Abstract
Background
A better definition of biomarkers and biological processes related to local recurrence and disease progression is highly warranted for ductal breast carcinoma in situ (DCIS). Stromal–epithelial interactions are likely of major importance for the biological, clinical, and pathological distinctions between high- and low-risk DCIS cases.
Methods
Stromal platelet derived growth factor receptor (PDGFR) was immunohistochemically assessed in two DCIS patient cohorts (n = 458 and n = 80). Cox proportional hazards models were used to calculate the hazard ratios of recurrence. The molecular mechanisms regulating stromal PDGFR expression were investigated in experimental in vitro co-culture systems of DCIS cells and fibroblasts and analyzed using immunoblot and quantitative real-time PCR. Knock-out of JAG1 in DCIS cells and NOTCH2 in fibroblasts was obtained through CRISPR/Cas9. Experimental data were validated by mammary fat pad injection of DCIS and DCIS-JAG1 knock-out cells (10 mice per group). All statistical tests were two-sided.
Results
PDGFRα(low)/PDGFRβ(high) fibroblasts were associated with increased risk for recurrence in DCIS (univariate hazard ratio = 1.59, 95% confidence interval [CI] = 1.02 to 2.46; P = .04 Wald test; multivariable hazard ratio = 1.78, 95% CI = 1.07 to 2.97; P = .03). Tissue culture and mouse model studies indicated that this fibroblast phenotype is induced by DCIS cells in a cell contact-dependent manner. Epithelial Jagged1 and fibroblast Notch2 were identified through loss-of-function studies as key juxtacrine signaling components driving the formation of the poor prognosis-associated fibroblast phenotype.
Conclusions
A PDGFRα(low)/PDGFRβ(high) fibroblast subset was identified as a marker for high-risk DCIS. The Jagged-1/Notch2/PDGFR stroma–epithelial pathway was described as a novel signaling mechanism regulating this poor prognosis-associated fibroblast subset. In general terms, the study highlights epithelial–stromal crosstalk in DCIS and contributes to ongoing efforts to define clinically relevant fibroblast subsets and their etiology.
Ductal breast carcinoma in situ (DCIS), a neoplastic proliferation of epithelial cells within the mammary ducts, can progress to invasive ductal carcinoma (IDC) (1,2). The introduction of mammography has led to a four- to sevenfold increase in the detection of DCIS (3,4). After breast conserving surgery, most patients receive additional radiotherapy and or hormonal treatment (1,3,5). The majority of patients would not recur, even without radiotherapy (6). Therefore, novel biomarkers are warranted to prevent overtreatment (1,4,5).
Invasion can be caused by DCIS-intrinsic factors (7,8). Tumor microenvironment-derived factors have also been implied as drivers of progression (2,4,9). Fibroblasts regulate progression of many solid tumors [reviewed in (10–12)], and an emerging concept regarding fibroblast tumor biology is the notion of functionally distinct subsets (13–15). Fibroblasts can support DCIS growth by suppressing myoepithelial-derived tumor-inhibitory signals (16). Other proposed mechanisms include metabolic crosstalk (17), induction of cyclooxygenase-2 in DCIS cells (18), and production of lysyl oxidases (19,20). Correlative analysis of fibroblasts in DCIS, however, remains sparse, although a loss of fibroblast Caveolin-1 has been associated with progression to IDC (21).
Platelet derived growth factor receptor-alpha and -beta (PDGFRα and PDGFRβ) are potent regulators of fibroblasts (22,23). Stromal PDGFRβ expression has been linked to poor prognosis in different solid tumors (24–29). Mechanistic studies have demonstrated pro-invasive effects of PDGF-activated fibroblasts (30) and a connection to estrogen receptor (ER)-expression in breast cancer cells (31,32).
The integrity of the basement membrane has also been suggested to determine progression of DCIS (33,34). According to electron microscopy studies, DCIS cells protrude into gaps of the basement membrane, and fibroblasts surrounding these gaps show myofibroblastic differentiation (33,35). The potential role of basement membrane breakdown in DCIS or stromal cell communication remains poorly understood.
This report uses clinical samples, tissue cultures, and mouse models to analyze signaling between DCIS and stroma cells. The study aims to identify if PDGFR-defined subsets of stromal fibroblasts affect the prognosis of DCIS and further addresses the question on how these fibroblast subsets are induced during DCIS development and progression.
Materials and Methods
Detailed materials and methods are provided within the Supplementary Material (available online). Primers and probes are listed in Supplementary Table 1 (available online).
Patient Samples
The tissue microarray (TMA) of the population-based DCIS cohort included cores from 458 women diagnosed with primary DCIS between 1986 and 2004 in Sweden (36). The TMA of the DCIS_Nation cohort (n = 80) used for validation includes samples from DCIS patients who all developed recurrences (37,38). TMAs included two cores per patient. Analysis was approved by the Ethics Committees of Uppsala University Hospital (Dnr.99422 and Dnr.2005: 118) and Umeå University (Dnr.2014–230-321M), and no written informed consent was needed. Core needle biopsies of normal, cancer-free breast tissue were provided within the Karolinska Mammography Project for Risk Prediction of Breast Cancer study (Dnr.2011/1464–31/1; ethical board Karolinska Institutet) (39). Staining and analysis of the invasive breast cancer cases are described in (24).
Immunohistochemistry (IHC)
PDGFR IHC was performed using the DAKO Techmate Horizon30 (DAKO, Glostrup, Denmark). The human-specific PDGFRα antibody (clone D13C6) was diluted 1:150, the mouse reactive PDGFRα antibody (clone D1E1E) 1:100, and the PDGFRβ antibody (clone 28E1) 1:75 (all CellSignaling Technology, Danvers, MA). Staining of porcine aortic endothelial cells (PAE)/PDGFRα and porcine aortic endothelial cells (PAE)/PDGFRβ cells excluded antibody cross-reactivity (Supplementary Figure 1A, available online).
Scoring was guided by a breast pathologist. The PDGFRα and -β staining was scored as the positive stroma fraction (0 = negative, 1= low, 2 = moderate, or 3 = high) as previously described (24) and dichotomized by defining scores 0 and 1 as low and scores 2 and 3 as high (Supplementary Figure 1B, available online). Scoring was performed independently and blinded by three individuals. In cases of disagreement (13.9%), a consensus score was established.
Animal Studies
Seven- to 8-week-old female CB17/Icr-Prkdc(scid)/IcrIcoCrl mice (Scanbur, Karlslunde, Denmark) were used for xenograft experiments with MCF10DCIS cells (Asterand, Detroit, MI) with 10 animals per experimental group. The mouse mammary tumor virus-polyoma middle tumor-antigen (MMTV-PyMT) in Friend Virus B-Type/NIH (FVB/N) background was used to collect mammary tumors at different ages. Animal experiments were approved by Jordbruksverket (N220/14; N96/11).
Cluster and Pathway Analysis
Publicly available, normalized gene expression data from Ma et al. (40) (GEO14548) was used for hierarchical clustering analysis of normal and DCIS stroma samples. Cluster analysis (median centred by feature or gene, Pearson correlation, average linkage) was performed in a semisupervised manner with the heatmap3 package in R version 3.4.0. Pathway analysis was performed using the Generally Applicable Gene-set Enrichment (GAGE) package (41).
Statistical Analysis
Associations between PDGFR expression and clinico-pathological parameters were analyzed with contingency tables and Fishers’ exact test (two-sided). The Kaplan-Meier plots and log-rank test were used to compare risk to develop local recurrence (in situ or invasive) or generalized disease (IBM SPSS Statistics Version 22, SPSS Inc). A Cox proportional hazards model was used for estimation of hazard ratios in univariate and multivariable analyses including relevant risk factors. Proportional hazard assumption was verified graphically through evaluation of parallelism of the log(-log(S(t))) vs time plot as well as statistically through the Schoenfeld Residuals Test. A weak interaction with time was observed after 240 months of follow-up, when patient numbers became low. This interaction completely disappears when dropping these last follow-up times from the analysis. P values for Cox regression are based on a Wald test.
