Abstract
Alterations in gut bacterial homeostasis result in changes in intestinal metabolites. To investigate the effects of alcohol on fecal metabolites and the role of cathelicidin-related antimicrobial peptide (CRAMP) in alcoholic liver disease (ALD), CRAMP knockout (KO) and their control wild type (WT) mice were fed a Lieber-DeCarli liquid diet with or without alcohol. Polar metabolites in mouse feces were analyzed by GC × GC-MS and 2DLC-MS, and the concentrations of short chain fatty acids (SCFAs) were measured by GC-MS. A total of 95 and 190 metabolites were detected by GC × GC-MS and 2DLC-MS, respectively. Among the significantly changed metabolites, taurine and nicotinic acid were decreased in WT mice fed alcohol, which were also down-regulated in KO mice fed without alcohol. Interestingly, these two metabolites were increased in KO mice fed alcohol compared to them in WT controls. Additionally, SCFAs were significantly decreased in WT mice fed alcohol and in KO mice fed without alcohol, whereas two branched-chain SCFAs were increased by alcohol treatment in KO mice. In summary, the analytical platforms employed in this study successfully dissected the alterations of polar metabolites and SCFAs in fecal samples, which helped understand the effects of alcohol consumption and CRAMP in intestinal metabolism and alcohol-induced liver injury.
Keywords: short chain fatty acids, alcoholic liver disease, cathelicidin-related antimicrobial peptide, taurine, nicotinic acid, metabolomics
Graphical Abstract
1. INTRODUCTION
Alcoholic liver disease (ALD) is a major cause of chronic liver disease and encompasses a broad spectrum of disorders including steatosis, steatohepatitis, fibrosis, and cirrhosis. ALD is characterized by gut dysbiosis and increased intestinal permeability, which lead to increased translocation of bacteria and their products, such as lipopolysaccharide (LPS), into the portal circulation and consequent activation of NFκB signaling pathways in the liver. The activation of NFκB causes an increased production of reactive oxygen species and proinflammatory factors that contribute to the development of liver injury.1,2 Accumulating evidence has shown that intestinal microbiota play a key role in gut metabolite regulation. As an example, short chain fatty acids (SCFAs) are critical in epithelial energy metabolism and host cell epigenetic regulation.3 Studies have shown that alcohol feeding decreased fecal SCFAs in mice.4
Antimicrobial peptides (AMPs) are endogenous polypeptides produced by a variety of organisms with the majority found in animals. They serve as an essential component of innate immune system of the host to counter microbial infections.5–7 Cathelicidin-related antimicrobial peptide (CRAMP, encoded by the CAMP gene) is an antimicrobial peptide in mice and rats produced by macrophages,8 neutrophils,9 and epithelial cells10 in response to invasive or inflammatory stimuli. CRAMP deficient mice are more sensitive to bacterial infection compared to wild type mice.10 CRAMP binds directly to bacterial cell membranes, kills bacteria by disrupting the membrane structure, and causes cytoplasm leakage.8,11 CRAMP also neutralizes microbial-derived endotoxins such as LPS and then suppresses inflammation.12–14 Studies also identified CRAMP as immune modulators in metabolic disease. CRAMP produced by β-cell in the pancreatic islets induces regulatory macrophages and dendritic cells (DCs), generating the pancreatic regulatory T cells; whereas in the absence or deficient of CRAMP, the macrophags and dendritic cells exhibit an inflammatory profile thus generating diabetogenic T cells, which attacks the pancrease and destroys the insulin-secreting cells, leading to decreased insulin production and type I diabetes. Notably, the production of CRAMP is controlled by gut derived SCFAs, which again demonstrated the importance of gut microbiota in the CRAMP regulation.15 Studies also demonstrated metabolic effects of CRAMP in the development of fatty liver in diabetic mice. CRAMP has been shown to suppress lipid accumulation and hepatic steatosis in high-fat diet (HFD) treated diabetic mice,16 suggesting that CRAMP may play a role in alcohol-induced hepatic steatosis. Our previous work showed that the intestinal CAMP mRNA expression was decreased in mice with ALD,17 indicating that CRAMP may play an important role in ALD. Further study showed that deficiency of CRAMP exacerbated ALD in mice (to be published separately). Therefore, examining the gut metabolite alterations will add valuable information for understanding the role of CRAMP in alcohol exposure and ALD.
The aim of this study was to analyze the fecal metabolite changes induced by alcohol consumption and CRAMP depletion in mice. Wild type and CRAMP knockout mice were treated with or without chronic-binge alcohol exposure. The feces of all mice were collected and analyzed. To increase metabolite coverage, the polar metabolites extracted from each fecal sample were analyzed by comprehensive two-dimensional gas chromatography mass spectrometry (GC × GC-MS) and parallel two-dimensional liquid chromatography mass spectrometry (2DLC-MS), respectively. SCFAs in each fecal sample were quantified by gas chromatography mass spectrometry (GC-MS) via selected ion monitoring (SIM).
2. MATERIALS AND METHODS
2.1. Chemicals and Reagents
A total of 205 polar metabolite standards were purchased from Sigma-Aldrich Corp. (St. Louis, MO, U.S.A.), Fisher Scientific (Loughborough, U.K.), and Cayman Chemical (Ann Arbor, MI, U.S.A.). Eight SCFA standards (sodium formate, sodium acetate, sodium propionate, sodium butyrate, isobutyric acid, sodium pentanoate, 2-methylbutyric acid, and isovaleric acid), 2,3,4,5,6-pentafluorobenzyl bromide (PFBBr), methoxyamine hydrochloride (MH), and N-(tert-butyldimethylsilyl)-N-methyltrifluoroacetamide (MTBSTFA) were purchased from Sigma-Aldrich Corp.
2.2. Animal and Biological Sample Preparation
Eight-week-old male C57BL/6J wild type (WT) mice were purchased from The Jackson Laboratory (Bar Harbor, ME, U.S.A.). The CRAMP knockout (KO) mice were crossed back to C57BL/6J background before establishing the colony. Mice were housed in a pathogen-free barrier facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care. The procedures of animal care were approved by the University of Louisville Institutional Animal Care and Use Committee. Mice were subjected to chronic-binge alcohol feeding as described previously18 with modification. Mice were fed Lieber DeCarli liquid diet containing 5% alcohol (alcohol-fed, AF) or isocaloric maltose dextrin (pair-fed, PF) for 24 days. At day 24, mice were gavaged with a bolus of alcohol at 5 g/kg 6 h before sacrificing. Four animal groups, PF-WT (n = 5), AF-WT (n = 9), PF-KO (n = 5), and AF-KO (n = 5) were, therefore, included in this study. After 24-day dietary intervention, mice were anesthetized with ketamine/xylazine (100/15 mg/kg i.m.), feces were collected from mouse colons, immediately frozen in liquid nitrogen, and stored in a 1.5 mL Eppendorf tube at −80 °C.
