ABSTRACT
T cell redirection mediated by bispecific antibodies (BsAbs) is a promising cancer therapy. Dual antigen binding is necessary for potent T cell redirection and is influenced by the structural characteristics of a BsAb, which are dependent on its IgG subclass. In this study, model BsAbs targeting CD19xCD3 were generated in variants of IgG1, IgG2, and IgG4 carrying Fc mutations that reduce FcγR interaction, and two chimeric IgG subclasses termed IgG1:2 and IgG4:2, in which the IgG1- or IgG4-F(ab)2 are grafted on an IgG2 Fc. Molecules containing an IgG2 or IgG4-F(ab)2 domain were confirmed to be the most structurally compact molecules. All BsAbs were shown to bind both of their target proteins (and corresponding cells) equally well. However, CD19xCD3 IgG2 did not bind both antigens simultaneously as measured by the absence of cellular clustering of T cells with target cells. This translated to a reduced potency of IgG2 BsAbs in T-cell redirection assays. The activity of IgG2 BsAbs was fully restored in the chimeric subclasses IgG4:2 and IgG1:2. This confirmed the major contribution of the F(ab)2 region to the BsAb’s functional activity and demonstrated that function of BsAbs can be modulated by engineering molecules combining different Fc and F(ab)2 domains.
Abbreviations: ADCC: Antibody-dependent cellular cytotoxicity; AlphaScreenTM: Amplified Luminescent Proximity Homogeneous Assay Screening; ANOVA: Analysis of variance; BiTE: bispecific T-cell engager; BSA: bovine serum albumin; BsAb: bispecific antibody; cFAE: controlled Fab-arm exchange; CDC: complement-dependent cellular cytotoxicity; CIEX: cation-exchange; CIR: chimeric immune receptor; DPBS: Dulbecco’s phosphate-buffered saline; EC50 value: effective concentration to reach half-maximum effect; EGFR: epidermal growth factor receptor; EI: expansion index (RAt=x/RAt=0); FACS: fluorescence-activated cell sorting; FVD: fixable viability dye; HI-HPLC: hydrophobic interaction HPLC; HI-FBS: heat-inactivated fetal bovine serum; HPLC: high-pressure liquid chromatography; IC50 value: effective concentration to reach half-maximum inhibition; IQ: Inhibition Quotient; IS: immunological synapse; MES: 2-(N-morpholino)ethanesulfonic acid; R-PE: recombinant phycoerythrin; RA: red area in μm2/well; RD: receptor density; RFP: red fluorescent protein; Rg: radius of gyration; RSV: respiratory syncytial virus; SAXS: small-angle x-ray scattering; scFv: single-chain variable fragment; SD: standard deviation; SPR: surface plasmon resonance; WT: wild-type
KEYWORDS: T cell redirection, bispecific antibody structure, antibody engineering, CD19xCD3
Introduction
T cell redirection using bispecific molecules to specifically eliminate tumor cells is a highly promising therapy for cancer malignancies.1,2 The first generation of bispecific molecules includes the CD3-targeting bispecific antibody (BsAb) catumaxomab (Removab®, Epcam x CD3, Trion Research GmbH, approved in the European Union in 2009),3 and the bispecific T-cell engager (BiTE) blinatumomab (Blincyto®, CD19xCD3, Micromet/Amgen, approved by the US Food and Drug Administration in 2015).4 The 50-kDa BiTE is designed to have two single-chain variable fragment (scFv) domains in one polypeptide chain.5 In addition, multiple formats ranging from Nanobodies (25 kDa) to IgG-fusions (≥150 kDa) are being investigated.6–8 While smaller molecules have the advantage of bridging shorter intercellular distances and can demonstrate better tumor penetration, IgG-based formats permit Fc engineering for controlled serum half-life via FcRn-mediated recycling and modulation of the Fc activity.2
T-cell killing activity requires the formation of immunological synapses (ISs), which are highly ordered structures connecting the T cell receptor to the peptide-MHC complex presented on the target cell. Formation of an active IS results in the release of cytotoxic granules and cytokines.9,10 Because of the spatial proximity of the T cell and target cell during the formation of ISs, large molecules such as CD45 are excluded from the intermembrane space.11,12 This allows an intercellular distance of ~ 15 nm,13 which is approximately the same distance as the Fab arm span of an IgG1.14 T cell redirection using BsAbs requires mimicry of the IS, which is achieved by bridging the T-cell receptor complex via CD3 binding to the target cell via binding to a target cell surface antigen. Multiple factors can affect T-cell redirection activity, such as differential affinities of the targeting arms, as shown by the example of a CLL1xCD3 knob-in-hole IgG1 BsAb.15 The distance of epitope engagement from the membrane is also important, as shown for the MCSPxCD3 BiTE and FcRH5xCD3 BsAb.16,17 The insertion of an Fc-based linker into the extracellular portion of a chimeric immune receptor (CIR) on T cells targeting a membrane-proximal domain of 5T4 also enhances T cell activation and target cell killing.18 In contrast, T cells targeting CD19 were activated to a higher level when expressing the CIR without the Fc-based linker but did not kill target cells more efficiently.18 Thus, structural characteristics, such as accessibility and distance of the epitope from the membrane and BsAb flexibility, can contribute to efficient T cell redirection.
The IgG subclass of a BsAb can also influence its functional activity due to the changes in the variable region presentation, hinge sequences, and disulfide-bond structures.19 The hinge region of IgG connects the Fc domain with the Fab domains, and varies in length, sequence, and disulfide bond architecture per IgG subclass.20 IgG1 has the longer hinge region with two disulfide bonds. IgG2 has the shortest and most rigid hinge region because of the four disulfide bonds that exist in three major isoforms.20 The hinge of IgG4 has the same amino acid length as IgG2 (12 amino acids), but contains more proline than cysteine residues and has only one disulfide-bond structure like IgG1 (Figure 1(a)). These structural variations influence the flexibility of the IgG hinge from IgG1 > IgG4 > IgG2 (IgG1 being the most flexible), as determined by electron microscopy.19 The conformations of the F(ab)2 domains of the different IgG subclasses were also analyzed by small-angle x-ray scattering (SAXS).21 The IgG1 molecules were present as an equal mixture of closed Y, intermediate Y/T, and widely open T conformations. The IgG4 molecules had primarily Y/T conformations, and the IgG2 molecules existed predominantly in Y conformations (Figure 1(a)). Although IgG1, IgG2, and IgG4 subclasses have been used as frameworks for therapeutic biologics, the influence of the IgG subclass on T-cell redirection activity has not been investigated in detail.22–24
Figure 1.
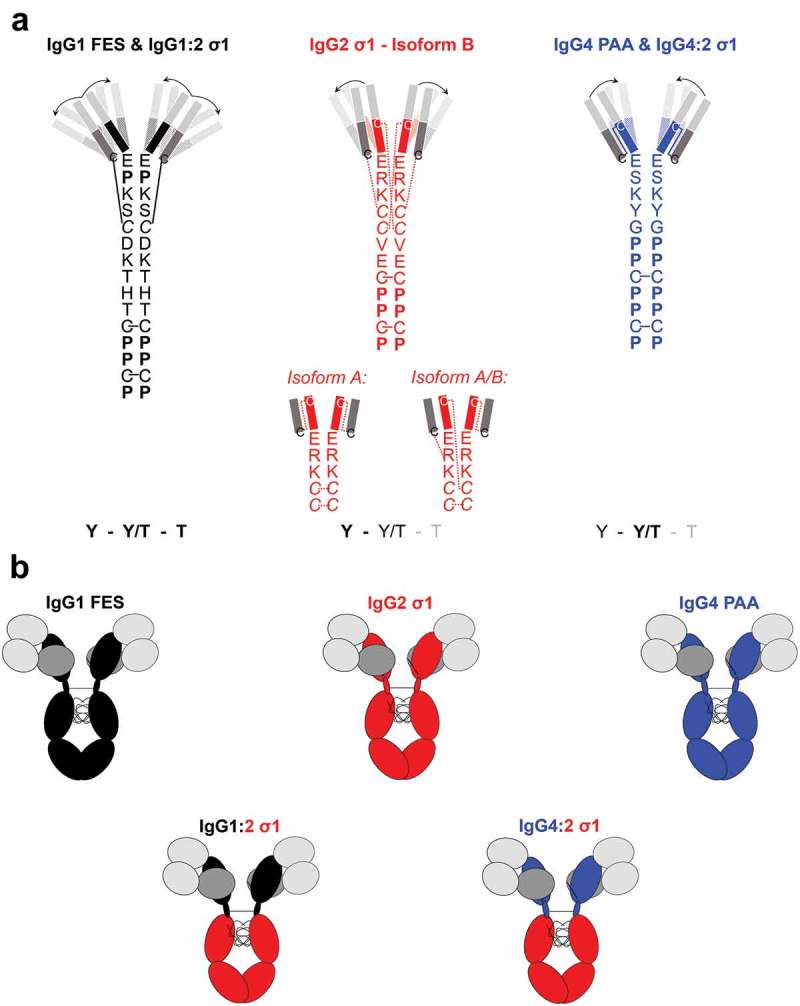
Structural characteristics of investigated IgG subclasses.
