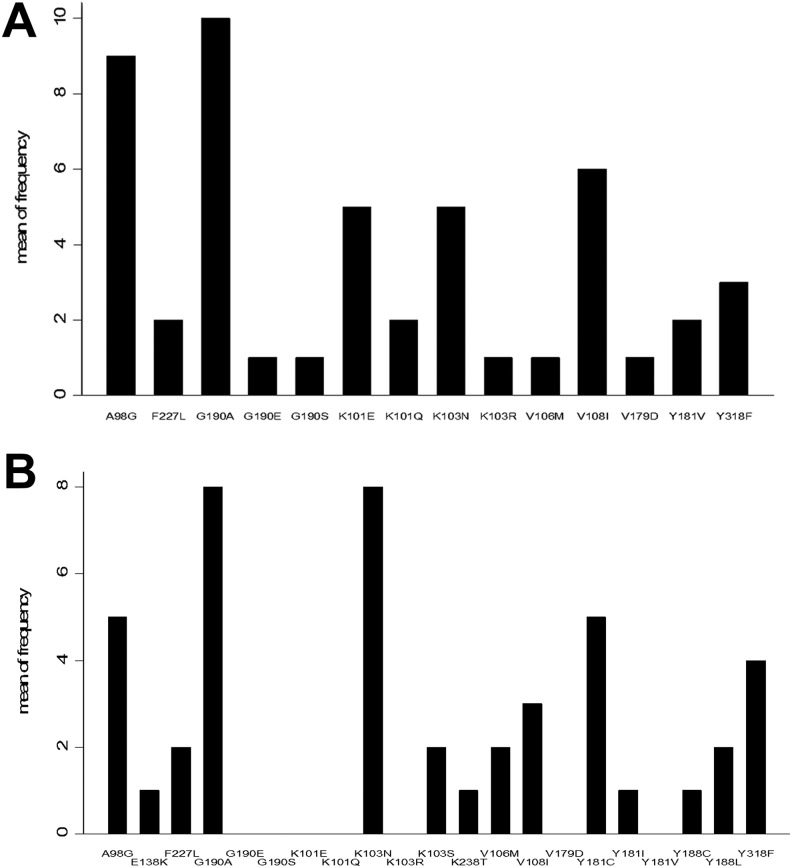
| NNRTI DRMs | 23 (92) | ||
| A98G | 9 (36) | V179D | 1 (4) |
| K101E | 5 (20) | Y181V | 2 (8) |
| K101Q | 2 (8) | G190A | 10 (40) |
| K103N | 5 (20) | G190E | 1 (4) |
| K103R | 1 (4) | G190S | 1 (4) |
| V106M | 1 (4) | F227L | 2 (8) |
| V108I | 6 (24) | Y318F | 3 (12) |
| PI—major DRMs | 1 (4) | ||
| G48M | 1 (4) | ||
| PI—minor DRMs | 2 (8) | ||
| T74S | 2 (8) |
Abbreviations: DRMs, drug-resistant mutations; NRTI, nucleoside reverse transcriptase inhibitors; NNRTI, non-nucleoside reverse transcriptase; PI, protease inhibitor.
aData are presented as frequency (percent).
Figure 1.
Frequency and type of non-nucleoside reverse transcriptase inhibitor (NNRTI) drug resistant mutation in patients with HIV and tuberculosis (A) and HIV alone (B).
Figure 2.
Frequency and type of nucleoside reverse transcriptase inhibitor (NRTI) drug-resistant mutation in patients with HIV and tuberculosis (A) and HIV alone (B).
In patients with HIV alone (n = 25), all patients had DRMs to at least 1 drug class. Both NRTI and NNRTI DRMs were found in 21 (84%) patients; 21 (84%) patients had DRMs to NRTIs, and all 25 patients had mutations to NNRTIs. These mutations are summarized in Table 4 and Figures 1B and 2B. Thymidine analog mutations were found in 18 patients; 8 (32%) patients had more than 3 mutations.
Table 4.
