Abstract
Background
Ewing sarcoma (EWS) manifests one of the lowest somatic mutation rates of any cancer, leading to a scarcity of druggable mutations and neoantigens. Immunotherapeutics targeting differentially expressed cell surface antigens could provide therapeutic benefit for such tumors. Pregnancy-associated plasma protein A (PAPP-A) is a cell membrane-associated proteinase produced by the placenta that promotes fetal growth by inducing insulinlike growth factor (IGF) signaling.
Methods
By comparing RNA expression of cell surface proteins in EWS (n = 120) versus normal tissues (n = 42), we comprehensively characterized the surfaceome of EWS to identify highly differentially expressed molecules. Using CRISPR/Cas-9 and anti-PAPP-A antibodies, we investigated biological roles for PAPP-A in EWS in vitro and in vivo in NSG xenograft models and performed RNA-sequencing on PAPPA knockout clones (n = 5) and controls (n = 3). All statistical tests were two-sided.
Results
EWS surfaceome analysis identified 11 highly differentially overexpressed genes, with PAPPA ranking second in differential expression. In EWS cell lines, genetic knockout of PAPPA and treatment with anti-PAPP-A antibodies revealed an essential survival role by regulating local IGF-1 bioavailability. MAb-mediated PAPPA inhibition diminished EWS growth in orthotopic xenografts (leg area mm2 at day 49 IgG2a control (CTRL) [n = 14], mean = 397.0, SD = 86.1 vs anti-PAPP-A [n = 14], mean = 311.7, SD = 155.0; P = .03; median OS anti-PAPP-A = 52.5 days, 95% CI = 46.0 to 63.0 days vs IgG2a = 45.0 days, 95% CI = 42.0 to 52.0 days; P = .02) and improved the efficacy of anti-IGF-1R treatment (leg area mm2 at day 49 anti-PAPP-A + anti-IGF-1R [n = 15], mean = 217.9, SD = 148.5 vs IgG2a-CTRL; P < .001; median OS anti-PAPP-A + anti-IGF1R = 63.0 days, 95% CI = 52.0 to 67.0 days vs IgG2a-CTRL; P < .001). Unexpectedly, PAPPA knockout in EWS cell lines induced interferon (IFN)-response genes, including proteins associated with antigen processing/presentation. Consistently, gene expression profiles in PAPPA-low EWS tumors were enriched for immune response pathways.
Conclusion
This work provides a comprehensive characterization of the surfaceome of EWS, credentials PAPP-A as a highly differentially expressed therapeutic target, and discovers a novel link between IGF-1 signaling and immune evasion in cancer, thus implicating shared mechanisms of immune evasion between EWS and the placenta.
Overall survival for patients with nonmetastatic Ewing sarcoma (EWS) has plateaued at approximately 70%, and less than 30% of patients with metastatic EWS are long-term survivors (1). Standard therapy incorporates dose-intensive cytotoxic chemotherapy, resulting in substantial risks of lifelong late effects, including a high rate of second malignant neoplasms (2,3). Decades of research have enhanced understanding of EWS biology and its oncogenic drivers (4,5), but these insights have not yet generated effective new therapies (6–8).
EWS shows one of the lowest rates of somatic mutations of any cancer (9,10), resulting in a dearth of druggable kinase mutations and a low rate of somatic, nonsynonymous mutations that could serve as neoantigens for checkpoint-based immunotherapies (11). Immunotherapeutics targeting differentially expressed surface antigens (12) can be effective against such tumors, but the surfaceome of EWS has been incompletely catalogued (13,14). We identify differentially expressed cell surface antigens in EWS, including pregnancy-associated plasma protein-A (PAPP-A), a molecule expressed at the maternal-fetal interface where it promotes fetal growth by inducing high levels of free insulinlike growth factor 1 (IGF-1) (15). In EWS, PAPP-A mediates essential growth pathways and unexpectedly regulates expression of HLA molecules and other genes involved in antigen processing/presentation. Together these findings identify previously unrecognized commonalities between the placenta and EWS, identify PAPP-A as a potential target for novel therapeutics in EWS, and provide the first evidence that IGF-1 signaling contributes to immune evasion.
Methods
All standard assays not detailed here are described in the Supplementary Methods (available online).
Tissue Processing and RNA-Seq of Pediatric Solid Tumors and Normal Tissue Samples and Differential Gene Expression Analysis in Ewing Sarcoma
Tumors were prepared and sequenced as previously described (10). The Cancer Genome Atlas (TCGA) RNAseq dataset was queried from the ISB Cancer Genomics Cloud Pilot (ISB-CGC). Gene expression in transcripts per million (TPM) of PAPPA was obtained for all 10 661 cancer samples. For further details, see Supplementary Methods (available online).
Cell Lines and In Vitro Proliferation Assays
Cell lines (NCI-EWS-925, NCI-EWS-95, NCI-EWS-021, NCI-EWS-981, NCI-EWS-022, TC71, 5838, EW8, TC32, RD-ES, 6647, CHLA258, A673, SKNMC, Kelly, and 293T) were cultured according to standard protocols as previously described (16). Proliferation assays utilized an IncuCyte Live Cell Analysis System (IncuCyte ZOOM, Essen Bioscience, Ann Arbor, MI, USA). For further details, see Supplementary Methods (available online).
In Vivo Experiments
All procedures were approved by the Animal Care and Use Committee of the NCI and Stanford University. Male 6- to 8-weeks-old NSG mice (NOD.Cg-Prkdcscid ILrgtm1Wjl/SzJ, Jackson Laborator, Bar Harbor, ME) were xenografted with 2 x 106 EW8 cells into the right M. gastrocnemius (4–5 mice/group) and received weekly intraperitoneal (IP) injections with 800 μg/mouse anti-PAPP-A 1/41 (AnshLabs, Webster, TX), IgG2a-control (CTRL) (#BE0085, BioXCell, West Lebanon, NH), 333 μg/mouse anti-IGF1R h7C10 (Merck, Kenilworth, NJ), PBS/human IgG1-CTRL (#C0001, Crown Bioscience, Santa Clara, CA), or both starting 3–4 days after tumor engraftment. Combined data of n = 3 individual experiments presented (n = 14–15 mice/group).
RNAsequencing of PAPPA-Targeted Knockout Clones
RNA extraction was performed using the RNeasy-Plus Mini Kit (Qiagen, Germantown, MD, USA). Subsequent processing and RNA sequencing analysis was carried out at the Stanford Functional Genomics Facility using standard methodology further specified in the Supplementary Methods (available online).
Statistical Analyses
All statistical tests were two-sided and P < .05 was considered statistically significant. TPM gene expression in tumors (n = 120) and normal tissues (n = 42) were log-transformed and standardized (Z-score) to calculate differences in gene expression between the two groups, and EdgeR was used to generate a list of differentially expressed genes. Differences in PAPP-A levels in patients and HLA-B/HLA-C expression in EWS/normal were calculated by Wilcoxon-Mann-Whitney U test. Pearson’s correlation was used for correlation analysis between two groups. Kaplan–Meier survival curves of vivo experiments were analyzed using log-rank. Effects on tumor growth curves in vitro and in vivo were calculated using two-way repeated measures analysis of variance (RM-ANOVA). P values of enrichment analysis were calculated using gene set enrichment analysis (GSEA) software. For all other experiments, a student’s t-test was used. Figures represent three replicates unless otherwise stated. For additional details, see Supplementary Methods (available online).