A: Hinge-region amino acid sequence, disulfide-bond structure20 and predominant F(ab)2-conformations of each natural IgG subclasses as determined by Tian et al. using SAXS-based structural modeling.21 Bold letters indicate the predominant conformation of each IgG subclass. The predominant F(ab)2 conformation is shown (opaque) with arrows indicating the flexibility towards the less-predominant F(ab)2 conformations (transparent). IgG1 consists of an equilibrium of Y, Y/T, and T conformations, IgG2 predominantly forms closed Y-conformation (53%) or intermediate Y/T conformation (40%) while IgG4 shows a predominant structural cluster with intermediate Y/T conformation (53%) and less frequent Y/T conformational clusters (36%). The disulfide bond structure of IgG2 hinge exists in three major isoforms which also influence the disulfide bond structure in the Fab domains.20 B: Overview of the three human IgG subclasses (carrying Fc-mutations, Supplementary Table 1) IgG1 FES, IgG2 σ1, IgG4 PAA, and the two engineered chimeras IgG1:2 σ1 and IgG4:2 σ1. The chimeras were generated by fusing the F(ab)2 domains of IgG1 FES or IgG4 PAA, respectively, onto an IgG2 σ1 Fc.Light grey boxes: VH/VL regions; the same sequences were used for each molecule. Dark grey boxes: CL regions; the same sequences were used for each molecule.Black boxes, black hinge regions: CH1, hinge, CH2 & CH3 sequences of the IgG1 subclass. Red boxes, red hinge region: CH1, hinge, CH2 & CH3 sequences of IgG2 subclass. Blue boxed, blue hinge regions: CH1, hinge CH2 & CH3 sequences of the IgG4 subclass. Note that the S228P mutation of IgG4 PAA in the hinge region prevents spontaneous Fab-arm exchange.25
To assess the impact of the IgG subclass on efficient T cell redirection, BsAbs targeting CD19 and CD3 were generated by the controlled Fab-arm exchange (cFAE) in three human IgG subclasses IgG1, IgG2 and IgG4.25–27 The parental antibody that bound the membrane-distal domain of CD19 was based on a humanized variant of blinatumomab.28 The parental antibody that bound to the CD3ϵ subunit of the T cell receptor was based on sequences of the variable regions of OKT3.29 To minimize the effects of Fcγ receptor (FcγR) engagement, constructs contained Fc mutations that reduced FcγR interaction: IgG1 FES, IgG2 σ1 and IgG4 PAA (Supplementary Table 1; Figure 1(b), upper panel).30–32 In addition, two chimeric IgG subclasses were designed: F(ab)2 including the hinge regions of IgG1 or IgG4 were fused to a human IgG2 σ1-Fc (named IgG1:2 σ1 and IgG4:2 σ1, respectively, Figure 1(b), lower panel). Protein and cell binding of all generated IgG subclasses were confirmed by surface plasmon resonance (SPR) and flow cytometry. In vitro activity was assessed by the ability of BsAbs to bridge T cells to their respective target cells and to inhibit target cell proliferation. This study provides a comprehensive analysis of how the binding of model CD19xCD3 BsAbs harboring IgG subclasses with differing molecular flexibilities can affect in vitro T cell activity.
Results
Expression, cFAE, and structural characterization of model antibodies in different IgG subclasses
Parental antibodies targeting CD19, CD3, and respiratory syncytial virus (RSV) as specificity control were transiently expressed in HEK293 ExpiTM cells with yields of 50 mg/L (CD3) to 300 mg/L (CD19, RSV). After purification by MabSelectTM SuReTM chromatography (routinely > 90% monodisperse and monomeric as defined by the elution of a single peak at the expected retention time for a monomeric antibody, and 100% purity (Supplementary Table 2), BsAbs were generated by cFAE and were > 95% monomeric by size-exclusion high-pressure liquid chromatography (SE-HPLC) and > 93% bispecific via hydrophobic interaction (HI)- or cation-exchange (CIEX)-HPLC except for CD19xRSV IgG2 σ1 (85%; 12% residual CD19 parental Ab). Endotoxin levels were < 0.4 EU/mg for each BsAb (Table 1).
Table 1.
Quality control of bispecific antibodies (BsAbs).
| BsAb | Recovery after cFAE (%) | Endotoxin (EU/mg) | Monodispersity (%) | Bispecificity (%)* |
|---|---|---|---|---|
| CD19xCD3 IgG1 FES | 92.36 | 0.29 | 95.69 | 94.34 |
| CD19xRSV IgG1 FES | 94.55 | 0.32 | 97.62 | 100.00 |
| RSVxCD3 IgG1 FES | 94.45 | 0.27 | 94.14 | 93.64 |
| CD19xCD3 IgG2 σ1 | 100.67 | 0.27 | 98.87 | 94.33 |
| CD19xRSV IgG2 σ1 | 100.87 | 0.23 | 100.00 | 85.07* |
| RSVxCD3 IgG2 σ1 | 94.95 | 0.26 | 97.93 | 93.43 |
| CD19xCD3 IgG4 PAA | 92.06 | 0.1 | 98.80 | 95.06 |
| CD19xRSV IgG4 PAA | 90.48 | 0.09 | 95.42 | 93.07* |
| RSVxCD3 IgG4 PAA | 91.76 | 0.09 | 96.24 | 96.57 |
| CD19xCD3 IgG1:2 σ1 | 97.94 | 0.04 | 97.94 | 96.66 |
| CD19xRSV IgG1:2 σ1 | 97.19 | 0.08 | 97.19 | 97.60* |
| RSVxCD3 IgG1:2 σ1 | 96.04 | 0.07 | 96.04 | 97.60 |
| CD19xCD3 IgG4:2 σ1 | 97.25 | 0.03 | 97.25 | 100.00 |
| CD19xRSV IgG4:2 σ1 | 98.29 | 0.03 | 98.29 | 95.14* |
| RSVxCD3 IgG4:2 σ1 | 96.71 | 0.03 | 96.71 | 96.04* |
*Molecules whose percentage of bispecificity was characterized by HI-HPLC are indicated with a star. Unmarked molecules were analyzed by CIEX-HPLC.
The radius of gyration (Rg) is an indicator of protein compactness and a measure for protein flexibility.21,33 We confirmed the Rg of each IgG subclass by analyzing CD19 parental antibodies as representative molecules by SAXS (Table 2). IgG2 and IgG4 had a tighter Rg value than IgG1. Furthermore, we demonstrated that the chimeric IgG subclasses showed similar Rg values as the natural IgG subclass values from which its F(ab)2 domains were derived.
Table 2.
The radius of gyration Rg (from Guinier)58 by IgG subclass compared to literature data. Rg values for investigated IgG subclasses were determined by SAXS and are presented in comparison to literature data.21
| IgG1 (FES) | IgG1:2 σ1 | IgG2 (σ1) | IgG4 (PAA) | IgG4:2 σ1 | |
|---|---|---|---|---|---|
| Experimental Rg (Å) | 47.74 ± 2.87 | 49.43 ± 0.97 | 46.85 ± 2.18 | 46.85 ± 1.08 | 46.86 ± 0.93 |
| Literature Rg (Å)21 | 49.4 ± 0.38 | n. d. | 47.6 ± 0.48 | 47.6 ± 0.49 | n. d. |
n. d. = not determined.
Binding of model BsABs to Fc receptors
The binding of model BsAbs of the different IgG subclasses containing mutations to reduce FcγR interaction to FcRs was assessed by Amplified Luminescent Proximity Homogeneous Assay Screening (AlphaScreenTM) competition assay. None of the applied Fc mutations affected binding of CD19xCD3 BsAbs and their RSV-controls to FcRn at pH 6.0 compared to wild-type (WT) IgG1 (Figure 2(a,b)). Likewise, the Fab domains had no effect on Fc-interaction properties: Competition for binding to the high-affinity FcγRI was ~ 100X weaker for IgG1 FES and IgG4 PAA than for WT IgG1 independent from the individual BsAb’s Fab domains. All BsAbs harboring an IgG2 σ1 Fc did not compete for binding to FcγRI at the highest applied concentration of 2,666 nM (Figure 2(c,d)). This finding was confirmed when testing competition for binding of concentrated CD19 parental antibodies in the different IgG subclasses to the low-affinity receptors FcγRIIa, IIb/c, and IIIa: IgG2 σ1 competed 10X (FcγRIIa, IIb/c) to 100X (FcγRIII) weaker than IgG1 FES and IgG4 PAA (Supplementary Figure 1).
Figure 2.
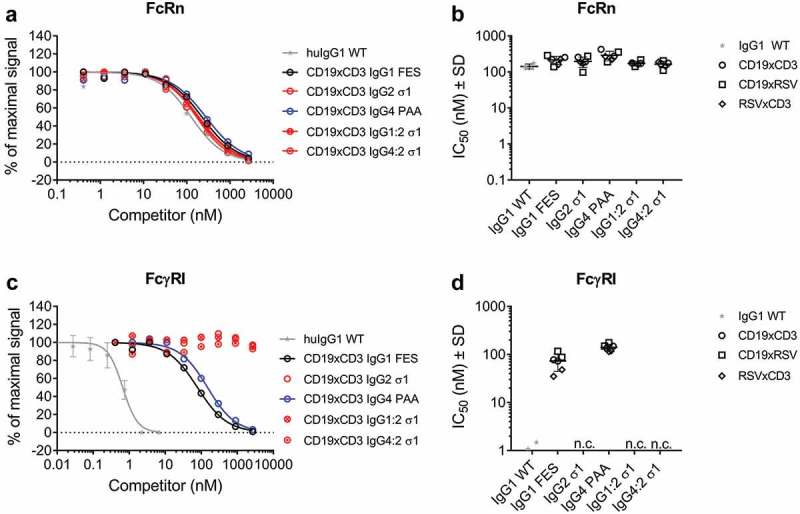
Binding of model BsAbs of different IgG subclasses to Fc receptors.
The binding to FcRn and FcγRI of was determined by AlphaScreenTM competition assay in comparison to a WT huIgG1 (grey stars). In such assays, a higher IC50 value translates to a weaker binding to the tested FcR. A: Dose-response of FcRn binding. Curves show the competition of CD19xCD3 BsAbs of one representative experiment (single measurement per concentration). The error bars represent the SD of the WT-control which was measured on multiple plates.B: IC50 values of FcRn binding. CD19xCD3 (open circles), CD19xRSV (open squares) and RSVxCD3 (open diamonds) BsAbs were grouped by Fc and individual values are plotted from two independent experiments. Lines represent the mean IC50 ± standard deviation (SD) through all tested molecules per Fc.C: Dose-response of FcγRI binding. Curves show the competition of CD19xCD3 BsAbs of one representative experiment (single measurement per concentration). The error bars represent the SD of the WT-control which was measured on multiple plates.D: IC50 values of FcγRI binding. CD19xCD3 (open circles), CD19xRSV (open squares) and RSVxCD3 (open diamonds) BsAbs were grouped by Fc and individual values are plotted from two independent experiments. Lines represent the mean IC50 ± standard deviation (SD) through all tested molecules per Fc.n. c.: no competition for binding to FcγRI was observed at the highest applied concentration (2666 nM) for BsAbs containing an IgG2 σ1 Fc.
