ABSTRACT
HIV-exposed but uninfected (HEU) children represent a growing population and show a significantly higher number of infectious diseases, several immune alterations, compromised growth, and increased mortality rates when compared to HIV-unexposed children. Considering the impact that the gut microbiota has on general host homeostasis and immune system development and modulation, we hypothesized that HEU children present altered gut microbiota that is linked to the increased morbidity and the immune system disorders faced by them. Our experiments revealed no differences in beta and alpha diversity of the gut microbiota between HEU and unexposed children or between HIV-infected and uninfected mothers. However, there were differences in the abundance of several taxa from the gut microbiota between HEU and unexposed children and between HIV-infected and uninfected mothers. Functional prediction based on 16S rRNA sequences also indicated differences between HEU and unexposed children and between infected and uninfected mothers. In addition, we detected no differences between HEU and unexposed children in relation to weight, weight-for-age z scores, albumin serum levels, or microbial translocation and inflammation markers. In summary, HIV-infected mothers and their HIV-exposed children present alterations in the abundance of several taxa in the gut microbiome and the predicted functional metagenome when compared to uninfected mothers and unexposed children. Knowledge about the gut microbiome of HEU children in different settings is essential in order to determine better treatments for this susceptible population.
KEYWORDS: HIV, children, exposed uninfected, gut microbiota, 16S rRNA, microbial translocation, inflammation
Introduction
From 2010 to 2016, the number of annual vertical HIV transmissions worldwide decreased by 60%, and it is estimated that more than 1 million new HIV infections among children were averted during this period.1 These numbers were achieved primarily due to the development of prevention of mother-to-child transmission programs, which had a rapid scale-up since 2010.1 With less HIV infections among children every year, health concerns are diverted towards the growing population of HIV-exposed but uninfected (HEU) children. According to several studies, HEU children show increased morbidity, mortality, growth problems, and immune system alterations when compared to unexposed and uninfected children.2-4 However, the causes and mechanisms underlying this are still far from being fully understood.
Considering the pivotal role that intestinal microbiota has in the development of the immune system and general host homeostasis, it has been hypothesized that the complications observed in HEU children can be related to alterations in the gut microbiota community.5,6 It is already well accepted that, compared to uninfected ones, HIV-infected individuals present an altered (dysbiotic) gut microbiota, in both treated and untreated individuals.7-9 The dysbiosis of HIV-infected patients includes increased richness,10,11 higher abundance of bacteria from the Proteobacteria phylum,11-13 an increased proportion of opportunistic pathogens,13-15 and lower abundance of commensal bacteria.13-15 Besides, dysbiosis may also lead to the disruption of the intestinal barrier, allowing the translocation of entire microbes or their components from the intestinal lumen to the systemic circulation.16-18 This process, known as microbial translocation, has been linked to increased immune activation in HIV-infected individuals.19
However, much less is understood about HEU children. Several factors have been associated to the immune alterations and higher morbidity rates exhibited by these children, with an emphasis on the factors directly related to mothers’ HIV infection, such as in utero exposure to HIV, exposure to antiretroviral therapy before and after birth, maternal immune compromise, and a maternal pro-inflammatory state.20 Also, it is known that the microbiota of mothers has an impact on the development of newborns’ microbiota.21-23 Considering that HIV-infected women may have altered microbiota,24 it is reasonable to hypothesize that HIV-exposed children also present dysbiosis. However, the intestinal microbiota of HEU children has not yet been thoroughly analyzed, with very few studies published to date.6,25 A previous study evaluating the gut microbial composition of HEU children was based on culture techniques and thus focused on a few components of the microbiome.25 With the recent development of next-generation sequencing techniques, the microbiome of HEU children can be studied in significantly more detail, revealing alterations in the microbial community structure among HEU and unexposed infants, while relating these alterations to the composition of oligosaccharides in mothers' breast milk.6
Moreover, considering that the human gut microbiota is widely variable, susceptible to alteration according to life and nutritional habits, and linked to environmental particularities of different geographic regions,26 it is essential to evaluate the gut microbiome of HEU children from different regions and with different nutritional and life habits. Furthermore, there are almost no data from HEU children gut microbiota beyond infancy (i.e., >1 y of age). Therefore, in order to contribute to a better understanding of the health and immune system alterations presented by HEU children, we performed a cross-sectional study of Brazilian mother–child pairs including HEU children and unexposed uninfected children. The analysis included microbiome characterization by 16S sequencing, evaluation of translocation, and inflammation markers of the participants’ diet. Our data confirmed the absence of inter-individual diversity in the gut microbiota of HEU children and their mothers compared to unexposed children and their mothers. It shows, for the first time, that several microbiota components differ between HEU children who are not breastfed and uninfected children.
Results
Characteristics of mother–child pairs
A total of 19 mother and child pairs (38 samples) were recruited in Florianópolis, Santa Catarina state, southern Brazil, from October 2016 to May 2017. Twelve children were HEU, and seven were unexposed and uninfected. No children were breastfeeding at the time of recruiting and had not breastfed within the previous eight months. There were no statistically significant differences for gender, skin color, and time or type of delivery between HEU and unexposed children (Table 1). Four HEU and two unexposed children consumed antibiotics within three months of recruitment, as declared by mothers. Of the 12 HIV-infected mothers, 11 reported to be under antiretroviral treatment and presented undetectable viral loads at the time of recruitment (Table 1). One HIV-infected mother was a smoker and smoked during pregnancy.
Table 1.
Clinical and demographic characteristics of the cohort of HIV-unexposed and HIV-exposed but uninfected (HEU) children and their HIV-negative and HIV-positive mothers from Florianópolis, Brazil.
| Children |
Mothers |
|||||
|---|---|---|---|---|---|---|
| Unexposed | HEU | p | HIV negative | HIV positive | p | |
| Number of individuals | 7 | 12 | - | 7 | 12 | - |
| Age (years, median (IQR)) | - | - | - | 29 (27 to 33) | 34 (24 to 35) | 1.00 |
| BMI (kg/m2, median (IQR)) | - | - | - | 24.86 (22.07 to 32.27) | 27.78 (22.44 to 36.91) | 0.4082 |
| Age (months, median (IQR)) | 24 (21 to 25) | 18 (16.75 to 22.25) | 0.02416 | - | - | - |
| Female sex (%) | 4 (57.14) | 6 (50) | 1.00 | - | - | - |
| Vaginal delivery (%) | 4 (57.14) | 6 (50) | 1.00 | - | - | - |
| Term delivery (%) | 5 (71.43) | 9 (75) | 1.00 | - | - | - |
| Weight (g, median (IQR)) | 12,426 (12,200 to 13,870) |
11,200 (10,700 to 11,850) |
0.1471 | - | - | - |
| Weight-for-age z score (median (IQR)) | 0.50 (−0.040 to 1.44) |
0.68 (−0.3150 to 0.9650) |
0.7914 | - | - | - |
| TCD4+ T cell count (cells/mm3, median (IQR)) | - | 2,636 (2,212 to 3,016) |
- | - | 749.5 (600.2 to 816.0) | - |
| TCD8+ T cell count (cells/mm3, median (IQR)) | - | 1,000 (792.5 to 1,486.8) |
- | - | 792.0 (725.5 to 1,132.0) | - |
| CD4/CD8+ T cell ratio (median (IQR)) | - | 2.575 (2.143 to 3.132) |
- | - | 0.65 (0.52 to 1.10) | - |
| CD45+ (cells/mm3, median (IQR)) | - | 5,614 (4,518 to 8,207) |
- | - | 2,056 (1,750 to 2,664) | - |
| On antiretroviral treatment (%) | - | - | - | - | 11 (91.67%) | - |
| Detectable viral load (%) | - | - | - | - | 1 (8.33%) | - |
Health and growth of HEU and unexposed children
To evaluate the frequency of infections in the two groups of children, mothers were asked to complete a survey to identify the incidence of disease in their children, with results indicating an increased frequency of diseases in HEU children when compared to unexposed ones. Mothers were asked to consider as “disease” any signs of infections such as fever, coughing, sneezing, and sore throat. For HIV-infected mothers, 25% reported that their children showed signs of disease at least once a month and 58.3% reported signs once in every six months. In the case of uninfected mothers, only one reported that her child showed signs of the disease every month and one reported signs of the disease every six months. Conversely, a large proportion of uninfected mothers reported that their children showed signs of disease yearly (42.9%) or less than once a year (28.6%). When inquired about the time since the child’s last infection, 83.3% of HIV-positive mothers reported that the last time they remembered their child being sick was at least within the previous six months, in contrast to the 57.1% of HIV-negative mothers responding the same.
