ABSTRACT
Based on modern microbiology, we propose a major revision in current space exploration philosophy and planetary protection policy, especially regarding microorganisms in space. Mainly, microbial introduction should not be considered accidental but inevitable. We hypothesize the near impossibility of exploring new planets without carrying and/or delivering any microbial travelers. In addition, although we highlight the importance of controlling and tracking such contaminations—to explore the existence of extraterrestrial microorganisms—we also believe that we must discuss the role of microbes as primary colonists and assets, rather than serendipitous accidents, for future plans of extraterrestrial colonization. This paradigm shift stems partly from the overwhelming evidence of microorganisms’ diverse roles in sustaining life on Earth, such as symbioses and ecosystem services (decomposition, atmosphere effects, nitrogen fixation, etc.). Therefore, we propose a framework for new discussion based on the scientific implications of future colonization and terraforming: (i) focus on methods to track and avoid accidental delivery of Earth's harmful microorganisms and genes to extraterrestrial areas; (ii) begin a rigorous program to develop and explore ‘Proactive Inoculation Protocols’. We outline a rationale and solicit feedback to drive a public and private research agenda that optimizes diverse organisms for potential space colonization.
Keywords: microorganisms, solar system, colonization, Mars, planetary protection policy
Discourse for scientists and space administrators on how and why to elevate microorganisms as essential components of future colonization strategies of our solar system.
INTRODUCTION—HISTORY AND PAST MICROBIAL POLICY IN SPACE
It should still be wondrous to realize that people took to the air only slightly more than a century ago with the Wright brothers and Santos-Dumont's inventive tenacity culminating in early 1900s. Soon after airplanes took off, space flight advanced to leave Earth's orbit, carrying people (and likely other organisms) beyond our gaseous atmosphere. Tsiolkovsky and science fiction writers from Jules Verne forward have notably dreamed of planetary escape and the expansion of Homo sapiens to outer space (Grant 2017). The general and early concepts of ‘extraterrestrial’ journeys tended toward the sensational and glamorous. In reality, of course, we must consider the untidy details—where will the liquid water and breathable air come from, what are the energy sources, how will waste be recycled, etc.
Since those halcyon days, humanity's quest to explore and study space has been relentless, albeit with waxing and waning moments based on fluctuating national budgets and resolve. Many know of the US Apollo and Space Shuttle programs, but in recent years, attention on space travel and study has again skyrocketed. Examples include plans for increased militarization of space, a new Martian rover and a Chinese landing on the far side of the moon (Castelvecchi 2019; Shammas and Holen 2019). Indeed private enterprises intending to ferry the public into space (e.g. SpaceX, Blue Origin, Virgin Galactic) not only yield infusions of new nongovernment funding but also introduce the ambition of eventual colonization of the solar system (Lee 2019). These activities highlight the need for a broader discussion and new policy for the roles of microbes in space.
International space microbial policy started with the 1967 United Nations Outer Space Treaty (OST), and especially Article IX, which has this statement (Rummel and Billings 2004)—‘States shall pursue studies of outer space, including the moon and other celestial bodies, and conduct exploration of them so as to avoid their harmful contamination…’ This mandate is further elaborated upon in the Committee for Space Research's (COSPAR) Planetary Protection Guidelines (http://w.astro.berkeley.edu/∼kalas/ethics/documents/environment/COSPAR%20Planetary%20Protection%20Policy.pdf; COSPAR 2003). A major tenet of these guidelines includes,
'COSPAR maintains and promulgates planetary protection policy for the reference of spacefaring nations, both as an international standard on procedures to avoid organic-constituent and biological contamination in space exploration, and to provide accepted guidelines in this area to guide compliance with the wording of this UN Space Treaty and other relevant international agreements.'
These conservative guidelines comprise a well-intentioned set of actions meant to avoid unintentional contamination of extraterrestrial habitats during the exploratory phase of the solar system with earthly organisms (e.g. microbes). This planetary protection policy was a noble and logical tact, since space exploration in the last century was pushing quickly for new boundaries of the unknown, and discovery of a new life form would probably be very different from that of Earth (Kminek and Rummel 2015). Humanity had to verify that no extraterrestrial life existed prior to human contact. Current COSPAR policy remains fairly consistent with these early tenets, and wisely states that policy should enable exploration and use of the solar system, not prohibit it (https://cosparhq.cnes.fr/scientific-structure/ppp).
A basic tenet of most space exploration has been to sterilize all spacecrafts in order to avoid potential contamination of space from Earth. A relevant incident occurred as a public faux pas in July 2017. The US Vice President was touring the Kennedy Space Center, and passed some equipment destined for space travel. A sign said ‘Critical space flight hardware—Do not touch’, but he accidentally did just that while on television. This story links to our essay's primary theme and the tacit message of the signage: ‘Do not touch to avoid contamination’. To the contrary, we suggest that it is now time to rethink this ‘contamination’ policy, including plans and protocols to track accidental introduction and, in parallel, developing a protocol for 'controlled colonizing' of another planetary body, if this is decided to be humanity's long-term goal. Our message in this paper intends to convince that substituting the more forward-looking term microbial ‘introductions or release’ into space would be more realistic than using the negative term ‘contamination’. Also, the current planetary protection policy is not consistent with future plans to ultimately colonize space.
The definition of ‘sterilize’ in a biological sense means the complete elimination of all living organisms, including microbes, their vegetative spores and viruses. Sterilization has merit in limiting what space exploration may deliver to an extraterrestrial site. However, we posit major flaws in this initial approach that requires corrections and adjustments. First, obsessing about microbes in space is not practical because they are essential components of life with a majority of microbes being beneficial and nonpathogenic (Gilbert and Knight 2017). Second, it seems unnecessary, costly and futile to strive for complete sterility of every nook and cranny on all space vessels for every mission. Humans have created a ‘built environment’ which teems with microbes (Lax, Nagler and Gilbert 2015; Blaustein et al. 2019). Microbes live everywhere from the lithosphere to dust particles in the stratosphere (Smith et al. 2011). Third, assuming that launching fully sterile space vessels were possible, we still could not sterilize the human crew with their own associated microbial communities or microbiomes. Moreover, humans and most living eukaryotes should be viewed as ‘metaorganisms’ i.e. composed of the host and its associated microbiome (Bosch and McFall-Ngai 2011). We still need to better understand the diversity of microbes and niche occupancy by microbes living inside and on our bodies. Some bacteria identified by culture-independent methods from the surface of prosthetic hip joints were identified as iron- oxidizing lithotroph ES-1 and hydrothermal vent eubacterium (Dempsey et al.2007).