Results
In Silico Identification of Differentially Expressed Cell Surface Antigens in Ewing Sarcoma
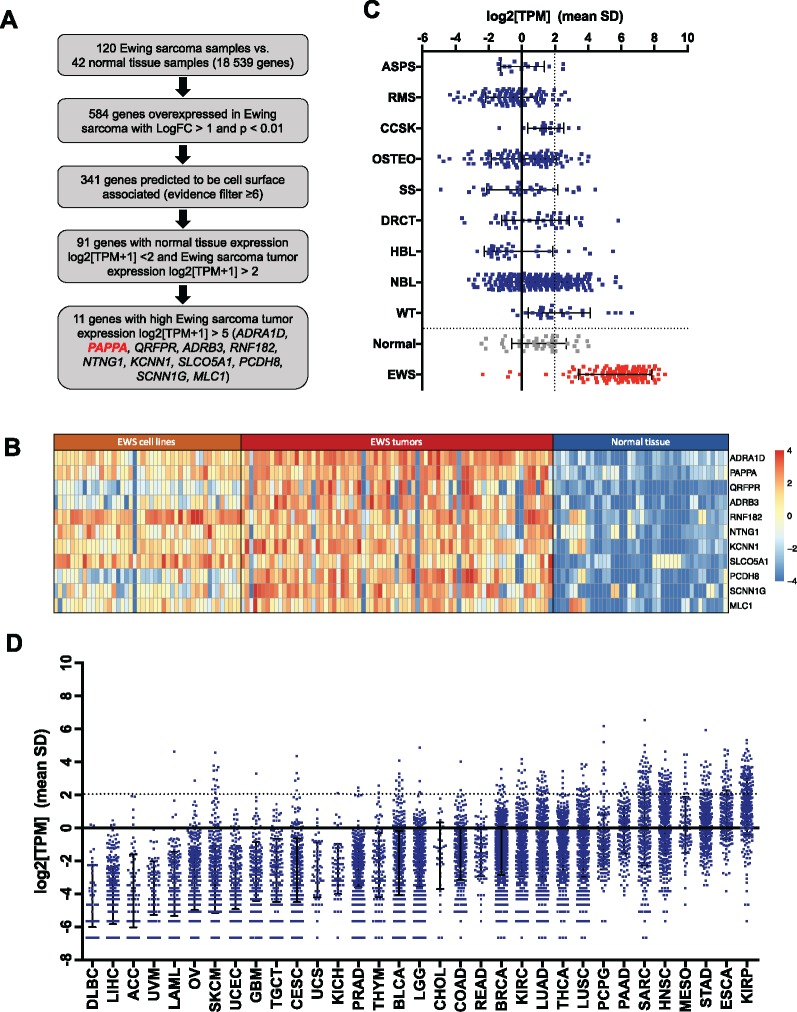
To identify cell surface proteins with potential to serve as immunotherapeutic targets for EWS (Figure 1A), RNA-sequencing data from 120 EWS samples (10) was compared to data from 42 normal tissue samples. This identified 584 genes overexpressed in EWS (LogFC [fold-change] > 1, P < .01), including numerous genes reported previously, such as CCND1 (17), CD99 (18), ALK (19), and STEAP1 (14). Using gene ontology and 11 cell-surface annotation datasets, we identified 341 genes predicted to be cell surface-associated. Excluding genes with substantial normal tissue, expression (AVElog2[TPM + 1] > 2) and insufficient expression in EWS (AVElog2[TPM + 1] < 2) ultimately identified 91 cell surface genes overexpressed on EWS compared to normal tissues (Supplementary Table 1, available online).
Figure 1.
In silico analysis of highly differentially expressed cell surface antigens in Ewing sarcoma compared to normal tissue. A) Prioritization strategy applied to RNAsequencing data to identify cell surface proteins overexpressed in Ewing sarcoma (EWS) compared to normal tissue. B) Heatmap of the 11 genes most differentially expressed in EWS (log2[TPM+1]>5) compared to normal tissue samples (log2[TPM+1]<2) ranked by P value, calculated using EdgeR. C) RNA PAPPA expression in primary tumor samples of children with alveolar soft part sarcoma (ASPS, n = 16), rhabdomyosarcoma (RMS, n = 99), desmoplastic round cell tumor (DRCT, n = 40), clear cell sarcoma of the kidney (CCSK, n = 16), synovial sarcoma (SS, n=31), hepatoblastoma (HBL, n= 30), Wilms’ tumor (WT, n = 27), osteosarcoma (OSTEO, n = 95), neuroblastoma (NBL, n = 206), normal tissue (n = 42), and EWS (n = 75) and EWS cell lines (n = 45). D) RNA PAPPA expression in a panel of adult tumors queried from the Cancer Genome Atlas (TCGA). Definition of tumor type abbreviations are provided in Supplementary Table 2 (available online). TPM = transcripts per million.
Among the top 11 genes with very high differential expression in EWS (AVElog2[TPM + 1] > 5) shown in Figure 1B, PAPPA was ranked second. PAPPA showed absent/very low expression in the vast majority of normal tissues (mean ± SD log2[TPM] = 1.04 ± 1.64, n = 42) (Figure 1C), which was validated in the Human Protein Atlas (www.proteinatlas.org) (20) (Supplementary Figure 1A, available online), demonstrating predominant PAPPA expression in the placenta. Two additional datasets from the Pediatric cBioPortal for Integrated Childhood Cancer Genomics (www.pedcbioportal.org) validated the differential PAPPA expression (Supplementary Figure 1B, available online).
PAPPA gene expression in non-EWS pediatric primary solid tumor samples is shown in Figure 1C. Using a cutoff for high PAPPA expression of log2(TPM) > 2, 95.0% EWS samples overexpressed PAPPA, whereas 5.1%–40.7% of other pediatric tumors showed high PAPPA expression. PAPP-A has been implicated in mediating oncogenic effects in adult cancers (21,22), thus we also evaluated PAPPA gene expression in adult malignancies and matched normal tissue samples from TCGA. Among 33 adult histotypes analyzed (TU n = 10 023/N n = 728, Figure 1D, Supplementary Table 2, available online), the highest and most consistent PAPPA expression was observed in renal papillary cell carcinoma. This dataset also confirmed low/absent PAPPA expression in all normal tissues with the exception of low PAPPA expression in matched normal kidney (Supplementary Table 2, available online). In summary, PAPPA is overexpressed in a substantial fraction of cancers, both pediatric and adult, but the frequency and level of PAPPA overexpression is higher in EWS than any other histologic subtype analyzed.
PAPP-A-mediated cleavage of IGF-1 from its binding proteins is essential for fetal and postnatal growth, and IGF-1 has been identified as a critical growth factor in EWS. Analysis of IGF signaling components in the EWS RNAseq dataset revealed greater expression of IGF-1 compared to IGF-2 (P = .004) (Supplementary Figure 1C, available online), as well as expression of IGF-1R, IR-A (the predominant splice variant of the insulin receptor in fetal tissues/cancer [23]), IGF-BP2–6, and stanniocalcin 1 and 2 (STC1/STC2), which are purported to play regulatory roles in IGF signaling (24,25). Unlike the placenta, EWS tumors do not express proMBP (PRG2), the major circulating binding partner of PAPP-A in pregnancy, which inhibits PAPP-A function (26,27).
EWS-FLI1-driven PAPPA expression has recently been validated (28). Using published datasets, binding of EWS-FLI1 to the PAPPA promoter (29) was confirmed in A673 and SKNMC cells (Supplementary Figure 1D, available online), and EWS-FLI1 silencing decreased gene expression of PAPPA (Supplementary Figure 1E, available online). Together, these data demonstrate that EWS tumors express all components necessary to sustain a local, autocrine IGF signaling loop and that the oncogenic driver in EWS induces expression of PAPPA.
Analysis of PAPP-A Expression within the EWS Tumor Microenvironment and in Plasma from EWS Patients
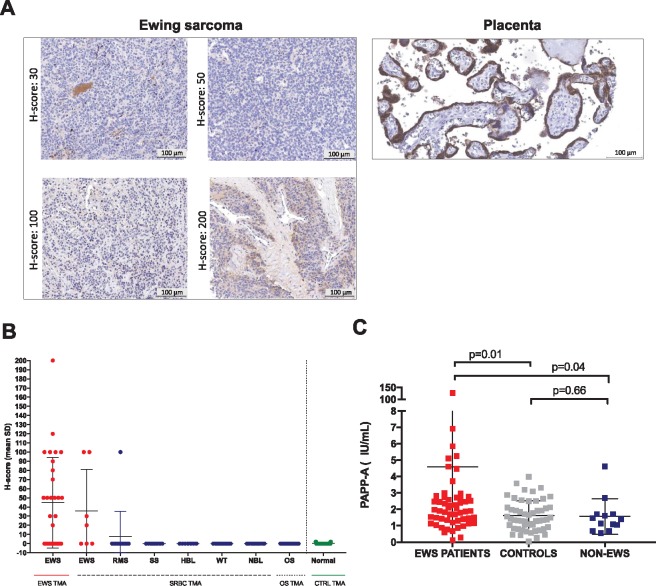
Immunohistochemistry (IHC) of pediatric solid tumor tissue microarrays (TMAs), placenta, and normal tissues (Figure 2,A and B, Supplementary Figure 2, A and B, available online) revealed that 57.1% (4/7) of EWS tumors on a SRBC TMA and 60.0% (18/30) of EWS samples on another TMA demonstrated anti-PAPP-A immunoreactivity (H-score 20–200). Overall, a subset of 21.6% (8/37) of EWS tumors demonstrated H-scores of greater than or equal to 100. Numerous other pediatric tumor histotypes, including synovial sarcoma, hepatoblastoma, Wilms' tumor, osteosarcoma, and neuroblastoma, were negative, and only 1/13 rhabdomyosarcoma samples tested positive. No PAPP-A staining was observed in normal tissues (Figure 2B, Supplementary Figure 2A, available online), including the kidney, except very weak staining in 1/3 tonsil samples (2% of the cells, intensity = 1, H-score 2) and strong staining in placental syncytiotrophoblasts (100% of cells, intensity = 3, H-score 300). Although we cannot rule out differences in the sensitivity of detection between IHC and RNAseq, our results show that PAPP-A protein expression is more restricted than RNA expression and confirm protein expression of PAPP-A in EWS tumors.