Moreover, three HEU children had bronchitis, and one unexposed child had hypothyroidism. In order to estimate the children’s nutritional state, albumin serum levels were evaluated, but no statistically significant differences were found between the two groups of children (data not shown), and the same was found for weight and weight-for-age z scores (Table 1). Taken together, these data indicate that, although HEU children seemed healthy at the time of the study, they display signs of infections more frequently than children born from HIV-negative mothers.
HEU children present differences in gut microbiome abundance
Microbiome composition was evaluated through 16S rRNA sequencing of stool samples obtained from HEU and unexposed children as well as their mothers. Bacterial DNA was sequenced and generated approximately 14 million reads, with an average of 375,182.7 (±92,706.08) reads per sample. These were processed and classified into a total of 1,246 operational taxonomic units (OTUs). To analyze inter-individual differences in the gut microbiome, the Bray Curtis (BC) dissimilarity index was used and showed that microbiome composition of mothers and children differed among each other (Adonis with 999 permutations, p < 0.01) (Figure 1A). However, there were no statistically significant differences in alpha and beta diversity between the two groups of children or the two groups of mothers (p > 0.05), indicating no difference in diversity either in between or within individuals (Figure 1A and B). However, in terms of microbial composition at the bacterial family level, the most abundant bacteria detected in the gut microbiome belonged to Ruminococcaceae in HIV-negative mothers (25.9%) and Prevotellaceae in HIV-positive mothers (37.4%). Lachnospiraceae was the most abundant bacteria family in both groups of children (33.7% in HEU and 26.7% in unexposed) (Figure 1C). At the genus level, the most abundant bacteria belonged to Bacteroides in both groups of children (25.2% in HEU and 48.4% in unexposed) and Prevotella in both groups of mothers (49.5% in HIV infected and 38.4% in uninfected).
Figure 1.
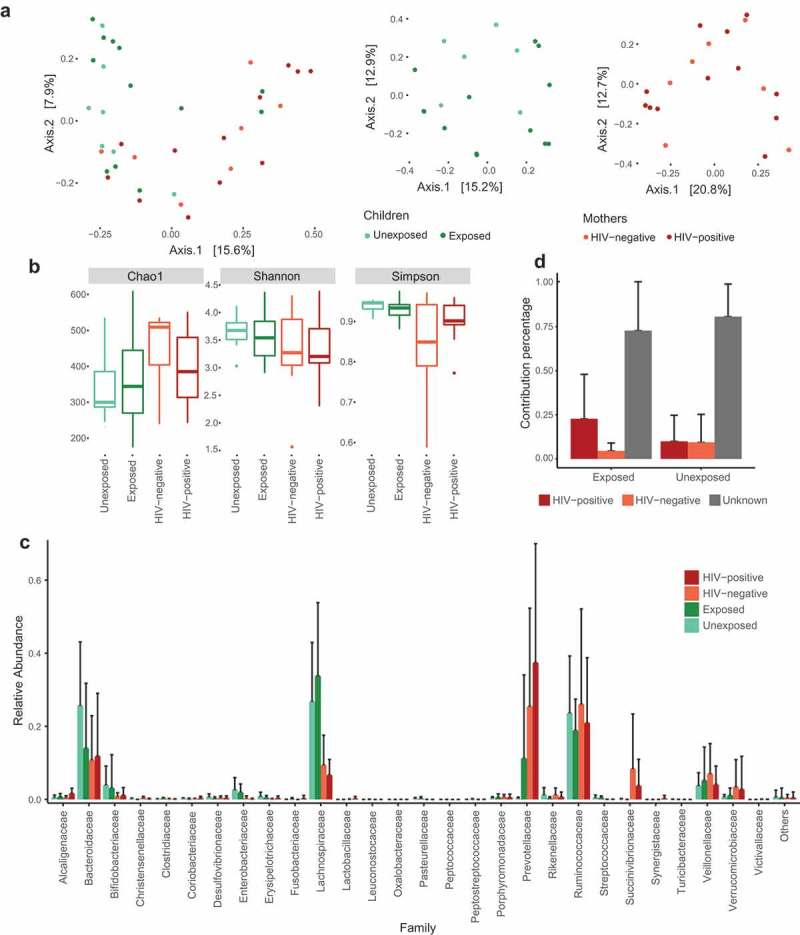
Gut microbiome of mothers and children. Principal coordinate analysis (PCoA) of Bray Curtis distances among groups of children and mothers (a); Richness (Chao1) and diversity (Shannon and Simpson) indexes (b); Relative abundance at the family level for groups of mothers and children (c); Mean source proportions of bacteria from the children’s gut microbiota (d).
To evaluate differential abundance of OTUs between groups, we used a negative binomial distribution model with the R package DEseq2. It was possible to identify 35 OTUs that were differentially abundant between HEU and unexposed children (false discovery rate (FDR)-adjusted p-value <0.05) (Figure 2A). Three of these OTUs were identified at the genus level and corresponded to Bacteroides uniformis, which was increased in unexposed children, and to Faecalibacterium prausnitzii and Prevotella copri, which were increased in HEU children. HEU children also showed increased proportions of other OTUs belonging to the genera Bacteroides, Dialister, Dorea, Klebsiella, Lachnobacterium, Lactococcus, Phascolarctobacterium, Roseburia, and unidentified OTUs from the families Clostridiaceae, Enterobacteriaceae, Erysipelotrichaceae, Lachnospiraceae, Ruminococcaceae, and the order Clostridiales. On the other hand, unexposed children showed an increase in the genus Paraprevotella. No significant correlations (FDR-adjusted p-value > 0.05) were observed for diversity estimates and weight, weight-for-age z scores and TCD4+, TCD8+, or CD45+ cells. In addition, no significant correlations (FDR-adjusted p-value >0.05) were found for individual OTU abundance and all previously mentioned clinical markers. For the two groups of mothers, we were able to identify 16 differentially abundant OTUs (FDR-adjusted p-value <0.05), with HIV-infected mothers showing increased proportions of P. copri, Prevotella stercorea, and other OTUs belonging to Prevotella, Dialister, Ruminococcaceae, and Bacteroidales. Conversely, HIV-negative mothers showed increased proportions of OTUs belonging to Bacteroides, Lachnospira, Oscillospira, Clostridiaceae, Lachnospiraceae, Rikenellaceae, and Clostridiales (Figure 2B).
