ABSTRACT
Intestinal damage driven by unrestricted immune responses against the intestinal microbiota can lead to the development of inflammatory diseases including inflammatory bowel disease. How such breakdown in tolerance occurs alongside the mechanisms to reinforce homeostasis with the microbiota are a focus of many studies. Our recent work demonstrates coordinated interactions between intact microbiota and CX3CR1 expressing intestinal antigen presenting cells (APCs) that limits T helper 1 cell responses and promotes differentiation of regulatory T cells (Treg) against intestinal antigens including pathogens, soluble proteins and the microbiota itself. We find a microbial attachment to intestinal epithelial cells is necessary to support these anti-inflammatory immune functions. In this addendum, we discuss how our findings enhance understanding of microbiota-directed homeostatic functions of the intestinal immune system and implications of modulating this interaction in ameliorating inflammatory disease.
KEYWORDS: Intestinal immunity, microbiota, Th1 cell responses, Treg responses, IL-10, CX3CR1 mononuclear phagocytes
Introduction
In the human body, dynamic interactions between the host and microbiota underlie many critical homeostatic functions. The human intestine provides a hospitable environment for the microbes while the microbiota provides numerous benefits to the host, including facilitating nutrient breakdown and absorption.1 Interactions between the host and microbiota shapes the development and responsiveness of both the mucosal and systemic immune systems, allowing for the induction of protective immunity against pathogens while also limiting aberrant inflammatory responses against the microbiota and self-antigens. Studies in germ-free mice demonstrate the importance of the microbiota in shaping host immunity as germ-free mice have reduced immune cellularity as well as a lack of organized structures such as B cell germinal centers.1-3 Further, these mice display reductions in effector molecules such as antimicrobial peptides and IgA that lead to reduced barrier function.1 Thus germ-free mice are more susceptible to opportunistic infections, consistent with loss of colonization resistance and reduced mucosal barrier function.4
It has become increasingly clear that alterations in the microbiota, also known as dysbiosis, develop along with inflammatory diseases in both humans and mouse models. Compositional changes in the microbiota are observed in inflammatory bowel disease (IBD),5 including Crohn’s disease and ulcerative colitis,6,7 as well as other diseases such as diabetes,8 asthma9, and rheumatoid arthritis.10 In IBD patients, reductions in potentially anti-inflammatory microbes such as Bacteroidetes, Lachnospiraceae6, and Faecalibacterium prausnitzii7,11 have been observed alongside increases in potentially inflammatory microbes such as Proteobacteria.12-14 Further, mucosa-associated bacteria is increased including Enterobacteriaceae, Pasteurellaceae, Veillonellaceae, and Fusobacteriaceae.5,12–16 This includes finding of increased adherent-invasive E. coli (AIEC). Similar changes have been observed in mouse colitis models.17,18 Given the importance of the microbiota in directing development and functions of the immune system, it is likely that these compositional changes contribute to the pathogenesis of associated disease. Supporting a role for the microbiota in inflammatory disease, germ-free mice are resistant to a number of models of colitis, including in IL-10 deficient mice19 and T cell transfer colitis.20 These results are consistent with studies demonstrating that mice defective in pathways of microbial recognition have reduced sensitivity in colitis models.21,22
While it is clear that the microbiota helps shape local and systemic immunity it is not completely understood how it does this. Further, it is unclear the impact of select members of the microbiota and how they can regulate distinct immunological outcomes.
Microbiota limits pro-inflammatory Th1 responses against intestinal pathogens
In our recent work,23 we set out to understand how microbiota shapes the balance between pro- and anti-inflammatory T cell responses. We utilized the enteric pathogen, Salmonella Typhimurium to determine the role of the microbiota in the regulation of pathogen-specific T cell responses. T helper 1 (Th1) cell responses are required to protect from Salmonella infection.24,25 While Th1 responses are crucial for clearance of intracellular pathogens, they are also associated with inflammatory disorders and autoimmune disease.26 Interferon-γ (IFN-γ), the prototypic Th1 cell effector cytokine, can induce tissue pathology associated with infectious disease.27 IFN-γ directly increases epithelial permeability both in vivo and in vitro28-30 resulting in the potential for increased food antigen, bacteria and bacterial products entering the mucosa alongside elevated local immune responses.31 Therefore, IFN-γ must be tightly regulated to mediate pathogen clearance while limiting unintended tissue damage.
