Summary
Tricellular adherens junctions are points of high tension that are central to the rearrangement of epithelial cells. However, the molecular composition of these junctions is unknown, making it difficult to assess their role in morphogenesis. Here we show that Sidekick, an immunoglobulin family cell adhesion protein, is highly enriched at tricellular adherens junctions in Drosophila. This localization is modulated by tension, and Sidekick is itself necessary to maintain normal levels of cell bond tension. Loss of Sidekick causes defects in cell and junctional rearrangements in actively remodeling epithelial tissues like the retina and tracheal system. The adaptor proteins Polychaetoid and Canoe are enriched at tricellular adherens junctions in a Sidekick-dependent manner; Sidekick functionally interacts with both proteins and directly binds to Polychaetoid. We suggest that Polychaetoid and Canoe link Sidekick to the actin cytoskeleton to enable tricellular adherens junctions to maintain or transmit cell bond tension during epithelial cell rearrangements.
Keywords: tricellular adherens junction, Sidekick, tension, epithelium, cell rearrangements, trachea, retina, Afadin, ZO-1, actin
Graphical Abstract
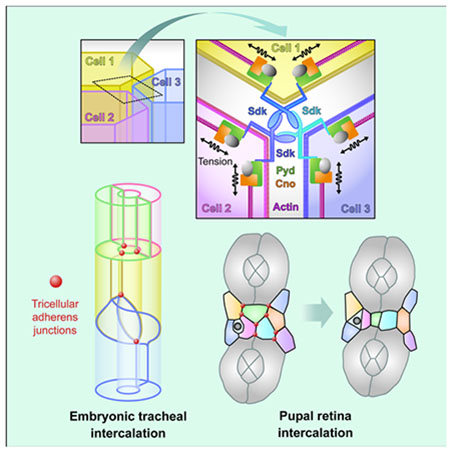
eTOC Blurb
Letizia et al. characterize the nature and function of tricellular adherens junctions. The transmembrane protein Sidekick is highly enriched at these junctions and connects to the actin cytoskeleton through Canoe and Polychaetoid. This complex can sense and modulate tension to promote normal cell rearrangements during developmental remodeling.
Introduction
Epithelial cells are linked together by multiple types of junctions that must be dynamically rearranged as the cells change their relative positions during tissue remodeling (Takeichi, 2014). Cadherin-based adherens junctions (AJs) are essential for cells to sense and respond to the mechanical forces that drive such rearrangements (Pinheiro and Bellaiche, 2018), while tight junctions (TJs) in vertebrates and septate junctions (SJs) in invertebrates seal the epithelium to form a permeability barrier. A specialized class of junctions is found at points where three or more cell membranes meet. Here, tricellular tight junctions (tTJs) or tricellular septate junctions (tSJs) maintain the integrity of the barrier. The molecular components that are specifically localized or enriched at these junctions include transmembrane proteins with sealing extracellular domains, such as angulins and tricellulins in vertebrates, and Gliotactin (Gli), Anakonda (Aka) and M6 in Drosophila (Byri et al., 2015; Dunn et al., 2018; Higashi and Miller, 2017; Schulte et al., 2003).
Tricellular adherens junctions (tAJs) are thought to be points of high tension, at which the ends of actin filaments must be anchored to the cell surface (Choi et al., 2016; Del Signore et al., 2018; Higashi et al., 2016; Higashi and Miller, 2017; Vanderleest et al., 2018; Yonemura, 2011). They are much less characterized than tTJs and tSJs, and no molecular components specific to tAJs have yet been identified. A few intracellular proteins are known to be enriched at tAJs, although they also localize continuously along bicellular adherens junctions (bAJs). One of them is the adaptor protein Afadin/Canoe (Cno), which links actin filaments to the junctional proteins E-cadherin (Ecad) and Echinoid (Ed) (Bonello et al., 2018; Choi et al., 2016; Sawyer et al., 2009; Wei et al., 2005). In the early Drosophila embryo, Cno enrichment at tAJs requires Rap1 activation by the guanine nucleotide exchange factor (GEF) Dizzy (Dzy) (Bonello et al., 2018), and in cultured MDCK cells tAJ localization of Afadin is enhanced by knocking down Zonula occludens 1 (ZO-1) family proteins (Choi et al., 2016). ZO-1 and Afadin physically interact (Takahashi et al., 1998; Yamamoto et al., 1997) and the single Drosophila ZO-1 homologue Polychaetoid (Pyd) has embryonic functions very similar to those of Cno (Choi et al., 2011), suggesting that the two proteins act together. As Ed is not necessary for the enrichment of Cno at tAJs (Sawyer et al., 2009), the protein that organizes tAJs by physically linking Cno to the cell surface at these positions remains unknown.
Sidekick (Sdk) proteins are members of the immunoglobulin (Ig) superfamily that can mediate homophilic adhesion through their four N-terminal Ig domains (Goodman et al., 2016). Their 13 Fibronectin type III (FnIII) domains interact with lipid membranes, allowing Sdks to facilitate tight cell-cell adhesion (Tang et al., 2018). Sdks are best known for their role in promoting interactions between specific neuronal processes in the retina (Astigarraga et al., 2018; Krishnaswamy et al., 2015; Yamagata and Sanes, 2008; Yamagata et al., 2002). However, the identification of the original Drosophila sdk gene was based on defects in eye development in sdk mutants, including abnormalities in the pigment cell lattice that results from epithelial cell rearrangements and shape changes at the pupal stage (Carthew, 2007; Nguyen et al., 1997). A Sdk protein trap accumulates at cell vertices in the epithelium of early embryos (Lye et al., 2014). Sdks are also expressed during branching morphogenesis in the developing mouse and human kidney, and their upregulation in glomeruli induces podocyte dysfunction (Kaufman et al., 2004; Kaufman et al., 2010). Sdk-1 controls actin cytoskeleton organization in prostate cancer cells and is required for their migration (Verone et al., 2013). The cytoplasmic domain of all Sdks ends in a conserved PDZ-binding motif that can interact with multiple PDZ-domain proteins (Meyer et al., 2004), including members of the MAGI family that control Sdk localization and function in the retina and kidney (Kaufman et al., 2010; Yamagata and Sanes, 2010).
Here, we show that Sdk accumulates at tAJs through a mechanism that is influenced by mechanical tension. Sdk is in turn necessary to maintain normal levels of cell bond tension, and in its absence cell and junctional rearrangements are perturbed. We find that Sdk assembles a tAJ complex by directly interacting with Pyd and enriching both Pyd and Cno at tAJs. These results suggest that Sdk links Cno and Pyd to the plasma membrane at tAJs, where they anchor the ends of actin filaments to maintain cellular tension and allow vertex remodeling during tissue rearrangements.
Results
Sdk is highly enriched at tricellular adherens junctions
A Sdk protein trap was previously reported to localize to tricellular vertices in the embryonic epidermis (Lye et al., 2014). Using an antibody to Sdk (Astigarraga et al., 2018), we confirmed that in several epithelia, including the epidermis and tracheae and the pupal retina, Sdk strongly accumulated at vertices (Fig. 1A, B, D). It was 14 to 35-fold more enriched at these positions than at bicellular junctions, in contrast to Ecad which showed only a slight enrichment at vertices (Fig. 1C). We observed that in embryos at the stage of germ band extension, Sdk was also present at high levels at bAJs in a planar polarized pattern in ventral cells that were undergoing intercalation (Fig. S1A, B). However, in this study we have focused on the role of Sdk in tissues in which it is strongly enriched at vertices.
Figure 1: Sdk localizes to tAJs.
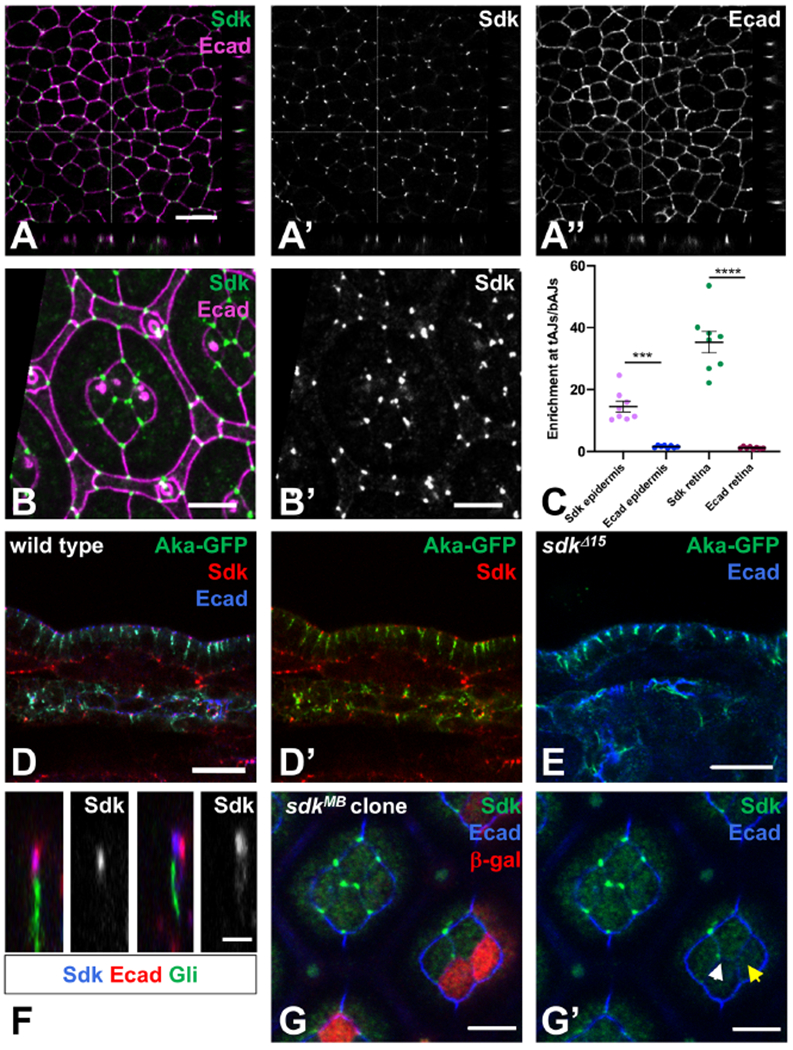
(A) Single confocal section of a wild type stage 10 embryo stained for Sdk (A’, green) and Ecad (A’’, magenta) showing the apical surface of the epidermis and xz and yz sections. (B) Apical surface of a 42 h pupal retina, stained for Ecad (magenta) and Sdk (B’, green). Four central cone cells are surrounded by two primary pigment cells and a lattice of secondary and tertiary pigment cells and mechanosensory bristles. (C) Quantification of Sdk and Ecad enrichment at tAJs relative to adjacent bAJs in stage 9-11 embryonic epidermis and in cone cells in 42h pupal retina. n=8 samples of each tissue, with each point being the mean of 10 cells from that sample. Bars show mean ± SEM. ****, p<0.0001, ***, p=0.0001, unpaired t-test with Welch’s correction. (D, E) cross-sections of the epidermis and the dorsal tracheal trunk of wild type (D) and sdkΔ15 (E) stage 15 embryos, stained for Sdk (red), Ecad (blue) and Aka-GFP (green). n=18 (D), n=8 control and 14 sdk (E). (F) xz sections of wild type 42 h pupal retina stained for Sdk (blue and single channels), Ecad (red) and Gli (green). n=15. Sdk is strongly enriched at tricellular vertices, at the same apical-basal level as and usually overlapping with Ecad, and apical to Gli and Aka. Aka localization is not affected in sdk mutants. (G) a sdkMB05054 clone marked with anti-β-galactosidase in red, stained for Sdk (green) and Ecad (blue), in a focal plane at the level of cone cell apical junctions. Sdk is absent from the tAJ when two of the three cells are sdk mutant (yellow arrow in G’) but present when only one cell is mutant (white arrow). Scale bars, 10 μm (A, D, E) 5 μm (B, G), or 2 μm (F). See also Figure S1.
Sdk localization was clearly apical to tSJ markers such as Gli and Aka (Byri et al., 2015; Schulte et al., 2003) (Fig. 1D, F, Fig. S1D); the center of Sdk staining was 2.4 ± 0.4 μm apical to the center of Gli staining (n=35 tAJs from 9 retinas). In sdk null mutants (Astigarraga et al., 2018), the tricellular junctional localization of Gli and Aka was unaffected (Fig. 1E; Fig. S1E, F), and dye injected into the embryo was unable to penetrate the trachea (Fig. S1G, H), indicating that the epithelial permeability barrier formed by tSJs (Schulte et al., 2003) is intact. The extracellular domain of Aka is thought to form a trimolecular complex that promotes its localization to tricellular junctions (Byri et al., 2015). Such a mechanism is unlikely to explain Sdk localization, as the presence of Sdk in two of three neighboring cells was sometimes sufficient for its enrichment at the vertex (Fig. 1G, Fig. S1I); 57% of vertices with one sdk mutant cell (n=248) and 7% of vertices with two mutant cells (n=117) showed detectable Sdk localization. These results show that Sdk is not a component of tSJs.
We found that Sdk at vertices was at the same apical-basal level as Ecad, or N-cadherin (Ncad) in retinal cone cells, where it usually colocalized with these cadherins (Fig. 1A, D, F, Fig. S1C, D) (Lye et al., 2014). Of 100 retinal pigment cell vertices examined in xz-projections, we observed complete overlap between Sdk and Ecad staining at 62, partial overlap at 27, and no overlap at 11. The non-overlapping cases may reflect vertices with lower or no accumulation of Ecad (Lye et al., 2014). In summary, in epithelial tissues from mid-embryogenesis onwards Sdk accumulates almost exclusively at vertices in the same plane as AJs. Specialized protein complexes have been postulated to assemble at tAJs (Higashi and Miller, 2017). Our results suggest that Sdk could be the first described transmembrane protein component of such a tAJ complex, and show that it does not localize to tSJs or contribute to the epithelial permeability barrier.