Figure 2.
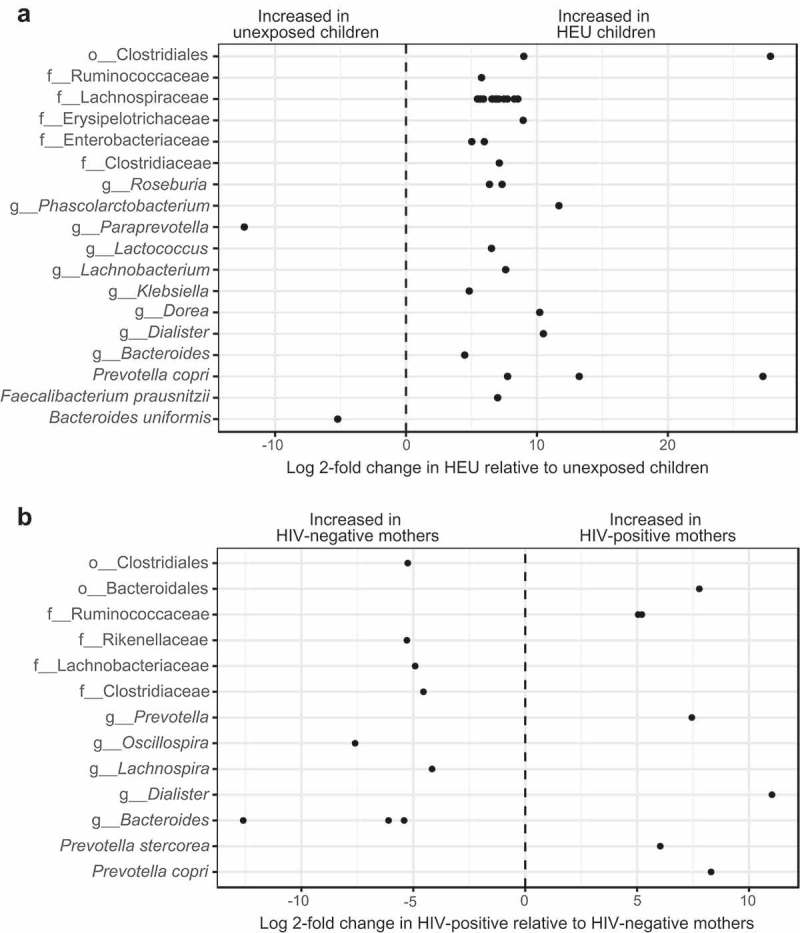
Differentially abundant OTUs in the gut microbiome of mothers and children. Log 2-fold change of differentially abundant OTUs between children (a) and mothers (b) categorized by the highest taxonomic level identified. Each dot corresponds to an OTU. “o” = Order; “f” = Family; “g” = Genera.
To evaluate the relationships between mother–child gut microbiota, we used SourceTracker,27 a tool to perform estimations of source proportions using Bayesian modeling of uncertainty based on known and unknown source environments. Although the majority of infant microbiota was considered from unknown sources, we observed that a relatively large proportion of the microbiota of HIV-exposed children could be tracked to HIV-infected mothers, with a smaller proportion of the microbiota from unexposed children being traceable to their respective mothers. These results could indicate a stronger similarity in gut microbiota between HEU children and HIV-infected mothers compared to unexposed children and their mothers. However, those differences did not meet the threshold of statistical significance (p > 0.05) due to the high contribution of the unknown sources to the overall gut microbiota (Figure 1D).
To further analyze the interactions involving the gut microbiome, functional profiles were inferred based on 16S data using PICRUst. It was possible to identify nine KEGG orthologs that differed between HEU and unexposed children and 13 that differed between HIV-positive and HIV-negative mothers (FDR-adjusted p-value <0.05). This indicates that the alterations in the gut microbiome of HEU children occur not only in microbial abundance but may also occur at the functional level.
Mothers and children present similar diet independent of HIV exposure
Differences in food consumption among the groups were evaluated according to answers to the food frequency questionnaire (FFQ). Food consumption pattern was similar between the two groups of children and the two groups of mothers (Figure 3A and B). The only statistically significant difference was in alcohol consumption (wine and beer excluded) between HIV-positive and HIV-negative mothers (p = 0.035), with HIV-positive mothers reporting increased consumption of alcohol (Figure 3B).
Figure 3.
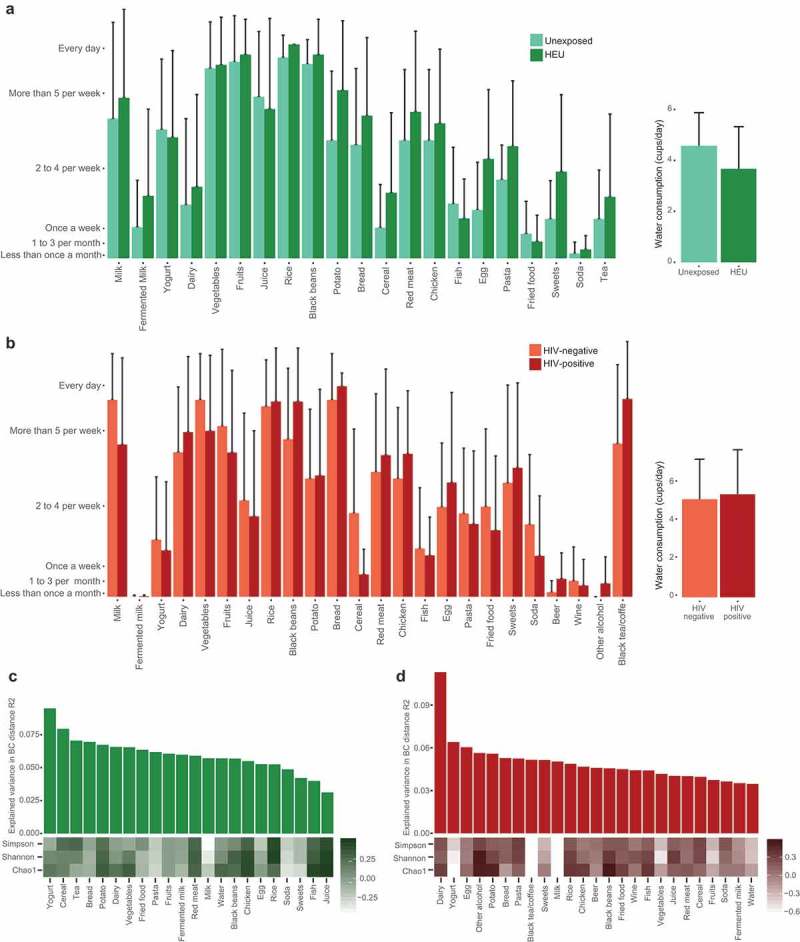
Food and microbiota diversity relationship. Weekly frequency of food consumption for children (a) and mothers (b). Relationship between consumption of individual food items and richness and diversity estimates for children (c) and mothers (d). Bar plots represent the Bray Curtis beta diversity explained variation for each food item from the food frequency questionnaire. The heatmap represents the correlation coefficients of food items and richness (Chao1) and diversity (Shannon and Simpson) estimates.