To understand the impact of the microbiota on induction of Th1 cell responses against Salmonella, we utilized antibiotic treatment that could reduce the intestinal microbial load to the limit of detection. We utilized antibiotics instead of performing our analysis in germ-free mice because germ-free mice have severe intestinal and immune defects including altered intestinal villus structure, loss of immune cellularity and organization as well as loss of intestinal antigen presenting cells.3,32 When we compared the Salmonella-specific T cell response in the mesenteric (MLN) from animals with an intact or antibiotic-depleted microbiota, we found an increase in IFN-γ producing, Salmonella-specific T cells in animals with a disrupted microbiota. Importantly, we did not find increased Salmonella in the MLN that could be driving the enhanced response. Further, we did not find global alterations in T cell responses further supporting the antigen specificity of this regulation. These findings strongly suggest that signals from the microbiota regulates host immunity to control intestinal inflammation after pathogen infection.
Anti-inflammatory function of CX3CR1+ APCs depends on the microbiota
We then sought to identify the intestinal antigen presenting cell (APC) that could drive this enhanced response. Within the intestine exist a number of APCs which can detect and respond to luminal antigens. APCs can be subdivided based on expression of CD103 or CX3CR1 (also known as fractalkine receptor), and each group has distinct roles in maintaining intestinal homeostasis.33 CX3CR1+ APCs arise from monocyte precursors34 which differentiate within the intestine into effector cell populations with characteristics of dendritic cells and macrophages.35-37 While the microbiota likely regulates a number of signaling pathways and immune cell populations, we previously found CX3CR1 expressing APCs are an essential target of regulation by the microbiota.37,38 Using mice where we could selectively deplete CX3CR1+ APCs,37,38 we demonstrated the enhanced Th1 cell response after antibiotic treatment was lost after depletion of CX3CR1+ APCs. Further, antigen presentation by CX3CR1+ APCs was required for this enhanced Salmonella-specific T cell response. Interestingly, in the presence of the intact microbiota, depletion of CX3CR1+ APCs resulted in an enhanced Th1 cell response, as did loss of antigen presentation by CX3CR1+ APCs, indicating these cells normally function to limit inflammatory T cell responses. This indicates the requirement of intact microbiota for CX3CR1+ APCs to limit Th1 cell responses.
We next asked if CX3CR1+ APCs could promote anti-inflammatory responses to soluble antigens, an important mechanism to limit immunity against antigens derived from food. Using a model of oral tolerance to ovalbumin, we found that tolerance could not be induced when CX3CR1+ APCs were depleted. This was due to reduced induction of ovalbumin-specific regulatory T cells (Tregs). As with the Salmonella responses, this also depended on antigen presentation by CX3CR1+ APCs. Interestingly, we found oral tolerance to ovalbumin was defective in animals treated with antibiotics. Similarly, it has been reported that germ-free mice have defective oral tolerance.39 In the context of an intact microbiota, ovalbumin-specific T cells differentiated into FoxP3+ single-positive Treg cells. After antibiotic treatment, however, we additionally found differentiation to FoxP3+RORγt+ double-positive Treg cells or RORγt+ Th17 cells. These findings suggest that the intact microbiota promotes Treg cells while suppressing inflammatory Th17 cell responses against food antigen and this regulation depends on CX3CR1+ APCs.
Within the intestine, it is also important to limit T cell reactivity to the microbiota itself. To ask if CX3CR1+ APCs also had an anti-inflammatory role in limiting microbiota-specific T cells, we depleted CX3CR1+ APCs in the T cell transfer model of colitis. We found that animals depleted of CX3CR1+ APCs had increased weight loss and intestinal pathology alongside reduced numbers of Treg cells and increased Th1 and Th17 cells. Overall, our data show that CX3CR1+ APCs, in the context of the normal microbiota function to limit inflammatory T cell responses and promote regulatory T cell responses.