The relationship between continuous variables was assessed through Spearman rank correlation, stating the correlation-coefficient rho and the P value. Group differences were evaluated by using Student t test for two-group comparisons and one-way ANOVA with Bonferroni post hoc test for multiple group comparisons. P values derived from multiple Student t test comparisons were adjusted by a 5% false discovery rate with Benjamini and Hochberg correction and referred to as q values. All statistical tests were two-sided and P values less than .05 were considered statistically significant.
Results
Alterations in Stromal PDGF Receptor Expression in DCIS
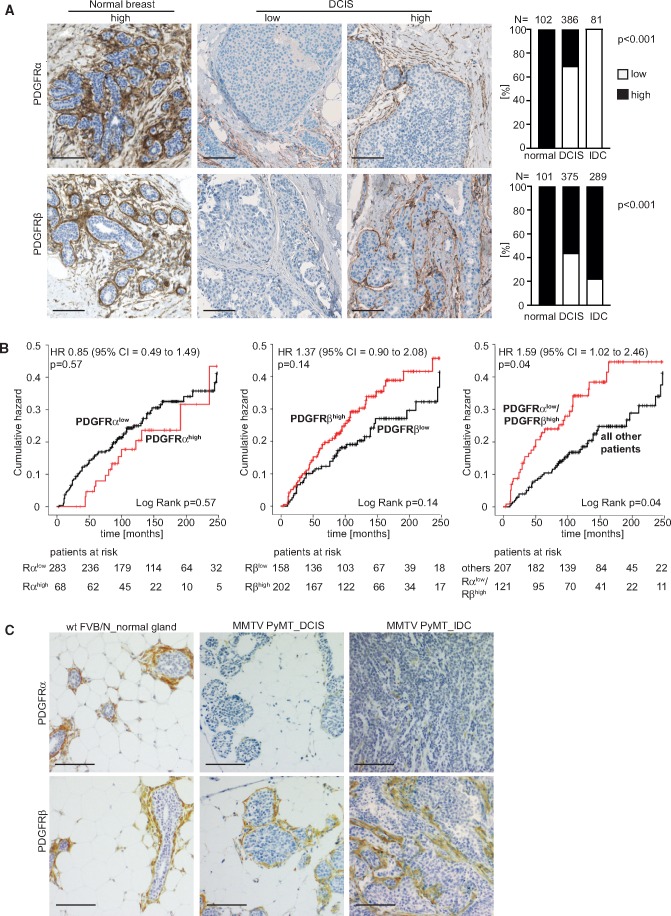
Stromal PDGFRα expression was analyzed in an IDC collection, and both PDGFRα and PDGFRβ were analyzed in a TMA of DCIS and a set of normal breast specimens (see Materials and Methods and Supplementary Figure 1, A and B, available online for staining and scoring information). PDGFR expression was predominantly observed in stromal cells. When specimens were co-stained with antibodies to PDGFRα and the myoepithelial marker CK14, no expression of PDGFRα in myoepithelial cells was observed (Supplementary Figure 1C, available online). All of the normal breast tissues displayed high stromal expression of both PDGFRs (Figure 1A). The PDGFRα expression was reduced in DCIS, with only 30.8% (119 of 386 patients) of cases displaying high PDGFRα, and no IDC cases displayed high PDGFRα. PDGFRβ expression remained high in the majority of DCIS and IDC cases.
Figure 1.
Differential platelet derived growth factor receptor (PDGFR)α and -β expression in stroma of human ductal breast carcinoma in situ (DCIS). A) Microphotographs of immunohistochemistry (IHC)-detected PDGFRα and PDGFRβ in normal human breast tissue and DCIS, together with results from analysis of differences in PDGFR status in normal breast tissue, DCIS, and invasive ductal cancer (IDC). P values were derived from Fisher’s exact test, two-sided. B) Kaplan-Meier plots showing relationships between PDGFR status and the risk for local recurrence (in situ or invasive) or metastasis in DCIS with P values derived from a log-rank test. Graphs also present hazard ratios, including confidence intervals as determined by univariate Cox proportional hazards regression analyses with P values derived from Wald test. Corresponding tables indicate the number of patients at risk. C) Microphotographs of IHC-detected PDGFRα and PDGFRβ in normal breast tissue of wild-type mice and in DCIS lesions (6 weeks) and invasive ductal carcinoma (12 weeks) of the MMTV-PyMT mouse breast cancer model. Images were adjusted for presentation. Size bars in A and C are 100 μm. FVB/N=Friend Virus B-Type/NIH mouse; MMTV-PyMT = mouse mammary tumor virus-polyoma middle tumor-antigen; wt = wild type.
High stromal PDGFRα expression was statistically significantly associated with ER positivity (P = .005, Fisher’s exact test) (Table 1; Supplementary Table 2, available online). High stromal PDGFRβ expression was instead linked to ER negativity and positively associated with high EORTC grade (P = .007 and P = .03, respectively; Fisher’s exact test). The combined PDGFRα(low)/PDGFRβ(high) metric showed strong associations with ER negativity and higher EORTC grade (P = .002 and P = .001, respectively; Fisher’s exact test). PDGFR status was not associated with tumor size.
Table 1.
Associations between stromal PDGFRα(low)/PDGFRβ(high) expression and clinicopathological parameters
| PDGFR expression, No. (%)* |
|||
|---|---|---|---|
| Clinicopathological parameter | PDGFRαlow/ PDGFRβhigh | All other patients | P † |
| Total | 121 | 207 | |
| Age, y | |||
| <50 | 38 (31.4) | 59 (28.5) | .90 |
| 50–64 | 47 (38.8) | 82 (39.6) | |
| ≥65 | 36 (29.8) | 66 (31.9) | |
| Size, mm | |||
| <15 | 48 (39.7) | 91 (44.0) | .62 |
| ≥15 | 47 (38.8) | 68 (32.8) | |
| Multifocal | 11 (9.1) | 25 (12.1) | |
| Missing | 15 (12.4) | 23 (11.1) | |
| ER status | |||
| ER negative | 47 (38.9) | 43 (20.8) | .002 |
| ER positive | 69 (57.0) | 152 (73.4) | |
| Missing | 5 (4.1) | 12 (5.8) | |
| EORTC grade | |||
| I | 6 (5.0) | 18 (8.7) | .001 |
| II | 40 (33.1) | 106 (51.2) | |
| III | 74 (61.1) | 83 (40.1) | |
| Missing | 1 (0.8) | 0 (0.0) | |
| Surgery | |||
| Mastectomy | 27 (22.3) | 46 (22.2) | 1.00 |
| Breast conserving | 94 (77.7) | 161 (77.8) | |
| Postoperative radiotherapy | |||
| No | 48 (39.7) | 75 (36.2) | .56 |
| Yes | 73 (60.3) | 132 (63.8) | |
Percentages are calculated within columns. EORTC = European Organization for Research and Treatment of Cancer; ER = estrogen receptor.
Associations were calculated with two-sided Fisher’s exact test.
Kaplan Meier analyses revealed an increased risk for local recurrences or metastasis in the cases with low PDGFRα or high PDGFRβ expression, although not statistically significant (Figure 1B). Interestingly, the PDGFRα(low)/PDGFRβ(high) subset showed a statistically significantly worse prognosis (P = .04, log-rank test) (Figure 1B). This association was also detected in univariate Cox regression analysis (hazard ratio [HR] = 1.59, 95% confidence interval [CI] = 1.02 to 2.46; P = .04 Wald test). The PDGFRα(low)/PDGFRβ(high) metric was a statistically significant independent marker for increased risk of local recurrence or metastasis in multivariable analysis (HR = 1.78, 95% CI = 1.07 to 2.97; P = .03) (Table 2; Supplementary Table 3, available online). Multivariable analysis also confirmed benefit of postoperative radiotherapy and increased recurrence risk associated with breast-conserving surgery.
Table 2.