Important issues that must be increasingly studied and used to establish new hypotheses are ‘How can our bacteria survive in unfamiliar space environments?’ and ‘How can we track accidental microbial introductions into space environments?’ Previous research has focused primarily on extremophiles and spore formers. Another important landmark issue is the very definition of what is considered extreme, as we are now expanding our knowledge about niche occupation. For example, the subsurface may support up to 23 billion tons of carbon, living biomass, which is four times higher than previous estimates (Magnabosco et al. 2018). A fundamental unanswered question for the robotic and human exploration of Mars is whether terrestrial microorganisms can adapt, possess active metabolism and replication capacity, at atmospheric pressure of ∼0.7 kPa at the Martian surface.
Different microorganisms, mainly extremophiles and spore formers, were recovered from spacecraft surfaces and processing facilities prior to the launching of Mars spacecraft (Ghosh et al. 2010; Venkateswaran, La Duc and Horneck 2014), and the survival of some of these species like Deinococcus spp. during the interplanetary transit between Earth and Mars has been shown to be quite likely (Paulino-Lima et al. 2011; Cheptsov et al. 2017). However, when landing on Mars, at least 17 or more separate biocidal factors are likely to be present on the Martian surface (Schuerger et al. 2013). At present, it is still unclear how many terrestrial microorganisms can overcome these potentially biocidal factors and interactions in order to develop replication capacity on Mars.
Alternative policy and science
A practical consideration for jettisoning strict no-contamination guidelines for our immediate solar system is cost. Protocols to keep every component of space hardware free of microbial contamination are wasteful and add extra layers of regulation, personnel expertise and time (Moissl-Eichinger, Cockell and Rettberg 2016; Moissl-Eichenger 2012; Rummel and Conley 2017). The potential introduction by the crew's microbiomes remains a concern that has only been recently addressed (Coil et al. 2016; Lang et al. 2017). The microbial communities that live in and within human bodies—the human microbiome— are often inseparable from the host and thus contain unique fingerprints that underpin identity and reveal the microbiome of each individual (Franzosa et al. 2015; Lloyd-Price et al. 2017). It is important to highlight that it is not yet possible to fully determine how a dysbiotic microbial community operates and that the definitions for beneficial and harmful microorganisms are still unclear and under debate. However, in the future, with ever-increasing developments in DNA, RNA and protein identification and sequencing techniques, as well as with more polyphasic approaches being developed to explore this topic, we may have sufficient capability to predict changes that indicate dysbiosis in humans, including, of course, astronauts (Gilbert and Knight 2017; Wilson 2019). It can be anticipated that savings from dropping the strict ‘no microbe’ mandate and focusing on removing only known harmful microorganisms (or their genes) could be estimated to save space projects millions of dollars.
Moreover, there has never been any rigorous follow up to determining microbial survival at the extraterrestrial sites already explored. Viable bacteria and fungi have been found on dust particles in our upper atmosphere and the International Space Station (ISS) (Smith et al. 2011), and these microbes could have already been accidentally delivered to extraterrestrial sites. Current efforts focus on characterizing microbes in the nonextreme conditions of vehicle interiors, such as the ISS (Coil et al. 2016; Lang et al. 2017), where most of the tested bacteria appeared unaffected by the space station conditions (Coil et al. 2016). In addition, the ISS was described having a diverse microbial community, including Archaea representatives, associated with the evaluated space station, which was more closely related to home surfaces on Earth than the human microbiome (Lang et al. 2017). Modern molecular forensics methods can be brought to bear for better identification and tracking of human-associated microbiomes in the built environment (Lax, Nagler and Gilbert 2015). However, these characterizations do not have space colonization in mind, and are mostly post hoc to actual launches, and yet are key tools for tracking of microbial introductions.
Instead of the old COSPAR policy (COSPAR 2003), we advocate and elaborate below a serious re-evaluation away from excessive obsession with total microbial sterilization to a more deliberate consideration of microbiological concepts and procedures, including targeted identification and prohibition of known pathogenic features to be transported/delivered. Recent articles by Fairen et al. (2018, 2019) parallel some of the ideas we advocate here, such as calls for a loosening of COSPAR policy, and the futility of ‘sterilization’ of spacecraft. They also correctly ascertain in their section 1.4 that ‘treating every bacterial species as a potential growing pathogen for Mars…. is a flawed approach’.
However, our current proposal will go several steps further, and differs by a stronger emphasis on realizing the inevitable microorganisms as pioneers or at least as ‘co-colonizers’.
DIFFERENTIATING SPACE EXPLORATION FROM COLONIZATION
One problem with the ‘no contamination’ policy is that a tacit dichotomy between exploration and colonization has not been acknowledged. However, we believe it is time to clearly differentiate society's intention to either (i) continue exploration or (ii) commit to extraterrestrial colonization (while still exploring). In the present context, we assert that humanity in genere stands at a precipice where people can rationally debate whether the initial ‘exploration’ phase of space travel in this solar system will soon culminate, and can be superseded by deliberate efforts at extraterrestrial colonization with its full implications. This rubicon should be clearly demarcated at both societal and scientific levels. For example, the lack of any discovery or evidence of life from any of the past 70+ space missions and probes which have left Earth's orbit points to only one unique presence of life in our immediate solar system.