2.3. Polar Metabolite Extraction and Derivatization
All sample processing procedures were performed at 4 °C to minimize the loss of volatile metabolites, unless stated otherwise. To extract polar metabolites for GC × GC-MS and 2DLC-MS analysis, fecal samples were thawed on ice. About 60 mg of mouse feces was weighed and ground in a glass vial. After adding 80% methanol at a ratio of 1:10 (g feces: mL solution), the mixture was sonicated for 20 min in an ultrasonic cleaner followed by centrifugation at 15,300 g for 20 min. The supernatants were collected and divided into two aliquots. One aliquot was lyophilized and redissolved in 20% acetonitrile. After centrifugation, the supernatant was used for 2DLC-MS analysis. A pooled sample was prepared for each group by mixing a small portion of the supernatant from each mouse sample in that group. A total of four pooled samples were prepared.
The other aliquot was derivatized by N-(tert-butyldimethylsilyl)-N-methyltrifluoroacetamide (MTBSTFA) and analyzed by GC × GC-MS. Two hundred microliters of the aliquot of each fecal sample was aspirated into a plastic tube and lyophilized. The metabolite extract was then dissolved in 20 μL of methoxyamine hydrochloride solution (30 mg/mL) and vigorously vortex-mixed for 1 min. Methoxymation was carried out at 60 °C for 1 h. After adding 30 μL of MTBSTFA mixed with 1% tert-butyldimethylchlorosilane (TBDMSCI), derivatization was carried out at 60 °C for 1 h. The derivatized sample was then transferred to a GC vial for analysis. The methoxymation and derivatization were prepared just before GC × GC-MS analysis. A pooled sample was also prepared by mixing a small portion of each derivatized sample.
To extract SCFAs from each fecal sample, about 30 mg of mouse feces was weighed and ground in a 1.5 mL Eppendorf tube. After adding water at a ratio of 1:10 (g feces: mL solution), the mixture was sonicated for 20 min and centrifuged at 15 300g for 20 min. The supernatant was collected and derivatized by pentafluorobenzyl bromide (PFBBr).19 Briefly, 150 μL of supernatant, 100 mM PFBBr in acetone, and 0.5 M phosphate buffer (pH 7.0) were mixed at a ratio of 5:14:2 (v:v:v) to make the acetone:water 14:7 (v:v) in a 2 mL glass tube. After 1 min of vigorous vortex-mixing, the mixture was incubated in a water bath at 60 °C for1.5 h. A total of 150 μL of hexane were added after the mixture cooled to room temperature. The mixture was then vortexed for 3 min followed by centrifugation in a SpeedVac at 3500g for 5 min, and 100 μL of supernatant (hexane phase) was transferred to a 200-μL GC vial for immediate GC-MS analysis. A blank sample prepared using distilled Milli-Q water was also derivatized as a reference for quality control.
2.4. GC × GC-MS Analysis
The LECO Pegasus 4D GC × GC-MS (St. Joseph, MI, U.S.A.) instrument was equipped with a Gerstel MPS2 autosampler and an Agilent 6890 gas chromatograph featuring a LECO two-stage cryogenic modulator and a secondary oven. A DB-5 ms (phenyl arylene polymer virtually equivalent to a (5%-phenyl)-methylpolysiloxane) 60.0 m × 0.25 mm 1dc ×0.25 μm 1df column was used as the first dimension column, and a DB-17 ms ((50%-phenyl)-methylpolysiloxane) 1.0 m × 0.1 mm 2dc ×0.1 μm 2df column was used as the second dimension column. Both columns were purchased from Agilent Technologies (Santa Clara, CA, U.S.A.), and were connected by means of a press-fit connector before the thermal modulator. The flow rate of ultrahigh purity helium carrier gas (99.999%) was set to 2.0 mL/min at a corrected constant flow via pressure ramps with an initial head pressure of 36.9 psi. One microliter sample was injected into the inlet chamber of the GC × GC-MS system and the split ratio was set to 5:1. The inlet temperature was 280 °C. The secondary oven and the thermal modulator were respectively set to +10 °C and +20 °C relative to the primary oven. The MS parameters were: MS range 45–800 m/z; data acquisition rate 200 spectra/s; temperature of ion source chamber 230 °C; temperature of MS transfer line 280 °C; detector voltage 1600 V; and electron energy 70 eV. The acceleration voltage was turned on after a solvent delay of 220 s. A mixture of C6–C24 n-alkanes was also analyzed for retention index calculation.
2.5. 2DLC-MS Analysis
All samples were analyzed on a Thermo Q Exactive HF Hybrid Quadrupole-Orbitrap Mass Spectrometer coupled with a Thermo DIONEX UltiMate 3000 HPLC system (Thermo Fisher Scientific, Waltham, MA, U.S.A.). The UltiMate 3000 HPLC system was equipped with a hydrophilic interaction chromatography (HILIC) and a reversed-phase chromatography (RPC) column. The HILIC column was a SeQuant ZIC-cHILIC HPLC column (150 × 2.1 mm i.d., 3 μm) purchased from Phenomenex (Torrance, CA, U.S.A.). The RPC column was an ACQUITY UPLC HSS T3 column (150 × 2.1 mm i.d., 1.8 μm) purchased from Waters (Milford, MA, U.S.A.).
The HILIC and RPC columns were configured in parallel mode.20 Each column was connected with a 2-μL sample loop. The temperature of these two columns was set to 40 °C. For separation on the HILIC column, 10 mM ammonium acetate (pH 3.25) was used as mobile phase A, and 100% acetonitrile was used as mobile phase B. The gradient was as follows: 0 min, 95% B; 0 to 5 min, 95% B to 35% B; 5 to 6 min, 35% B; 6.0 to 6.1 min, 35% B to 5% B; 6.1 to 23 min, 5% B; 23.0 to 23.1 min, 5% to 95%. The flow rate was 0.3 mL/min. For separation on the RPC column, water with 0.1% formic acid was used as mobile phase A, and 100% acetonitrile with 0.1% formic acid was used as mobile phase B. The gradient was as follows: 0 min, 5% B; 0 to 5 min, 5% B; 5 to 6.1 min, 5 to 15% B; 6.1 to 10 min, 15 to 60% B; 10 to 12 min, 60% B; 12 to 14 min, 60% to 100% B; 14 to 27.0 min, 100% to 5% B; 27.0 to 27.1 min, 5% B. The flow rate was 0.4 mL/min.
The parameters for mass spectrometry were set up exactly in the same way as our previous study.21 The full scan range from 60 to 900 (m/z), maximum injection time was 50 ms, and the resolution was set to 30 000 for both positive and negative modes. Each biological sample was analyzed by 2DLC-MS in both positive mode (+) and negative mode (−) to obtain the full MS data. Four pooled samples were made for each group and these pooled samples were analyzed by 2DLC-MS/MS in positive and negative mode to acquire MS/MS spectra at different collision energies (10, 20, 40, and 60 eV) for metabolite identification.
2.6. SCFAs Quantification by GC-MS
For GC-MS analysis, a Thermo ITQ 1100 GC-Ion Trap MS instrument was coupled with a 1310 autosampler and a Thermo Trace 1310 gas chromatography system (Thermo Scientific, Waltham, MA, U.S.A.). The instrument settings were identical to our previous study,19 except that the column temperature was programmed with an initial temperature of 40 °C to better focus the sample on the column head. Briefly, two GC columns, DB-225 ms (30 m × 0.25 mm 1dc ×0.25 μm 1dp, (50%-cyanopropylphenyl)-methylpolysiloxane) and DB-5 ms (30 m × 0.25 mm 1dc ×0.25 μm 1dp, (5%-phenyl)-methylpolysiloxane) were purchased from Agilent Technologies J&W (Santa Clara, CA, U.S.A.). The two columns were connected by a column connector purchased from Restek Corporation (Bellefonte, PA, U.S.A.), where the DB-225 ms column was the first column and the DB-5 ms column was the second column. The helium carrier gas (99.999% purity) flow rate was set to 1.0 mL/min. The temperatures of inlet, ion source, and transfer line were all set to 220 °C. The column temperature was programmed as follows: initial temperature 40 °C for 0.5 min; 10 °C/min to 170 °C; holding at 170 °C for 0.5 min; 5 °C/min to 220 °C; and holding at 220 °C for 5 min. The energy of electron ionization (EI) was set to 70 eV.