Binding of model antibodies and BsABs to target antigen and cells
T-cell redirection activity can be influenced by changes in the BsAb affinity to the target antigens. Single-arm affinities to CD19 and CD3 were determined by SPR spectroscopy for parental antibodies and BsAbs (Table 3, left panel). All molecules had similar affinity values for binding to CD19 (2.3–5.6 nM) or CD3δϵ-Fc (14.4–42.2 nM) regardless of their IgG subclass.
Table 3.
Binding of model antibodies and BsAbs to antigen and HBL-1 and Daudi cells.
| SPR: KD in nM (± range) |
Flow Cytometry: EC50 in nM (± range) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Molecule | CD19 | CD3 | HBL-1 | Daudi | HBL-1 RFP | Daudi RFP | human T cells | |
| CD19 parental Ab | IgG1 FES | 2.52 (0.15) | n. b. | n. d. | n. d. | 0.22 (0.05) | 0.30 (0.00) | n. b. |
| IgG2 σ1 | 5.21 (0.08) | n. b. | n. d. | n. d. | 0.38 (0.07) | 0.68 (0.07) | n. b. | |
| IgG4 PAA | 3.07 (0.14) | n. b. | n. d. | n. d. | 0.41 (0.10) | 0.47 (0.01) | n. b. | |
| IgG1:2 σ1 | 4.91 (0.40) | n. b. | n. d. | n. d. | 0.21 (0.03) | 0.41 (0.00) | n. b. | |
| IgG4:2 σ1 | 4.59 (0.01) | n. b. | n. d. | n. d. | 0.31 (0.04) | 0.41 (0.06) | n. b. | |
| CD3 parental Ab | IgG1 FES | n. b. | 28.39 (4.23) | n. d. | n. d. | n. b. | n. b. | 0.34 (0.04) |
| IgG2 σ1 | n. b. | 28.4 (1.27) | n. d. | n. d. | n. b. | n. b. | 2.00 (0.49) | |
| IgG4 PAA | n. b. | 42.24 (5.49) | n. d. | n. d. | n. b. | n. b. | 0.37 (0.05) | |
| IgG1:2 σ1 | n. b. | 36.72 (1.10) | n. d. | n. d. | n. b. | n. b. | 0.18 (0.11) | |
| IgG4:2 σ1 | n. b. | 37.78 (1.84) | n. d. | n. d. | n. b. | n. b. | 0.24 (0.14) | |
| IgG1 FES BsAb | CD19xCD3 | 3.02 (0.22) | 21.31 (8.61) | 0.73 (0.13) | 1.20 (0.10) | 0.70 (0.07) | 0.82 (0.28) | 7.79 (1.05) |
| CD19xRSV | 2.60 (0.17) | n. b. | 0.64 (0.05) | 0.97 (0.19) | 0.65 (0.17) | 0.91 (0.14) | n. b. | |
| RSVxCD3 | n. b. | 36.66 (0.17) | n. b. | n. b. | n. b. | n. b. | 4.47 (0.79) | |
| IgG2 σ1 BsAb | CD19xCD3 | 4.42 (0.05) | 14.40 (1.14) | 0.55 (0.01) | 0.85 (0.07) | 0.68 (0.06) | 0.90 (0.12) | 54.48 (10.86) |
| CD19xRSV | 3.58 (0.14) | n. b. | 0.67 (0.00) | 0.92 (0.02) | 0.81 (0.13) | 1.00 (0.12) | n. b. | |
| RSVxCD3 | n. b. | 18.68 (4.81) | n. b. | n. b. | n. b. | n. b. | 39.39 (2.42) | |
| IgG4 PAA BsAb | CD19xCD3 | 2.29 (0.21) | 22.80 (0.98) | 0.39 (0.01) | 0.76 (0.02) | 0.64 (0.07) | 1.01 (0.10) | 25.31 (3.05) |
| CD19xRSV | 3.34 (0.07) | n. b. | 0.57 (0.06) | 0.77 (0.04) | 0.60 (0.05) | 0.67 (0.20) | n. b. | |
| RSVxCD3 | n. b. | 22.73 (0.61) | n. b. | n. b. | n. b. | n. b. | 12.23 (0.19) | |
| IgG1:2 σ1 BsAb | CD19xCD3 | 5.55 (0.09) | 34.49 (7.10) | 0.54 (0.05) | 0.79 (0.01) | 0.60 (0.03) | 0.85 (0.05) | 29.36 (4.2) |
| CD19xRSV | 4.76 (0.16) | n. b. | 0.51 (0.09) | 0.87 (0.11) | 0.62 (0.03) | 0.89 (0.05) | n. b. | |
| RSVxCD3 | n. b. | 30.83 (0.31) | n. b. | n. b. | n. b. | n. b. | 12.05 (2.16) | |
| IgG4:2 σ1 BsAb | CD19xCD3 | 4.52 (0.08) | 26.26 (4.91) | 0.60 (0.11) | 0.95 (0.16) | 0.62 (0.06) | 1.05 (0.05) | 34.65 (0.34) |
| CD19xRSV | 4.35 (0.01) | n. b. | 0.51 (0.04) | 0.77 (0.12) | 0.62 (0.01) | 0.82 (0.16) | n. b. | |
| RSVxCD3 | n. b. | 21.79 (4.51) | n. b. | n. b. | n. b. | n. b. | 10.54 (0.98) | |
SPR spectroscopy and flow cytometry were performed as outlined in the methods. The single-arm affinity values of parental antibodies and BsAbs for binding to CD19 and CD3 are presented as KD (left panel), binding of BsAbs to CD3+ T cells and CD19+ cell lines as EC50 values (right panel). No binding was detected to the CD19- and CD3-negative cell lines HLY-1 (RFP) (not shown).
All binding data are shown as mean (± range) in nM of two independent experiments per data point. n. b.: no binding. n. d.: not determined.
Association and dissociation constant values are reported in Supplementary Table 4.
Next, the binding of model BsAbs to their targets was assessed by flow cytometry in a cellular context (Table 3, right panels). Measurements were conducted on CD19-expressing B-cell lymphoma lines HBL-1 (low CD19 receptor density (RD)) and Daudi (high CD19 RD). WT and red fluorescent protein (RFP)-transduced cell lines (used in kinetic CD3 killing assays) were used to ensure that the RFP-transduction did not alter CD19-recognition. The parental CD3 Abs did not bind to CD19-expressing cell lines, while the parental CD19 Abs did not bind to the human pan T cells, demonstrating specificity of the antibody binding. BsAbs bound both CD19-expressing cell lines with a similar effective concentration to reach half-maximum effect (EC50 value) across IgG subclasses, independent of CD19 RD (Table 4) and RFP-transduction (Table 3), though 2-3X weaker than their parental antibodies. In contrast, the bivalent CD3 IgG2 σ1 antibody bound T cells 6-10X weaker than CD3 antibodies of the other IgG subclasses. Furthermore, monovalent BsAbs bound CD3 at least 10X weaker than their parental bivalent antibodies. RSVxCD3 BsAbs bound to T cells with a 2-3X slightly tighter EC50 value than their CD19xCD3 counterparts across the other IgG subclasses despite their similar single-arm affinity. CD19xCD3 IgG1 FES bound tightest among the BsAb subclasses, with an EC50 value of 8 nM. CD19xCD3 IgG2 σ1 bound 7X weaker than CD19xCD3 IgG1 FES (EC50 value of 55 nM). The other three subclasses, IgG4 PAA, IgG1:2 σ1, and IgG4:2 σ1, bound intermediately, with EC50 values of ~ 30 nM (3X weaker than CD19xCD3 IgG1 FES).
Table 4.
CD19 receptor density (RD) of HBL-1 and Daudi cells (WT and RFP transduced).
| HBL-1 | HBL-1 RFP | Daudi | Daudi RFP | |
|---|---|---|---|---|
| CD19 RD (SD) in molecules/cell | 49,054 (1,300) | 40,596 (8,250) | 258,063 (36,374) | 322,212 (15,325) |
CD19 RD was determined by quantitative flow cytometry. The mean RD ± standard deviation (SD) were derived from three independent experiments as outlined in the methods, each in technical duplicates.
Cellular clustering as a function of the IgG subclass
Cellular association between effector and target cells is required for potent effector T cell activity. To distinguish the two cell types, target cells were labeled with CellTrace Violet, and the T cells were labeled with CellTrace FarRed. To measure BsAb-mediated cell clustering, WT target cells were pre-incubated with 20 μg/mL BsAbs and incubated with effector T cells. This concentration was determined to be at saturating levels for both CD19 and CD3 monovalent binding. At this concentration, the relative binding level to CD19 on both target cell lines was not affected by the IgG subclass. CD19xCD3 IgG2 σ1, however, bound 25% weaker than all other IgG subclasses to CD3 on T cells (Supplementary Table 4). Cellular clustering was demonstrated by the appearance of violet-red double-positive events by flow cytometry. These double-positive events appeared at an increased size and granularity, thus confirming their cellular cluster nature.
No clustering was observed for the CD19− cell line HLY-1 (Figure 3(a)). Low levels of clustering were observed for all IgG subclasses except for IgG2 σ1 on HBL-1 cells (max. 2X of untreated, Figure 3(b)), while these BsAbs mediated a ~ 30X increased clustering over untreated controls on Daudi cells (Figure 3(c)). The low CD19 RD present on HBL-1 cells represented the detection limit for cellular clusters in this assay. Despite this, the observed clustering reached levels of statistical significance on both target cell lines. Dual antigen binding was required to cluster both cell types, as demonstrated by the lack of activity of the CD19xRSV control BsAbs on both cell lines. Strikingly, CD19xCD3 IgG2 σ1 BsAb was not able to facilitate cellular clustering despite its similar single-arm affinity towards CD19 and CD3 and its similar binding towards CD19+ target cells.
Figure 3.