The US National Aeronautic and Space Agency (NASA) seemed to follow this idea by announcing concrete plans to colonize our nearest, most hospitable planetary neighbor, Mars (NASA 2015). If this new strategy is to be seriously pursued, then the concept of ‘terraforming’ (e.g. transforming (a planet) so as to resemble Earth, especially so that it can support human/Earth-based life) should likewise be extensively discussed in the future. To date, only a few scholarly works have regarded this activity with scientific rigor (Moissl-Eichinger, Cockell and Rettberg 2016). This exercise will involve the identification and deliberate or random introduction of beneficial microbes, desirably preceded by simulations and tests in micro- and mesocosms on Earth.
Our alternative perspective to current space science policy can be considered a paradigm shift. It recommends a focus on microorganisms, which should actually represent the first prerequisite wave of earthly pioneers for any successful colonization of the solar system by humans from Earth. This idea acknowledges current policy for exploration, but challenges it when colonization becomes the primary goal on the horizon. This view also stems from our improved understanding of microbiology and corresponding ecosystem services, and a scientific consensus that symbiotic microbes comprise essential elements for life on Earth (McFall-Ngai et al. 2013; Thompson et al. 2017). Assuming that a colonization plan aims for eventual permanence, the first colonists should consist of microbial species, not human, paralleling what likely happened on primordial Earth. The paradigm shift we now advocate is that a deliberate seeding of microbes would ultimately promote colonization goals—e.g terraforming. This will not be easy by any means, and many hurdles to successful terraforming exist (Lage et al. 2012; Jakosky and Edwards 2018). Yet, to date, there has been insufficient incorporation of microbiology principles in previous plans, or a lack of coordinated networking, which could possibly pave the way for the subsequent successful colonization of macroorganisms.
We make this provocative paradigm shift suggestion based on a foundation of microbial ecology, evolution and planetary science. Biologists understand that there can be no life on Earth without the ecosystem services of various microbes (bacteria, Archaea, some fungi, algae, protozoans) (McFall-Ngai et al. 2013; Stolz 2017). The first life forms and ‘colonists’ of terrestrial Earth were not amphibians, or even plants but rather single-celled microorganisms (Pikuta, Hoover and Tang 2007). Microbial ancestors conditioned ancient Earth atmosphere billions of years ago, adding more oxygen via photosynthesis (De Marais 2000). If humanity is seriously contemplating colonizing Mars, another planet or one of the nearby moons in the future, then people need to identify, understand and send the most competitive and beneficial pioneers. Choosing or developing the most durable microbial taxa or communities may be done with deliberation, systematic research and current data, rather than sending random bacteria serendipitously hitchhiking on space stations (Coil et al. 2016; Lang et al. 2017).
Moreover, there is an operational understanding that referring to the term symbiotic also means beneficial. Most microbial activity and associations can be viewed more as a continuum of beneficial (mutualistic), neutral or harmful effects for the host (Bjork et al. 2018), depending on host and environmental factors. Symbiology in itself is a rapidly growing field, which spans almost every organismal system and habitat (Bosch and McFall-Ngai 2011). The beneficial products that some microbial symbionts provide holobionts (e.g. hosts + microbes) include nitrogen fixation for plants, defensive natural products, competitive exclusion to foreign and potentially pathogenic invaders, probiotic mechanisms, as well as commensal interactions to other microbiome members (McFall-Ngai et al. 2013; Peixoto et al. 2017; Lopez 2019). For nonsymbionts, free living microbes beneficially condition the atmosphere here on Earth by affecting levels of CO2, oxygen, methane and nitrogen (Stolz 2017).
In this regard, a clear definition of ‘beneficial’ should also include characterizing the genotypes and phenotypes to be promoted, in the context of expected extraterrestrial environmental conditions. The current environmental conditions and stability of Earth, including its habitability, stem from complex interactions between biosphere, atmosphere, hydrosphere and lithosphere components. These generated a unique chemical composition, constantly supported and driven by a Gaia hypothesis, and by extension, its diverse microbiomes (Stolz 2017; Thompson et al. 2017). The knowledge and manipulation of specific Gaian microbiome capacities could lead to potential beneficial mechanisms or candidate taxa for testing, in order to explore and reproduce our unique environmental conditions on extraterrestrial areas. Such studies can also support the improvement of Earth's stability in the light of global changes. Several experimental efforts such as EXPOSE and space exposure biology have been applied but need further expansion and support (Schulte et al. 2006; Rabbow et al. 2015). (As a footnote, the authors do not actually endorse expensive interplanetary colonization [at this time], but would rather see scarce resources go more towards better conserving and characterizing the still relatively unknown biological diversity that is increasingly threatened on our own planet. For example the Earth BioGenome Project fits in this mold (Lewin et al.2018).
One can rightly argue that microbes released on Mars will represent invasive species that are being introduced into an unexplored and possibly pristine ecosystem (Rummel and Conley 2017). How should we control releases, or protect any unique system from harm? Of course, these are issues that will require extensive policy debate and scientific experimentation on Earth before actual extraterrestrial terraforming. Yet colonization efforts should eventually integrate not only the latest technologies but also basic principles such as ecological succession and exotic invasions (Simberloff and Von Holle 1999). Examples of uncontrolled spreading of fecund organisms into new habitats are abound, but we have no room to fully discuss them here (Albins and Hixon 2013; Hess-Erga et al. 2019). As in any colonization scenario, the most extreme conditions must be achieved by confronting and harnessing dangerous, unknown habitats, which can possibly be transformed for the survival of Earth-sourced organisms. Microbes can carry out these essential large-scale transformations of an environment (not instantaneously), which is another reason to keep them in the forefront. We propose future research platforms which would allow microbes to compete. However, tracking microbes with current methods (microscopy or even gene probes) remains very difficult. Nor do we advocate rushing microbial introductions without thorough research on Earth. Instead, we envision a deliberate and measured program of research into microbial colonization, realizing the limits of current technologies (https://mars.nasa.gov/news/8358/mars-terraforming-not-possible-using-present-day-technology/). Thus, we advocate a conservative schedule of microbial introductions into space, while also realizing that human colonization cannot be separate from microbial introductions.