Next, 1 μL of PFBBr derivatives was injected into GC-MS in splitless mode with a splitless time of 1.0 min. Solvent delay time was set to 13.8 min. The mass spectral data were collected in selected ion monitoring (SIM) mode.
2.7. RNA Isolation and Real-Time PCR
The mRNA levels were assessed by real-time PCR. In brief, the total RNA was extracted with Trizol according to manufacturer’s protocol (Invitrogen, Carlsbad, CA, U.S.A.). GenAmp RNA PCR kit (Applied Biosystems, Foster City, CA, U.S.A.) was then used to reverse-transcribe RNA to cDNA. Prepared cDNA samples were analyzed on an ABI 7500 real-time PCR thermocycler for quantitative real-time PCR analysis using SYBR green PCR Master Mix (Applied Biosystems). The relative quantities of target transcripts were calculated from duplicate samples after normalization of the data against the housekeeping gene, 18S. Dissociation curve analysis was performed after PCR amplification to confirm the specificity of the primers. Relative mRNA expression was calculated using the Ct method. The following primer pairs were used: Taut forward 5′GCACACGGCCTGAAGATGA3′, Taut reverse 5′ATTTTTGTAGCAGAGGTACGGG3′, Csd forward 5′CCAGGACGTGTTTGGGATTGT3′, Csd reverse 5′ACCAGTCTTGACACTGTAGTGA3′, Cdo forward 5′GGGGACGAAGTCAACGTGG3′, Cdo reverse 5′ACCCCAGCACAGAATCATCAG3′, 18S forward 5′ GTAACCCGTTGAACCCCATT 3′, and 18S reverse 5′ CCATCCAATCGGTAGTAGCG 3′.
2.8. Hematoxylin and Eosin Staining
Formalin fixed, paraffin embedded liver tissue was sliced at 5 μm and mounted on positively charged glass slides. The sections were then deparaffinized with Citrisolv and rehydrated by immersing in graded ethanol solutions. Hematoxylin and eosin (H&E) was then used to stain the nuclear and cytoplasm components, respectively. After staining, sections were dehydrated through graded alcohol, cleared in Citrisolv, and mounted with Cytoseal XYL (Thermo Scientific, Waltham, MA, U.S.A.). Liver tissue morphology was observed by microscopy.
2.9. Data Analysis
The LECO ChromaTOF software (version 4.51), equipped with the National Institute of Standards and Technology MS database (NIST 2014), was used to analyze the GC × GC-MS data for spectrum deconvolution and tentative metabolite identification.22 MetPP software was used for retention index matching, cross-sample peak list alignment, normalization, and statistical analysis.23 For metabolite tentative identification, the threshold of spectral similarity score was set as S ≥ 600 with a maximum spectral similarity score of 1000. The p-value threshold was set as p ≤ 0.001 for retention index matching in MetPP. A tentative metabolite identification was considered as a correct identification only if the experimental information on the authentic metabolite agreed with the corresponding information on the chromatographic peak in the biological samples; that is, difference of the first dimension retention time Δ1tR ≤ 10 s, difference of the second dimension retention time Δ2R ≤ 0.05 s, and the spectral similarity score S ≥ 600.
For 2DLC-MS data analysis, MetSign software was used for spectrum deconvolution, metabolite identification, cross-sample peak list alignment, normalization, and statistical analysis.24–27 To identify metabolites, the 2DLC-MS/MS data of the four pooled samples were first matched to an in-house database that contains the parent ion m/z, MS/MS spectra, and retention time of 205 metabolite standards. The threshold for the spectral similarity of the MS/MS spectra of a metabolite standard and a spectrum of the pooled sample was set as ≥0.4 with a maximum score of one, while the thresholds of retention time difference and m/z variation window were respectively set to ≤0.15 min and ≤4 ppm. The 2DLC-MS/MS data without a match in the in-house database were then analyzed using Compound Discoverer software (version 2.0, Thermo Fisher Scientific, Inc., Germany), where the MS/MS spectra similarity score threshold was set as ≥40 with a maximum score of 100.
For GC-MS data, Thermo Xcalibur software Quan (version2.2 SP1.48) was used for peak picking. The calibration curves were determined by analyzing a series of SCFA solutions with different concentrations as described in our previous study.19 The signal-to-noise ratio (S/N) was set to S/N = 3.
2.10. Statistic Analysis
Statistical analyses were performed using the software MetPP, MetSign, and R v.3.5.1. Results are expressed as mean ± standard error of mean (SEM). Statistical comparisons were performed using an unpaired two-tailed t test. To investigate metabolic profiling difference between groups, metabolites identified by GC × GC-MS and 2DLC-MS/MS in each sample were merged based on the name of native metabolites, i.e., the names of metabolites without derivatization. Partial least-squares discriminant analysis (PLS-DA), a supervised technique that uses the PLS algorithm to explain and predict the membership of samples to groups, was performed to give an overview on the metabolic profile difference between groups. For purpose of comparison, principal component analysis (PCA), an unsupervised pattern recognition method, was also performed. PCA seeks a linear combination of variables such that the maximum variance is extracted from the variables for each component.
3. RESULTS AND DISCUSSION
3.1. Fecal Metabolic Profiling
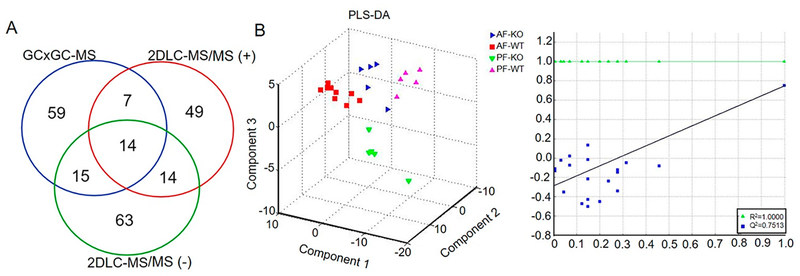
A total of 95 metabolites were identified by GC × GC-MS, while 106 metabolites were identified by 2DLC-MS/MS (−) and 84 metabolites by 2DLC-MS/MS (+). Among those metabolites, 14 were identified by all three platforms and 36 were identified by two platforms (Figure 1A). The identified metabolites included fatty acids, organic acids, amino acids, and other metabolites.
Figure 1.
Metabolic profiling of mouse fecal metabolites by three platforms. (A) Overlap of identified metabolites by GC × GC-MS, 2DLC-MS/MS (−), and 2DLC-MS/MS (+). (B) Result of PLS-DA. All data measured by the three platforms GC × GC-MS, 2DLC-MS (−), and 2DLC-MS (+) were merged and used as input for PLS-DA. The left is PLS-DA 3-D score plot and the right is the performance plot of PLS-DA model. AF, alcohol-fed; PF, pair-fed; WT, wild type; KO, CRAMP knockout.