Sdk localization is regulated by tension
Since interactions between Sdk molecules on two adjacent cells are sufficient for its localization, it is unclear how it is restricted to tAJs. Sdk localizes to vertices earlier in embryogenesis than Gli and Aka (Byri et al., 2015; Schulte et al., 2003), so its localization cannot depend on these proteins. tAJs are thought to be the points of highest tension in an epithelium based on the localization of tension-sensitive proteins such as Vinculin (Choi et al., 2016; Hara et al., 2016; Higashi and Miller, 2017; Kale et al., 2018; Trichas et al., 2012), suggesting the possibility that Sdk localization could be regulated by mechanical tension. Myosin II, which promotes contractility, is activated primarily through phosphorylation of its regulatory light chain by Rho kinase, and inactivated through dephosphorylation by Myosin light chain phosphatase (Somlyo and Somlyo, 2000). We analyzed Sdk accumulation in tissues in which the mechanical properties of cells have been shown to change upon Myosin II perturbation (Del Signore et al., 2018; Fischer et al., 2014; Warner and Longmore 2009a, b).
When we reduced tension in the cells of the embryonic amnioserosa by expressing a constitutively active form of the Myosin-binding subunit (Mbs) of Myosin light chain phosphatase (Lee and Treisman, 2004) to inhibit Myosin II, Sdk became less enriched at tAJs and more diffusely expanded at bicellular adherens junctions (bAJs) along cell edges (Fig. 2A, B, D). A similar expansion of Sdk was observed surrounding clones of cells in the pupal retina that lacked the myosin regulatory light chain encoded by spaghetti squash (sqh) (Karess et al., 1991) (Fig. 2E, F). These cells had enlarged apical areas, demonstrating a failure of tension-mediated apical constriction (Fig. 2E) (Del Signore et al., 2018; Warner and Longmore 2009a, b). Conversely, increasing tension in the amnioserosa by overexpressing myosin light chain kinase (MLCK) resulted in increased enrichment of Sdk at tAJs (Fig. 2C, D). In the wing disc, an actomyosin cable has been shown to assemble at the border of ed mutant clones and contract to constrict the mutant cells and extrude them from the epithelium (Chang et al., 2011). We found that Sdk levels at tAJs were elevated within and at the border of these clones (Fig. 2G, I). All these results are consistent with differential localization of Sdk to regions experiencing relatively higher tension, implying that Sdk can respond to contractility. Sdk localization to tAJs was disrupted in clones in which Ecad was depleted by RNAi (Fig. 2H, I), suggesting that the Ecad complex may enable Sdk to respond to tension or maintain adhesion that is necessary for Sdk localization.
Figure 2: Tension promotes Sdk localization to tAJs.
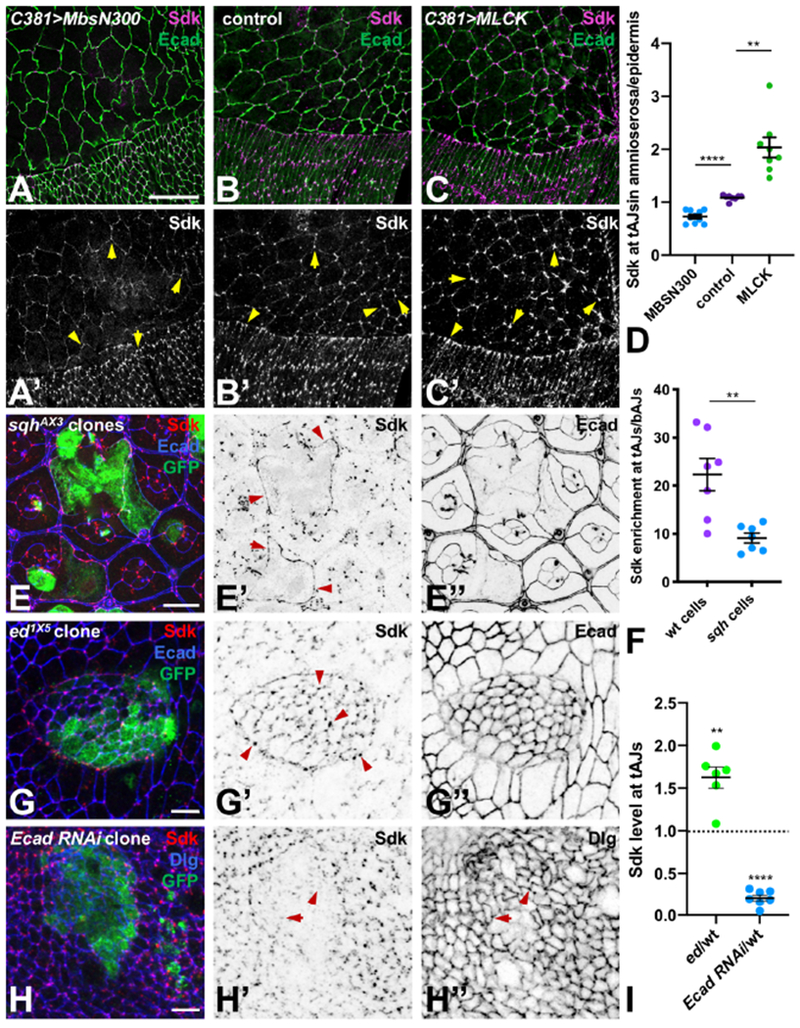
(A-C) show the dorsal epidermis and amnioserosa of stage 14 embryos during dorsal closure stained for Sdk (A’-C’, magenta) and Ecad (green). C381-GAL4 drives UAS-MbsN300 (A), no transgene (B) or UAS-MLCK (C) in the amnioserosa. Sdk is less enriched at tAJs when myosin activity is reduced (A) and more enriched when it is increased (C) (arrows). (D) Quantification of Sdk levels at tAJs in the amnioserosa relative to the epidermis in these three conditions. n=10 (MbsN300) or 8 embryos (control and MLCK), and points are the ratio of the means of 20 cells of each type per embryo. ****, p<0.0001; **, p=0.0015, unpaired t-test with Welch’s correction. (E) sqhAX3 clones in a 48 h APF pupal retina marked with GFP (green) and stained for Sdk (E’, red) and Ecad (E’’, blue). Mutant cells have expanded apical areas and show diffuse localization of Sdk along bAJs (arrows). (F) Quantification of Sdk enrichment at tAJs relative to bAJs in cells at the border of sqh clones compared to wild type lattice cells in the same retina. n=7 retinas, and points are the means of 10 cells of each genotype. **, p=0.007, unpaired t-test with Welch’s correction. (G, H) ed1X5 (G) and Ecad RNAi (H) clones in third instar wing imaginal discs, marked with GFP (green) and stained for Sdk (G’, H’, red) and Ecad (G’’, blue in G) or Discs-large (Dlg, H’’, blue in H), a marker of septate junctions. Sdk levels are increased within and at the border of the ed clone, where an actomyosin cable constricts the mutant cells (arrows), and Sdk is not correctly localized at tAJs within the Ecad RNAi clone (arrows). (I) Quantification of Sdk levels at tAJs in ed mutant or Ecad RNAi-expressing cells relative to wild type cells in the same wing disc. n=6 (ed) or 7 (Ecad RNAi), and points are the ratio of the means of 15-20 cells of each genotype per disc. **, p=0.004, ****, p<0.0001 for difference from wild type by one sample t-test. All graphs show mean ± SEM. Scale bars, 25 μm (A-C), 10 μm (E), 5 μm (G, H).
Sdk is required for normal cell rearrangements
tAJs have been proposed to represent tension hot spots in dynamic epithelial tissues (Higashi and Miller, 2017); however, the lack of information about their molecular composition has hindered the analysis of their functions in morphogenesis. The localization of Sdk primarily to tAJs gave us an opportunity to investigate their importance in developing epithelia. To do this, we examined the consequences of removing Sdk function in two different tissues that undergo extensive remodeling. We first analyzed the embryonic tracheal system during the phase of cell rearrangements, in which several branches, including the dorsal branches, undergo cell intercalation. Intercalation is a stepwise process in which cells originally positioned in pairs become positioned end-to-end, generating a lumen surrounded by a single cell (Ribeiro et al., 2004). It involves a major remodeling of the junctional region, as a straight-line autocellular junction gradually replaces the ring-shaped intercellular junctions that initially connect paired cells (Fig. S2A, B) (Ribeiro et al., 2004). Although only two cells contact each other during this “zipping up” step (stage 14), vertices still form at connections between the autocellular junction and the intercellular junction with the neighboring cell, due to the tubular topology (Fig. S2B). We found that Sdk strongly accumulated at cell-cell vertices during intercalation (Fig. S2C, D). In sdk null mutants, the transformation of intercellular junctions into autocellular ones was blocked; many branches still maintained intercellular junctions at stage 15-16, when branches are fully elongated and cell intercalation should be complete (Fig. 3A, B,E, S2E, Movie S1). The defects could be rescued by expressing a full-length UAS-HA-Sdk transgene, but not by a transgene that lacks the C-terminal PDZ-binding motif (UAS-HA-SdkΔBD, Fig. 3C–E), showing the importance of this motif for Sdk function. Intercalation is thought to be driven by the tension generated by migration of the tip cells of the branch (Ochoa-Espinosa et al., 2017), suggesting that Sdk acts at autocellular-intercellular vertices to transmit this tension and ensure that vertex remodeling proceeds correctly.
Figure 3: sdk is required for cell and junctional rearrangements.
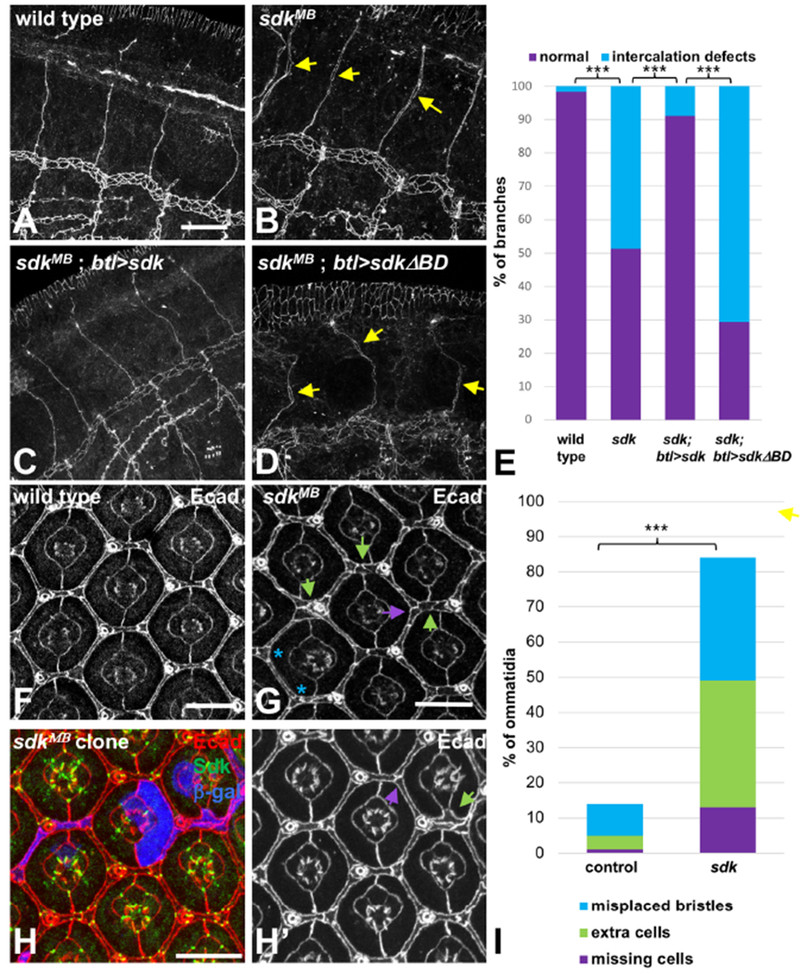
(A-D) show stage 15-16 embryos stained for Ecad to mark AJs in the tracheal branches. (A) wild type; (B) sdkMB05054; (C) sdkMB05054; btl-GAL4>UAS-sdk; (D) sdkMB05054; btl-GAL4>UAS-sdkΔBD. Intercellular junctions are completely converted to autocellular junctions in wild type embryos at this stage, but some remain in sdk mutants (arrows). This phenotype is rescued by expressing wild-type Sdk, but not SdkΔBD, in the tracheal system. (E) is a quantification of the percentage of branches with intercalation defects in stage 15-16 embryos of each genotype. n=120 branches from 30 embryos (wild type), 201 branches from 42 embryos (sdk), 90 branches from 16 embryos (sdk; btl>sdk), or 51 branches from 11 embryos (sdk; btl>sdkΔBD). (F-H) show 42 h APF pupal retinas stained with Ecad (F, G, H’, red in H) and Sdk (green in H). (F) wild type; (G) sdkMB05054; (H) sdkMB05054 clone marked with anti-β-galactosidase (blue). sdk mutant retinas or clones have extra lattice cells (green arrows) or missing cells (purple arrows). (I) is a quantification of the percentage of ommatidia showing these phenotypes or misplaced bristles (two bristles or two tertiary pigment cells at adjacent vertices; blue asterisks in G). n=274 ommatidia in 8 retinas (wild type) or 180 ommatidia in 6 retinas (sdk). Scale bars, 10 μm. ***, p<0.001, Fisher’s exact test. See also Figures S2 and S3 and Movies S1–S5.
