Abstract
The glutamate pyruvate transaminase 2 (GPT2) gene produces a nuclear-encoded mitochondrial enzyme that catalyzes the reversible transfer of an amino group from glutamate to pyruvate, generating alanine and alpha-ketoglutarate. Recessive mutations in GPT2 have been recently identified in a new syndrome involving intellectual and developmental disability (IDD), postnatal microcephaly, and spastic paraplegia. We have identified additional families with recessive GPT2 mutations and expanded the phenotype to include small stature. GPT2 loss-of-function mutations were identified in four families, nine patients total, including: a homozygous mutation in one child [c.775T>C (p.C259R)]; compound heterozygous mutations in two siblings [c.812A>C (p.N271T)/c.1432_1433delGT (p.V478Rfs*73)]; a novel homozygous, putative splicing mutation [c.1035C>T (p.G345=)]; and finally, a recurrent mutation, previously identified in a distinct family [c.1210C>T (p.R404*)]. All patients were diagnosed with IDD. A majority of patients had remarkably small stature throughout development, many <1st percentile for height and weight. Given the potential biological function of GPT2 in cellular growth, this phenotype is strongly suggestive of a newly-identified clinical susceptibility. Further, homozygous GPT2 mutations manifested in at least 2 of 176 families with IDD (approximately 1.1%) in a Pakistani cohort, thereby representing a relatively common cause of recessive IDD in this population, with recurrence of the p.R404* mutation in this population. Based on variants in the ExAC database, we estimated that approximately 1 in 248 individuals are carriers of moderately or severely deleterious variants in GPT2.
Keywords: Genetics, Intellectual disability, Mitochondrial metabolism, Development, Neurology, GPT2
INTRODUCTION
Intellectual and developmental disability (IDD) is common (~1–2% of the population) and carries an average lifetime cost per person of ~$1,000,000 (in 2003 dollars), which makes it a substantial public health concern (CDC 2004). The etiologic heterogeneity challenges screening and development of effective treatments (CDC 2013). Successes with conditions such as in the case of phenylketonuria (PKU) provide the model for newborn screening and subsequent prevention of progressive cognitive disability (Rose and Wick 2016). IDD manifests secondary to genetic and/or environmental factors and commonly co-occurs with other central nervous system conditions such as autism spectrum disorder, epilepsy, motor abnormalities (e.g., spastic paraplegia), and microcephaly (Jamra 2018). More than 800 genes have been implicated in the pathogenesis of IDD, with approximately one third (28–34%) of patients possessing genetic mutations discoverable with available clinical genetic testing (Iglesias et al. 2014; Lee et al. 2014; Nambot et al. 2018; Rump et al. 2016; Thevenon et al. 2016). With the advent of genome-wide sequencing, future promise for newborn screening and preventative treatments for some forms of IDD is now a realistic future.
One gene recently associated with IDD is glutamate pyruvate transaminase 2 (GPT2) (Celis et al. 2015; Ouyang et al. 2016; Yang et al. 2002). The GPT2 protein represents a nuclear-encoded mitochondrial enzyme that catalyzes the reversible addition of an amino group from glutamate to pyruvate, yielding alanine and alpha-ketoglutarate. GPT2 is specifically localized to mitochondria, within which it is responsible for the regulation of metabolic processes such as amino acid metabolism and the tricarboxylic acid (TCA) cycle. It may also be involved in neurotransmitter metabolism, as glutamate is the major excitatory neurotransmitter in brain and the inhibitory neurotransmitter gamma-aminobutyric acid is synthesized from glutamate. Consistent with a role in synapse biology, GPT2 transcription is upregulated in the postnatal developing brain (Ouyang et al. 2016).
A recently developed mouse model of loss-of-function GPT2 mutations identified specific neural abnormalities, including reduced overall brain growth and metabolic abnormalities (Ouyang et al. 2016). Ouyang et al. (2016) also specifically implicated this enzyme in the process of anaplerosis, the replenishment of TCA cycle intermediates. Anaplerosis balances cataplerosis, which is the utilization of TCA cycle intermediates for biosynthesis; it is a critically needed homeostatic process during rapid cell growth (Owen et al. 2002). Interestingly, GPT2 has been studied recently in the context of cancer, wherein this enzyme may be a target to limit cancer growth due the role of this enzyme in anaplerosis (Cao et al. 2017; Kim et al. 2019).
In patients, homozygous loss-of-function mutations in GPT2 lead to a novel, neurodevelopmental phenotype characterized by intellectual disability, postnatal microcephaly, and motor abnormalities, as well as commonly, but not uniformly identified, epilepsy, mild dysmorphic features, and slowly progressive spastic diplegia/paraplegia (Celis et al. 2015; Hengel et al. 2018; Kaymakcalan et al. 2018; Lobo-Prada et al. 2017; Ouyang et al. 2016). In this report, we present our discovery of several new, important aspects of GPT2-related disease. First, we report new mutations and expand the phenotype to include unusually small stature, which was notably prominent in two siblings with previously unreported compound heterozygous mutations. Susceptibility to small stature, above and beyond the prior reports of isolated postnatal microcephaly, is highly consistent with a role of the enzyme in growth. Additionally, we report estimates of prevalence of mutations based on a control dataset (the Exome Aggregation Consortium, ExAC; http://exac.broadinstitute.org) (Lek et al. 2016) and a sample of Pakistani pedigrees with autosomal recessive intellectual disability (Harripaul et al. 2018). Surprisingly, the measured prevalence of GPT2 mutations in the Pakistani cohort is higher than predicted, suggesting a prominent role for this novel GPT2-related genetic syndrome in Pakistan.
MATERIALS AND METHODS
Human subjects
Institutional review boards at Brown University and Lifespan Healthcare approved this study. Written informed consent was obtained from all participants or their legal guardians. Clinical information was collected by direct parental interview and neurological/neuropsychological exam of participant and from available medical records.
DNA sequence analysis
All patient mutations were verified by Sanger DNA sequencing. Sequence alignments are based on human reference genome GrCH37/hg19. References for mutation coordinates are based on NCBI Reference Sequences NM_133443.2 and NM_133443.3 (cDNA) and NP 597700.1 (protein).
DNA constructs and antibodies
The human GPT2 gene (GenBank: BC062555.1) was cloned into the pcDNA3.1/V5-His-TOPO mammalian expression vector to generate C-terminal V5-His-tagged human GPT2 (hGPT2-V5His). GPT2 mutants Q80E, C259R, N271T, and V478Rfs*73 were constructed using QuickChange site-directed mutagenesis (Agilent Technologies, Santa Clara, CA, USA). All vectors were verified by Sanger DNA sequencing. Antibodies used were mouse anti-V5 antibody (Life Technologies, Thermo Fisher Scientific, Carlsbad, CA, USA), mouse anti-tubulin antibody (abcam, Cambridge, MA, USA), and goat anti-mouse Alexa Fluor 488 (Life Technologies, Thermo Fisher Scientific, Carlsbad, CA, USA).
GPT enzymatic activity assay
GPT transaminase enzymatic activity was analyzed using the alanine aminotransferase (ALT) Assay Kit (Sigma-Aldrich, St. Louis, MO, USA). HeLa cells transfected with constructs encoding for wild-type or mutant hGPT2-V5His were homogenized in ice-cold ALT assay buffer and centrifuged. Protein concentrations of supernatants were determined by bicinchoninic acid (BCA) assay. GPT enzymatic activity was detected using 3.5 μg of cell extracts. Data are presented as means ± SEMs.
Immunocytochemistry
Prior to fixation, HeLa cells exogenously expressing wild-type or mutant hGPT2-V5His were incubated with MitoTracker Red (Life Technologies, Thermo Fisher Scientific, Carlsbad, CA, USA) for 30 min. Cells were then rinsed twice with 1X PBS, fixed in 4% (w/v) paraformaldehyde for 15 min, and permeabilized with 0.25% (w/v) Triton X-100 in 1X PBS for 15 min at room temperature. Cells were then blocked with 10% normal goat serum in 1X PBS + 0.1% Triton X-100 (PBST) for 1 h at room temperature, followed by incubation overnight at 4°C with primary antibodies diluted in PBST containing 2% normal goat serum. After rinsing 3 × 5 min in PBST, cells were incubated for 1 h at room temperature with secondary antibody diluted in PBST containing 2% normal goat serum. Nuclei were counter stained with Hoechst dye (Life Technologies, Thermo Fisher Scientific, Carlsbad, CA, USA) at the same time as incubation with secondary antibody. Cells were again rinsed 3 × 5 min in PBST and then mounted in Fluoromount-G (SouthernBiotech, Birmingham, AL, USA).
Microscopy
Structured illumination microscopy (SIM) images were collected using a DeltaVision OMX SR microscope (GE Healthcare Life Sciences, USA). Z-series images were collected using a 60X oil objective (refractive index immersion oil-1.516) and light path-SI under sequential acquisition mode. Z-series images were processed by performing OMX SI reconstruction, alignment, and maximal projection sequentially using softWoRx software. Images were analyzed using ImageJ software (NIH, USA).