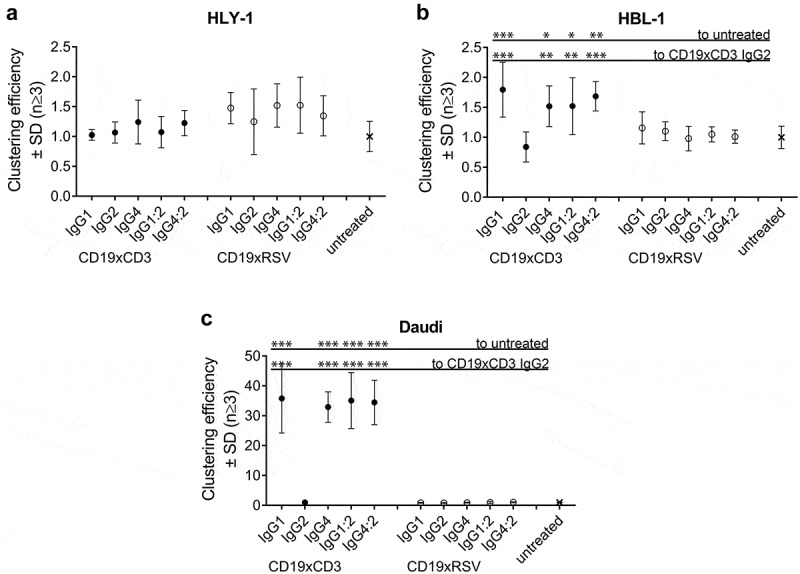
Efficiency of cellular clustering efficiency by IgG subclass.
The efficiency of CD19xCD3 BsAb IgG subclasses to facilitate clustering of target cells and T cells was assessed by flow cytometry using 20 μg/mL BsAb and a 1:1 effector cell number: target cell number ratio. Cell clustering efficiency was the percentage of double-positive events (red-labeled effector T cells and violet-labeled target cells) out of a live-cell gate. Data are presented as mean ± SD of 3–5 independent experiments, normalized to the clustering percentage in the absence of BsAb per cell line and replicate. A: CD19-negative HLY-1 cells.B: CD19-positive HBL-1 target cells (low CD19 RD).C: CD19-positive Daudi target cells (high CD19 RD).Applied Fc mutations (Supplementary Table 1) have not been included in the x-axis label for better legibility. Statistical significance was analyzed by ANOVA, and level of significance of a BsAb in comparison to untreated or IgG2 σ1 is shown as: *p-value ≤ 0.05; **p-value ≤ 0.001; ***p-value <0.0001. Not shown comparisons did not reach significance.
T-cell redirection activity of CD19xCD3 BsABs of different IgG subclasses
The T-cell redirection activity of CD19xCD3 BsAbs of different IgG subclasses was assessed in kinetic T-cell redirection assays. Upon bridging T cells with their target cells, T cells are activated and secrete cytotoxic granules.9,10 The resulting kinetics of target cell killing were measured as growth inhibition resulting from reduced proliferation over the course of 4–5 days. The activity of the CD19xCD3 BsAbs was evaluated by two parameters: efficacy (maximum inhibitory effect), and potency (effective concentration to reach half-maximum inhibition (IC50 value) in inhibition of proliferation). No BsAb inhibited the growth of the CD19-negative cell line HLY-1 RFP at relevant concentrations (Supplementary Figure 2).
The activity of CD19xCD3 BsAbs was dependent on the IgG subclass and the target cell line. CD19xCD3 IgG4 PAA was the most efficacious IgG subclass (= 100%) on HBL-1 RFP cells. IgG1 FES reached about 75% efficacy while displaying the same potency (Figure 4(a,c,e)). IgG2 σ1 showed an intermediate efficacy of 90%, but a clear, statistically significant 10X weaker potency than the other IgG subclasses. Strikingly, the chimeric CD19xCD3 IgG4:2 σ1 showed the same activity (efficacy and potency) as IgG4 PAA. Similarly, IgG1:2 σ1 behaved like IgG1 FES (Figure 4(a,c,e)). RSV-controls only showed minor inhibitory effects at saturating concentrations.
Figure 4.
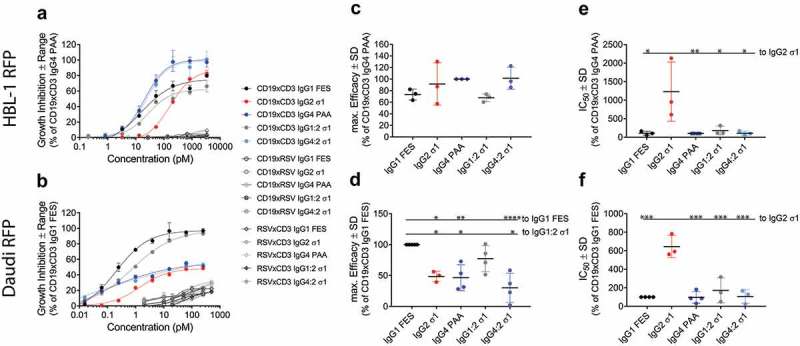
Inhibition of proliferation by IgG subclass.
The inhibition of proliferation as function of BsAb concentration was assessed in an IncuCyte ZOOM® system using a 5:1 effector: target ratio for 4–5 days. Data are normalized to the most potent IgG subclass per cell line, i.e. CD19xCD3 IgG4 PAA on HBL-1 RFP cells, or CD19xCD3 IgG1 FES on Daudi RFP cells.A,B: Representative results for the growth inhibition as function of BsAb concentrationA: on HBL-1 RFP cells.B: on Daudi RFP cells.Data are plotted as mean ± range of technical duplicates.C,D: Maximum efficacy (mean ± SD of ≥3 independent experiments) of CD19xCD3 BsAbsC: on HBL-1 RFP cells.D: on Daudi RFP cells.E,F: Potency (IC50 values, mean ± SD of ≥3 independent experiments) of CD19xCD3 BsAbsE: on HBL-1 RFP cells.F: on Daudi RFP cells.IgG1 FES: black; IgG1:2 σ1: grey; IgG2 σ1: red; IgG4 PAA: blue and IgG4:2 σ1: light blue. Statistical significance was analyzed by ANOVA, and level of significance of a BsAb in comparison to IgG1 FES or IgG2 σ1 is shown as: *: p-value ≤ 0.05; **: p-value ≤ 0.001; ***: p-value <0.0001. Not shown comparisons did not reach significance.
In contrast to HBL-1 RFP cells, the growth of Daudi RFP cells was inhibited best by CD19xCD3 IgG1 FES. CD19xCD3 IgG4 PAA reached only 50% maximum efficacy (Figure 4(b,d)). Here, CD19xCD3 IgG2 σ1 reached a similar maximum efficacy to IgG4 PAA but was again clearly less potent (Figure 4(d,f)). Despite the significant difference in maximum efficacy between the IgG4 PAA to IgG1 FES subclasses, the chimeric IgG subclasses IgG1:2 σ1 and IgG4:2 σ1 again fully restored their F(ab)2-parental BsAbs’ activity.
We preferred this kinetic assay because it provides a better all-encompassing analysis of a BsAb’s ability to promote tumor growth inhibition than a setup that seeks to analyze one single time point. However, we confirmed our results in a flow-cytometric end-point assay analyzing target cell viability (Supplementary Figure 3(a,b)). CD19xCD3 BsAbs harboring an IgG1 or IgG4 F(ab)2 domain mediated the best target cell killing using HBL-1 cells, while CD19xCD3 IgG2 σ1 was ~ 10X less potent. Daudi cells were killed best by CD19xCD3 IgG1 and its chimera IgG1:2 σ1, while there was no difference between IgG2 σ1, IgG4 PAA and IgG4:2 σ1 at the tested concentration range in this assay. However, CD19xCD3 IgG2 σ1-activated T cells were the least potent on both target cell lines (Supplementary Figure 3(b,d)).
To analyze whether these results were unique to the chosen targeting pair, we generated CD20xCD3 molecules based on the well-characterized public-domain CD20-targeting antibody rituximab.34 BsAbs were generated and analyzed similarly to CD19xCD3 BsAbs, keeping OKT3 to target CD3. The CD20xCD3 BsAb in IgG2 σ1 subclass was again the least active molecule on both HBL-1 RFP and Daudi RFP cells (Supplementary Figure 4), underlining the general nature of the presented findings.
Discussion
The influence of the IgG subclass on T cell redirection by BsAbs has not been studied in detail before. While T-cell redirecting BsAbs have been generated in different IgG subclasses, no side-by-side comparisons of killing activity have been reported.35,36 In this study, we evaluated the implications of different structural characteristics, which are inherent to different IgG subclasses, on T-cell redirection activity. To this end, BsAbs targeting CD19xCD3 were successfully generated by cFAE with high quality in the three human IgG subclasses with Fc mutations reducing FcγR interaction: IgG1 FES, IgG2 σ1 and IgG4 PAA, and two chimeric subclasses termed IgG1:2 σ1 and IgG4:2 σ1. There was no evidence that these mutations had effects on the generation of the parental antibodies or the cFAE process to generate the respective BsAbs.
We confirmed the Rg that had previously been determined for the three natural human IgG subclasses as initial measure for protein compactness.21,33 We further demonstrated that the chimeric subclasses IgG1:2 σ1 and IgG4:2 σ1 displayed the same Rg as the IgG subclass from which their F(ab)2 domains were obtained. In their study, Tian et al. further derived maximal molecular dimensions per IgG subclass from their SAXS data and modeled predominant structural clusters of each IgG subclass.21 IgG1 comprised the most structural clusters with an equal distribution of Y, Y/T or T conformations, indicating maximum flexibility for this subclass. In contrast, IgG2 and IgG4 had a tighter Rg in both this study and pre-existing literature.21 However, Tian et al. attributed a higher flexibility to IgG4 over IgG2 due to IgG4’s broader maximum molecular dimensions, and IgG2’s more similar and more compact clusters. This compactness can likely be attributed to the rigid hinge containing the highest number of disulfide bonds.20 We substantiated the structural compactness with a different monoclonal antibody than that used by Tian et al.; Consequently, these findings were extrapolated to the investigated CD19xCD3 BsAbs used in the further functional studies.