To study the metabolic profile difference among the four groups, the GC × GC-MS data and the full MS data of 2DLC-MS (−) and 2DLC-MS (+) were merged. The merged data were then clustered using PLS-DA. Figure 1B depicts the PLS-DA results. Distinct separation of the metabolic profiles of the identified metabolites suggests a remarkable alteration in mouse fecal metabolites caused by alcohol-feeding and CRAMP knockout. The large values of R2 = 1.00 and Q2 = 0.75 demonstrate the PLS-DA model has good discrimination and high predictability for clustering the metabolic profile data acquired in this study. Figure S-1 depicts the PLS-DA results of respectively classifying the four groups using data acquired by each analytical platform. The single platform-based PLS-DA models have worse discrimination and lower predictability than the merged data, suggesting that the merged data have more metabolite information and are more close to represent the mouse fecal metabolome. Similarly, the merged data provide a better discrimination than the single platform data when PCA was employed (Figure S-2).
3.2. Alcohol-Feeding Changed Metabolites in Feces of Wild Type Mice
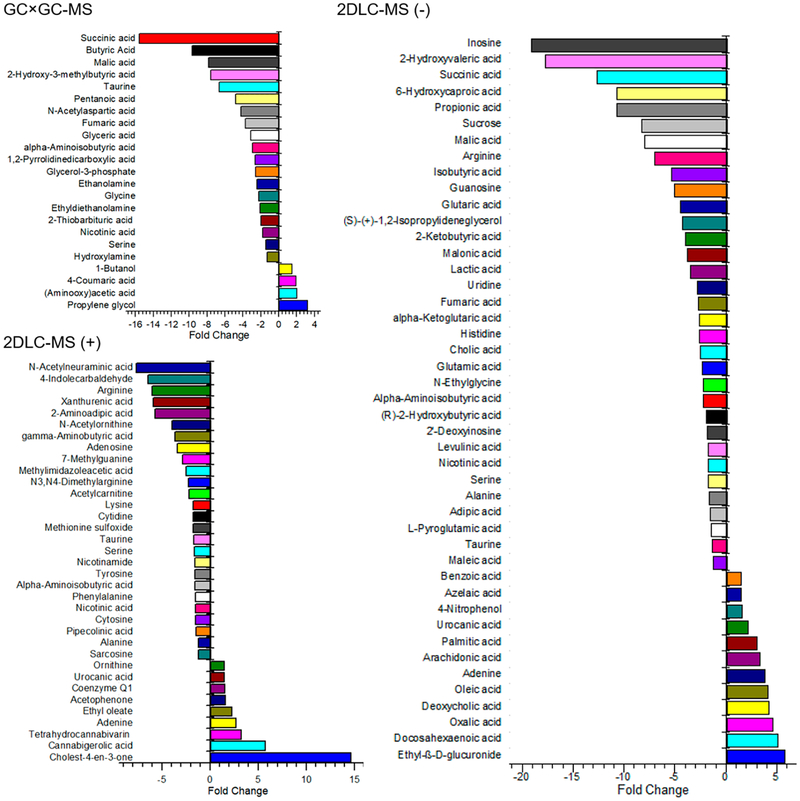
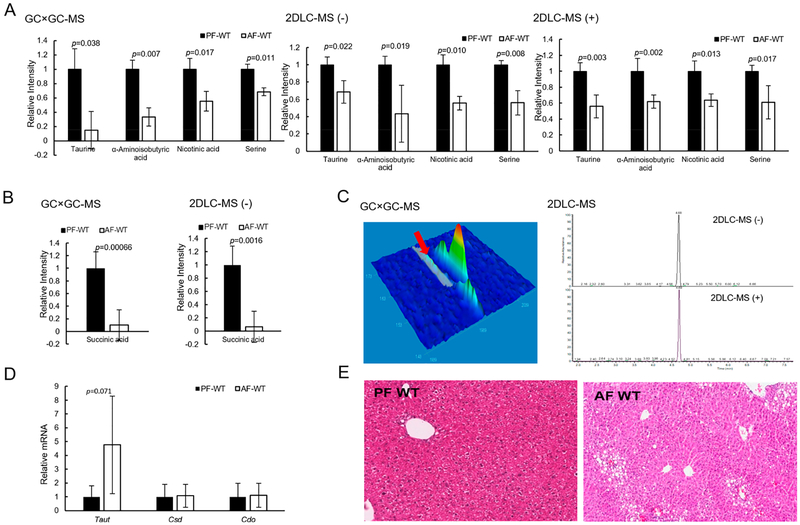
The abundance of metabolites detected in groups PF-WT and AF-WT were used to study the influence of alcohol in the fecal metabolome in wild type mice. Figure 2 shows that 23, 45, and 35 metabolites were detected with significant changes in their abundance levels by GC × GC-MS, 2DLC-MS (−), and 2DLC-MS (+), respectively. Among those metabolites, four and seven metabolites were detected in three and two platforms, respectively. Figure 3A shows the abundances of the four metabolites (taurine, α-aminoisobutyric acid, nicotinic acid, and serine) that were detected in all three platforms. The abundance levels of those four metabolites were lower in AF mice than those in PF mice, and were consistently demonstrated by all three analytical platforms, showing the high accuracy and high stability of the three platforms in analyzing complex biological samples.
Figure 2.
Metabolites with significant abundance changes in feces of wild type mice after PF or AF treatment for 24 days. The polar metabolites extracted from each sample were analyzed by GC × GC-MS, 2DLC-MS (−), and 2DLC-MS (+), respectively.
Figure 3.
Alcohol-feeding changed the amount of fecal metabolites and increased liver fat. (A) Abundance levels of the four metabolites detected by all three platforms. (B) Abundance levels of succinic acid in PF-WT mice and AF-WT mice detected by GC × GC-MS and 2DLC-MS (−). (C) shows the chromatographic peak shapes of taurine measured by GC × GC-MS, 2DLC-MS (−) and 2DLC-MS (+). (D) is gene expression of Taut, Csd, and Cdo in the liver tissues of PF-WT mice and AP-WT mice. (E) H&E staining of the liver tissues of PF-WT mice and AP-WT mice. AF, alcohol-fed; PF, pair-fed; WT, wild type.
Succinic acid was detected with significant abundance changes by GC × GC-MS and 2DLC-MS (−). In fact, it has the largest fold change in the GC × GC-MS data and the third largest fold change in the 2DLC-MS (−) data (Figure 2). Figure 3B shows that levels of succinic acid were significantly decreased by alcohol feeding. It has been reported that Parabacteroides distasonis could increase the production of succinic acid in gut, and oral gavage of Parabacteroides distasonis to ob/ob mice could alleviate nonalcoholic fatty liver disease and metabolic disorders by increasing succinic acid in gut and upregulating the intestinal gluconeogenesis pathway.28 The reduction of succinic acid in fecal samples suggests a decrease of succinic acid-producing bacteria, such as Parabacteroides distasonis in the intestine in response to alcohol exposure. However, this notion needs further investigation.