Image quantification and analysis
Co-localization of exogenously expressed wild-type or mutant hGPT2-V5His with MitoTracker Red was quantified based on images collected using SIM. Images were analyzed using the co-localization application of MetaMorph software (San Jose, CA, USA), with either: (1) the V5 and MitoTracker Red co-stained area in the numerator and the total V5 stained area in the denominator or (2) the V5 and MitoTracker Red co-stained area in the numerator and the total MitoTracker Red stained area in the denominator. Thresholds were set for each channel to exclude background staining, and the percent co-localization was calculated for each cell analyzed. Data are presented as means ± SEMs.
Structural analysis
GPT2 variants were visualized using PyMOL visualization software, with GPT2 mutants generated using the PyRosetta framework (PDB reference 3IHJ) (Chaudhury et al. 2010). Homology analysis was performed by analyzing the multiple sequence alignment from the aspartate aminotransferase (AAT)-like family of proteins using the NCBI tool Conserved Protein Domain Family and NCBI Reference Sequence NP_597700.1 (Marchler-Bauer et al. 2017; Sayers et al. 2011). Model refinement was performed using PyRosetta and a combination of sampling methods during every cycle. Two hundred independent trajectories were run for each model. The top five conformations with lowest scores, as indicated by Rosetta Energy Units (REU), were visualized, and the lowest scoring conformation that was representative of the group of top proposed models was selected for presentation (DeLuca et al. 2015; Moretti et al. 2018).
Analysis of the effects of GPT2 mutations on the energy of the pyridoxal 5’-phosphate (PLP) binding site was performed using the Ligand Docking protocol on the ROSIE web server (Lyskov et al. 2013), which is hosted by the Gray Lab at Johns Hopkins University (https://rosie.graylab.jhu.edu/ligand_docking). Rosetta software was used to predict the interface energy of the ligand and protein, and the conformation space of the binding pocket and the ligand was thoroughly sampled. Eight hundred independent trajectories were used for each model. To model the docked complexes, the top five conformations with lowest scores, as indicated by REU, were visualized, and the lowest scoring conformation that was representative of the group of top proposed models was selected for presentation (DeLuca et al. 2015; Moretti et al. 2018).
Exon trapping assays
Exon trapping assays were performed for GPT2 with respect to exon 2 and exon 8 and mutations in these exons. For generation of vectors used in these assays, either exon 2 or exon 8 of human GPT2, as well as portions of the flanking introns (102 bp of intron 1 and 288 bp of intron 2; 141 bp of intron 7 and 290 bp of intron 8), was PCR-amplified from the genomic sample of a control patient, after which the amplicon was digested with SalI and BamHI or SalI alone and ligated into the multiple cloning site of the pET01 vector (MoBiTec GmbH, Goettingen, Germany). The pET01 multiple cloning site is located between two bordering exons, thereby allowing for analysis of the effects on splicing of sequences ligated into this site. Site-directed mutagenesis was performed to generate pET01-GPT2 vectors with the missense mutation in exon 2 (c.238C>G) and the synonymous mutation in exon 8 (c.1035C>T). Briefly, the base substitution was induced by PCR using mutagenic primers, followed by digestion of the parental template using DpnI, phosphorylation of the mutagenic amplicon by T4 Polynucleotide Kinase (New England Biolabs, Ipswich, MA, USA), and circularization using Quick T4 DNA Ligase (New England Biolabs, Ipswich, MA, USA). All vectors were verified by Sanger DNA sequencing. Primers used for generating the pET01-GPT2 vectors were as follows, with the SalI and BamHI recognition sequences in lower-case and the substituted bases underlined. GPT2_Exon2_SalI_F: 5’-CACTGgtcgacGGCACCGCTCGCTGAAAG-3’; GPT2_Exon2_BamHI_R: 5’-CACTGggatccGAGAGTAGGGCAGACACACA-3’; GPT2_Exon8_SalI_F: 5’-GACTgtcgacGCTCATCCCTGGAATGGCA-3’; GPT2_Exon8_SalI_R: 5’-GACTgtcgacACCTCGGAACAAGTGACAGA-3; GPT2_Exon2_Mutagenesis_F: 5’-GCTCGAGCTGGAGCGGGTGAG-3’; GPT2_Exon2_Mutagenesis_R: 5’-TCGATCTCGCCGGCCTTGAG-3’; GPT2_Exon8_Mutagenesis_F: 5’-GGGCTACATGGGTGAGTACGTGGGC-3’; and GPT2 Exon8 Mutagenesis R: 5’-GCCCACGTACTCACCCATGTAGCCC-3’.
For performance of exon trapping assays, wild-type, mutant, and control (empty pET01) vectors were separately transfected into HEK293T cells using Lipofectamine 3000 (Life Technologies, Thermo Fisher Scientific, Carlsbad, CA, USA). Twenty-four hours post-transfection, RNA was extracted from cells, and RT - PCR were performed using primers complementary to the pET01 exons bordering the multiple cloning site. PCR amplicons were separated by electrophoresis on agarose gels, and bands were excised, purified, and analyzed by Sanger DNA sequencing. Primers used for analyzing splicing between the multiple cloning site and bordering exons of the pET01 vector were as follows: pET01_cDNA_PCR_F: 5’-ATCGATCTGCTTCCTGGC-3’ and pET01_cDNA_PCR_R: 5’-GGGCCACCTCCAGTGCC-3’.
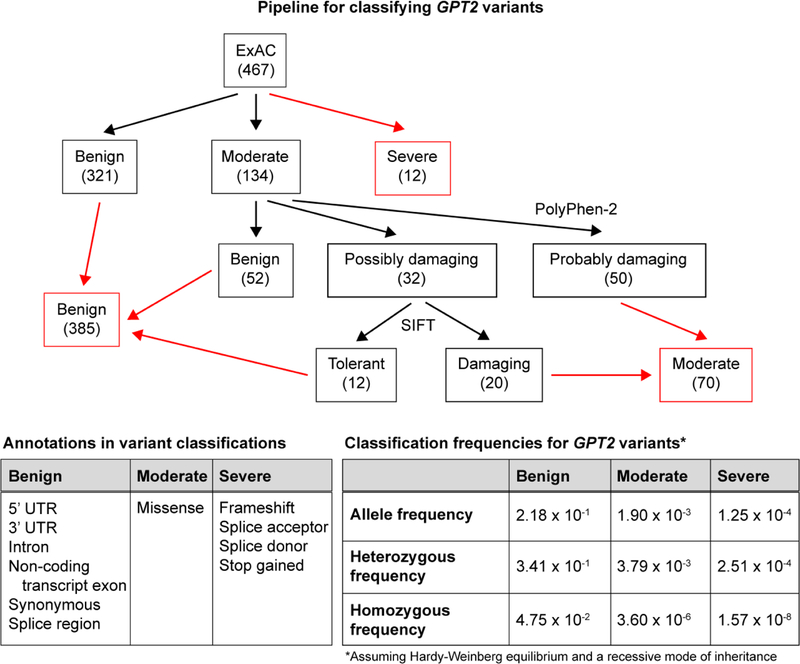
Gene variant analysis pipeline and allele frequency calculations
GPT2 variants, as reported in the ExAC database (467 variants at time of access), were sorted into three categories based on the annotation provided by ExAC: benign (321 variants), moderate (134 variants), and severe (12 variants). Missense variants falling under the moderate category were then further differentiated, first using ANNOVAR-PolyPhen-2 and then using ANNOVAR-SIFT (Wang et al. 2010). The annotation table was set with a reference of hg19 dbnsfp35a for both tools. Missense variants processed using PolyPhen-2 were split into three categories: benign (52 variants), possibly damaging (32 variants), and probably damaging (50 variants). Variants classified as possibly damaging were then further processed using SIFT and split into two categories: tolerant (12 variants) and damaging (20 variants). (See Fig. 6 for a summary of the analysis pipeline and the final number of variants in each category.)
Fig 6.
Pipeline for the classification of GPT2 variants and prediction of allele frequencies. GPT2 variants were characterized as benign, moderate, or severe based on variant annotations provided by ExAC (bottom left table) and use of PolyPhen-2 and SIFT (see Materials and Methods for details). Final variant counts in each category are shown in red boxes. Total allele frequency was computed across all variants in each category (bottom right table). The frequencies of heterozygous alleles and homozygous alleles in each variant category were predicted based on calculations assuming Hardy-Weinberg equilibrium and a recessive mode of inheritance
The total variant allele frequency for each of the three categories was calculated by summing the adjusted frequencies of the individual variant alleles within each category, using data reported in the ExAC web browser. The population carrier frequency (i.e., heterozygous frequency) and the predicted affected frequency (i.e., homozygous frequency) for each of the categories were calculated using the equations below, based on assumption of Hardy-Weinberg equilibrium and a recessive mode of inheritance.
Heterozygous frequency: 2(variant allele frequency)(1 - variant allele frequency) Homozygous frequency: (variant allele frequency)^2
RESULTS
Clinical genetics and phenotypes
We established strong genetic and functional evidence for association of the GPT2 mutations with the clinical phenotype for five distinct mutations in four families. Notably, we identified one family with compound heterozygous mutations (i.e., two mutations in this one family) (Fig. 1). A fifth family, family 5, with a missense change (c.238C>G, p.Q80E) was identified, yet we were not able to establish functional evidence that this GPT2 variant is pathogenic. Clinical features are summarized in Table 1.