All molecules were generated on Fc domains containing mutations reducing FcγR interaction to exclusively study the effect of the hinge-mediated molecular flexibility on T cell redirection. Residues involved in FcRn-binding (T250, M252, S254, T256, M428) were conserved among the Fc-mutated subclasses used in our study.37,38 As expected, the implemented silencing mutations did not affect FcRn binding compared to a human WT IgG1, suggesting that a proper serum half-life can be expected. The implemented Fc mutations, however, reduced the binding to FcγRs. In line with literature, all molecules comprising an IgG2 σ1 Fc interacted the least with all FcγRs.39 It has been demonstrated that the limited interaction of IgG2 σ1 with FcγRs in vitro translated to low or no activity in several in vitro functional assays, such as antibody-dependent cell-mediated cytotoxicity (ADCC), complement-mediated cytotoxicity (CDC), and antibody-dependent cellular phagocytosis.39 This observation confirmed that T cell activity would not likely be mediated via Fc because applied Fc mutations minimized binding to FcγR at the functional concentration ranges of BsAb used in killing experiments although subsets of T cells can express FcγRs.40
Single-arm affinities to CD19 and CD3 were recorded within a range of < 3X for all molecules, revealing that neither the IgG subclass nor molecular format (antibody vs. BsAb) significantly affected affinity in vitro. Similarly, CD19 was bound equally on cells across IgG subclasses, and independent from CD19 RD and RFP-transduction. Because the bivalent IgGs bound only 2-3X tighter than BsAbs, these observations indicated only little avidity of the molecules. In contrast, CD3-specific BsAbs bound T cells up to 10X weaker than their bivalent parental antibodies, suggesting stronger avidity effects for CD3 than observed for CD19. This could be driven by the antigens themselves, in particular the specific epitopes, which either allow for bivalent binding in the case of CD3, but not of CD19. Molecules of the IgG2 σ1 subclass bound T cells weakest in both molecular formats. This indicated that the more rigid hinge region and higher structural compactness of the IgG2 σ1 subclass may have impaired binding of CD3 on cells. Across IgG subclasses, RSVxCD3 BsAbs bound T cells slightly stronger than CD19xCD3 BsAbs, indicating that the pairing Fab arm may also influence CD3 recognition. It is known that the CD3: T-cell receptor complex undergoes a conformational change upon OKT3-binding to CD3ϵ.41 The structure of the different IgG subclasses could therefore affect the binding to CD3 in a cellular context. In line with these observations, changing the orientation and stabilization of disulfide bonds of the GD2-targeting scFv in a GD2xCD3 bs-scFv affected the binding of OKT3 to CD3.42 In general, tight binding to the B-cell lymphoma lines appears in the low-nM range at which none of the tested Fc-mutated IgG subclasses competed for binding to any FcγR. Thus, any effect of the Fc domain on binding activity, and consequently further contribution to killing activity, can be excluded, even though HBL-1 and Daudi cells express FcγRIIb.43,44
The IgG subclass influences antigen binding by a bivalent antibody.45–47 For example, changing the IgG subclass from IgG1 to IgG4 and adding hinge modifications resulted in enhancement of the agonistic activity of an anti-thrombopoietin-receptor antibody.48 Similarly, the F-VIII-mimetic activity was optimal if the FIXa x FX BsAb was used as IgG4 instead of IgG2 or IgG1.49 We observed similar effects of the IgG subclass change on the dual cell binding. CD19xCD3 model BsAbs harboring an IgG1 or IgG4 F(ab)2 domain, which mediate at least an intermediate flexibility, clustered T cells with target cells equally well. The abundance of clusters increased with a higher CD19 RD. In contrast, CD19xCD3 IgG2 σ1 was not able to efficiently cluster T cells with their target cells despite similar single-arm affinity. Its relative saturated binding level to CD3 on T cells was ~ 25% lower than that of the other IgG subclasses, while binding levels to CD19 on target cells were independent from the IgG subclass. However, the cluster formation was completely abrogated for CD19xCD3 IgG2 σ1 and is thus likely affected more by the structural characteristics than the monovalent binding level. Otherwise, partial clustering would have been expected to be recorded. Clustering of effector and target cells, however, is a prerequisite for the formation of active ISs. Higher flexibility not only means a wider angle between the F(ab)2 domains, but also a higher degree of F(ab)2 domain bending or rotation. In the case of IgG1 and IgG4 F(ab)2 domains, this leads to an optimal architecture to induce active ISs. IgG2, however, displays a similar structural compactness to IgG4 F(ab)2, but is predominantly present in a closed Y-shaped conformation.21 Additionally, IgG2 has a rigid disulfide-bond connection limiting free movement of the Fab domains.20 These characteristics probably decreased the capability of dual antigen binding to cluster T cells with their target cells, and less formed ISs, less activated T cells, and consequently less potent target cell killing. Further experiments evaluating T cell activation profiles and cytokine secretion at several time points should be addressed in a follow-up study to gain full mechanistical insight into the observations of this study.
Several previous studies gave evidence that a bispecific molecule’s structure contributes significantly to its activity: Modulating the size and amino-acid composition of the linker in a CD19xCD3 tandem diabody indicated that an optimal length and flexibility is required for efficient cytotoxic activity.50 In contrast, insertion of a green fluorescent protein-tag between the two targeting scFvs did not significantly reduce the activity of CD33xCD3 and CD3xPSCA single-chain tandem-Fvs (VHA-VLA – VHB-VLB in one polypeptide chain, similar to the BiTE format) at a fixed amount of 10–20 pmol.51 In a previous publication, the authors demonstrated that the domain-orientation in a CD3xPSMA bispecific scFv affected its performance.52 We observed that either IgG1- or IgG4-F(ab)2-molecules are most active dependent on the targeted cell line: The clustering efficiency was predictive of T-cell redirection potency, but was not an indicator for the maximum efficacy. Potentially, the extracellular matrix differs per cell line in amount, size, or glycosylation of proteins. Thus, an optimal BsAb flexibility and size would be required to most efficiently bridge the intermembrane distance between the T and target cells. Additionally, expression of co-stimulatory/inhibitory proteins, downstream signaling pathways, spatial determinants and activation of T cells could be further investigated to fully resolve this question.
In summary, we demonstrated that CD19xCD3 IgG2 σ1 BsAbs are less suitable for T cell redirection than their IgG1- or IgG4 counterparts. IgG2 is currently used to reduce effector functions of a therapeutic antibody to neutralize soluble ligands or to inhibit receptor-ligand interactions, such as the anti-epidermal growth factor receptor (EGFR) antibody panitumumab.23,24,53 Further, the CD40 antibody CDX-1140 in IgG2 subclass has superior Fc-independent agonist activity than its IgG1 counterpart.54 Recently, it has been demonstrated that EGFR-specific IgG2 can induce CDC and ADCC.55 This underlines the necessity of using Fc mutations to reduce FcγR interaction also for this supposedly less Fc-competent IgG subclass. In this study, IgG2 displayed a clearly inferior cellular clustering capacity, which is likely caused by reduced flexibility. We have demonstrated that structural and functional components can easily be combined to engineer molecules with desired efficacy by the example of the two chimeric BsAbs consisting of either the IgG1- or IgG4(Fab)2 domain on an IgG2 σ1 Fc domain: The superior T cell redirection conferred via the IgG1- or IgG4-hinge-region as part of the F(ab)2 was combined with the decreased FcγR interaction, conferred by the IgG2 σ1 Fc. Combining natural components of an antibody to improve two desired characteristics of a molecule (preferably without introducing further mutations) may be beneficial from a clinical point of view because fewer potentially immunogenic epitopes are introduced into the therapeutic molecule.56 In this aspect, a less stimulatory Fc domain is of special interest in T-cell targeting immunotherapy to avoid overstimulation of FcγR-positive effector cells in vivo: The overstimulation of Kupffer cells in a target-independent manner led to the termination of a clinical study investigating intravenous application of Fc-competent catumaxomab.57 Whether the applied Fc mutations are required for optimal in vivo functionality, or whether other mutations or a combination of a WT Fc with a mutated Fc to reduce FcγR interaction may be sufficient, could be addressed in further studies.
Material and methods
Kits and reagents used in the methods were used as per manufacturer’s instructions.
Generation of bispecific antibodies (BsABs) by cFAE
The parental antibodies were based on the variable regions encoding for OKT3 (CD3) and CD19 (humanized version of blinatumomab).22,29 As a null-arm control, a sequence encoding for an antibody targeting RSV was used. This targeted protein is neither expressed on target nor T cells and thus the RSV arm served as specificity control. The DNA constructs were ordered from Integrated DNA Technologies and cloned (In-Fusion® HD Cloning Kit, Clontech Laboratories, Inc.) into mammalian expression vectors.
HEK Expi293TM cells were transfected with the appropriate expression vectors (ExpiFectamineTM 293 transfection kit, ThermoFisher Scientific). Supernatants were harvested after 4–6 days by centrifugation (4,000 g, 15 min), passed through a 0.45-μm filter, and purified at 4°C by MabSelectTM SuReTM chromatography on an ÄKTAxpress system (both GE Healthcare) using Dulbecco’s phosphate-buffered saline (DPBS) (Sigma) as running buffer and 0.1 M sodium acetate, pH 3.5 as elution buffer. Elutions were immediately neutralized using 25% (v/v) 2 M Tris-HCl pH 7.0, dialyzed to DPBS, sterilized by 0.22-μm filtration and stored at 4°C. Concentrations were determined by absorbance at 280 nm on a Nanodrop ND-1000 spectrophotometer (ThermoFisher Scientific).
cFAE reactions were performed by reduction of a 1.05:1 molar mixture of CD19 and CD3 parental monoclonal antibodies using 75 mM cysteamine-hydrochloride (Sigma) essentially as described previously.27 An excess of 5% of CD19 parental antibody was used to minimize the presence of bivalent CD3-parental antibody in the BsAb preparations to minimize side-effects of residual bivalent CD3 in the IgG molecule format in the preparation.