Taurine levels were decreased in AF-WT mice compared with that in PF-WT mice with fold-changes of 0.15, 0.68, and 0.56 in GC × GC-MS, 2DLC-MS (−), and 2DLC-MS (+), respectively (Table S-1). While the data of all three platforms showed that the abundance level of taurine was decreased in AF-WT, the magnitude of the fold-change calculated from the GC × GC-MS data has a larger variation than those calculated from the 2DLC-MS (−) and 2DLC-MS (+) data. Manual analysis showed that the low abundance taurine has a poor chromatographic peak shape in GC × GC-MS with a significant tailing in the second dimension GC. However, taurine had a large instrument response with a symmetric chromatographic peak shape in 2DLC-MS (−) and 2DLC-MS (+) (Figure 3C). The low instrument response and the poor chromatographic peak shape of taurine resulted in a degree of variation in quantifying its peak area in GC × GC-MS data.
Taurine is a sulfur-containing amino acid that exists naturally in the diet. It is absorbed by ileal intestine through transporters such as Taut. The absorbed taurine enters into the liver through the portal circulation, and conjugates bile acids. Conjugated bile acids flow back to the intestine and help with food digestion. The taurine-conjugated bile acids are deconjugated by gut bacteria that have bile salt hydrolase activity and the taurine is then released.29 Therefore, fecal taurine concentrations are composed of two important parts: the unabsorbed and the deconjugation-freed taurine. The detected level reflects the balance of the two contributions. Interestingly, alcohol feeding tended to increase Taut mRNA, while Csd and Cdo, two enzymes responsible for taurine synthesis from cysteine, remained unchanged (Figure 3D). In our study, the mass spectrometry data of taurine acquired by three independent analytical platforms demonstrated that taurine was decreased in AF-WT mice. It is likely that AF-decreased fecal taurine levels are, at least in part, due to the increased intestinal transport.
Nicotinic acid (NA) is an organic compound that affects lipid metabolism in multiple organs by preventing lipolysis in adipose tissue, decreasing triglyceride synthesis in liver, and reducing oxidative damage.30 Our results showed that the level of NA was decreased in AF-WT mice with fold-changes of 0.55, 0.56, and 0.64 in GC × GC-MS, 2DLC-MS (−), and 2DLC-MS (+), respectively (Table S-1). The decreased NA in AF-WT mice indicates that lipolysis might be increased in alcohol-fed mice. In fact, the level of fatty acids such as oleic acid, palmitic acid, docosahexaenoic acid, and arachidonic acid detected by 2DLC-MS (−) were significantly increased in AF-WT mice (Table S-1). Levels of linoelaidic acid, oleic acid, palmitic acid, stearic acid, and pentadecanoic acid detected by GC × GC-MS were also increased (Table S-1). The increased levels of free fatty acids in fecal samples indicate a likely high level of lipolysis in adipose tissue. Indeed, alcohol feeding significantly decreased white fat mass in WT mice.31 In addition, a previous study showed that dietary NA supplementation increased hepatic fatty acid oxidation and decreased hepatic de novo lipogenesis.32 Therefore, the decreased levels of NA may be a contributing factor for the increased fatty liver induced by alcohol (Figure 3E).
3.3. Levels of SCFAs Were Decreased in Alcohol-Fed Wild Type Mice
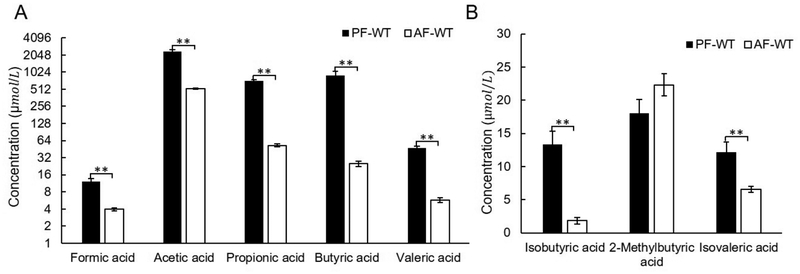
SCFAs (volatile organic compounds) are produced in the intestine by gut bacteria. Recent studies have demonstrated that SCFAs mediate multiple mechanisms in ALD.33 Butyric acid is a major nutrient of enterocytes and serves as a histone deacetylase (HDAC) inhibitor to regulate gene expression.34 Propionic acid and isobutyric acid are involved in intestinal epithelial cell homeostasis and gut barrier integrity. The decreased abundance levels of these two metabolites in the feces of alcoholics compared to healthy individuals indicates that alcohol consumption might decrease the levels of those two metabolites in intestine.35
The concentrations of SCFAs were quantified by GC-MS via SIM in this study. Figures 4A,B show that the abundance levels of seven SCFAs significantly decreased in the alcohol fed mice, i.e., AF-WT mice. However, the level of 2-methylbutyric acid was not significantly altered by alcohol (Figure 4B). The increased ratio of colonic propionic acid:acetic acid could prevent the accumulation of liver fat by impairing hepatic acetic acid metabolism.36 In this study, the ratio of colonic propionic acid:acetic acid in AF-WT mouse was 1:9.9, which is notably decreased compared with that in PF-WT mouse (1:3.3) in mouse feces (Table 1). Such a dramatic change indicates an increased fat accumulation in alcohol-fed mice (Figure 3E), which is consistent with the previous report.37
Figure 4.
Alcohol-feeding affected the amount of fecal SCFAs in wild type mice. (A) Alcohol-feeding decreased the concentrations of straight-chain SCFAs in feces of the wild type mice. (B) The concentrations of two branched-chain SCFAs in mouse feces were affected by alcohol-fed. ** p < 0.01. AF, alcohol-fed; PF, pair-fed; WT, wild type.
Table 1.
