Abstract
Motivation
Interest in text mining full-text biomedical research articles is growing. To facilitate automated processing of nearly 3 million full-text articles (in PubMed Central® Open Access and Author Manuscript subsets) and to improve interoperability, we convert these articles to BioC, a community-driven simple data structure in either XML or JavaScript Object Notation format for conveniently sharing text and annotations.
Results
The resultant articles can be downloaded via both File Transfer Protocol for bulk access and a Web API for updates or a more focused collection. Since the availability of the Web API in 2017, our BioC collection has been widely used by the research community.
Availability and implementation
https://www.ncbi.nlm.nih.gov/research/bionlp/APIs/BioC-PMC/.
1 Introduction
Text mining of the full-text biomedical research literature is growing in importance (Bada et al., 2012; Cejuela et al., 2014; Czarnecki and Shepherd, 2014; Gyori et al., 2017; Islamaj Dogan et al., 2014; Kim et al., 2015). There is a significant amount of information available in the full text that is not available in the abstract (Tudor et al., 2015; Van Landeghem et al., 2013; Westergaard et al., 2018). Even when the major conclusion or discovery appears in the abstract, the data needed by curators to confirm or verify that discovery appears in the full text (Van Auken et al., 2014). Often that confirmation appears in figures or figure captions (Islamaj Dogan et al., 2017; Liechti et al., 2017).
This interest demonstrates the desire and need for increased access to full text for text mining. PubMed Central® (PMC), built and maintained by the US National Library of Medicine®, is a collection of biomedical research literature available to read on the web. The PMC Open Access Subset is a well-known portion of the PMC articles under a Creative Commons or similar license that allows more liberal reuse than traditional copyright (https://www.ncbi.nlm.nih.gov/pmc/tools/openftlist/). Less well-known is the Author Manuscript Collection. These articles have been made available in compliance with the NIH Public Access Policy or similar policies of other funders (https://www.ncbi.nlm.nih.gov/pmc/about/mscollection/). Figure 1 shows that the proportion of PMC articles available for text and data mining (in the Open Access Subset and the Author Manuscript Collection) is steadily growing (∼55% overall and over 80% in recent years).
Fig. 1.
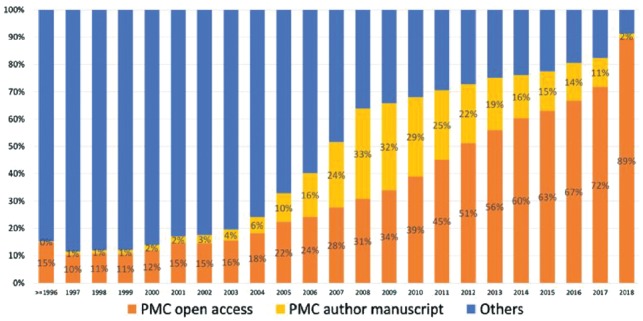
Proportion of PMC articles published each year available in the Open Access Subset and the Author Manuscript Collection, as of the end of December 2018. (The Others portion of the 2018 bar is smaller than might be expected because of the 1-year embargo period enforced upon some publications in this category.)
PMC articles are encoded using the Journal Article Tag Suite (JATS) (https://jats.nlm.nih.gov). This is a powerful, but complicated system that tracks all available meta-information and allows articles to be displayed in a manner very similar to their appearance in a printed journal. One sign of this flexible system is that 277 XML elements are defined. But JATS is not designed to aid text processing. Text appears at different levels and in various structures. The JATS-XML includes display markup, which while visually informative, is another complication to be addressed when text mining.
The BioCreative community has previously developed BioC, which is a simple, straightforward data structure for dealing with text and annotations in order to achieve interoperability (Comeau et al., 2013). Unlike some other formats such as PubAnnotation (Kim and Wang, 2012) and BRAT (http://brat.nlplab.org/index.html), BioC easily handles documents of various lengths, whether it is an abstract or full-text article. Even with additions for annotations and relations, BioC XML only uses 15 elements. Even more valuable, for BioC XML there are dedicated libraries that populate and preserve native BioC classes, or data structures, in a number of different languages (C++, Go, Java, Python, Perl, Ruby) (Comeau et al., 2014; Liu et al., 2014). With these libraries, no knowledge of internal or external XML format is required. As a result, text and annotations can be painlessly shared between multiple tools.