One of the hallmark cytokines secreted by CX3CR1+ APCs is the anti-inflammatory cytokine IL-10.40,41 We found that CX3CR1+ APCs from animals with a disrupted microbiota expressed significantly reduced levels of IL-10. Further, when we infected mice whose CX3CR1+ APCs lack IL-10 production with Salmonella, we found an increased Salmonella specific Th1 response. If these mice were treated with antibiotics first, however, we did not observe an altered Salmonella specific Th1 cell response. These data suggest that the intact microbiota drives IL-10 production by CX3CR1+ APCs which provides a critical signal to suppress pathogen-induced inflammatory T cell responses. In line with significant role for IL-10 induced by the intact microbiota, we also found reduced generation of ovalbumin-specific Treg cells in ovalbumin fed mice who lacked IL-10 production by CX3CR1+ APCs. Overall, our data show that IL-10 production of CX3CR1+ APCs is a key molecular mechanism of microbiota regulation of anti-inflammatory T cell responses.
Microbial attachment can drive host anti-inflammatory response
The intestine and intestinal immune system must detect and properly respond to complex microbiota-derived signals including a large number of pathogen-associated molecular patterns (PAMPs) and metabolites. Identifying specific microbiota signals with the capacity to limit intestinal inflammatory responses is a key for treating inflammatory diseases. PAMPs such as LPS can directly induce IL-10 production by macrophages42 but we found in vivo treatment with LPS was insufficient to induce IL-10 production by CX3CR1+ APCs in antibiotic-treated mice. Instead, we found that colonizing antibiotic-treated mice with human mucosa-derived AIEC12,16 was sufficient to induce IL-10 production and limit Th1 cell responses against Salmonella in antibiotic-treated animals. Animals depleted of CX3CR1+ APCs lost this effect. Importantly, AIEC with reduced epithelial attachment15 was unable to induce IL-10 production or reduce Th1 responses. Overall, our data show that microbial attachment to intestinal epithelium leads to anti-inflammatory function of CX3CR1+ APCs (Figure 1(a)).
Figure 1.
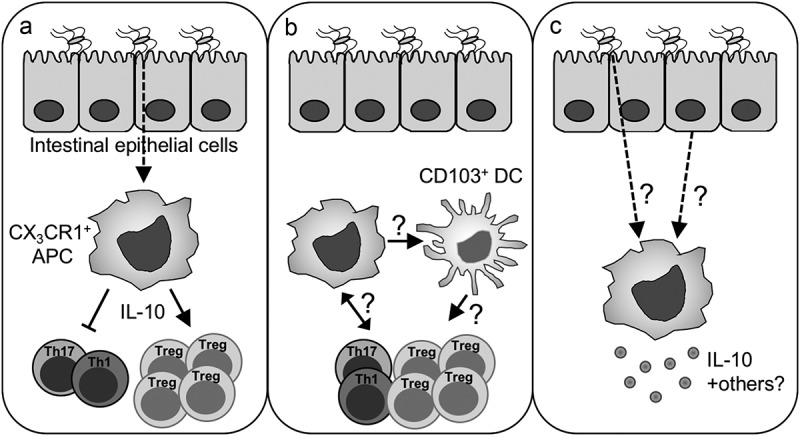
Potential mechanisms of anti-inflammatory intestinal immune responses. (a) Attachment of microbes to the intestinal epithelia leads to IL-10 production by CX3CR1+ APCs. IL-10 suppresses Th1 and Th17 cell responses and induces Treg cells. (b) CX3CR1+ APC-derived signals could modulate the T effector/Treg cell balance directly or indirectly through other intestinal APCs such as CD103+ dendritic cells. Crosstalk between T cells and CX3CR1+ APC could further direct the functions of CX3CR1+ APCs. (c) Modulation of CX3CR1+ APC function by intestinal microbes may depend on direct recognition of microbes or microbial products or microbial signals may be relayed by intestinal epithelial cells.