Secondary Drug Resistance Mutations in Patients with HIV Alone.a
| Mutations (n = 25) | |||
|---|---|---|---|
| NRTIs DRMs | 21 (84) | ||
| M41L | 12 (24) | Y115F | 1 (4) |
| A62V | 1 (4) | F116Y | 1 (4) |
| K65R | 2 (8) | V118I | 2 (8) |
| D67E | 1 (4) | Q151M | 2 (8) |
| D67N | 6 (26) | M184V | 19 (76) |
| T69D | 2 (8) | L210W | 1 (4) |
| K70R | 9 (36) | T215F | 9 (36) |
| L74V | 1 (4) | T215Y | 6 (24) |
| V75M | 4 (16) | T215S | 1 (4) |
| V75I | 1 (4) | K219E | 3 (12) |
| K219Q | 5 (20) | ||
| NNRTIs DRMs | 25 (100) | ||
| A98G | 5 (20) | Y188L | 2 (8) |
| K103N | 8 (32) | G190A | 8 (32) |
| K103S | 2 (8) | F227L | 2 (8) |
| V106M | 2 (8) | K238T | 1 (4) |
| V108I | 3 (12) | Y318F | 4 (16) |
| E138K | 1 (4) | Y188C | 1 (4) |
| Y181C | 5 (20) | Y181I | 1 (4) |
| PI—major DRMs | 1 (4) | ||
| M46I | 1 (4) |
Abbreviations: DRMs, drug-resistant mutations; NRTI, nucleoside reverse transcriptase inhibitors; NNRTI, non-nucleoside reverse transcriptase; PI, protease inhibitor.
aData are presented as frequency (percent).
Drug Resistance Profile According to Stanford Resistance Score
In patients with HIV-TB coinfection, among the NRTIs, 80% of patients had high-level resistance to zidovudine (ZDV) and stavudine (d4T), 72% to abacavir (ABC), and 68% to 3TC. For NNRTIs, 84% of patients had high-level resistance to nevirapine (NVP) and 72% to EFV.
In patients with HIV alone, among the NRTIs, 52% of patients had high-level resistance to zidovudine, 64% to d4T, 48% to ABC, and 76% to 3TC. For NNRTIs, 92% and 60% of patients had high-level resistance to NVP and EFV, respectively.
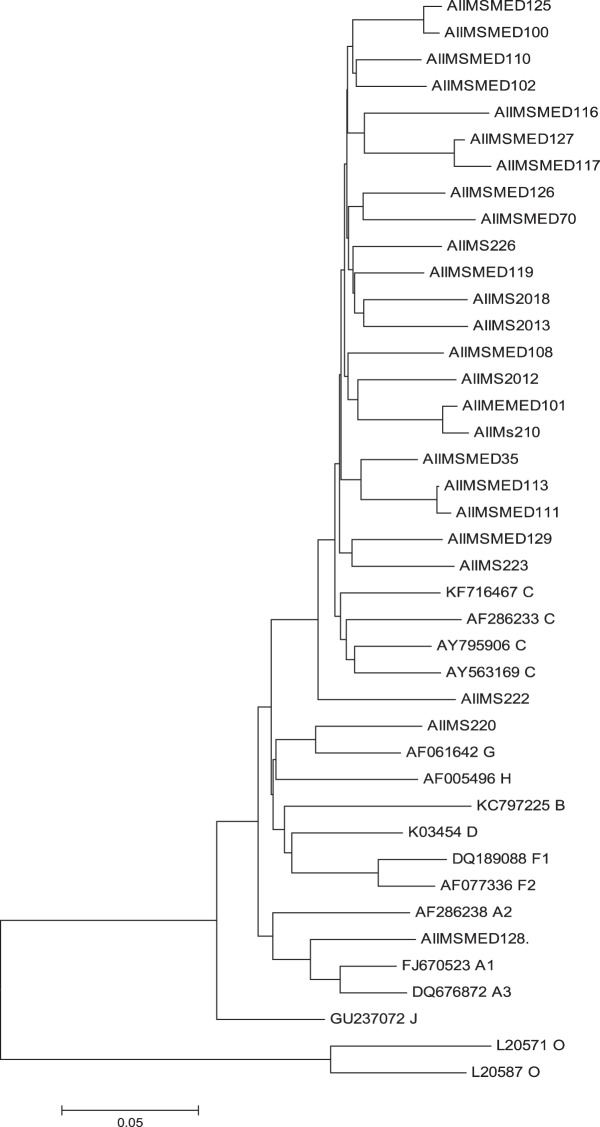
Clade Typing and Phylogenetic Tree
Phylogenetic analysis of sequences obtained showed that the majority of the sequences are subtype C in both groups (Figures 3 and 4).
Figure 3.
Phylogenetic tree of HIV-1 subtype in patients with HIV and tuberculosis.
Figure 4.
Phylogenetic tree of HIV-1 subtype in patients with HIV alone.
Discussion
Almost all patients in both groups with suspected ART failure were found to have DRMs to at least one of the ART drug classes. Emergence of DRMs decimates the goal of suppressing viral replication to break the chain of transmission across the globe.