On the other the hand, the benefits of rapid growth of the introduced species can represent a desired goal and hallmark of successful ecological colonization in a novel extraterrestrial context. Moreover, molecular genetics, phylogenetics and multidisciplinary methods have advanced far in the last five decades to include high throughput DNA sequencing, MALDI-TOF (matrix-assisted laser desorption ionization–time of flight) mass spectrophotometry and other types of diagnostics. Hence, humans now possess a strong capacity to classify and differentiate different life forms after colonization. Therefore, we could safely assume that any accidental co-mingling of Earth life is unlikely (e.g. no hybridization) and could also be distinguished from any extraterrestrial life form (NASEM 2018). Recent scientific advances in the culture-free genomic diagnostics and identifications would allow the distinction between earthly and extraterrestrial life forms, assuming that there was the possibility that a chance encounter could lead to exchanging genetic information.
On another level, there are rapid developments in microbiology, robotics, astrobiology and artificial intelligence that will enable future, more precise ‘within-field’ type of terraforming, in which space agencies and scientists will be able to act as space farmers and regularly monitor the modifications needed to have a more ‘friendly’ environment to a possible Mars colonization by humans. Whereas this development will someday allow manipulating microbial communities by this ‘Interplanetary Microbiome Engineering (IME)’, inclusion of quality parameters will be a necessary next step.
To foster the quality of Mars soils at lower scale, the concept of ‘smart’ or ‘precision’ farming that has been recently introduced to terrestrial agriculture could be used as a first step. Smart farming proposes the use of advanced interdisciplinary methods to assess and foster soil quality at fine levels, in order to improve agricultural production within a field (Wolfert et al. 2017). Its central premises are targeted and site-specific interventions, with on-the-spot highly automated (robots and drones) agents that monitor crops—via advanced imaging techniques—at individual plant level and intervene at this level in case of possible problems in the crop. Speculating on interplanetary missions, the observational agents (rovers and sensors) could yield massive data that will be provided to machine and deep learning algorithms, so as to provide robust algorithms that direct on-planet management. Unfortunately, the knowledge and technology necessary for robotics-driven management, microbiome manipulation and transplantation, features and interactions among microbes and modeling the effects of evolutionary forces on introduced terrestrial microbes on Mars are still in their infancy.
Choosing microorganisms for an extraterrestrial journey and their role in ecosystems
If we assume humanity intends to eventually colonize parts of our solar system, then the ‘contamination’ of these new areas with terrestrial microorganisms, by our expeditions, will also be inevitable and possibly desirable. In this context, we should begin to systematically discard ‘contamination’ terminology and instead determine the criteria for ‘selecting’ which microbes to be introduced as pioneering colonists on a Martian or extraterrestrial landscape. Recent microbiology research has provided insights to determine the correct criteria. An important primary need is the generation of habitable atmosphere with decreased CO2 and more oxygen, which some microbes can produce. Thereafter, another benefit would be to support growth of sustainable food supplies through symbiosis—e.g. nitrogen or carbon fixation to generate organic materials—and other ‘agriculture-beneficial’ mechanisms, to be further explored.
Beneficial mechanisms and the efficiency of the use of probiotics or environmental probiotics have been well described and/or proposed for different organisms, such as plants, humans (Lax, Nagler and Gilbert 2015), fishes (Dawooda and Koshio 2016) and corals (Peixoto et al. 2017). What microbes accomplish on Earth can benefit human colonization of Mars or other planets.
Extremophiles
An extremophile is an organism that is tolerant or even dependent and thrives in environmental extremes and that has evolved to grow optimally under one or more of these extreme conditions. Earth's habitat was likely very inhospitable more than 4 billion years ago, but microbial life arose and evolved over time. The first microbial colonists of extraplanetary bodies will likely derive from extremophiles.
Many studies, including Horneck et al. (2010; 2012), have characterized the conditions that earthly microbes could encounter in space, starting at low Earth orbit. The extreme conditions include increased UVA/UVB exposure, dessication, low pressure and freezing temperatures and most water on present-day Mars is frozen in the regolith. The surface environment of Mars is composed of 95% carbon dioxide: 2.7% nitrogen and only 0.13% oxygen, and has an average temperature of −63°C (−81°F) with a maximum temperature of 20°C (68°F) and a minimum of −140°C (−220°F) measured at Viking landers’ sites. Thus, on Mars, the surface can be a very hostile environment and the subsurface can be a good choice for seeding the microbiome (Nimmo and Tanaka, 2005). We can get an idea of what can happen by using the subsurface of the Earth as an example since a great number of the bacteria and archaea on Earth are found in subsurface environments. Actually, a recent updating on subsurface cellular estimates that the total global prokaryotic biomass is ∼23 to 31 Pg of carbon C (PgC), roughly 4 to 10 times less than previous estimates (Magnabosco et al. 2018). Microbial cells in these stable and oligotrophic settings catabolize 104- to 106-fold more slowly than model organisms and subsist with energy fluxes that are 1000-fold lower than the typical culture-based estimates of maintenance requirements (Hoehler and Jørgensen 2013). Thus, subsurface microbes could be candidates for terraforming.
For example, to handle the anoxic conditions, perhaps microbes related to lithotrophic hypersaline anaerobes should be proposed for testing (Pikuta et al. 2017). Microbes (fungi and pigmented bacteria) have been found and cultured from as high as 77 km and below the frozen ice sheets of Antarctica. The possibility of liquid water exists but only with high salt content lowering freezing points or if subsurface geothermal warming existed. If the coldest conditions prevail, they will likely not welcome even the hardiest of Earth's known organisms. The second most hospitable body in our solar system, the sixth-largest moon of Jupiter, Europa, appears to have oxygen within its atmosphere (Hall et al. 1995). Europa's water remains mostly frozen due to its distance from the Sun, but underneath the surface could be thawed by superheated plumes. And water is actually not an uncommon molecule in space, so conditions exist for survival, which are consistent with the current proposal (Mora et al. 2016). Indeed, Hand and German (2018) argue for more exploration of extraterrestrial oceans, which may be a platform for life.