Concluding remarks
Understanding the signals that limit intestinal inflammation and promote homeostasis are a major focus in the field of mucosal immunology. In our work, we have demonstrated that important anti-inflammatory signals are relayed through direct contact of the microbiota with the intestinal epithelium and highlight potential epithelial contribution to mucosal responses to intestinal microbes. These signals, which could be microbe or epithelial derived, are integrated by CX3CR1+ intestinal APCs. Defining these signals is an area of active investigation. CX3CR1+ APCs, through antigen presentation and cytokine secretion limit the expansion of antigen-specific Th1 cells and promote the differentiation of antigen-specific Treg cells. As the balance between T effector and Treg cells supports intestinal homeostasis, this cellular pathway likely limits inflammatory conditions such as IBD.43
Critical questions remain regarding how microbial signals are relayed between distinct immune populations in the intestine. For example, other intestinal APCs such as CD103+ dendritic cells (DCs) are also thought to promote Treg cell responses in the intestine.44 It will be important to understand if the function of these cells is regulated by CX3CR1+ APCs or directly influenced by intestinal microbes (Figure 1(b)). It remains an open question which antigen presenting cell is driving the enhanced Th1 cell response after depletion of CX3CR1+ APCs. Further, other intestinal cell populations are likely part of a feedback loop within the intestine. Recent work has shown that intestinal LAG3+ Treg cells restrain inflammatory cytokine production by CX3CR1+ APCs during models of colitis.45 It will be important to understand the role of microbial signals in this loop as well as the cellular network of communication between individual intestinal immune cell populations (Figure 1(b)).
Additional critical questions remain regarding the regulation of intestinal immunity by the microbiota. First, is microbial attachment to the epithelium sufficient to induce IL-10 and limit inflammatory responses to intestinal antigens? Further, how is the signal from attached microbes being relayed to the immune system (Figure 1(c)). Attachment could allow for increased local concentrations of microbial products or epithelial signaling could relay information about the attached microbe. We know intestinal epithelial cells secrete a number of mediators such as serum amyloid A (SAA) which activates inflammatory Th17 cell responses.46 It will be important to define the epithelial mediators that activate anti-inflammatory pathways along with the upstream microbial drivers. One of the AIEC, we utilized to induce IL-10 enhances intestinal pathology in IL-10 deficient animals.16 The balanced response to such pathosymbionts will determine if a microbe can be contained within the lumen or induce tissue pathology. Such microbes could be more pathogenic depending on the genetic makeup of the host who might have defects in anti-inflammatory signals. This underscores the importance in host genetics or alterations in microbial responsiveness in linking the microbiota to intestinal disease. Additionally, for pathogenic organisms, the ability to limit inflammatory responses and clearance would enhance colonization. Of direct human relevance, increased colonization with AIEC has been found by a number of groups when comparing IBD patients with controls.12,13,16 As we find epithelial adhesion is a critical for microbial induction of anti-inflammatory host immune cell responses, it will be interesting to understand the selective pressures of microbes to adopt this phenotype.
In our system, we were only looking at outcomes after colonization with a single AIEC. A number of other microbes or microbial products have been identified which can also induce anti-inflammatory responses, including increased Treg cells. This includes microbes such as Helicobacter hepaticus47 and Clostridia species48 or microbial products or metabolites such as outer membrane proteins49 and short chain fatty acids.50-52 Integration of signals from multiple types of microorganisms and/or their metabolites likely occurs and dictates the overall immune tone of the tissue. It remains to be determined if the activated anti-inflammatory pathways overlap or are distinct. Further, the genetic makeup of the host will determine whether colonization with an organism increases or limits intestinal inflammation. Together, it will be important to assess the overall balance between pro- and anti-inflammatory immune responses after colonization with a more complex microbial community.
