Abstract
This editorial refers to ‘Calmodulin mutations and life-threatening cardiac arrhythmias: insights from the International Calmodulinopathy Registry’†, by L. Crotti et al., on page 2964.
Calmodulin is the primary sensor of intracellular calcium concentration shifts. On binding calcium, calmodulin undergoes large conformational changes, allowing rapid transmission of signals to ≥300 target proteins. Calmodulin integrity is so crucial that the human genome contains three independent calmodulin-encoding genes (CALM1–CALM3), all functional and encoding the same protein, which further is 100% conserved across vertebrates.1
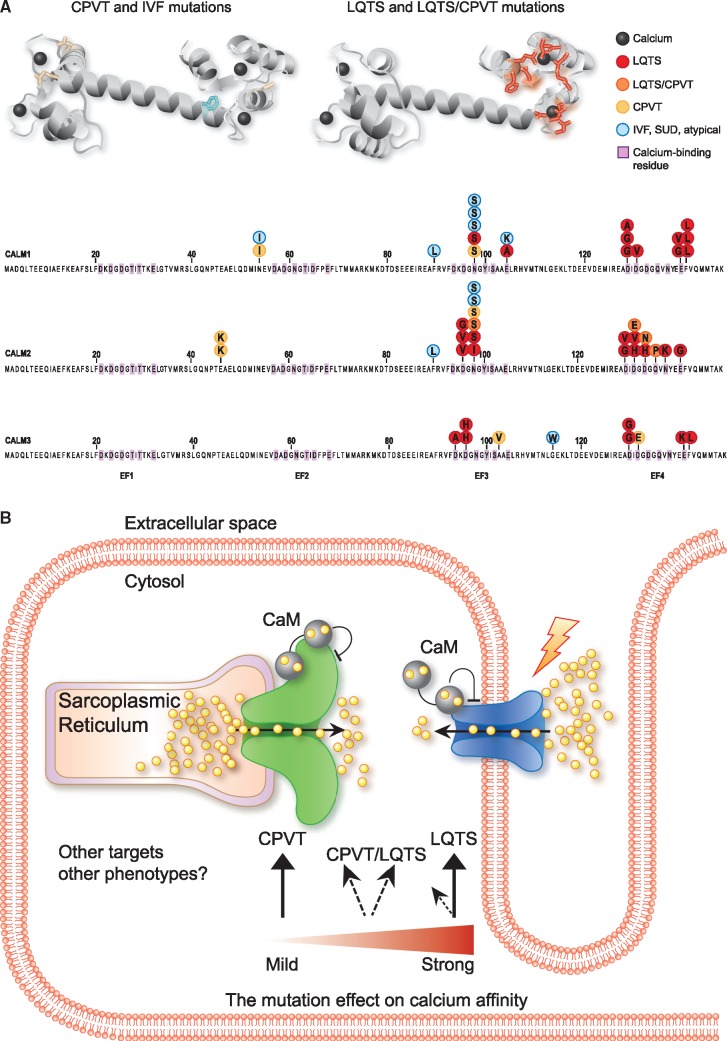
Given this level of conservation, calmodulin mutations were long thought to be incompatible with life. It therefore came as a big surprise when a missense mutation was identified in a genetic linkage study of a large Swedish family with catecholaminergic polymorphic ventricular tachycardia (CPVT).2 All affected family members carried a p.Asn54Ile substitution in CALM1 (Take home figure A). The mutated residue is in the calmodulin N-terminal domain but induces only minimal changes in calcium binding.3
Take home figure.
Position and main mechanism of arrhythmogenic calmodulin mutations. (A) Upper: three-dimensional structure of calmodulin in calcium-bound form with mutated residues shown in stick representation (PDB-ID: 1CLL). Lower: individual founder mutations shown on the protein encoded from the representative CALM gene. SUD, sudden unexpected death. (B) Schematic illustration of a cardiomyocyte and the main molecular mechanism for LQTS, CPVT, and mixed phenotypes, indicating main targets (RyR2 in green, CaV1.2 in blue) and correlation with impact on calmodulin C-terminal domain calcium affinity loss. Yellow spheres, calcium ions; CaM, calmodulin.
In the following year, three more CALM missense mutations were identified among four unrelated infants with long QT syndrome (LQTS) and recurrent cardiac arrest.4 These de novo missense mutations in either CALM1 or CALM2 expanded the phenotypic spectrum to include a mechanistically entirely different disease. More reports followed, with novel missense mutations also identified in CALM3, and including patients with a diagnosis of idiopathic ventricular fibrillation (IVF).5,6 It thus became clear that calmodulin mutations underlie a complex phenotypic spectrum, today known as calmodulinopathies.7
In this issue of the European Heart Journal, Crotti et al. describe the International Calmodulinopathy Register (ICalmR), a large collaborative effort to systematically register individuals carrying pathogenic calmodulin mutations.8 Of 74 included subjects carrying a pathogenic missense variant in calmodulin, 58 were ascertained from collaborating centres and 16 from published literature. For the ascertained cases, the authors used a registry-specific case report file and meticulously recorded the clinical picture, genetic variant, and treatment response. This effort has more than doubled the number of known carriers and brought the count of distinct pathogenic amino acid changes to 28. The predominant phenotype is LQTS (49%), followed by CPVT (28%). Other phenotypes include overlapping features of LQTS/CPVT, IVF, sudden unexplained death (SUD), and neurological impairments. Most pathogenic mutations are de novo, in line with the severe phenotypes, with median onset ages as low as 1.5 years for LQTS and 6 years for CPVT.