Next, we examined the developing retina, as sdk mutations were previously reported to affect the number and arrangement of retinal pigment cells (Nguyen et al., 1997). During pupal stages, the precursors of these cells change their shapes and relative positions and reduce their numbers by cell death to generate a final hexagonal lattice in which one elongated secondary pigment cell occupies each edge, and tertiary pigment cells and mechanosensory bristles occupy alternating vertices (Carthew, 2007) (Fig. 3F). In sdk mutants this arrangement was irregular; some extra cells remained within the lattice, while at other positions cells were missing or bristles were misplaced (Fig. 3G, I). These defects could be observed even within small sdk mutant clones (Fig. 3H), indicating that sdk has a cell-autonomous effect on the junctional rearrangements that enable cells to change their relative positions and suggesting a role for tAJs in this process. In summary, Sdk is required for regulated cell rearrangements in two different tissues that undergo dynamic remodeling.
Sdk regulates cell bond tension
To determine how the defects observed in sdk mutants arise, we carried out live imaging of retinas in which apical junctions were labeled with GFP-tagged α-catenin. Between 24 and 28 h after puparium formation (APF), lattice cells form a single file by cell intercalation, which involves contraction of some bicellular junctions and expansion of others through tension exerted on the actomyosin network (Bardet et al., 2013; Bertet et al., 2004; Blankenship et al., 2006; Chan et al., 2017; Del Signore et al., 2018; Yu and Fernandez-Gonzalez, 2016). In the retina, we observed accumulation of Myosin II along shrinking junctions and F-actin at their vertices, consistent with a requirement for tension exerted on shrinking junctions (Movie S2, Fig. S3A, B). In sdk mutants, intercalation was often incomplete (Movie S3, Fig. S3C,D), indicating a failure of junctional rearrangements. We also carried out live imaging of the dorsolateral region of stage 7-8 embryos using GFP-tagged E-cad, which revealed that in this epithelium cell boundaries were less straight in sdk mutants than in wild-type controls (Movies S4, S5). These findings, together with the defects observed in tracheal intercalation, suggest that Sdk is required to maintain or transmit the appropriate level of tension on cell contacts. However, we could not detect gross differences in myosin localization or phosphorylation in sdk mutant cells or embryos (Fig. S3E–G). Interestingly, Sdk and Myosin II often showed a complementary localization pattern (Fig. S3H, I), consistent with the presence of myosin on bicellular bonds undergoing contraction.
To test more directly whether cell bond tension was affected in sdk mutants, we estimated the relative forces acting at cell edges by using a laser to sever cell-cell junctions (Farhadifar et al., 2007; Hutson et al., 2003; Rauzi and Lenne, 2015; Rauzi et al., 2008). We induced laser cuts in bicellular bonds in the epidermis of control and sdk mutant embryos and followed the displacement of vertices over time (Fig. 4A, B). We measured the initial recoil velocity of the vertices of the cut bonds and the amplitude of vertex displacement, and found that both were significantly decreased in sdk mutants compared to controls (Fig. 4C, D), indicating that sdk mutant bonds are under lower tension. Sdk thus not only responds to tension, but is also necessary to sustain that tension, making it a key component of the tension-regulating function of tAJs.
Figure 4: Sdk modulates cell bond tension.
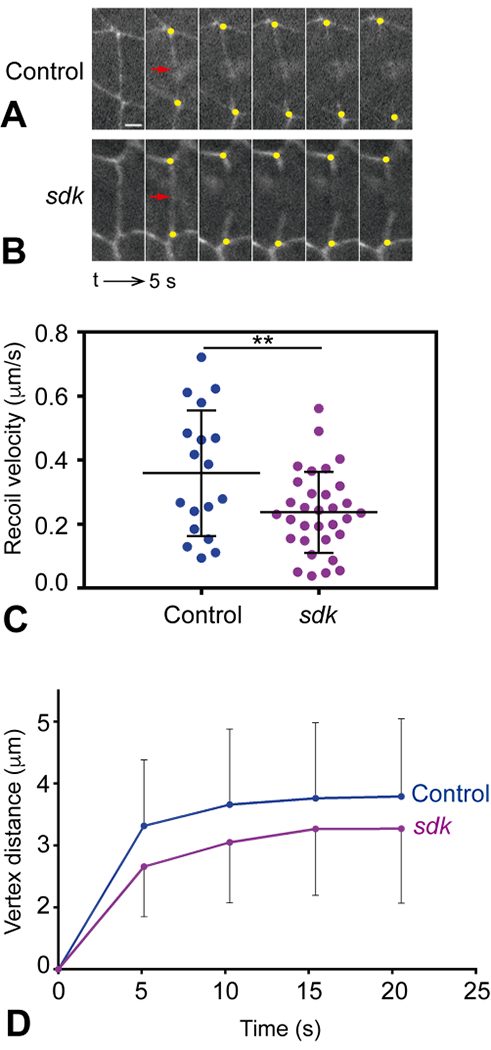
(A, B) Still images showing junction retraction after laser ablations in control ECad-GFP (A) and sdkMB05054; ECad-GFP mutant embryos (B). Cell junctions are visualized by the presence of ECad-GFP. The site of the cut is indicated by a red arrow and the recoiling vertices by yellow dots. Note the decreased recoil of vertices in sdk mutants. The sequence interval is 5 s. Scale bar, 2 μm. (C) Quantification of the recoil speed upon laser ablation in control and sdk mutant embryos. The recoil velocity in sdk mutants is lower, indicating reduced tension (Student’s t test, **p = 0.0098). (D) Quantification of vertex distance increase over time upon laser ablation in control and sdk mutant embryos. Vertex distance was normalized to the initial distance before ablation. Vertex displacement in sdk mutants is lower than in control embryos (Mann Whitney test, *p= 0.0181). n = 18 control embryos, n = 32 sdk mutants. Means ± SDs are shown in (C, D).
Sdk localizes Cno and Pyd to tAJs
We next wanted to understand the mechanism by which Sdk regulates tension at tAJs. The actin cytoskeleton plays key roles in regulating tension, cell rearrangements and tissue architecture (Pinheiro and Bellaiche, 2018). We found that Sdk colocalized with a pool of actin at tAJs in the embryonic epidermis, trachea and retina (Fig. 5A, B, S4C, D), suggesting that it may control tension through interactions with the actin cytoskeleton. The short cytoplasmic domain of Sdk terminates in a predicted PDZ-binding motif which is necessary for its function (Fig. 3D, E); we therefore investigated the possibility that one or more PDZ domain proteins could link Sdk to actin filaments. In cultured mammalian epithelial cells, the PDZ domain-containing proteins Afadin and ZO-1 control cell bond tension by anchoring the ends of actin filaments to vertices (Choi et al., 2016). The Drosophila Afadin homologue Cno colocalizes with the Ecad complex along all AJs. In addition, Cno, but not the Ecad complex, is strongly enriched at vertices slightly basal to the apical junction (Bonello et al., 2018; Sawyer et al., 2009). We found that Cno perfectly colocalized with Sdk and actin in these regions (Fig. 5A, B), suggesting that Cno is also a component of the Sdk complex at tAJs. In agreement with this, we found that in sdk mutant embryos, the enrichment of Cno and actin at tAJs was absent, but their uniform localization at bAJs was unaffected (Fig. 5C, F). A similar enrichment of GFP-tagged endogenous Cno to tAJs was observed in the third instar wing imaginal disc, and this enrichment was lost in sdk mutant clones (Fig. 5D, G). The ZO-1 homologue Polychaetoid (Pyd) was likewise enriched at tAJs, where it colocalized with Sdk, and this enrichment was also lost in sdk mutant cells (Fig. 5E–G, S4A, B, D). These results show that Sdk is required for the accumulation of Cno, Pyd and actin at tAJs, suggesting that sdk mutant phenotypes may reflect the loss of this specific pool of the three proteins.
Figure 5: Sdk localizes Cno and Pyd to tAJs.
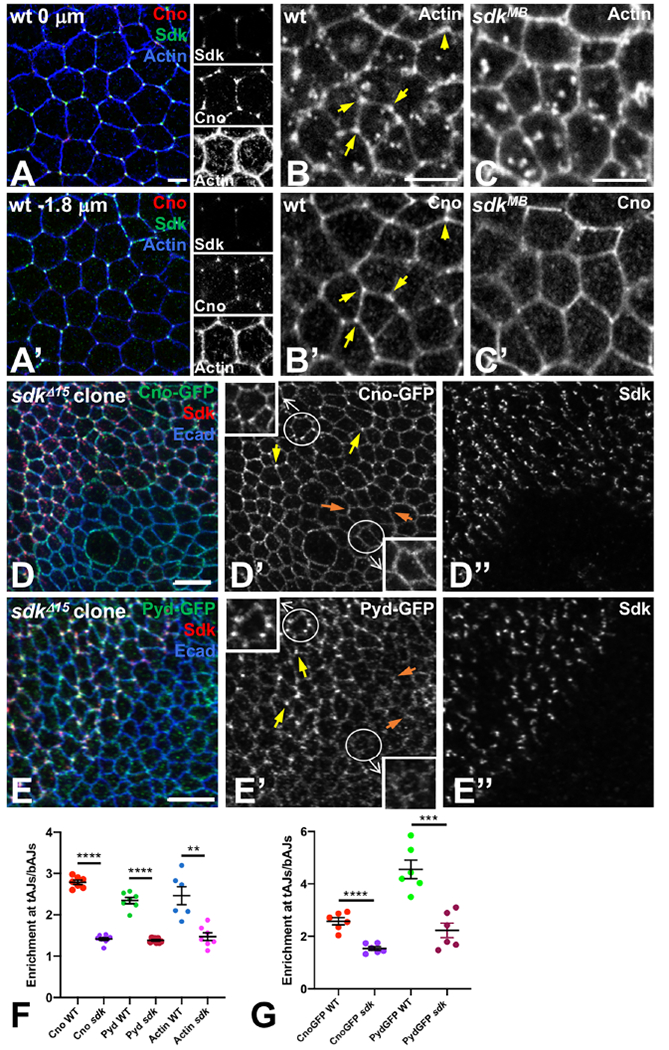
(A) lateral view of a wild type stage 6/7 embryo, stained for Cno (red), Sdk (green) and actin (blue). Single channels are shown in the right panels. (A) shows the apical surface and (A’) is 1.8 μm below the apical surface. Cno is enriched at tAJs, especially subapically, where it colocalizes with Sdk and a pool of actin filaments. (B, C) wild type (B) and sdkMB05054 (C) stage 9 embryos showing actin (B, C) and Cno (B’, C’) staining 1.8 μm subapically. Actin and Cno are enriched at tAJs in control embryos (yellow arrows in B, B’) but not in sdk mutants. (D, E) wing discs with sdkΔ15 clones marked by the absence of Sdk (D’’, E’’, red in D, E), stained for Ecad (blue), Cno-GFP (D’, green in D) or Pyd-GFP (E’, green in E). Cno and Pyd are enriched at tAJs in wild type regions (yellow arrows) but not in sdk mutant regions (orange arrows). The circled cells are enlarged in the insets. (F) Quantification of enrichment of Cno, Pyd and Actin at tAJs relative to adjacent bAJs in wild type and sdkMB05054 stage 9-11 embryos. n=7 embryos (Cno WT, Pyd WT, Actin sdk), 9 embryos (Cno sdk), 8 embryos (Pyd sdk), or 6 embryos (Actin WT). Each point is the mean of 10 cells from that embryo. (G) Quantification of enrichment of Cno-GFP and Pyd-GFP at tAJs relative to adjacent bAJs in wild type cells and sdkΔ15 clones in third instar wing discs. n=6 discs from each genotype, and each point is the mean of 10-15 cells from that disc. For both graphs, bars show mean ± SEM. ****, p<0.0001, ***, p<0.001, **, p=0.001, unpaired t-test (with Welch’s correction for Pyd in wt v. Pyd in sdk). Scale bars, 5 μm. See also Figures S4 and S5.
We next examined whether any of these proteins were required for the maintenance of Sdk localization at tAJs. To test the role of actin, we treated embryos with Latrunculin A, which binds actin monomers and inhibits polymerization of F-actin (Spector et al., 1983). Junctions at which F-actin was lost also lost Cno and Sdk, although Ecad was maintained (Fig. S4E, F). In regions in which F-actin was less disrupted, Sdk was still present at vertices but also expanded onto bAJs (Fig. S4E). This indicates that interactions with the cytoskeleton are important to maintain Sdk enrichment at tAJs. We found that Sdk could still localize to tAJs in cno or pyd mutant cells or in cells expressing pyd RNAi (Fig. S5). However, in later embryos and wing discs loss of cno reduced Sdk enrichment at tAJs, concomitant with loss of Ecad from the AJs (Fig. S5B, E, H). Cno and Pyd are thus not necessary for the initial localization of Sdk at tAJs, although Cno may help to maintain and reinforce its accumulation there through its effects on Ecad. Taken together, our results show that Sdk is the key component that promotes the recruitment to tAJs of a Pyd/Cno complex associated with actin filaments.