To better understand the impact of diet on the microbial community structure in different groups, correlation analyses were performed between diversity and richness estimates and the frequency of consumption of each food item. However, no significant correlations were found for children and mothers (FDR-adjusted p-value >0.1). To analyze the impact of diet on the interindividual variance of the microbiota, we next sought to evaluate the association of interindividual variation and the frequency of food consumption through the explained variation (R2), according to the BC distance for each food from the FFQ. The food items that presented the highest association to microbiome interindividual variance were yogurt for children and dairy for mothers (Figure 3C and D). These analyses indicate that diet did not have a strong influence in gut microbiota diversity in the studied groups, possibly due to the low variability in the food consumption pattern among individuals.
We also evaluated the relationship between the frequency of food consumption and abundance of individual OTUs, with results revealing a negative correlation between Bacteroides ovatus and fruit consumption in children (FDR-adjusted p < 0.05). In mothers, OTUs belonging to Veillonella dispar and Ruminococcus sp. and an OTU from the Order Bacteroidales correlated negatively with bread consumption, while OTUs from Oscillospira sp. and Anaerostipes sp. and two OTUs belonging to the family Ruminococcaceae correlated positively with alcohol consumption (FDR-adjusted p < 0.05). However, none of the OTUs which were found to be statistically different between the two groups of children or the two groups of mothers presented correlation with food consumption, indicating that the differences observed among the groups were not caused by differences in food consumption.
HEU children do not present alterations in microbial translocation and inflammation markers in plasma
Since alterations in gut microbiota are related to microbial translocation and immune activation, plasmatic markers of microbial translocation and inflammation were also evaluated. Although HEU children showed increased levels of IL-6, IL-8, and IL-10 compared to unexposed children, these differences were not statistically significant (Figure 4A). An equivalent result was observed for C-reactive protein (p = 0.08) (assessed for seven HEU and four unexposed children) (Figure 4B), and no statistical differences were also observed for lipopolysaccharide-binding protein (LBP) and the monocyte activation marker soluble CD14 (Figure 4D and E). Spearman correlation tests were performed in order to evaluate the relationship between each of the plasmatic markers assessed and: (i) individual OTU abundance, (ii) abundance at the family level, and (iii) alpha diversity estimates. However, no statistically significant correlations (FDR-adjusted p-value >0.05) were observed. HIV-infected and uninfected mothers showed similar levels of inflammatory cytokines and LBP (Figure 4C and D). However, HIV-infected mothers showed increased sCD14 plasma levels when compared to uninfected mothers (p = 0.02) (Figure 4E).
Figure 4.
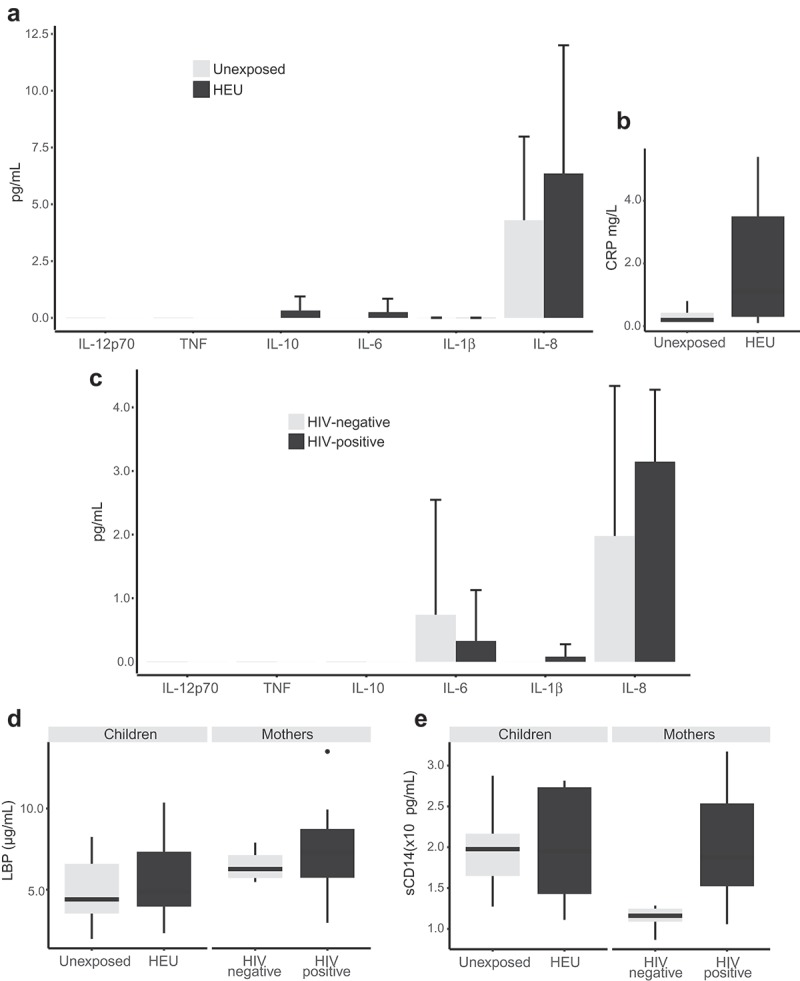
Determination of microbial translocation and inflammation markers in plasma from children and mothers. Inflammatory cytokines (a) and C-reactive protein (b) in children; inflammatory cytokines (c) in mothers; lipopolysaccharide-binding protein (LBP) (d) and soluble CD14 (sCD14) (e) for mothers and children.
Discussion
Here, we evaluated the gut microbiota composition, diet habits, and microbial translocation markers in peripheral circulation in HEU children and their mothers and compared them to unexposed children and their mothers. Although we could not find differences in microbiota alpha and beta diversity or in microbial translocation and inflammation markers, we did find several OTUs that were differentially abundant between the studied groups, indicating that these individuals show compositional alterations in the gut microbiota.
Several studies report an increased rate of infections and hospital admissions in HEU children.4,28,29 In this study, the rate of infections was evaluated through mother’s answers to a survey. Although this is a general way to evaluate the rate of infections and mothers may differ in their perception, the reports indicate an increased frequency of diseases in HEU children compared to unexposed children, in accordance with the literature. We did not observe statistically significant differences between HEU and unexposed children with regard to weight and weight-for-age z score. Although previous studies show that HEU children present lower weight and weight-for-age z scores when compared to unexposed children,30-32 there are still uncertainties whether these differences are due to HIV exposure or if there are other causes such as socioeconomic or behavioral factors that explain the reduced physical development of HEU children.28,31
Analysis of the gut microbiome composition showed that the most abundant bacteria family in the two groups of children was Lachnospiraceae, while the most abundant genus was Bacteroides, similarly to a previously reported study on children’s gut microbiota.33 Moreover, no differences were observed between HEU and unexposed children regarding alpha and beta diversity. However, analysis of the differentially abundant OTUs between the two groups of children in our study revealed that HEU children present increased proportions of OTUs belonging to the families Lachnospiraceae, Prevotellaceae, Ruminococcaceae, Enterobacteriaceae, Veillonellaceae, Clostridiaceae, Streptococcaceae, and Erysipelotrichaceae, with the majority of differentially abundant OTUs belonging to the family Lachnospiraceae. A decrease in the proportion of bacteria from the Lachnospiraceae family has been previously reported in HIV-infected individuals when compared to uninfected controls.12,15,16 However, an increased proportion of bacteria from the Lachnospiraceae family has previously been observed by Bender et al. (2016)6 in HEU children when compared to unexposed children, but this difference was not considered the statistically significant difference. It has been described that several members of the Lachnospiraceae family produce short-chain fatty acids, especially butyric acid34, which is essential to the maintenance of the gut epithelial barrier and related to the modulation of immune responses, contributing to gut microbiota homeostasis35. Therefore, increased proportions of OTUs belonging to Lachnospiraceae could be an indication that alterations in the gut microbiota of HEU children are not necessarily indicators of a pathogenic state. The fact that HEU children in our study did not show alterations in microbial translocation markers also corroborates this hypothesis.