With access to this impressive collection of mutation carriers, the CALM genes are now irrevocably established as major genes for cardiac arrhythmias. Their missense mutations can cause LQTS, CPVT, overlapping features of the two, and IVF (Take home figure A). With the continued collection of detailed clinical information, the ICalmR will become invaluable for variant interpretation of new CALM mutations associated with cardiac arrhythmia.
Interpretation of missense mutations in asymptomatic individuals is less clear. Ascertainment of primarily symptomatic and familial cases is expected to create an upwards bias in estimated penetrance of identified disease-causing mutations.9 Capturing the full unbiased phenotypic spectrum and associated penetrance of CALM mutations will therefore require sequencing of large population-based cohorts. Of interest, six carriers had neurological impairments unrelated to cardiac arrest, suggesting that some calmodulinopathies may involve the brain.
Impaired regulation of calcium entry into the cytosol has been suggested to explain how calmodulin mutations can lead to both CPVT and LQTS (reviewed in Jensen et al.10). For early-onset severe LQTS, aberrant calcium influx through compromised voltage-gated L-type calcium channels (CaV1.2) leads to increased action potential duration. CPVT, in contrast, arise from aberrant regulation of the intracellular sarcoplasmic reticulum calcium release channel, RyR2. For both targets, calmodulin inhibits calcium flux in a calcium-dependent manner, providing a negative feedback loop for appropriate regulation of magnitude and duration of calcium transients.
The question remains of how a mutation in only one of six alleles encoding identical calmodulin proteins dominantly leads to fatal cardiac arrhythmias. One possible explanation comes from the ability of calmodulin to pre-associate with both CaV1.2 and RyR2 at resting calcium concentrations. Calmodulin mutations that do not interfere with calmodulin–target binding at low calcium concentrations can compete with normal calmodulin for binding and impair channel response to increased calcium concentrations. This results in loss of negative feedback and excessive calcium release. It is likely because of these features, shared between CaV1.2 and RyR2 calcium channels, that one of six alleles can exert a strong dominant negative effect. Moreover, the mutations may differ in how they affect target binding affinity, setting the stage for a complex genotype–phenotype correlation.
Despite this complexity, the emerging picture is an association between clinical presentation and a mutation’s effect on the calcium binding affinity of the calmodulin C-terminal domain. According to a number of reports,10,11 the reduction in calcium affinity correlates with loss of calcium-dependent inactivation of CaV1.2. The effect on RyR2 is more complicated and not directly related to calcium affinity. Thus, in simplified terms, if a mutation has low impact on calcium binding, CaV1.2 dysregulation and LQTS may be avoided. However, this will probably unmask CPVT features from aberrant RyR2 regulation (Take home figure B). Mutations among LQTS patients in ICalmR indeed cluster in C-terminal domain residues directly involved in calcium binding. In contrast, mutations in patients with CPVT occur in both N- and C-terminal domain residues that do not bind calcium (Take home figure B). However, neither calcium nor target binding impact of the mutation can fully explain the phenotypic outcome. At least 10 patients in ICalmR carry the p.Asn98Ser mutation, which induces a moderate ∼4-fold reduction in C-terminal domain calcium affinity. Yet this group represents the entire phenotypic spectrum, including LQTS, CPVT, IVF, and overlapping features of LQTS/CPVT (Take home figure A), so environmental factors or modifier genes must also have an influence.
Curiously, the first missense mutation identified in calmodulin (p.Asn54Ile), in the large Swedish family with CPVT, had an almost undetectable impact on calcium binding.3 It is now clear that this feature is exactly why the mutation could segregate across four generations. Related individuals carrying the same inherited variant are now part of ICalmR, offering a chance to identify potential modifier genes.
Importantly, ICalmR underscores that current treatments for calmodulinopathies are inadequate. Possibly because of calmodulin’s role as a calcium signalling hub for a number of important cardiac targets,12 the full mutation impact is complex and more difficult to treat than with ‘classical’ arrhythmia genes. A potentially promising option is a CRISPR- (clustered regularly interspaced short palindromic repeats) based strategy to remove the mutated allele, as shown for a severe LQTS CALM2 mutation (p.Asp130Gly).13
Overall, the establishment of ICalmR offers an unprecedented overview of the remarkable spectrum of phenotypes associated with calmodulin mutations. It also highlights the serious inadequacies of current treatments and the need for new approaches. We undoubtedly are only at the beginning of unravelling calmodulinopathy prevalence and mechanisms.