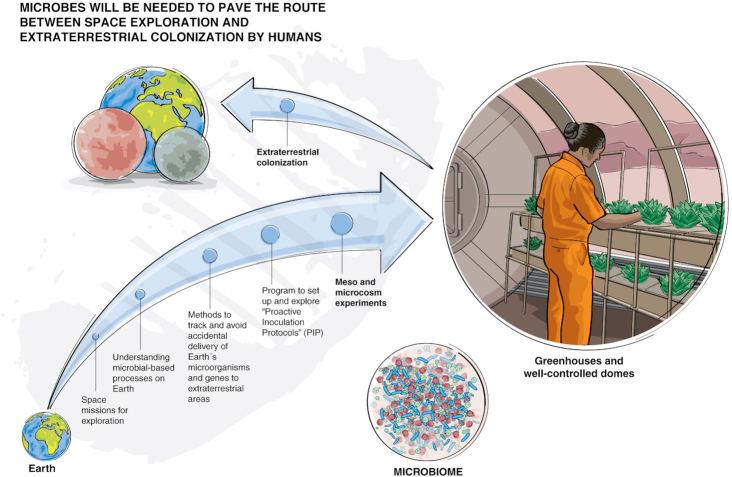
Selecting microbes for potential space travel should be evaluated scientifically and objectively with the latest technologies, perhaps with consensus through a proposed program we tentatively entitle as ‘Proactive Inoculation Protocols (PIPs)’. This approach promotes both (i) a lowering of contamination alarm levels; (ii) tracking accidental contamination; (iii) systemically studying, choosing or engineering types of beneficial microbes that could pre- or co-colonize a new extraterrestrial site (e.g. Mars), and/or support the development of life in the new sites (i.e. plant growth promoting rhizobacteria [Kloepper and Schroth 1978] to support plant development; selected microorganisms to provide a healthier environment according to the microbiome of the built environment [NASEM 2017] future discoveries and directions, etc.). Alternatively, astro-microbiology focuses on whether Earth bacteria may survive unique and extreme space conditions (Zea et al. 2016). Many have speculated on potential Human Assisted Panspermia (HPA) parameters, and the factors that may enable the viable transportation of microorganisms from space to Earth, and vice versa. For example, Mileikowsky et al. (2000) list hurdles to space transport as 'microbe survival in a vacuum; central meteorite temperatures at launch, orbiting, and arrival; pressure and acceleration at launch; spontaneous DNA decay; metal ion migration, etc.' Moissl-Eichinger, Cockell and Rettberg (2016) has provided an excellent review of microbial studies for space but these studies represent only a beginning, and well-funded PIP would advance the field. One underlying thesis of this paper is to acknowledge the stochastic nature of microbial evolution. We cannot fully control for all aspects of microbial introduction into space. The ideal situation will be the formation of a ‘more hospitable’ environment to facilitate the colonization of other planetary bodies. It should be noted that even if this process takes a long time (and perhaps may not ever be reached), there are currently several projects and plans of the different space agencies for greenhouses and more controlled sites (such as Biosphere 2 and Domes—see Fig. 1), which seem to be more feasible in a relatively imminent future than the process of planetary transformation. Still, such initiatives will need to rely heavily on microorganisms if they are to succeed.
Figure 1.
Potential trajectory for how terraforming, PIPs and other related microbiologically focused methods can be applied in a concerted effort to colonize the solar system. A long period of rigorous study and experimentation on Earth prior to extraterrestrial releases is thus expected.
For the immediate future, PIP would provide systematic and controlled approaches to microbial pioneering. The serendipitous survival on vehicles such as the ISS (Coil et al. 2016) could be related with the microbial genomic capacity to adapt and colonize extreme conditions. In this regard, it is important to highlight that each and all attempts to colonize extraterrestrial areas including the use of Gaia's microbiome as first colonizers should be first evaluated in well-controlled experiments on micro- and mesocosms. In addition, of course we would want to leave out deadly and harmful known pathogens from any colonizing vehicle. But is this realistic? It is important to consider our inability to unequivocally predict whether some of colonizers can become harmful in a different environment, or if and how some yet undiscovered extraterrestrial life form could be affected. Many of our most common pathogens (hemorrhagic Escherichia coli) obtain their pathogenicity islands through lateral gene transfer. This is almost impossible to prevent or predict. Perhaps microbes slated for new space colonies could be genetically screened. Using modern bioinformatics methods, genes known to code for pathogenic phenotypes (toxins, adhesins, antibiotic resistance) could be detected and used to remove them or their bacterial carriers. Candidate microorganisms could also be modified or synthetically engineered, using genomic data from currently known extremophiles, such as Antarctic Chlamydomonas (Szyszka, Ivanov and Hüner 2017) or Carnobacteria(Nicholson et al. 2013).
Our arguments still emphasize the need for balanced discussions, in order to avoid unwanted results (yet to be defined). Concurrent with experimental PIPs, we stress that public ethical and philosophical discussions should be raised and encouraged so that experimental parameters can be clearly delineated. One model program that the astrobiology community can use to explore the ethical dimensions of extraterrestrial microbial introductions would be molecular biologists’ self-moratorium on recombinant DNA research in the early 1970s or the NIH Human Genome Project ethics panel (Swazey, Sorenson and Wong 1977; Knoppers and Chadwick 2005).
A recent example where cautionary measures were not upheld is with the application of CRISPR. This very effective cutting-edge technology (to edit and change the human genome) appears to be outpacing any ethical, moral and philosophical guidelines. Recently, an apparently isolated, rogue scientist worked in a vacuum, without guidance from the wider community of scientists (Normile 2018; Brokowski and Adli 2019; Daley 2019). We realize that either accidental or deliberate inoculation of planetary bodies is a nontrivial, sensitive issue. Yet in the same context, few policy makers, scientists and entrepreneurs have raised similar cautionary alarms and standards pertaining to populating space with humans (and their many inseparable microbial symbionts) in the future (Fairen and Schulze-Makuch 2013; Fairen et al. 2018; Schwendner et al. 2017). The stated aims of commercial enterprises and NASA are to eventually colonize Mars or other bodies that could support human existence, preventing our extinction. Thus, our proposal is consistent with current aspirations. Yet we do aim to avoid previous technological missteps by making any future microbial inoculation protocols safe, transparent, and based on the latest symbiosis concepts and peer-reviewed microbiological methods.