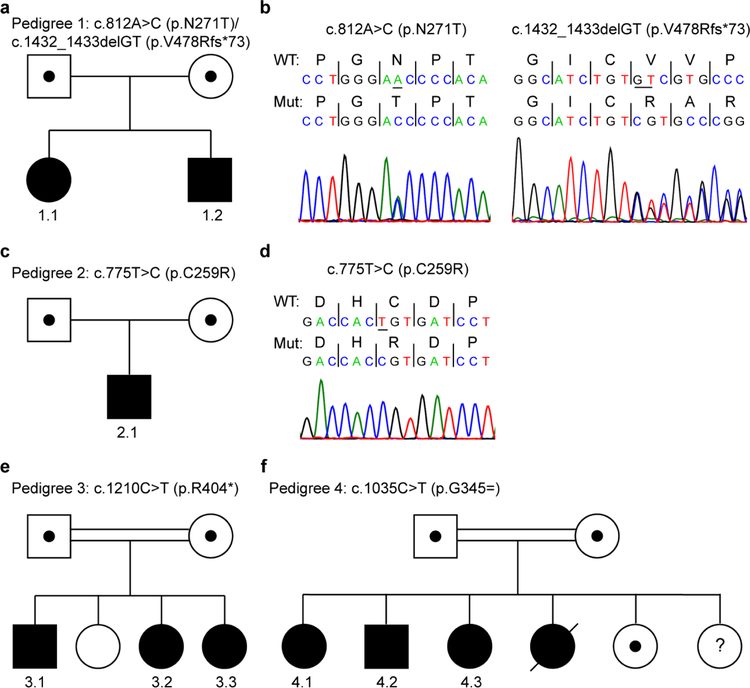
Fig 1.
Pedigrees and segregation of GPT2 mutations in the current study. a-d Pedigrees and corresponding sequence chromatograms for the compound heterozygous GPT2 mutations identified in Pedigree 1 (a, b) and homozygous mutation identified in Pedigree 2 (c, d). e,f Pedigrees of GPT2 mutations identified in a Pakistani cohort of individuals with autosomal recessive intellectual disability. Note, some pedigrees do not show all siblings to aid in maintaining anonymity of the patient and family. Positions of mutations are according to transcripts NM_133443.3 (a-e) and NM_133443.2 (f) and protein NP_597700.1
Table 1.
Summary of clinical and neurological findings for six probands with GPT2 mutations
| Patient | 1.1 | 1.2 | 2.1 | 3.1 | 3.2 | 3.3 |
|---|---|---|---|---|---|---|
| Consanguinity (parental relatedness) | No | No | No | Yes | Yes | Yes |
| Gender | Female | Male | Male | Male | Female | Female |
| Age at examination | 15 y | 13 y | 17 y | 17 y | 14 y | 10 y |
| Intellectual disability | Moderate | Moderate | Moderate | Severe to profound | Severe to profound | Severe to profound |
| Weight at birth | 54th percentile | 59th percentile | 2nd percentile | NA | NA | NA |
| Head circumference at examination | 1st percentile | 1st percentile | <1st percentile | <1st percentile | <1st percentile | <1st percentile |
| Height at examination | NA; <5th percentile throughout childhood | NA; <1st percentile based on most recent clinical note | 21st percentile | 1st percentile | <1st percentile | <1st percentile |
| Weight at examination | NA; <5th percentile throughout childhood | NA; <1st percentile based on most recent clinical note | 10th percentile | 8th percentile | 1st percentile | 2nd percentile |
| Motor development | Delayed motor signs by 17 mo; apraxia | Delayed motor signs by 9–12 mo; head holding at 2–3 mo; standing at 14 mo | Normal gross motor development | Delayed motor development | Delayed motor development | Delayed motor development |
| Age of being able to walk | 4 y | 2 y | 13–14 mo | 3 y | 4 y | 6 y |
| Subsequent deterioration in walking | No | No | No | Yes, after fall at 4 y | No | No |
| Speech | Dysarthria; globally impaired | Dysarthria; speech apraxia; globally delayed | Delayed | Non-verbal; communicates with gestures | Monotone speech; some dysarthria | Monotone speech; some dysarthria |
| Oral-motor function | Drooling; vomiting | Drooling; vomiting | Normal | Severe impairment in oral-motor skills; drooling | Severe impairment in oral-motor skills; drooling | Severe impairment in oral-motor skills; drooling |
| Motor examination | Spasticity; increased tone in legs in terms of rotation; hyperreflexia; bilateral coordination impaired; wide-based gait; mild scissoring | Spasticity; increased tone in legs in terms of rotation; hyperreflexia in all joints, with 4–5 beats of clonus bilaterally in ankles | Strength and tone normal; abnormal gait | Hyperreflexia at all joints; severely impaired coordination | Strength reduced (power 3/5); tone normal; no spasticity of upper or lower limbs; hyperreflexia at all joints; poor motor coordination | Strength reduced (power 4/5); tone normal; no spasticity of upper or lower limb; hyperreflexia at all joints; severely impaired coordination |
| Data supporting seizures/epilepsy | 1st seizure at 9 y; EEG in 2012 showed bursts of generalized 3-Hz spike and wave activity | 1st seizure around 9 y; medically refractory epilepsy; currently prescribed lamotrigine and cannabidiol oil | No seizures observed | Equivocal falling behavior, but seizure workup unavailable | No seizures observed | Equivocal falling behavior, but seizure workup unavailable |
| Imaging | CT/MRI-MR spectroscopy in 2008 normal | MRI showed bilateral prominent perivascular spaces at centrum semiovale | CT of head normal | Low resolution MRI normal | Low resolution MRI normal | Low resolution MRI normal |
| GPT2 mutationa | Compound heterozygous c.812A>C (p.N271T)/c.1432_1433delGT (p.V478Rfs*73) | Compound heterozygous c.812A>C (p.N271T)/c.1432_1433delGT (p.V478Rfs*73) | Homozygous c.775T>C (p.C259R) | Homozygous c.1210C>T (p.R404*) | Homozygous c.1210C>T (p.R404*) | Homozygous c.1210C>T (p.R404*) |
Positions of mutations are according to transcript NM_133443.3.
y years, mo months, NA not available
Pedigree 1 – Patient 1.1 (Female)
This patient is a 15-year-old Caucasian female of English and French-Canadian descent. Both she and her 13-year-old brother (Patient 1.2) were found to have compound heterozygous GPT2 mutations by clinical exome sequencing: c.812A>C (p.N271T)/c.1432_1433delGT (p.V478Rfs*73) (Fig. 1a,b). Her parents are not related, and each parent is heterozygous for one of the two mutations. For the female patient, mother’s pregnancy was full-term and unremarkable. Patient’s birth weight was at the 54th percentile (z-score = .1).
The patient experienced a febrile seizure at approximately 21 months of age, although her first non-febrile seizure occurred at 9 years old. She has had 4–5 total non-febrile seizures in her lifetime, with the last seizure occurring approximately 4–5 years ago. Seizures were previously described as episodes involving staring, unresponsiveness, and eyelid quivering or flickering movements. Also, in at least one of these episodes, the patient was noted to have wandered around her school gymnasium while exhibiting the staring and eyelid movements. These episodes were characterized as atypical absence seizures by prior clinicians. Of note, at the time of these episodes, parents reported to providers that the patient had experienced multiple staring episodes per week for multiple years. An EEG at 9 years old demonstrated bursts of generalized 3-Hz spike and wave activity with drowsiness. The patient has never been prescribed anti-epileptic drugs. An MRI-MR spectroscopy at 9 years old and a head CT scan at 4 years old were read as normal.
The family first noted concerns about delayed development of the patient at approximately 17 months old. She took her first steps at 2 years old and walked at 4 years old. The patient has a history of clinically diagnosed apraxia and global developmental delay. She also has a history of cyclic vomiting, gastroesophageal reflux disease (GERD), excessive drooling (with Botox injection), and early choking/gagging, which were associated with a failure to thrive. Notably, after normal birth weights, she has been less than 5th percentile (z-score <-1.5) for height and weight throughout development. The patient’s mother and father are of normal stature. The patient had negative celiac serology.
On general exam, the patient was noted to have a head circumference of 51.5 cm (1st percentile for age; z-score = −2.4). Other than hypersalivation, the remainder of the general exam was normal. On neurological exam, the patient was alert and oriented to name and place. She could follow one-step directions, with some difficulty staying focused, and she did not follow directions that involve crossing the midline. Her cranial nerves were intact except for significant dysarthria. Her pupils were equal, visual fields full, extra-ocular movements intact, face symmetric, sensation intact, tongue midline, and her palate elevated symmetrically. Sternocleidomastoid/trapezius muscles appeared to be full strength. Her motor exam showed normal bulk, no atrophy, no tremor, and full and symmetric strength in arms and legs. Tone was increased in legs. Her sensory exam was normal to light touch and vibration. Coordination was mildly impaired. Finger-nose-finger was mildly ataxic bilaterally, and she had slight clumsiness of her fine finger movements; however, rapid alternating movements were intact. Her reflexes were symmetric in biceps, triceps, brachioradialis, patellar, and Achilles tendons. These were brisk without evidence of clonus and spreading. Her gait was wide-based, with mild scissoring and turned-in knees, showing evidence of spasticity. Her Romberg test was negative.