Figure 2.
Analysis of pregnancy-associated plasma protein A (PAPP-A) expression within the Ewing sarcoma (EWS) tumor microenvironment and circulating PAPP-A levels EWS patients. A) Representative immunohistochemistry (IHC) images of PAPP-A expression in the placenta and EWS tissue microarray samples. Scale bars represent 100 µm. B) IHC H-score values of pediatric tissue microarrays analyzed for PAPP-A via IHC. C) Circulating PAPP-A levels in EWS patients (n = 56), healthy control donors (n = 50), and non-EWS sarcoma patients (n = 13). Levels represent mean ± SD, P value according to two-sided Wilcoxon-Mann-Whitney U test. Definition of tumor type abbreviations are provided in Supplementary Table 2 (available online). TMA = tissue microarray, EWS = Ewing sarcoma; RMS = Rhabdomyosarcoma; SS = Synovial Sarcoma; HBL = Hepatoblastoma; WT = Wilms' Tumor; NBL = Neuroblastoma; OS = Osteosarcoma.
Circulating PAPP-A from 56 EWS patients (Supplementary Table 3, Supplementary Figure 2C, available online) and 53 healthy controls (Figure 2C) demonstrated that PAPP-A levels were increased in EWS patients compared to controls (EWS mean [SD]= 4.6[16.7] vs CTRL mean[SD] = 1.6[0.9]; P = .01) and compared to patients with non-EWS sarcomas (non-EWS mean[SD] = 1.6[1.1]; EWS vs non-EWS; P = .04), despite overlap between the groups. A total of 16.1% EWS patients demonstrated PAPP-A levels above the 95% reference range of the controls. The relatively low levels of circulating PAPP-A in EWS compared to pregnancy (30) likely derives from diminished expression in EWS compared to the placenta as well as the absence of proform of eosinophil major basic protein (proMBP) (no PRG2 gene expression, Supplementary Figure 1C, available online) (26,27,31) and support a model wherein EWS-derived PAPP-A is primarily localized to the tumor microenvironment.
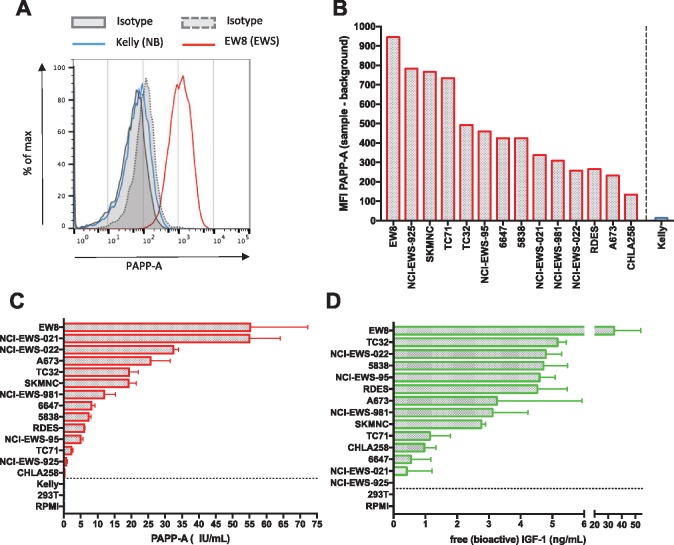
Analysis of Proteolytic Activity of PAPP-A and the Effect on IGF-1 Signaling and Growth in EWS Cell Lines
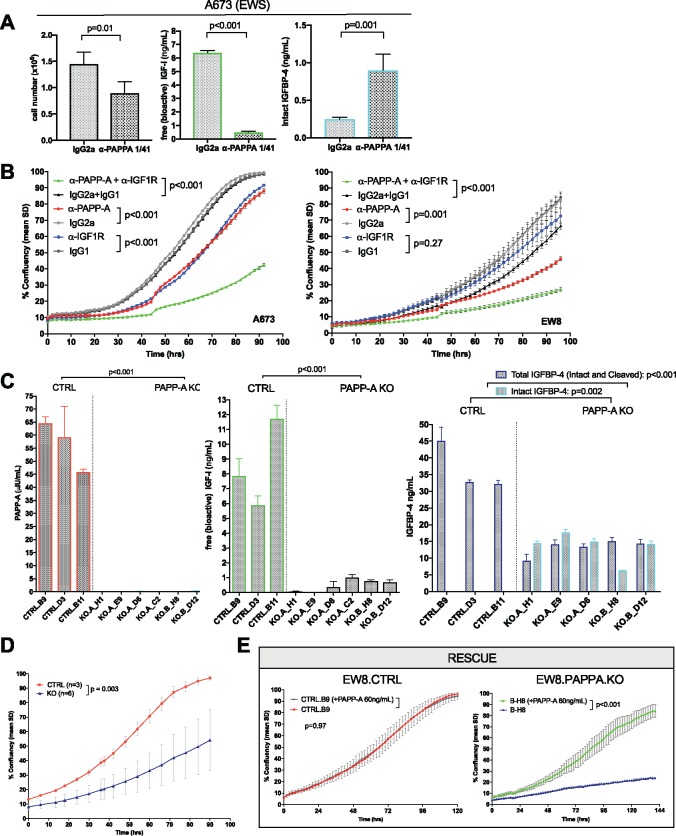
PAPP-A was present on the surface of 14 EWS cell lines analyzed by flow cytometry (Figure 3, A and B), and proteolytically active PAPP-A was secreted by all EWS cell lines examined (Figure 3C). Free, bioactive IGF-1 was detected in the supernatant of 13/14 EWS cell lines (Figure 3D), and PAPP-A levels correlated with the abundance of free, bioactive IGF-1 (Pearson 0.574, P = .03). To evaluate whether PAPP-A plays a functional role in autocrine IGF signaling in EWS, we used a monoclonal antibody (mAb 1/41) to inhibit its proteolytic activity (32). Anti-PAPP-A decreased proliferation of A673 EWS cells; diminished levels of free, bioactive IGF-1; and increased intact IGBP-4, indicating complexed IGFs (Figure 4A). MAb-mediated inhibition of PAPP-A proteolytic activity also induced antiproliferative effects in EW8 cells (measured as % confluency upon treatment), which was statistically significantly enhanced upon combination with anti-IGF-1R mAb h7C10 (endpoint: anti-PAPP-A mean[SD] = 46.1[3.1]% vs IgG2a mean[SD] = 82.8[11.5]%; P = .001; anti-IGF-1R mean[SD] = 72.6[11.4]% vs IgG1 mean[SD] = 84.1[6.6]%; P = .27; anti-PAPP-A + anti-IGF-1R mean[SD] = 27.1[3.0]% vs IgG2a + IgG mean[SD] = 66.5[6.0]%; P < .001 according to two-sided two-way RM-ANOVA). Similar results were observed for the A673 cell line (endpoint: anti-PAPP-A mean[SD] = 88.1[3.30]% vs IgG2a mean[SD] = 99.5[0.5]%; P < .001 according to two-sided two-way RM-ANOVA; anti-IGF-1R mean[SD] = 91.6[1.4]% vs IgG1 mean[SD] = 98.4[1.8]%; P < .001 according to two-sided two-way RM-ANOVA; anti-PAPP-A + anti-IGF-1R mean[SD] = 42.6[2.4]% vs IgG2a + IgG mean[SD] = 98.6[0.9]%; P < .001 according to two-sided two-way RM-ANOVA) (Figure 4B).
