Abstract
Motivation
Traditional glycan microarray data is typically presented as excel files with limited visualization and interactivity. Thus, comparisons and analysis of glycan array data have been difficult, and there is need for a tool to facilitate data mining of glycan array data.
Results
GLAD (GLycan Array Dashboard) is a web-based tool to visualize, analyze, present and mine glycan microarray data. GLAD allows users to input multiple data files to create comparisons. GLAD extends the capability of the microarray data to produce more comparative visualizations in the form of grouped bar charts, heatmaps, calendar heatmaps, force graphs and correlation maps in order to analyze broad sets of samples. Additionally, it allows users to filter, sort and normalize the data and view glycan structures in an interactive manner, to facilitate faster visual data mining.
Availability and implementation
GLAD is freely available for use on the Web at https://glycotoolkit.com/Tools/GLAD/ with all major modern browsers (Edge, Firefox, Chrome, Safari).
Supplementary information
Full documentation and video tutorials for GLAD can be found on https://glycotoolkit.com/GLAD.
1 Introduction
Out of the four major macromolecules within human cells, glycans (oli-gosaccharides) are by far the most complicated as they can be made up of multiple monosaccharide units, having 3–5 possible sites of attachments, with 2 anomeric configurations (α or β) for glycosidic bonds and range in size from one- to >20-mers long (Seeberger, 2015). Thus, it is not just the diversity in the molecules and length of the glycan, but also the variety of linkages and possibility of branching, which increases the molecular complexity of glycans orders of magnitudes higher than other macromolecules. Glycans play crucial roles in a number of biological functions and pathologies such as immunity, host-pathogen interactions, adhesion and cell signaling via binding to glycan binding proteins (Cummings and Pierce, 2014; Varki, 2017; Varki et al., 2015). Studying interactions that involve glycans can potentially lead to a better understanding of biology for potential diagnostics and therapeutics.
Defined glycan arrays such as those produced by the Consortium for Functional Glycomics (CFG) and the National Center for Functional Glycomics (NCFG) contain synthetic glycans of known structure and are printed on functionalized glass microarray surfaces (Heimburg-Molinaro et al., 2011). Such arrays can be utilized to screen Glycan Binding Proteins (GBPs) and biological samples (e.g. serum) using very small sample volumes to define their binding specificities (Liang and Wu, 2009). To date, data for >3900 experiments has been made public by the CFG, with more data being added (http://www.functionalglycomics.org/).
Data visualization is an important step in exploratory data analysis, and should be made comprehensive but less tedious (Shelly, 1996). Tools to visualize glycan array data and binding patterns are limited. There have been a few glycan array database tools created previously, for example, CFG database (Venkataraman et al., 2015), GlycoPattern (Agravat et al., 2014), GlycanBinder and GlycoSearch (Kletter et al., 2015; Porter et al., 2010). These resources provide large amounts of raw data or automated presentation of binding motifs. However, the software have limited data visualization features (bar graphs and heatmaps) and are restrictive with respect to usage (need login or special access), thus limiting their use in visual analytics. To facilitate the data visualization and visual comparison of glycan array data, we present here a new tool, GLAD (GLycan Array Dashboard), which is a browser-based tool to visualize and mine glycan array data. GLAD was developed using a combination of JavaScript (JS) libraries including D3.js, jQuery, Lodash, Bootstrap and Select2js. As GLAD is mainly coded in JS, it removes the need for users to upload their data to any server, and hence keeps user data and analysis private on their local site. This significantly reduces the need to maintain a high-performance server. Furthermore, the data visualization includes not only bar graphs and heatmaps, but also calendar heatmaps, force graphs and correlation maps. All visualizations are interactive and in SVG format for the best user experience. GLAD also allows users to filter, sort and normalize data to accentuate key data and binding relationships. Overall, GLAD helps to make glycan array data analysis simpler and more easily accessible to a larger community.
2 Materials and methods
The general workflow for GLAD is as follows (Fig. 1): 1. Input data as tab-delimited text files in the correct format. 2. Visualize and select data for display using bar charts (Stage-1), or, alternatively add the entire data file to the selections. 3. Save/Load the selections from multiple experiments. This allows the user to perform their analysis locally and resume at any time. Filter, Sort or Normalize the selections options allow the user to mine the data to analyze binding patterns. 4. Visualize the data selected from various experiments in one of the Stage-2 visualizations, i.e.:
Grouped Bar Charts- useful to compare small datasets.
Heatmaps- useful to compare data for large numbers of samples across small numbers of glycans.