As IBD results from dysregulated interactions between the immune system and the microbiota1,3 it is important to understand how normal homeostasis is achieved and maintained. By defining a global anti-inflammatory role for the microbiota in limiting inflammatory T cell responses and/or promoting regulatory T cells against pathogen, soluble antigens and the microbiota, our studies demonstrate how intestinal microbes set the stage for intestinal homeostasis. Understanding how these signals are relayed to underlying immune cells such as CX3CR1+ APCs will define numerous therapeutic opportunities to limit pathology in IBD.
Funding Statement
This work was supported by the NIH AI123945 (G.E.D.), NIH AI125264 (G.E.D.) institutional NRSA T32AI053831_Corry (A.A.H), AAI Careers in Immunology Fellowship (M.H.K.).
References
- 1.Sommer F, Bäckhed F.. The gut microbiota–masters of host development and physiology. Nat Rev Immunol. 2013;11:227–238. [DOI] [PubMed] [Google Scholar]
- 2.Thompson GR, Trexler PC. Gastrointestinal structure and function in germ-free or gnotobiotic animals. Gut. 1971;12:230–235. doi: 10.1136/gut.12.3.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamada N, Seo S-U, Chen GY, Núñez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 5.Dalal SR, Chang EB. The microbial basis of inflammatory bowel diseases. J Clin Invest. 2014;124:4190–4196. doi: 10.1172/JCI72330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frank DN, Amand ALS, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peterson DA, Frank DN, Pace NR, Gordon JI. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host and Microbe. 2008;3:417–427. doi: 10.1016/j.chom.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 9.Abrahamsson TR, Jakobsson HE, Andersson AF, Björkstén B, Engstrand L, Jenmalm MC. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin Exp Allergy. 2014;44:842–850. doi: 10.1111/cea.12322. [DOI] [PubMed] [Google Scholar]
- 10.Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, Rostron T, Cerundolo V, Pamer EG, Abramson SB, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux -J-J, Blugeon S, Bridonneau C, Furet J-P, Corthier G, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baumgart M, Dogan B, Rishniw M, Weitzman G, Bosworth B, Yantiss R, Orsi RH, Wiedmann M, McDonough P, Kim SG, et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. ISME J. 2007;1:403–418. doi: 10.1038/ismej.2007.39. [DOI] [PubMed] [Google Scholar]
- 13.Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frank DN, Robertson CE, Hamm CM, Kpadeh Z, Zhang T, Chen H, Zhu W, Sartor RB, Boedeker EC, Harpaz N, et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm Bowel Dis. 2011;17:179–184. doi: 10.1002/ibd.21339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dogan B, Suzuki H, Herlekar D, Sartor RB, Campbell BJ, Roberts CL, Stewart K, Scherl EJ, Araz Y, Bitar PP, et al. Inflammation-associated adherent-invasive Escherichia coli are enriched in pathways for use of propanediol and iron and M-cell translocation. Inflamm Bowel Dis. 2014;20:1919–1932. doi: 10.1097/MIB.0000000000000183. [DOI] [PubMed] [Google Scholar]
- 16.Viladomiu M, Kivolowitz C, Abdulhamid A, Dogan B, Victorio D, Castellanos JG, Woo V, Teng F, Tran NL, Sczesnak A, et al. IgA-coated E. coli enriched in Crohn’s disease spondyloarthritis promote TH17-dependent inflammation. Sci Transl Med. 2017;9:eaaf9655. doi: 10.1126/scitranslmed.aaf9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaser A, Zeissig S, Blumberg RS. Inflammatory Bowel Disease. Annu Rev Immunol. 2010;28:573–621. doi: 10.1146/annurev-immunol-030409-101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elson CO, Cong Y. Host-microbiota interactions in inflammatory bowel disease. Gut Microbes. 