Multivariable analysis of risk for local recurrence (in situ or invasive) and metastasis
| Clinicopathological parameter | Events/total, No. | HR (95% CI)* | P † |
|---|---|---|---|
| Age, y | |||
| <50 | 37/121 | 1.00 (Reference) | .10 |
| 50–64 | 39/187 | 0.58 (0.33 to 1.01) | |
| ≥65 | 40/150 | 0.58 (0.31 to 1.09) | |
| Size‡, mm | |||
| <15 mm | 52/197 | 1.00 (Reference) | .07 |
| ≥15 mm | 52/158 | 0.94 (0.50 to 1.75) | |
| Multifocal | 24/49 | 2.04 (1.05 to 3.97) | |
| ER status§ | |||
| Negative | 31/123 | 1.00 (Reference) | .82 |
| Positive | 75/292 | 1.07 (0.60 to 1.92) | |
| EORTC grade‖ | |||
| I | 11/37 | 1.00 (Reference) | .93 |
| II | 58/203 | 0.97 (0.40 to 2.35) | |
| III | 47/215 | 0.88 (0.36 to 2.14) | |
| Surgery¶ | |||
| Mastectomy | 11/104 | 1.00 (Reference) | .02 |
| Breast conserving | 103/350 | 2.93 (1.22 to 7.07) | |
| Postoperative radiotherapy | |||
| No | 84/297 | 1.00 (Reference) | .008 |
| Yes | 32/161 | 0.46 (0.26 to 0.82) | |
| PDGFR# | |||
| All other patients | 45/207 | 1.00 (Reference) | .03 |
| PDGFRαlow/PDGFRβhigh | 36/121 | 1.78 (1.07 to 2.97) | |
| Total No. | 116/458 | — | — |
A Cox proportional hazard model was applied including all variables (n = 272). HRs are estimated using proportional hazards regression with event defined as local recurrence or metastasis. CI = confidence interval; ER = estrogen receptor; HR = hazard ratio.
P values are based on a two-sided Wald test.
Data missing for 54 patients.
Data missing for 43 patients.
Data missing for 3 patients.
Data missing for 4 patients.
Data missing for 130 patients.
These results were confirmed on a validation cohort of DCIS patients who all developed local recurrences (Supplementary Figure 2A, upper row, available online). The PDGFRα(low)/PDGFRβ(high) subset showed a statistically significantly worse prognosis in both univariate and multivariable analysis. Further, a digital analysis approach was applied to the validation cohort, leading to similar results (Supplementary Figure 2A, lower row, available online). The data of the digital analysis were also used to investigate if intratumoral heterogeneity affected the scoring outcome. A strong statistically significant correlation between the PDGFR status in cores derived from same patients was noted (Supplementary Figure 2B, available online).
Analysis of The Cancer Genome Atlas gene expression datasets demonstrated in many solid tumor types lower PDGFRα expression in tumors than in corresponding normal tissue (Supplementary Figure 3A, available online). No distinct pattern was found for PDGFRβ expression. Fresh-frozen sections from four normal, four DCIS, and eight invasive cancers were analyzed for PDGFR transcript abundance with padlock probes (Supplementary Figure 3B, available online). Within DCIS stroma, the fraction of PDGFRA of total PDGFR transcript abundance was statistically significantly reduced.
Analyses of the MMTV-PyMT mouse breast cancer model also revealed a progression-associated loss of stromal PDGFRα expression, occurring with maintained stromal PDGFRβ expression (Figure 1C). Together, these data demonstrate clinically relevant alterations in the stroma of DCIS, and identify the combined PDGFRα(low)/PDGFRβ(high) metric as a novel marker for increased risk to develop local recurrence or metastasis.
Analysis of Stromal PDGFRα Expression and Peri-Epithelial Laminin-γ2 Deposition
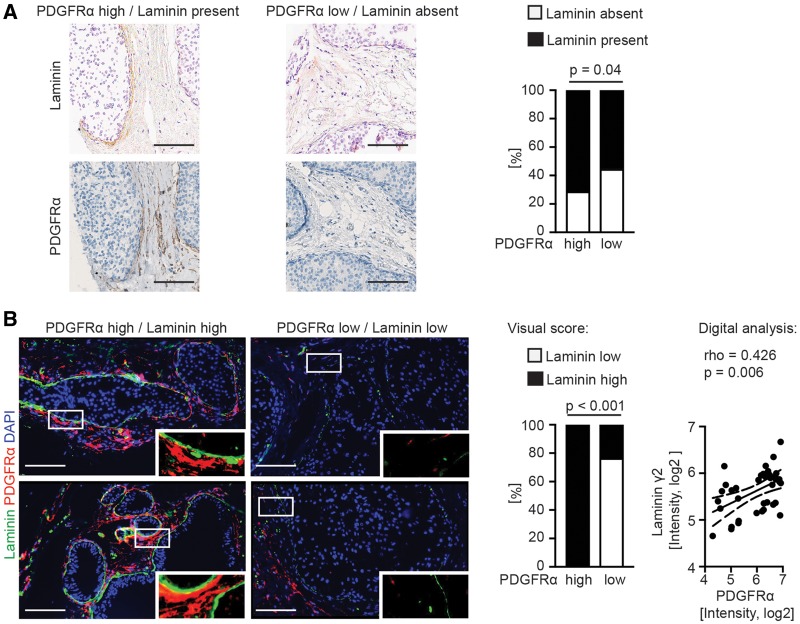
To investigate possible links between PDGFR expression and the basement membrane status, associations between stromal PDGFRα expression and laminin-γ2 levels, a key component of epithelial basement membranes (42), were analyzed. As shown in Figure 2A, a statistically significant association was detected between low laminin-γ2 level and the PDGFRα(low) phenotype (P = .04 Fisher’s exact test). Furthermore, 40 lesions from four different DCIS cases were subjected to combined laminin-γ2 and PDGFRα immunofluorescence analysis. There was a strong positive correlation between low laminin-γ2 levels and low PDGFRα expression in analyses using visual scoring (P < .001 Fisher’s exact test) or digital image analysis (P = .006, rho = 0.426, Spearman rank correlation (Figure 2B).
Figure 2.
Analysis of stromal platelet derived growth factor receptor (PDGFR)α expression and the basement membrane component laminin-γ2. A) Microphotographs of immunohistochemistry (IHC)-detected PDGFRα and laminin-γ2 of selected cases from the PDGFR-defined human ductal breast carcinoma in situ (DCIS) patient cohort, together with the results from patient-based correlation analysis. Laminin-γ2 was scored as present or absent. P value was derived from Fisher’s exact test, two-sided. B) Microphotographs of immunofluorescence-detected PDGFRα and laminin-γ2 in selected lesions of human DCIS. White boxes indicate regions shown at higher magnification. The correlation analysis of the visual scoring was based on the distribution of laminin (high) and laminin (low) within a total of 40 different PDGFR-defined lesions selected from four different DCIS sections. P value was derived from Fisher’s exact test, two-sided. The analysis of the digital scoring was based on the mean fluorescent intensity of the laminin-γ2 staining and the PDGFRα staining in the corresponding stroma region. The data were log2-transformed for presentation. Rho and P values were derived from Spearman rank correlation, two-sided. The linear regression line with 95% confidence interval is indicated. Images were adjusted for presentation. Size bars are 100 μm.
Impact of DCIS Cells on PDGF Receptor Expression in Fibroblasts
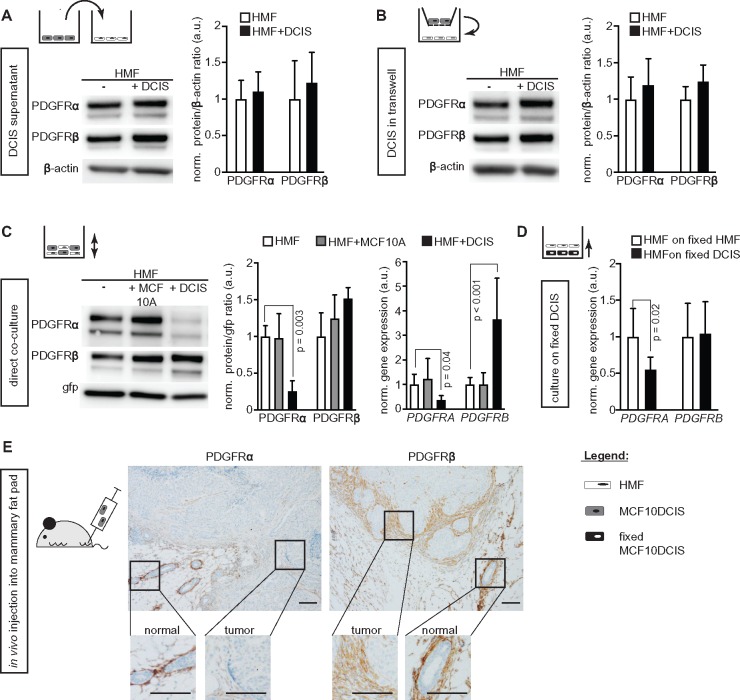
To address if the PDGFRα(low)/PDGFRβ(high) phenotype signature was induced by paracrine signaling from DCIS cells, the MCF10A.DCIS cell model (16) was used in co-cultures with immortalized human mammary fibroblasts (HMF) positive for both PDGFRα and β (Figure 3, A–D), immortalized skin fibroblast, or primary mammary fibroblasts (Supplementary Figure 3C, available online). The DCIS cells are negative for both PDGFRs and did not show an induction of PDGFR expression after co-culture (Supplementary Figure 3, C and D, available online). MCF10A normal breast epithelial cells were included as control cells (Figure 3C).