On neuropsychological examination, intellectual testing (Wechsler Intelligence Scale for Children, Fifth Edition [WISC-V]) indicated moderately impaired overall IQ (FSIQ = 41). Receptive vocabulary (Receptive One-Word Picture Vocabulary Test, Fourth Edition [ROWPVT-4]) was mildly impaired (standard score = 65) and expressive vocabulary (Expressive One-Word Picture Vocabulary Test, Fourth Edition [EOWPVT-4]) was moderately impaired (standard score <55). Overall adaptive functioning (Vineland Adaptive Behavior Scales, Second Edition [VABS-II]) was mildly impaired (standard score = 56).
Pedigree 1 – Patient 1.2 (Male)
This patient is a 13-year-old Caucasian male and a full brother of Patient 1.1. He harbored the same compound heterozygous mutations as his sister: c.812A>C (p.N271T)/c.1432_1433delGT (p.V478Rfs*73) (Fig. 1a,b). Pregnancy was full-term and unremarkable. His birth weight was at the 59th percentile (z-score = .3).
The patient experienced his first seizure at 9 years old. He continues to experience seizures, primarily complex generalized seizures, particularly in 4–5-day clusters. His seizures are described as two distinct types. One type involves unresponsiveness, wandering, clicking sound with his tongue, backward head extension, eyebrow twitching, and “throaty” voice, while the other type involves whole-body shaking, followed by entire body stiffening and falling to the ground. Both types are accompanied by a post-ictal state of approximately 20 minutes. Seizures persist despite current prescription of lamotrigine and cannabidiol oil (prior lorazepam and topiramate). EEG at 9 years old was read as abnormal due to rare epileptiform discharges from right centrotemporal region during awake state and intermittent epileptiform discharges from right centrotemporal region during sleep. Subsequent EEGs, at 10 years old, were read as normal. Brain MRI at 9 years old was only notable for bilateral prominent perivascular spaces at centrum semiovale.
The family first noted concerns about development of the patient at an age of 9–12 months. He was standing by 14 months and walked at 2 years old. The patient has a history of speech apraxia, excessive drooling, GERD, and frequent vomiting (up to 2–3 times per day). He has some history of aggressive or dysregulated behaviors. His height and weight at clinical appointments have consistently been less than 1st percentile (z-score <-2.5) for age. A clinical bone age evaluation completed when the patient was 6.5 years old indicated a bone age of 3.5–4 years old.
On physical examination, the patient’s head circumference was 50.5 cm (1st percentile; z-score ≤-2.5) at the time of the examination. He also had evidence of hypersalivation, but remainder of the general exam was otherwise normal. This patient had medical evaluation for short stature, with evidence of delayed bone age as indicated above. He had normal thyroid function test results and IGF-1 levels that were slightly below the lower limit of the reference range for pubertal stage (Tanner Stage 1).
On neurological examination, the patient was alert and oriented to name and date. He knew that he was in the hospital. The patient was noted as not being cooperative with examination, but able to understand questions (e.g., “What would you like to eat?”) and answer appropriately (e.g., “Cheeseburger.”). Cranial nerve exam was largely normal except for significant dysarthria. Pupils were equal, visual fields full, extra-ocular movements intact, face symmetric, and his tongue protruded in the midline. The patient had full strength in the upper and lower extremities by motor exam. However, his tone was increased in legs. His sensation was normal to light touch and vibration. His coordination appeared largely normal without evidence of ataxia. He had brisk reflexes throughout, but in his ankles, he also demonstrated 4–5 beats of clonus bilaterally. In contrast to his sister, the patient had a normal-based gait. However, he had the tendency to walk on his toes with inward rotation of his knees.
Upon neuropsychological examination, intellectual testing (WISC-V) indicated moderately impaired overall IQ (FSIQ = 40). Receptive (ROWPVT-4) and expressive (EOWPVT-4) vocabulary were moderately impaired (standard score <55). Overall adaptive functioning (VABS-II) was moderately impaired (standard score = 50).
Pedigree 2 – Patient 2.1 (Male)
Patient 2.1 presented at 15 years of age to neurology for a developmental evaluation to determine the etiology of his delays (17 years old at current report). Chromosomal microarray analysis and Fragile × testing were negative. A commercial gene panel for IDD/autism revealed a homozygous GPT2 variant of unknown significance, c.775T>C (p.C259R) (Fig. 1c,d). This variant was found to cause loss of enzyme activity (see below).
The patient was born at term after an unremarkable fraternal twin pregnancy. His weight at birth was at the 2nd percentile (z-score = −2.0). The patient crawled at 1 year, walked at 13–14 months, talked at 2 years, and was toilet trained by 3.5 years. He was diagnosed with intellectual disability at 4 years old. A prior head CT scan was read as normal. At age 15, it was noted that the patient needed assistance with bathing and dressing, and he had issues with nocturnal enuresis. He attended school, which focused on activities of daily living. Ongoing challenges with adaptive functioning have persisted into adolescence (e.g., nocturnal enuresis, assistance with bathing and dressing), and his education focuses on developing independence.
On examination, the patient was microcephalic (<1st percentile; z-score <-2.5), and his height and weight were at the 21st (z-score = −.75) and 10th (z-score = −1.25) percentiles, respectively. General physical examination was otherwise unremarkable. Neurological examination revealed that the patient was easily distracted, with limited expressive language. He could not identify colors, shapes, letters, or numbers. Cranial nerves were all intact. He had normal strength and tone. He had a shuffling gait.
Pedigrees 3 and 4
One hundred seventy-six families from Pakistan (plus 16 from Iran) with cousin marriage and IDD were prospectively recruited (author JBV) (Harripaul et al. 2018). Of the 176 Pakistani families, 3 of the families harbored homozygous, putative mutations in GPT2. Notably, this would reflect an approximately 1.7% frequency of GPT2 disease in this population. However, we were not able to establish functional evidence for the variant in family 5, i.e., c.238C>G (p.Q80E); thus, at present, this variant reflects a variant of unknown significance.
Pedigree 3 – Patient 3.1 (Male)
For Pedigree 3, the parents come from a rural community and are first cousins. Their oldest daughter is married to one of her cousins and has two unaffected children. Based on linkage analysis and whole-exome sequencing analysis, the three other siblings in this pedigree were found to be homozygous for the GPT2 variant c.1210C>T (p.R404*) (Fig. 1e) (Harripaul et al. 2018). Notably, this is the same mutation as was found by Ouyang et al. in a distinct family, also from Pakistan but from a geographically distinct region (Ouyang et al. 2016).
Patient 3.1 is a 17-year-old male. He was born after full-term pregnancy and with normal delivery. His milestones were delayed. He started walking at 3 years of age. He had a fall when he was 4 years old, which resulted in further deterioration in walking. He can recognize his family members and people who mostly visit his house. He can eat and clean himself without any help. He does not use any words but communicates with gestures. His motor examination was normal apart from hyperreflexia bilaterally. His coordination is significantly impaired. His oral-motor skills are severely impaired. At examination, head circumference was at <1st percentile at 48.3 cm (z-score = −4.6), height was at 1st percentile at 154.9 cm (z-score = −2.4), and weight was at 8th percentile at 51 kg (z-score = −1.4).
Pedigree 3 – Patient 3.2 (Female)
Patient 3.2 is a 14-year-old female. Birth of the patient was reportedly normal, although delayed milestones were noted early in development. She started walking at the age of 4 years with no further deterioration in walking. Her motor coordination is poor. She can perform routine and repetitive activities without any help. She has trouble in remembering the names of her family members. She feels uneasy and upset if anyone visits her house; however, she can recognize her family members and relatives. She can count up to ten. She has no history of epilepsy. Her most recent physical exam identified head circumference, height, and weight at <1st (43.2 cm; z-score = −9.6), <1st (119.4 cm; z-score = −6.2), and the 1st (34 kg; z-score = −2.3) percentiles, respectively.
Pedigree 3 – Patient 3.3 (Female)
Patient 3.3 is a 10-year-old female. Delivery of the patient was reportedly normal, although delayed milestones were noted early in development. She started walking at the age of 6 years with no further deterioration in walking. She experiences intermittent rigidity and irritability. She insists to eat the same meal every day and becomes very angry if served something different. She can recognize her family members and people who mostly visit her house. She can eat and clean herself without any help. She can count up to twenty. Her most recent physical exam identified head circumference, height, and weight at <1st (40.6 cm; z-score = −9.1), <1st (106.7 cm; z-score = −4.8), and 2nd (23 kg; z-score = −2.1) percentiles, respectively. Her sensory and motor examination was normal apart from hyperreflexia bilaterally. Her coordination is significantly impaired.
Pedigree 4
For Pedigree 4, the parents are also cousins. Four siblings (one deceased) were found to be homozygous for the GPT2 variant c.1035C>T (p.G345=) (Fig. 1f), which reflects a likely splicing mutation (see below). Two unaffected siblings also exist in this pedigree, one of which was confirmed to be heterozygous for the variant and the other for which information concerning the presence or absence of the variant was unavailable. Unfortunately, very limited clinical information is available from this pedigree based on examination from neurological and medical specialists. All affected patients were definitively diagnosed with intellectual disabilities in the mild to moderate range. Two of the three living patients, a female age 40 years and a male age 36 years, were described as having a jerky gait and compact size. Limited information is available on the third affected sibling, a 32-year-old female at time of examination, beyond the clear diagnosis of intellectual disability.