Concentration of SCFAs in Fecal Samples Detected by GC-MS
| compound name | PF-WT | AF-WT | PF-KO | AF-KO | |||||
|---|---|---|---|---|---|---|---|---|---|
| concentration (μmol/L) | SEM | concentration (μmol/L) | SEM | concentration μmol/L) | SEM | concentration (μmol/L) | SEM | ||
| formic acid | 12.1 | 1.5 | 3.94 | 0.25 | 8.76 | 0.64 | 4.88 | 1.00 | |
| acetic acid | 2380 | 126 | 521 | 13 | 1060 | 119 | 694 | 192 | |
| propionic acid | 717 | 39 | 52.5 | 3.0 | 197 | 22 | 63.3 | 19.8 | |
| butyric acid | 893 | 2 | 24.9 | 2.6 | 74.0 | 10.7 | 32.9 | 10.0 | |
| valeric acid | 47.8 | 3.1 | 5.75 | 0.51 | 8.73 | 1.15 | 6.11 | 1.25 | |
| isobutyric acid | 13.3 | 2.0 | 1.82 | 0.53 | 3.20 | 0.69 | 3.93 | 0.76 | |
| 2-methylbutyric acid | 18.1 | 2.0 | 22.3 | 1.7 | 18.7 | 2.1 | 24.4 | 2.6 | |
| isovaleric acid | 12.1 | 1.5 | 6.57 | 0.46 | 5.58 | 0.53 | 10.9 | 0.7 | |
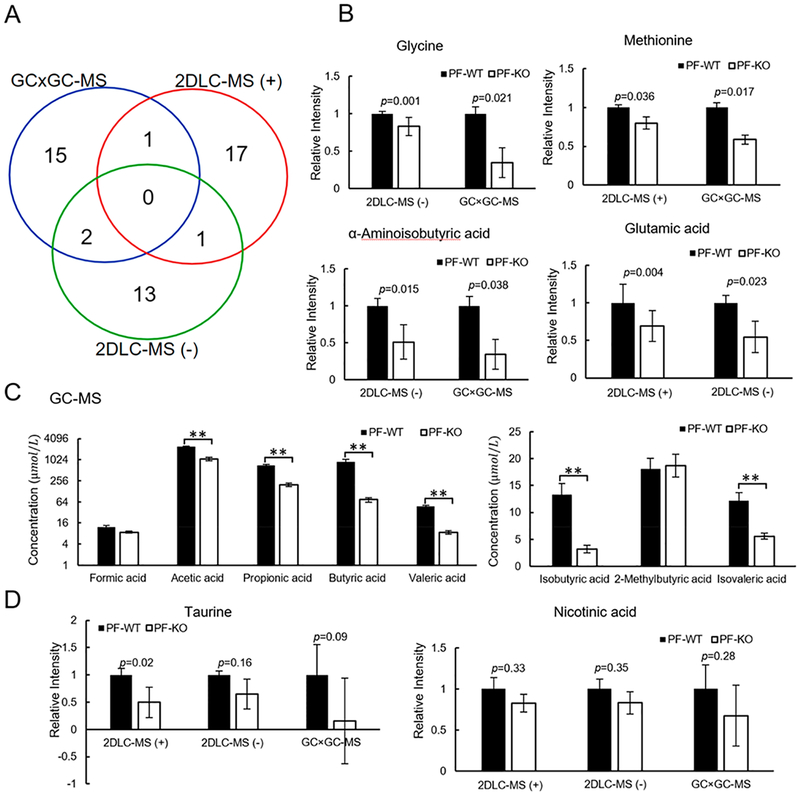
3.4. Role of CRAMP in Mouse Fecal Metabolism
CRAMP plays an important role in intestinal microbiota and immune system. In order to investigate the role of CRAMP in mouse fecal metabolism, polar metabolites in feces of PF-KO mice were compared with those in feces of PF-WT mice. Statistical significance tests showed that 18, 16, and 19 metabolites had significant changes in their abundance levels detected by GC × GC-MS, 2DLC-MS (−), and 2DLC-MS (+), respectively (Figure 5A). Four metabolites (methionine, glycine, alpha-aminoisobutyric acid, and glutamic acid) were detected with significant abundance changes between PF-KO mice and PF-WT mice by two platforms (Figure 5B). While measured by different platforms, the levels of these four metabolites were consistently lower in the KO mice compared to WT mice under pair-feeding conditions, demonstrating the high accuracy of the three platforms in quantifying these metabolites (Figure 5B and Table S-2). Interestingly, the changes of SCFAs in PF-KO mice mirrored their changes in AF-WT mice (Figure 5C), suggesting that the composition change of gut microbiota has a certain degree of similarity between alcohol feeding and CRAMP deficiency.
Figure 5.
Fecal metabolite levels in PF-KO mice compared to that in PF-WT mice. (A) Overlap of metabolites with significant changes between PF-KO mice and PF-WT mice detected by GC × GC-MS, 2DLC-MS (−), and 2DLC-MS (+). (B) Relative abundance of four metabolites detected by two platforms. (C) Concentrations of SCFAs in feces of PF-KO mice and PF-WT mice. ** p < 0.01. (D) Relative abundance of taurine and NA. PF, pair-fed; WT, wild type; KO, CRAMP knockout.
Taurine and NA were detected in all three platforms, but it was not considered with statistical significant changes between PF-KO mice and PF-WT mice. Notably, taurine levels were decreased by 2.0-fold in 2DLC-MS (+) data, and were decreased by 1.5- and 6.3-fold in the 2DLC-MS (−) and GC × GC-MS data, respectively (Figure 5D). Manual investigation of the data shows that the variation in the magnitude of the fold-change of taurine was mainly caused by the low instrument response and poor chromatographic peak shape like illustrated in Figure 3C.
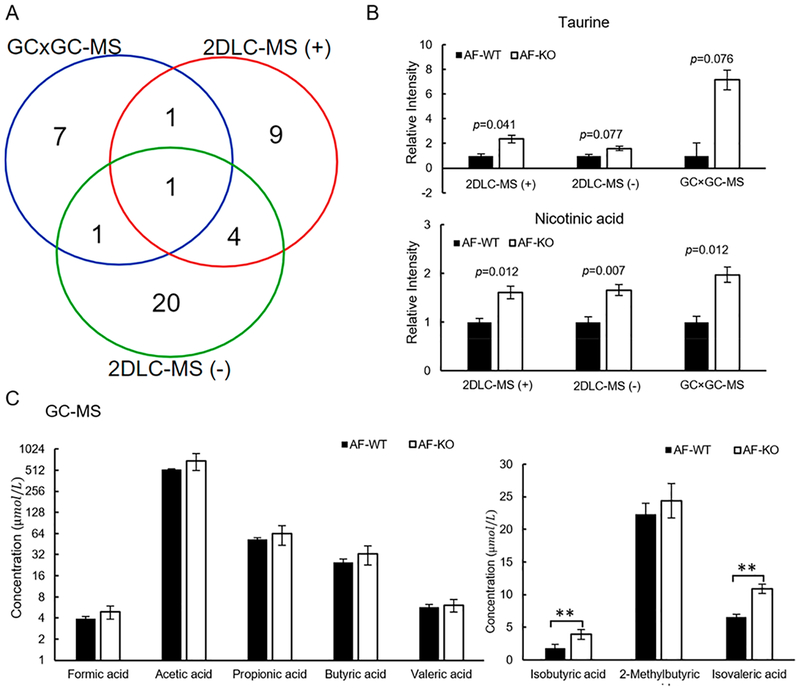
In order to determine the role of alcohol on fecal metabolism in CRAMP deficiency mice, the metabolite differences between AF-WT mice and AF-KO mice were analyzed. Figure 6A shows that 43 metabolites had significant changes in their abundance levels in feces between the two groups. NA is the only metabolite that was detected with significant changes in its abundance level by the three platforms. Six metabolites (α-aminoisobutyric acid, malic acid, alpha-aminoadipic acid, tyrosine, citrulline, and N-acetylneuraminic acid) were detected with significant abundance changes between groups by two platforms. Compared with AF-WT mice, NA in AF-KO mice was increased by 1.6-, 1.7-, and 2.0-fold in AF-WT mice as detected by 2DLC-MS (+), 2DLC-MS (−), and GC × GC-MS, respectively (Figure 6B and Table S-3).
Figure 6.
CRAMP KO increased the amount of taurine, NA, and SCFAs in AF mice. (A) Overlap of significantly changed metabolites between AF-WT mice and AF-KO mice detected by GC × GC-MS, 2DLC-MS (−), and 2DLC-MS (+). (B) Changes of taurine and NA detected by all three platforms in feces of AF-WT mice and AF-KO mice. (C) Quantification results of SCFAs in feces of AF-WT mice and AF-KO mice. ** p < 0.01. AF, alcohol-fed; WT, wild type; KO, CRAMP knockout.