Figure 3.
Analysis of proteolytically active pregnancy-associated plasma protein A (PAPP-A) expression in Ewing sarcoma (EWS) cell lines. A) PAPP-A expression in a representative EWS cell line (EW8), which is absent on a representative neuroblastoma cell line (Kelly). B) Range of PAPP-A cell surface antigen density as measured by mean fluorescent intensity (MFI) in a panel of EWS cell lines. C, D) PAPPA levels and free, bioactive insulinlike growth factor 1 (IGF-1) measured in cell culture supernatant obtained from a panel of EWS cell lines vs non-EWS cell lines (separated by dotted line). Values represent mean ± SD. Results are representative of three individual experiments.
Figure 4.
Role of pregnancy-associated plasma protein A (PAPP-A) on growth of Ewing sarcoma (EWS) cell lines ex vivo. A) Impact of neutralizing anti-PAPPA mAb (1/41 or isotype control (CTRL) IgG2a) (20 µg/mL) on cell number, free (bioactive) insulinlike growth factor 1 (IGF-1), total IGFBP-4, and intact (complexed) IGFBP-4. Levels represent mean ± SD, 4 replicates/condition, representative of n = 3 experiments. P value according to two-sided Student’s t-test. B) Impact of neutralizing anti-PAPP-A (mAb 1/41) and/or anti-IGF-1R (mAb h7C10) or isotype controls on cell growth over time as measured by confluency in an IncuCyte ZOOM Live Cell Analysis System (6 replicates/condition, representative of n= 3 experiments), values represent mean % confluency ± SD, P value according to two-sided two-way repeated measures analysis of variance (RM-ANOVA). C) Cell culture supernatants (24 h incubation, 100 000 cells/well) of PAPPA targeted CRISPR/Cas9 knockout and control clones were analyzed by ELISA for PAPP-A secretion, free (bioactive) IGF-1, and intact (complexed) IGFBP-4. Values represent mean ± SD. Representative of n = 3 experiments, 3 replicates per condition. P value according to two-sided Student’s t-test. D) Growth of EW8 in CTRL clones (n = 3) vs PAPPA.KO clones (n=6). E) Growth of CTRL vs PAPPA.KO clones upon culture +/- 60 ng/mL soluble PAPP-A (3 replicates/condition, representative of n = 3 experiments). Values represent mean % confluency± SD, P value according to two-sided two-way RM-ANOVA.
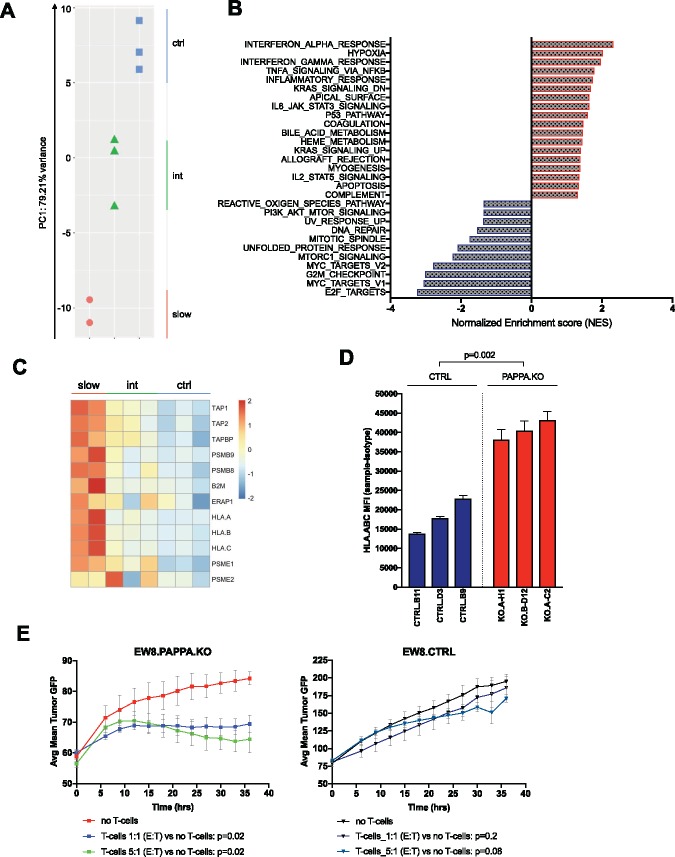
To better understand the role of PAPP-A in EWS, we utilized CRISPR/Cas9 to delete PAPPA in the EW8 cell line, creating EW8.PAPPA.KO clones, which demonstrated a complete absence of PAPP-A expression and secretion (Figure 4C). Supernatant from EW8.CTRL clones contained substantial levels of bioactive IGF-1, whereas levels were essentially absent in EW8.PAPPA.KO (ng/mL in control mean[SD] = 8.51[2.97] vs knockout mean[SD] = 0.48[0.42]; P < .001). Intact IGFBP-4 (IGFBP-4/IGF complexes) was present in supernatants from EW8.PAPPA.KO clones, but not from EW8.CTRLs (ng/mL in control mean = below limit of detection vs knockout mean[SD] = 13.6[4.26]; P = .002), confirming loss of PAPP-A proteolytic activity toward IGFBP-4 (Figure 4C). Because PAPPA gene editing could potentially lead to a truncated protein undetectable by ELISA, but still able to cleave IGFBP-5, absent metalloproteinase activity toward IGFBP-5 in EW8.PAPPA.KO cultures was confirmed in a proteinase cleavage assay using 125I-labeled IGFBP-5 (Supplementary Figure 3A, available online).
EW8.PAPPA.KO clones demonstrated statistically significantly diminished growth rates (assessed by mean % confluency) compared to control clones (control mean[SD] = 97.07[1.26]% vs knockout mean[SD] = 54.18[20.82]%; P = .003) (Figure 4D), albeit with heterogeneity (Supplementary Figure 3B, available online). Daily addition of 60 ng/mL of soluble PAPP-A had no effect on EWS.CTRL clones, but rescued growth of PAPPA.KO clones (Figure 4E, Supplementary Figure 3C, available online), confirming that diminished growth was due to PAPP-A deficiency. Together, these results identify an essential role for PAPP-A in cleaving IGF-1 from IGFBP-4/5 within the EWS microenvironment and demonstrate that PAPP-A is required to supply bioactive IGF-1, an essential growth factor in EWS.
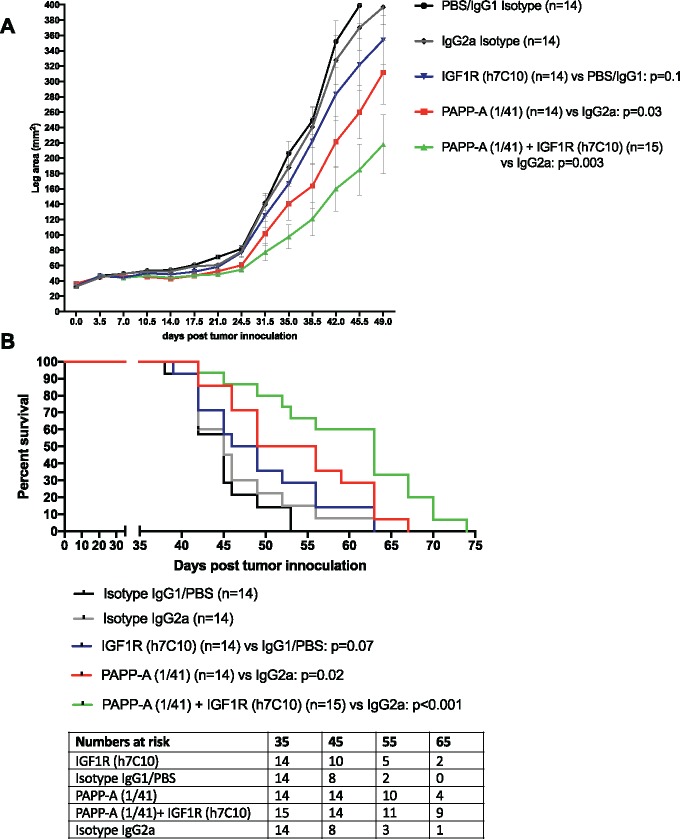
We next analyzed the effects of anti-PAPP-A mAb as single agent and in combination with anti-IGF-1R mAb in vivo, in an EW8 orthotopic xenograft model (Figure 5, A and B). Treatment with anti-IGF-1R alone had no effect but treatment with anti-PAPP-A statistically significantly delayed tumor growth (leg area mm2 at day 49 IgG2a-CTRL mean[SD] = 397.0[86.1] vs anti-PAPP-A mean[SD] = 311.7[155.0]; P = .03 according to two-sided two-way RM-ANOVA) in vivo and prolonged survival of mice (median OS anti-PAPP-A = 52.5 days, 95% CI = 46.0 to 63.0 days vs IgG2a = 45.0 days, 95% CI = 42.0 to 52.0 days; P = .02). This effect was enhanced when anti-PAPP-A was combined with anti-IGF-1R (leg area mm2 at day 49 anti-PAPP-A + anti-IGF-1R mean = 217.9, SD = 148.5 vs IgG2a-CTRL, P < .001; median OS anti-PAPP-A + anti-IGF-1R = 63.0 days, 95% CI = 52.0 to 67.0 days vs IgG2a-CTRL, P < .001).