Sdk functionally interacts with Cno and Pyd
To determine whether normal morphogenesis requires Sdk to localize Cno to tAJs , we compared the effects of misexpressing Cno in wild type and sdk mutant cells. In the retina, Cno misexpression dramatically disrupted the pigment cell lattice, leading to excessive numbers of lattice cells at some edges and loss of cells at others (Fig. 6A, C). Sdk localization was also expanded in cells that overexpressed Cno (Fig. 6A). In contrast, Cno had a much weaker effect when it was expressed in sdk mutant cells; most lattice edges had a single secondary pigment cell, as in sdk mutant and wild type retinas (Fig. 6B, C). This indicates that Sdk is necessary for Cno to affect cell rearrangements. We also observed a genetic interaction between sdk and cno in the tracheal system; sdk/+; cno/+ double heterozygotes had intercalation defects similar to sdk homozygotes (23% of transheterozygous embryos, n=35) (Fig. 6D). A similar genetic interaction occurred in sdk/+; pyd/+ double heterozygotes (35% of transheterozygotes showed intercalation defects, n=37) (Fig. 6E).
Figure 6: Sdk interacts with Cno and Pyd.
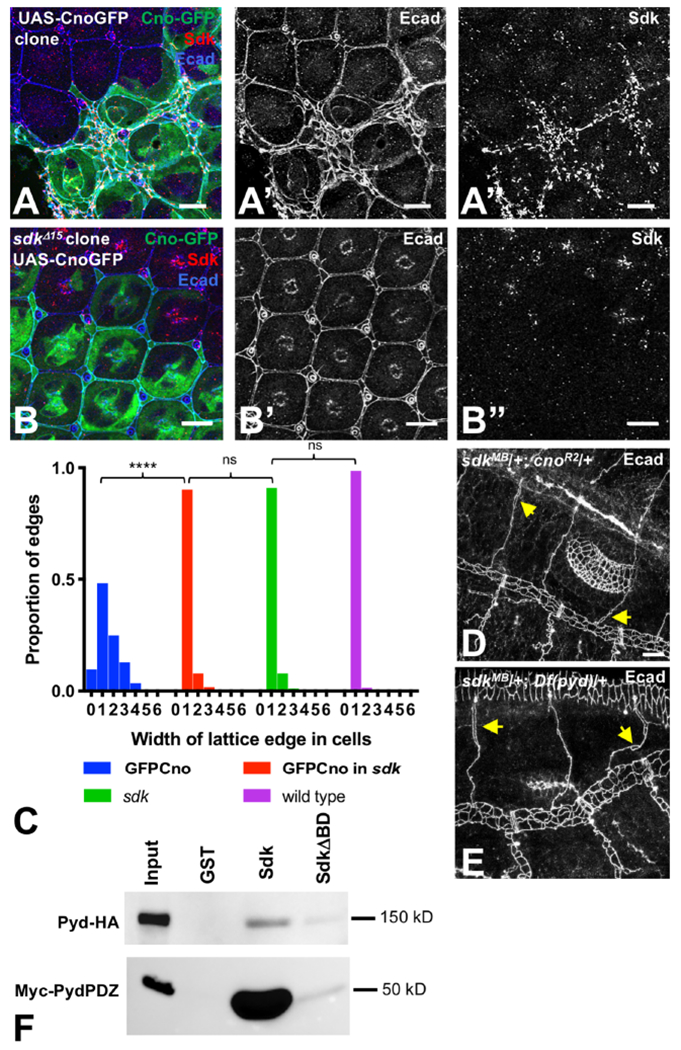
(A, B) 48 h APF pupal retinas with wild type (A) or sdkΔ15 (B) clones expressing UAS-CnoGFP (green), stained for Ecad (A’, B’, blue) and Sdk (A’’, B’’, red). Cno overexpression expands Sdk expression and dramatically disrupts the pigment cell lattice, with some edges containing many extra cells and others missing cells. When Cno is expressed in sdk mutant cells, the phenotypes are much milder. (C) shows a quantification of the distribution of the number of cells spanning each lattice edge. ****, p<0.0001, ns, not significant, chi-squared test. n=344 ommatidia of each genotype in 11 retinas (UAS-CnoGFP and UAS-CnoGFP in sdk), 8 retinas (sdk) or 15 retinas (wild-type ommatidia taken from the same images). (D,E) stage 15-16 sdkMB05054/+; cnoR2/+ (D) and sdkMB05054/+; Df(pyd)B12/+ (E) embryos stained for Ecad to mark AJs in the tracheal branches, showing open intercellular junctions (arrows). (F) pulldowns of full-length HA-tagged Pyd or the myc-tagged PDZ domains of Pyd with GST, GST fused to the cytoplasmic domain of Sdk, or GST fused to the cytoplasmic domain of SdkΔBD. n=4 (full-length) or 5 (PDZ). Sdk directly interacts with Pyd; the Pyd PDZ domains are sufficient for this interaction, and the PDZ-binding motif of Sdk is required. Scale bars, 10 μm. See also Figure S6.
To test whether the functional interactions between Sdk, Cno and Pyd result from direct physical interactions, we attempted to pull down tagged Cno or Pyd proteins from lysates of transfected S2 cells using a fusion of glutathione-S-transferase (GST) to the cytoplasmic domain of Sdk. We also used a similar GST fusion lacking the PDZ-binding motif (SdkΔBD) to determine whether the interaction was dependent on this motif. GST-Sdk pulled down both full-length HA-tagged Pyd and a Myc-tagged construct with the three PDZ domains of Pyd, and these interactions were strongly reduced by deleting the PDZ-binding motif of Sdk (Fig. 6F, S6A). Consistent with this result, the Sdk PDZ-binding motif is predicted to bind to the first PDZ domain of Pyd, but not to the PDZ domain of Cno (http://pow.baderlab.org/). Although we could not demonstrate a direct interaction with Cno, the requirement for Sdk to localize Cno to tAJs suggests that it is in a complex with Cno. As Pyd and its homologues have been reported to directly bind to Cno and its homologues (Takahashi et al., 1998; Yamamoto et al., 1997), Pyd could potentially recruit Cno to this complex. However, Cno still localized to tAJs in pyd mutant embryos and in wing disc cells from which Pyd was depleted by RNAi (Fig. S6B–D), arguing that its interaction with Sdk does not depend on Pyd. Additional components of the Sdk intracellular complex may contribute to the recruitment of Cno. Overall, our results are consistent with the hypothesis that Sdk maintains or transmits cell bond tension by anchoring the ends of actin filaments to tricellular adherens junctions through Pyd, Cno and potentially other proteins (Fig. 7).
Figure 7: Sdk acts at tAJs to control cell bond tension.
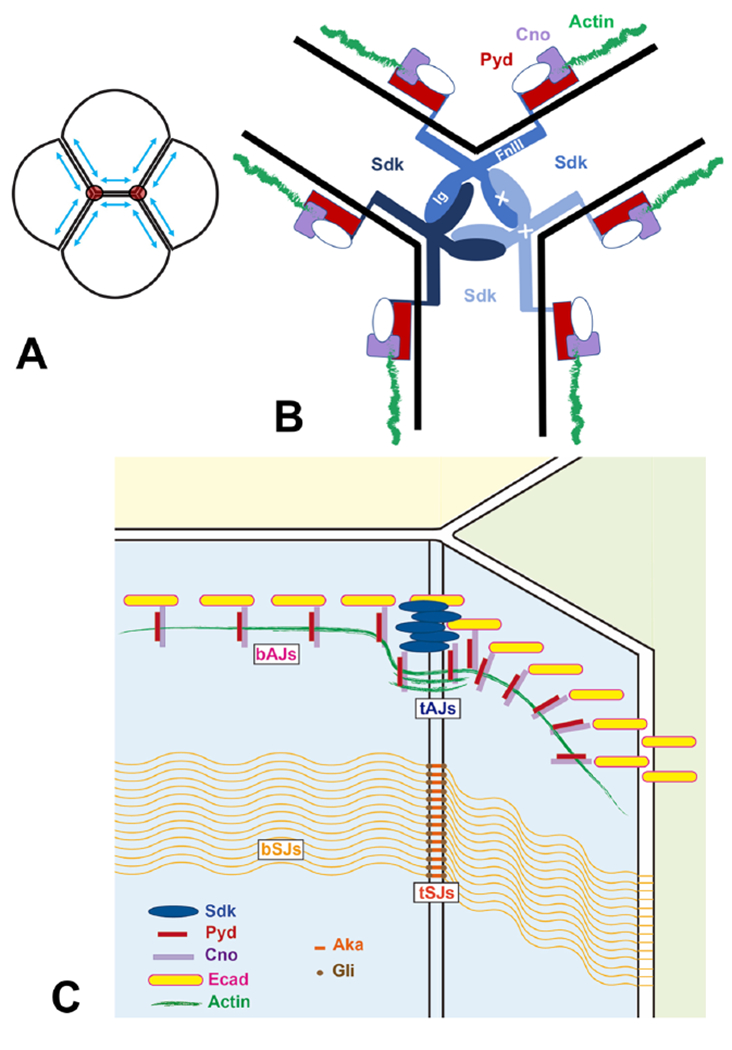
(A) diagram of four cells indicating how tension (blue arrows) is highest at tricellular junctions (red circles). (B) model showing how the structure of Sdk might favor interactions at cell contacts with the geometry of tAJs and how Sdk could anchor the ends of actin filaments through Pyd and Cno. Potential trans interactions between Sdk Ig domains and cis interactions between FnIII domains are indicated by white crosses. (C) Diagram showing the position of tAJs relative to bAJs, bSJs and tSJs.
Discussion
Sdk acts as a hub to anchor the cytoskeleton to tAJs
Sdk is the only protein so far shown to be specifically localized to tAJs and almost excluded from bAJs in most epithelia. Although several proteins were known to be enriched at tricellular contacts at the level of AJs (Beati et al., 2018; Bonello et al., 2018; Lye et al., 2014; Rauskolb et al., 2014; Razzell et al., 2018; Sawyer et al., 2009), most of them are also present along the whole bicellular junction. We find that Sdk is required for the specific enrichment of Cno, Pyd and actin at tAJs, but not their localization at bAJs. Consistent with this, cno and pyd mutants have stronger phenotypes than sdk mutants (Choi et al., 2011; Matsuo et al., 1999; Seppa et al., 2008), indicating that these proteins have functions independent of Sdk. In contrast, Sdk localizes to tAJs even in the absence of Cno and Pyd. We propose that Sdk is the hub that organizes a protein complex specifically at tAJs to modulate the actin cytoskeleton.
We have shown that the C-terminal predicted PDZ-binding motif is required for Sdk function, and directly interacts with Pyd. The requirement for Sdk to recruit Cno to tAJs and the ability of misexpressed Cno to expand Sdk suggest that Cno is also in a complex with Sdk. As the vertebrate homologues of Cno and Pyd can bind to each other (Takahashi et al., 1998; Yamamoto et al., 1997), all three proteins may be present in the same complex. However, knocking down ZO-1 proteins in MDCK cells increases the recruitment of Afadin to tAJs (Choi et al., 2016), so it is also possible that Cno and Pyd compete for the same binding site on Sdk. Although we could not detect a direct interaction between Sdk and Cno, our experiments did not exclude this possibility. Alternatively, Sdk might recruit Cno indirectly, for example by binding to a GEF that increases Rap activity (Bonello et al., 2018). Cno is known to interact with Ed, which is necessary for Ecad recruitment to bAJs (Wei et al., 2005); it is not yet clear whether Ed is in the same complex as Sdk or whether Cno interacts with the two proteins in a mutually exclusive manner. In addition to anchoring the ends of actin filaments (Choi et al., 2016), Cno might recruit other regulators of tension such as the LIM domain protein Smallish (Beati et al., 2018). pyd mutant embryos show defects in tracheal intercalation similar to sdk mutants (Jung et al., 2006), and pyd and cno genetically interact with sdk in this context, supporting a general role for these factors downstream of Sdk.
Although Cno and Pyd are likely to be important mediators of the effects of Sdk on tension and junctional stability (Choi et al., 2016), other partners may also contribute to Sdk function. The cytoplasmic domain of Sdk has several regions of strong evolutionary conservation, which could serve as interaction domains for non-PDZ proteins. Elucidating the nature of the complex that has Sdk as its hub will provide important clues to the structure and function of tAJs. The expression of mouse Sdks in tissues other than the nervous system, such as the ureteric bud during branching morphogenesis of the kidney (Kaufman et al., 2004), suggests that the role of Sdk at tAJs may be conserved, although it is not yet known whether Sdks accumulate at tAJs in epithelia in other organisms.
Sdk localization to tAJs may depend on both geometry and tension
While Sdk can mediate homophilic interactions at bicellular contacts in cultured cells (Tang et al., 2018), in epithelial tissues it localizes almost exclusively to vertices. Several mechanisms might contribute to this localization pattern. It is possible that the topology of membranes at vertices imposes a particular structural conformation on Sdk that promotes its clustering there. The FnIII domains of Sdk molecules can interact with lipids and are thought to lie flat along the plasma membrane at bicellular contacts between parallel membranes, while the first four Ig domains mediate homophilic adhesion; the fourth and fifth Ig domains meet at a relatively rigid angle of 126° (Goodman et al., 2016; Tang et al., 2018). This might make Sdk-mediated adhesion at tricellular interfaces, at which cells meet at an average angle of 120°, more energetically favorable (Fig. 7B). Alternatively or additionally, this angle might favor cis-interactions between Sdk molecules through their FnIII domains over FnIII-membrane interactions, contributing to Sdk clustering (Kunz et al., 2002). Such a geometric model could explain why Sdk is still enriched at tAJs when it is present on only two of the three cells that are in contact. However, this cannot be the only mechanism for Sdk localization, as tricellular vertices can form at a range of angles.