Figure 3.
Ductal breast carcinoma in situ (DCIS) cell-dependent regulation of platelet derived growth factor receptor (PDGFR) expression in fibroblasts. Representative immunoblots of PDGFR expression in human mammary fibroblasts (HMF) and results from corresponding quantifications of (A–C) PDGFR protein and (C and D) PDGFR mRNA under conditions of (A) HMF exposure to control medium or DCIS cell medium (n = 4), (B) trans-well HMF cultures with or without DCIS cells (n = 3), (C) mono-culture or co-cultures of green fluorescent protein (gfp)-labeled HMF with MCF10A normal breast epithelial cells or breast MCF10A.DCIS cells (n = 4), and (D) HMF-gfp cultured either on fixed HMF or fixed DCIS cells (n = 7). Protein levels (immunoblot data) were determined using β-actin (A and B) or gfp (C) as loading control and further normalized to the mean of the control group (set to 1.0 arbitrary units [a.u.]). Data are presented as average with standard deviation (SD). For PDGFRα and -β detection, the membranes were stripped and reprobed. β-Actin or gfp was detected on the same membrane. Changes in gene expression (quantitative real-time PCR data; C and D) were calculated by the comparative ΔΔCT-method with GFP as housekeeping gene for input control. Expression values were further normalized to the mean of the control monoculture HMF (set to 1 a.u.) and are represented as average with SD. P values were derived from ANOVA with Bonferroni post hoc test. (E) Microphotographs of immunohistochemistry-detected PDGFR expression in orthotopic tumors formed after injection of DCIS cells into the mammary fat pad of SCID mice. Images were adjusted for presentation. Size bars are 100 μm.
DCIS supernatant or DCIS co-culture in trans-well plates did not affect PDGFR expression in fibroblasts (Figure 3, A and B). In contrast, a statistically significant, DCIS-induced down-regulation of PDGFRα protein and mRNA was detected in fibroblasts upon direct co-culture with MCF10A.DCIS cells (protein: 0.26-fold of the control monoculture (0.14 SD), P = .003; mRNA: 0.36-fold (0.18), P = .04; Figure 3C). Conversely, PDGFRβ expression was up-regulated at the mRNA level (3.65-fold of control monoculture (1.68), P < .001) with an associated slight up-regulation of PDGFRβ protein. A similar pattern was also observed in co-culture experiments with other fibroblasts (Supplementary Figure 3C, available online). Formaldehyde-fixed DCIS cells also elicited the effect (Figure 3D).
DCIS cells injected into the mammary fat pad of SCID mice (Figure 3E) formed lesions, which were associated with fibroblasts with lower PDGFRα expression than fibroblasts surrounding the normal glands. In contrast, stromal PDGFRβ expression was retained.
These studies imply cell contact-dependent paracrine signaling as the mechanism underlying the formation of the poor prognosis-associated PDGFRα(low)/PDGFRβ(high) fibroblasts detected in human DCIS.
Gene Expression Changes in Fibroblasts upon Co-Culture with DCIS Cells
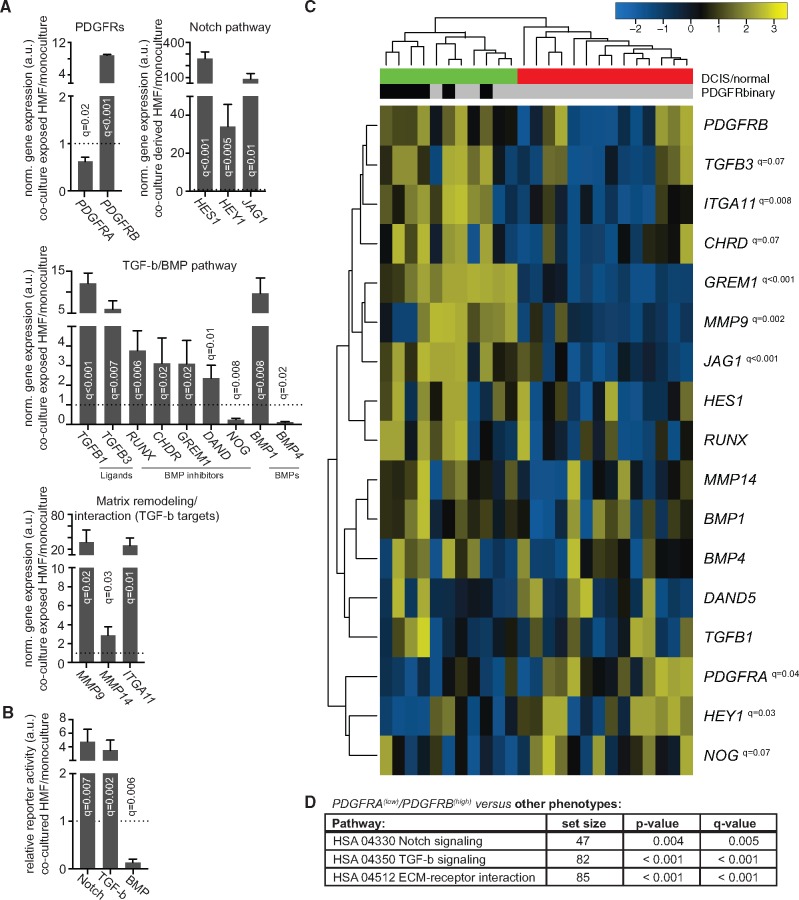
The next steps aimed to identify signaling pathways involved in the DCIS-induced modulation of fibroblast PDGFR expression. Initial qRT-PCR array–based screening detected co-culture–induced changes of genes related to Notch and transforming growth factor (TGF) and bone morphogenetic protein (BMP) signaling (Supplementary Table 4, available online). QRT-PCR validation confirmed a robust up-regulation of Notch-induced genes (HES1, HEY1, JAG1), TGF-beta signaling components (ligands TGFB1 and TGFB3 and targets MMP9 and IGA11) and BMP inhibitors (RUNX1, CHRD, GREM1) in sorted, co-culture–exposed fibroblasts (Figure 4A).
Figure 4.
Gene expression analysis of platelet derived growth factor receptor (PDGFR)A(low)/PDGFRB(high) fibroblasts. A) Gene expression analysis of sorted, co-culture–exposed human mammary fibroblast (HMF) (n = 4). The expression values were calculated by the comparative ΔΔCT-method with GAPDH as housekeeping gene for input control. Expression values were further normalized to the mean of the control monoculture HMF (set to 1 arbitrary unit [a.u.] and indicated by dotted line) and are represented as average with standard deviation (SD). B) Pathway-specific signaling activity was determined in HMF under conditions of monoculture or co-culture with MCF10A.DCIS cells. Transforming growth factor (TGF)-beta signaling was measured with the pGL3-(CAGA)12-Luciferase, bone morphogenetic protein (BMP) signaling with the pGL3-BRE2-Luciferase, and Notch signaling with the 12xCSL-Luciferase vector. Quantifications were based on at least four experiments. Data were normalized to the mean of the control monoculture HMF (set to 1.0 a.u. and indicated by dotted line) and are presented as average with SD. C) Semisupervised hierarchical cluster analysis of genes of interest within normal (n = 14) and ductal breast carcinoma in situ (DCIS) (n = 11) stroma fractions. The gene expression data were derived from GEO14548 (40). Hierarchical clustering was performed by Pearson correlation with average linkage. Red bar represents stroma from normal cases, green bar from DCIS cases. The bad prognosis–associated PDGFRA(low)/PDGFRB(high) phenotype is marked with black bars vs the other phenotypes indicated by grey bars. Median PDGFRA and PDGFRB expression was used to define samples as high or low. Multiple t test comparisons in A–C were adjusted by 5% false discovery rate with Benjamini and Hochberg correction and q values are indicated. D) Gene ontology analysis for changes of indicated pathways for the PDGFRA(low)/PDGFRB(high) samples vs all others as defined in (C). Global P values were based on t statistics and derived by testing whether the mean fold change in the Notch, TGF-beta, and ECM gene sets were different from the mean fold change in the other background genes in the microarray experiment, as is default for the Generally Applicable Gene-set Enrichment (GAGE) package. Q values were based on adjustment of the global P value using a Benjamini and Hochberg correction. ECM = extracellular matrix; HSA = homo sapiens.