An original PMC article is encoded in UTF-8 Unicode. Our Web API provides both this original Unicode encoding, as well as an ASCII version, to facilitate tool interoperability. Many tools were developed before Unicode was widely used. Other tools can handle UTF-8 encoded texts with an occasional accented character, but do not address the many space characters, or the rich variety of punctuation marks. By treating accented and unaccented versions of the same character identically, ASCII provides convenient effective synonymy. Random access to Unicode code points requires more memory/time. For these reasons, articles are available in ASCII, using a Unicode to ASCII translation that we have found useful and convenient (http://bioc.sourceforge.net). However, if your tool chain can handle Unicode well, that is recommended.
In recent years, interest in JavaScript Object Notation (JSON) APIs is more than four times higher than XML APIs, as shown by Google Trends (https://trends.google.com/trends/explore?date=all&q=json%20api,xml%20api). Because of this increase in popularity, BioC articles are now available in JSON, in addition to XML.
When PMC Open Access articles were first made available in BioC, they were only available as large File Transfer Protocol (FTP) files. This is convenient for obtaining a large number of articles. As a complement to this FTP access, a RESTful web service is now available, making it possible to download exactly the articles needed. This is much more convenient for small collections or for updating an existing large collection.
When all of these reasons are taken together, there are multiple advantages to text miners to using BioC over PMC XML.
2 Materials and methods
PMC XML files are converted to BioC XML format via an in-house program described by Islamaj Dogan et al. (2014). The PMC XML files are obtained from the PMC Open Access Subset (https://www.ncbi.nlm.nih.gov/pmc/tools/openftlist/). Now articles are also obtained from the 500 000+ members of the Author Manuscript collection (https://www.ncbi.nlm.nih.gov/pmc/about/authorms/).
The original PMC XML files and thus the first set of BioC files are in Unicode. These Unicode files are converted to ASCII using a sample program released with the C++ BioC library (http://bioc.sourceforge.net). This tool uses a simple lookup table to replace Unicode characters with strings of zero or more ASCII characters. Both the source and lookup table are available.
The XML files from the previous two steps are converted to JSON using a BioC-JSON tool in Python (https://github.com/ncbi-nlp/BioC-JSON). The JSON data structure has the same BioC internal data structures as used by BioC libraries, except stored in JSON. That means objects are typically available as dictionaries, as is expected for JSON. The output of JSON parsers is usually convenient to work with, unlike an XML DOM. For this reason, in contrast to the multiple BioC XML libraries, only a Python BioC library is available to convert between JSON and internal BioC classes.
3 Results
As of the end of December 2018, there are over 2.8 million full-text articles from the two combined subsets in PMC: Open Access and Author Manuscript. These collections continue to grow. PMC Open Access articles were previously available in BioC XML through FTP (Islamaj Dogan et al., 2014). In this work, we significantly improve the utility and accessibility of this collection by a number of enrichments, including new Web APIs, an alternative JSON format and ASCII encoding and the inclusion of 500 000+ additional articles in Author Manuscript subset. All of these files are available via a Web API (https://www.ncbi.nlm.nih.gov/research/bionlp/APIs/BioC-PMC/). In addition to PMC, the entire set of 29 million PubMed articles is also available via our RESTful webservice (https://www.ncbi.nlm.nih.gov/research/bionlp/APIs/BioC-PubMed/).
PMC Open Access articles available via the API are updated daily. The Author Manuscript Collection is updated twice weekly. This is the pace that NCBI updates these collections. The BioC FTP files are updated quarterly. These articles are all available in BioC XML and BioC JSON. They are also available in both Unicode (UTF-8) and ASCII encodings.