In his comparison of the gut microbiome composition between HEU and unexposed children, Bender et al. (2016)6 reported abundance and diversity results that slightly differ from ours. However, the alterations in the microbiome of HEU children reported by him were related to an altered composition of human milk oligosaccharides (HMOs) in the breast milk of HIV-positive mothers.6 HEU children in our study were not breastfed, which may be the reason for these discrepant results. It is also relevant to notice note that, when compared to the results observed by Bender et al., our data indicate that HMOs are not the only cause of dysbiosis in HEU children. Therefore, it is possible that the alterations in the microbiota of these children are related to alterations in their mothers. Our results on the source proportions estimative with SourceTracker27 further indicate that the HEU children’s gut microbiota suffer, at least in part, some influence from their mother’s altered gut microbiota.
We also observed differences between HIV-infected and uninfected mothers in the abundance of some taxa, but not in richness or alpha and beta diversity. This was unexpected since alterations in gut microbiome richness and diversity of HIV-infected individuals have been described in several cases.11,15,36 However, the impact of antiretroviral therapy on the gut microbiota is still not entirely clear, with some reports showing that diversity alterations in the gut microbiota can remain after antiretroviral treatment37,38 and others showing that treatment can revert these changes to a point where diversity indexe of the microbiota of HIV-infected individuals is similar to the one of uninfected individuals.14 Therefore, it is possible that the absence of alterations in diversity indexes in this report is related to the fact that the majority of HIV-infected women in our study had been receiving antiretroviral therapy for an extended period and presented undetectable viral loads.
We did observe differences in OTU abundance between HIV-infected and uninfected mothers, with OTUs classified in the families Rickenellaceae, Lachnospiraceae, Clostridiaceae, and Bacteroidaceae increased in HIV-negative mothers and OTUs from Prevotellaceae and Paraprevotellaceae increased in HIV-positive mothers. Moreover, the most abundant family in mothers’ gut microbiota differed between those who are HIV positive and HIV negative, with Ruminococcaceae more abundant in HIV negative and Prevotellaceae in HIV positive. In accordance, an increased proportion of Prevotellaceae and a decrease in the proportion of Bacteroidaceae have been described in HIV-positive individuals multiple times.16,36,39
We also observed an increased proportion of P. copri in both HIV-infected mothers and HEU children, which could indicate a contribution from the dysbiotic microbiota of HIV-infected mothers to the alterations in their children’s microbiota. It is tempting to speculate that increased P. copri could represent an adaptation of HEU children’s microbiota to ensure Th17 cell development, since this bacterium has been postulated to be the human equivalent of Segmented Filamentous Bacteria (SFB), which is pivotal for murine Th17 T cell development in the gut lamina propria.40,41 The increased presence of Th17-inducer bacteria in the microbiota of HEU children could be associated to the hyper-responsiveness of T-cell-mediated immunity observed in HEU children2 or even interact with host genetics to promote subsequent development of autoimmune disorders, like rheumatoid arthritis.41,42 Further studies will be needed to confirm these observations and to determine the impact of P. copri on HEU children immune system development. Analysis of the predicted functional metagenome further indicates that there are alterations in the gut microbiome of HEU children and HIV-infected mothers, although it is important to mention that results represent predictions based on the 16S rRNA data and thus should be interpreted only as an indication of alteration rather than conclusive.
Our study did not reveal a substantial impact of diet on the gut microbiome. There were no significant differences between the diet habits of HEU and unexposed children, and few differences were observed between the two groups of mothers. We also evaluated the impact of diet on microbiome composition of our groups, and it revealed a low impact of the food items evaluated in both alpha and beta diversity estimates. Although diet is a well-recognized and important factor that modulates gut microbiota,43 the fact that we did not observe a major influence may be due to low variability in diet among individuals of the studied groups and their homogeneity regarding the origin and socioeconomic profile.
Even though some studies report alterations in the immune response of HEU children,2,20 HEU and unexposed children in our evaluations did not present significant differences regarding the presence of soluble markers in plasma. A previous study with HEU children from Brazil also revealed similar levels of microbial translocation and inflammation markers in plasma of HEU and unexposed children.44 In HIV-infected individuals, alterations in gut microbiota are generally linked to increased microbial translocation.18 Although we show that HEU children present differences in gut microbiota composition, there were no signs of microbial translocation, indicating that the microbiota alterations observed in HEU children are not directly linked to microbial translocation.
There were also no significant differences between HIV-infected and uninfected mothers in the concentration of plasmatic inflammatory cytokines. Increased immune activation and inflammation are a characteristic of HIV infection, but it has been reported that viral load suppression with prolonged antiretroviral therapy can partially or even entirely reverse pro-inflammatory pathways in HIV-infected individuals.45 The mothers who participated in this study were in good health, including the HIV-infected ones who presented undetectable viral loads and high TCD4+ and TCD8+ cell counts, possibly explaining the equivalent levels of inflammatory cytokines in plasma relative to HIV-negative mothers. However, immune activation was not entirely suppressed by antiretroviral therapy as HIV-infected mothers still showed increased soluble CD14 in plasma. Both results, normal T cell counts and increased sCD14 in plasma in HIV+ mothers with undetectable viral loads and under retroviral therapy, were also observed by Villanueva-Millan et al.46
We recognize that our study presents limitations. This was a cross-sectional study with a small number of participants in a single location. Moreover, some important characteristics such as age, mode of delivery, and breastfeeding were variable between the groups. Nonetheless, efforts were made to recruit unexposed children who were not breastfed for an extended period, but they were still breastfed during the first months of life. HEU children, in contrast, received supplement formula since birth, following guidelines of the Brazilian Ministry of Health making it difficult to match the characteristics between HEU and unexposed children.
Previous studies have compared the gut microbiota of HEU children to that of unexposed children.6,25 However, to the best of our knowledge, these studies were not conducted in settings where children were not breastfed. The results presented here are relevant as they show that even in a region where breastfeeding is not recommended for HIV-infected mothers, their children still show a slightly altered microbiota. This warrants further evaluations, especially of extensive prospective studies, with regard to the implications for HEU children’s health given the differences observed in their microbiota composition. Based on these results, it could also be interesting to plan the introduction of probiotics or prebiotics as supplementation to infant formulas. The use of probiotics in infant formulas for HIV-exposed infants has already been tested and demonstrated to be safe.47 A better understanding of the gut microbial composition may point to new bacteria that could be used as a supplement with the goal to improve the health of HEU children.
Methods
Study population and inclusion criteria
This was a cross-sectional study in which the study population was comprised of 19 women and their children with age between 16 and 26 months. Mother–child pairs were grouped according to maternal HIV infection status as follows: (i) HIV-exposed uninfected children and their HIV-positive mothers (n = 12) and (ii) HIV-unexposed uninfected children and their HIV-negative mothers (n = 7). Following the Brazilian guidelines that HIV-exposed children should not be breastfed,48 efforts were made to recruit only unexposed uninfected children that had not been breastfed for at least 1 y, in order to minimize bias in the gut microbiota evaluation. The Hospital Infantil Joana de Gusmão Ethics Committee approved this study under the protocol number 2.022.007, and all volunteers provided written informed consent before enrollment.