Pathway-specific reporter gene assays demonstrated statistically significantly increased Notch and TGF-beta signaling (Notch: 3.5-fold of the control monoculture [0.54 SD], q = 0.002; TGF-beta: 4.8-fold [0.92], q = 0.007), and decreased BMP signaling (0.1-fold of the control monoculture [0.03], q = 0.006), in fibroblasts during co-culture with DCIS cells (Figure 4B).
Semisupervised cluster analysis was done on a gene expression dataset from microdissected stroma of normal breast tissue and DCIS (40). The PDGFRA(low)/PDGFRB(high) phenotype clustered exclusively within the DCIS cases (Figure 4C). Furthermore, the PDGFRA(low)/PDGFRB(high) cluster showed a very similar gene expression pattern as the co-culture–exposed fibroblasts, with prominent changes of Notch target genes and TGF-beta or BMP signaling components (Figure 4, C and D).
Molecular Analysis of the DCIS-Induced Modulation of Fibroblast PDGFR Status
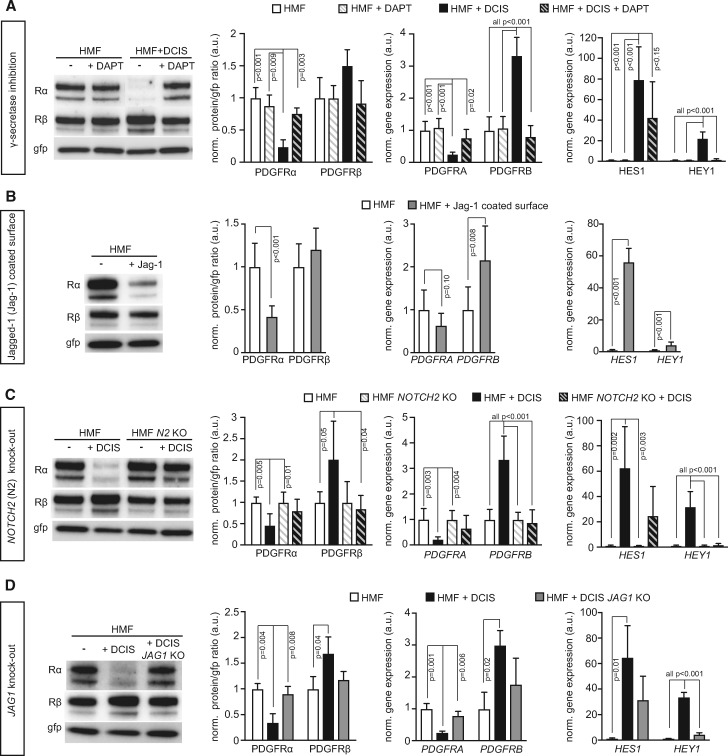
The presented data motivated further analysis of the Notch pathway, a cell-cell interaction signaling mechanism, leading to Notch receptor cleavage by γ-secretase and transcriptional activation (43). Treatment of co-cultures with the γ-secretase inhibitor DAPT blocked DCIS-induced down-regulation of PDGFRα and up-regulation of PDGFRβ (Figure 5A). On the contrary, a down-regulation of PDGFRα, accompanied by an increase in PDGFRB mRNA expression, was observed in fibroblasts after exposure to immobilized Jagged-1 protein (Figure 5B).
Figure 5.
Involvement of Notch-signaling components in the regulation of platelet derived growth factor receptor (PDGFR) expression in fibroblasts. Representative immunoblots of PDGFR expression in fibroblasts (human mammary fibroblasts [HMF]) and results from corresponding quantifications of PDGFR protein as well as mRNA levels of PDGFR, HES1, and HEY1 under conditions of (A) HMF-green fluorescent protein (gfp) mono-cultures or MCF10A.DCIS co-cultures in the absence or presence of the Notch-signaling inhibitor DAPT, (B) HMF-gfp mono-culture under control conditions or on immobilized Jagged-1, (C) HMF-gfp control or HMF-gfp with CRISPR/Cas9-mediated NOTCH2 deletion in mono-cultures or co-cultures with MCF10DCIS cells, and (D) HMF-gfp mono-cultures or co-cultures with control MCF10DCIS cells or MCF10DCIS cells with CRISPR/Cas-mediated JAG1 deletion. Protein levels (immunoblot data) were determined using gfp as loading control and further normalized to the mean of the control group (set to 1.0 a.u.). Data are presented as average with standard deviation (SD). For PDGFRα and -β detection, the membranes were stripped and reprobed. Gfp was detected on the same membrane. Changes in gene expression (quantitative real-time PCR data) were calculated by the comparative ΔΔCT-method with GFP as housekeeping gene for input control. Expression values were further normalized to the mean of the control monoculture HMF (set to 1.0 a.u.) and are represented with SD. P values were derived from ANOVA with Bonferroni post hoc test. KO = knock-out.
JAG1 and NOTCH2 were the most highly expressed Notch ligands and receptors on the DCIS cells and fibroblasts, respectively, as determined by gene expression analysis of different Notch ligand and receptor paralogs (Supplementary Figure 4A, available online). To assess their specific roles, CRISPR/Cas9 gene editing was used to inactivate the JAG1 gene in DCIS cells and the NOTCH2 gene in fibroblasts (Supplementary Figure 4C, available online). The statistically significant PDGFRα down-regulation observed in co-culture experiments with wild-type fibroblasts was abrogated both at the protein and mRNA levels when using NOTCH2-deficient fibroblasts (Figure 5C; Supplementary Figure 4, D and E, available online). Similarly, the PDGFRβ up-regulation was obliterated in co-culture experiments with NOTCH2-deficient fibroblasts (Figure 5C). SiRNA-based knock-down of NOTCH2 expression in fibroblasts yielded similar results (Supplementary Figure 4, G–J, available online). The paracrine modulation of fibroblast PDGFR status was also lost in fibroblasts from co-cultures with JAG1 knock-out DCIS cells (Figure 5D; Supplementary Figure 4F, available online). The efficacy of JAG1/NOTCH2 manipulation in these experiments was confirmed by reduced induction of HES1 and HEY1 (Figure 5, A–D right panel; Supplementary Figure 4, B and J, available online).