All purified parental antibodies and recombined BsAbs were analyzed by analytical SE-HPLC on an Agilent 2600 BioInert system using a TSKgel BioAssist G3SWxl column (Tosoh) at a flow rate of 0.8 mL/min. A monomeric and monodisperse antibody elutes at a single peak at the expected retention time of ~ 10 min at these settings. Monodispersity is the presence of a single species. A monomeric antibody is a correctly assembled antibody consisting of two heavy and two light chains, without aggregates or free unmatched light or heavy chains. Successful recombination of BsAbs by cFAE was verified by CIEX-HPLC on a ProPac™ SCX-10 LC Column (ThermoFisher Scientific) using 20 mM 2-(N-morpholino)ethanesulfonic acid (MES), pH 5.8 as binding buffer and a linear 0–100% gradient elution over 30 min using 20 mM MES, 1 M NaCl, pH 5.8 at a flow rate of 1 mL/min. Each parental antibody elutes at a defined retention time based on its isoelectric point. A successfully recombined BsAb elutes at a retention time that was between those of the parental antibodies. In the case that the parental antibodies could not be separated by CIEX-HPLC, HI-HPLC was applied as quality control. Molecules were bound to a TSKgel butyl-NPR column (Tosoh) using 20 mM MES, pH 6.0, 1.5 M (NH4)2SO4, and eluted with 20 mM MES, pH 6.0 in a linear 20–100% gradient elution over 40 min at a flow rate of 0.8 mL/min. Analytical HPLCs were run with 30-40 μg loaded protein. Samples were verified as low endotoxin levels using a chromogenic LAL kit (Lonza).
Structural characterization by small angle x-ray scattering
SAXS-data were generated and analyzed at the Illinois Institute of Technology. CD19-targeting parental antibodies in the five generated IgG subclasses (IgG1, IgG2, IgG4, IgG1:2, and IgG4:2) were used as representative model antibodies for structural studies. Samples (250 μL at 12 mg/ml) were purified through a Wyatt SEC Analytical Column (5 μm, 300 Å, 7.8 mm) using an Agilent Infinity II HPLC system. After being eluted from the column, the sample passed through a multiple wavelength UV detector, followed by a DAWN HELEOS II (18 angle research grade light scattering photometer), a Wyatt QELS (Integrated into the multi-angle light scattering detector to perform simultaneous QELS (dynamic light scattering) measurements in the same flow cell or cuvette), an Optilab TrEX (Refractive Index detector for measurement of Differential and Absolute refractive index measurement) and finally to the SAXS flow cell. The beam was attenuated to 23–30% of full intensity, every 3 s for 0.5 s. The Rg value was derived from the SAXS data.58
Binding studies by SPR spectroscopy
SPR spectroscopy to determine single-arm affinities of parental antibodies or recombined BsAbs targeting CD19 was performed on a Biacore 4000 (GE Healthcare) using HBS-EP (0.01 M HEPES, 0.15 M NaCl, 3 mM EDTA, 0.05% v/v Surfactant P20) as running buffer. CD19 was recombinantly expressed as a hexahistidine-tagged extracellular domain in HEK Expi293TM cells (Biologics Research, Janssen R&D, Spring House, PA, US). Antibodies were bound at 10 μg/mL on Protein A sensor chips for 220 s at 10 µL/min. Monomeric CD19 antigen was injected for 300 s at a flow rate of 30 µL/min in 5-point 1:2 serial dilutions starting at 500 nM. Dissociation followed for 800 s. Between each concentration, the Protein A chip was regenerated using 10 mM Glycine-HCl pH 1.5 for at least 30 s. In this setup, no avidity-effect is measured and resulting values represent single-arm affinity. Fits were performed with a 1:1 binding model.
SPR spectroscopy to determine single-arm affinity values towards CD3 was performed on a Biacore T200 (GE Healthcare) using HBS-EP as running buffer. CD3 was recombinantly expressed as an Avi-tagged CD3δϵ-huFc fusion protein in HEK Expi293TM cells and site-directedly biotinylated post purification (Biologics Research, Janssen R&D, Spring House, PA, US). bt-CD3δϵ-Fc antigen was bound at 0.2 μg/mL on streptavidin (SA) sensor chips for 15–40 s resulting in 30–40 RU. This low immobilization level was applied to avoid avidity effects. Parental antibodies or BsAbs were injected for 200 s at a flow rate of 60 µL/min in 5-point 1:2 serial dilutions starting at 100 nM. Dissociation followed for 800 s. Between each kinetic run, the chip was regenerated 1 M NaCl in 50 mM NaOH for 30 s (leaving the bt-CD3δϵ-huFc bound to the SA-chip). Fits were performed with a steady-state binding model.
Cell culture
Daudi cells (CD19+) were obtained from ATCC (CCL-213). HBL-1 (CD19+) and HLY-1 (CD19−) were obtained from Dr. Miguel A. Piris (Hospital Universitario Marques de Valdecilla, Santander, Spain).59 Cell lines (WT and RFP-transduced) were sub-cultured thrice a week in RPMI1640 (Sigma) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (HI-FBS, BioWest) and 1X GlutaMaxTM (Gibco) (= complete medium) between 1.2 (HBL-1, HLY-1) or 4 (Daudi) and 20 × 105 cells/mL. Viability was assessed using the MoxiFlow cell counter (Orflo Technologies). Cells were transduced with RFP by lentiviral transduction using the CellPlayer™ NucLight Red reagent (Lenti, EF-1α; Essen BioScience). Transduced cells were sorted by fluorescence-activated cell sorting (FACS) on a BD FACS AriaIITM cell sorter. All cell lines were tested mycoplasma-free by the MycoAlert™ mycoplasma detection kit (Lonza).
Human pan T cells were isolated by Ficoll density gradient centrifugation (GE Healthcare) from 1-day-old buffy coats purchased from the Belgian Red Cross. T cells were enriched from the isolated peripheral blood mononuclear cell layer using a pan T cell isolation kit (CD3 negative selection, MACS Miltenyi) and stored at 15 × 106 cells/mL in Recovery™ Cell Culture Freezing Medium (Gibco) in the vapor phase of liquid N2. Typically, the T-cell purity was >85% as assessed by quantifying CD3+ cells (anti-huCD3APC, BD Pharmingen, clone UCHT1) on a BD FACSVerseTM flow cytometer. Before use, human pan T cells were thawed for 3 min at 37°C, washed twice (400 g, 4 min) and resuspended in complete or IncuCyte medium (phenol-red free RPMI1640 (Sigma) supplemented with 1X GlutaMax® (Gibco), 10% (v/v) HI-FBS (BioWest) and 25 mM HEPES (Sigma)).
FcR binding by AlphaScreenTM competition assay
AlphaScreenTM competition assays were performed in a total volume of 50 μL in 1X DPBS (Sigma) supplemented with 0.05% (w/v) bovine serum albumin (BSA, Sigma) and 0.01% (v/v) Tween-20 at a final pH-value of 7.2 for all FcγRs, or 6.0 for FcRn, in white 96-well half-area assay plates (Greiner). His-tagged FcγRs (R&D Systems) or FcRn (Sino Biological) bound to Ni2+-acceptor beads (PerkinElmer) were crosslinked by a biotinylated reference antibody bound to SA-donor beads. Model BsAbs or parental antibodies were titered in a 9-point 1:3 serial dilution to compete for binding to the FcRs with the biotinylated reference antibody. Specific concentrations, volumes and incubation times are provided in the supplements (Supplementary Table 5).
As reference antibody, a WT human IgG1 or human IgG1 containing Fc mutations reducing FcγR interaction (L234A, L235A)30 (both Biologics Research, Janssen R&D, Spring House, PA, US) was biotinylated using the SureLINKTM Chromophoric Biotin Labeling Kit (KPL, Inc.). Wells without biotinylated crosslinking antibody and competitor served as background. After the appropriate incubation times, plates were read on an EnVision plate reader (PerkinElmer) using the default AlphaScreenTM protocol. IC50 values were calculated using GraphPad Prism by applying a four-parameter logarithmic fitting model on a % of maximum signal over logarithmic protein concentration plot.
Cell binding by flow cytometry
Thawed T cells or cultured cell lines were harvested by centrifugation (400 g, 4 min), counted (Moxi Flow, Orflo), resuspended to 1 × 107 cells/mL, and FcγRs were blocked using a human Fc domain (Janssen) for 30 min at room temperature in FACS stain buffer (BSA, BD Pharmingen). Cells with blocked FcγRs were incubated for 30 min with a serial dilution of antibodies or BsAbs at 4°C, followed by a 30-min incubation at 4°C with 1:100-diluted anti-human IgGFcAlexaFluor®647 (Biolegend, clone HP6017). Ten thousand cells were counted on a BD FACS VerseTM flow cytometer and analysis was performed using BD FACSSuiteTM. EC50 values were calculated using GraphPad Prism by applying 4-parameter logarithmic fitting model on a median fluorescence intensity over logarithmic protein concentration plot.
Quantification of CD19 RD
To determine CD19 RD, CD19xRSV IgG1 FES and RSV IgG1 FES K409R as isotype control molecules were unimolarly labeled with recombinant phycoerythrin (R-PE) as previously described.60
FcγRs were blocked as described for cell binding. Subsequently, 100,000 cells/well were stained in a 100-μl volume with 9 μg/mL CD19xRSV IgG1 FESPE or its isotype control. These amounts were previously titrated to be at saturating concentration. Ten thousand cells were read on a BD FACS VerseTM flow cytometer and analysis was performed using BD FACSSuiteTM. Prior to readings, a calibration curve using the Quantum™ R-PE MESF kit (Bangs Laboratories, Inc.) was run on the same day with the same photomultiplier tube voltage and gain settings for calibration.
Cellular clustering by flow cytometry
The ability of CD19xCD3 IgG subclasses to cluster target and effector cells was assessed by flow cytometry. One lakh target cells, previously labeled with CellTrace Violet (Invitrogen) and treated with FcγR-blocking reagent as described for cell binding, were incubated for 45 min at 4°C with 20 μg/mL CD19xCD3 or CD19xRSV (negative control) and washed once. Subsequently, thawed T cells, previously labeled with CellTrace Far Red (Invitrogen), were added in a 1:1 ratio and cluster formation was allowed for 6 h at 4°C. Subsequently, cells were stained with a green fixable viability dye (FVD eFluor® 520; eBioScience) and fixed with a final concentration of 4% (w/v) paraformaldehyde (Pierce). Plates were stored at 4°C until analysis. Thirty-thousand FVD eFluor® 520− cells were counted on a BD FACS VerseTM flow cytometer and analysis was performed using BD FACSSuiteTM software. Clustering efficiency of the BsAbs was defined as the percentage of double-positive events of the live-cell gate in a violet over red scatter plot, and was normalized to the untreated condition per cell line and biological replicate.