Figure 5.
Effect of pregnancy-associated plasma protein A (PAPP-A) inhibition ± IGF-1R blockade on tumor growth and survival in a Ewing sarcoma (EWS) xenograft model. In vivo antitumor effects on tumor growth (A) and survival (B) following administration of anti-PAPP-A neutralization and/or anti-IGF-1R mAbs. Male 6- to 8-weeks-old NSG mice were xenografted with 2 x 106 EW8 cells into the left M. gastrocnemius. Mice received weekly intraperitoneal (IP) injections with 800 μg/mouse PAPP-A mAB 1/41, 333 μg/mouse anti-IGF1R h7C10, or both (n = 4–5/group/experiment), combined data of n = 3 shown. Treatment effect: P value according to two-way repeated measures analysis of variance (RM-ANOVA). Kaplan–Meier survival according to log-rank. All statistical tests were two-sided.
Effect of Disruption of IGF-1 Signaling via PAPP-A Inhibition on Immune Response Gene Expression and Antigen Presentation by EWS
To further investigate molecular pathways modulated by PAPP-A in EWS, we performed RNA sequencing on EW8.PAPPA.KO clones (n = 5) and EW8.CTRL clones (n = 3). Principal component analysis suggested that transcriptional profiles clustered based on the lack of PAPPA expression and proliferation rates (Figure 6A). GSEA to identify hallmark gene set collections altered upon PAPPA knockout (nominal P < .05) (33) (Figure 6B and Supplementary Table 4, available online) identified downregulation of IGF signaling, such as PI3K/Akt/mTOR and mTORC1 signaling, and cell cycle/proliferation, such as E2F targets, MYC targets, and G2M checkpoint, while an apoptosis transcriptional profile was enriched, consistent with well-described anti-apoptotic effects of IGF signaling (34–36).
Figure 6.
Effect of disruption of insulinlike growth factor 1 (IGF-1) signaling via pregnancy-associated plasma protein A (PAPP-A) inhibition on expression of immune response genes and immune recognition of Ewing sarcoma (EWS). A) Principal component analysis of PAPPA knockout clones and controls. B) Gene set enrichment analysis of RNAseq data in PAPPA knockout clones (n = 5) and CTRL clones (n = 3) according to Molecular Signatures Database (MSigDB). Y-axis reflects all hallmark pathways in which nominal P < .05. X-axis reflects normalized enrichment score. C) Heatmap of genes associated with antigen processing and presentation in PAPPA.KO and CTRL clones. D) Mean fluorescence intensity (MFI) of HLA.ABC expression on EW8.PAPPA.KO and CTRL clones assessed by flow cytometric analysis using a mAb recognizing a public, monomorphic epitope on HLA-A, HLA-B, and HLA-C. Values represent mean ± SD. Samples were run in 3 replicates/condition, representative of n = 3 experiments, P value according to two-sided Student’s t-test. E) Alloreactivity of activated T-cells against green fluorescent protein (GFP) expressing EW8.PAPPA.KO (clone B-D12) and CTRL (clone B11) cell lines assessed in an IncuCyte ZOOM live cell analysis system (3 replicates/condition, representative of n = 3 experiments). Values represent mean % confluency ± SD, P value according to two-sided two-way repeated measures analysis of variance (RM-ANOVA). int = intermediate.
Surprisingly, gene sets induced upon PAPPA knockout were largely immune related, such as complement system, allograft rejection, inflammatory response, acute-phase response (IL-6_JAK_STAT3), tumor necrosis factor (TNF)-α signaling, and interferon (IFN)-α/IFN-γ response. Enrichment of key genes in the antigen processing/presentation pathway, such as the peptide loading complex TAP-1/TAP-2/TAPBP, the proteasome components LMP2 (PSMB9) and LMP7 (PSMB8), proteasome activator complex subunits PSME1/PSME2, the endoplasmic reticulum aminopeptidase ERAP1, and in particular the subunits of MHC class I molecules β2-microglobulin (B2M) and classical MHC class I HLA-A, HLA-B, HLA-C was observed in PAPPA KO clones. Notably, the greatest enrichment occured in clones wherein PAPPA.KO had the greatest impact on cell growth (Figure 6C). These findings were confirmed by increased cell surface protein expression of HLA-ABC on EW8.PAPPA.KO clones (Figure 6D). To determine whether this led to an enhanced capacity to present antigens, we tested the killing capability of allogeneic T cells toward EW8 isogenic cell lines with varying PAPPA expression. As shown in Figure 6E, activated, alloreactive T cells demonstrated increased killing of EW8.PAPPA.KO compared to EW8.CTRL cell lines in vitro.
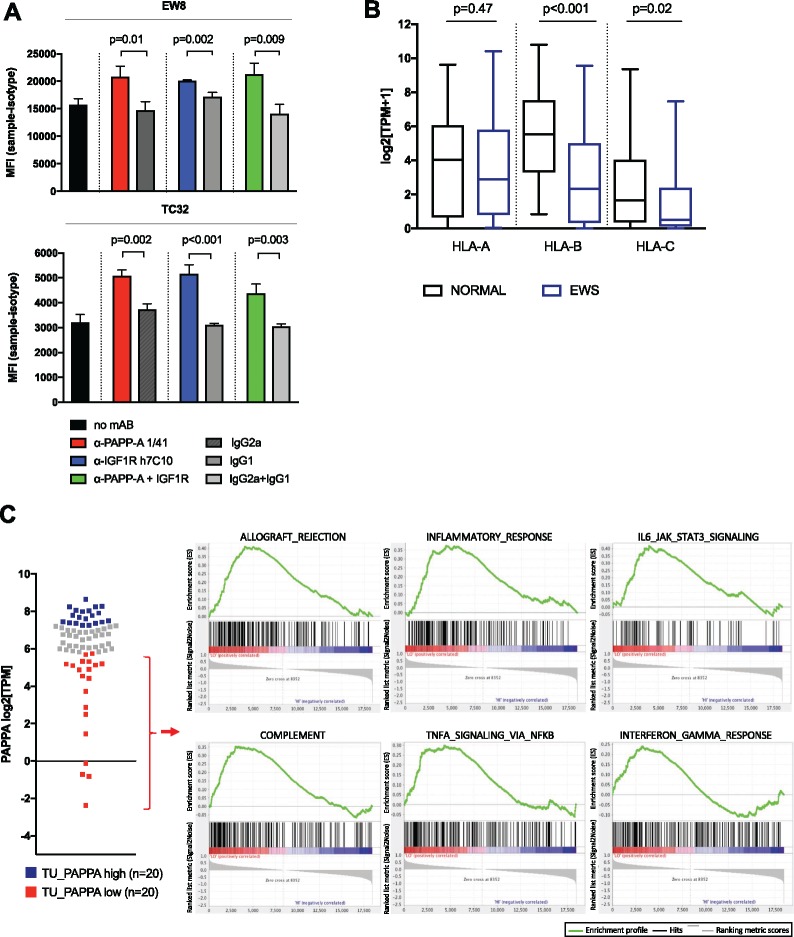
To confirm that these observations were a result of disrupting IGF-1 signaling, we assessed changes in HLA-I expression following treatment of two EWS cells lines with anti-PAPP-A and/or anti-IGF-1R mAbs. In response, both cell lines upregulated HLA-ABC molecules, as measured by mean fluorescence intensity (MFI) (Figure 7A). Together these results demonstrate that IGF-1 signaling and PAPPA expression in EWS is associated with changes in antigen processing/presentation that diminish the capacity for EWS to present antigens recognized by T cells.