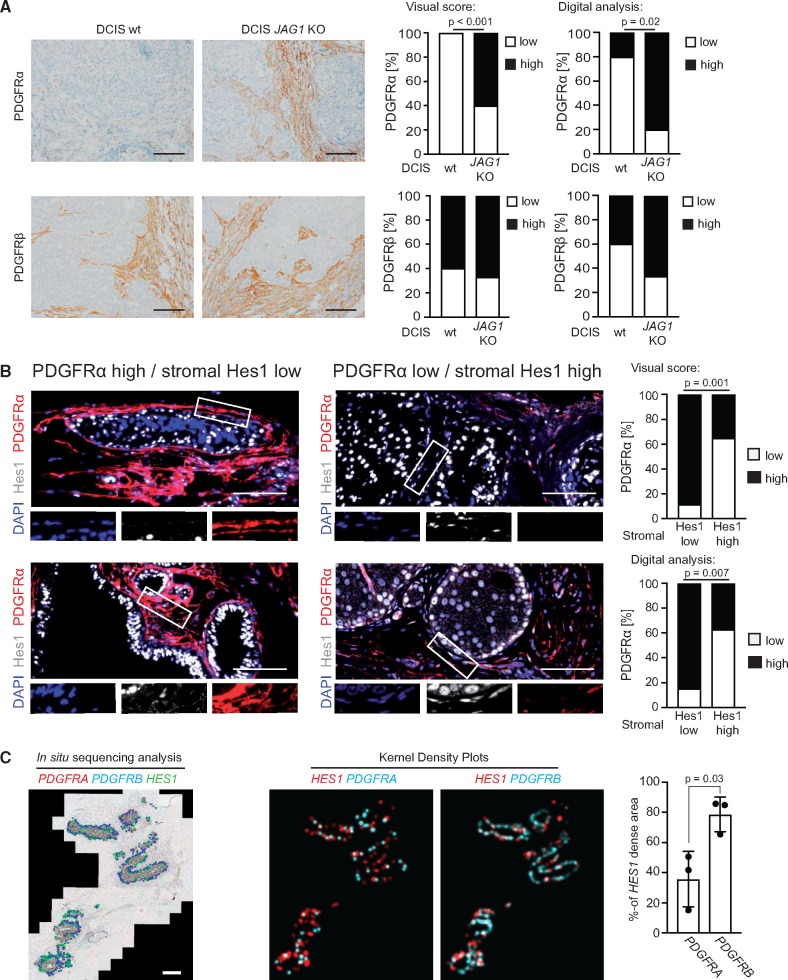
To test the role of JAG1/NOTCH2 in vivo, DCIS cells deficient for JAG1 were used for mammary fat pad injection. Lesions from JAG1-deficient DCIS cells displayed a statistically significantly higher PDGFRα expression in the tumor-associated stroma compared with lesions formed by wild-type cells, as analyzed by visual scoring and digital analysis (P < .001 and P = .02, respectively, Fisher’s exact test) (Figure 6A).
Figure 6.
Correlation analysis of platelet derived growth factor receptor (PDGFR)α down-regulation and Notch signaling in stromal fibroblasts of experimental and human ductal breast carcinoma in situ (DCIS) samples. A) Microphotographs and quantifications of immunohistochemistry (IHC)-detected stromal PDGFR expression in orthotopic tumors formed after injection of wild-type MCF10DCIS cells (left panel) or MCF10DCIS cells with CRISPR/Cas-mediated JAG1 deletion (DCIS JAG1 knock-out, right panel) into the mammary gland of SCID mice. Correlation analysis of the dichotomized visual scoring (right graphs) and the digital image analysis of the mean intensity (left graphs) was based on Fisher’s exact test, two-sided. For the digital analysis, the PDGFR expression was defined as low or high by splitting the datasets at the median. Results were derived from 10 tumors per group, except for PDGFRβ analysis of the DCIS JAG1 depleted group, where only nine tumor sections could be analyzed due to tissue detachment in one case. Only the tumor-associated stroma was included in the analyses. Images were adjusted for presentation. Size bars are 100 μm. B) Microphotographs of immunofluorescence-detected PDGFRα and Hes1 in selected lesions of human DCIS cases. The white boxes indicate regions displayed at higher magnification. The correlation analysis was based on the presence of nuclear Hes1 in tumor-associated stroma cells with cases being defined as “Hes1 high” when more than one-quarter of cells showed nuclear Hes1 positivity. Correlation analyses of the dichotomized visual scoring (upper graph) and the digital image analysis of the mean intensity (lower graph) were based on Fisher’s exact test, two-sided. For the digital analysis, dichotomization of the PDGFRα mean intensity into high and low was based on a histogram-guided cutoff value. Results were derived from analysis of 40 different regions within four different DCIS sections. Images were adjusted for presentation. Size bars are 100 μm. C) Padlock probe–based in situ sequencing to analyze areas of high mRNA density for PDGFRA (blue dots), PDGFRB (red dots), and HES1 (green dots). The analysis was restricted to the tumor-associated stroma (representative image on the left). Three different DCIS cases were analyzed. Kernel density estimation plots (representative images on the right) were generated to analyze overlap of regions with high abundance of HES1 (red) or PDGFR (cyan) transcripts. Quantification is given as the percentage overlap with regions of high HES1 density. The P value was calculated by Student’s t test, two-sided. Data are presented as average with standard deviation (SD). Size bar is 1000 μm.
Collectively, these data provide evidence for paracrine Jagged-1/Notch2-signaling as the driver of the PDGFRα(low)/PDGFRβ(high) fibroblast phenotype.
Correlative Analysis of Loss of Stromal PDGFRα in Human DCIS and the Expression of the Notch Target Gene HES1 in Fibroblasts
To validate the connection between Notch signaling and the PDGFRα(low)/PDGFRβ(high) phenotype in clinical samples, the relationship between PDGFRα and Hes1 expression was analyzed using double-immunofluorescence on different DCIS sections. Statistical analysis indicated a statistically significant inverse correlation between stromal PDGFRα and Hes1 in tumor-associated stroma cells as determined by visual scoring and digital analysis (P = .001 and P = .007, respectively; Fisher’s exact test) (Figure 6B).
Padlock probe–based in situ sequencing was used to analyze the tumor-associated stroma for mRNA transcript density of PDGFRA, PDGFRB, and HES1 (Figure 6C). Only 35.9% (10.7% SD) of the area with high HES1 transcript density showed high PDGFRΑ transcript density, whereas a 78.7% (6.6% SD) overlap was observed between areas of high HES1 and high PDGFRΒ transcript density (P = .03; Student's t test).
Together, these data support the notion that activation of Notch signaling in stroma cells coincides with a loss of PDGFRα expression.
Discussion
This study identifies a novel interplay between DCIS and stroma cells involving Notch signaling and demonstrates that PDGFRα(low)/PDGFRβ(high) fibroblasts are statistically significantly associated with a higher risk for local recurrence in DCIS. Importantly, this marker remained an independent prognostic factor in multivariable analysis, suggesting stromal PDGFR status as a potential marker for DCIS stratification. These findings add to ongoing efforts to identify highly warranted prognostic markers for DCIS disease.
Concerning limitations of this study, it is noted that the results rely on retrospective analyses of population-based cohorts. Therefore, firm conclusions cannot be made regarding the impact of the marker on the natural course of the disease or benefit to treatment. This topic merits attention in future studies.
High stromal PDGFRβ expression is linked to poor prognosis for several different malignancies including breast (24,25), prostate (26,27), and ovarian cancer (28) as well as rhabdomyosarcoma (29). Mechanistic studies have demonstrated pro-tumoral and -metastatic effects of PDGFRβ-activated fibroblasts (30,44,45). This study is the first report to our knowledge on regulatory and prognostic roles of a marker-defined fibroblast subset in DCIS as well as the first analysis that implicates low PDGFRα as a marker associated with poor prognosis.
Our data demonstrate that elevated Notch signaling in the fibroblasts is a critical regulator of the PDGFRα(low)/PDGFRβ(high) phenotype. The up-regulation of PDGFRβ expression is likely directly mediated by the Notch intracellular domain/CSL (CBF1, Suppressor of Hairless, Lag-1) transcriptional complex, because the PDGFRB promoter contains CSL binding sites to which Notch intracellular domain has been shown to bind in vascular smooth muscle cells (46). The Notch-mediated down-regulation of PDGFRα expression may instead be explained through the induction of the Notch downstream transcriptional repressors HES1 and HEY1. Keeping with this notion, the 5000-bp promoter region upstream and 1000-bp promoter region downstream of the initiating ATG of the PDGFRΑ gene contains 13 putative Hes1 and five putative Hey1 binding sites (http://epd.vital-it.ch/).
The laminin-γ2 analyses suggest partial disruption of the basement membrane in DCIS. A discontinuous basement membrane in pure DCIS lesions was earlier detected through type IV collagen IHC analysis and correlated positively with higher nuclear grade (47). Similarly, a correlation between disintegration of the laminin components within the basement membrane and increasing anaplastic appearance was found (48). Disruption of basement membrane enables direct interactions between stroma cells and tumor cells. In DCIS, tumor cells were indeed found to protrude into gaps of the basement membrane, and the fibroblasts surrounding these gaps show an altered phenotype (33). The loss of the epithelial basement membrane component laminin-γ2 observed here may therefore facilitate Notch signaling by allowing direct contact between DCIS and stromal cells. The DCIS-induced expression of MMPs in fibroblasts supports a combined role for DCIS and the stroma in initiating this process. Several studies also imply a role for Notch in MMP regulation (49,50).