Inhibition of proliferation assays using incucyte ZOOM® technology
4–5-day proliferation assays were carried out using an IncuCyte ZOOM® Continuous Live-cell Imaging & Analysis System (Essen BioScience) in a 96-well format using IncuCyte medium at 37°C/5% CO2. RFP-transduced target cells were incubated with thawed T cells using a 5:1 effector: target cell ratio in the presence or absence (= untreated condition) of BsAbs. Each well was imaged in 4-h intervals by quantifying the red area (RA) in μm2/well. Image analysis was terminated when untreated control wells reached their growth plateau. Per time point, RAt=x was first normalized to the initial RAt=0, resulting in the expansion index (EI = RAt=x/RAt=0). This measure for normalized growth in treated conditions (EItreated) was set in relation to EIuntreated, resulting in the Inhibition Quotient (IQ = EIuntreated/EItreated). The IQ was plotted over time, and the area under the curve (AUC) was determined for each condition. This AUC represents the growth inhibition per BsAb concentration and was plotted over the BsAb concentration. IC50 values were calculated from these plots using GraphPad Prism using a four-parameter logarithmic fitting model. The maximum efficiency was defined as the upper asymptote.
Funding Statement
This work was supported by the Flemish Agency for Innovation and Entrepreneurship [IWT.145054 – ImmunoTherapy].
Acknowledgments
We acknowledge Nele Vloemans (Oncology Biology & Discovery Janssen Research and Development, Belgium) for the RFP-transduction of cell lines; Renouard Sanders (Janssen Diagnostics, Janssen Research and Development, Huntingdon Valley, PA, USA) for unimolar PE-labelling of molecules for receptor density experiments; Luc Gabriels (Discovery Sciences, Screening BE, Janssen Research and Development, Beerse, Belgium) for mycoplasma testing of cell lines, Anthony Armstrong (Biologics Research, Janssen Research and Development, Spring House, PA, USA) for coordinating structural SAXS measurements with the Illinois Institute of Technology. This work was supported by the Flemish Agency for Innovation and Entrepreneurship (IWT.145054 – ImmunoTherapy).
Disclosure of interest
Janssen Research and Development provided funding for the research. All authors are/were employees of Janssen Research and Development.
Supplemental material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Thakur A, Lum LG.. “NextGen” biologics: bispecific antibodies and emerging clinical results. Expert Opin Biol Ther. 2016;16:675–88. doi: 10.1517/14712598.2016.1150996. [DOI] [PubMed] [Google Scholar]
- 2.Kontermann RE, Brinkmann U.. Bispecific antibodies. Drug Discov Today. 2015;20:838–47. doi: 10.1016/j.drudis.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Seimetz D, Lindhofer H, Bokemeyer C. Development and approval of the trifunctional antibody catumaxomab (anti-EpCAM x anti-CD3) as a targeted cancer immunotherapy. Cancer Treat Rev. 2010;36:458–67. doi: 10.1016/j.ctrv.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Przepiorka D, Ko CW, Deisseroth A, Yancey CL, Candau-Chacon R, Chiu HJ, Gehrke BJ, Gomez-Broughton C, Kane RC, Kirshner S, et al. FDA approval: blinatumomab. Clin Cancer Res. 2015;21:4035–39. doi: 10.1158/1078-0432.CCR-15-0612. [DOI] [PubMed] [Google Scholar]
- 5.Batlevi CL, Matsuki E, Brentjens RJ, Younes A. Novel immunotherapies in lymphoid malignancies. Nat Rev Clin Oncol. 2016;13:25–40. doi: 10.1038/nrclinonc.2015.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith EJ, Olson K, Haber LJ, Varghese B, Duramad P, Tustian AD, Oyejide A, Kirshner JR, Canova L, Menon J, et al. A novel, native-format bispecific antibody triggering T-cell killing of B-cells is robustly active in mouse tumor models and cynomolgus monkeys. Sci Rep. 2015;5:17943. doi: 10.1038/srep17943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spiess C, Zhai Q, Carter PJ. Alternative molecular formats and therapeutic applications for bispecific antibodies. Mol Immunol. 2015;67:95–106. doi: 10.1016/j.molimm.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Schaefer W, Regula JT, Bahner M, Schanzer J, Croasdale R, Durr H, Gassner C, Georges G, Kettenberger H, Imhof-Jung S, et al. Immunoglobulin domain crossover as a generic approach for the production of bispecific IgG antibodies. Proc Natl Acad Sci U S A. 2011;108:11187–92. doi: 10.1073/pnas.1019002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis DM, Dustin ML. What is the importance of the immunological synapse? Trends Immunol. 2004;25:323–27. doi: 10.1016/j.it.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 10.Dustin ML, Depoil D. New insights into the T cell synapse from single molecule techniques. Nat Rev Immunol. 2011;11:672–84. doi: 10.1038/nri3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.James JR, Vale RD. Biophysical mechanism of T-cell receptor triggering in a reconstituted system. Nature. 2012;487:64–69. doi: 10.1038/nature11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Merwe PA, Dushek O. Mechanisms for T cell receptor triggering. Nat Rev Immunol. 2011;11:47–55. doi: 10.1038/nri2887. [DOI] [PubMed] [Google Scholar]
- 13.Alakoskela JM, Koner AL, Rudnicka D, Kohler K, Howarth M, Davis DM. Mechanisms for size-dependent protein segregation at immune synapses assessed with molecular rulers. Biophys J. 2011;100:2865–74. doi: 10.1016/j.bpj.2011.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klein JS, Gnanapragasam PN, Galimidi RP, Foglesong CP, West AP Jr., Bjorkman PJ. Examination of the contributions of size and avidity to the neutralization mechanisms of the anti-HIV antibodies b12 and 4E10. Proc Natl Acad Sci U S A. 2009;106:7385–90. doi: 10.1073/pnas.0811427106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leong SR, Sukumaran S, Hristopoulos M, Totpal K, Stainton S, Lu E, Wong A, Tam L, Newman R, Vuillemenot BR, et al. An anti-CD3/anti-CLL-1 bispecific antibody for the treatment of acute myeloid leukemia. Blood. 2017;129:609–18. doi: 10.1182/blood-2016-08-735365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bluemel C, Hausmann S, Fluhr P, Sriskandarajah M, Stallcup WB, Baeuerle PA, Kufer P. Epitope distance to the target cell membrane and antigen size determine the potency of T cell-mediated lysis by BiTE antibodies specific for a large melanoma surface antigen. Cancer Immunol Immunother. 2010;59:1197–209. doi: 10.1007/s00262-010-0844-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Stagg NJ, Johnston J, Harris MJ, Menzies SA, DiCara D, Clark V, Hristopoulos M, Cook R, Slaga D, et al. Membrane-proximal epitope facilitates efficient T cell synapse formation by anti-FcRH5/CD3 and is a requirement for myeloma cell killing. Cancer Cell. 2017. doi: 10.1016/j.ccell.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guest RD, Hawkins RE, Kirillova N, Cheadle EJ, Arnold J, O’Neill A, Irlam J, Chester KA, Kemshead JT, Shaw DM, et al. The role of extracellular spacer regions in the optimal design of chimeric immune receptors: evaluation of four different scFvs and antigens. J Immunother. 2005;28:203–11. [DOI] [PubMed] [Google Scholar]
- 19.Roux KH, Strelets L, Michaelsen TE. Flexibility of human IgG subclasses. J Immunol. 1997;159:3372–82. [PubMed] [Google Scholar]
- 20.Liu H, May K. Disulfide bond structures of IgG molecules: structural variations, chemical modifications and possible impacts to stability and biological function. MAbs. 2012;4:17–23. doi: 10.4161/mabs.4.1.18347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian X, Vestergaard B, Thorolfsson M, Yang Z, Rasmussen HB, Langkilde AE. In-depth analysis of subclass-specific conformational preferences of IgG antibodies. IUCrJ. 2015;2:9–18. doi: 10.1107/S205225251402209X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brinkmann U, Kontermann RE. The making of bispecific antibodies. MAbs. 2017;9:182–212. doi: 10.1080/19420862.2016.1268307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carter PJ. Potent antibody therapeutics by design. Nat Rev Immunol. 2006;6:343–57. doi: 10.1038/nri1837. [DOI] [PubMed] [Google Scholar]
- 24.Kretschmer A, Schwanbeck R, Valerius T, Rosner T. Antibody isotypes for tumor immunotherapy. Transfus Med Hemother. 2017;44:320–26. doi: 10.1159/000479240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Labrijn AF, Buijsse AO, van Den Bremer ET, Verwilligen AY, Bleeker WK, Thorpe SJ, Killestein J, Polman CH, Aalberse RC, Schuurman J, et al. Therapeutic IgG4 antibodies engage in Fab-arm exchange with endogenous human IgG4 in vivo. Nat Biotechnol. 2009;27:767–71. doi: 10.1038/nbt.1553. [DOI] [PubMed] [Google Scholar]
- 26.Labrijn AF, Meesters JI, Priem P, de Jong RN, van Den Bremer ET, van Kampen MD, Gerritsen AF, Schuurman J, Parren PW. Controlled Fab-arm exchange for the generation of stable bispecific IgG1. Nat Protoc. 2014;9:2450–63. doi: 10.1038/nprot.2014.169. [DOI] [PubMed] [Google Scholar]
- 27.Labrijn AF, Meesters JI, de Goeij BE, van Den Bremer ET, Neijssen J, van Kampen MD, Strumane K, Verploegen S, Kundu A, Gramer MJ, et al. Efficient generation of stable bispecific IgG1 by controlled Fab-arm exchange. Proc Natl Acad Sci U S A. 2013;110:5145–50. doi: 10.1073/pnas.1220145110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loffler A, Kufer P, Lutterbuse R, Zettl F, Daniel PT, Schwenkenbecher JM, Riethmuller G, Dorken B, Bargou RC. A recombinant bispecific single-chain antibody, CD19 x CD3, induces rapid and high lymphoma-directed cytotoxicity by unstimulated T lymphocytes. Blood. 2000;95:2098–103. [PubMed] [Google Scholar]