Functional and structural characterization of putative GPT2 mutants
The functional impact of the newly discovered mutations on GPT2 protein stability, enzymatic function (Fig. 2), and subcellular localization (Fig. 3) was tested. To do so, vectors encoding for V5-tagged versions of the C259R, V478Rfs*73, N271T, and Q80E GPT2 mutants were generated, which were then used to transfect HeLa cells for the functional studies described below.
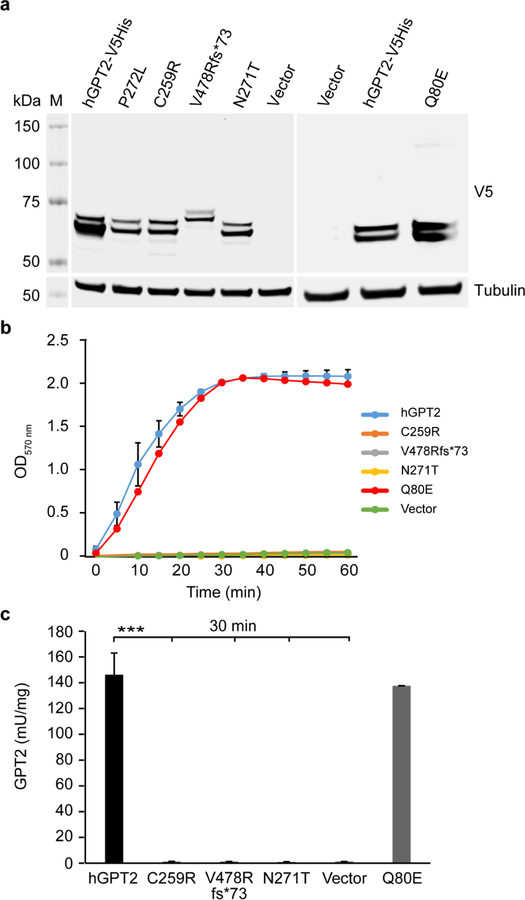
Fig 2.
Exogenous expression and transaminase enzymatic activity of GPT2 mutants. a Western blot results of lysates from HeLa cells transfected to express C-terminal V5-His-tagged human GPT2 (hGPT2-V5His) or one of the GPT2 mutants P272L (previously identified loss-of-function mutation), C259R, V478Rfs*73, N271T, or Q80E. Lysates from cells transfected with empty vector were used as a control. Blots were probed with an anti-V5 antibody to detect exogenously expressed hGPT2 protein; blotting for tubulin served as a loading control. b,c Results of measurement of transaminase enzymatic activity of wild-type hGPT2 and hGPT2 mutants using lysates from HeLa cells as described in (a). Shown are a time course of hGPT enzymatic activity over a 60-min incubation period with substrate (b) and hGPT enzymatic activity measured in cell lysates after incubating with substrate for 30 min (c). n = 3 (V478Rfs*73, N271T, and Q80E), n = 6 (hGPT2, C259R, and vector control). Data are presented as means ± SEMs. ***p <0.001
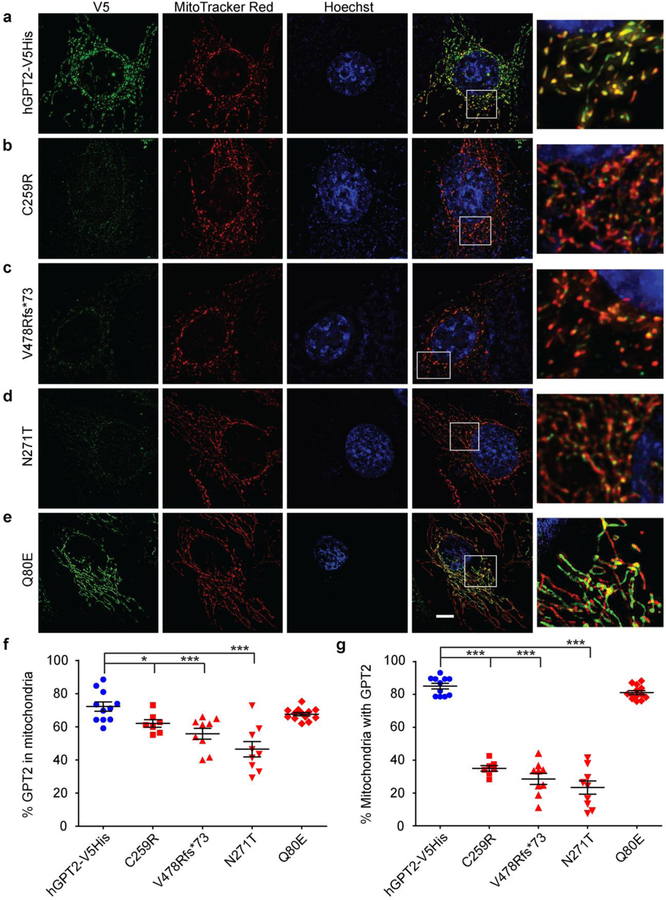
Fig 3.
Subcellular localization of GPT2 mutants in cultured cells. a-e Structured illumination microscopy (SIM) images of HeLa cells exogenously expressing C-terminal V5-His-tagged human GPT2 (hGPT2-V5His) (a) or one of the hGPT2 mutants C259R (b), V478Rfs*73 (c), N271T (d), or Q80E (e) and immunostained with an anti-V5 antibody to detect hGPT2 (green). Prior to fixation, cells were incubated with MitoTracker Red to label mitochondria (red). Nuclei were labeled with Hoechst dye (blue). Boxed areas are magnified on the right. All images were acquired using the same imaging parameters. Scale bar, 5 μm. f,g Quantification of the percent co-localization of exogenously expressed wild-type or mutant hGPT2 with MitoTracker Red based on images similar to those shown in (a-e). Co-localization was calculated as a percent of total hGPT2 stained area, as indicated by V5 staining (f), and as a percent of total MitoTracker Red stained area (g). n = 11 (hGPT2-V5His), n = 7 (C259R), n = 9 (V478Rfs*73), n = 9 (N271T), n = 13 (Q80E). Data are presented as means ± SEMs. *p ≤0.05, ***p ≤0.001
Protein stability of GPT2 coding variants
The molecular weight of V5-tagged wild-type hGPT2 and of the V5-tagged C259R, N271T, and Q80E GPT2 mutants is 57.9 kDa. The molecular weight of the V5-tagged V478Rfs*73 GPT2 mutant, however, is 65.7 kDa. This is due to a 2-bp deletion (GT) and subsequent frameshift, which leads to read through of the original stop codon and extension of the C-terminus of GPT2 by 26 amino acids. Western blotting with an anti-V5 antibody detected double bands in the 58–66 kDa range for wild-type and mutant hGPT2, suggesting similar post-translational modification of all proteins (Fig. 2a). However, other than the Q80E GPT2 mutant, the mutant proteins showed reduced expression levels, which is potentially suggestive of protein instability (Fig. 2a).
Enzymatic activity of GPT2 coding variants
Transaminase enzymatic activity of wild-type and mutant hGPT2 was tested by ALT assay (see Materials and Methods). To do so, total protein was extracted from transfected HeLa cells, and lysates of equal protein amounts for each population of transfected cells were used for assaying enzymatic activity. In the cell lines expressing either the C259R, V478Rfs*73, or N271T GPT2 mutant, enzymatic activity was substantially diminished such that it was comparable to cells transfected with empty vector (Fig. 2b,c). By contrast, the Q80E GPT2 mutant showed GPT2 enzymatic activity comparable to wild-type hGPT2 (Fig. 2b,c).
Mitochondrial localization of GPT2 coding variants
The N-terminus of GPT2 contains a strong mitochondrial localization peptide sequence. Based on PSORT II predictions, none of the four mutant proteins appears to interfere with the peptide sequence predicted to mediate mitochondrial localization. Mitochondrial localization was tested directly by way of immunocytochemistry on transfected HeLa cells followed by imaging using SIM; mitochondrial localization of wild-type vs. mutant hGPT2-V5His was also quantified using images similar to those shown (Fig. 3). The mitochondrial marker MitoTracker Red showed a high level of co-staining with exogenously expressed wild-type hGPT2 (Fig. 3a), as well as with the Q80E hGPT2 mutant (Fig. 3e). When quantified as a percent of total hGPT2 staining, as indicated by V5 staining, 72.2% ± 2.8% and 67.6% ± 1.0% of wild-type and Q80E hGPT2, respectively, co-stained with MitoTracker Red (Fig. 3f). When quantified as a percent of total MitoTracker Red staining, 85.0% ± 1.7% and 81.1% ± 1.1% of MitoTracker Red co-stained with wild-type and Q80E hGPT2, respectively (Fig. 3g). By comparison, cells expressing any of the hGPT2 mutants C259R, V478Rfs*73, or N271T showed statistically significant reductions in levels of hGPT2 staining in mitochondria relative to wild-type hGPT2 (Fig. 3b–d). That is, the percent of hGPT2 that localized to mitochondria was reduced, as was the percent of mitochondrial area containing hGPT2 (Fig. 3f,g). When quantified as a percent of total hGPT2 staining, co-staining with MitoTracker Red was as follows: 62.0% ± 2.3%, p = 0.02 for C259R; 55.8% ± 3.3%, p = 0.001 for V478Rfs*73; and 46.5% ± 4.6%, p = 0.0001 for N271T (Fig. 3f). When quantified as a percent of total MitoTracker Red staining, co-staining with hGPT2 was as follows: 35.0% ± 1.7%, p = 1.03 × 10−12 for C259R; 28.5% ± 3.4%, p = 4.79 × 10−12 for V478Rfs*73; and 23.3% ± 4.1%, p = 1.15 × 10−11 for N271T (Fig. 3g). These results indicate that the C259R, V478Rfs*73, and N271T mutations cause a profound reduction in the localization of GPT2 to mitochondria, in addition to loss of enzymatic activity. Thus, the C259R, V478Rfs*73, and N271T mutations in GPT2 are associated with reductions in protein levels and loss of enzymatic activity, whereas the Q80E missense change appears to be benign at the protein level, with little effect on protein stability, localization, and enzymatic activity.