Patients with HIV-TB coinfection were older (47 versus 37 years) and had a lower body mass index. Higher age in HIV-TB coinfection suggests longer duration of HIV infection and a higher viral load. There was a trend toward lower CD4 counts and higher viral copies in HIV-TB coinfected group, which could not reach statistical significance. There was a significant difference in the metabolic panel of the 2 groups as well, with HIV-TB coinfected patients having a higher erythrocyte sedimentation rate, aspartate transaminase, alanine transaminase, and serum bilirubin. This could be due to the infection itself and the treatment with anti-TB drugs (rifampicin, isoniazid, pyrazinamide), which are known to raise liver enzymes and bilirubin.
The high prevalence of DRMs reported in our study supports results from other parts of India.11,17–19 M184V was the most commonly observed NRTI-associated mutation in both groups (64% in HIV-TB patients and 76% in HIV-positive patients). This mutation causes high-level resistance to 3TC and emtricitabine (FTC) but makes the virus more susceptible to AZT, d4T, and TDF. This mutation is associated with increased ABC resistance when associated with thymidine analog mutations.20,21
Thymidine analog mutations are commonly observed in low-income countries where fixed-dose combinations containing thymidine analogs are the backbone of the therapy. Patients with HIV and TB (88%) had more thymidine analog mutations than those with HIV alone (72%). The high prevalence of thymidine analog mutations in the patients included in this study might explain the increased DRMs in HIV and HIV-TB patients despite the known protective role of the M184V mutation.22
There were 2 uncommon mutations observed. The T69Ins mutation was observed in 1 patient from the HIV-TB coinfected group. This patient had 3 thymidine analog mutations, and the M184V mutation, which makes it resistant to all NRTIs. The Q151M mutation was observed in 2 patients with HIV alone. Q151M mutations cause intermediate-/high-level resistance to AZT, ddI, d4T, and ABC with low-level resistance to TDF, 3TC, and FTC. Also, in combination with accessory mutations at positions 62, 75, 77, and 116, Q151M confers high-level resistance to AZT, ddI, d4T, and ABC and intermediate-level resistance to TDF, 3TC, and FTC. These patients had mutations at these other sites also, thereby rendering a state of multidrug resistance.23
There is predominance of HIV-1 subtype C in both HIV and HIV-TB coinfected patients, which is in agreement with our previously published study.11 We have been able to demonstrate a high prevalence of DRMs in patients with HIV-TB and HIV alone. There were a few limitations in our study. A sample size of convenience was taken. All the drug mutations listed and tested may not be clinically relevant, and more data are required to find their significance. We did not follow up the patients to look for treatment outcome.
Worldwide, there is greater emphasis than ever before on virological suppression in all patients on ART to break the chain of transmission. The present study shows that there is a high prevalence of DRMs in patients with suspected ART failure. It is imperative to study the pattern of these DRMs to guide policy changes in future to change ART regimens. Although genotyping for DRMs in all patients is currently not a cost-effective measure, it may help in personalizing HIV treatment based on individual variations in drug susceptibility.
Conclusion
There is a high prevalence of DRMs in HIV and HIV-TB coinfected patients with first-line failure.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Sanjeev Sinha, MD  https://orcid.org/0000-0001-8060-4231
https://orcid.org/0000-0001-8060-4231
References
- 1. Global AIDS Update 2016. 2016. http://www.unaids.org/sites/default/files/media_asset/global-AIDS-update-2016_en.pdf. Accessed March 28, 2018.
- 2. Larder BA, Darby G, Richman DD. HIV with reduced sensitivity to zidovudine (AZT) isolated during prolonged therapy. Science. 1989;243(4899):1731–1734. [DOI] [PubMed] [Google Scholar]
- 3. Haile-Selassie H. World Health Organization. WHO HIV Drug Resistance Report 2017. 2017. http://apps.who.int/iris/bitstream/10665/255896/1/9789241512831-eng.pdf. Accessed March 28, 2018.
- 4. India | UNAIDS. 2017. http://www.unaids.org/en/regionscountries/countries/india. Accessed May 31, 2018.
- 5. Ministry of Health and Family Welfare. Antiretroviral therapy guidelines for HIV-infected adults and adolescents. 2013. http://naco.gov.in/sites/default/files/Antiretroviral%20Therapy%20Guidelines%20for%20HIVInfected%20Adults%20and%20Adolescents%20May%202013%281%29_0.pdf. Accessed August 8, 2017.
- 6. OM-Revised guidelines in initiation of ART. 2017. http://www.indiahivinfo.naco.gov.in/naco/resource/om-revised-guidelines-initation-art. Accessed August 8, 2017.