Figure 7.
Effect of inhibition of insulinlike growth factor (IGF)-signaling on HLA expression in Ewing sarcoma (EWS) cell lines and association of diminished PAPPA expression with MHC Class I and immune response gene expression in EWS tumors. A) Mean fluorescence intensity (MFI) of HLA.ABC expression in EW8 and TC32 Ewing sarcoma cells treated with anti-PAPP-A 1/41, anti-IGF1R h7C10, both, or according controls. Values represent mean ± SD, samples and isotype control samples were run in 3 replicates/condition, and MFI of isotype samples was subtracted. Representative of n = 3 experiments, P value according to two-sided Student’s t-test. B) Classical HLA class I expression in EWS compared to normal tissue, boxplot of gene expression values of HLA-A, HLA-B, and HLA-C in primary EWS tumor samples (n = 75) and normal tissue (n = 42), P values according to two-sided Wilcoxon-Mann-Whitney U test. C) Gene set enrichment analysis of Hallmark pathways in primary Ewing sarcoma tumor samples comparing a cohort of lowest (n = 20) vs highest (n = 20) PAPPA expression.
Effect of Diminished PAPPA Expression on Immune Response Gene Expression and Immune Cell Infiltration in EWS Tumors
We assessed HLA-I expression in our RNA-sequencing dataset of primary EWS tumors and cell lines (n = 120). Compared to normal tissue samples (n = 42), MHC class I gene expression is downregulated for HLA-B and HLA-C (Figure 7B). To validate the link between expression of genes associated with immune response and PAPPA expression, we stratified our 75 EWS tumor samples into PAPPA-high (n = 20) and PAPPA-low (n = 20) and performed GSEA analysis using all genes interrogated (n = 18 539). Hallmark pathways associated with immune responses were upregulated in the cohort of EWS patient samples showing the lowest PAPPA expression (Figure 7C, Supplementary Table 5, available online), and these findings were validated using a previously published EWS RNA expression dataset (n = 117, GSE34620, Delattre et al., Supplementary Figure 3D, available online).
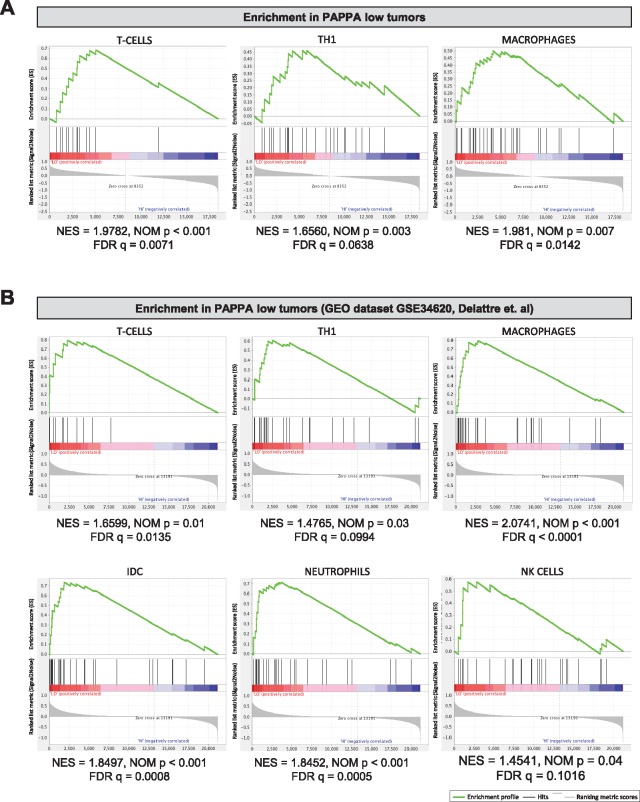
To investigate whether the increased expression of genes associated with immune responses in PAPPA-low tumors was associated with differences in tumor-infiltrating immune cells, we assessed the immune landscape in PAPPA low vs high EWS tumors by GSEA based on previously described immune cell gene expression signatures (37). Using a total of 16 immune cell signatures, we found enrichment for T cell, Th1, and macrophage signatures in PAPPA-low (n = 20) tumors, while no enrichment was seen in PAPPA-high (n = 20) tumors (Figure 8A). To validate these findings, we performed the same analysis in the GSE34620 dataset and, again, found enrichment of immune cell signatures in the PAPPA-low group (Figure 8B).
Figure 8.
Association of PAPPA expression in Ewing sarcoma (EWS) with the abundance of tumor-infiltrating immune cells. A) Gene set enrichment analysis of gene signatures of various immune cells based on Bindea et al. (38) in our RNAseq data of PAPPA-low (n=20) compared to PAPPA-high (n = 20) primary EWS tumors and in (B) the GEO dataset Delattre et al. (GSE34620). Normalized enrichment score (NES), FDR, and P values were calculated using gene set enrichment analysis (GSEA) software. FDR = false discovery rate; NOM p = Nominal P Value.
Discussion
Placental trophoblasts and cancer cells share numerous features, including expression of proto-oncogenes, growth factors, hormones, proteases, and tumor-associated antigens that enable rapid growth, tissue invasion, angiogenesis, and immune evasion (reviewed in [39–42]). We demonstrate that similar to the placenta, EWS expresses high levels of PAPP-A, as previously reported (14), and that PAPP-A expression in EWS ensures availability of free, bioactive IGF-1 to promote cell growth. Although there are clear distinctions between placental and EWS-associated PAPP-A, including higher levels of PAPP-A in placental trophoblasts and higher circulating PAPP-A levels during pregnancy, our results are consistent with the placenta and EWS sharing a common growth pathway. Furthermore, we discovered that PAPP-A, via augmentation of IGF-1 signaling, contributes to EWS immune evasion, because genetic deletion or neutralization of PAPP-A induces expression of interferon response genes and enhances HLA expression and immune recognition of EWS. These results raise the prospect that PAPP-A and IGF-1 signaling may serve a heretofore unappreciated role in placental and tumor immune tolerance.
The findings presented here have several potential implications for EWS therapy. Although IGF-1 signaling drives EWS growth (38), clinical trials using mAbs to inhibit IGF1R signaling demonstrated response rates of only 10%–14% (43–45). Our data demonstrate that combinatorial targeting of PAPP-A and IGF-1R was more effective in EWS, a result that may be explained by the fact that resistance of EWS to anti-IGF-1R mAbs can involve signaling of IGF-1 and/or IGF-2 via insulin receptor (IR)-A homodimers (46,47). Consistent with this, high IR expression correlates with resistance to anti-IGF-1R treatment in pediatric solid tumor models (48). PAPP-A inhibition limits the availability of ligands, which would also prevent signaling through IR-A. Nevertheless, inhibition of only proximal proteins will likely be insufficient for sustained clinical responses because of the development of bypass resistance, which will necessitate simultaneous inhibition of downstream signaling pathways (49).
We also provide a comprehensive analysis of the EWS surfaceome, identifying numerous differentially expressed surface proteins that could serve as targets for mAb-derived immunotherapeutics. The strikingly favorable tumor-to-normal tissue expression of PAPPA, and the fact that PAPP-A anchors to cell surface heparan sulfate proteoglycans (50,51), raises the prospect of EWS tumor-specific targeting via PAPP-A-directed radioconjugate antibodies, antibody drug conjugates, bispecific antibodies, or chimeric antigen receptors. Furthermore, a recent report demonstrated that a T cell receptor (TCR) targeting a peptide derived from PAPP-A expressed in the context of HLA-A2 mediated antitumor activity (52). Future studies are warranted to determine whether immunotherapeutics targeting PAPP-A could mediate antitumor effects in EWS.
Downregulation of pathways involved in antigen processing and presentation is a well-known strategy utilized by tumors to evade the immune response (53), and low HLA expression has been linked to inferior outcomes in numerous cancers (54–58). In EWS, HLA class I expression was previously reported to be absent or low in the vast majority of tumors and of prognostic significance, as it correlated with decreased tumor infiltrating CD8+ T cells and diminished long-term overall survival (59). We present evidence that PAPPA expression and IGF signaling play a role in diminishing classical HLA class I expression and other proteins involved in antigen presentation in EWS, consistent with a model wherein immune evasion is linked to canonical growth signaling pathways. Similarly, previous studies demonstrate that MYC mediates immune privilege (60,61), albeit via distinct mechanisms from those described here.