Structural analysis of two deleterious GPT2 missense mutants, C259R and N271T
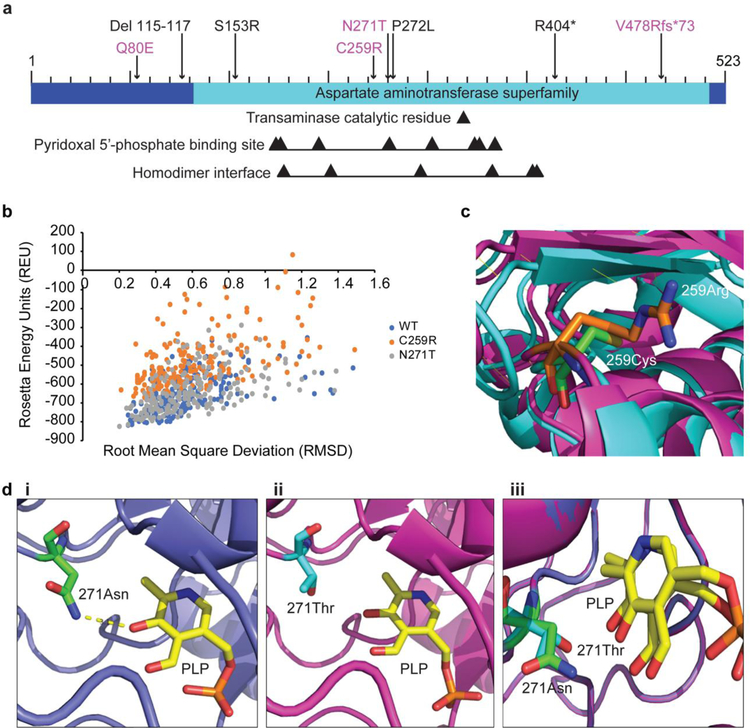
Structural analysis based on computational modeling was next performed to aid in understanding the structural basis by which the C259R and N271T GPT2 mutants might confer loss of enzymatic activity. Notably, N271 is a binding site for PLP, the coenzyme pyridoxal 5’-phosphate. C259 is in the homologous region for both the PLP binding site and the homodimer interface, but it is not a catalytic residue in either interaction (Fig. 4a). For the structural analysis studies, the software PyRosetta was used. PyRosetta is a Python interface for the Rosetta software suite that allows for the computational modeling and analysis of protein structures using a statistical energy function and a nuanced Bayes network (Chaudhury et al. 2010) (see Materials and Methods).
Fig 4.
Structural analysis of two deleterious GPT2 variants. a Bar diagram of the GPT2 protein, with mutations newly characterized in the current study indicated in magenta. Black arrows point to the transaminase catalytic residue, residues involved in binding of the coenzyme pyridoxal 5’-phosphate (PLP), and residues at the homodimer interface. b Stability analysis of wild-type (WT) GPT2 (blue) and the two GPT2 mutants C259R (orange) and N271T (gray), as reflected by Rosetta Energy Units (REU). REU is correlated with energy, which is an indicator of how stable the structure is (lower REU score = higher structure stability). c Visualization of the C259R GPT2 mutant. Wild-type GPT2 is shown in cyan, with C259 in green; mutant GPT2 is shown in magenta, with R259 in orange. di-iii Visualization of the N271T GPT2 mutant and PLP binding. Wild-type GPT2 is shown in blue, with N271 in green (i, iii); mutant GPT2 is shown in magenta, with T271 in cyan (ii, iii); PLP is shown in yellow (i-iii). A hydrogen bond between PLP and N271 is present in wild-type GPT2 (i, dashed yellow line), but is lacking in the N271T mutant (ii)
The measurement of REU (Rosetta Energy Units) is a function of many aspects of a protein’s structure. It is a strong indicator of structure stability and “native-ness” of conformation, although REU is not thermodynamically translatable to energy. In plotting REU, wild-type GPT2 and the N271T GPT2 mutant were found to have similar lowest-energy scores (−817.25 REU and −822.12 REU, respectively), whereas the C259R GPT2 mutant had a lowest-energy score of −670.03 REU (Fig. 4b). This large increase in REU supports that the C259R GPT2 mutant is less stable than wild-type GPT2.
Modeling of the GPT2 protein shows that residue C259 is located in a loop region on the exterior of GPT2, between multiple alpha helices and beta sheets (Berman et al. 2000) (Fig. 4c). Replacing cysteine with the larger, charged arginine could lead to both steric and electrostatic clashes with the surrounding residues. The C259R mutant might also favor an alternate folding conformation that provides a basis for decreases in protein stability and enzymatic activity.
PLP is a coenzyme involved in transamination reactions, and it binds to GPT2 (Rebhan et al. 1997). Based on the Conserved Domain Database, N271 is a very conserved PLP binding site (Fig. 4a). Also, homology analysis revealed that this residue is highly conserved across species (data not shown). N271 is located in a loop structure near the center of the protein, and the importance of this residue to GPT2 function is supported by ligand docking simulations (Fig. 4di-iii). The docking simulations to both wild-type GPT2 and the N271T GPT2 mutant resulted in lowest-energy complexes with PLP in the same location and orientation as observed in crystal structure (data not shown). However, the substitution of threonine for asparagine eliminates a hydrogen bond between threonine and the docked PLP, which could decrease the binding affinity (Fig. 4di,ii). Indeed, the binding energy score for wild-type GPT2 vs. the N271T mutant increases from −13.712 REU to −13.051 REU (Table 2). A similar effect is not observed for the C259R mutant, which is not predicted to affect PLP binding.
Table 2.
Interface energies of wild-type and mutant GPT2a
| Wild-type | C259R | N271T |
|---|---|---|
| −13.712 | −14.109 | −13.051 |
| −12.819 | −13.904 | −12.797 |
| −12.596 | −13.64 | −12.796 |
| −12.457 | −12.916 | −12.637 |
| −12.123 | −12.905 | −12.384 |
| −12.101 | −12.888 | −12.207 |
| −12.002 | −12.711 | −12.09 |
| −11.694 | −12.486 | −12.045 |
| −11.551 | −12.337 | −12.021 |
| −11.488 | −12.21 | −11.962 |
| −11.457 | −12.158 | −11.533 |
Interface energy is defined as the contribution of the ligand and any surrounding residues to the overall energy score. Shown are the interface energy scores for the ten lowest-scoring conformations for wild-type GPT2 and two GPT2 mutants included in this study, C259R and N271T
Evaluation of putative splicing mutations by exon trapping
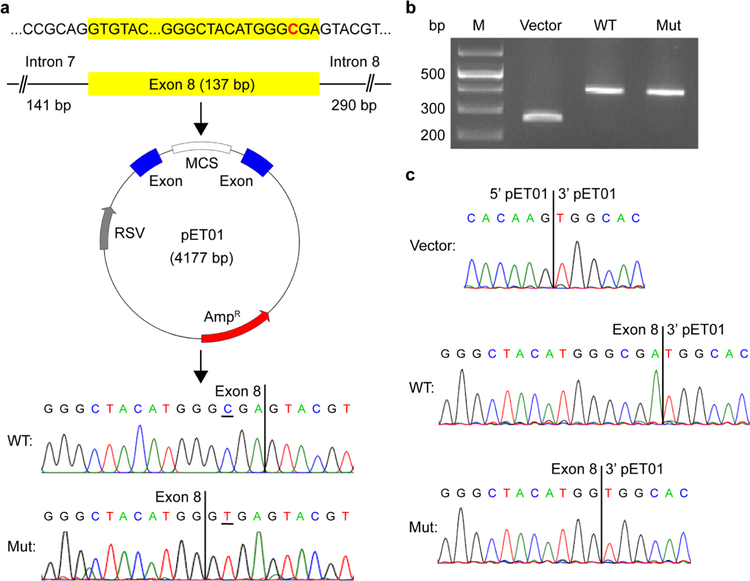
The putative GPT2 mutation in Pedigree 4, c.1035C>T (p.G345=), is a synonymous mutation. Nonetheless, given strong linkage data to chromosome 16q11.2 specifically at the GPT2 locus and the proximity of the mutation to an exon-intron boundary, we further evaluated this mutation for potential effects on splicing. Importantly, we discerned that the C-to-T change would introduce a strong splice donor site 4 bp upstream of the reference splice donor at the end of exon 8. If this mutant splice donor were to engage the splice acceptor on exon 9, then we would predict a 4-bp deletion and subsequent frameshift, resulting in a nonsense mutation.