Calendar Heatmaps- useful to compare data for large numbers of glycans across multiple experiments.
Force Graphs- useful to visualize any network-like binding patterns and identify common binding glycans among samples.
Correlation plots- useful to compare and quantify similarities or differences in binding patterns for a set of experiments, based on the generated Pearson Coefficient (r) values.
Bubble Box plots- allows users to get statistical values as box plots and mean values with scatter-plot like data.
Fig. 1.
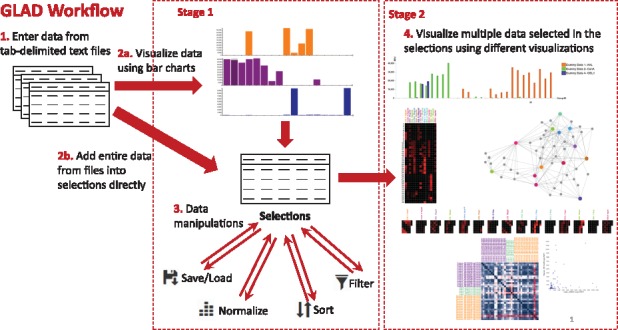
GLAD workflow showing how the user can input, manipulate and visualize glycan array data
The users can save and export the figures as SVG files. Users can view the glycan structure in the Glycan Structure drawer at any time by hovering the mouse over data points. Full documentation can be found in the GLAD documentation online along with video snippets and tutorials to guide the user on how to use the tools. In addition, we have put up an example demonstrating the ability to use GLAD to differentiate between Galectin-3, Galectin-3C and Galectin-7 online.
3 Results and discussion
GLAD is a unique tool offered to glycoscientists to assist in mining and comparing glycan array data in an easy and private manner. GLAD can be extremely useful in uncovering hidden relationships between array datasets, which was previously unexplored. The ability to visualize glycan array data, alongside glycan structure and the ability to filter the data based on user-defined criteria enables the user to mine data in an efficient and thorough manner. In addition, the selection files which are saved in the tool can be attached to manuscripts as Supplementary Information for reviewers and readers to have a more interactive view of the data. The tool is constantly being updated to add new features as requested by internal users of the NCFG, which provides glycan microarray assays as a service to the scientific community, and is open to suggestions from the glycoscience community. Additionally, newer data visualizations are constantly being investigated. In the future, we intend to host a database, which could feed data that is public into the GLAD interface for visualization. This will allow users to analyze and compare their data along with publicly available data.
Acknowledgements
The authors would like to thank all members of the National Center for Functional Glycomics for their valuable feedback, especially Chao Gao, Lauren Byrd-Leotis, Tanya McKitrick, Jamie Heimburg-Molinaro and Alyssa McQuillan.
Funding
This work was supported by National Institutes of Health Grants P41GM103694 and U01GM125267.
Conflict of Interest: none declared.
References
- Agravat S.B. et al. (2014) GlycoPattern: a web platform for glycan array mining. Bioinformatics, 30, 3417–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings R.D., Pierce J.M. (2014) The challenge and promise of glycomics. Chem. Biol., 21, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimburg-Molinaro J. et al. (2011) Preparation and analysis of glycan microarrays. Curr. Protoc. Protein Sci., 10, 12.10.1–12.10.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kletter D. et al. (eds.) (2015) Glycoscience: Biology and Medicine. Springer, Tokyo, Japan, pp. 61–68. [Google Scholar]
- Liang C.H., Wu C.Y. (2009) Glycan array: a powerful tool for glycomics studies. Exp. Rev. Proteomics, 6, 631–645. [DOI] [PubMed] [Google Scholar]
- Porter A. et al. (2010) A motif-based analysis of glycan array data to determine the specificities of glycan-binding proteins. Glycobiology, 20, 369–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeberger P.H. (2015) Monosaccharide diversity In: Varki A.et al. (eds.) Essentials of Glycobiology. Cold Spring Harbor, New York, pp. 19–30. [Google Scholar]
- Shelly M.A. (1996) Exploratory data analysis: data visualization or torture? Infect. Control Hosp. Epidemiol., 17, 605–612. [DOI] [PubMed] [Google Scholar]
- Varki A. (2017) Biological roles of glycans. Glycobiology, 27, 3–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A. et al. (eds.) (2015) Essentials of Glycobiology. Cold Spring Harbor, New York: pp. 77–88. [Google Scholar]
- Venkataraman M. et al. (2015) Glycan array data management at consortium for functional glycomics. Methods Mol. Biol., 1273, 181–190. [DOI] [PubMed] [Google Scholar]