Figure 6B and Table S-3 show that taurine level was increased in AF-WT mice by 2.4-fold in 2DLC-MS (+) data (p = 0.041), 1.6-fold in 2DLC-MS (−) data (p = 0.077), 7.1-fold in GC × GC-MS data (p = 0.076). Taurine was not significantly changed between the two groups in GC × GC-MS and 2DLC-MS (–) data because of the large variations in its abundance data. We have shown that both alcohol and CRAMP deficiency decreased fecal taurine levels compared to their respective controls (Figures 3A and 5D), and the fecal taurine concentration may be a result of the balance of absorption and deconjugation of bile acids. Therefore, the increased taurine in the CRAMP KO mice induced by alcohol feeding may reflect the disturbance of that balance. Alcohol treatment did not significantly increase intestinal Taut mRNA levels, which might indicate an unchanged taurine absorption between the KO and WT mice by alcohol. The increased taurine levels in feces may be due to the increased deconjugation of taurine-conjugated bile acid by gut micro-biota, which were altered by alcohol and CRAMP deficiency (data not shown).
Compared with AF-WT mice, the abundance levels of branched-chain fatty acids, isobutyric acid and isovalric acid, were significantly increased in AF-KO mice, while the levels of straight-chain SCFAs were not changed in AF-KO mice (Figure 6C). Branched-chain SCFAs are mainly derived from the catabolism of branched-chain amino acids such as valine, leucine, and isoleucine.38 Thus, it is likely that upon the treatment by alcohol, CRAMP deficiency does not further change the gut microbiota that favor SCFAs production, but does increase branched-chain amino acids catabolism that favor isobutyric acid and isovalric acid production.
4. CONCLUSIONS
Profiling of polar metabolites extracted from mouse feces by three analytical platforms and quantification of SCFAs by GC-MS showed a significant metabolic profile difference between the four animal groups. Among the significantly changed metabolites that were detected by all three platforms, the levels of taurine and NA were decreased by alcohol in wild type mice and in pair fed mice by CRAMP depletion. Taurine and NA levels were increased when CRAMP KO mice were exposed to alcohol compared to the AF-WT mice. Compared with PF-WT mice, the abundance levels of all straight-chain SCFAs were significantly decreased in AF-WT mice and PF-KO mice. However, the abundance levels of branched-chain SCFAs were increased in AF-KO mice when compared with AF-WT mice. These results provide further evidence demonstrating the roles of alcohol and CRAMP in intestinal metabolism in ALD, and the utility of analytical platforms optimized to detect metabolic changes in ALD and other diseases.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Mrs. Marion McClain for review of this manuscript. This work was supported by NIH [S10OD020106 (X.Z.); 1R01AA23190 (W.F.); 1P20GM113226 (C.J.M.); 1P50AA024337 (C.J.M.); 1U01AA021893 (C.J.M.); 1U01AA021901 (C.J.M.); 1U01AA022489–01A1 (C.J.M.); and 1R01AA023681 (C.J.M.)]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work was also supported by the Department of Veterans Affairs 1I01BX002996–01A2 (C.J.M.).
Footnotes
Notes
The authors declare no competing financial interest.
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.9b00181.
Figure S-1: The results of individual PLS-DA analysis; Figure S-2: The results of PCA analysis; Table S-1: Metabolites changed in feces by alcohol detected by three platforms; Table S-2: Metabolites changed in feces by CRAMP knockout detected by three platforms; Table S-3: Metabolites changed in feces by CRAMP knockout exposed to alcohol detected by three platforms (PDF)
REFERENCES
- (1).Thurman RG; Bradford BU; Iimuro Y; Knecht KT; Arteel GE; Yin M; Connor HD; Wall C; Raleigh JA; Frankenberg MV; Adachi Y; Forman DT; Brenner D; Kadiiska M; Mason RP The role of gut-derived bacterial toxins and free radicals in alcohol-induced liver injury. J. Gastroenterol. Hepatol 1998, 13 (Suppl), S39–50. [DOI] [PubMed] [Google Scholar]
- (2).Beier JI; Arteel GE; McClain CJ Advances in alcoholic liver disease. Current gastroenterology reports 2011, 13, 56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Cummings JH Short chain fatty acids in the human colon. Gut 1981, 22, 763–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Xie G; Zhong W; Zheng X; Li Q; Qiu Y; Li H; Chen H; Zhou Z; Jia W Chronic ethanol consumption alters mammalian gastrointestinal content metabolites. J. Proteome Res 2013, 12, 3297–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Zasloff M Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [DOI] [PubMed] [Google Scholar]
- (6).Kang HK; Kim C; Seo CH; Park Y The therapeutic applications of antimicrobial peptides (AMPs): a patent review. J. Microbiol 2017, 55, 1–12. [DOI] [PubMed] [Google Scholar]
- (7).Seo MD; Won HS; Kim JH; Mishig-Ochir T; Lee BJ Antimicrobial peptides for therapeutic applications: a review. Molecules 2012, 17, 12276–12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Rosenberger CM; Gallo RL; Finlay BB Interplay between antibacterial effectors: a macrophage antimicrobial peptide impairs intracellular Salmonella replication. Proc. Natl. Acad. Sci. U. S. A 2004, 101, 2422–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Soehnlein O; Wantha S; Simsekyilmaz S; Doring Y; Megens RT; Mause SF; Drechsler M; Smeets R; Weinandy S; Schreiber F; Gries T; Jockenhoevel S; Moller M; Vijayan S; van Zandvoort MA; Agerberth B; Pham CT; Gallo RL; Hackeng TM; Liehn EA; Zernecke A; Klee D; Weber C Neutrophil-derived cathelicidin protects from neointimal hyperplasia. Sci. Transl. Med 2011, 3, 103ra98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Chromek M; Slamova Z; Bergman P; Kovacs L; Podracka L; Ehren I; Hokfelt T; Gudmundsson GH; Gallo RL; Agerberth B; Brauner A The antimicrobial peptide cathelicidin protects the urinary tract against invasive bacterial infection. Nat. Med 2006, 12, 636–641. [DOI] [PubMed] [Google Scholar]
- (11).Kovach MA; Ballinger MN; Newstead MW; Zeng X; Bhan U; Yu FS; Moore BB; Gallo RL; Standiford TJ Cathelicidin-related antimicrobial peptide is required for effective lung mucosal immunity in Gram-negative bacterial pneumonia. J. Immunol 2012, 189, 304–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Rosenfeld Y; Papo N; Shai Y Endotoxin (lipopolysaccharide) neutralization by innate immunity host-defense peptides. Peptide properties and plausible modes of action. J. Biol. Chem 2006, 281, 1636–1643. [DOI] [PubMed] [Google Scholar]
- (13).Kandler K; Shaykhiev R; Kleemann P; Klescz F; Lohoff M; Vogelmeier C; Bals R The anti-microbial peptide LL-37 inhibits the activation of dendritic cells by TLR ligands. Int. Immunol 2006, 18, 1729–1736. [DOI] [PubMed] [Google Scholar]