To test for effects on splicing, an exon trapping assay was performed using the exon trapping vector pET01 and by cloning the mutant and reference exon 8 into the multiple cloning site (Fig. 5a) (see also Materials and Methods). Wild-type, mutant, and control (empty pET01) vectors were then separately transfected into HEK293T cells. Following extraction of RNA from transfected cells, RT-PCR were performed using primers complementary to the pET01 exons bordering the multiple cloning site, and PCR amplicons were separated by electrophoresis on an agarose gel (Fig. 5b) and analyzed by Sanger DNA sequencing (Fig. 5c). As predicted, the C-to-T change in exon 8 led to a 4-bp deletion in comparing the wild-type vs. mutant splicing reaction. These data thereby provide strong support that the c.1035C>T (p.G345=) GPT2 mutation is a loss-of-function mutation.
Fig 5.
The GPT2 synonymous mutation c.1035C>T (p.G345=) results in aberrant use of a donor splice site. a Generation of an exon trapping vector for testing of the effects of the c.1035C>T (p.G345=) mutation on splicing. Exon 8 of the human wild-type GPT2 gene, as well as portions of the flanking introns (141 bp of intron 7 and 290 bp of intron 8), was ligated into the multiple cloning site (MCS) of the pET01 vector (mutated base in red). Site-directed mutagenesis was performed to generate a pET01-GPT2 vector with the c.1035C>T mutation in exon 8. Wild-type (WT) and mutant (Mut) vectors were confirmed by Sanger DNA sequencing (bottom chromatograms). The mutated base is underlined. b,c Results of exon trapping assays. RT-PCR were performed with cDNA generated from HEK293T cells transfected with either the wild-type (WT), mutant (Mut), or control (empty pET01) vector. Amplicons were separated by agarose gel electrophoresis (b) and analyzed by Sanger DNA sequencing (c). The c.1035C>T mutation in exon 8 results in a 4-bp deletion due to creation of a new splice donor site (compare bottom two chromatograms)
The GPT2 missense variant identified in family 5, c.238C>G (p.Q80E), was also tested using the exon trapping assay. Here, splice-site algorithms (http://www.fruitfly.org/seq_tools/splice.html) predicted a reduction in efficiency of the splice donor at the exon 1/intron 1 boundary, with a reduction in score from 0.78 to 0.58. No change in splice-site usage was observed, however, using the exon trapping assay (data not shown). Whether more subtle effects on splicing might actually occur in vivo is at present unclear, and this possibility cannot be excluded.
Frequency of GPT2 variants in a population-based sample without intellectual disability
The estimate of GPT2 mutations in the Pakistani cohort was approximately 2 of 176, or 1.1% of autosomal recessive intellectual disability. We set out to estimate the population prevalence of heterozygous GPT2 mutations in a control dataset, that is, the frequency of the carrier state. To this end, the ExAC database, a cohort of approximately 70,000 people, presumably without developmental disability and largely from Northern European geographic ancestry, was utilized (Lek et al. 2016). At the time of access, ExAC reported 467 different variants in GPT2, with each variant annotated for effect on protein sequence and overall frequency in the population. Using the protein sequence effect annotation, together with effects predicted by PolyPhen-2 and SIFT, variants were sorted into three categories: benign, moderate, and severe (Fig. 6). The total variant allele frequency for each of the three categories was calculated by summing the adjusted frequencies of the individual variant alleles within each category. The population carrier frequency (i.e., heterozygous frequency) and the predicted affected frequency (i.e., homozygous frequency) for each category was then calculated, assuming Hardy-Weinberg equilibrium and a recessive mode of inheritance (Fig. 6) (see also Materials and Methods).
Carrier frequency for severe GPT2 mutations is estimated at approximately 1 in 3,991 individuals and for moderate mutations at 1 in 264 individuals. Thereby, the carrier frequency of severe or moderate mutations is approximately 1 in 248 individuals, which leads to a prevalence estimate of 1 in 246,016 people with homozygous or compound heterozygous mutations. Interestingly, the c.1210C>T (p.R404*) variant has thus far been reported as homozygous in two distinct pedigrees with IDD, both in Pakistan (Harripaul et al. 2018; Ouyang et al. 2016). This highly deleterious variant was found in heterozygosity in the ExAC database in four different individuals. Three of four individuals were of South Asian descent, and the remaining individual was of non-Finnish, European descent. These data suggest that this variant may occur at a relatively higher than expected frequency in the South Asian population.
DISCUSSION
We present new pedigrees for individuals with IDD with mutations in GPT2: novel compound heterozygous mutations in two siblings, c.812A>C (p.N271T)/c.1432_1433delGT (p.V478Rfs*73); two novel homozygous mutations, c.775T>C (p.C259R) and c.1035C>T (p.G345=); and a recurrent mutation identified in a new family, c.1210C>T (p.R404*). These mutations reflect two new missense mutations and a novel synonymous mutation with effects on splicing. The clinical phenotype of these patients is characterized by IDD and motor delays/abnormalities, consistent with the previously documented GPT2 mutation phenotype, including spastic paraplegia in some patients (Celis et al. 2015; Hengel et al. 2018; Kaymakcalan et al. 2018; Lobo-Prada et al. 2017; Ouyang et al. 2016). However, there are several important new findings that extend the phenotype and also provide additional data regarding the frequency of prior observations. We also demonstrate the functional impact of the newly identified GPT2 mutations using a transaminase enzymatic activity assay, immunocytochemistry and super-resolution microscopy (SIM), and structural modeling.
One of the most striking features is the small stature of the majority of patients. For example, for the two sibling patients with the compound heterozygous mutations documented height and weight was less than 5th percentile throughout development for the female, and less than the 1st percentile for the male (within the context of parents of typical height and weight). The small stature was documented over a decade of their lives and into adolescence. Additionally, the three patients in Pedigree 3 had small stature, at the 1st percentile or less. In retrospect, all three of the patients reported by Celis et al. (Celis et al. 2015) were noted to have small stature (i.e., height and/or weight that was <5th percentile) during at least one time point in development. However, small stature was only noted in one of the patients by the most recent exam in that prior report.
Small stature has not been previously described as a core component of the GPT2 phenotype; however, susceptibility to small stature is highly plausible given the biological function of the enzyme in anaplerosis. First, there are several studies recently that have implicated GPT2 in cancer cell proliferation and growth (Cao et al. 2017; Feng et al. 2016; Hao et al. 2016; Itkonen et al. 2016; Kim et al. 2019; Smith et al. 2016; Xu et al. 2016). Smith et al. highlight the importance of GPT2’s role in coupling the Warburg effect to glutamine catabolism toward tumorigenesis (Smith et al. 2016). Hao et al. have shown that oncogenic PIK3CA mutations upregulate GPT2 levels in colorectal cancer, suggesting GPT2-mediated glutamine dependency in cancer progression (Hao et al. 2016). The feature of small stature noted in our study, paired with at least one patient reported by Celis et al. (Celis et al. 2015), suggests that small stature may manifest as a component of the GPT2 mutation phenotype.
Notably, in the absence of small stature, the vast majority of patients have prominent postnatal microcephaly, which is an attenuation of brain growth in the postnatal period. This phenotype perhaps reflects an increased susceptibility of the brain to limitations in tissue growth and development. The biological underpinnings of this brain growth remain to be defined in mechanistic detail; however, it is plausible to hypothesize a role for GPT2 in growth of cell size. The siblings in our study here with the compound heterozygous mutations (Pedigree 1) were also noted as experiencing challenges with vomiting, which raises the possibility that nutrition could in part contribute to the phenotype of small stature. Nonetheless, given known metabolic functions of GPT2 in anaplerosis, and thereby cell growth, it is important for clinicians to consider the possibility of decreased growth and stature in patients with GPT2 mutations.
In patients presented to date, intellectual disability in patients with GPT2 mutations has been noted as “severe” in status in at least 81% of patients (Hengel et al. 2018). Two of the currently reported patients (again in Pedigree 1) were diagnosed as having moderate intellectual disability status, based on formally assessed overall intelligence (standard scores = 40 and 41) and adaptive functioning (standard scores = 50 and 56). Expressive and receptive language skills were similarly impaired, and these two children (aged 13 and 15 years) were estimated to be functioning at a level comparable to a typically developing 1- to 6-year-old child. These findings reiterate that intellectual disability is a universal component of the phenotype, although there may be some degree of variability in the overall severity.
With regard to the neurological phenotype, motor abnormalities, particularly spastic paraplegia, is described as a core characteristic of the phenotype (Hengel et al. 2018; Ouyang et al. 2016). A majority of patients for whom we had reliable motor exam results showed symptoms consistent with spastic paraplegia, including lower extremity spasticity, hyperreflexia, abnormal gait, or motor apraxia. Approximately 38% of prior patients with GPT2 mutations have a history of seizures (Hengel et al. 2018), with a history of epilepsy documented in at least two of the patients reported here.