2012;3:332–344. doi: 10.4161/gmic.20228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sellon RK, Tonkonogy S, Schultz M, Dieleman LA, Grenther W, Balish E, Rennick DM, Sartor RB. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun. 1998;66:5224–5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kieper WC, Troy A, Burghardt JT, Ramsey C, Lee JY, Jiang H-Q, Dummer W, Shen H, Cebra JJ, Surh CD. Recent immune status determines the source of antigens that drive homeostatic T cell expansion. J Immunol. 2005;174:3158–3163. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi M, Kweon M-N, Kuwata H, Schreiber RD, Kiyono H, Takeda K, Akira S. Toll-like receptor-dependent production of IL-12p40 causes chronic enterocolitis in myeloid cell-specific Stat3-deficient mice. J Clin Invest. 2003;111:1297–1308. doi: 10.1172/JCI17085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rakoff-Nahoum S, Hao L, Medzhitov R. Role of toll-like receptors in spontaneous commensal-dependent colitis.Immunity.2006;25:319–329. doi: 10.1016/j.immuni.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 23.Kim M, Galan C, Hill AA, Wu W-J, Fehlner-Peach H, Song HW, Schady D, Bettini ML, Simpson KW, Longman RS, et al. Critical role for the microbiota in CX3CR1+ intestinal mononuclear phagocyte regulation of intestinal T cell responses. Immunity. 2018;49:151–155. doi: 10.1016/j.immuni.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hess J, Ladel C, Miko D, Kaufmann SH. Salmonella typhimurium aroA- infection in gene-targeted immunodeficient mice: major role of CD4+ TCR-alpha beta cells and IFN-gamma in bacterial clearance independent of intracellular location. J Immunol. 1996;156:3321–3326. [PubMed] [Google Scholar]
- 25.Ravindran R, Foley J, Stoklasek T, Glimcher LH, McSorley SJ. Expression of T-bet by CD4 T cells is essential for resistance to Salmonella infection. J Immunol. 2005;175:4603–4610. doi: 10.4049/jimmunol.175.7.4603. [DOI] [PubMed] [Google Scholar]
- 26.Cope A, Le Friec G, Cardone J, Kemper C. The Th1 life cycle: molecular control of IFN-γ to IL-10 switching. Trends Immunol. 2011;32:278–286. doi: 10.1016/j.it.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 27.Dolowschiak T, Mueller AA, Pisan LJ, Feigelman R, Felmy B, Sellin ME, Namineni S, Nguyen BD, Wotzka SY, Heikenwalder M, et al. IFN-γ hinders recovery from mucosal inflammation during antibiotic therapy for salmonella gut infection. Cell Host and Microbe. 2016;20:238–249. doi: 10.1016/j.chom.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 28.Madara JL, Stafford J. Interferon-gamma directly affects barrier function of cultured intestinal epithelial monolayers. J Clin Invest. 1989;83:724–727. doi: 10.1172/JCI113938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferrier L, Mazelin L, Cenac N, Desreumaux P, Janin A, Emilie D, Colombel J-F, Garcia-Villar R, Fioramonti J, Bueno L. Stress-induced disruption of colonic epithelial barrier: role of interferon-gamma and myosin light chain kinase in mice. Ygast. 2003;125:795–804. [DOI] [PubMed] [Google Scholar]
- 30.Beaurepaire C, Smyth D, McKay DM. Interferon-gamma regulation of intestinal epithelial permeability. J Interferon Cytokine Res. 2009;29:133–144. doi: 10.1089/jir.2008.0057. [DOI] [PubMed] [Google Scholar]
- 31.Sartor RB. Bacteria in Crohn’s disease: mechanisms of inflammation and therapeutic implications. J Clin Gastroenterol. 2007;41(Suppl 1):S37–43. [DOI] [PubMed] [Google Scholar]
- 32.Bain CC, Bravo-Blas A, Scott CL, Gomez Perdiguero E, Geissmann F, Henri S, Malissen B, Osborne LC, Artis D, Mowat AM. Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat Immunol. 2014;15:929–937. doi: 10.1038/ni.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chow A, Brown BD, Merad M. Studying the mononuclear phagocyte system in the molecular age. Nat Rev Immunol. 2011;11:788–798. doi: 10.1038/nri3087. [DOI] [PubMed] [Google Scholar]