Notably, Notch signaling in other contexts has also been shown to operate between cells separated by a basement membrane, for example between endothelial and vascular smooth muscle cells in the adult vasculature (51), in manners possibly involving ligand expression on dynamic filopodia (52) or on exosomes (53,54). Collectively, this interplay gives credence to the overall concept that tumor fibroblasts are active participants in the tumor process rather than idle bystanders (55).
As Notch signaling is now emerging as a critical mediator for the induction of a poor prognosis-associated fibroblast subset, it may be interesting to try to intervene with Notch signaling in DCIS (43,56). Notch-based therapies have not yet reached the clinic. However, considerable efforts are made to develop Notch-modulating therapies (43). It will be important to consider at what level Notch pathway signaling should be blocked. Interestingly, genetic ablation of CSL, both in tumors and stroma, produced growth-promoting rather than growth-inhibiting effects (57,58), indicating that blockade should be better executed at the ligand-receptor level.
In conclusion, our data identify a novel, clinically relevant Notch-mediated mechanism of fibroblast activation in early stages of breast cancer development. Activation of Notch, occurring in association with basement membrane disruption, induces fibroblasts with a PDGFRα(low)/PDGFRβ(high) phenotype together with increased expression of matrix-remodeling enzymes and TGF-beta ligands. The identification of this novel and strong marker combination supports an important regulatory role of fibroblasts in the transition of DCIS to invasive cancer. The identification of the PDGFRα(low)/PDGFRβ(high) fibroblasts subset also contributes to ongoing efforts to better define clinically relevant and functionally distinct fibroblast subsets.
Funding
The AÖ group received support from the Swedish Cancer Society (grant no. 150895), Swedish Research Council (Diarienr 349–2006-160), STARGET Linné grant, Radiumhemmets forskningsfonder, EU Caffein ITN network (grant no. 3047/12), BRECT network of Karolinska Institutet, and Stockholm County Council. The authors further thank Uppsala-Umeå Comprehensive Cancer Consortium (U-CAN) for financial support.
Notes
Affiliations of authors: Department of Oncology-Pathology, Cancer Center Karolinska (CCK) (CS, JP, NPT, AM, CM, HJ, SMW, JB, AÖ), Department of Cell and Molecular Biology (SBJ, UL), Division of Vascular Biology, Department of Medical Biochemistry and Biophysics (PR), and Department of Neuroscience, Science for Life Laboratory (NM, JM), Karolinska Institutet, Stockholm, Sweden; Science for Life Laboratory, Department of Biochemistry and Biophysics, Stockholm University, Solna, Sweden (CS, JS, MN); Science for Life Laboratory, Department of Immunology, Genetics and Pathology, Uppsala University, Uppsala, Sweden (AM, MN); Department of Medical Epidemiology & Biostatistics, Karolinska Institutet, Solna, Sweden (PH); Department of Oncology, Södersjukhuset, Stockholm, Sweden (PH); Department of Cancer Biology, Mayo Clinic Comprehensive Cancer Center, Jacksonville, FL (DCR); Division of Translational Cancer Research, Department of Laboratory Medicine, Lund University, Lund, Sweden (KP); Radiumhemmet, Karolinska University Hospital, Stockholm, Sweden (JB); Department of Surgical Sciences, Uppsala University, Uppsala Academic Hospital, Uppsala, Sweden (FW).
The funders had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication. The authors have no conflicts of interest to disclose.
Members of the AÖ group are acknowledged for support throughout the study. Special thanks to Christer Betsholtz for valuable support and contribution. The authors furthermore thank all members of the STARGET and BRECT consortia at Karolinska Institutet for constructive discussion and comments throughout the study. All staff at the MTC animal facility are acknowledged for excellent assistance within animal experiments, and Rhys Fox is especially acknowledged for technical support. The pGL3-(CAGA)12-Luc and the pGL3-BRE2-Luc reporter vectors were both a generous gift of Prof. Dr Aristidis Moustakas (Ludwig Institute for Cancer Research, Uppsala University, Uppsala, Sweden).
Supplementary Material
References
- 1. Mardekian SK, Bombonati A, Palazzo JP.. Ductal carcinoma in situ of the breast: the importance of morphologic and molecular interactions. Hum Pathol. 2016;49:114–123. [DOI] [PubMed] [Google Scholar]
- 2. Yeong J, Thike AA, Tan PH, Iqbal J.. Identifying progression predictors of breast ductal carcinoma in situ. J Clin Pathol. 2017;702:102–108. [DOI] [PubMed] [Google Scholar]
- 3. Virnig BA, Tuttle TM, Shamliyan T, Kane RL.. Ductal carcinoma in situ of the breast: a systematic review of incidence, treatment, and outcomes. J Natl Cancer Inst. 2010;1023:170–178. [DOI] [PubMed] [Google Scholar]
- 4. Cowell CF, Weigelt B, Sakr RA, et al. Progression from ductal carcinoma in situ to invasive breast cancer: revisited. Mol Oncol. 2013;75:859–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Falk RS, Hofvind S, Skaane P, Haldorsen T.. Second events following ductal carcinoma in situ of the breast: a register-based cohort study. Breast Cancer Res Treat. 2011;1293:929–938. [DOI] [PubMed] [Google Scholar]
- 6. Wadsten C, Heyman H, Holmqvist M, et al. A validation of DCIS registration in a population-based breast cancer quality register and a study of treatment and prognosis for DCIS during 20 years. Acta Oncol Stockh Swed. 2016;5511:1338–1343. [DOI] [PubMed] [Google Scholar]
- 7. Vincent-Salomon A, Lucchesi C, Gruel N, et al. Integrated genomic and transcriptomic analysis of ductal carcinoma in situ of the breast. Clin Cancer Res. 2008;147:1956–1965. [DOI] [PubMed] [Google Scholar]
- 8. Johnson CE, Gorringe KL, Thompson ER, et al. Identification of copy number alterations associated with the progression of DCIS to invasive ductal carcinoma. Breast Cancer Res Treat. 2012;1333:889–898. [DOI] [PubMed] [Google Scholar]
- 9. Gil Del Alcazar CR, Huh SJ, Ekram MB, et al. Immune escape in breast cancer during in situ to invasive carcinoma transition. Cancer Discov. 2017;710:1098–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer. 2016;169:582–598. [DOI] [PubMed] [Google Scholar]
- 11. Marsh T, Pietras K, McAllister SS.. Fibroblasts as architects of cancer pathogenesis. Biochim Biophys Acta. 2013;18327:1070–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McAllister SS, Weinberg RA.. The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat Cell Biol. 2014;168:717–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rhim AD, Oberstein PE, Thomas DH, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell. 2014;256:735–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Öhlund D, Handly-Santana A, Biffi G, et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med. 2017;2143:579–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Özdemir BC, Pentcheva-Hoang T, Carstens JL, et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2014;256:719–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hu M, Yao J, Carroll DK, et al. Regulation of in situ to invasive breast carcinoma transition. Cancer Cell. 2008;135:394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Martins D, Beça FF, Sousa B, Baltazar F, Paredes J, Schmitt F.. Loss of caveolin-1 and gain of MCT4 expression in the tumor stroma: key events in the progression from an in situ to an invasive breast carcinoma. Cell Cycle Georget Tex. 2013;1216:2684–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hu M, Peluffo G, Chen H, Gelman R, Schnitt S, Polyak K.. Role of COX-2 in epithelial–stromal cell interactions and progression of ductal carcinoma in situ of the breast. Proc Natl Acad Sci USA. 2009;1069:3372–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cox TR, Erler JT.. Remodeling and homeostasis of the extracellular matrix: implications for fibrotic diseases and cancer. Dis Model Mech. 2011;42:165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cox TR, Bird D, Baker A-M, et al. LOX-mediated collagen crosslinking is responsible for fibrosis-enhanced metastasis. Cancer Res. 2013;736:1721–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Witkiewicz AK, Dasgupta A, Nguyen KH, et al. Stromal caveolin-1 levels predict early DCIS progression to invasive breast cancer. Cancer Biol Ther. 2009;811:1071–1079. [DOI] [PubMed] [Google Scholar]