Sample collection and processing
Mothers were recruited during children’s routine appointments at the Hospital Infantil Joana de Gusmão in Florianópolis, SC, Brazil. All mothers signed a consent form, answered a survey about life and nutritional habits, and received containers and instructions to perform the stool collection from themselves and their children. Samples were then collected either at the volunteers’ residence or at the hospital and kept frozen at −20°C until arrival at the laboratory, where DNA extraction was performed with the commercial kit QIAamp DNA Stool Mini Kit (Qiagen, 51504) and stored at −20°C. A sample of sterile water was used as negative control for the DNA extraction and submitted to the same procedures as the biological samples. For microbial translocation markers and inflammatory cytokine assessment, blood samples were also collected from mothers and children at the time of recruitment, and plasma was separated and stored at −80°C. Information about height, T CD4+ and T CD8+ cell counts, viral load, and albumin plasma levels were obtained from patients’ hospital records. For the evaluation of participants’ diet, a short FFQ was created based on literature covering foods that might influence on the microbiota49 and typical and most consumed food in the region of study. Weight-for-age z scores were calculated according to WHO child growth standards.50
Library preparation and sequencing
DNA was quantified with the Qubit dsDNA HS kit (Thermo Fisher Scientific, Q32851) and amplified with the Illumina primers S-D-Bact-0341-b-S-17 (forward) and S-D-Bact-0785-a-A-21 (reverse)51 targeting the V3 and V4 regions of the 16S rRNA. Polymerase chain reactions were performed with 10 µL of the polymerase GoTaq Colorless Master Mix 2x (Promega, M7143), 1.0 μL of the forward primer, 1.0 μL of the reverse primer, 40 ng of the extracted DNA, and ultrapure sterile water as necessary to reach the final volume of 20 μL. Amplification conditions consisted in an initial denaturation at 94°C for 3 min, followed by 28 cycles of denaturation at 94°C for 30 s, annealing at 53°C for 1 min, and extension at 72°C for 2 min, with a final extension period at 72°C for 6 min. Amplification was performed with positive and negative controls and confirmed through electrophoresis in agarose gel, with resulting amplicons in the size of approximately 500 base-pairs on positive and sample reactions. For library preparation, PCR product was purified with AMPure XP (Beckman Coulter, A63881), adaptors for sequence identification were added with the commercial kit Nextera XT Index (Illumina, FC-131–2001), and samples were quantified through qPCR with the KAPA Universal Library Quantification Kit (Illumina, KK4824) commercial kit. After quantification, a 3 nM equimolar pool was prepared and sequencing was performed with all samples multiplexed in a single run, using a V2 reagent kit 2 × 250 bp (500 cycles) (Illumina, MS-102–2003) at the Illumina MiSeq platform.
Bioinformatics analysis
Low-quality sequences were filtered with Trimmomatic v0.3652 according to sequence size and Phred score. Nucleotides with a Phred score under 20 at the beginning and end of each sequence, or groups of nucleotides that showed a mean Phred score every five nucleotides inferior to 20, were considered low quality and thus removed. Sequences shorter than 100 nucleotides were also removed. Paired-end sequences were merged with PEAR v0.9.8 (2015-04-09),53 and the resulting contigs were filtered to maintain only sequences with between 435 and 468 bp, which corresponded to the most frequent sequence length, in order to remove sequences that were too short or that had issues during pairing. Chimera removal, determination of OTUs, and taxonomic assignment were performed as described elsewhere.54 Briefly, to search for and remove chimeric sequences, we used a script developed by Comeau, Douglas, and Langille54 that uses VSEARCH v1.11.1 (13 April 2016)55 (implementing the UCHIME56 algorithm) to filter out chimeric reads from multiple files. We classified the reads into OTUs at 97% identity using a QIIME 1 v1.9.157 pipeline for open-reference OTU picking which includes the open-source methods SortMeRNA v2.0-dev (2014-10-31)58 for reference picking and SUMACLUST v1.0.0059 for the de novo OTU picking. The resulting OTU table was filtered to remove unwanted OTUs, including those resulting from unfiltered chimeras, clusters containing only one sequence (singletons), and low-confidence OTUs, using a dynamic cutoff of less than 0.1% of the total number of sequences. Finally, the OTU table was normalized per sample by subsampling so that all samples have the same depth (15,722 reads). Taxonomy determination was performed using the 13.8 release of the Greengenes database. The functional prediction was based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) and performed with PICRUSt v1.1.2 (30 August 2017)60 using data normalized considering 16S rRNA gene copy number to predict KEGG orthologs and KEGG pathways.
Microbial translocation markers and inflammatory cytokine assessment
To estimate microbial translocation, plasma soluble CD14 (sCD14) and LBP were quantified from plasma samples in triplicate with commercially available kits ELISA-sCD14 DuoSet (R&D Systems, Minneapolis, USA) and Human LBP DuoSet ELISA (R&D Systems), respectively. Inflammatory cytokines IL-12p70, TNF, IL-10, IL-6, IL-1β, and IL-8 were assessed through cytometric bead array with a Human Inflammatory Cytokines Kit (BD Biosciences, Franklin Lakes, USA). Information about C-reactive protein levels was obtained from hospital records.
Statistical analysis
Statistical analyses were performed in R.61 The significance of quantitative demographic data was tested with Mann–Whitney–Wilcoxon and Student's t-tests for nonparametric and parametric variables, respectively. For qualitative variables, Fisher’s exact test was applied. Diversity estimates were performed with the phyloseq R package.62 To test the statistical significance of alpha diversity estimates among groups of mothers and children, a Student’s t-test was applied, and to analyze beta diversity statistical significance, an Adonis test with 999 permutations was applied. All correlations were evaluated through Spearman rank correlation. To identify taxa that were differentially abundant between groups, an OTU table not normalized was used. OTUs with zero counts in more than 80% of the samples were removed, and a differential analysis test based on the negative binomial distribution was performed with the R package DESeq2.63 OTUs with an adjusted p-value inferior to 0.05 according to Wald binomial significance test with Benjamini–Hochberg correction were considered as statistically different. Differences in the predicted functional metagenome were also analyzed with DESeq2, under the same parameters described above. To evaluate the relationship between mother–child microbiome, we applied a Bayesian model using SourceTracker,27 and differences in the mean proportions between groups were evaluated through Kruskal–Wallis with Turkey post-hoc test.
Funding Statement
No direct funding for the execution of this study was received. AM received a CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) student fellowship. ARP is a CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) scholar.
Abbreviations
- BMI
Body mass index
- HEU
HIV-exposed but uninfected
- HIV
Human immunodeficiency virus
- FFQ
Food frequency questionnaire
- HMOs
Human milk oligosaccharides
- IL
Interleukin
- KEGG
Kyoto encyclopedia of genes and genomes
- LBP
Lipopolysaccharide-binding protein
- OTU
Operational taxonomic unit
- PICRUSt
Phylotypic investigation of communities by reconstruction of unobserved states
- QIIME
Quantitative insights into microbial ecology
- PCR
Polymerase chain reaction
- PEAR
Paired-end read merger
- SFB
Segmented filamentous bacteria
- TNF
Tumor necrosis factor
- FDR
false discovery rate
Acknowledgments
The authors wish to thank all mothers that volunteered to participate in this study and the staff from Hospital Infantil Joana de Gusmão who collected samples from patients. The authors also thank Dr Juliano Bordignon and D Oscar Bruna-Romero for providing reagents and Dr Nadim Ajami for helpful discussion on data analysis.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Ethics statement
This study was approved by the Hospital Infantil Joana de Gusmão Ethics Committee under the protocol number 2.022.007. All volunteers were informed about the study and provided written consent prior to enrollment.