- 34.Varol C, Zigmond E, Jung S. Securing the immune tightrope: mononuclear phagocytes in the intestinal lamina propria. Nat Rev Immunol. 2010;10:415–426. doi: 10.1038/nri2778. [DOI] [PubMed] [Google Scholar]
- 35.Zigmond E, Varol C, Farache J, Elmaliah E, Satpathy AT, Friedlander G, Mack M, Shpigel N, Boneca IG, Murphy KM, et al. Ly6Chi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity. 2012;37(6):1076–1090. doi: 10.1016/j.immuni.2012.08.026. [DOI] [PubMed] [Google Scholar]
- 36.Varol C, Vallon-Eberhard A, Elinav E, Aychek T, Shapira Y, Luche H, Fehling H-J, Hardt W-D, Shakhar G, Jung S. Intestinal lamina propria dendritic cell subsets have different origin and functions. Immunity. 2009;31:502–512. doi: 10.1016/j.immuni.2009.06.025. [DOI] [PubMed] [Google Scholar]
- 37.Diehl GE, Longman RS, Zhang J-X, Breart B, Galan C, Cuesta A, Schwab SR, Littman DR. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX(3)CR1(hi) cells. Nature. 2013;494:116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Longman RS, Diehl GE, Victorio DA, Huh JR, Galan C, Miraldi ER, Swaminath A, Bonneau R, Scherl EJ, Littman DR. CX₃CR1+ mononuclear phagocytes support colitis-associated innate lymphoid cell production of IL-22. J Exp Med. 2014;211:1571–1583. doi: 10.1084/jem.20140678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murai M, Turovskaya O, Kim G, Madan R, Karp CL, Cheroutre H, Kronenberg M. Interleukin 10 acts on regulatory T cells to maintain expression of the transcription factor Foxp3 and suppressive function in mice with colitis. Nat Immunol. 2009;10:1178–1184. doi: 10.1038/ni.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Denning TL, Wang Y-C, Patel SR, Williams IR, Pulendran B. Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17-producing T cell responses. Nat Immunol. 2007;8:1086–1094. doi: 10.1038/ni1511. [DOI] [PubMed] [Google Scholar]
- 42.Dobrovolskaia MA, Vogel SN. Toll receptors, CD14, and macrophage activation and deactivation by LPS. Microbes Infect. 2002;4:903–914. doi: 10.1016/S1286-4579(02)01613-1. [DOI] [PubMed] [Google Scholar]
- 43.Shevach EM. Mechanisms of foxp3+ T regulatory cell-mediated suppression. Immunity. 2009;30:636–645. doi: 10.1016/j.immuni.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 44.Sun CM, Hall JA, Blank RB, Bouladoux N, Oukka M, Mora JR, Belkaid Y. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med. 2007;204:1775–1785. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bauché D, Joyce-Shaikh B, Jain R, Grein J, Ku KS, Blumenschein WM, Ganal-Vonarburg SC, Wilson DC, McClanahan TK, Malefyt RDW, et al. LAG3+ regulatory T cells restrain Interleukin-23-Producing CX3CR1+ gut-resident macrophages during group 3 innate lymphoid cell-driven colitis. Immunity. 2018;49:342–345. doi: 10.1016/j.immuni.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 46.Sano T, Huang W, Hall JA, Yang Y, Chen A, Gavzy SJ, Lee J-Y, Ziel JW, Miraldi ER, Domingos AI, et al. An IL-23R/IL-22 circuit regulates epithelial serum amyloid a to promote local effector Th17 responses. Cell. 2015;163:381–393. doi: 10.1016/j.cell.2015.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chai JN, Peng Y, Rengarajan S, Solomon BD, Ai TL, Shen Z, Perry JSA, Knoop KA, Tanoue T, Narushima S, et al. Helicobacter species are potent drivers of colonic T cell responses in homeostasis and inflammation. Sci Immunol. 2017;2(13). pii: eaal5068. doi: 10.1126/sciimmunol.aal5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 49.Shen Y, Torchia MLG, Lawson GW, Karp CL, Ashwell JD, Mazmanian SK. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host and Microbe. 2012;12:509–520. doi: 10.1016/j.chom.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]