BioC has been used in various studies such as corpus annotation and information extraction from literature in the past. In terms of API uses, there have been 11.6 million external calls since July 2017 where over 97.8% of the requests have been for the XML format. This is not surprising since the XML format has been available earlier in FTP files, so that is what most tools use. We expect that JSON will see more use as new tools start to use it. We also noticed about 66% of the requests are for ASCII versus 34% for Unicode. This shows the value of providing both encodings. Finally, section type identifiers have been added based on the labels and regular expressions found in Kafkas et al. (2015).
4 Summary
To date, more than 2.8 million PMC articles are freely available for text mining. This is a large and growing proportion of the total. These articles are provided in both BioC XML and BioC JSON formats that allow easy storing and sharing of annotations. They can be accessed via either a Web API or FTP. Unicode and ASCII encodings are both available. Convenient and growing programmatic access to full-text biomedical research articles will facilitate new developments and improvements in biomedical information retrieval and knowledge extraction, accelerating discovery.
Funding
This work was supported by the Intramural Research Program of the National Institutes of Health, National Library of Medicine.
Conflict of Interest: none declared.
References
- Bada M., et al. (2012) Concept annotation in the CRAFT corpus. BMC Bioinformatics, 13, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejuela J.M., et al. (2014) tagtog: interactive and text-mining-assisted annotation of gene mentions in PLOS full-text articles. Database (Oxford), 2014, bau033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeau D.C., et al. (2013) BioC: a minimalist approach to interoperability for biomedical text processing. Database (Oxford), 2013, bat064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeau D.C., et al. (2014) Natural language processing pipelines to annotate BioC collections with an application to the NCBI disease corpus. Database (Oxford), 2014, bau056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarnecki J., Shepherd A.J. (2014) Mining biological networks from full-text articles. Methods Mol. Biol., 1159, 135–145. [DOI] [PubMed] [Google Scholar]
- Gyori B.M., et al. (2017) From word models to executable models of signaling networks using automated assembly. Mol. Syst. Biol., 13, 954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islamaj Dogan R., et al. (2014) BioC and Simplified Use of the PMC Open Access Dataset for Biomedical Text Mining. In: Proceedings of the 4th Workshop on Building and Evaluating Resources for Health and Biomedical Text Processing. LREC, Reykjavik, Iceland, 2014. [Google Scholar]
- Islamaj Dogan R., et al. (2017) The BioC-BioGRID corpus: full text articles annotated for curation of protein-protein and genetic interactions. Database (Oxford), 2017, baw147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafkas S., et al. (2015) Section level search functionality in Europe PMC. J. Biomed. Semantics, 6, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.D., et al. (2015) Extending the evaluation of Genia Event task toward knowledge base construction and comparison to Gene Regulation Ontology task. BMC Bioinformatics, 16 (Suppl. 10), S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.D., Wang Y. (2012) PubAnnotation: a persistent and sharable corpus and annotation repository. In: Proceedings of the 2012 Workshop on Biomedical Natural Language Processing, pp. 202–205. [Google Scholar]
- Liechti R., et al. (2017) SourceData: a semantic platform for curating and searching figures. Nat. Methods, 14, 1021–1022. [DOI] [PubMed] [Google Scholar]
- Liu W., et al. (2014) BioC implementations in Go, Perl, Python and Ruby. Database (Oxford), 2014, bau059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudor C.O., et al. (2015) Construction of phosphorylation interaction networks by text mining of full-length articles using the eFIP system. Database (Oxford), 2015, bav020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Auken K., et al. (2014) BC4GO: a full-text corpus for the BioCreative IV GO task. Database (Oxford), 2014, bau074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Landeghem S., et al. (2013) Large-scale event extraction from literature with multi-level gene normalization. PLoS One, 8, e55814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergaard D., et al. (2018) A comprehensive and quantitative comparison of text-mining in 15 million full-text articles versus their corresponding abstracts. PLoS Comput. Biol., 14, e1005962. [DOI] [PMC free article] [PubMed] [Google Scholar]