For future space expeditions, we may one day allow natural selection to act upon microbes that fortuitously or deliberately travel on the landers and probes. As they did during ancient Earth evolution, a lucky microbe may find a way to adapt to the extremes and then pave the way for future colonists. Although a heretically sounding idea on first mention, openly discussing the pros and cons of deliberately sending a community of hand-picked microbial taxa to specific (non-APEX [Astrobiology Priority Exploration]) sites, and monitoring their survival, as starter colonies at controlled extraterrestrial mesocosms could be beneficial to a long-term colonization program.
A caveat in PIPs is that total control or a full inventory of microbial taxa and their genomes sent into space can never be realistically achieved. Also, retrieval of microbes once sent may be impossible, and stochastic processes will be similarly in play. In parallel, controlled micro- and mesocosm experiments, selecting and inoculating beneficial microbiomes in the ISS or simulated extraterrestrial conditions (such as rejuvenated ‘Biosphere 2’ experiments) should be encouraged and supported (Lage et al. 2012; Brandt et al. 2015; Schwendner et al. 2017), in order to understand, develop and predict transformations prompted by manipulation and potential colonization. Such approaches must also indicate the most competitive strains to be tested. Eventually, this can lead to the most practical and optimized microbial inoculum to be sent for extraterrestrial colonization long before humans intend to colonize.
Regarding IME, microbial consortia can harbor complex microbial communities, which can potentially serve as models for studies of microbial ecology and biotechnological processes in mixed culture, as well as systems biology. Integrated molecular analyses (metagenomics, metatranscriptomics, metaproteomics and metabolomics) are gaining more importance as they can provide a better understanding of the structure, functioning and dynamics of the in situ community, as well as offer the potential to discover new biological functionalities within the scope of (eco)systems biology (Morrison et al. 2019). The integration of information from the genome to the metabolome allows the establishment of associations between the genetic potential and the final phenotype. This information is only obtained through an integrated and systemic approach, and not when only one of these tools is used on an individual basis. According to Narayanasamy et al. (2015), the systemic approach integrating the different ‘omics’ should be the future standard for large-scale characterization of microbial consortia. The data obtained in an integrated way can allow the ‘deconvolution’ of structure–function relations, identifying the main members and functions. This knowledge can establish the basis for discovering new genes on a much larger scale compared to previous efforts. In a broader sense, the knowledge obtained through systems and synthetic biology disciplines could allow the optimization of microbial processes, either through a better control of the processes in mixed culture, through the use of more efficient enzymes in bioengineering applications, or even genome recoding (Ben Said and Or 2017; Fredens et al. 2019).
With a plethora of modern microbiology aspects still being studied (Thompson et al. 2017; Magnabosco et al. 2018; Wilson 2019), and considering the inevitable transport of microbial organisms in spacecraft, we re-emphasize that a new attitude toward space should be based on evolutionary and microbiological principles and Earth history. Within our solar system we can envision future life stretch as a continuum connecting certain planetary bodies, with microbes doing the work of pioneer habitat conditioning. This allowance will require many systematic and controlled experimental studies with an ethical platform developed on Earth, prior to releases. Since we know that at least a billion years may have been needed for primordial life to arise on this planet, we should consider the best method to plant seeds, initiate systematic and scientifically based PIPs or some derivation, and find ways to manipulate beneficial microorganisms. These could advance natural transformation processes and eventual terraforming, if humanity truly wishes to colonize our solar system.
ACKNOWLEDGEMENTS
We thank Drs Stefan Kautsch, Stephen J. O'Brien, Diego Castano, Robert Smith and Hidetoshi Urakawa for very helpful feedback on early drafts of the manuscript. We also thank the reviewers for their stimulating comments. The original solar system graphic was kindly provided by NASA Jet Propulsion Laboratory Caltech.
Conflict of interests. None declared.
REFERENCES
- Albins MA, Hixon MA. Worst case scenario: potential long-term effects of invasive predatory lionfish (Pteroisvolitans) on Atlantic and Caribbean coral-reef communities. Environ Biol Fishes. 2013;96:1151–7. [Google Scholar]
- Ben Said S, Or D. Synthetic microbial ecology: engineering habitats for modular consortia. Front Microbiol. 2017;8:1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjork JR, O'Hara R, Ribes M et al.. The dynamic core microbiome: structure, dynamics and stability. 2018: Preprint at bioRxiv: 10.1101/137885-2018. [DOI]
- Blaustein RA, McFarland AG, Maamar SB et al.. Pangenomic approach to understanding microbial adaptations within a model built environment, the International Space Station, relative to human hosts and soil. MSystems. 2019;4:e00281–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch TCG, McFall-Ngai MJ. Metaorganisms as the new frontier. Zoology. 2011;114:185–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt A, Vera JP, Onofri S et al.. Viability of the lichen Xanthoriaelegans and its symbionts after 18 months of space exposure and simulated Mars conditions on the ISS. Int J Astrobiol. 2015;14:441–25. [Google Scholar]
- Brokowski C, Adli M. CRISPR ethics: moral considerations for applications of a powerful tool. J Mol Biol. 2019;431:88–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelvecchi D. China becomes first nation to land on the Moon's far side. Nature. 2019;565:146–7. [DOI] [PubMed] [Google Scholar]