In addition, we find that tension enhances Sdk recruitment to tAJs. Although the Pyd homologue ZO-1 is stretched by tension, which controls its interactions with some of its binding partners (Spadaro et al., 2017), neither Pyd nor Cno is required for the initial localization of Sdk to tAJs. This argues that Sdk can detect tension by a mechanism independent of and prior to its own role in anchoring actin filaments through Cno and Pyd. Sdk might itself be mechanosensitive, as FNIII domains can stretch in response to force (Ohashi et al., 1999). Force exerted on the plasma membrane to pull it away from Sdk-Sdk adhesions could extend the FNIII domains to open up more interaction sites. Alternatively, as Ecad is necessary for Sdk localization, Sdk could detect tension through the cadherin-catenin complex, perhaps by interacting with α-catenin or another protein that is deformed by mechanical force.
Tricellular adherens junctions modulate tension during cell rearrangements
Mechanical forces can regulate tissue morphogenesis by promoting cell intercalation, oriented cell division or cell extrusion (Pinheiro and Bellaiche, 2018). Adherens junctions are known to transduce mechanical force, with one prominent mechanism being the force-dependent unfolding of α-catenin, which exposes binding sites for vinculin and other proteins (Yonemura et al., 2010). Vinculin is enriched at tAJs, indicating that tension is highest at these positions (Choi et al., 2016; Higashi et al., 2016)(Fig. 7A) and suggesting that tAJs might sense and regulate tension (Higashi and Miller, 2017). At bAJs, the cadherin-catenin complex resists tension by linking cell adhesions to the actin cytoskeleton through catch bonds (Pinheiro and Bellaiche, 2018). tAJs may require additional mechanisms to resist contractility because they anchor the ends of actin filaments (Choi et al., 2016; Yonemura et al., 2010), generating tension in an actomyosin network both along the cell cortex and in radially directed filaments (Vanderleest et al., 2018). The requirement for Sdk to maintain normal levels of tension along cell bonds makes it a good candidate to mediate this tension-modulating function of tAJs. The homophilic adhesive properties of the extracellular domain of Sdk and its interaction with the actin cytoskeleton could allow it to transmit tension between cells or organize supracellular contractile networks that facilitate mechanical coordination across a tissue. Sdks mediate strong and compact adhesions with tightly packed molecules (Tang et al., 2018), potentially allowing them to resist force. However, the Ecad complex is sufficient to maintain adhesion at tAJs in the absence of Sdk.
Cell shape changes and cell rearrangements often involve oscillations in apical area driven by pulses of actomyosin contraction (Del Signore et al., 2018; Fernandez-Gonzalez and Zallen, 2011; Gorfinkiel and Blanchard, 2011; Martin et al., 2009; Rauzi et al., 2010). Both reduced tension on cell bonds in the embryo and defects in cell intercalation in the retina in sdk mutants could result from failure to connect the oscillatory network to tAJs. A lack of mechanical coupling of tensile actomyosin networks to tAJs would disrupt cell intercalation by reducing the efficiency of T1 transitions mediated by changes in junction length. In the trachea, Sdk accumulates at autocellular-intercellular vertices and is required for the replacement of intercellular junctions by autocellular ones. Tracheal cell intercalation does not require myosin contractility (Ochoa-Espinosa et al., 2017), suggesting that other actin regulators provide local forces to resolve cell rearrangements. Sdk may mediate the replacement of intercellular junctions by homophilic adhesion and/or by modulating the actin cytoskeleton to provide polarized tension or polarized membrane growth that allows vertex displacement during the zipping process. In general, vertex displacements in complex cell rearrangements may rely on the Sdk complex to transmit tension.
As tTJs can also contribute to actin organization and tension (Oda et al., 2014), it will be important to investigate the interactions between the two types of tricellular junctions. Such studies may shed light on the role of tricellular junctions in regulating cell proliferation, stem cell homeostasis, and tissue integrity (Bosveld et al., 2018).
STAR Methods
Contact for reagent and resource sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jessica Treisman (Jessica.Treisman@nyulangone.org).
Experimental model and subject details
Drosophila melanogaster strains were maintained on cornmeal/agar molasses fly food at room temperature (20°C). Embryos, third instar larvae and pupae of both sexes were used for the experiments described, except that sdk and sqh mutant clones could only be generated in females, as these genes are on the X chromosome. The stocks used were sdkMB05054; sdk315 (Astigarraga et al., 2018); sdkCPTI000337 (Lye et al., 2014); akaYD1046 (Aka-GFP) (Byri et al., 2015); cnoCPTI000590(Cno-GFP, Flybase); pydMI01205-GFSTF.1 (Pyd-GFP, Flybase); UAS-LifeAct-Ruby, UAS-LifeAct-GFP (Flybase); IGMR-GAL4 (Flybase); C381-GAL4 (Manseau et al., 1997); UAS-MbsN300 (Lee and Treisman, 2004); UAS-ctMLCK (Kim et al., 2002); sqhAX3 (Jordan and Karess, 1997); ed1X5 (Bai et al., 2001); btl-GAL4 (Vincent et al., 1997); UAS-CnoGFP (Perez-Gomez et al., 2013); cnoR2 (Sawyer et al., 2009); Df(pyd)B12 (Choi et al., 2011); UAS-pyd RNAiTRiPHM05131 (Flybase); Ecad-GFP (Huang et al., 2009); α-catenin-GFP (Venken et al., 2011); and apmd544-GAL4 (O’Keefe et al., 1998). UAS-Sdk-HA (Astigarraga et al., 2018) and UAS-SdkΔBD-HA were injected by Genetivision into flies containing the VK31 attP site at 62E1. Stocks used to make clones were (1) eyFLP, tub-GAL80, FRT19A; tub-GAL4, UAS-GFP/CyO, P(Tb, Ubi-RFP) (2) eyFLP, tub-GAL80, FRT19A; tub-GAL4/ CyO, P(Tb, Ubi-RFP) (3) sdkMB05054, FRT19A; UAS-lacZ (4) UbxFLP, UAS-GFP; tub-GAL80, FRT40A; tub-GAL4/TM6B (5) FRT82B, UAS-CnoGFP (6) eyFLP; tub-GAL4, FRT82B, tub-GAL80/TM6B (7) sdkΔ15, FRT19A; UAS-CnoGFP (8) FRT82B, cnoR2/TM6B (9) hsFLP122; FRT82B, ovoD1/TM3 (10) FRT19A, sqhAX3/FM6, P(Tb, Ubi-RFP) (11) FRT40A, ed1X5/SM6-TM6B (12) FRT82, UAS-pyd RNAi. S2 cells were maintained at 25°C in Schneider’s medium supplemented with 10% fetal calf serum. We believe this cell line to be male based on unpublished RNA-Seq experiments in which we detected expression of a gene on the Y chromosome.
Method details
Immunohistochemistry
Embryos were staged as described previously (Campos-Ortega and Hartenstein, 1985) and stained following standard protocols. Immunostainings were performed on embryos fixed in 4% formaldehyde in PBS-Heptane (1:1) for 20 minutes, except for DCAD2 staining, for which the embryos were fixed for 10 minutes. Pupal retinas and wing imaginal discs were dissected in 0.1M sodium phosphate buffer pH 7.2 or in PBS and fixed for 30 min on ice in 4% formaldehyde in 0.1 M PIPES pH 7.0/2 mM EGTA/1mM MgSO4. Samples were washed for 15 min in phosphate buffer/0.2% Triton X-100 and incubated in primary antibody in phosphate buffer/0.2% Triton X-100/10% donkey serum overnight at 4°C. After three 5-minute washes in phosphate buffer/0.2% Triton X-100, they were incubated for 2-3 h at 4°C in secondary antibody in phosphate buffer/0.2% Triton X-100/10% donkey serum, washed again three times, and mounted in 80% glycerol in PBS. Embryos were mounted in Fluoromount-G (Southern Biotech). Antibodies used were rat anti-Ecad (1: 10; DCAD2, Developmental Studies Hybridoma Bank (DSHB)); rat anti-Ncad (1:10; DN-Ex #8, DSHB); guinea pig anti-Sdk (1:300; (Astigarraga et al., 2018)); mouse anti-Gli 1F6 (1:10; (Auld et al., 1995)); chicken anti-GFP (1:400; Life Technologies); rabbit anti-phospho-myosin light chain 2 (Ser19) (Cell Signaling #3675); mouse anti-actin (clone C4, 1:1000, MP Biomedicals); rabbit anti-Zip (gift from T. Ohshiro); anti-phospho-Sqh (SqhP1; (Zhang and Ward, 2011)); rabbit anti-β-galactosidase (1:5000; Cappel); rabbit anti-Cno (1:400; (Speicher et al., 2008)); mouse anti-Dlg (1:10, DSHB) and mouse anti-Pyd (1:10; PYD2, DSHB). Rhodamine-labelled 10 KD dextran injections were used for an in vivo permeability assay in stage 17 embryos and were performed as described (Tsarouhas et al., 2007).
Live imaging
Live imaging of pupal eyes was performed as previously described (Del Signore et al., 2018), using α-catenin::GFP (Venken et al., 2011) to follow cellular behavior, and both UAS-Lifeact::GFP (Riedl et al., 2008) and sgh-Sqh::mCherry (Martin et al., 2009) to follow actin and myosin dynamics simultaneously. An image stack containing 15 sections was obtained every minute with optimal pinhole using a 63X, 1.4 NA, plan Apochromat oil immersion objective, 0.7 μm per optical section with a 50 % overlap between sections, at a scan speed of 7, averaging of 1 with an overall pixel dwell time of ~1. Image stacks were then projected along the Z axis using the maximal intensity projection plugin, processed using the Gaussian blur filter (sigma=1.3) and registered in time using the Correct 3D Drift plugin.
Embryos for live imaging were dechorionated and lined up on Menzel-Glaser cover slips with 10-S Voltalef oil (VWR) and covered with a stretch membrane (YSI membrane kit). Live imaging was performed on a Spinning disk confocal microscope (Andor Revolution system equipped with a EMCCD camera) using a 100X, 1.4 NA oil immersion objective. Stacks of 0.5 μm confocal planes were acquired every 15 seconds, imported to ImageJ and projected along the Z axis using the maximal intensity projection option.
Laser ablation
Laser nanosurgery was performed with a standing beam of a 355-nm pulsed (470 ps)-laser (Teemphotonics, France) with average exposure per cut of 20 to 30 pulses at 100 Hz and 250 nJ energy per pulse, coupled through the epi-port of an AxioVert 200 M (Carl Zeiss, Germany) and focused onto cell junctions through a Zeiss C-Apo 63x/1.2 W lens, as described (Colombelli and Solon, 2013). Fluorescence confocal images were acquired through a custom spinning disk unit coupled to the inverted microscope and with a Flash4.0 SCMOS camera (Hamamatsu, Japan), with the excitation line at 488 nm of a solid-state laser (LuxX series, Omicron, Germany). Fluorescence imaging was done at the rate of 0.642 sec/frame. GFP fluorescence of a knock-in ECad-GFP allele (Huang et al., 2009) was used to visualise cell-cell junctions. Experiments were performed using as the control condition ECad-GFP embryos (n=18 cuts from 16 embryos, acquired from two independent experiments) and as mutant condition sdkMB805054; ECad-GFP embryos (n= 32 cuts from 18 embryos, acquired from two independent experiments). Stage 10-11 embryos were used in all experiments. Cell-cell junctions of the central region of the epidermis not involved in cell divisions were selected for laser ablation (avoiding the most anterior and posterior regions and the most dorsal and ventral regions). Time-lapse images were then analysed using ImageJ software (version Fiji Lifeline, June 2014) (Schindelin et al., 2012). MTrackJ plugin was used to measure the displacement of the vertices in contact with the dissected junctions. To extract initial retraction velocity, the profile of spatial relaxation was automatically calculated from the path length of the tracks and fitted with a single exponential curve using the Igor software (Wavemetrics, Portland, OR). The initial retraction velocity V0 was calculated from the fitting function f(t) as follows: f(t) = A · (1-e-t/τ ) and V0 = df(0)/dt = A/τ where A is the total retraction amplitude and τthe decay rate constant of the exponential. The fluorescence recoil velocity of control and mutant embryos was compared using the Scatter Plot tool of GraphPad Prism. Error bars indicate standard deviation (SD). p-values were obtained with an unpaired two-tailed Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001.
Cloning
The UAS-SdkΔBD-HA plasmid was made by replacing the 3’ Xbal-Nhel fragment of UAS-Sdk (Astigarraga et al., 2018) with an Xbal-Nhel PCR fragment from the 3’ end of sdk in which a stop codon replaced the last six amino acids. UAS-SdkΔBD-HA also has a change of Ser 2213 to Ala, which was introduced inadvertently but is present in some Sdk homologues from other species. sdk cDNA fragments encoding the intracellular portion of Sdk (amino acids 2025 to 2224) or SdkΔBD (amino acids 2025 to 2220) were cloned into the EcoRI and Notl sites of the pGEX-4T-1 vector by PCR, using UAS-Sdk as the template. UAS-Myc-PydPDZ was generated by PCR using UAS-Pyd-FLAG-HA (UFO07742, Drosophila Genomics Resource Center) as a template to amplify the three PDZ domains of Pyd with a Myc tag in the 5’ primer, and cloned into the EcoRI and Xhol sites of pUAST-attB.
GST pulldowns
GST-Sdk, GST-SdkΔBD and pGEX-4T-1 were transformed into BL21 E. coli competent cells for translation upon induction with 0.1 mM IPTG. Bacteria were sonicated in PBS/1% Triton X-100/cOmplete Protease Inhibitor Cocktail (Roche) and GST fusion proteins were purified by incubating the supernatant with glutathione-sepharose beads and centrifuging 1 min at 750 g. Beads were washed and resuspended in PBS/ cOmplete Protease Inhibitor Cocktail.