Limitations of our study include the absence of an immunocompetent EWS model, which would allow for a more comprehensive analysis of immunomodulating effects of anti-PAPP-A.
The impact of the PAPP-A/IGF-1R axis on immune evasion suggest that combinatorial therapies that simultaneously target oncogene-mediated growth pathways and augment immune responses may prove synergistic. Specifically, it raises the question whether combined targeting of anti-PAPP-A/anti-IGF-1R therapies with amplifiers of the natural tumor immune response should be tested in future studies. Antibody-based therapeutics targeting PAPP-A could theoretically induce anti-EWS immune responses as a result of enhanced antigen presentation of PAPP-A-deficient EWS tumors. In summary, our work credentials PAPP-A as a tumor-specific, cell surface antigen with properties favorable for immunotherapeutic targeting. We further discovered a previously unrecognized role for PAPP-A and IGF-1 signaling in immune evasion in EWS, which provides a rationale for combining T cell-directed immunotherapies with agents to inhibit PAPP-A and IGF-1 signaling in EWS.
Funding
This work was in part supported by the Intramural Research Program of the National Cancer Institute. It was also supported by a SU2C-St. Baldrick’s Pediatric Cancer Dream Team Translational Research Grant (SU2CAACR-DT1113). Stand Up To Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research.
Notes
Affiliations of authors: Stanford Cancer Institute, Stanford University School of Medicine, Stanford, CA (SH, ES, PX, RJ, CLM); Pediatric Oncology Branch, National Cancer Center, National Institutes of Health, Bethesda, MD (JFS, AA); Cancer Genetics Branch, National Cancer Center, National Institutes of Health, Bethesda, MD (SS, PM, JK); Lady Davis Institute, SMBD Jewish General Hospital/Department of Oncology, McGill University/Department of Experimental Medicine, McGill University, Montreal, QC, Canada (MP); Department of Molecular Biology and Genetics, Aarhus University, Aarhus, Denmark (PRN, CO); Department of Pathology and Laboratory Medicine, University of British Columbia, Vancouver, BC, Canada (JL, PHS); Vancouver Prostate Centre, University of British Columbia, Vancouver, BC, Canada (LF); Division of Hematology-Oncology, Connecticut Children’s Medical Center and Jackson Laboratory for Genomic Medicine, CT (CL); Endocrine Research Unit, Mayo Clinic, Rochester, MN (CAC); Children’s Hospital of Philadelphia and Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA (JMM); Department of Pediatrics, Stanford University School of Medicine, Stanford, CA (CLM)
Present affiliations: University of California, Los Angeles, CA (AA).
The funders had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication.
Conflict of interest disclaimer: CO and CAC are inventors on patent or patent application relating to the use of pregnancy-associated plasma protein-A (PCT/EP2009/050796, CO; US 7115382). All other authors do not have disclosures related to the work presented in this manuscript.
The authors thank AnshLabs (Webster, TX) for providing the PAPP-A neutralizing antibody 1/41 and Michael C. Jensen (Seattle Children’s Hospital, Seattle, WE) for providing annotation datasets that were utilized for generation of our cell surface prioritization strategy.
CLM is a member of the Parker Institute for Cancer Immunotherapy, which supports the Stanford University Cancer Immunotherapy Program.
Supplementary Material
References
- 1. Stahl M, Ranft A, Paulussen M, et al. Risk of recurrence and survival after relapse in patients with Ewing sarcoma. Pediatr Blood Cancer. 2011;574:549–553. [DOI] [PubMed] [Google Scholar]
- 2. Fuchs B, Valenzuela RG, Petersen IA, et al. Ewing’s sarcoma and the development of secondary malignancies. Clin Orthop Relat Res. 2003;415:82–89. [DOI] [PubMed] [Google Scholar]
- 3. Goldsby R, Burke C, Nagarajan R, et al. Second solid malignancies among children, adolescents, and young adults diagnosed with malignant bone tumors after 1976: follow-up of a Children's Oncology Group cohort. Cancer. 2008;1139:2597–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tomazou EM, Sheffield NC, Schmidl C, et al. Epigenome mapping reveals distinct modes of gene regulation and widespread enhancer reprogramming by the oncogenic fusion protein EWS-FLI1. Cell Rep. 2015;107:1082–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sheffield NC, Pierron G, Klughammer J, et al. DNA methylation heterogeneity defines a disease spectrum in Ewing sarcoma. Nat Med 2017;23(3):386–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grohar PJ, Glod J, Peer CJ, et al. A phase I/II trial and pharmacokinetic study of mithramycin in children and adults with refractory Ewing sarcoma and EWS-FLI1 fusion transcript. Cancer Chemother Pharmacol 2017;80(3):645–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Choy E, Butrynski JE, Harmon DC, et al. Phase II study of olaparib in patients with refractory Ewing sarcoma following failure of standard chemotherapy. BMC Cancer. 2014;14:813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Subbiah V, Hess KR, Khawaja MR, et al. Evaluation of novel targeted therapies in aggressive biology sarcoma patients after progression from US FDA approved therapies. Sci Rep. 2016;6:35448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;4997457:214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brohl AS, Solomon DA, Chang W, et al. The genomic landscape of the Ewing Sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS Genet. 2014;107:E1004475.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vornicova O, Bar-Sela G.. Investigational therapies for Ewing sarcoma: a search without a clear finding. Expert Opin Investig Drugs. 2016;256:679–686. [DOI] [PubMed] [Google Scholar]
- 12. Majzner RG, Heitzeneder S, Mackall CL.. Harnessing the immunotherapy revolution for the treatment of childhood cancers. Cancer Cell. 2017;314:476–485. [DOI] [PubMed] [Google Scholar]
- 13. Town J, Pais H, Harrison S, et al. Exploring the surfaceome of Ewing sarcoma identifies a new and unique therapeutic target. Proc Natl Acad Sci USA. 2016;11313:3603–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Staege MS, Hutter C, Neumann I, et al. DNA microarrays reveal relationship of Ewing family tumors to both endothelial and fetal neural crest-derived cells and define novel targets. Cancer Res. 2004;6422:8213–8221. [DOI] [PubMed] [Google Scholar]
- 15. Conover CA. Key questions and answers about pregnancy-associated plasma protein-A. Trends Endocrinol Metab. 2012;235:242–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Merchant MS, Woo C-W, Mackall CL, et al. Potential use of imatinib in Ewing’s sarcoma: evidence for in vitro and in vivo activity. J Natl Cancer Inst. 2002;9422:1673–1679. [DOI] [PubMed] [Google Scholar]
- 17. Sanchez G, Delattre O, Auboeuf D, et al. Coupled alteration of transcription and splicing by a single oncogene: boosting the effect on cyclin D1 activity. Cell Cycle. 2008;715:2299–2305. [DOI] [PubMed] [Google Scholar]
- 18. Kovar H, Dworzak M, Strehl S, et al. Overexpression of the pseudoautosomal gene MIC2 in Ewing’s sarcoma and peripheral primitive neuroectodermal tumor. Oncogene. 1990;57:1067–1070. [PubMed] [Google Scholar]
- 19. Fleuren EDG, Roeffen MHS, Leenders WP, et al. Expression and clinical relevance of MET and ALK in Ewing sarcomas. Int J Cancer. 2013;1332:427–436. [DOI] [PubMed] [Google Scholar]
- 20. Uhlen M, Fagerberg L, Hallstrom BM, et al. Proteomics: tissue-based map of the human proteome. Science. 2015;3476220:1260419. [DOI] [PubMed] [Google Scholar]
- 21. Becker MA, Haluska P, Bale LK, et al. A novel neutralizing antibody targeting pregnancy-associated plasma protein-a inhibits ovarian cancer growth and ascites accumulation in patient mouse tumorgrafts. Mol Cancer Ther. 2015;144:973–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Takabatake Y, Oxvig C, Nagi C, et al. Lactation opposes pappalysin-1-driven pregnancy-associated breast cancer. EMBO Mol Med. 2016;84:388–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Frasca F, Pandini G, Scalia P, et al. Insulin receptor isoform A, a newly recognized, high-affinity insulin-like growth factor II receptor in fetal and cancer cells. Mol Cell Biol. 1999;195:3278–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kloverpris S, Mikkelsen JH, Pedersen JH, et al. Stanniocalcin-1 potently inhibits the proteolytic activity of the metalloproteinase pregnancy-associated plasma protein-A. J Biol Chem. 2015;29036:21915–21924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jepsen MR, Kløverpris S, Mikkelsen JH, et al. Stanniocalcin-2 inhibits mammalian growth by proteolytic inhibition of the insulin-like growth factor axis. J Biol Chem. 2015;2906:3430–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Overgaard MT, Haaning J, Boldt HB, et al. Expression of recombinant human pregnancy-associated plasma protein-A and identification of the proform of eosinophil major basic protein as its physiological inhibitor. J Biol Chem. 2000;27540:31128–31133. [DOI] [PubMed] [Google Scholar]