- 7. Baxter JD, Mayers DL, Wentworth DN, et al. A randomized study of antiretroviral management based on plasma genotypic antiretroviral resistance testing in patients failing therapy. CPCRA 046 study team for the Terry Beirn Community Programs for Clinical Research on AIDS. AIDS Lond Engl. 2000;14(9):F83–F93. [DOI] [PubMed] [Google Scholar]
- 8. Palella FJ, Armon C, Buchacz K, et al. The association of HIV susceptibility testing with survival among HIV-infected patients receiving antiretroviral therapy: a cohort study. Ann Intern Med. 2009;151(2):73–84. [DOI] [PubMed] [Google Scholar]
- 9. Saini S, Bhalla P, Gautam H, Baveja UK, Pasha ST, Dewan R. Resistance-associated mutations in HIV-1 among patients failing first-line antiretroviral therapy. J Int Assoc Physicians AIDS Care (Chic). 2012;11(3):203–209. doi:10.1177/1545109711421217. [DOI] [PubMed] [Google Scholar]
- 10. Sungkanuparph S, Win MM, Kiertiburanakul S, Phonrat B, Maek-a-nantawat W. HIV-1 drug resistance at virological failure versus immunological failure among patients failing first-line antiretroviral therapy in a resource-limited setting. Int J STD AIDS. 2012;23(5):316–318. doi:10.1258/ijsa.2011.011337. [DOI] [PubMed] [Google Scholar]
- 11. Sinha S, Shekhar RC, Ahmad H, et al. Prevalence of HIV drug resistance mutation in the northern Indian population after failure of the first line antiretroviral therapy. Curr HIV Res. 2012;10(6):532–538. [DOI] [PubMed] [Google Scholar]
- 12. tbhiv_factsheet_2015.pdf. 2016. http://www.who.int/tb/challenges/hiv/tbhiv_factsheet_2015.pdf. Accessed July 4, 2018.
- 13. Eshleman SH, Hackett J, Swanson P, et al. Performance of the Celera Diagnostics ViroSeq HIV-1 Genotyping System for sequence-based analysis of diverse human immunodeficiency virus type 1 strains. J Clin Microbiol. 2004;42(6):2711–2717. doi:10.1128/JCM.42.6.2711-2717.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. HIV Drug Resistance Database. https://hivdb.stanford.edu/. Accessed July 5, 2018.
- 15. Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi:10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang KJ, Alter GM, Wooley DP. The reverse transcriptase sequence of human immunodeficiency virus type 1 is under positive evolutionary selection within the central nervous system. J Neurovirol. 2002;8(4):281–294. doi:10.1080/13550280290100716. [DOI] [PubMed] [Google Scholar]
- 17. Sen S, Tripathy SP, Chimanpure VM, Patil AA, Bagul RD, Paranjape RS. Human immunodeficiency virus type 1 drug resistance mutations in peripheral blood mononuclear cell proviral DNA among antiretroviral treatment-naive and treatment-experienced patients from Pune, India. AIDS Res Hum Retroviruses. 2007;23(4):489–497. doi:10.1089/aid.2006.0221. [DOI] [PubMed] [Google Scholar]
- 18. Deshpande A, Recordon-Pinson P, Deshmukh R, et al. Molecular characterization of HIV type 1 isolates from untreated patients of Mumbai (Bombay), India, and detection of rare resistance mutations. AIDS research and human retroviruses. 2004;20(9):1032–1035. [DOI] [PubMed] [Google Scholar]
- 19. Gupta R, Hill A, Sawyer AW, Pillay D. Emergence of drug resistance in HIV type 1-infected patients after receipt of first-line highly active antiretroviral therapy: a systematic review of clinical trials. Clin Infect Dis. 2008;47(5):712–722. doi:10.1086/590943. [DOI] [PubMed] [Google Scholar]
- 20. Lanier ER, Ait-Khaled M, Scott J, et al. Antiviral efficacy of abacavir in antiretroviral therapy-experienced adults harbouring HIV-1 with specific patterns of resistance to nucleoside reverse transcriptase inhibitors. Antivir Ther. 2004;9(1):37–45. [DOI] [PubMed] [Google Scholar]
- 21. Marcelin AG, Flandre P, Pavie J, et al. Clinically relevant genotype interpretation of resistance to didanosine. Antimicrob Agents Chemother. 2005;49(5):1739–1744. doi:10.1128/AAC.49.5.1739-1744.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wainberg MA, Moisi D, Oliveira M, Toni TD, Brenner BG. Transmission dynamics of the M184V drug resistance mutation in primary HIV infection. J Antimicrob Chemother. 2011;66(10):2346–2349. doi:10.1093/jac/dkr291. [DOI] [PubMed] [Google Scholar]
- 23. Wensing AM, Calvez V, Günthard HF, et al. 2017 Update of the drug resistance mutations in HIV-1. Top Antivir Med. 2017;24(4):132–133. [PMC free article] [PubMed] [Google Scholar]