S2 cells were transfected with full-length UAS-Pyd-FLAG-HA or UAS-Myc-PydPDZ, using Effectene (Qiagen) according to the manufacturer’s protocol. After 3 days, cells were lysed in 75mM NaCl/ 50mM Tris-HCl pH7.5/0.2% NP-40/1mM EDTA/ cOmplete Protease Inhibitor Cocktail/5mM NaF/1mM Na3VO4, centrifuged to remove debris, pre-cleared with glutathione-sepharose beads for 90 min, centrifuged and incubated with GST protein-glutathione-sepharose beads for 2 h. The beads were pelleted at 750 g, washed 3 times in lysis buffer, boiled in Laemmli buffer and run on a 15% SDS-PAGE gel. Aliquots of the GST proteins run on a separate gel were stained with Coomassie blue to compare the amounts of protein used for the pulldowns.
Western blotting
For Western blotting, gels were transferred to nitrocellulose membranes (Biorad) and were blocked overnight with TBST (20 mM Tris pH 7.5/150 mM NaCl/0.2% Tween-20) supplemented with 10% low-fat milk. Membranes were incubated with TBST + 10% milk supplemented with antibodies for 1 hour at room temperature. Blots were washed with TBST for an hour and incubated with HRP-conjugated secondary antibodies (1:2000, Jackson Laboratories) for another hour. Blots were developed with enhanced chemiluminescence (Pierce). Antibodies used were rat anti-HA 3F10 (Roche) and mouse anti-Myc (Cell Signaling).
Latrunculin A treatment
Embryos expressing the Cno-GFP or Ecad-GFP protein trap were dechorionated and rinsed several times with 0.9% NaCl. Embryos were incubated in 1:1 octane/5mM Latrunculin A (Enzo) in 0.9% NaCl for 40 min at RT with rocking. After removing both phases, embryos were rinsed twice with heptane and fixed immediately. The working solution of Latrunculin A was prepared from a 1 mM solution in 10% DMSO. For controls, the embryos were treated with the DMSO carrier alone. Embryos were then stained with anti-GFP, anti-Sdk and anti-actin.
Quantification and statistical analysis
All quantifications of fluorescence intensity at tAJs were done on maximum projections of several sections from confocal z-stacks stained for Ecad to mark cell boundaries. To quantify protein enrichments of Sdk, Ecad, Cno, actin and Pyd at tAJs, we used ImageJ to measure the fluorescence intensity of antibody staining in identical circular ROIs (just large enough to encompass the staining at tAJs) at one tAJ per cell, an adjacent bAJ, and the center of the cell (used as a measure of the background fluorescence). Enrichment at each tAJ was calculated as the background-subtracted intensity at the tAJ divided by the background-subtracted intensity at the adjacent bAJ. To quantify the enrichment of Sdk upon manipulations of myosin activity, we used identical circular ROIs for each embryo to measure the intensity of Sdk antibody fluorescence, after subtracting background, at 20 tAJs in the amnioserosa and 20 tAJs in the dorsal epidermis in each embryo analyzed. The enrichment of Sdk in the amnioserosa was defined as the ratio of the mean intensity at tAJs in the amnioserosa over the mean intensity at tAJs in the epidermis for each embryo. To determine the relative intensity of Sdk fluorescence at tAJs including sdk mutant cells, we used identical circular ROIs to measure fluorescence intensity at tAJs with 0, 1 or 2 mutant cells, and to measure background fluorescence at at least 10 locations in the same image. The mean background fluorescence intensity was subtracted from each measurement, and the intensity at tAJs that included mutant cells was normalized to the mean intensity at tAJs with only wild type cells in the same image. To count the number of tAJs that had detectable Sdk fluorescence, we first increased the gain so that even low levels of fluorescence were visible. Similarly, Sdk intensity at tAJs in ed mutant or Ecad RNAi expressing clones was normalized to the mean intensity at tAJs in wild type regions from the same image, after subtracting background from each measurement. Significance was calculated by unpaired t-test, with Welch’s correction if variances were significantly different, or by one-sample t-test when comparing the ratio of Sdk in ed mutant or Ecad RNAi expressing cells to wild type cells in the same wing disc to an expected value of 1.
We used ImageJ to measure the length of a line connecting the apical-basal center of anti-Sdk staining to the apical-basal center of anti-Gli staining on xz-projections of confocal stacks from wild type retinas. We also counted the number of vertices in xz projections in which Sdk staining fully or partially overlapped Ecad staining or was adjacent to it. We used xy projections of several confocal sections to count the percentage of vertices showing Sdk localization that was complementary to the pattern of Zip or p-Sqh, partial colocalization of Sdk with Zip or p-Sqh (including weak accumulation of myosin at the tAJ) or full colocalization of Sdk with a distinct spot of myosin. To quantify actin and myosin levels at contacts during peak expansion or contraction, we used ImageJ to measure LifeAct-GFP or Sqh::mCherry fluorescence along the length of the contact, with a line width of 1.7 μm. We used Fiji to measure the straightness of the junctions in movies S4 and S5. We selected a comparable time frame of control and mutant embryos and analyzed cells of the dorsal epidermis which were not in direct contact with the amnioserosa. Straightness was calculated as a ratio of the shortest distance between junction vertices to the path length of the junction. We counted the number of tracheal branches in stage 15-16 embryos that had no defects or that showed remaining intercellular junctions (Shaye et al., 2008). To quantify retinal defects, we counted the number of missing lattice cells, extra lattice cells, and adjacent bristle pairs or adjacent tertiary pigment cell pairs and the total number of ommatidia in the field in wild type and sdkMB05054 42 h pupal retinas. Significance was calculated by Fisher’s exact test. To compare the effects of Cno overexpression in wild type or sdk mutant cells, we measured the number of interommatidial cells separating the primary pigment cells on each lattice edge within the GFP-labeled clone. Wild-type controls were non-GFP-labeled ommatidia from the same images. Significance was calculated using a chi-squared test. Sample numbers and definitions of error bars are given in the figure legends.
Supplementary Material
Movie S1, related to Figure 3: sdk is required for tracheal intercalation. sdkMB05054; Ecad-GFP embryo imaged for 114 min, focused on two dorsal branches during the process of cell intercalation (from early stage 14 to stage 15). In normal conditions (as in the left dorsal branch, marked with a yellow arrow), the zipping up process replaces ring-shaped intercellular junctions with straight-line autocellular junctions. This junctional remodelling accompanies the repositioning of the cells from a side-by-side to an end-to-end position. At the end of the intercalation phase the dorsal branches are sealed by autocellular junctions interrupted by small rings of intercellular junctions (which connect the cells at their ends). In sdk mutant embryos the zipping up process is perturbed in many branches (right dorsal branch marked with a blue arrow). This perturbation generates fully extended dorsal branches that still maintain intercellular junctions, indicating that the cells have not repositioned into a row and maintain a side-by-side position. Scale bar, 10 μm.
Movie S2, related to Figure 3: Actomyosin is recruited to shrinking junctions during retinal lattice cell intercalation. LifeAct-GFP (green, A’); Sqh-mCherry (red, A’’) pupal retina imaged at 26-27 h APF, showing dynamic recruitment of actin and myosin to shrinking junctions during cell intercalation and their loss from expanding junctions.
Movie S3, related to Figure 3: sdk is required for cell intercalation in the retina. sdkA15; GFP-α-catenin pupal retina imaged at 28-32h APF. (A) shows an ommatidium with normal cell intercalation, followed by apoptosis of the extra cells. (B) shows an ommatidium with defective cell intercalation in the same eye resulting in misplacement of bristles and lattice cells. Note the apoptosis of cells that failed to intercalate. (A’, B’) show enlargements of ommatidial edges that are demarcated in boxes in upper panels. Scale bars, 10 μm.
Movie S4, related to Figure 3: Cell bonds are under high tension in wild type embryos. Wild type stage 7 Ecad-GFP embryo imaged for 39.5 min at 29°C, focused on the dorsolateral region of the epidermis, showing the amnioserosa (larger cells) and the dorsal epidermis (smaller cells) in contact. Cell bonds are straight (junction straightness is 1.0 ± 0.05, n=30 cells of the dorsal epidermis), reflecting high levels of tension. Scale bar 15μm.
Movie S5, related to Figure 3: Cell bond tension is reduced in sdk mutant embryos. sdkMB05054; Ecad-GFP stage 8 embryo imaged for 52 min at 29°C, focused on the dorsolateral region of the epidermis, showing the amnioserosa (larger cells) and the dorsal epidermis (smaller cells) in contact. The presence of wiggly bonds indicates reduced tension. Junction straightness of the dorsal epidermis cells is 0.87 ± 0.10 (n=30 cells), which is significantly different from wildtype (p<0.0001, unpaired t-test). Scale bar 10μm.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken polyclonal anti-GFP | Invitrogen | Cat # A10262 |
| Rat monoclonal anti-N-cadherin | Developmental Studies Hybridoma Bank | Cat # DN-Ex #8; RRID AB_528121 |
| Guinea pig polyclonal anti-Sidekick | Astigarraga et al., 2018 | N/A |
| Rat monoclonal anti-E-cadherin | Developmental Studies Hybridoma Bank | Cat # DCAD2; RRID AB_528120 |
| Mouse monoclonal anti-Gliotactin | Auld et al., 1995 | RRID AB_2566915 |
| Rabbit monoclonal anti-myosin light chain 2 (Ser19) | Cell Signaling | Cat # 3675; RRID AB_2250969 |
| Mouse monoclonal anti-actin | MP Biomedicals | Clone C4; RRID AB_2335127 |
| Rabbit polyclonal anti-β-galactosidase | MP Biomedicals | Cat # 0855976; AB_2334934 |
| Rabbit polyclonal anti-Canoe | Speicher et al., 2008 | N/A |
| Mouse monoclonal anti-Discs large | Developmental Studies Hybridoma Bank | Cat # 4F3; RRID AB_528203 |
| Mouse monoclonal anti-Polychaetoid | Developmental Studies Hybridoma Bank | Cat # PYD2; RRID AB_2618043 |
| Rabbit polyclonal anti-Zipper | Tomokazu Ohshiro | N/A |
| Rat monoclonal anti-HA 3F10 | Roche | Cat # 11815016001; RRID AB_390914 |
| Mouse monoclonal anti-Myc tag 9B11 | Cell Signaling | Cat # 2040; RRID AB_2148465 |
| GFP-Trap A | Chromotek | Cat # gta-10; RRID AB_2631357 |
| Alexa Fluor 555-conjugated phalloidin | Invitrogen | Cat # A34055 |
| Guinea pig anti-Sqh1P | Zhang and Ward, 2011 | N/A |
| Rabbit polyclonal anti-GFP | Molecular Probes - Thermo Fisher Scientific | Cat # A11122; RRID: AB_221569 |
| Bacterial and Virus Strains | ||
| E.coli BL21 | Sigma | Cat # B2685-10X50μl |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Effectene Transfection Reagent | Qiagen | Cat # 301425 |
| Dextran, Rhodamine B, 10,000MW, Neutral | ThermoFisher Scientific | Cat # 1824 |
| Gibco® Schneider’s Drosophila medium | Invitrogen | Cat # 21720001 |
| Gibco® Fetal bovine serum, qualified, heat inactivated | Invitrogen | Cat # 16140-071 |
| Shields and Sang M3 insect medium | Sigma | Cat # S3652 |
| Latrunculin A | Enzo | Cat # BML-T119 |
| Octane | Sigma-Aldrich | Cat # 657042 |
| GST-Sdk | This paper | N/A |
| GST-SdkΔBD | This paper | N/A |
| cOmplete Protease Inhibitor Cocktail | Roche | Cat # 11873580001 |
| Glutathione-sepharose beads | Thermo-Fisher | Cat # 17075601 |
| 10-S Voltalef oil | VWR Chemicals | Cat # 24627.188 |
| Experimental Models: Cell Lines | ||
| D. melanogaster: Cell line S2: S2-DRSC | Laboratory of Ruth Lehmann | FlyBase: FBtc0000181 |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster: sdkMB05054 | Bloomington Drosophila Stock Center | BDSC; 24603; Flybase: FBal0198242 |
| D. melanogaster: sdkΔ15 | Astigarraga et al., 2018 | BDSC; 24603; Flybase: FBal0340184 |
| D. melanogaster: sdkCPTI00037 | Kyoto Drosophila Genomics and Genetic Resources | Kyoto; 115107; Flybase: FBal0262454 |
| D. melanogaster: sdkMI101498-GFSTF.1 | Bloomington Drosophila Stock Center | BDSC; 60169; Flybase; FBti0178565 |
| D. melanogaster: akaYD1046 | Bloomington Drosophila Stock Center | BDSC; 51179; Flybase: FBal0212250 |
| D. melanogaster: pydMI01205-GFSTF.1 | Bloomington Drosophila Stock Center | BDSC; 60166; Flybase: FBal0314561 |
| D. melanogaster: UAS-LifeAct-Ruby | Bloomington Drosophila Stock Center | BDSC; 35545; Flybase: FBti0143328 |
| D. melanogaster: P{ubi-GFP.D} | Bloomington Drosophila Stock Center | BDSC; 1681; Flybase: FBti0004735 |
| D. melanogaster: longGMR-GAL4 | Bloomington Drosophila Stock Center | BDSC; 8605; Flybase: FBti0058798 |
| D. melanogaster: C381-GAL4 | Manseau et al., 1997 | N/A |
| D. melanogaster: UAS-MbsN300 | Lee and Treisman, 2004 | Flybase: FBal0182792 |
| D. melanogaster: UAS-MLCK.ct | Bloomington Drosophila Stock Center | BDSC; 37527; Flybase: FBti0026705 |
| D. melanogaster: sqhAX3 | Bloomington Drosophila Stock Center | BDSC; 25712; Flybase: FBal0035707 |
| D. melanogaster: ed1X5 | Bai et al., 2001 | Flybase: FBal0086650 |
| D. melanogaster: btl-GAL4 | Bloomington Drosophila Stock Center | BDSC; 78328; Flybase: FBtp0001208 |
| D. melanogaster: UAS-CnoGFP | Perez-Gomez et al., 2013 | Flybase: FBal0294979 |
| D. melanogaster: cnoR2 | Bloomington Drosophila Stock Center | BDSC; 51328; Flybase: FBal0243335 |
| D. melanogaster: Df(pyd)B12 | Bloomington Drosophila Stock Center | BDSC; 51329; Flybase: FBal0285508 |
| D. melanogaster: UAS-pydRNAiHM05131 | Bloomington Drosophila Stock Center | BDSC; 28920; Flybase: FBtp0051218 |
| D. melanogaster: Ecad-GFP | Huang et al., 2009 | Flybase: FBtp0247909 |
| D. melanogaster: α-Cat::GFP | Bloomington Drosophila Stock Center | BDSC; 59405; Flybase; FBti0168861 |
| D. melanogaster: apmd544-GAL4 | Bloomington Drosophila Stock Center | BDSC; 3041; Flybase: FBal0051787 |
| D. melanogaster: UAS-Lifeact::GFP; sqh-Sqh::mCherry, GMR-GAL4 | Del Signore et al., 2018 | N/A |
| D. melanogaster: sqh-UtrABD::Cherry | Vazquez et al., 2014 | Flybase: FBal0298059 |
| Oligonucleotides | ||
| Sdk XbaI 5’ primer AGACTCTAGAGGAATCTATGGCCATG |
This paper | N/A |
| Sdk NheI 3’ primer CAGCTAGCGGCAGTGGTGCCCTC |
This paper | N/A |
| Sdk EcoRI 5’ primer ATCGGAATTCAAGAGCAAGAGCTATAAA |
This paper | N/A |
| Sdk NotI 3’ primer ATCGGCGGCCGCTCAGACGAATGACGAGAAGC |
This paper | N/A |
| SdkΔBD NotI 3’ primer ATCGGCGGCCGCTCAGAAGCCGG |
This paper | N/A |
| Pyd Myc EcoRI 5’ primer ACTGGAATTCCAAACATGGAGCAGAAGCTGATCTCCGAGGAGGACCTGGCTAGCGAGTACCACACGGTGGCGGTGA | This paper | N/A |
| Pyd XhoI 3’ primer ATCGCTCGAGTCAGCAGTACTGCACAATCAGATCGATGC |
This paper | N/A |
| Recombinant DNA | ||
| pUAS-Pyd-FLAGHA | Drosophila Genomics Resource Center | Cat # UFO07742 |
| pUAS-Myc-PydPDZ | This paper | N/A |
| pGEX-4T-Sdk | This paper | N/A |
| pGEX-4T-SdkΔBD | This paper | N/A |
| Software and Algorithms | ||
| ImageJ 1.50d | National Institutes of Health; Schindelin et al., 2012 | http://imagej.nih.gov.ij |
| GraphPad Prism 7.0c | GraphPad Software, Inc. | https://www.graphpad.com |
| Igor Pro 6.04 | WaveMetrics, Inc. | http://wavemetrics.com |
Highlights.