While the frequency of mutation carriers in the ExAC database is reflective of a rare status, the frequency in the Pakistani autosomal recessive intellectual disability convenience sample was determined to be surprisingly high. Namely, for the Pakistani cohort, approximately 1.1% of intellectual disabilities were secondary to autosomal recessive GPT2 mutations. However, it is also conceivable that our estimate reflects an underestimate in this given convenience sample, as GPT2 was sequenced only in those pedigrees that mapped to the chromosome 16 locus. There were other pedigrees in the cohort that were not sufficiently powered to reveal a locus and were not sequenced. Additionally, one pedigree with GPT2 mutation reported by Ouyang et al. (2016) was from Pakistan. Based on available estimates, a frequency of 1.1% is high. Extrapolating from X-linked intellectual disability (XLID) wherein 88 of approximately 800 genes may cause XLID, one arrives at approximately 10% of genes as being causal. With regard to autosomal recessive intellectual disability, we might estimate nearly 2,000 genes. An estimate of 500–1,000 genes would suggest that the frequency of 1.1% is five to ten times expected. Of course, we have to interpret this finding with caution given the small sample size of the cohort. Such a study needs to be studied in a robustly powered replication cohort with the same geographic ancestry.
In summary, GPT2-related autosomal recessive intellectual disability represents an important new neurogenetic syndrome involving developmental as well as progressive features, notably motor disabilities. We present evidence to extend the phenotype to potentially involving susceptibility to small stature, which is plausible given the metabolic function of GPT2. Additionally, we present population prevalence estimates reflecting populations from largely Northern European ancestry, as well as from Pakistan. In the Pakistani cohort, the prevalence of GPT2 mutations makes this new syndrome among the most common causes of autosomal recessive intellectual disability.
ACKNOWLEDGEMENTS
The authors would like to thank the families for their participation in the current study.
FUNDING
Research reported in this publication was supported in part by the following: National Institute of Mental Health grant numbers R01MH102418 and R01MH105442 and the Hassenfeld Child Health Innovation Institute at Brown University (EMM); Canadian Institutes of Health Research grant numbers MOP-102758 and PJT-156402 (JBV); and a Carney Institute for Brain Science and Suna Kıraç Fellowship Graduate Award in Brain Science (OB).
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
DATA AVAILABILITY
All data generated or analyzed as part of this study are included herein or are available from the corresponding author on reasonable request.
REFERENCES
- Berman HM et al. (2000) The Protein Data Bank. Nucleic Acids Res 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Lin SH, Wang Y, Chin YE, Kang L, Mi J (2017) Glutamic Pyruvate Transaminase GPT2 Promotes Tumorigenesis of Breast Cancer Cells by Activating Sonic Hedgehog Signaling. Theranostics 7:3021–3033. doi: 10.7150/thno.18992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celis K, Shuldiner S, Haverfield EV, Cappell J, Yang R, Gong DW, Chung WK (2015) Loss of function mutation in glutamic pyruvate transaminase 2 (GPT2) causes developmental encephalopathy. J Inherit Metab Dis 38:941–948. doi: 10.1007/s10545-015-9824-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (2004) Economic costs associated with mental retardation, cerebral palsy, hearing loss, and vision impairment--United States, 2003. MMWR Morb Mortal Wkly Rep 53:57–59. [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (2013) Intellectual Disability Among Children https://www.cdc.gov/ncbddd/developmentaldisabilities/documents/IntellectualDisabilities.pdf. Accessed on 10 October 2018
- Chaudhury S, Lyskov S, Gray JJ (2010) PyRosetta: a script-based interface for implementing molecular modeling algorithms using Rosetta. Bioinformatics 26:689–691. doi: 10.1093/bioinformatics/btq007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca S, Khar K, Meiler J (2015) Fully Flexible Docking of Medium Sized Ligand Libraries with RosettaLigand. PLoS One 10:e0132508. doi: 10.1371/journal.pone.0132508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Hao Y, Wang Z (2016) Targeting glutamine metabolism in PIK3CA mutant colorectal cancers. Genes Dis 3:241–243. doi: 10.1016/j.gendis.2016.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y et al. (2016) Oncogenic PIK3CA mutations reprogram glutamine metabolism in colorectal cancer. Nat Commun 7:11971. doi: 10.1038/ncomms11971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harripaul R et al. (2018) Mapping autosomal recessive intellectual disability: combined microarray and exome sequencing identifies 26 novel candidate genes in 192 consanguineous families. Mol Psychiatry 23:973–984. doi: 10.1038/mp.2017.60 [DOI] [PubMed] [Google Scholar]
- Hengel H et al. (2018) GPT2 mutations cause developmental encephalopathy with microcephaly and features of complicated hereditary spastic paraplegia. Clin Genet 94:356–361. doi: 10.1111/cge.13390 [DOI] [PubMed] [Google Scholar]
- Iglesias A et al. (2014) The usefulness of whole-exome sequencing in routine clinical practice. Genet Med 16:922–931. doi: 10.1038/gim.2014.58 [DOI] [PubMed] [Google Scholar]
- Itkonen HM et al. (2016) Inhibition of O-GlcNAc transferase activity reprograms prostate cancer cell metabolism. Oncotarget 7:12464–12476. doi: 10.18632/oncotarget.7039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamra R (2018) Genetics of autosomal recessive intellectual disability. Med Genet 30:323–327. doi: 10.1007/s11825-018-0209-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaymakcalan H, Yarman Y, Goc N, Toy F, Meral C, Ercan-Sencicek AG, Gunel M (2018) Novel compound heterozygous mutations in GPT2 linked to microcephaly, and intellectual developmental disability with or without spastic paraplegia. Am J Med Genet A 176:421–425. doi: 10.1002/ajmg.a.38558 [DOI] [PubMed] [Google Scholar]
- Kim M, Gwak J, Hwang S, Yang S, Jeong SM (2019) Mitochondrial GPT2 plays a pivotal role in metabolic adaptation to the perturbation of mitochondrial glutamine metabolism. Oncogene doi: 10.1038/s41388-019-0751-4 [DOI] [PubMed]
- Lee H et al. (2014) Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA 312:1880–1887. doi: 10.1001/jama.2014.14604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M et al. (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature 536:285–291. doi: 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo-Prada T, Sticht H, Bogantes-Ledezma S, Ekici A, Uebe S, Reis A, Leal A (2017) A Homozygous Mutation in GPT2 Associated with Nonsyndromic Intellectual Disability in a Consanguineous Family from Costa Rica. JIMD Rep 36:59–66. doi: 10.1007/8904_2016_40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyskov S et al. (2013) Serverification of molecular modeling applications: the Rosetta Online Server that Includes Everyone (ROSIE). PLoS One 8:e63906. doi: 10.1371/journal.pone.0063906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A et al. (2017) CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45:D200–D203. doi: 10.1093/nar/gkw1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti R, Lyskov S, Das R, Meiler J, Gray JJ (2018) Web-accessible molecular modeling with Rosetta: The Rosetta Online Server that Includes Everyone (ROSIE). Protein Sci 27:259–268. doi: 10.1002/pro.3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambot S et al. (2018) Clinical whole-exome sequencing for the diagnosis of rare disorders with congenital anomalies and/or intellectual disability: substantial interest of prospective annual reanalysis. Genet Med 20:645–654. doi: 10.1038/gim.2017.162 [DOI] [PubMed] [Google Scholar]
- Ouyang Q et al. (2016) Mutations in mitochondrial enzyme GPT2 cause metabolic dysfunction and neurological disease with developmental and progressive features. Proc Natl Acad Sci U S A 113:E5598–5607. doi: 10.1073/pnas.1609221113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen OE, Kalhan SC, Hanson RW (2002) The key role of anaplerosis and cataplerosis for citric acid cycle function. J Biol Chem 277:30409–30412. doi: 10.1074/jbc.R200006200 [DOI] [PubMed] [Google Scholar]
- Rebhan M, Chalifa-Caspi V, Prilusky J, Lancet D (1997) GeneCards: integrating information about genes, proteins and diseases. Trends Genet 13:163. [DOI] [PubMed] [Google Scholar]
- Rose NC, Wick M (2016) Current recommendations: Screening for Mendelian disorders. Semin Perinatol 40:23–28. doi: 10.1053/j.semperi.2015.11.004 [DOI] [PubMed] [Google Scholar]
- Rump P et al. (2016) Whole-exome sequencing is a powerful approach for establishing the etiological diagnosis in patients with intellectual disability and microcephaly. BMC Med Genomics 9:7. doi: 10.1186/s12920-016-0167-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers EW et al. (2011) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 39:D38–51. doi: 10.1093/nar/gkq1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B, Schafer XL, Ambeskovic A, Spencer CM, Land H, Munger J (2016) Addiction to Coupling of the Warburg Effect with Glutamine Catabolism in Cancer Cells. Cell Rep 17:821–836. doi: 10.1016/j.celrep.2016.09.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevenon J et al. (2016) Diagnostic odyssey in severe neurodevelopmental disorders: toward clinical whole-exome sequencing as a first-line diagnostic test. Clin Genet 89:700–707. doi: 10.1111/cge.12732 [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38:e164. doi: 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P et al. (2016) LRH-1-dependent programming of mitochondrial glutamine processing drives liver cancer. Genes Dev 30:1255–1260. doi: 10.1101/gad.277483.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang RZ, Blaileanu G, Hansen BC, Shuldiner AR, Gong DW (2002) cDNA cloning, genomic structure, chromosomal mapping, and functional expression of a novel human alanine aminotransferase. Genomics 79:445–450. doi: 10.1006/geno.2002.6722 [DOI] [PubMed] [Google Scholar]