- Cheptsov VS, Vorobyova EA, Manucharova NA et al.. 100 kGy gamma-affected microbial communities within the ancient Arctic permafrost under simulated Martian conditions. Extremophiles. 2017;21:1057. [DOI] [PubMed] [Google Scholar]
- Coil DA, Neches RY, Lang JM et al.. Growth of 48 built environment bacterial isolates on board the International Space Station (ISS) PeerJ. 2016;4:e1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COSPAR (Committee on Space Research) Planetary protection policy. COSPAR Inform Bull. 2003;156:67–74. [Google Scholar]
- Daley J. China's ‘CRISPR Babies’ May Be More Likely to Die Young. 2009 https://www.smithsonianmag.com/smart-news/crispr-babies-face-higher-risk-premature-death-180972345/ (9 September 2019, date last accessed). [Google Scholar]
- Dawooda MAD, Koshio S. Recent advances in the role of probiotics and prebiotics in carp aquaculture: areview. Aquaculture. 2016;14:243–51. [Google Scholar]
- De Marais DJ. When did photosynthesis emerge on Earth? Science. 2000;289:1703–5. [PubMed] [Google Scholar]
- Dempsey KE, Riggio MP, Lennon A et al.. Identification of bacteria on the surface of clinically infected and non-infected prosthetic hip joints removed during revision arthroplasties by 16S rRNA gene sequencing and by microbiological culture. Arthritis Res Ther. 2007;9:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairen AG, Parro V, Schulze-Makuch D et al.. Is searching for Martian life a priority for the Mars community? Astrobiology. 2018;18:101–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairen AG, Schulze-Makuch D, Whyte L et al.. Planetary protection and the astrobiological exploration of Mars: proactive steps in moving forward. Adv Space Res. 2019;63:1491–7. [Google Scholar]
- Fairén AG, Schulze-Makuch D. The overprotection of Mars. Nat Geosci. 2013;6:510. [Google Scholar]
- Franzosa EA, Huang KH, Meadow JF et al.. Identifying personal microbiomes using metagenomic codes. Proc Natl Acad Sci USA. 2015;112:E2930–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredens J, Wang K, de la Torre D et al.. Total synthesis of Escherichia coli with a recoded genome. Nature. 2019;569:514–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Osman S, Vaishampayan P et al.. Recurrent isolation of extremotolerant bacteria from the clean room where Phoenix spacecraft components were assembled. Astrobiology. 2010;10:325–35. [DOI] [PubMed] [Google Scholar]
- Gilbert J, Knight R. Dirt is Good. New York: St. Martin's Press, 2017. [Google Scholar]
- Grant RG. Flight: The Complete History of Aviation. New York: DK Publishers, 2017. [Google Scholar]
- Hall DT, Strobel DF, Feldman PD et al.. Detection of an oxygen atmosphere on Jupiter's moon Europa. Nature. 1995;373:677–81. [DOI] [PubMed] [Google Scholar]
- Hand KP, German CR. Exploring ocean worlds on Earth and beyond. Nat Geosci. 2018;11:2–4. [Google Scholar]
- Hess-Erga OK, Moreno-Andrés J, Enger Ø et al.. Microorganisms in ballast water: disinfection, community dynamics, and implications for management. Sci Total Environ. 2019;657:704–16. [DOI] [PubMed] [Google Scholar]
- Hoehler TM, Jørgensen BB. Microbial life under extreme energy limitation. Nat Rev Microbiol. 2013;11:83–94. [DOI] [PubMed] [Google Scholar]
- Horneck G, Klaus DM, Mancinelli RL. Space microbiology. Microbiol Mol Biol Rev. 2010;74:121–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horneck G, Moeller R, Cadet J et al.. Resistance of bacterial endospores to outer space for planetary protection purposes—experiment PROTECT of the EXPOSE-E mission. Astrobiology. 2012;12:445–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakosky BM, Edwards CS. Inventory of CO 2 available for terraforming Mars. Nat Astron. 2018;2:634. [Google Scholar]
- Kloepper JW, Schroth M. Plant growth-promoting rhizobacteria on radishes. In Proceedings of the 4th International Conference on Plant Pathogenic Bacteria, Vol. 2. INRA: Gilbert-Clarey Tours 1978, 879–82. [Google Scholar]
- Kminek G, Rummel JD. COSPAR's planetary protection policy. Space Res Today. 2015;193:7–17. [Google Scholar]
- Knoppers BM, Chadwick R. Human genetic research: emerging trends in ethics. Nat Rev Genet. 2005;6:75. [DOI] [PubMed] [Google Scholar]
- Lage CAS, Dalmaso GZL, Teixeira LCRS et al.. Mini-review: probing the limits of extremophilic life in extraterrestrial environment-simulated experiments. Int J Astrobiol. 2012;11:251–6. [Google Scholar]
- Lang JM, Coil DA, Neches RY et al.. A microbial survey of the International Space Station (ISS). PeerJ. 2017;5:e4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lax S, Nagler CR, Gilbert JA. Our interface with the built environment: immunity and the indoor microbiota. Trends Immunol. 2015;36:121–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. Jeff Bezos Says He Wants to See ‘a Mark Zuckerberg of Space’ and Thinks Blue Origin Will Lead to ‘Dorm-Room’ Entrepreneurship off Earth. http://businessinsider.com/jeff-bezos-blue-origin-space-entreprenuers-mark-zuckerberg-2019-2?smid=nytcore-ios-share (9 September 2019, date last accessed). [Google Scholar]
- Lewin HA, Robinson GE, Kress WJ et al.. Earth BioGenome Project: Sequencing life for the future of life. Proceedings of the National Academy of Sciences. 2018;115:4325–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Price J, Mahurkar A, Rahnavard G et al.. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature. 2017;550:61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez JV. After the taxonomic identification phase: addressing the functions of symbiotic communities within marine invertebrates. In Symbionts of Marine Sponges and Corals. Li Z.(ed). Dordrecht, The Netherlands: Springer, 2019, 105–44. [Google Scholar]
- Magnabosco C, Lin LH, Dong H et al.. The biomass and biodiversity of the continental subsurface. Nat Geosci. 2018;11:707–17. [Google Scholar]
- McFall-Ngai M, Hadfield MG, Bosch TC et al.. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA. 2013;110:3229–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mileikowsky C, Cucinotta FA, Wilson JW et al.. Natural transfer of viable microbes in space: 1. From Mars to Earth and Earth to Mars. Icarus. 2000;145:391–427. [DOI] [PubMed] [Google Scholar]