Sidekick is the hub of a complex specific to tricellular adherens junctions
Sidekick can sense and modulate cell bond tension
Sidekick is necessary for normal cell and junctional rearrangements
Sidekick links actin filaments to vertices through Canoe and Polychaetoid
Acknowledgements
We thank Vanessa Auld, Ana Carmena, Yang Hong, Jui-Chou Hsu, Adam Martin, Tomokazu Ohshiro, Mar Ruiz-Gómez, the Bloomington Drosophila stock center, the Drosophila Genomics Resource Center and the Developmental Studies Hybridoma Bank for fly stocks and reagents, and Flybase for essential information. We also thank A. D’Angelo, A. Godeau and J. Solon (CRG, Barcelona) for discussions, technical advice and help. The manuscript was improved by the critical comments of Jordi Casanova, Ariel Hairston, Cheuk Hei Ho and Hongsu Wang. This work was supported by the National Institutes of Health (grants EY025540 to J.E.T. and GM129151 to V.H.) and by Ministerio de Economía y Competitividad of the Spanish Government (BFU2012-39509-C02, BFU2015-68098-P to M.L.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests: The authors declare no competing interests.
References
- Astigarraga S, Douthit J, Tarnogorska D, Creamer MS, Mano O, Clark DA, Meinertzhagen IA, and Treisman JE (2018). Drosophila Sidekick is required in developing photoreceptors to enable visual motion detection. Development 145, pii: dev158246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auld VJ, Fetter RD, Broadie K, and Goodman CS (1995). Gliotactin, a novel transmembrane protein on peripheral glia, is required to form the blood-nerve barrier in Drosophila. Cell 81, 757–767. [DOI] [PubMed] [Google Scholar]
- Bai J, Chiu W, Wang J, Tzeng T, Perrimon N, and Hsu J (2001). The cell adhesion molecule Echinoid defines a new pathway that antagonizes the Drosophila EGF receptor signaling pathway. Development 128, 591–601. [DOI] [PubMed] [Google Scholar]
- Bardet PL, Guirao B, Paoletti C, Serman F, Leopold V, Bosveld F, Goya Y, Mirouse V, Graner F, and Bellaiche Y (2013). PTEN controls junction lengthening and stability during cell rearrangement in epithelial tissue. Dev Cell 25, 534–546. [DOI] [PubMed] [Google Scholar]
- Beati H, Peek I, Hordowska P, Honemann-Capito M, Glashauser J, Renschler FA, Kakanj P, Ramrath A, Leptin M, Luschnig S, et al. (2018). The adherens junction-associated LIM domain protein Smallish regulates epithelial morphogenesis. J Cell Biol 217, 1079–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertet C, Sulak L, and Lecuit T (2004). Myosin-dependent junction remodelling controls planar cell intercalation and axis elongation. Nature 429, 667–671. [DOI] [PubMed] [Google Scholar]
- Blankenship JT, Backovic ST, Sanny JS, Weitz O, and Zallen JA (2006). Multicellular rosette formation links planar cell polarity to tissue morphogenesis. Dev Cell 11, 459–470. [DOI] [PubMed] [Google Scholar]
- Bonello TT, Perez-Vale KZ, Sumigray KD, and Peifer M (2018). Rap1 acts via multiple mechanisms to position Canoe and adherens junctions and mediate apical-basal polarity establishment. Development 145, pii: dev157941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosveld F, Wang Z, and Bellaiche Y (2018). Tricellular junctions: a hot corner of epithelial biology. Curr Opin Cell Biol 54, 80–88. [DOI] [PubMed] [Google Scholar]
- Byri S, Misra T, Syed ZA, Batz T, Shah J, Boril L, Glashauser J, Aegerter-Wilmsen T, Matzat T, Moussian B, et al. (2015). The triple-repeat protein Anakonda controls epithelial tricellular junction formation in Drosophila. Dev Cell 33, 535–548. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega AJ, and Hartenstein V (1985). The embryonic development of Drosophila melanogaster. Springer-Verlag New York, p. 10–84. [Google Scholar]
- Carthew RW (2007). Pattern formation in the Drosophila eye. Curr Opin Genet Dev 17, 309–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EH, Chavadimane Shivakumar P, Clement R, Laugier E, and Lenne PF (2017). Patterned cortical tension mediated by N-cadherin controls cell geometric order in the Drosophila eye. Elife 6, pii: e22796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang LH, Chen P, Lien MT, Ho YH, Lin CM, Pan YT, Wei SY, and Hsu JC (2011). Differential adhesion and actomyosin cable collaborate to drive Echinoid-mediated cell sorting. Development 138, 3803–3812. [DOI] [PubMed] [Google Scholar]
- Choi W, Acharya BR, Peyret G, Fardin MA, Mege RM, Ladoux B, Yap AS, Fanning AS, and Peifer M (2016). Remodeling the zonula adherens in response to tension and the role of afadin in this response. J Cell Biol 213, 243–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W, Jung KC, Nelson KS, Bhat MA, Beitel GJ, Peifer M, and Fanning AS (2011). The single Drosophila ZO-1 protein Polychaetoid regulates embryonic morphogenesis in coordination with Canoe/afadin and Enabled. Mol Biol Cell 22, 2010–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombelli J, and Solon J (2013). Force communication in multicellular tissues addressed by laser nanosurgery. Cell Tissue Res 352, 133–147. [DOI] [PubMed] [Google Scholar]
- Del Signore SJ, Cilla R, and Hatini V (2018). The WAVE regulatory complex and branched F-actin counterbalance contractile force to control cell shape and packing in the Drosophila eye. Dev Cell 44, 471–483 e474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn BS, Rush L, Lu JY, and Xu T (2018). Mutations in the Drosophila tricellular junction protein M6 synergize with RasV12 to induce apical cell delamination and invasion. Proc Natl Acad Sci U S A 115, 8358–8363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhadifar R, Roper JC, Aigouy B, Eaton S, and Julicher F (2007). The influence of cell mechanics, cell-cell interactions, and proliferation on epithelial packing. Curr Biol 17, 2095–2104. [DOI] [PubMed] [Google Scholar]
- Fernandez-Gonzalez R, and Zallen JA (2011). Oscillatory behaviors and hierarchical assembly of contractile structures in intercalating cells. Phys Biol 8, 045005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer SC, Blanchard GB, Duque J, Adams RJ, Arias AM, Guest SD, and Gorfinkiel N (2014). Contractile and mechanical properties of epithelia with perturbed actomyosin dynamics. PLoS One 9, e95695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman KM, Yamagata M, Jin X, Mannepalli S, Katsamba PS, Ahlsen G, Sergeeva AP, Honig B, Sanes JR, and Shapiro L (2016). Molecular basis of Sidekick-mediated cell-cell adhesion and specificity. Elife 5, pii: e19058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorfinkiel N, and Blanchard GB (2011). Dynamics of actomyosin contractile activity during epithelial morphogenesis. Curr Opin Cell Biol 23, 531–539. [DOI] [PubMed] [Google Scholar]
- Hara Y, Shagirov M, and Toyama Y (2016). Cell boundary elongation by non-autonomous contractility in cell oscillation. Curr Biol 26, 2388–2396. [DOI] [PubMed] [Google Scholar]
- Higashi T, Arnold TR, Stephenson RE, Dinshaw KM, and Miller AL (2016). Maintenance of the epithelial barrier and remodeling of cell-cell junctions during cytokinesis. Curr Biol 26, 1829–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi T, and Miller AL (2017). Tricellular junctions: how to build junctions at the TRICkiest points of epithelial cells. Mol Biol Cell 28, 2023–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Zhou W, Dong W, Watson AM, and Hong Y (2009). Directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering. Proc Natl Acad Sci U S A 106, 8284–8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutson MS, Tokutake Y, Chang MS, Bloor JW, Venakides S, Kiehart DP, and Edwards GS (2003). Forces for morphogenesis investigated with laser microsurgery and quantitative modeling. Science 300, 145–149. [DOI] [PubMed] [Google Scholar]
- Jordan P, and Karess R (1997). Myosin light chain-activating phosphorylation sites are required for oogenesis in Drosophila. J Cell Biol 139, 1805–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung AC, Ribeiro C, Michaut L, Certa U, and Affolter M (2006). Polychaetoid/ZO-1 is required for cell specification and rearrangement during Drosophila tracheal morphogenesis. Curr Biol 16, 1224–1231. [DOI] [PubMed] [Google Scholar]
- Kale GR, Yang X, Philippe JM, Mani M, Lenne PF, and Lecuit T (2018). Distinct contributions of tensile and shear stress on E-cadherin levels during morphogenesis. Nat Commun 9, 5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karess RE, Chang XJ, Edwards KA, Kulkarni S, Aguilera I, and Kiehart DP (1991). The regulatory light chain of nonmuscle myosin is encoded by spaghetti-squash, a gene required for cytokinesis in Drosophila. Cell 65, 1177–1189. [DOI] [PubMed] [Google Scholar]
- Kaufman L, Hayashi K, Ross MJ, Ross MD, and Klotman PE (2004). Sidekick-1 is upregulated in glomeruli in HIV-associated nephropathy. J Am Soc Nephrol 15, 1721–1730. [DOI] [PubMed] [Google Scholar]
- Kaufman L, Potla U, Coleman S, Dikiy S, Hata Y, Kurihara H, He JC, D’Agati VD, and Klotman PE (2010). Up-regulation of the homophilic adhesion molecule Sidekick-1 in podocytes contributes to glomerulosclerosis. J Biol Chem 285, 25677–25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Fritz JL, Seneviratne AK, and VanBerkum MF (2002). Constitutively active myosin light chain kinase alters axon guidance decisions in Drosophila embryos. Dev Biol 249, 367–381. [DOI] [PubMed] [Google Scholar]
- Krishnaswamy A, Yamagata M, Duan X, Hong YK, and Sanes JR (2015). Sidekick 2 directs formation of a retinal circuit that detects differential motion. Nature 524, 466–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz B, Lierheimer R, Rader C, Spirig M, Ziegler U, and Sonderegger P (2002). Axonin-1/TAG-1 mediates cell-cell adhesion by a cis-assisted trans-interaction. J Biol Chem 277, 4551–4557. [DOI] [PubMed] [Google Scholar]
- Lee A, and Treisman JE (2004). Excessive Myosin activity in Mbs mutants causes photoreceptor movement out of the Drosophila eye disc epithelium. Mol Biol Cell 15, 3285–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lye CM, Naylor HW, and Sanson B (2014). Subcellular localisations of the CPTI collection of YFP-tagged proteins in Drosophila embryos. Development 141, 4006–4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manseau L, Baradaran A, Brower D, Budhu A, Elefant F, Phan H, Philp AV, Yang M, Glover D, Kaiser K et al. (1997). GAL4 enhancer traps expressed in the embryo, larval brain, imaginal discs, and ovary of Drosophila. Dev Dyn 209, 310–322. [DOI] [PubMed] [Google Scholar]
- Martin AC, Kaschube M, and Wieschaus EF (2009). Pulsed contractions of an actin-myosin network drive apical constriction. Nature 457, 495–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T, Takahashi K, Suzuki E, and Yamamoto D (1999). The Canoe protein is necessary in adherens junctions for development of ommatidial architecture in the Drosophila compound eye. Cell Tissue Res 298, 397–404. [DOI] [PubMed] [Google Scholar]
- Meyer G, Varoqueaux F, Neeb A, Oschlies M, and Brose N (2004). The complexity of PDZ domain-mediated interactions at glutamatergic synapses: a case study on neuroligin. Neuropharmacology 47, 724–733. [DOI] [PubMed] [Google Scholar]
- Nguyen DN, Liu Y, Litsky ML, and Reinke R (1997). The sidekick gene, a member of the immunoglobulin superfamily, is required for pattern formation in the Drosophila eye. Development 124, 3303–3312. [DOI] [PubMed] [Google Scholar]
- O’Keefe DD, Thor S, and Thomas JB (1998). Function and specificity of LIM domains in Drosophila nervous system and wing development. Development 125, 3915–3923. [DOI] [PubMed] [Google Scholar]
- Ochoa-Espinosa A, Harmansa S, Caussinus E, and Affolter M (2017). Myosin II is not required for Drosophila tracheal branch elongation and cell intercalation. Development 144, 2961–2968. [DOI] [PubMed] [Google Scholar]