- 22. Heldin C-H, Lennartsson J, Westermark B.. Involvement of platelet-derived growth factor ligands and receptors in tumorigenesis. J Intern Med. 2018;2831:16–44. [DOI] [PubMed] [Google Scholar]
- 23. Östman A. PDGF receptors in tumor stroma: biological effects and associations with prognosis and response to treatment. Adv Drug Deliv Rev. 2017;121:117–123. [DOI] [PubMed] [Google Scholar]
- 24. Paulsson J, Sjöblom T, Micke P, et al. Prognostic significance of stromal platelet-derived growth factor β-receptor expression in human breast cancer. Am J Pathol. 2009;1751:334.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Paulsson J, Rydén L, Strell C, et al. High expression of stromal PDGFRβ is associated with reduced benefit of tamoxifen in breast cancer. J Pathol Clin Res. 2017;31:38–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hägglöf C, Hammarsten P, Josefsson A, et al. Stromal PDGFRbeta expression in prostate tumors and non-malignant prostate tissue predicts prostate cancer survival. PLoS One. 2010;55:e10747.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nordby Y, Richardsen E, Rakaee M, et al. High expression of PDGFR-β in prostate cancer stroma is independently associated with clinical and biochemical prostate cancer recurrence. Sci Rep. 2017;7:43378.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Corvigno S, Wisman GBA, Mezheyeuski A, et al. Markers of fibroblast-rich tumor stroma and perivascular cells in serous ovarian cancer: inter- and intra-patient heterogeneity and impact on survival. Oncotarget. 2016;714:18573–18584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ehnman M, Missiaglia E, Folestad E, et al. Distinct effects of ligand-induced PDGFRα and PDGFRβ signaling in the human rhabdomyosarcoma tumor cell and stroma cell compartments. Cancer Res. 2013;737:2139–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Peña C, Céspedes MV, Lindh MB, et al. STC1 expression by cancer-associated fibroblasts drives metastasis of colorectal cancer. Cancer Res. 2013;734:1287–1297. [DOI] [PubMed] [Google Scholar]
- 31. Roswall P, Bocci M, Bartoschek M, et al. Microenvironmental control of breast cancer subtype elicited through paracrine platelet-derived growth factor-CC signaling. Nat Med. 2018;244:463–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jansson S, Aaltonen K, Bendahl P-O, et al. The PDGF pathway in breast cancer is linked to tumour aggressiveness, triple-negative subtype and early recurrence. Breast Cancer Res Treat. 2018;1692:231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tulusan AH, Grünsteidel W, Ramming I, Egger H.. A contribution to the natural history of breast cancer. III. Changes in the basement membranes in breast cancers with stromal microinvasion. Arch Gynecol. 1982;2313:209–218. [DOI] [PubMed] [Google Scholar]
- 34. Rosen PP. Rosen’s Breast Pathology. Philadelphia, PA: Lippincott Williams & Wilkins; 2001. [Google Scholar]
- 35. Giannelli G, Falk MJ, Schiraldi O, Stetler SWG, Quaranta V.. Induction of cell migration by matrix metalloprotease-2 cleavage of laminin-5. Science. 1997;2775323:225–228. [DOI] [PubMed] [Google Scholar]
- 36. Zhou W, Jirström K, Johansson C, et al. Long-term survival of women with basal-like ductal carcinoma in situ of the breast: a population-based cohort study. BMC Cancer. 2010;10:653.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhou W, Johansson C, Jirström K, et al. A comparison of tumor biology in primary ductal carcinoma in situ recurring as invasive carcinoma versus a new in situ. Int J Breast Cancer. 2013;2013:1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Karlsson E, Sandelin K, Appelgren J, et al. Clonal alteration of breast cancer receptors between primary ductal carcinoma in situ (DCIS) and corresponding local events. Eur J Cancer. 2014;503:517–524. [DOI] [PubMed] [Google Scholar]
- 39. Gabrielson M, Eriksson M, Hammarström M, et al. Cohort profile: the Karolinska Mammography Project for Risk Prediction of Breast Cancer (KARMA). Int J Epidemiol. 2017; 466:1740–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ma X-J, Dahiya S, Richardson E, Erlander M, Sgroi DC.. Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res BCR. 2009;111:R7.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ.. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics. 2009;101:161.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mizushima H, Koshikawa N, Moriyama K, et al. Wide distribution of laminin-5 gamma 2 chain in basement membranes of various human tissues. Horm Res. 1998;50(suppl 2):7–14. [DOI] [PubMed] [Google Scholar]
- 43. Andersson ER, Lendahl U.. Therapeutic modulation of Notch signalling—are we there yet? Nat Rev Drug Discov. 2014;135:357–378. [DOI] [PubMed] [Google Scholar]
- 44. Pietras K, Rubin K, Sjöblom T, et al. Inhibition of PDGF receptor signaling in tumor stroma enhances antitumor effect of chemotherapy. Cancer Res. 2002;6219:5476–5484. [PubMed] [Google Scholar]
- 45. Furuhashi M, Sjöblom T, Abramsson A, et al. Platelet-derived growth factor production by B16 melanoma cells leads to increased pericyte abundance in tumors and an associated increase in tumor growth rate. Cancer Res. 2004;648:2725–2733. [DOI] [PubMed] [Google Scholar]
- 46. Jin S, Hansson EM, Tikka S, et al. Notch signaling regulates platelet-derived growth factor receptor-beta expression in vascular smooth muscle cells. Circ Res. 2008;10212:1483–1491. [DOI] [PubMed] [Google Scholar]
- 47. Rajan PB, Perry RH.. A quantitative study of patterns of basement membrane in ductal carcinoma in situ (DCIS) of the breast. Breast J. 1995;15:315–321. [Google Scholar]
- 48. Albrechtsen R, Nielsen M, Wewer U, Engvall E, Ruoslahti E.. Basement membrane changes in breast cancer detected by immunohistochemical staining for laminin. Cancer Res. 1981;41(12)(pt 1):5076–5081. [PubMed] [Google Scholar]
- 49. Blaise R, Mahjoub M, Salvat C, et al. Involvement of the Notch pathway in the regulation of matrix metalloproteinase 13 and the dedifferentiation of articular chondrocytes in murine cartilage. Arthritis Rheum. 2009;602:428–439. [DOI] [PubMed] [Google Scholar]
- 50. Funahashi Y, Shawber CJ, Sharma A, Kanamaru E, Choi YK, Kitajewski J.. Notch modulates VEGF action in endothelial cells by inducing Matrix Metalloprotease activity. Vasc Cell. 2011;31:2.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liu H, Kennard S, Lilly B.. NOTCH3 expression is induced in mural cells through an autoregulatory loop that requires endothelial-expressed JAGGED1. Circ Res. 2009;1044:466–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cohen M, Georgiou M, Stevenson NL, Miodownik M, Baum B.. Dynamic filopodia transmit intermittent Delta-Notch signaling to drive pattern refinement during lateral inhibition. Dev Cell. 2010;191:78–89. [DOI] [PubMed] [Google Scholar]
- 53. Sheldon H, Heikamp E, Turley H, et al. New mechanism for Notch signaling to endothelium at a distance by Delta-like 4 incorporation into exosomes. Blood. 2010;11613:2385–2394. [DOI] [PubMed] [Google Scholar]
- 54. Boelens MC, Wu TJ, Nabet BY, et al. Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways. Cell. 2014;1593:499–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ostman A, Augsten M.. Cancer-associated fibroblasts and tumor growth—bystanders turning into key players. Curr Opin Genet Dev. 2009;191:67–73. [DOI] [PubMed] [Google Scholar]
- 56. Previs RA, Coleman RL, Harris AL, Sood AK.. Molecular pathways: translational and therapeutic implications of the Notch signaling pathway in cancer. Clin Cancer Res. 2015;215:955–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Procopio M-G, Laszlo C, Al Labban D, et al. Combined CSL and p53 downregulation promotes cancer-associated fibroblast activation. Nat Cell Biol. 2015;179:1193–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Braune E-B, Tsoi YL, Phoon YP, et al. Loss of CSL unlocks a hypoxic response and enhanced tumor growth potential in breast cancer cells. Stem Cell Rep. 2016;65:643–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.