- 27. Oxvig C, Sand O, Kristensen T, et al. Circulating human pregnancy-associated plasma protein-A is disulfide-bridged to the proform of eosinophil major basic protein. J Biol Chem. 1993;26817:12243–12246. [PubMed] [Google Scholar]
- 28. Jayabal P, Houghton PJ, Shiio Y.. EWS-FLI-1 creates a cell surface microenvironment conducive to IGF signaling by inducing pappalysin-1. Genes Cancer. 2017;8(11–12):762–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Riggi N, Knoechel B, Gillespie SM, et al. EWS-FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma. Cancer Cell. 2014;265:668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Leguy M, Brun S, Pidoux G, et al. Pattern of secretion of pregnancy-associated plasma protein-A (PAPP-A) during pregnancies complicated by fetal aneuploidy, in vivo and in vitro. Reprod Biol Endocrinol. 2014;121:129.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Glerup S, Kløverpris S, Laursen LS, et al. Cell surface detachment of pregnancy-associated plasma protein-A requires the formation of intermolecular proteinase-inhibitor disulfide bonds and glycosaminoglycan covalently bound to the inhibitor. J Biol Chem. 2007;2823:1769–1778. [DOI] [PubMed] [Google Scholar]
- 32. Mikkelsen JH, Resch ZT, Kalra B, et al. Indirect targeting of IGF receptor signaling in vivo by substrate-selective inhibition of PAPP-A proteolytic activity. Oncotarget. 2014;54:1014–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liberzon A, Birger C, Thorvaldsdóttir H, et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;16:417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dudek H, Datta SR, Franke TF, et al. Regulation of neuronal survival by the serine-threonine protein kinase Akt. Science. 1997;2755300:661–665. [DOI] [PubMed] [Google Scholar]
- 35. Galvan V, Logvinova A, Sperandio S, et al. Type 1 insulin-like growth factor receptor (IGF-IR) signaling inhibits apoptosis signal-regulating kinase 1 (ASK1). J Biol Chem. 2003;27815:13325–13332. [DOI] [PubMed] [Google Scholar]
- 36. Peruzzi F, Prisco M, Morrione A, et al. Anti-apoptotic signaling of the insulin-like growth factor-I receptor through mitochondrial translocation of c-Raf and Nedd4. J Biol Chem. 2001;27628:25990–25996. [DOI] [PubMed] [Google Scholar]
- 37. Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;394:782–795. [DOI] [PubMed] [Google Scholar]
- 38. Toretsky JA, Kalebic T, Blakesley V, et al. The insulin-like growth factor-I receptor is required for EWS/FLI-1 transformation of fibroblasts. J Biol Chem. 1997;27249:30822–30827. [DOI] [PubMed] [Google Scholar]
- 39. Ferretti C, Bruni L, Dangles-Marie V, et al. Molecular circuits shared by placental and cancer cells, and their implications in the proliferative, invasive and migratory capacities of trophoblasts. Hum Reprod Update. 2007;132:121–141. [DOI] [PubMed] [Google Scholar]
- 40. Holtan SG, Creedon DJ, Haluska P, et al. Cancer and pregnancy: parallels in growth, invasion, and immune modulation and implications for cancer therapeutic agents. Mayo Clin Proc. 2009;8411:985–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Reeves E, James E.. Tumour and placenta establishment: the importance of antigen processing and presentation. Placenta. 2017;56:34–39. [DOI] [PubMed] [Google Scholar]
- 42. Salanti A, Clausen TM, Agerbæk MØ, et al. Targeting human cancer by a glycosaminoglycan binding malaria protein. Cancer Cell. 2015;284:500–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pappo AS, Patel SR, Crowley J, et al. R1507, a monoclonal antibody to the insulin-like growth factor 1 receptor, in patients with recurrent or refractory Ewing sarcoma family of tumors: results of a phase II Sarcoma Alliance for Research through Collaboration study. J Clin Oncol. 2011;2934:4541–4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Anderson PM, Bielack SS, Gorlick RG, et al. A phase II study of clinical activity of SCH 717454 (robatumumab) in patients with relapsed osteosarcoma and Ewing sarcoma. Pediatr Blood Cancer. 2016;6310:1761–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Juergens H, Daw NC, Geoerger B, et al. Preliminary efficacy of the anti-insulin-like growth factor type 1 receptor antibody figitumumab in patients with refractory Ewing sarcoma. J Clin Oncol. 2011;2934:4534–4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Scotlandi K, Manara MC, Serra M, et al. Expression of insulin-like growth factor system components in Ewing’s sarcoma and their association with survival. Eur J Cancer. 2011;478:1258–1266. [DOI] [PubMed] [Google Scholar]
- 47. Buck E, Gokhale PC, Koujak S, et al. Compensatory insulin receptor (IR) activation on inhibition of insulin-like growth factor-1 receptor (IGF-1R): rationale for cotargeting IGF-1R and IR in cancer. Mol Cancer Ther. 2010;910:2652–2664. [DOI] [PubMed] [Google Scholar]
- 48. Forest A, Amatulli M, Ludwig DL, et al. Intrinsic resistance to cixutumumab is conferred by distinct isoforms of the insulin receptor. Mol Cancer Res. 2015;1312:1615–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lamhamedi-Cherradi SE, Menegaz BA, Ramamoorthy V, et al. IGF-1R and mTOR blockade: novel resistance mechanisms and synergistic drug combinations for Ewing sarcoma. J Natl Cancer Inst. 2016;10812:djw182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Weyer K, Overgaard MT, Laursen LS, et al. Cell surface adhesion of pregnancy-associated plasma protein-A is mediated by four clusters of basic residues located in its third and fourth CCP module. Eur J Biochem. 2004;2718:1525–1535. [DOI] [PubMed] [Google Scholar]
- 51. Laursen LS, Overgaard MT, Weyer K, et al. Cell surface targeting of pregnancy-associated plasma protein A proteolytic activity. J Biol Chem. 2002;27749:47225–47234. [DOI] [PubMed] [Google Scholar]
- 52. Kirschner A, Thiede M, Grünewald TGP, et al. Pappalysin-1 T cell receptor transgenic allo-restricted T cells kill Ewing sarcoma in vitro and in vivo. Oncoimmunology. 2017;62:e1273301.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. McGranahan N, Rosenthal R, Hiley CT, et al. Allele-Specific HLA loss and immune escape in lung cancer evolution. Cell. 2017;1716:1259–1271.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Campoli M, Ferrone S.. HLA antigen changes in malignant cells: epigenetic mechanisms and biologic significance. Oncogene. 2008;2745:5869–5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kageshita T, Hirai S, Ono T, et al. Down-regulation of HLA class I antigen-processing molecules in malignant melanoma: association with disease progression. Am J Pathol. 1999;1543:745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mehta AM, Jordanova ES, Kenter GG, et al. Association of antigen processing machinery and HLA class I defects with clinicopathological outcome in cervical carcinoma. Cancer Immunol Immunother. 2008;572:197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pedersen MH, Hood BL, Beck HC, et al. Downregulation of antigen presentation-associated pathway proteins is linked to poor outcome in triple-negative breast cancer patient tumors. Oncoimmunology. 2017;65:e1305531.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Haworth KB, Leddon JL, Chen C-Y, et al. Going back to class I: MHC and immunotherapies for childhood cancer. Pediatr Blood Cancer. 2015;624:571–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yabe H, Tsukahara T, Kawaguchi S, et al. Prognostic significance of HLA class I expression in Ewing’s sarcoma family of tumors. J Surg Oncol. 2011;1035:380–385. [DOI] [PubMed] [Google Scholar]
- 60. Casey SC, Baylot V, Felsher DW.. MYC: master regulator of immune privilege. Trends Immunol. 2017;384:298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Casey SC, Tong L, Li Y, et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science. 2016;3526282:227–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.