- Oda Y, Otani T, Ikenouchi J, and Furuse M (2014). Tricellulin regulates junctional tension of epithelial cells at tricellular contacts through Cdc42. J Cell Sci 127, 4201–4212. [DOI] [PubMed] [Google Scholar]
- Ohashi T, Kiehart DP, and Erickson HP (1999). Dynamics and elasticity of the fibronectin matrix in living cell culture visualized by fibronectin-green fluorescent protein. Proc Natl Acad Sci U S A 96, 2153–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Gomez R, Slovakova J, Rives-Quinto N, Krejci A, and Carmena A (2013). A Serrate-Notch-Canoe complex mediates essential interactions between glia and neuroepithelial cells during Drosophila optic lobe development. J Cell Sci 126, 4873–4884. [DOI] [PubMed] [Google Scholar]
- Pinheiro D, and Bellaiche Y (2018). Mechanical force-driven adherens junction remodeling and epithelial dynamics. Dev Cell 47, 3–19. [DOI] [PubMed] [Google Scholar]
- Rauskolb C, Sun S, Sun G, Pan Y, and Irvine KD (2014). Cytoskeletal tension inhibits Hippo signaling through an Ajuba-Warts complex. Cell 158, 143–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauzi M, and Lenne PF (2015). Probing cell mechanics with subcellular laser dissection of actomyosin networks in the early developing Drosophila embryo. Methods Mol Biol 1189, 209–218. [DOI] [PubMed] [Google Scholar]
- Rauzi M, Lenne PF, and Lecuit T (2010). Planar polarized actomyosin contractile flows control epithelial junction remodelling. Nature 468, 1110–1114. [DOI] [PubMed] [Google Scholar]
- Rauzi M, Verant P, Lecuit T, and Lenne PF (2008). Nature and anisotropy of cortical forces orienting Drosophila tissue morphogenesis. Nat Cell Biol 10, 1401–1410. [DOI] [PubMed] [Google Scholar]
- Razzell W, Bustillo ME, and Zallen JA (2018). The force-sensitive protein Ajuba regulates cell adhesion during epithelial morphogenesis. J Cell Biol 217, 3715–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro C, Neumann M, and Affolter M (2004). Genetic control of cell intercalation during tracheal morphogenesis in Drosophila. Curr Biol 14, 2197–2207. [DOI] [PubMed] [Google Scholar]
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, et al. (2008). Lifeact: a versatile marker to visualize F-actin. Nat Methods 5, 605–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer JK, Harris NJ, Slep KC, Gaul U, and Peifer M (2009). The Drosophila afadin homologue Canoe regulates linkage of the actin cytoskeleton to adherens junctions during apical constriction. J Cell Biol 186, 57–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte J, Tepass U, and Auld VJ (2003). Gliotactin, a novel marker of tricellular junctions, is necessary for septate junction development in Drosophila. J Cell Biol 161, 991–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppa MJ, Johnson RI, Bao S, and Cagan RL (2008). Polychaetoid controls patterning by modulating adhesion in the Drosophila pupal retina. Dev Biol 318, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaye DD, Casanova J, and Llimargas M (2008). Modulation of intracellular trafficking regulates cell intercalation in the Drosophila trachea. Nat Cell Biol 10, 964–970. [DOI] [PubMed] [Google Scholar]
- Somlyo AP, and Somlyo AV (2000). Signal transduction by G-proteins, rho-kinase and protein phosphatase to smooth muscle and non-muscle myosin II. J Physiol 522, 177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spadaro D, Le S, Laroche T, Mean I, Jond L, Yan J, and Citi S (2017). Tension-dependent stretching activates ZO-1 to control the junctional localization of its interactors. Curr Biol 27, 3783–3795. [DOI] [PubMed] [Google Scholar]
- Spector I, Schochet NR, Kashman Y, and Groweiss A (1983). Latrunculins: novel marine toxins that disrupt microfilament organization in cultured cells. Science 219, 493–495. [DOI] [PubMed] [Google Scholar]
- Speicher S, Fischer A, Knoblich J, and Carmena A (2008). The PDZ protein Canoe regulates the asymmetric division of Drosophila neuroblasts and muscle progenitors. Curr Biol 18, 831–837. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Matsuo T, Katsube T, Ueda R, and Yamamoto D (1998). Direct binding between two PDZ domain proteins Canoe and ZO-1 and their roles in regulation of the jun N-terminal kinase pathway in Drosophila morphogenesis. Mech Dev 78, 97–111. [DOI] [PubMed] [Google Scholar]
- Takeichi M (2014). Dynamic contacts: rearranging adherens junctions to drive epithelial remodelling. Nat Rev Mol Cell Biol 15, 397–410. [DOI] [PubMed] [Google Scholar]
- Tang H, Chang H, Dong Y, Guo L, Shi X, Wu Y, Huang Y, and He Y (2018). Architecture of cell-cell adhesion mediated by Sidekicks. Proc Natl Acad Sci U S A 115, 9246–9251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trichas G, Smith AM, White N, Wilkins V, Watanabe T, Moore A, Joyce B, Sugnaseelan J, Rodriguez TA, Kay D et al. (2012). Multi-cellular rosettes in the mouse visceral endoderm facilitate the ordered migration of anterior visceral endoderm cells. PLoS Biol 10, e1001256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsarouhas V, Senti KA, Jayaram SA, Tiklova K, Hemphala J, Adler J, and Samakovlis C. (2007). Sequential pulses of apical epithelial secretion and endocytosis drive airway maturation in Drosophila. Dev Cell 13, 214–225. [DOI] [PubMed] [Google Scholar]
- Vanderleest TE, Smits CM, Xie Y, Jewett CE, Blankenship JT, and Loerke D (2018). Vertex sliding drives intercalation by radial coupling of adhesion and actomyosin networks during Drosophila germband extension. Elife 7, pii: e34586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez CG, Tworoger M,and Martin AC (2014). Dynamic myosin phosphorylation regulates contractile pulses and tissue integrity during epithelial morphogenesis. J Cell Biol 206, 435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJ, Schulze KL, Haelterman NA, Pan H, He Y, Evans-Holm M, Carlson JW,Levis RW, Spradling AC, Hoskins RA, et al. (2011). MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat Methods 8, 737–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verone AR, Duncan K, Godoy A, Yadav N, Bakin A, Koochekpour S, Jin JP, and Heemers HV (2013). Androgen-responsive serum response factor target genes regulate prostate cancer cell migration. Carcinogenesis 34, 1737–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent S, Ruberte E, Grieder NC, Chen CK, Haerry T, Schuh R, and Affolter M (1997). DPP controls tracheal cell migration along the dorsoventral body axis of the Drosophila embryo. Development 124, 2741–2750. [DOI] [PubMed] [Google Scholar]
- Warner SJ, and Longmore GD (2009a). Distinct functions for Rho1 in maintaining adherens junctions and apical tension in remodeling epithelia. J. Cell Biol. 185, 1111–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner SJ, and Longmore GD (2009b). Cdc42 antagonizes Rho1 activity at adherens junctions to limit epithelial cell apical tension. J. Cell Biol. 187, 119–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei SY, Escudero LM, Yu F, Chang LH, Chen LY, Ho YH, Lin CM, Chou CS, Chia W, Modolell J, et al. (2005). Echinoid is a component of adherens junctions that cooperates with DE-Cadherin to mediate cell adhesion. Dev Cell 8, 493–504. [DOI] [PubMed] [Google Scholar]
- Yamagata M, and Sanes JR (2008). Dscam and Sidekick proteins direct lamina-specific synaptic connections in vertebrate retina. Nature 451, 465–469. [DOI] [PubMed] [Google Scholar]
- Yamagata M, and Sanes JR (2010). Synaptic localization and function of Sidekick recognition molecules require MAGI scaffolding proteins. J Neurosci 30, 3579–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata M, Weiner JA, and Sanes JR (2002). Sidekicks: synaptic adhesion molecules that promote lamina-specific connectivity in the retina. Cell 110, 649–660. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Harada N, Kano K, Taya S, Canaani E, Matsuura Y, Mizoguchi A, Ide C, and Kaibuchi K (1997). The Ras target AF-6 interacts with ZO-1 and serves as a peripheral component of tight junctions in epithelial cells. J Cell Biol 139, 785–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura S (2011). Cadherin-actin interactions at adherens junctions. Curr Opin Cell Biol 23, 515–522. [DOI] [PubMed] [Google Scholar]
- Yonemura S, Wada Y, Watanabe T, Nagafuchi A, and Shibata M (2010). alpha-Catenin as a tension transducer that induces adherens junction development. Nat Cell Biol 12, 533–542. [DOI] [PubMed] [Google Scholar]
- Yu JC, and Fernandez-Gonzalez R (2016). Local mechanical forces promote polarized junctional assembly and axis elongation in Drosophila. Elife 5, pii:e10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, and Ward R.E.t. (2011). Distinct tissue distributions and subcellular localizations of differently phosphorylated forms of the myosin regulatory light chain in Drosophila. Gene Expr Patterns 11, 93–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1, related to Figure 3: sdk is required for tracheal intercalation. sdkMB05054; Ecad-GFP embryo imaged for 114 min, focused on two dorsal branches during the process of cell intercalation (from early stage 14 to stage 15). In normal conditions (as in the left dorsal branch, marked with a yellow arrow), the zipping up process replaces ring-shaped intercellular junctions with straight-line autocellular junctions. This junctional remodelling accompanies the repositioning of the cells from a side-by-side to an end-to-end position. At the end of the intercalation phase the dorsal branches are sealed by autocellular junctions interrupted by small rings of intercellular junctions (which connect the cells at their ends). In sdk mutant embryos the zipping up process is perturbed in many branches (right dorsal branch marked with a blue arrow). This perturbation generates fully extended dorsal branches that still maintain intercellular junctions, indicating that the cells have not repositioned into a row and maintain a side-by-side position. Scale bar, 10 μm.
Movie S2, related to Figure 3: Actomyosin is recruited to shrinking junctions during retinal lattice cell intercalation. LifeAct-GFP (green, A’); Sqh-mCherry (red, A’’) pupal retina imaged at 26-27 h APF, showing dynamic recruitment of actin and myosin to shrinking junctions during cell intercalation and their loss from expanding junctions.
Movie S3, related to Figure 3: sdk is required for cell intercalation in the retina. sdkA15; GFP-α-catenin pupal retina imaged at 28-32h APF. (A) shows an ommatidium with normal cell intercalation, followed by apoptosis of the extra cells. (B) shows an ommatidium with defective cell intercalation in the same eye resulting in misplacement of bristles and lattice cells. Note the apoptosis of cells that failed to intercalate. (A’, B’) show enlargements of ommatidial edges that are demarcated in boxes in upper panels. Scale bars, 10 μm.
Movie S4, related to Figure 3: Cell bonds are under high tension in wild type embryos. Wild type stage 7 Ecad-GFP embryo imaged for 39.5 min at 29°C, focused on the dorsolateral region of the epidermis, showing the amnioserosa (larger cells) and the dorsal epidermis (smaller cells) in contact. Cell bonds are straight (junction straightness is 1.0 ± 0.05, n=30 cells of the dorsal epidermis), reflecting high levels of tension. Scale bar 15μm.
Movie S5, related to Figure 3: Cell bond tension is reduced in sdk mutant embryos. sdkMB05054; Ecad-GFP stage 8 embryo imaged for 52 min at 29°C, focused on the dorsolateral region of the epidermis, showing the amnioserosa (larger cells) and the dorsal epidermis (smaller cells) in contact. The presence of wiggly bonds indicates reduced tension. Junction straightness of the dorsal epidermis cells is 0.87 ± 0.10 (n=30 cells), which is significantly different from wildtype (p<0.0001, unpaired t-test). Scale bar 10μm.