- (14).Mookherjee N; Brown KL; Bowdish DM; Doria S; Falsafi R; Hokamp K; Roche FM; Mu R; Doho GH; Pistolic J; Powers JP; Bryan J; Brinkman FS; Hancock RE Modulation of the TLR-mediated inflammatory response by the endogenous human host defense peptide LL-37. J. Immunol 2006, 176, 2455–2464. [DOI] [PubMed] [Google Scholar]
- (15).Sun J; Furio L; Mecheri R; van der Does AM; Lundeberg E; Saveanu L; Chen Y; van Endert P; Agerberth B; Diana J Pancreatic beta-Cells Limit Autoimmune Diabetes via an Immunoregulatory Antimicrobial Peptide Expressed under the Influence of the Gut Microbiota. Immunity 2015, 43, 304–317. [DOI] [PubMed] [Google Scholar]
- (16).Hoang-Yen Tran D; Hoang-Ngoc Tran D; Mattai SA; Sallam T; Ortiz C; Lee EC; Robbins L; Ho S; Lee JE; Fisseha E; Shieh C; Sideri A; Shih DQ; Fleshner P; McGovern DP; Vu M; Hing TC; Bakirtzi K; Cheng M; Su B; Law I; Karagiannides I; Targan SR; Gallo RL; Li Z; Koon HW Cathelicidin suppresses lipid accumulation and hepatic steatosis by inhibition of the CD36 receptor. Int. J. Obes 2016, 40, 1424–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Shao T; Zhao C; Li F; Gu Z; Liu L; Zhang L; Wang Y; He L; Liu Y; Liu Q; Chen Y; Donde H; Wang R; Jala VR; Barve S; Chen SY; Zhang X; Chen Y; McClain CJ; Feng W Intestinal HIF-1alpha deletion exacerbates alcoholic liver disease by inducing intestinal dysbiosis and barrier dysfunction. J. Hepatol 2018, 69, 886–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Bertola A; Mathews S; Ki SH; Wang H; Gao B Mouse model of chronic and binge ethanol feeding (the NIAAA model). Nat. Protoc 2013, 8, 627–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).He L; Prodhan MAI; Yuan F; Yin X; Lorkiewicz PK; Wei X; Feng W; McClain C; Zhang X Simultaneous quantification of straight-chain and branched-chain short chain fatty acids by gas chromatography mass spectrometry. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci 2018, 1092, 359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Klavins K; Drexler H; Hann S; Koellensperger G Quantitative metabolite profiling utilizing parallel column analysis for simultaneous reversed-phase and hydrophilic interaction liquid chromatography separations combined with tandem mass spectrometry. Anal. Chem 2014, 86, 4145–4150. [DOI] [PubMed] [Google Scholar]
- (21).He L; Wei X; Ma X; Yin X; Song M; Donninger H; Yaddanapudi K; McClain CJ; Zhang X Simultaneous Quantification of Nucleosides and Nucleotides from Biological Samples. J. Am. Soc. Mass Spectrom 2019, 30, 987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Shi X; Wei X; Yin X; Wang Y; Zhang M; Zhao C; Zhao H; McClain CJ; Feng W; Zhang X Hepatic and fecal metabolomic analysis of the effects of Lactobacillus rhamnosus GG on alcoholic fatty liver disease in mice. J. Proteome Res 2015, 14, 1174–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Wei X; Shi X; Koo I; Kim S; Schmidt RH; Arteel GE; Watson WH; McClain C; Zhang X MetPP: a computational platform for comprehensive two-dimensional gas chromatography time-of-flight mass spectrometry-based metabolomics. Bioinformatics 2013, 29, 1786–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Wei X; Sun W; Shi X; Koo I; Wang B; Zhang J; Yin X; Tang Y; Bogdanov B; Kim S; Zhou Z; McClain C; Zhang X MetSign: a computational platform for high-resolution mass spectrometry-based metabolomics. Anal. Chem 2011, 83, 7668–7675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Wei X; Shi X; Kim S; S K; Patrick JS; Binkley J; Kong M; McClain C; Zhang X Data dependent chromatographic peak model-based spectrum deconvolution for analysis of LC-MS data. Anal. Chem 2014, 86, 2156–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Wei X; Shi X; Kim S; Zhang L; Patrick JS; Binkley J; McClain C; Zhang X Data preprocessing method for liquid chromatography-mass spectrometry based metabolomics. Anal. Chem 2012, 84, 7963–7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Wei X; Lorkiewicz P; Salabei JK; Shi B; Hill BG; Kim S; McClain CJ; Zhang X Analysis of stable isotope assisted metabolomics data acquired by high resolution mass spectrometry. Anal. Methods 2017, 9, 2275–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Wang K; Liao M; Zhou N; Bao L; Ma K; Zheng Z; Wang Y; Liu C; Wang W; Wang J; Liu SJ; Liu H Parabacteroides distasonis Alleviates Obesity and Metabolic Dysfunctions via Production of Succinate and Secondary Bile Acids. Cell Rep 2019, 26 (222), No. e5. [DOI] [PubMed] [Google Scholar]
- (29).Chikai T; Nakao H; Uchida K Deconjugation of bile acids by human intestinal bacteria implanted in germ-free rats. Lipids 1987, 22, 669–671. [DOI] [PubMed] [Google Scholar]
- (30).Kharbanda KK Nicotinic acid supplementation in the context of alcoholic liver injury: friend or foe? Alcohol.: Clin. Exp. Res 2014, 38, 1829–1831. [DOI] [PubMed] [Google Scholar]
- (31).Crowell KT; Steiner JL; Coleman CS; Lang CH Decreased Whole-Body Fat Mass Produced by Chronic Alcohol Consumption is Associated with Activation of S6K1-Mediated Protein Synthesis and Increased Autophagy in Epididymal White Adipose Tissue. Alcohol.: Clin. Exp. Res 2016, 40, 1832–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Li Q; Xie G; Zhang W; Zhong W; Sun X; Tan X; Sun X; Jia W; Zhou Z Dietary nicotinic acid supplementation ameliorates chronic alcohol-induced fatty liver in rats. Alcohol.: Clin. Exp. Res 2014, 38, 1982–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Cassard AM; Ciocan D Microbiota, a key player in alcoholic liver disease. Clinical and molecular hepatology 2018, 24, 100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Nudelman A; Levovich I; Cutts SM; Phillips DR; Rephaeli A The role of intracellularly released formaldehyde and butyric acid in the anticancer activity of acyloxyalkyl esters. J. Med. Chem 2005, 48, 1042–1054. [DOI] [PubMed] [Google Scholar]
- (35).Couch RD; Dailey A; Zaidi F; Navarro K; Forsyth CB; Mutlu E; Engen PA; Keshavarzian A Alcohol induced alterations to the human fecal VOC metabolome. PLoS One 2015, 10, No. e0119362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Chambers ES; Byrne CS; Rugyendo A; Morrison DJ; Preston T; Tedford C; Bell JD; Thomas L; Akbar AN; Riddell NE; Sharma R; Thursz MR; Manousou P; Frost G The effects of dietary supplementation with inulin and inulin-propionate ester on hepatic steatosis in adults with non-alcoholic fatty liver disease. Diabetes, Obes. Metab 2019, 21, 372–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Sang WH; Zeng MC; Chen S; Chen R; Fan XF; Gong YS; Zhang HL; Zhang HY; Kong XX Effect of autophagy inhibitor chloroquine on acute alcoholinduced liver disease. Chinese journal of applied physiology 2018, 34, 102–105. [PubMed] [Google Scholar]
- (38).Macfarlane S; Macfarlane GT Regulation of short-chain fatty acid production. Proc. Nutr. Soc 2003, 62, 67–72. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.