Availability of data
The sequence data supporting the results of this study are available in NCBI sequence read archive (SRA) under accession SRP144334.
Authors’ contributions
AM designed the study, recruited volunteers, processed samples, analyzed the data, and wrote the manuscript. ARP and CRZB designed the study, supervised the work, and revised the manuscript. RD helped with data analysis and revised the manuscript. MMSP designed the study and recruited volunteers. All authors read and approved the final version of the manuscript.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
References
- 1.UNAIDS On the fast-track to an AIDS-free generation. Geneva: 2016. [accessed 2017 December18] http://www.unaids.org/sites/default/files/media_asset/GlobalPlan2016_en.pdf.
- 2.Abu-Raya B, Kollmann TR, Marchant A, MacGillivray DM.. The immune system of HIV-exposed uninfected infants. Front Immunol. 2016;7:1–10. doi: 10.3389/fimmu.2016.00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arikawa S, Rollins N, Newell ML, Becquet R. Mortality risk and associated factors in HIV-exposed, uninfected children. Trop Med Int Heal. 2016;21:720–734. doi: 10.1111/tmi.12695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slogrove AL, Goetghebuer T, Cotton MF, Singer J, Bettinger JA. Pattern of infectious morbidity in HIV-exposed uninfected infants and children. Front Immunol. 2016;7:1–8. doi: 10.3389/fimmu.2016.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans C, Humphrey JH, Ntozini R, Prendergast AJ. HIV-exposed uninfected infants in Zimbabwe: insights into health outcomes in the pre-antiretroviral therapy era. Front Immunol. 2016;7:190. doi: 10.3389/fimmu.2016.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bender JM, Li F, Martelly S, Byrt E, Rouzier V, Leo M, Tobin N, Pannaraj PS, Adisetiyo H, Rollie A, et al. Maternal HIV infection influences the microbiome of HIV-uninfected infants. Sci Transl Med. 2016;8:1–12. doi: 10.1126/scitranslmed.aaf0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lozupone CA, Rhodes ME, Neff CP, Fontenot AP, Campbell TB, Palmer BE. HIV-induced alteration in gut microbiota. Gut Microbes. 2014;5:562–570. doi: 10.4161/gmic.32132. [DOI] [PubMed] [Google Scholar]
- 8.Pinto-Cardoso S, Klatt NR, Reyes-Terán G. Impact of antiretroviral drugs on the microbiome: unknown answers to important questions. Curr Opin HIV AIDS. 2018;13:53–60. doi: 10.1097/COH.0000000000000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li S, Armstrong A, Neff C, Shaffer M, Lozupone C, Palmer B. Complexities of gut microbiome dysbiosis in the context of HIV infection and antiretroviral therapy. Clin Pharmacol Ther. 2016;99:600–611. doi: 10.1002/cpt.v99.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noguera-Julian M, Rocafort M, Guillén Y, Rivera J, Casadellà M, Nowak P, Hildebrand F, Zeller G, Parera M, Bellido R, et al. Gut microbiota linked to sexual preference and HIV infection. EBioMedicine. 2016;5:135–146. doi: 10.1016/j.ebiom.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nowak P, Troseid M, Avershina E, Barqasho B, Neogi U, Holm K, Hov JR, Noyan K, Vesterbacka J, Svärd J, et al. Gut microbiota diversity predicts immune status in HIV-1 infection. AIDS. 2015;29:2409–2418. doi: 10.1097/QAD.0000000000000869. [DOI] [PubMed] [Google Scholar]
- 12.Sun Y, Ma Y, Lin P, Tang Y-W, Yang L, Shen Y, Zhang R, Liu L, Cheng J, Shao J, et al. Fecal bacterial microbiome diversity in chronic HIV-infected patients in China. Emerg Microbes Infect. 2016;5:1–7. doi: 10.1038/emi.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang L, Poles MA, Fisch GS, Ma Y, Nossa C, Phelan JA, Pei Z. HIV-induced immunosuppression is associated with colonization of the proximal gut by environmental bacteria. AIDS. 2016;30:19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McHardy IH, Li X, Tong M, Ruegger P, Jacobs J, Borneman J, Anton P, Braun J. HIV Infection is associated with compositional and functional shifts in the rectal mucosal microbiota. Microbiome. 2013;1(1):12. doi: 10.1186/2049-2618-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mutlu EA, Keshavarzian A, Losurdo J, Swanson G, Siewe B, Forsyth C, French A, Demarais P, Sun Y, Koenig L, et al. A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS Pathog. 2014;10:1–18. doi: 10.1371/journal.ppat.1003829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dillon SM, Lee EJ, Kotter CV, Austin GL, Dong Z, Hecht DK, Gianella S, Siewe B, Smith DM, Landay AL, et al. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol. 2014;7:983–994. doi: 10.1038/mi.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dinh DM, Volpe GE, Duffalo C, Bhalchandra S, Tai AK, Kane AV, Wanke CA, Ward HD. Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J Infect Dis. 2015;211:19–27. doi: 10.1093/infdis/jiu409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zevin AS, Mckinnon L, Burgener A, Klatt NR. Microbial translocation and microbiome dysbiosis in HIV- associated immune activation. Curr Opin HIV AIDS. 2016;11:182–190. doi: 10.1097/COH.0000000000000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, Kazzaz Z, Bornstein E, Lambotte O, Altmann D, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 20.Afran L, Garcia Knight M, Nduati E, Urban BC, Heyderman RS, Rowland-Jones SL. HIV-exposed uninfected children: A growing population with a vulnerable immune system? Clin. Exp Immunol. 2014;176:11–22. doi: 10.1111/cei.12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jenmalm MC. The mother-offspring dyad: microbial transmission, immune interactions and allergy development. J Intern Med. 2017;282:484–495. doi: 10.1111/joim.12652. [DOI] [PubMed] [Google Scholar]
- 22.Romano-Keeler J, Weitkamp JH. Maternal influences on fetal microbial colonization and immune development. Pediatr Res. 2015;77:189–195. doi: 10.1038/pr.2014.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107:11971–11975. doi: 10.1073/pnas.0910097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams B, Landay A, Presti R. Microbiome alterations in HIV infection a review. Cell Microbiol. 2016;18:645–651. doi: 10.1111/cmi.12570. [DOI] [PubMed] [Google Scholar]
- 25.González R, Mandomando I, Fumadó V, Sacoor C, Macete E, Alonso PL, Menendez C, Uversky VN. Breast milk and gut microbiota in African mothers and infants from an area of high HIV prevalence. PLoS One. 2013;8:1–9. doi: 10.1371/journal.pone.0080299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta VK, Paul S, Dutta C. Geography, ethnicity or subsistence-specific variations in human microbiome composition and diversity. Front Microbiol. 2017;8. doi: 10.3389/fmicb.2017.01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG, Bushman FD, Knight R, Kelley ST. Bayesian community-wide culture-independent microbial source tracking. Nat Methods. 2011;8:761–763. doi: 10.1038/nmeth.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Roux SM, Abrams EJ, Nguyen K, Myer L. Clinical outcomes of HIV-exposed, HIV-uninfected children in sub-Saharan Africa. Trop Med Int Heal. 2016;21:829–845. doi: 10.1111/tmi.12716. [DOI] [PubMed] [Google Scholar]