- 29.Kung P, Goldstein G, Reinherz EL, Schlossman SF. Monoclonal antibodies defining distinctive human T cell surface antigens. Science 1979;206:347–49. [DOI] [PubMed] [Google Scholar]
- 30.Hezareh M, Hessell AJ, Jensen RC, van de Winkel JG, Parren PW. Effector function activities of a panel of mutants of a broadly neutralizing antibody against human immunodeficiency virus type 1. J Virol. 2001;75:12161–68. doi: 10.1128/JVI.75.24.12161-12168.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oganesyan V, Gao C, Shirinian L, Wu H, Dall’Acqua WF. Structural characterization of a human Fc fragment engineered for lack of effector functions. Acta Crystallogr D Biol Crystallogr. 2008;64:700–04. doi: 10.1107/S0907444908007877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vafa O, Gilliland GL, Brezski RJ, Strake B, Wilkinson T, Lacy ER, Scallon B, Teplyakov A, Malia TJ, Strohl WR. An engineered Fc variant of an IgG eliminates all immune effector functions via structural perturbations. Methods. 2014;65:114–26. doi: 10.1016/j.ymeth.2013.06.035. [DOI] [PubMed] [Google Scholar]
- 33.Lobanov M, Bogatyreva NS, Galzitskaia OV. [Radius of gyration is indicator of compactness of protein structure]. Mol Biol (Mosk). 2008;42:701–06. [PubMed] [Google Scholar]
- 34.Anderson DR, Grillo-Lopez A, Varns C, Chambers KS, Hanna N. Targeted anti-cancer therapy using rituximab, a chimaeric anti-CD20 antibody (IDEC-C2B8) in the treatment of non-hodgkin’s B-cell lymphoma. Biochem Soc Trans. 1997;25:705–08. [DOI] [PubMed] [Google Scholar]
- 35.Strop P, Ho WH, Boustany LM, Abdiche YN, Lindquist KC, Farias SE, Rickert M, Appah CT, Pascua E, Radcliffe T, et al. Generating bispecific human IgG1 and IgG2 antibodies from any antibody pair. J Mol Biol. 2012;420:204–19. doi: 10.1016/j.jmb.2012.04.020. [DOI] [PubMed] [Google Scholar]
- 36.Dillon M, Yin Y, Zhou J, McCarty L, Ellerman D, Slaga D, Junttila TT, Han G, Sandoval W, Ovacik MA, et al. Efficient production of bispecific IgG of different isotypes and species of origin in single mammalian cells. MAbs. 2017;9:213–30. doi: 10.1080/19420862.2016.1267089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dall’Acqua WF, Kiener PA, Wu H. Properties of human IgG1s engineered for enhanced binding to the neonatal Fc receptor (FcRn). J Biol Chem. 2006;281:23514–24. doi: 10.1074/jbc.M604292200. [DOI] [PubMed] [Google Scholar]
- 38.Dall’Acqua WF, Woods RM, Ward ES, Palaszynski SR, Patel NK, Brewah YA, Wu H, Kiener PA, Langermann S. Increasing the affinity of a human IgG1 for the neonatal Fc receptor: biological consequences. J Immunol. 2002;169:5171–80. doi: 10.4049/jimmunol.169.9.5171. [DOI] [PubMed] [Google Scholar]
- 39.Tam S, McCarthy S, Armstrong A, Somani S, Wu S-J, Liu X, Gervais A, Ernst R, Saro D, Decker R, et al. Functional, biophysical, and structural characterization of human IgG1 and IgG4 Fc variants with ablated immune functionality. Antibodies. 2017;6:12. doi: 10.3390/antib6030012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chauhan AK, Chen C, Moore TL, DiPaolo RJ. Induced expression of fcgammariiia (CD16a) on CD4+ T cells triggers generation of IFN-gammahigh subset. J Biol Chem. 2015;290:5127–40. doi: 10.1074/jbc.M114.599266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kjer-Nielsen L, Dunstone MA, Kostenko L, Ely LK, Beddoe T, Mifsud NA, Purcell AW, Brooks AG, McCluskey J, Rossjohn J. Crystal structure of the human T cell receptor CD3 epsilon gamma heterodimer complexed to the therapeutic mAb OKT3. Proc Natl Acad Sci U S A. 2004;101:7675–80. doi: 10.1073/pnas.0402295101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng M, Ahmed M, Xu H, Cheung NK. Structural design of disialoganglioside GD2 and CD3-bispecific antibodies to redirect T cells for tumor therapy. Int J Cancer. 2015;136:476–86. doi: 10.1002/ijc.29007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rankin CT, Veri MC, Gorlatov S, Tuaillon N, Burke S, Huang L, Inzunza HD, Li H, Thomas S, Johnson S, et al. CD32B, the human inhibitory Fc-gamma receptor IIB, as a target for monoclonal antibody therapy of B-cell lymphoma. Blood. 2006;108:2384–91. doi: 10.1182/blood-2006-05-020602. [DOI] [PubMed] [Google Scholar]
- 44.Psathas JN, Doonan PJ, Raman P, Freedman BD, Minn AJ, Thomas-Tikhonenko A. The Myc-miR-17-92 axis amplifies B-cell receptor signaling via inhibition of ITIM proteins: a novel lymphomagenic feed-forward loop. Blood. 2013;122:4220–29. doi: 10.1182/blood-2012-12-473090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hubbard MA, Thorkildson P, Kozel TR, AuCoin DP. Constant domains influence binding of mouse-human chimeric antibodies to the capsular polypeptide of bacillus anthracis. Virulence. 2013;4:483–88. doi: 10.4161/viru.25711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCloskey N, Turner MW, Steffner P, Owens R, Goldblatt D. Human constant regions influence the antibody binding characteristics of mouse-human chimeric IgG subclasses. Immunology. 1996;88:169–73. doi: 10.1111/j.1365-2567.1996.tb00001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morelock MM, Rothlein R, Bright SM, Robinson MK, Graham ET, Sabo JP, Owens R, King DJ, Norris SH, Scher DS, et al. Isotype choice for chimeric antibodies affects binding properties. J Biol Chem. 1994;269:13048–55. [PubMed] [Google Scholar]
- 48.Kai M, Motoki K, Yoshida H, Emuta C, Chisaka Y, Tsuruhata K, Endo C, Muto M, Shimabe M, Nishiyama U, et al. Switching constant domains enhances agonist activities of antibodies to a thrombopoietin receptor. Nat Biotechnol. 2008;26:209–11. doi: 10.1038/nbt1376. [DOI] [PubMed] [Google Scholar]
- 49.Sampei Z, Igawa T, Soeda T, Funaki M, Yoshihashi K, Kitazawa T, Muto A, Kojima T, Nakamura S, Hattori K. Non-antigen-contacting region of an asymmetric bispecific antibody to factors IXa/X significantly affects factor VIII-mimetic activity. MAbs. 2015;7:120–28. doi: 10.4161/19420862.2015.989028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Le Gall F, Reusch U, Little M, Kipriyanov SM. Effect of linker sequences between the antibody variable domains on the formation, stability and biological activity of a bispecific tandem diabody. Protein Eng Des Sel. 2004;17:357–66. doi: 10.1093/protein/gzh039. [DOI] [PubMed] [Google Scholar]
- 51.Stamova S, Feldmann A, Cartellieri M, Arndt C, Koristka S, Apel F, Wehner R, Schmitz M, Bornhauser M, von Bonin M, et al. Generation of single-chain bispecific green fluorescent protein fusion antibodies for imaging of antibody-induced T cell synapses. Anal Biochem. 2012;423:261–68. doi: 10.1016/j.ab.2011.12.042. [DOI] [PubMed] [Google Scholar]
- 52.Feldmann A, Stamova S, Bippes CC, Bartsch H, Wehner R, Schmitz M, Temme A, Cartellieri M, Bachmann M. Retargeting of T cells to prostate stem cell antigen expressing tumor cells: comparison of different antibody formats. Prostate. 2011;71:998–1011. doi: 10.1002/pros.21315. [DOI] [PubMed] [Google Scholar]
- 53.Wang X, Mathieu M, Brezski RJ. IgG Fc engineering to modulate antibody effector functions. Protein Cell. 2018;9:63–73. doi: 10.1007/s13238-017-0473-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vitale LA, Thomas LJ, He LZ, O’Neill T, Widger J, Crocker A, Sundarapandiyan K, Storey JR, Forsberg EM, Weidlick J, et al. Development of CDX-1140, an agonist CD40 antibody for cancer immunotherapy. Cancer Immunol Immunother. 2019;68:233–45. doi: 10.1007/s00262-018-2267-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rosner T, Kahle S, Montenegro F, Matlung HL, Jansen JHM, Evers M, Beurskens F, Leusen JHW, van Den Berg TK, Valerius T. Immune effector functions of human IgG2 antibodies against EGFR. Mol Cancer Ther. 2019;18:75–88. doi: 10.1158/1535-7163.MCT-18-0341. [DOI] [PubMed] [Google Scholar]
- 56.Kubota T, Niwa R, Satoh M, Akinaga S, Shitara K, Hanai N. Engineered therapeutic antibodies with improved effector functions. Cancer Sci. 2009;100:1566–72. doi: 10.1111/j.1349-7006.2009.01222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Borlak J, Langer F, Spanel R, Schondorfer G, Dittrich C. Immune-mediated liver injury of the cancer therapeutic antibody catumaxomab targeting EpCAM, CD3 and Fcgamma receptors. Oncotarget. 2016;7:28059–74. doi: 10.18632/oncotarget.8574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guinier A, Fournet G. Small-angle scatterring of X-rays. New York: Wiley; 1955. [Google Scholar]
- 59.Gomez-Abad C, Pisonero H, Blanco-Aparicio C, Roncador G, Gonzalez-Menchen A, Martinez-Climent JA, Mata E, Rodriguez ME, Munoz-Gonzalez G, Sanchez-Beato M, et al. PIM2 inhibition as a rational therapeutic approach in B-cell lymphoma. Blood. 2011;118:5517–27. doi: 10.1182/blood-2011-03-344374. [DOI] [PubMed] [Google Scholar]
- 60.Jarantow SW, Bushey BS, Pardinas JR, Boakye K, Lacy ER, Sanders R, Sepulveda MA, Moores SL, Chiu ML. Impact of cell-surface antigen expression on target engagement and function of an epidermal growth factor receptor x c-MET bispecific antibody. J Biol Chem. 2015;290:24689–704. doi: 10.1074/jbc.M115.651653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


