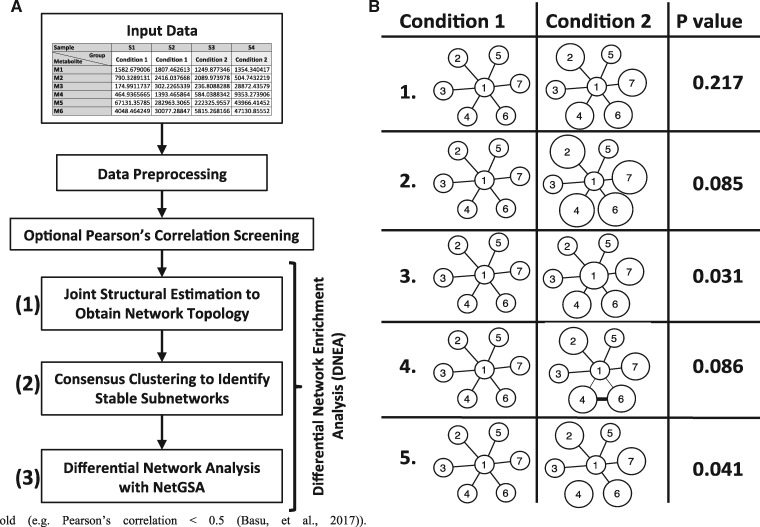
Fig. 1.
Differential Network Enrichment Analysis (DNEA) and Proof of Concept. (A) Workflow for DNEA. The workflow includes pre-processing of input data, followed by optional Pearson’s correlation. Main DNEA analysis includes joint structural network estimation, consensus clustering and differential network analysis. (B) NetGSA results using simulated data. The first three scenarios have DE nodes, whereas the last two scenarios have both DE nodes and differential edges