- Moissl-Eichinger C, Cockell C, Rettberg P. Venturing into new realms? Microorganisms in space. FEMS Microbiol Rev. 2016:40:722–37. [DOI] [PubMed] [Google Scholar]
- Moissl-Eichinger C. Extremophiles in Spacecraft Assembly Clean Rooms. Vienna: Springer, 2012. [Google Scholar]
- Mora M, Perras A, Alekhova TA et al.. Resilient microorganisms in dust samples of the International Space Station—survival of the adaptation specialists. Microbiome. 2016;4:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison MD, Fajardo-Cavazos P, Nicholson WL. Comparison of Bacillus subtilis transcriptome profiles from two separate missions to the International Space Station. NPJ Microgravity. 2019;5:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanasamy S, Muller EEL, Sheik AR et al.. Integrated omics for the identification of key functionalities in biological wastewater treatment microbial communities. Microb Biotechnol. 2015;8:363–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NASA. Journey to Mars. http://go.nasa.gov/1VHDXxg(9 September 2019, date last accessed). [Google Scholar]
- National Academies of Sciences, Engineering, and Medicine (NASEM). Microbiomes of the Built Environment: A Research Agenda for Indoor Microbiology, Human Health, and Buildings. Washington, DC: The National Academies Press, 2017. [PubMed] [Google Scholar]
- National Academies of Sciences, Engineering, and Medicine (NASEM). Review and Assessment of Planetary Protection Policy Development Processes. Washington, DC: The National Academies Press 3, 2018. [Google Scholar]
- Nicholson WL, Krivushin K, Gilichinsky D et al.. Growth of Carnobacterium spp. from permafrost under low pressure, temperature, and anoxic atmosphere has implications for Earth microbes on Mars. Proc Natl Acad Sci USA. 2013;110:666–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo F, Tanaka K. Early crustal evolution of Mars. Annu Rev Earth Planet Sci. 2005;33:133–61. [Google Scholar]
- Normile D. Shock greets claim of CRISPR-edited babies. Science. 2018;362:978–9. [DOI] [PubMed] [Google Scholar]
- Paulino-Lima IG, Janot-Pacheco E, Galante D et al.. Survival of Deinococcus radiodurans against laboratory-simulated solar wind charged particles. Astrobiology. 2011;11:875–82. [DOI] [PubMed] [Google Scholar]
- Peixoto RS, Rosado PM, Leite DC et al.. Beneficial microorganisms for corals (BMC): proposed mechanisms for coral health and resilience. Front Microbiol. 2017;8:341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikuta EV, Hoover RB, Tang J. Microbial extremophiles at the limits of life. Crit Rev Microbiol. 2007;33:183–209. [DOI] [PubMed] [Google Scholar]
- Pikuta EV, Lyu Z, Williams MD et al.. Sanguibacter gelidistatuariae sp. nov., a novel psychrotolerant anaerobe from an ice sculpture in Antarctica, an emendation of descriptions of the family Sanguibacteraceae, the genus Sanguibacter and species S. antarcticus, S inulinus, S. kedieii, S. marinus, S. soli and S. suarezii. International J Syst Evol Microbio. 2017;67:1442–50. [DOI] [PubMed] [Google Scholar]
- Rabbow E, Rettberg P, Barczyk S et al.. The astrobiological mission EXPOSE-R on board of the International Space Station. Int J Astrobiol. 2015;14:3–16. [Google Scholar]
- Rummel JD, Billings L. Issues in planetary protection: policy, protocol and implementation. Space Policy. 2004;20:49–54. [Google Scholar]
- Rummel JD, Conley CA. Four fallacies and an oversight: searching for martian life. Astrobiology. 2017;17:971–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuerger AC, Ulrich R, Berry BJ et al.. Growth of Serratia liquefaciens under 7 mbar, 0°C, and CO2-enriched anoxic atmospheres. Astrobiology. 2013;13:115–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte W, Demetes R, Baglioni P et al.. BIOPAN and ESPOSE: space exposure platforms for exo/astrobiological research in Earth orbit with relevance for Mars exploration. Geophys Res Abs. 2006;8:06643 [Google Scholar]
- Schwendner P, Mahnert A, Koskinen K et al.. Preparing for the crewed Mars journey: microbiota dynamics in the confined Mars500 habitat during simulated Mars flight and landing. Microbiome. 2017:5:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shammas VL, Holen TB. One giant leap for capitalist kind: private enterprise in outer space. Palgrave Commun. 2019;5:1 [Google Scholar]
- Simberloff D, Von Holle B. Positive interactions of nonindigenous species: invasional meltdown? Biol Invasions. 1999;1:21–32. [Google Scholar]
- Smith DJ, Griffin DW, McPeters RD et al.. Microbial survival in the stratosphere and implications for global dispersal. Aerobiologia. 2011;27:319–32. [Google Scholar]
- Stolz JF. Gaia and her microbiome. FEMS Microbiol Ecol. 2017;93:fiw247. [DOI] [PubMed] [Google Scholar]
- Swazey JP, Sorenson JR, Wong CB. Risks and benefits, rights and responsibilities: a history of the recombinant DNA research controversy. S Cal L Rev. 1977;51:1019. [PubMed] [Google Scholar]
- Szyszka B, Ivanov AG, Hüner NP. Adaptation to low temperature in a photoautotrophic antarctic psychrophile, Chlamydomonas sp. UWO 241. In Photosynthesis: Structures, Mechanisms, and Applications. Cham: Springer, 2017, 275–303. [Google Scholar]
- Thompson LR, Sanders JG, McDonald D et al.. A communal catalogue reveals Earth's multiscale microbial diversity. Nature. 2017;551:7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkateswaran K, La Duc MT, Horneck G. Microbial existence in controlled habitats and their resistance to space conditions. Microbes Environ. 2014;29:243–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson N. A microbial hitchhiker's guide to the galaxy: researchers race to understand effects of deep space on microbiome. Bioscience. 2019;69:5–11. [Google Scholar]
- Wolfert S, Ge L, Verdouw C et al.. Big data in smart farming—a review. Agric Syst. 2017;153:69–80. [Google Scholar]
- Zea L, Prasad N, Levy SE et al.. Molecular genetic basis explaining altered bacterial behavior in space. PLoS One. 2016;11:e0164359. [DOI] [PMC free article] [PubMed] [Google Scholar]