- 29.Rupérez M, González R, Maculuve S, Quintó L, López-Varela E, Augusto O, Vala A, Nhacolo A, Sevene E, Naniche D, et al. Maternal HIV infection is an important health determinant in non-HIV-infected infants. AIDS. 2017;31:1545–1553. doi: 10.1097/QAD.0000000000001499. [DOI] [PubMed] [Google Scholar]
- 30.Filteau S, Baisley K, Chisenga M, Kasonka L, Gibson RS. Provision of micronutrient-fortified food from 6 months of age does not permit HIV-exposed uninfected Zambian children to catch up in growth to HIV-unexposed children: a randomized controlled trial. JAIDS J Acquir Immune Defic Syndr. 2011;56:166–175. doi: 10.1097/QAI.0b013e318201f6c9. [DOI] [PubMed] [Google Scholar]
- 31.Muhangi L, Lule SA, Mpairwe H, Ndibazza J, Kizza M, Nampijja M, Nakazibwe E, Kihembo M, Elliott AM, Webb EL. Maternal HIV infection and other factors associated with growth outcomes of HIV-uninfected infants in Entebbe, Uganda. Public Health Nutr. 2013;16:1548–1557. doi: 10.1017/S1368980013000499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosala-Hallas A, Bartlett JW, Filteau S. Growth of HIV-exposed uninfected, compared with HIV-unexposed, Zambian children: a longitudinal analysis from infancy to school age. BMC Pediatr. 2017;17:1–9. doi: 10.1186/s12887-016-0759-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim ES, Zhou Y, Zhao G, Bauer IK, Droit L, Ndao IM, Warner BB, Tarr PI, Wang D, Holtz LR. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat Med. 2015;21:1228–1234. doi: 10.1038/nm.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meehan CJ, Beiko RG. A phylogenomic view of ecological specialization in the Lachnospiraceae, a family of digestive tract-associated bacteria. Genome Biol Evol. 2014;6:703–713. doi: 10.1093/gbe/evu037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature. 2016;535:65–74. doi: 10.1038/nature18847. [DOI] [PubMed] [Google Scholar]
- 36.Vázquez-Castellanos JF, Serrano-Villar S, Latorre A, Artacho A, Ferrús ML, Madrid N, Vallejo A, Sainz T, Martínez-Botas J, Ferrando-Martínez S, et al. Altered metabolism of gut microbiota contributes to chronic immune activation in HIV-infected individuals. Mucosal Immunol. 2014;8:760–772. doi: 10.1038/mi.2014.107. [DOI] [PubMed] [Google Scholar]
- 37.Lozupone CA, Li M, Campbell TB, Flores SC, Linderman D, Gebert MJ, Knight R, Fontenot AP, Palmer BE. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe. 2013;14:329–339. doi: 10.1016/j.chom.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Y, Ou Z, Tang X, Zhou Y, Xu H, Wang X, Li K, He J, Du Y, Wang H, et al. Alterations in the gut microbiota of patients with acquired immune deficiency syndrome. J Cell Mol Med. 2018;22:2263–2271. doi: 10.1111/jcmm.13508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paquin-Proulx D, Ching C, Vujkovic-Cvijin I, Fadrosh D, Loh L, Huang Y, Somsouk M, Lynch SV, Hunt PW, Nixon DF, et al. Bacteroides are associated with GALT iNKT cell function and reduction of microbial translocation in HIV-1 infection. Polym J. 2016;10:69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, Rostron T, Cerundolo V, Pamer EG, Abramson SB, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;2:1–20. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maeda Y, Kurakawa T, Umemoto E, Motooka D, Ito Y, Gotoh K, Hirota K, Matsushita M, Furuta Y, Narazaki M, et al. Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheumatol. 2016;68:2646–2661. doi: 10.1002/art.39783. [DOI] [PubMed] [Google Scholar]
- 43.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyamoto M, Ono E, Pahwa S, Moraes-Pinto MID. Immune development in HIV-exposed uninfected children born to HIV-infected women maristela. Rev Inst Med Trop Sao Paulo. 2017;59:1–9. doi: 10.1590/s1678-9946201759030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deeks SG, Tracy R, Douek DC. Systemic effects of inflammation on health during chronic HIV infection. Immunity. 2013;39:633–645. doi: 10.1016/j.immuni.2013.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Villanueva-Millán MJ, Pérez-Matute P, Recio-Fernández E, Lezana Rosales JM, Oteo JA. Differential effects of antiretrovirals on microbial translocation and gut microbiota composition of HIV-infected patients. J Int AIDS Soc. 2017;20:1–13. doi: 10.7448/IAS.20.1.21526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cooper P, Bolton KD, Velaphi S, De Groot N, Emady-Azar S, Pecquet S, Steenhout P. Early benefits of a starter formula enriched in prebiotics and probiotics on the gut microbiota of healthy infants born to HIV+ mothers: a randomized double-blind controlled trial. Clin Med Insights Pediatr. 2016;10:119–130. doi: 10.4137/CMPed.S40134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ministério da Saúde Protocolo clínico e diretrizes terapêuticas para manejo da infecção pelo HIV em crianças e adolescentes. 1st ed. Brasilia/DF, Brazil; 2017. [Google Scholar]
- 49.Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, Mujagic Z, Vila AV, Falony G, Vieira-Silva S, et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science (80-). 2016;352:565–569. doi: 10.1126/science.aad3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.World Health Organization WHO Child Growth Standards R igrowup package. 2011. [accessed 2018 June19] 1–12 http://www.who.int/childgrowth/software/readme_r.pdf?ua=1
- 51.Klindworth A, Pruesse E, Schweer T, Rg Peplies J, Quast C, Horn M, Glö Ckner FO. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:1–11. doi: 10.1093/nar/gks1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang J, Kobert K, Flouri TŠ, Stamatakis A. PEAR: a fast and accurate illumina paired-end read merger. Genome Anal. 2014;30:614–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Comeau AM, Douglas GM, Langille MGI. Microbiome helper: a custom and streamlined workflow for microbiome research. mSystems. 2017;2:e00127–16. [assessed 2018 June19] http://msystems.asm.org/content/msys/2/1/e00127-16.full.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pẽa AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kopylova E, Noé L, Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28:3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 59.Mercier C, Boyer F, Bonin A, Coissac E. SUMATRA and SUMACLUST : fast and exact comparison and clustering of sequences. 2013. [accessed 2018 June19] http://metabarcoding.org/sumatra/
- 60.Langille M, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes J, Clemente J, Burkepile D, Vega Thurber R, Knight R, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.R Core Team R: A language and environment for statistical computing. R Found Stat Comput. 2017;739:409 [assessed 2018 June19] https://www.r-project.org/. [Google Scholar]
- 62.McMurdie PJ, Holmes S, Watson M. An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:1–8. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- UNAIDS On the fast-track to an AIDS-free generation. Geneva: 2016. [accessed 2017 December18] http://www.unaids.org/sites/default/files/media_asset/GlobalPlan2016_en.pdf.
Data Availability Statement
The sequence data supporting the results of this study are available in NCBI sequence read archive (SRA) under accession SRP144334.