Conflict of interest: M.T.O. reports grants from The Lundbeck Foundation, The Novo Nordisk Foundation, and the Danish Council for Independent Research, Natural Sciences, during the conduct of the study. M.N. has no conflicts to declare.
Footnotes
† doi:10.1093/eurheartj/ehz311.
The opinions expressed in this article are not necessarily those of the Editors of the European Heart Journal or of the European Society of Cardiology.
References
- 1. Rhoads FF, Allen R.. Evolutionary aspects of calmodulin. IUBMB Life 2001;51:215–221. [DOI] [PubMed] [Google Scholar]
- 2. Nyegaard M, Overgaard MT, Søndergaard MT, Vranas M, Behr ER, Hildebrandt LL, Lund J, Hedley PL, Camm AJ, Wettrell G, Fosdal I, Christiansen M, Børglum AD.. Mutations in calmodulin cause ventricular tachycardia and sudden cardiac death. Am J Hum Genet 2012;91:703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Søndergaard MT, Sorensen AB, Skov LL, Kjaer-Sorensen K, Bauer MC, Nyegaard M, Linse S, Oxvig C, Overgaard MT.. Calmodulin mutations causing catecholaminergic polymorphic ventricular tachycardia confer opposing functional and biophysical molecular changes. FEBS J 2015;282:803–816. [DOI] [PubMed] [Google Scholar]
- 4. Crotti L, Johnson CN, Graf E, De Ferrari GM, Cuneo BF, Ovadia M, Papagiannis J, Feldkamp MD, Rathi SG, Kunic JD, Pedrazzini M, Wieland T, Lichtner P, Beckmann B-M, Clark T, Shaffer C, Benson DW, Kääb S, Meitinger T, Strom TM, Chazin WJ, Schwartz PJ, George AL.. Calmodulin mutations associated with recurrent cardiac arrest in infants. Circulation 2013;127:1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reed GJ, Boczek NJ, Etheridge S, Ackerman MJ.. CALM3 mutation associated with long QT syndrome. Heart Rhythm 2015;12:419–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Marsman RF, Barc J, Beekman L, Alders M, Dooijes D, Wijngaard A van den, Ratbi I, Sefiani A, Bhuiyan ZA, Wilde AAM, Bezzina CR.. A mutation in CALM1 encoding calmodulin in familial idiopathic ventricular fibrillation in childhood and adolescence. J Am Coll Cardiol 2014;63:259–266. [DOI] [PubMed] [Google Scholar]
- 7. George AL. Calmodulinopathy: a genetic trilogy. Heart Rhythm 2015;12:423–424. [DOI] [PubMed] [Google Scholar]
- 8. Crotti L, Spazzolini C, Tester DJ, Ghidoni A, Baruteau A-E, Beckmann B-M, Behr ER, Bennett JS, Bezzina CR, Bhuiyan ZA, Celiker A, Cerrone M, Dagradi F, De Ferrari GM, Etheridge SP, Fatah M, Garcia-Pavia P, Al-Ghamdi S, Hamilton RM, Al-Hassnan ZN, Horie M, Jimenez-Jaimez J, Kanter RJ, Kaski JP, Kotta M-C, Lahrouchi N, Makita N, Norrish G, Odland HH, Ohno S, Papagiannis J, Parati G, Sekarski N, Tveten K, Vatta M, Webster G, Wilde AAM, Wojciak J, George AL, Ackerman MJ, Schwartz PJ.. Calmodulin mutations and life-threatening cardiac arrhythmias: insights from the International Calmodulinopathy Registry. Eur Heart J 2019;40:2964–2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wright CF, West B, Tuke M, Jones SE, Patel K, Laver TW, Beaumont RN, Tyrrell J, Wood AR, Frayling TM, Hattersley AT, Weedon MN.. Assessing the pathogenicity, penetrance, and expressivity of putative disease-causing variants in a population setting. Am J Hum Genet 2019;104:275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jensen HH, Brohus M, Nyegaard M, Overgaard MT.. Human calmodulin mutations. Front Mol Neurosci 2018;11:396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kotta M-C, Sala L, Ghidoni A, Badone B, Ronchi C, Parati G, Zaza A, Crotti L.. Calmodulinopathy: a novel, life-threatening clinical entity affecting the young. Front Cardiovasc Med 2018;5:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sorensen AB, Søndergaard MT, Overgaard MT.. Calmodulin in a heartbeat. FEBS J 2013;280:5511–5532. [DOI] [PubMed] [Google Scholar]
- 13. Limpitikul WB, Dick IE, Tester DJ, Boczek NJ, Limphong P, Yang W, Choi MH, Babich J, DiSilvestre D, Kanter RJ, Tomaselli GF, Ackerman MJ, Yue DT.. A precision medicine approach to the rescue of function on malignant calmodulinopathic long-QT syndrome. Circ Res 2017;120:39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]